Abstract
During the life cycle of the transmissible gastroenteritis coronavirus (TGEV), two types of virus-related particles are detected in infected swine testis cells: large annular viruses and small dense viruses. We have studied the relationships between these two types of particles. Immunoelectron microscopy showed that they are closely related, since both large and small particles reacted equally with polyclonal and monoclonal antibodies specific for TGEV proteins. Monensin, a drug that selectively affects the Golgi complex, caused an accumulation of large annular viral particles in perinuclear elements of the endoplasmic reticulum-Golgi intermediate compartment. A partial reversion of the monensin blockade was obtained in both the absence and presence of cycloheximide, a drug that prevented the formation of new viral particles. After removal of monensin, the Golgi complex recovered its perinuclear location, and a decrease in the number of perinuclear large viral particles was observed. The release of small dense viral particles into secretory vesicles and the extracellular medium was also observed, as was a partial recovery of infectivity in culture supernatants. Small viral particles started to be seen between the third and the fourth Golgi cisternae of normally infected cells. All of these data strongly indicate that the large annular particles are the immature precursors of the small dense viruses, which are the infectious TGEV virions. The immature viral particles need to reach a particular location at the trans side of the Golgi stack to complete their morphological maturation.
Coronaviruses are members of a family of RNA viruses that cause different pathologies in humans and animals. An important aspect of their study is the finding of efficient antiviral strategies that could prevent the considerable economic losses caused by these infectious agents all over the world (29). The factors involved in coronavirus-receptor binding, virus entry, and uncoating are poorly characterized, as is how coronaviruses replicate their own large genome and transcribe subgenomic mRNAs in the cytoplasm of infected cells. Different proteins associated with viral RNA seem to participate in the replication of coronaviruses (25, 45). Recently, a cellular protein purified from the replicative complexes has been identified as the heterogeneous nuclear ribonucleoprotein (heterogeneous nuclear RNP) A1, which in the cell participates in the regulation of alternative RNA splicing (27).
The transmissible gastroenteritis coronavirus (TGEV) causes a serious illness in newborn piglets, with mortality rates approaching 100% (7). TGEV virions are constituted by basic elements present in most of the members of the Coronaviridae: three structural envelope proteins (S, which forms the peplomers; M, the integral membrane protein, a key factor in assembly; and E, a minor component of unknown function) and a fourth structural polypeptide, the nucleocapsid (N) protein (26). The N protein interacts with the viral genome, a long (around 30-kb) positive, single-stranded RNA molecule, forming a helical RNP (22, 31). A role for N protein in viral replication has been also proposed (5). The M and E proteins are key factors for assembly, as suggested by studies with virus-like particles and mutant viruses obtained by targeted recombination (3, 6, 9, 56). Infectious TGEV virions enter susceptible cells after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment (16). After completing replication and RNA transcription, coronaviruses exhibit a complex morphogenesis in which the constitutive secretory pathway of the infected cell plays a key role. The endoplasmic reticulum-Golgi intermediate compartment (ERGIC), which contains perinuclear and peripheral tubulo-vesicular elements (41, 43), constitutes the budding compartment for coronaviruses at early times postinfection (p.i.). Assembly of coronavirus particles starts with the association of the RNPs with membranes of the perinuclear ERGIC, where the envelope proteins are already inserted (23). A budding process originates viral particles that are transported through the Golgi complex and collected inside secretory vesicles, which release virions out of the cell (52, 53).
The study of viral assembly and maturation is intimately associated with the study of the cellular systems involved in the process. In recent years, the considerable advances in ultrastructural analysis provided by methods based on cryomicroscopy are giving us a new view of how viruses are built in infected cells (4, 10, 38, 39, 47, 49, 57). A large amount of new data is being obtained for different viral groups. This is the case for coronaviruses, the subject of recent detailed structural studies developed in our group and focused on TGEV (34, 36, 37). These studies have provided new concepts concerning the structure and assembly of coronaviruses (26): (i) extracellular infectious coronavirus particles contain an internal core shell, probably icosahedral, that encloses the RNP (36); (ii) a detectable number of molecules of the membrane (M) protein of TGEV expose both the amino- and carboxy-terminal domains on the external surface of the virions, which differs from the conventional N-exo, C-endo topology described for mouse hepatitis virus (MHV) M protein (34); (iii) two different types of virus-related particles form inside coronavirus-infected cells, with the larger viral particles being the first to be seen at the perinuclear area, while the smaller virions accumulate inside secretory vesicles and on the cell surface (37); and (iv) transport along the constitutive exocytic route, and in particular through a functional Golgi complex, seems to be necessary for morphological transformation (37). The complex structural reorganization of coronavirus particles during their transport along the exocytic route and the key participation of the Golgi complex are interesting subjects for analysis, due to the novelty of the processes involved. If small virions actually originate from the perinuclear large viral particles, the structural transformation taking place involves a major reorganization of all viral components, with the formation of the icosahedral core shell, and a dramatic change in volume (reduced to approximately 50%). To analyze these aspects in more detail, we have used monensin, a drug that reversibly disorganizes the Golgi complex and blocks transport along the exocytic route (50). We have also studied the effects of cycloheximide (an inhibitor of protein synthesis) on TGEV assembly and structural maturation. The effects of different treatments on the integrity of the exocytic route and the simultaneous changes to viral morphogenesis were analyzed. The results of this structural analysis, together with the immunodetection of five TGEV proteins, strongly support the precursor role for the large viral particles. We have also confirmed that these particles need to reach a particular location within the Golgi complex to complete their morphological maturation.
MATERIALS AND METHODS
Cells, viruses, antibodies, and drugs.
Culture of swine testis (ST) cells and virus infections were performed as previously described (19). Monolayers of ST cells were infected with TGEV (PUR 46-MAD strain) at a multiplicity of infection of 10 PFU/cell and collected at different times p.i. to be processed for immunofluorescence or electron microscopy. Virus infectivity in culture supernatants was calculated by the plaque lysis titration assay (19). The previously characterized murine monoclonal antibodies (MAbs) specific for TGEV N, M, and S proteins (13, 34, 40) and the hyperimmune rabbit serum against the PUR 46-MAD strain of TGEV were generously provided by L. Enjuanes (Centro Nacional de Biotecnología, Madrid, Spain). MAbs specific for TGEV E protein (14) were a kind gift of H. Laude (Centre de Recherches, Institut National de la Recherche Agronomique, Jouy-en Josas, France). The polyclonal antiserum specific for the TGEV hydrophobic protein (HP), the product of TGEV gene 7 (54), was kindly provided by D. Brian (University of Tennessee, Knoxville). The rabbit antiserum to human giantin, a Golgi-specific marker (44), and the mouse MAb G1/93, specific for human ERGIC-53 protein (42), were kindly provided by M. Renz (Institute of Immunology and Molecular Genetics, Karlsruhe, Germany) and H. P. Hauri (Biozentrum, University of Basel, Basel, Switzerland), respectively. Goat anti-rabbit and goat anti-mouse antibodies conjugated with fluorescein or rhodamine were purchased from Southern Biotechnology Associates, Inc. (Birmingham, Ala.). Colloidal gold conjugates of goat anti-mouse and goat anti-rabbit immunoglobulin G were provided by Biocell Research Laboratory (Cardiff, United Kingdom). Monensin and cycloheximide were purchased from Sigma (St. Louis, Mo.).
Treatment of ST cells with monensin and cycloheximide.
Confluent monolayers of uninfected ST cells were incubated with the carboxylic ionophore monensin at different concentrations (1, 2, and 5 μM) and for different times (2, 4, and 6 h). Once the treatment conditions that cause the described characteristic morphological effects on the Golgi complex (swelling, fragmentation, and spreading) were determined (by immunofluorescence and electron microscopy), they were used for infected ST cells. Monensin was added to cell cultures at a final concentration of 5 μM and at different times (1, 2, 3, and 4 h) p.i. Treatment with the drug was prolonged for 7, 6, 5, or 4 h (to complete 8 h p.i.). Some of the uninfected and infected cell cultures were further incubated for 0.5, 1, 1.5, or 2 h after monensin removal to study whether it was possible to revert the effects caused by the drug. To study the effects of cycloheximide on TGEV morphogenesis, the drug was added to infected ST cultures (8 h p.i.) at a final concentration of 100 μg/ml. Cells were incubated with the drug for 0.5, 1, 1.5, and 2 h. Cycloheximide was also used during reversion of the monensin blockade. Some of the infected cells were incubated for 0.5, 1, 1.5, or 2 h with cycloheximide after monensin removal. Control cells as well as cultures of cells subjected to the different treatments were processed for immunofluoresecence detection of cellular markers or TGEV proteins or for structural studies at the electron microscopy level.
Fluorescence microscopy.
ST cells were grown on 12-mm-diameter glass coverslips. For immunodetection of ERGIC-53 protein or the Golgi protein giantin, cells were briefly washed with phosphate-buffered saline (PBS) (pH 7.4) containing 0.1% bovine serum albumin (PBS-A) and then fixed with 3% paraformaldehyde in PBS for 30 min. After three washes with PBS-A, the cells were permeabilized for 3 min with pure methanol at −20°C. The coverslips were then washed two times with PBS and three times with PBS containing 2% bovine serum albumin (PBS-B) and incubated for 1 h with MAb G1/93 (specific for the human ERGIC-53 marker) previously diluted 1:50 in PBS-A or with the antigiantin antiserum diluted 1:50 in PBS-A. For TGEV detection, cells grown on coverslips were fixed in a mixture of methanol and acetone (1:1) for 20 min at −20°C. After being washed with PBS, the coverslips were incubated with PBS-B for 15 min. Cells were then incubated 1 h with MAb 6A.C3 (specific for TGEV S protein) or with a rabbit anti-TGEV polyclonal antiserum, diluted 1:200 in PBS-A. After treatment with the primary antibodies, samples were washed three times with PBS-A and incubated for 1 h with the secondary antibodies (conjugated with rhodamine or fluorescein) diluted in PBS-A. The coverslips were then washed three times with PBS-A and twice with PBS and finally mounted on glass slides with a mixture of glycerol and PBS (9:1). Samples were studied with a Zeiss Axiophot fluorescence microscope. Images were collected with a MicroMax digital camera system.
Electron microscopy. (i) Processing of infected ST cells for embedding in EML-812 for ultrastructural studies.
Monolayers of ST cells were fixed in situ with a mixture of 2% glutaraldehyde and 2% tannic acid in 0.4 M HEPES buffer (pH 7.4) for 1 h at room temperature. Fixed monolayers were removed from the dishes in the fixative and transferred to Eppendorf tubes. After centrifugation in a microcentrifuge and several washes with HEPES buffer, the pellets were processed for embedding in the epoxy resin EML-812 (Taab Laboratories, Berkshire, United Kingdom) by methods previously described in detail (37). Postfixation of cells was done with a mixture of 1% osmium tetroxide and 0.8% potassium ferricyanide in distilled water for 1 h at 4°C. After four washes with HEPES buffer, samples were treated with 2% uranyl acetate, washed again, and dehydrated in increasing concentrations of acetone (50, 70, 90, and 100%) for 10 min each at 4°C. Infiltration in the resin EML-812 was done at room temperature for 1 day. Polymerization of infiltrated samples was done at 60°C for 3 days. Ultrathin (20- to 30-nm-thick) sections of the samples were stained with saturated uranyl acetate and lead citrate by standard procedures.
(ii) Processing of ST cells for embedding in Lowicryl K4M for immunoelectron microscopy.
Cultures of ST cells were subjected to a mild fixation with a solution of 4% paraformaldehyde containing 0.1% glutaraldehyde in PBS at 4°C for 30 min. Fixed cells were removed from the culture dishes, washed with PBS, and dehydrated at −20°C in increasing concentrations of ethanol. Infiltration with the resin Lowicryl K4M (EML Laboratories, Berkshire, United Kingdom) at −30°C and polymerization with UV light were done as previously described (34). Ultrathin sections were processed for immunogold detection.
(iii) Quick freezing and freeze-substitution of ST cells.
Small pellets of chemically fixed cells were cryoprotected with glycerol, applied to small pieces of filter paper, blotted for 15 s, and quick frozen in liquid propane at an approximate speed of 104°C/s. Vitrified specimens were transferred to a Reichert-Jung AFS freeze-substitution unit (Leica, Vienna, Austria) and maintained for 24 h at −90°C in pure methanol or in a mixture of pure acetone and 0.5% (wt/vol) osmium tetroxide for complete substitution of the water of the sample (37, 38). After freeze-substitution in methanol, samples were infiltrated in Lowicryl HM23 (EML Laboratories) at −80°C and polymerized with UV light. Samples processed by freeze-substitution in acetone-osmium were subjected to a controlled increase of temperature before embedding in EML-812. Ultrathin sections of the samples were either stained or processed for immunogold labeling.
(iv) Immunoelectron microscopy.
Immunogold detection of TGEV proteins on ultrathin sections of infected ST cells was done at room temperature with hyperimmune anti-TGEV or anti-HP rabbit serum or MAbs specific for M, N, S, and E proteins by established procedures (34, 35). Sections collected on coated gold electron microscopy grids were incubated for 30 min with Tris buffer-gelatin and then floated on drops of diluted primary antibodies for 2 h. After jet washing with PBS, samples were incubated for 1 h with secondary antibodies conjugated with 5- or 10-nm-diameter gold particles and washed again with PBS and distilled water. Samples were then allowed to dry on filter paper before being stained with saturated uranyl acetate for 25 min. An identical procedure was used to label the Golgi complex or the ERGIC, using the antigiantin and anti-ERGIC-53 antibodies described above. All samples were studied with a JEOL 1200 EX II electron microscope.
RESULTS
Immunocytochemical characterization of the two types of TGEV viral particles.
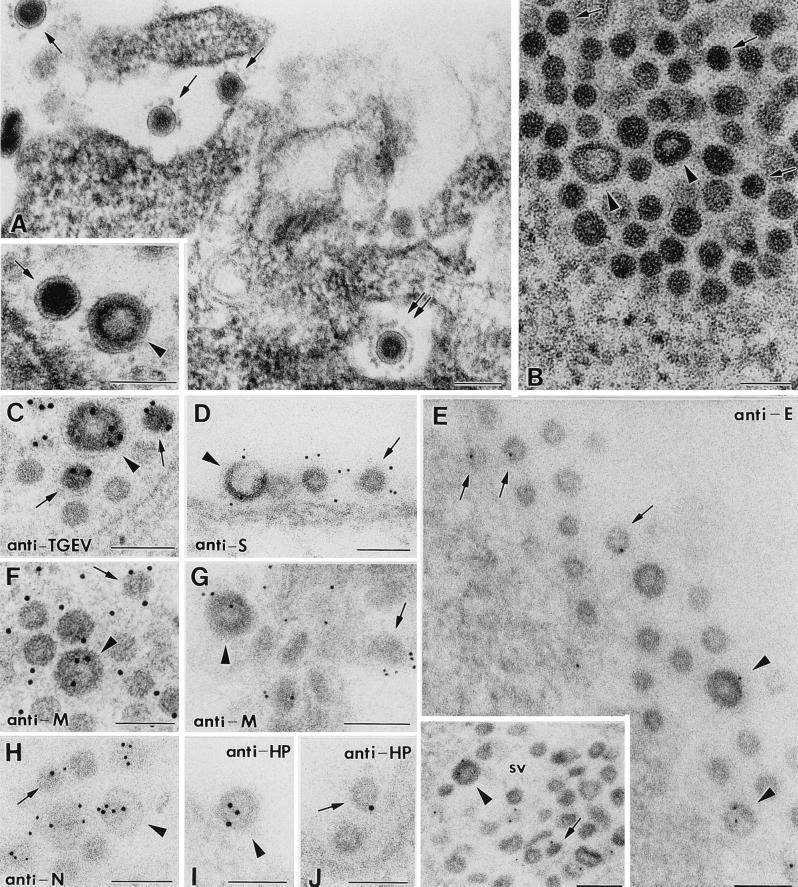
A detailed characterization of the large and small viral particles at the cytochemical level was performed by using ultrathin sections from infected cells processed by different methods (Fig. 1). Larger virus-like particles (marked with arrowheads in Fig. 1) are characterized by an annular stained region under the external envelope, while smaller virions (marked with arrows in Fig. 1) have a more dense core, which is heavily stained. Both types of virus-related particles are clearly distinguished in ultrathin sections of EML-812 and Lowicryl resins (Fig. 1A and B). The use of different processing procedures for the samples was an attempt to obtain signals with both polyclonal antisera and MAbs specific for the different TGEV proteins. Both large and small viruses from different cellular locations reacted similarly with an anti-TGEV polyclonal serum (Fig. 1C) and with several MAbs specific for TGEV S, E, M, and N structural proteins (Fig. 1D to H). MAbs specific for the minor structural component E protein provided a weak signal in approximately 20% of both intra- and extracellular large and small particles (Fig. 1E). A slightly lower percentage of viral particles (around 10%) were weakly labeled with a polyclonal antiserum specific for TGEV HP (Fig. 1I and J). This protein, of unknown function, is the product of gene 7, an open reading frame in the very 3′ end of the TGEV genome (26, 54). It has been suggested that HP could incorporate into virions and participate in the membrane association of replication complexes or in virion assembly (12, 54). From the immunocytochemical point of view, it is equally represented in both large and small viral particles.
FIG. 1.
Characterization of large and small TGEV viral particles by immunoelectron microscopy. All images show TGEV-infected ST cells at 8 h p.i. In all panels representative large annular viral particles are marked with arrowheads, while small dense viral particles are marked with arrows. (A) Both types of particles are clearly distinguished in sections of the epoxy resin EML-812. The main field shows several extracellular small viruses, while the inset shows a small dense particle on the left and a large annular particle on the right, for a direct comparison of size and morphology. Double arrows in the main field point to a small virus with a well defined corona of extended peplomers. (B) In low-contrast Lowicryl sections membranes exhibit a poor definition, but both types of viral particles are clearly distinguished due to the different structures of their cores. (C) A polyclonal anti-TGEV antiserum clearly reacts with both types of viral particles. (D) MAbs specific for S protein also react with large and small viral particles. (E) Anti-E MAbs show a weak specific signal in approximately 20% of extracellular (main field) and intracellular (inset) large and small viral particles. (F to H) MAbs specific for the carboxy-terminal domain of M protein react equally with both small and large viral particles (F and G), as do anti-N MAbs (H). (I and J) An antiserum specific for TGEV HP weakly labels approximately 10% of both large (I) and small (J) viral particles. Panel A shows a sample conventionally embedded in EML-812, panel C shows a sample processed by freeze-substitution in osmium-acetone, and panel G shows a sample freeze-substituted in methanol; the rest of panels show samples conventionally embedded in Lowicryl K4M at low temperature. Colloidal gold conjugates 10 nm in diameter were used for panels C, F, I, and J, while the rest of the panels show 5-nm-diameter gold particles. Panels A and D and the main field in panel E show viral particles at the cell surface, while the rest of the micrographs show viral particles inside secretory vesicles (sv). Bars, 100 nm.
Accumulation of large viral particles in pre-Golgi compartments.
In our previous studies, the distribution of TGEV large and small viral particles in different cellular compartments, as well as the effects of brefeldin A, suggested that transport along the exocytic pathway correlated with TGEV morphological transformation. In the present work we have studied in detail the progression of TGEV infection in ST cells and the effects of monensin, a drug more specific for the Golgi complex, since the effect of brefeldin A derives from the fusion between Golgi and pre-Golgi compartments. As shown in Fig. 2A and B, the immunofluorescence signal associated with TGEV starts to accumulate at the perinuclear area of cell islets at early times p.i. (5 h), extending all over the cells as a punctate signal at 8 h p.i. Almost 100% of the cells in the confluent monolayer were labeled at 8 h p.i. The two compartments of the exocytic route that play key roles in coronavirus morphogenesis (the ERGIC and the Golgi complex) were also localized by immunofluorescence with specific markers (Fig. 2C and D). TGEV infection did not cause any appreciable change in the organization of these compartments, as detected by immunofluorescence at 8 h p.i. (not shown). We then studied the effect of monensin on the integrity of the Golgi complex of ST cells. Monensin induced a fragmentation and dispersion of the Golgi stacks (Fig. 3A) and a retention of the signal associated with the virus in the perinuclear area, where the budding takes place (Fig. 3B). However, no significant changes in the ERGIC were detected in the presence of the drug (Fig. 3C). At the electron microscopy level, the punctate immunofluorescence pattern of infected cells at 8 h p.i. corresponds to the accumulation of secretory vesicles (Fig. 3D) filled with viruses, most of them small and dense. Normally infected cells have also large annular viruses at the perinuclear region (Fig. 3D). Many extracellular virions, most of them small and dense, are also seen (Fig. 3D). The effects of monensin on TGEV assembly were also monitored at the ultrastructural level after addition of the drug at different times p.i. At earlier times p.i., viral assembly was almost completely inhibited. When monensin was added at 3 h p.i. (after enough viral components have already accumulated) large viral particles accumulated inside perinuclear vesicles of smooth membranes and did not spread following the Golgi redistribution (Fig. 3E). Numerous smooth membrane vesicles filled with large annular viral particles accumulated at these perinuclear locations. In freeze-substituted samples (Fig. 3F and G) these large particles are indistinguishable from the large viral particles that are the first to assemble at the perinuclear area in normally infected cells (37). Numerous RNPs and characteristic budding profiles are also seen in these perinuclear vesicles (Fig. 3F and G). Freeze-substitution also allowed good immunolabeling signals with anti-ERGIC-53 and antigiantin antibodies (Fig. 3H and I). The vesicles with viruses reacted with the anti-ERGIC-53 antibody (Fig. 3H). Antigiantin labeled only empty vesicles (Fig. 3I), while vesicles with viruses did not react with the antigiantin serum (Fig. 3J).
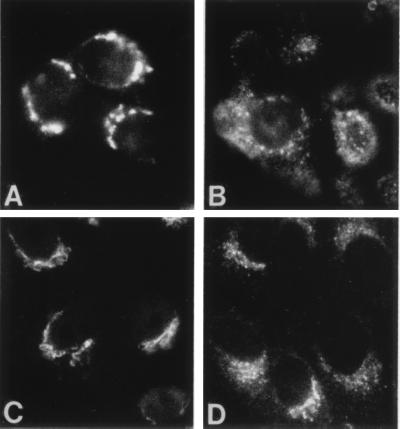
FIG. 2.
Immunofluorescence detection of cellular markers and TGEV in ST cells. (A and B) Detection of TGEV with MAb 6A.C3 (specific for the S protein) to monitor TGEV infection in confluent monolayers of ST cells. (A) At 5 h p.i., labeling concentrates at a perinuclear location, coincident with the described budding compartment. (B) At 8 h p.i., signal spreads throughout the cytoplasm, exhibiting a punctate pattern in most of the cells. (C and D) Detection of two cellular compartments of the exocytic route that function as key factors in TGEV morphogenesis. (C) Detection of the Golgi apparatus with an antibody specific for the peripheral protein giantin. (D) Organization of the ERGIC as seen with an antibody to the resident protein ERGIC-53.
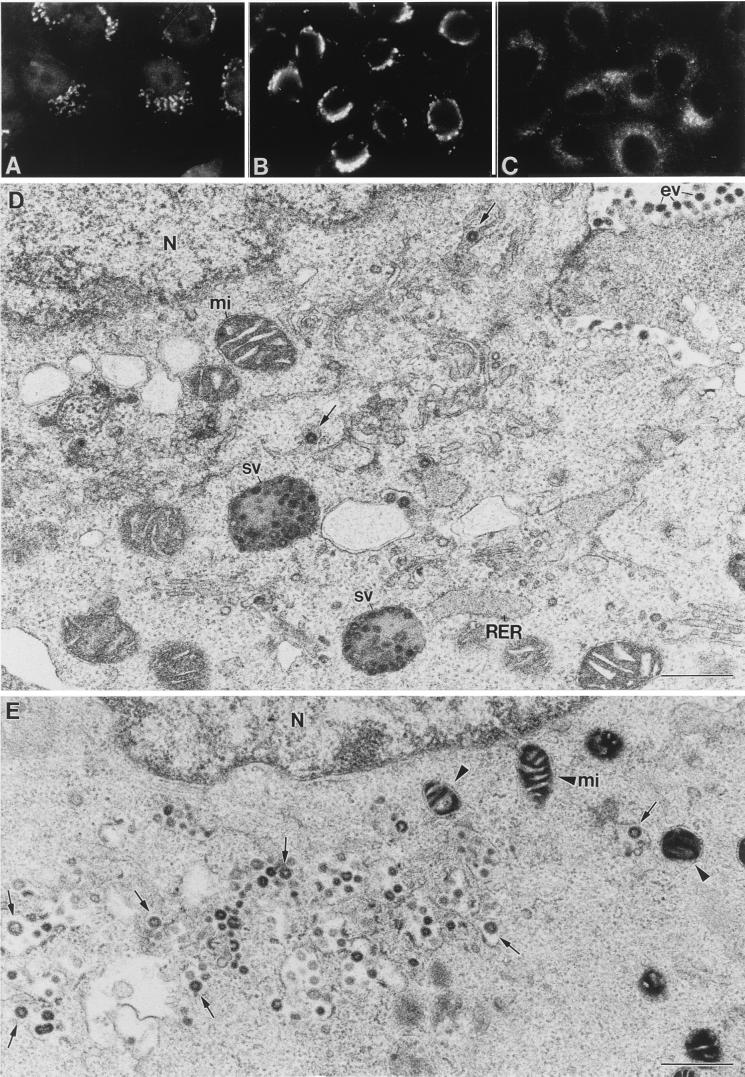
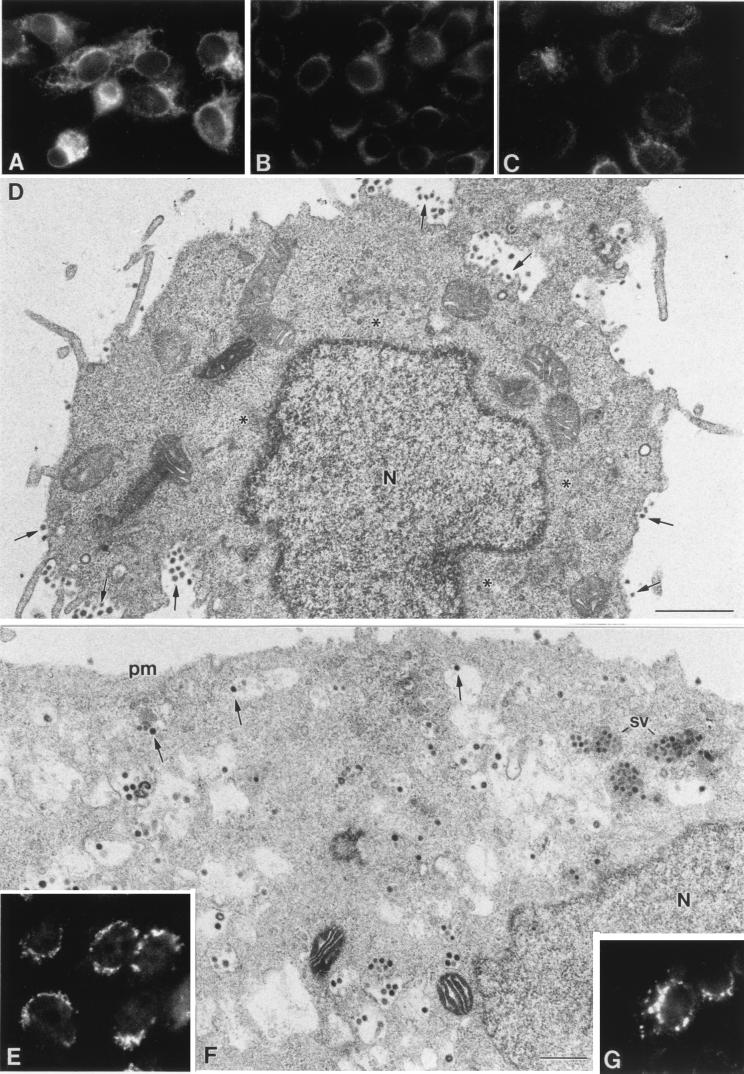
FIG. 3.
Effects of the Golgi-disrupting drug monensin on TGEV morphogenesis. (A to C) Immunofluorescence study of the effects of monensin on two components of the exocytic route and TGEV distribution in infected ST cells. (A) Monensin causes a generalized fragmentation and spreading of the Golgi complex of ST cells, as visualized with the antigiantin antibody (compare with control cells in Fig. 2C). (B) Under these conditions, signal associated with TGEV (at 8 h p.i.) is restricted to the perinuclear region, coincident with the budding compartment (compare to normally infected cells at 5 and 8 h p.i. in Fig. 2). (C) Monensin treatment does not induce any significant change in the organization of the intermediate compartment, as shown by immunofluorescence with the anti-ERGIC-53 antibody (compare with control cells in Fig. 2D). (D) Electron microscopy shows the characteristic situation of normally infected cells at 8 h p.i. These cells have large annular viruses (arrows) around the nucleus (N) and large secretory vesicles (sv) filled with viruses, which are most probably responsible for the punctate labeling seen by immunofluorescence. Most of the extracellular viruses (ev) are small and dense. mi, mitochondrion. (E) Cells infected and treated with monensin at 3 h p.i. exhibit a very different situation than those at 8 h p.i. The perinuclear signal observed by immunofluorescence when localizing TGEV (panel B) corresponds to large viral particles that accumulated inside dilated vesicles of smooth membranes (arrows). Arrowheads point to abnormal mitochondria characteristic of monensin treatment. (F and G) Structures of the large viral particles (arrowheads) and the abundant viral RNPs and budding profiles (double arrows) formed in monensin-treated cells, seen with more detail in higher-magnification fields from samples processed by freeze-substitution in osmium-acetone. (H and I) In these samples the membranes of vesicles filled with viruses (v) clearly react with the anti-ERGIC-53 antibody (arrows in panel H), while the Golgi-specific antigiantin antibody reacts with vesicles that do not contain any viral particles (asterisks in panel I). (J) Vesicles filled with viruses did not show any labeling with the Golgi-specific marker. (K) Viruses that have accumulated in the lumen of RER cisternae of normally infected cells at 16 h p.i. are large and annular. pm: plasma membrane. Bars, 0.5 μm in panels D and E and 100 nm in the rest of the panels.
TGEV is also able to assemble in the rough endoplasmic reticulum (RER) cisternae at late times p.i. (16 h p.i.), when viral components have accumulated in RER membranes (52). Large annular viral particles are seen inside this pre-ERGIC compartment (Fig. 3K).
Effects of cycloheximide and reversion of the monensin blockade.
Cycloheximide is an inhibitor of protein synthesis that is frequently used to inhibit viral assembly (23). Its effects on TGEV morphogenesis are shown in Fig. 4. The addition of the drug to a monolayer of infected ST cells at 8 h p.i. caused a progressive decrease in the intracellular signal associated with the virus, as observed by immunofluorescence after 30 or 60 min (Fig. 4A to C). At the electron microscopy level, the viral particles disappeared from the perinuclear region (Fig. 4D), and small dense virions were seen inside secretory vesicles (not shown) and almost exclusively on the cell surface 60 min after addition of the drug (Fig. 4D). Subsequently, when no new large viral particles were formed due to the inhibition of the protein synthesis, small viruses finally accumulated in infected cells.
FIG. 4.
Effects of cycloheximide on TGEV morphogenesis and reversion of monensin effects. (A to C) The immunofluorescence signal associated with TGEV in normally infected cells sometimes exhibits a reticular pattern at 8 h p.i. (A), which is indicative of accumulation of viruses in RER cisternae. This signal progressively decreases after treatment of the culture with cycloheximide for 30 (B) or 60 (C) min. (D) Electron microscopy images of cells after 60 min with cycloheximide show the disappearance of viral particles at the perinuclear region (asterisks), while numerous small viruses are seen on the cell surface (arrows). (E to G) A partial reversion of monensin effects is observed after removal of the drug in the presence of cycloheximide. (E) In 2 h, dispersed Golgi elements have returned to the perinuclear region, as observed by immunofluorescence detection of giantin. (F) At the electron microscopy level, many viruses are seen leaving the perinuclear area and approaching the cell surface (arrows). Secretory vesicles (sv) filled with small viruses and extracellular viral particles attached to the plasma membrane (pm) are seen again. (G) In these cells, immunofluorescence detection of TGEV shows a punctate pattern indicative of virus release in secretory vesicles. Bars, 1 μm in panel D and 0.5 μm in panel F.
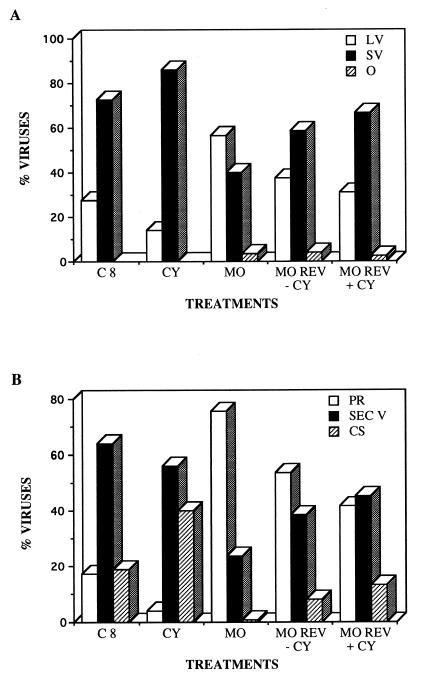
Reversion of the monensin blockade was also studied in both the absence and the presence of cycloheximide. The Golgi complex was able to recover its perinuclear location from the fragmented and dispersed state caused by monensin when the drug was removed from the cultures (Fig. 4E). This suggested that a reversion of the effects on TGEV morphogenesis caused by the monensin blockade could be also obtained. If the reversion also takes place in the presence of cycloheximide, we could then monitor the fate of the large annular viruses previously accumulated by use of monensin. During reversion of this blockade, we observed a progressive decrease in the number of large viruses inside dilated perinuclear vesicles, as well as the formation of secretory vesicles filled with small dense viruses, which were also seen on the cell surface (Fig. 4F). At the immunofluorescence level, the punctate signal characteristic of the formation of secretory vesicles with viruses was also recovered in many cells (Fig. 4G). The data obtained in two independent sets of treatments were also expressed in a quantitative form. The percentages of large and small viruses as well as the percentages of viruses in different cellular compartments with the treatments described are shown in Fig. 5. Control infected cells at 8 h p.i. exhibited a major population of small dense virions (Fig. 5A), most of them inside secretory vesicles and the extracellular cell surface, with a minor population of perinuclear large viral particles (Fig. 5B). Treatment with cycloheximide increased the percentage of small dense viruses on the extracellular cell surface, and a considerable decrease of large perinuclear viral particles was observed (Fig. 5). Disorganization of a functional Golgi by monensin treatment caused the opposite situation: large viral particles accumulated at the perinuclear region (Fig. 5A), while the amount of small viruses inside secretory vesicles significantly decreased, together with the disappearance of the viruses on the cell surface (Fig. 5B). During reversion of the monensin blockade, a recovery of the population of small dense viruses was observed even in the presence of cycloheximide (Fig. 5A). Correspondingly, the amount of perinuclear viral particles decreased, while a higher percentage of viruses accumulated inside secretory vesicles and on the cell surface (Fig. 5B).
FIG. 5.
Quantitative aspects of the effects of the different treatments on TGEV morphogenesis. Quantification included large viruses (LV), small viruses (SV), and those viruses that cannot be included in these two categories (other [O]), which could represent aberrant assemblies or new maturation intermediates. (A) Percentages of viruses with the different morphologies among normally infected (control) cells at 8 h p.i. (C8) and among infected cells subjected to different treatments: infected cells (8 h p.i.) treated with cycloheximide for 30 min (CY), infected cells (8 h p.i.) treated with monensin (MO) (the drug was added to the cultures at 3 h p.i.), and cells after reversion of the monensin blockade (MO REV) that were incubated for 2 h without monensin in the absence of cycloheximide (−CY) or without monensin in the presence of cycloheximide (+CY). (B) Percentages of viral particles in different cellular regions of normally infected cells and in infected cells subjected to the treatments described above. The different cellular locations studied were the perinuclear region (PR), the secretory vesicles (SEC V), and the extracellular cell surface (CS). A total of 4,531 viruses (at least 700 per treatment) were included in the quantification.
The values for viral infectivity in culture supernatants with the different treatments were also determined in duplicate titration assays. While in normally infected cells (8 h p.i.) titration of supernatants rendered a value of 8 × 107 PFU/ml, at the same time p.i. the supernatants from infected and monensin-treated cells had an infectivity of 2 × 105 PFU/ml. At 2 h after removal of monensin from the cultures, infectivity moderately recovered, and the values were 6 × 106 PFU/ml (without cycloheximide) and 4 × 106 PFU/ml (with cycloheximide). These results indicate that infectivity in culture supernatants sharply decreases with the monensin treatment compared with normally infected cells and partially recovers during reversion of the monensin blockade, when transport along the exocytic pathway is partially reestablished. It is also clear that recovery of infectivity is closely related to the presence of small dense virions on the cell surface.
TGEV structural transformation within the Golgi cisternae.
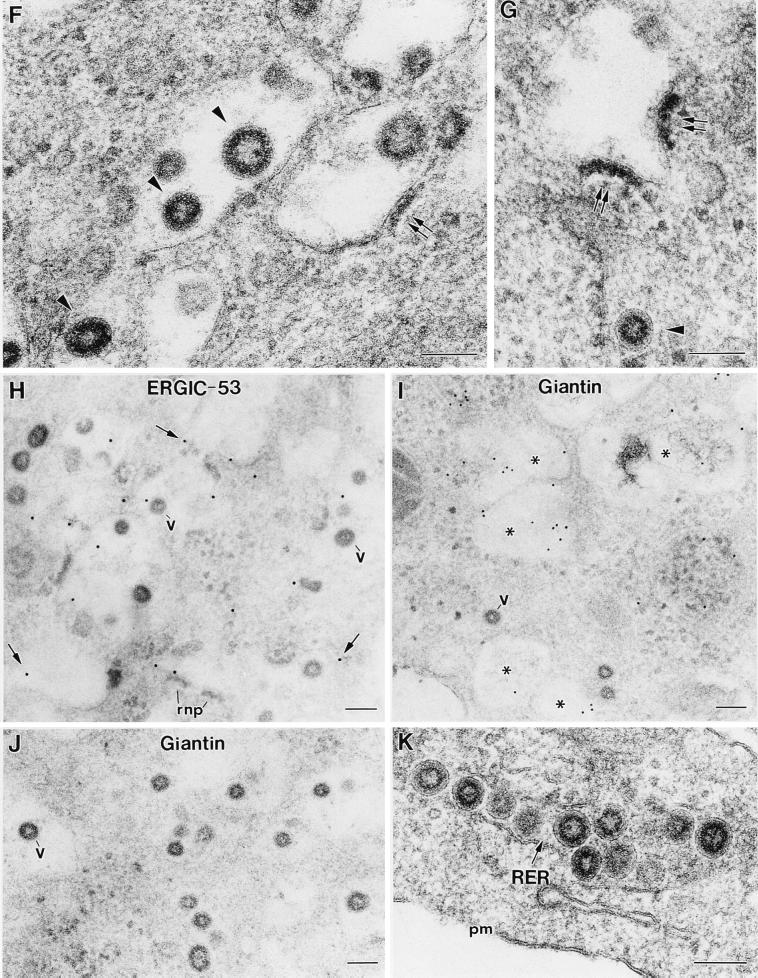
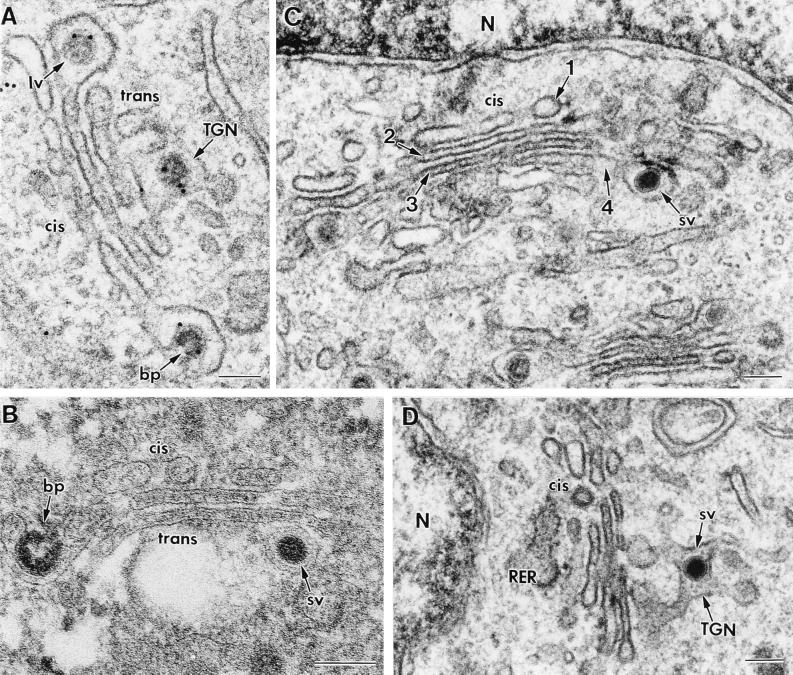
Evidence obtained in a previous study (37) and, mainly, in the present work has clearly shown that a functional (although not necessarily morphologically intact) Golgi complex is necessary for TGEV structural transformation from large to small viral particles. The two viral morphologies were equally represented in the Golgi complex of infected ST cells (37). Here, we have studied the distribution of large and small viruses within the different Golgi subcompartments to determine where the large particles start their transformation. Only Golgi complexes with a clear cis-trans morphology were considered. When many viruses had passed through the Golgi complex, a progressive loss of its fine organization in individual cisternae was observed (not shown). Thus, we studied Golgi complexes from cells infected at early times p.i. (5 and 6 h). After analyzing more than 100 Golgi complexes with viruses, we observed that at the cis side of the Golgi complex, only large annular viruses were seen (Fig. 6A), and very few small dense viral particles were seen inside the third Golgi cisterna (not shown). However, most of the viral particles inside the fourth cisterna and the trans-Golgi network (TGN) were small dense viruses (Fig. 6B to D). This suggests that the transformation from large to small viral particles takes place between the third and the fourth Golgi cisternae.
FIG. 6.
Different viral morphologies within Golgi subcompartments in normally infected cells. Panels A, C, and D show sections from conventionally embedded material, while panel B shows a freeze-substituted sample. All panels show TGEV-infected ST cells at 6 h p.i. In panel A immunogold labeling with the anti-TGEV polyclonal serum and a secondary antibody conjugated to 10-nm-diameter gold particles was also performed. Budding profiles (bp), large viruses (lv), and small viruses (sv) are indicated. The cis and trans sides of the Golgi complex and the TGN are indicated. (A and B) Budding profiles and large viruses are often seen at the cis side of the Golgi stack, where its first cisterna exhibits a characteristic fenestrated structure. Large viral particles are occasionally seen in the TGN (A), although the trans side of the stack usually contains small viruses (B). (C) The four Golgi cisternae are numbered from the cis to the trans side. A small virus is seen in the fourth cisterna. (D) Small viral particles are usually observed in the TGN. N, nucleus. Bars, 100 nm.
DISCUSSION
The characterization of the structure and morphogenesis of coronaviruses is one of the challenging fields in the study of this viral family (26). Previous studies have shown that coronaviruses assemble as large particles of annular morphology at the perinuclear region of the cell, changing their morphology into small dense virions during the transport along the exocytic route (37, 53). More direct evidence of the precursor-product relationship between large and small viral particles was needed, as was confirmation of the key role of the Golgi apparatus in the complex structural transformation of coronaviruses, a process that has not been described before for any other viral family.
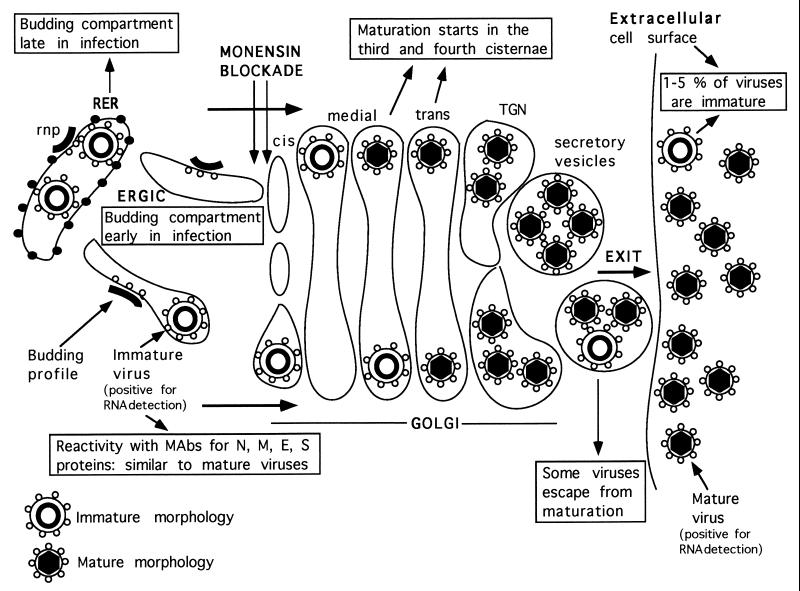
Figure 7 shows a summary of the experimental data obtained so far in the characterization of TGEV morphogenesis. (i) The two types of viral particles seen in TGEV-infected cells are closely related, because they react equally with MAbs specific for the four TGEV structural proteins. It has also been reported that both large and small viral particles studied in situ were positive for RNA detection (33, 37). (ii) TGEV budding takes place in pre-Golgi compartments (the RER or ERGIC, depending on the time p.i.) and creates large viral particles. (iii) These particles change their morphology between the third and fourth Golgi cisternae, creating small dense viruses whose internal cores exhibit polygonal contours (37). These viral particles are collected into secretory vesicles and released to the extracellular environment. (iv) A small percentage of viruses escape from this structural transformation and exit the cell.
FIG. 7.
TGEV morphogenesis in ST cells. This diagram is a summary of the experimental results on TGEV morphogenesis obtained in the present work and in a previous study by Risco et al. (37). The immature and mature morphologies of TGEV viral particles are shown at the bottom. The monensin blockade at a pre-Golgi level is also indicated. Reactivity with MAbs refers to immunolabeling experiments with thin sections of infected cells. Mature virions contain dense cores with polygonal contours, which have been observed in ultrathin sections of freeze-substituted samples (37). They suggest an icosahedral symmetry for these cores, which needs to be confirmed by high-resolution structural studies.
Monensin blockade of TGEV morphogenesis, which, as indicated in Fig. 7, takes place at the pre-Golgi level, has been very useful to obtain evidence for the two main aspects under study: the Golgi complex is necessary for TGEV structural transformation, and large viral assemblies transform into small viruses, as observed after reversion of the monensin blockade. The effects of monensin in different cell types have been extensively characterized. This drug is unique in terms of its specific action on the Golgi complex, while protein synthesis and early steps in intracellular transport proceed at normal rates (50, 51, 55). Monensin has been also used before in virus-infected cells to study several viruses that depend on processes related to the exocytic pathway for completing their morphogenesis. Budding of the Uukuniemi virus, which usually assembles in pre-Golgi and Golgi membranes (18), was effectively inhibited by monensin, although viral nucleocapsids were detected attached to intracellular membranes (24). The viral nucleocapsids of Sindbis virus, which usually assemble at the plasma membrane, change their budding location in the presence of monensin. They assemble at intracellular membranes, although their exit from the infected cell is blocked (20). In the case of herpes simplex virus, monensin does not affect the assembly and envelopment of nucleocapsids, but it drastically inhibits the transport of progeny viruses to the surfaces of the infected cells (21). TGEV, as well as other coronaviruses, assembles at pre-Golgi ERGIC membranes early in infection and in the RER at late times p.i. (Fig. 7). For cells infected with the mouse hepatitis coronavirus (MHVA59) at late times p.i., Niemann et al. (32) showed that monensin did not interfere with the budding of the virus from the membranes of the RER, but it inhibited virus release and fusion of infected cells. In the present work we have shown that TGEV budding and assembly were not inhibited by the drug at early times p.i. Instead, numerous large viral particles were retained in dilated pre-Golgi ERGIC elements of vacuolar morphology, since transport out of this compartment was blocked. A similar structural alteration of the ERGIC was also observed as a consequence of the blockade in transport caused by incubating the cells at 15°C (41). The few small viruses detected in our monensin-treated cells were probably produced before addition of the drug at 3 h p.i. Earlier treatments inhibited assembly almost completely, probably due to a lack of sufficient amounts of correctly processed viral components. Damage in the Golgi or mitochondria, which look abnormally dense in monensin-treated cells, could contribute to this deficiency. During reversion of the monensin blockade, although the Golgi complex recovers its perinuclear location as indicated by immunofluorescence, the Golgi stacks did not recover the fine ultrastructure of untreated cells, as seen by electron microscopy. However, transport of viral particles and release of small TGEV particles were at least partially reestablished. These results agree with previous observations for infected, nocodazole-treated ST cells. Fragmented, but functional, Golgi stacks formed with nocodazole were able to support TGEV morphological transformation (37). The reversion of the monensin blockade in the presence of cycloheximide indicates that the same population of large viral particles arrested in the ERGIC-associated vesicles later transforms into the small dense viruses. As a consequence, we can conclude that the large annular viral particles are immature precursors of the small dense virions and that the described structural transformation takes place in the Golgi complex.
Another interesting observation for monensin-treated cells is that the drug induces the accumulation of large amounts of membrane-associated RNPs and budding profiles at different stages. In normally infected cells these structures are seen in much smaller amounts, probably due to the higher speed of the normal assembly process. The monensin blockade, then, gives us the opportunity for a more detailed study of the very first steps of coronavirus assembly. Working with the reversion of the monensin blockade and with the help of cryomethods, we might be able to study potential intermediate viral forms between precursor and mature viruses.
Which factors associated with the Golgi could be potentially involved in the structural maturation of coronaviruses? Our data indicate that maturation takes place somewhere at the trans side of this compartment. The distributions of both viral morphologies within the different regions of the Golgi complex point to a change in morphology for TGEV between the third and the fourth cisternae (Fig. 7). In the TGN most of the viruses have a mature morphology, and, as seen inside secretory vesicles, a small percentage of large viral particles have escaped from the maturation process. In a previous study (53) small dense viral particles of MHV were seen exclusively in the TGN of infected AtT20 cells. Thus, something present in the trans-most side of the Golgi complex acts on immature coronavirus particles to trigger their structural transformation. Currently, the study of the functional compartmentalization of the Golgi complex is a very dynamic area of research among cell biologists. The trans-most Golgi cisterna and the TGN have exclusive characteristics, such as a low pH and specific enzymes (glyco- and sulfotransferases, phosphatases, and endopeptidases) (1, 15, 48). In particular, furin-type proteases (which are active at low pH) have been found to be involved in the proteolytic processing of alphavirus proteins during their transport through these Golgi subcompartments (30). Viral enzymes also could be involved in maturation. In this regard, the only enzymatic activity associated with coronavirus particles reported to date is a virion-associated protein kinase activity described for the murine coronavirus JHM (46). The origin and role of this kinase activity in the coronavirus life cycle have not been determined. Its potential participation in the phosphorylation of the nucleocapsid protein and virus assembly should be investigated, since phosphorylation and dephosphorylation are important events in the maturation of other viruses (28).
The extremely different organizations of large and small viruses made us think of potential differences in the reactivities of the two types of particles with antibodies specific for different TGEV components. Although these aspects will be analyzed more extensively with purified viruses (to obtain information about changes in the topology of the envelope proteins), the immunocytochemical data that we present here show that there are no significant differences between sectioned large immature and small mature viral particles regarding the reactivity with MAbs specific for the four TGEV structural proteins. E protein, for example, does not seem to be present in larger amounts within the immature viruses. Thus, this protein probably develops its role before the formation of these precursor viruses, where it could already be present in a low number of copies. E protein is a very minor component of the extracellular mature virion, and it is known that it plays a key role in assembly (3, 9, 58). In both MHV and TGEV, virus-like envelopes have been obtained (2, 3, 56). In these studies, it has been demonstrated that the E and M proteins are able to assemble those particles, in which E protein is present in a much higher number of copies (similar to M) than in native viruses (56). Higher-resolution structural studies are necessary to determine if molecules of E protein occupy strategic positions within the budding profiles, the immature viral particles, and the mature viruses. E protein could somehow prepare the membrane or other viral proteins, as proposed for small, acylated glycoproteins from other viruses, such as alphaviruses and orthomyxoviruses (11, 17, 59). The potential role of the incorporation in the viral particles of some other minor components, such as TGEV HP, should be also investigated.
Considerable progress on the knowledge of coronavirus biology has been made in the last decade. However, numerous aspects related to the assembly of coronaviruses remain to be defined. Some of these aspects are (i) the potential participation of proteins encoded by some poorly characterized open reading frames (8, 12, 26, 54), (ii) the role of the posttranslational modifications of viral structural proteins in morphogenesis, (iii) the molecular and structural characterization of the RNA encapsidation process, and (iv) the participation of cellular factors in coronavirus assembly and maturation. Within this context, the characterization of the immature viral assemblies, presently under way, will be fundamental in understanding the nature of coronavirus maturation.
ACKNOWLEDGMENTS
We express our gratitude to Luis Enjuanes for generously providing the MAbs specific for TGEV M, N, and S proteins and the hyperimmune serum against the PUR 46-MAD strain of TGEV. Special thanks go to Hubert Laude and David Brian for their support to this work by providing the anti-E MAbs and the anti-HP antiserum, respectively, and for their constructive comments. We are also grateful to Hans P. Hauri and Manfred Renz for kindly providing the anti-ERGIC-53 MAb and the antigiantin antiserum, respectively.
I.J.S. is the recipient of a fellowship for postgraduate students from the Gobierno Vasco. This work has been supported by grant PB96-0818 from the Comisión Interministerial de Ciencia y Tecnología of Spain (to J.L.C.).
REFERENCES
- 1.Anderson R G W, Pathak R K. Vesicles and cisternae in the trans Golgi apparatus of human fibroblasts are acidic compartments. Cell. 1985;40:635–643. doi: 10.1016/0092-8674(85)90212-0. [DOI] [PubMed] [Google Scholar]
- 2.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos E C W, Luytjes W, Van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castón J R, Trus B L, Booy F P, Wickner R B, Wall J S, Steven A C. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compton S R, Rogers D B, Holmes K V, Fertsch D, Remenick J, McGowan J J. In vitro replication of mouse hepatitis virus strain A59. J Virol. 1987;61:1814–1820. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Haan C A M, Kuo L, Masters P S, Vennema H, Rottier P J M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enjuanes L, Van der Zeijst B A M. Molecular basis of the transmissible gastroenteritis virus epidemiology. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 337–376. [Google Scholar]
- 8.Fischer F, Peng D, Hingley S T, Weiss S R, Masters P S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 11.Gaedigk-Nitschko K, Ding M, Levy M A, Schlesinger M J. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology. 1990;175:282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 12.Garwes D J, Stewart F, Britton P. The polypeptide of Mr 14000 of porcine transmissible gastroenteritis virus: gene assignment and intracellular location. J Gen Virol. 1989;70:2495–2499. doi: 10.1099/0022-1317-70-9-2495. [DOI] [PubMed] [Google Scholar]
- 13.Gebauer F, Posthumus W A P, Correa I, Suñé C, Sánchez C M, Smerdou C, Lenstra J A, Meloen R, Enjuanes L. Residues involved in the formation of the antigenic sites of the S protein of transmissible gastroenteritis coronavirus. Virology. 1991;183:225–238. doi: 10.1016/0042-6822(91)90135-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godet M, L’Haridon R, Vautherot J F, Laude H. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths G, Simons K. The trans Golgi network: sorting at the exit site of the Golgi complex. Science. 1986;234:438–443. doi: 10.1126/science.2945253. [DOI] [PubMed] [Google Scholar]
- 16.Hansen G H, Delmas B, Besnardeau L, Vogel L K, Laude H, Sjöström H, Norén O. The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment. J Virol. 1998;72:527–534. doi: 10.1128/jvi.72.1.527-534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova L, Lustig S, Schlesinger M J. A pseudo-revertant of a Sindbis virus 6K protein mutant, which corrects for aberrant particle formation, contains two new mutations that map to the ectodomain of the E2 glycoprotein. Virology. 1995;206:1027–1034. doi: 10.1006/viro.1995.1025. [DOI] [PubMed] [Google Scholar]
- 18.Jäntti J, Hildén P, Rönka H, Mäkiranta V, Keränen S, Kuismanen E. Immunocytochemical analysis of Uukuniemi virus budding compartments: role of the intermediate compartment and the Golgi stack in virus maturation. J Virol. 1997;71:1162–1172. doi: 10.1128/jvi.71.2.1162-1172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiménez G, Correa I, Melgosa M P, Bullido M J, Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson D C, Schlesinger M J. Vesicular stomatitis virus and Sindbis virus glycoprotein transport to the cell surface is inhibited by ionophores. Virology. 1980;103:407–424. doi: 10.1016/0042-6822(80)90200-7. [DOI] [PubMed] [Google Scholar]
- 21.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from the infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapke P A, Brian D A. Sequence analysis of the porcine transmissible coronavirus nucleocapsid protein gene. Virology. 1986;151:41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krijnse-Locker J, Ericsson M, Rottier P J, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuismanen E, Saraste J, Pettersson R F. Effect of monensin on the assembly of Uukuniemi virus in the Golgi complex. J Virol. 1985;55:813–822. doi: 10.1128/jvi.55.3.813-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai M M C. RNA-protein interactions in the regulation of coronavirus RNA replication and transcription. Biol Chem. 1997;378:477–481. doi: 10.1515/bchm.1997.378.6.477. [DOI] [PubMed] [Google Scholar]
- 26.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N, Brown D T. Phosphorylation and dephosphorylation events play critical roles in Sindbis virus maturation. Virology. 1993;196:703–711. doi: 10.1006/viro.1993.1527. [DOI] [PubMed] [Google Scholar]
- 29.MacIntosh K. Coronaviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1095–1133. [Google Scholar]
- 30.Moehring J M, Inocencio N M, Robertson B J, Moehring T J. Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and viruses complements the genetic lesion. J Biol Chem. 1993;268:2590–2594. [PubMed] [Google Scholar]
- 31.Molenkamp R, Spaan W J M. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemann H, Boschek B, Evans D, Rosing M, Tamura T, Klenk H-D. Post-translational glycosylation of coronavirus glycoprotein E1: inhibition by monensin. EMBO J. 1982;1:1499–1504. doi: 10.1002/j.1460-2075.1982.tb01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintana C, Marco S, Bonnet N, Risco C, Gutiérrez M L, Guerrero A, Carrascosa J L. Optimization of phosphorus localization by EFTEM of nucleic acid containing structures. Micron. 1998;29:297–307. doi: 10.1016/S0968-4328(98)00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risco C, Antón I M, Suñé C, Pedregosa A M, Martín-Alonso J M, Parra F, Carrascosa J L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risco C, Menéndez-Arias L, Copeland T D, Pinto da Silva P, Oroszlan S. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of the nucleocapsid protein and integrase. J Cell Sci. 1995;108:3039–3050. doi: 10.1242/jcs.108.9.3039. [DOI] [PubMed] [Google Scholar]
- 36.Risco C, Antón I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Risco C, Muntión M, Enjuanes L, Carrascosa J L. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J Virol. 1998;72:4022–4031. doi: 10.1128/jvi.72.5.4022-4031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risco C, Carrascosa J L. Visualization of viral assembly in the infected cell. Histol Histopathol. 1999;14:905–926. doi: 10.14670/HH-14.905. [DOI] [PubMed] [Google Scholar]
- 39.Ross N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffith G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 40.Sánchez C M, Jiménez G, Laviada M D, Correa I, Suñé C, Bullido M J, Gebauer F, Smerdou C, Callebaut P, Escribano J M, Enjuanes L. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saraste J, Kuismanen E. Pathways of protein sorting and membrane traffic between the rough endoplasmic reticulum and the Golgi complex. Semin Cell Biol. 1992;3:343–355. doi: 10.1016/1043-4682(92)90020-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer A, Fransen J A M, Baechi T, Ginsel L, Hauri H P. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer A, Fransen J A M, Matter K, Kreis T E, Ginsel L, Hauri H-P. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- 44.Seelig H P, Schranz P, Schroter H, Wiemann C, Griffiths G, Renz M. Molecular and genetic analysis of a 376-kilodalton Golgi complex membrane protein (giantin) Mol Cell Biol. 1994;14:2564–2576. doi: 10.1128/mcb.14.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddell S G, Barthel A, ter Meulen V. Coronavirus JHM: a virion-associated protein kinase. J Gen Virol. 1981;52:235–243. doi: 10.1099/0022-1317-52-2-235. [DOI] [PubMed] [Google Scholar]
- 47.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, vant’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sossin W S, Fisher J M, Scheller R H. Sorting within the regulated secretory pathway occurs in the trans-Golgi network. J Cell Biol. 1990;110:1–12. doi: 10.1083/jcb.110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steven A C, Trus B L, Booy F P, Cheng N, Zlotnick A, Castón J R, Conway J F. The making and breaking of symmetry in virus capsid assembly: glimpses of capsid biology from cryoelectron microscopy. FASEB J. 1997;11:733–742. doi: 10.1096/fasebj.11.10.9271358. [DOI] [PubMed] [Google Scholar]
- 50.Tartakoff A M. Perturbation of vesicular traffic with the carboxylic ionophore monensin. Cell. 1983;32:1026–1028. doi: 10.1016/0092-8674(83)90286-6. [DOI] [PubMed] [Google Scholar]
- 51.Tartakoff A M, Vassalli P. Plasma cell immunoglobulin secretion. Arrest is accompanied by alterations of the Golgi complex. J Exp Med. 1977;146:1332–1345. doi: 10.1084/jem.146.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 53.Tooze J, Tooze S A, Fuller S D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tung F Y T, Abraham S, Sethna M, Hung S-L, Sethna P, Hogue B G, Brian D A. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology. 1992;186:676–683. doi: 10.1016/0042-6822(92)90034-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchida N, Smilowitz H, Tanzer M L. Monovalent ionophores inhibit secretion of procollagen and fibronectin from cultured human fibroblasts. Proc Natl Acad Sci USA. 1979;76:1868–1872. doi: 10.1073/pnas.76.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opsteltein D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- 59.Zebedee S L, Lamb R A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]