Abstract
Theiler’s virus causes a persistent infection and a demyelinating disease of mice which is a model for multiple sclerosis. Susceptibility to viral persistence maps to several loci, including the interferon gamma locus. Inactivating the gene coding for the interferon gamma receptor makes 129/Sv mice susceptible to persistent infection and clinical disease, whereas inactivating the interferon gamma gene makes C57BL/6 mice susceptible to persistent infection but not to clinical disease. This difference in phenotype is due to the difference in genetic background. Clinical disease depends on high viral load and Tmevd5, a locus on chromosome 11. These results have consequences for the identification of viruses which might be implicated in multiple sclerosis.
The DA strain of Theiler’s virus, a picornavirus, is responsible for a chronic demyelinating disease in genetically susceptible mice (12, 22). This natural infection is one of the best models for multiple sclerosis. During the first 2 weeks following intracranial inoculation, the virus causes an encephalomyelitis regardless of the genetic background of the mouse. Neurons of the brain and spinal cord are the main cells infected at this stage. All mice recover, although some of them may present with sequellae, such as flaccid paralysis of the hind legs (38). In genetically susceptible strains of mice, such as the SJL/J strain, this early disease is followed by a lifelong persistent infection of the white matter of the spinal cord. At this stage, the virus infects predominantly macrophage and microglial cells but also oligodendrocytes and possibly astrocytes (1, 10, 30, 31, 33). Persistence is associated with chronic inflammation and primary demyelination, which may cause gait disorders and spastic paralysis, particularly when the mice are inoculated with a high dose of virus (105 or 2 × 106 PFU). In contrast, genetically resistant mouse strains, such as the C57BL/6 and 129/Sv strains, eliminate the virus after early gray matter encephalomyelitis. Inflammation and demyelination does not occur in these mice.
Persistence of infection and demyelination are under the control of several host genes (3, 4, 37). The haplotype of the major histocompatibility complex class I H-2D gene has a major effect on viral persistence (2, 6, 24, 31, 34). H-2Db strains, such as C57BL/6 and 129/Sv, are resistant, and their resistance is dominant. The SJL/J strain, however, is more susceptible to viral persistence than predicted by its H-2Ds haplotype (6). A locus on chromosome 10, close to the Ifng locus, explains most of the susceptibility of this strain (5). The role of the interferon gamma pathway in the persistence of the infection was tested by inoculating 129/Sv mice in which the gene coding for the interferon gamma receptor had been inactivated (129/Sv Ifngr−/− mice). These mice were highly susceptible to persistent infection and presented with very severe neurological symptoms (16). The role of this pathway can also be tested by using C57BL/6 Ifng−/− mice, which are now available. They offer the possibility of testing the role of the cytokine itself in the resistance of an H-2b mouse strain. In the present article, we report that these mice were persistently infected, but at a low level, and that they did not show any clinical signs. We also show that most of the difference of susceptibility between the 129/Sv Ifngr−/− and the C57BL/6 Ifng−/− strains is due to the difference of genetic background. A complete genome scan of two F2 crosses showed that clinically apparent disease results from two independent factors: high viral load and a locus on chromosome 11, which we called Tmevd5 (for Theiler’s murine encephalomyelitis virus demyelination locus 5). The data indicate that Tmevd5 modifies the risk of clinical disease in mice infected at a high level.
MATERIALS AND METHODS
Animals.
C57BL/6 mice were purchased from Janvier (Le Genest-St-Isle, France), and 129/Sv mice were purchased from the Institut Pasteur animal facility. C57BL/6 Ifng−/− mice were obtained from the Jackson Laboratory (Bar Harbor, Maine). 129/Sv Ifngr−/− mice were provided by Michel Aguet (Institut Suisse de Recherches Expérimentales sur le Cancer, Lausanne, Switzerland). These mice have been described previously (11, 19). F2 Ifng−/− and F2 Ifngr−/− mice (see Results for the definition of these crosses) were bred at the Institut Pasteur animal facility. The mice were genotyped for Ifng or Ifngr by PCR amplification with DNA extracted from tail biopsies. The following sets of primers were used to distinguish between Ifng−/−, Ifng+/−, and wild-type mice: the first (5′-AGAAGTAAGTGGAAGGGCCCAGAAG-3′ and 5′-AGGGAAACTGGGAGAGGAGAAATAT-3′) set of primers amplified a 220-bp product from the Ifng gene, and the second (5′-TCAGCGCAGGGGCGCCCGGTTCTTT-3′ and 5′-ATCGACAAGACCGGCTTCCATCCGA-3′) set of primers amplified a 375-bp product from the 2-kb neomycin resistance gene inserted into exon 2 of the Ifng gene. Amplification was performed with a Gene Amp kit (Bio-Rad Laboratories, Gaithersburg, Md.) and a 9600 reactor (Perkin-Elmer Cetus, Norwalk, Conn.). After denaturation at 94°C for 2 min, 30 cycles of DNA amplification were performed under the following conditions for the first set of primers: 94°C for 40 s, 60°C for 40 s, and 72°C for 15 s. For the second set of primers, the annealing temperature was 65°C instead of 60°C. The following primers were used to distinguish between Ifngr−/−, Ifngr+/−, and wild-type mice: 5′-CCCATTTAGATCCTACATACGAAACATACGG-3′ and 5′-TTTCTGTCATCATGGAAAGGAGGGATACAG-3′. These primers are located in the Ifngr gene, upstream and downstream, respectively, from the neomycin cassette. Amplification was carried out under the following conditions: 94°C for 2 min followed by 40 cycles at 94°C for 40 s, 55°C for 40 s, and 72°C for 2 min.
Viral inoculation.
The DA strain of Theiler’s virus was produced by transfection of BHK-21 cells with the pTMDA plasmid as described elsewhere (26). Three- to 4-week-old anesthetized mice were inoculated intracranially with 104 PFU of the DA strain in 40 μl of phosphate buffered saline (PBS). All mice were observed once or twice a week to record clinical signs and mortality. They were sacrificed at 6, 21, or 45 days postinoculation (p.i.), depending on the experiment.
Histological analysis.
Anesthetized mice were perfused through the left ventricle with PBS followed by 4% paraformaldehyde in PBS. The spinal cords were dissected out, postfixed, embedded in paraffin, and sectioned as previously described (1). Detection of Theiler’s virus capsid antigen in the paraffin sections was carried out with a primary rabbit hyperimmune anti-capsid serum, a secondary biotinylated goat anti-rabbit immunoglobulin, and the ABC peroxidase detection system (labelled streptavidin biotin [LSAB] peroxidase; Dako). Slides were counterstained with Mayer hematoxylin.
Extraction of RNA from the spinal cord and quantification of viral RNA.
The assay has been described in detail elsewhere (6). Briefly, mice were anesthetized and perfused with 20 ml of PBS. Their spinal cords were removed, and total RNA was extracted by the procedure of Chomczynski and Sacchi (8). For each mouse, series of fivefold dilutions, starting with 10 μg of total RNA, were dot blotted on Hybond C-extra filters (Amersham Corp., Arlington Heights, Ill.). The blots were hybridized with a 32P-labeled cDNA probe (specific activity, greater than 108 cpm/μg) specific for the 5′ extremity of the Theiler’s virus genome. The hybridized filters were analyzed with a phosphorimager. For each sample, the highest dilution which gave a positive hybridization signal was used to estimate the viral RNA content. The results were expressed as a dilution factor that ranged from 1 to 625 or, to perform statistical analyses, as a score ranging from 0 to 4 (see Fig. 1 and 3 for examples of the relationship between the dilution factor and the score). The data were normalized by two procedures. First, a duplicate blot was hybridized with a β-actin-specific probe to correct, if necessary, for variations in the amount of total RNA. Second, reference samples from previous blots were repeatedly analyzed to adjust for small variations of hybridization efficiency from blot to blot. In most cases, quantification was performed by visual observation of a phosphorimager printout. For the F2 Ifng−/− cross, quantification was also done by image analysis. Both methods gave essentially identical results.
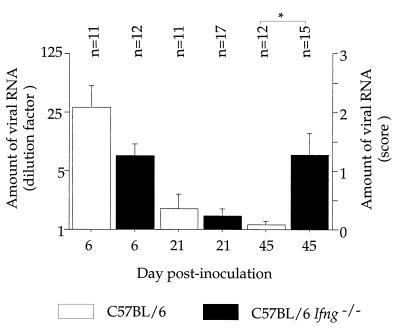
FIG. 1.
Amount of viral RNA in the central nervous system at different times p.i. The amount of viral RNA was measured in the brain on day 6 p.i. and in the spinal cord on days 21 and 45 p.i. This amount is expressed as the highest RNA dilution which gave a hybridization signal (left ordinate) or the score of viral persistence (right ordinate). n, number of animals per group. ∗, P < 0.05. The error bars indicate the SEM.
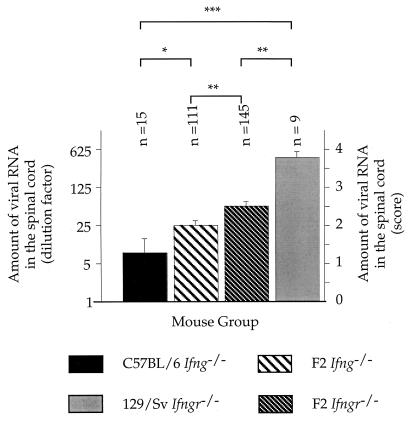
FIG. 3.
Amount of viral RNA 45 days p.i. in the spinal cord of the two F2 crosses and their parental strains. The amount of viral RNA is expressed as the highest RNA dilution (+ SEM) which gave a hybridization signal (left ordinate) or the score of viral persistence (right ordinate). n, number of animals per group. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
Genetic analysis.
DNA was extracted from tail biopsies by standard techniques (5). Mouse genotypes were determined with polymorphic microsatellite markers. The sequences of the PCR primers used are available online (13a). The primers were synthesized at Research Genetics, Genset, or the Institut Pasteur oligonucleotide facility. Amplifications were performed as previously described (28). The optimal annealing temperature varied from 50 to 55°C. The PCR products were visualized on 5% agarose gels stained with ethidium bromide. Genetic maps were constructed with the Mapmanager program (25). Depending on the F2 cross and the markers, 91 to 151 mice were used to calculate genetic distances. Exclusion mapping and localization of susceptibility loci were performed with the Mapmaker/QTL program (21), and the Kosambi function was used to calculate genetic distances.
Statistical analysis.
The mean and standard error of the mean (SEM) of the score of viral persistence were computed for the three genotypes at each locus. Means were compared by using analysis of variance from the Statview F-4.5 package. To account for deviation of the persistence score from the Laplace-Gauss distribution, an empirical significance level was obtained by a Monte Carlo method (data not shown). The F distribution was evaluated in two simulations of 20,000 random replicates, each under the assumption of no linkage between the genotype and the phenotype. For the simulations, persistence scores were those observed in the experiment and genotypes were randomly assigned to members of the F2 cross. For each of the two crosses, the simulated distribution was very close to the F distribution of the table, and this distribution was used during the remainder of the study. The linkage between clinical signs and each locus was tested by a χ2 test. Dates of appearance of clinical signs were studied as a function of genotypes with the Logrank test of the nonparametric Kaplan-Meier analysis from the Statview F-4.5 package. Statistical criteria for the genetic analysis were those from Lander and Kruglyak (20). Two levels of significance were used: suggestive linkage, when the probability of linkage by chance was less than 1.6 × 10−3 (decimal logarithm of the odds [LOD] score, >2.8), and significant linkage, when the probability of linkage by chance was less than 5.2 × 10−5 (LOD score, >4.3).
RESULTS
C57BL/6 mice become susceptible to persistent infection with Theiler’s virus after inactivation of the Ifng gene.
C57BL/6 Ifng−/− and wild-type C57BL/6 mice were inoculated intracranially with 104 PFU of the DA strain and observed for 45 days to record clinical signs and mortality. Some mice were sacrificed at 6, 21, or 45 days p.i. Their brains and spinal cords were dissected out to quantify viral persistence by a dot blot assay or to localize viral antigens by immunocytochemistry.
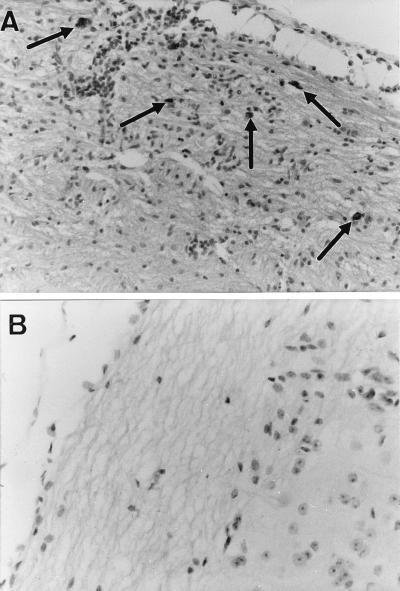
Six days p.i., the level of infection in the brain was lower in C57BL/6 Ifng−/− mice than in wild-type mice (mean value for B6Ifng−/− [mB6Ifng−/−] = 1.25 ± 0.2 [n = 12]; mB6 = 2.1 ± 0.4 [n = 11]), although the difference was not statistically significant (NS) (P = 0.0524) (Fig. 1). Twenty-one days p.i., the levels of viral RNA in the spinal cord were low and very similar in the two groups (mB6Ifng−/− = 0.2 ± 0.1 [n = 17]; mB6 = 0.4 ± 0.2 [n = 11]; NS) (Fig. 1). However, the locations of infected cells were different in the two groups. Infected cells were found in the white matter of the spinal cord, in the vicinity of the central canal, in three of seven C57BL/6 Ifng−/− mice examined. In contrast, a small number of infected cells were found in the gray matter of the spinal cord of two of nine C57BL/6 mice, close to the central canal (data not shown). Forty-five days p.i., the level of infection in the spinal cord was very low in the C57BL/6 control mice, as expected. In contrast, significant amounts of viral RNA were present in the spinal cords of C57BL/6 Ifng−/− mice (Fig. 1). The levels of infection were significantly different in the two strains (mB6Ifng−/− = 1.3 ± 0.4 [n = 15]; mB6 = 0.1 ± 0.1 [n = 12]; P = 0.0115). Immunocytochemistry did not detect viral antigens in the spinal cords of three wild-type mice examined, although there was mild inflammation in the gray matter (Fig. 2). In contrast, infected cells were found in the white matter of the spinal cord in 8 of 11 C57BL/6 Ifng−/− mice. Furthermore, this infection was associated with inflammation such as perivascular cuffs, meningitis, and diffuse parenchymal infiltration (Fig. 2). None of the C57BL/6 wild-type and C57BL/6 Ifng−/− mice showed clinical signs or died during this study.
FIG. 2.
Longitudinal sections of spinal cord from a C57BL/6 Ifng−/− mouse (A) and a wild-type C57BL/6 mouse (B) with Theiler’s virus 45 days p.i. The arrows indicate infected cells detected by immunocytochemistry. Magnification, ×200 (A) and ×320 (B).
In a previous work, we reported that 129/Sv Ifngr−/− mice were highly susceptible to persistent infection with Theiler’s virus and died from extensive white matter disease before 45 days p.i. (16). We repeated these experiments to compare C57BL/6 Ifng−/− and 129/Sv Ifngr−/− mice under the same conditions. Forty-five days p.i., the level of viral RNA was significantly lower in C57BL/6 Ifng−/− mice than in 129/Sv Ifngr−/− mice (mB6Ifng−/− = 1.3 ± 0.4 [n = 15]; m129Ifngr−/− = 3.8 ± 0.1 [n = 9]; P < 0.0001) (Fig. 3). At that time, seven of nine 129/Sv Ifngr−/− mice had developed hind-leg paralysis whereas no symptoms were observed in 26 C57BL/6 Ifng−/− mice (Table 1). No mortality was observed in either group of knockout mice.
TABLE 1.
Clinical disease for the different groups of null-mutant mice
| Mouse group | No. of mice | Morbidity [no. (%)] | Mortality [no. (%)] |
|---|---|---|---|
| 129/Sv Ifngr−/− | 9 | 7 (78) | 0 (0) |
| C57BL/6 Ifng−/− | 26 | 0 (0) | 0 (0) |
| F2Ifng−/−a | 117 | 17 (14.5) | 3 (2.5) |
| F2Ifngr−/−b | 151 | 39 (26) | 6 (4) |
F2 Ifng−/− is short for (C57BL/6 Ifng−/− × 129/Sv) F2 Ifng−/−.
F2 Ifngr−/− is short for (C57BL/6 × 129/Sv Ifngr−/−) F2 Ifngr−/−.
Modifier genes controlling virus load and clinical signs explain differences between C57BL/6 Ifng−/− and 129/Sv Ifngr−/− mice.
As discussed above, C57BL/6 Ifng−/− mice are less susceptible to persistent infection with Theiler’s virus and to the associated clinical symptoms than 129/Sv Ifngr−/− mice. This could be due to the fact that the gene which is inactivated codes for the cytokine in the former strain whereas it codes for the cytokine receptor in the latter. Alternatively, it could be due to the difference in genetic background between the two strains. Because null mutants with the same genetic background were not available, we tested these hypotheses by comparing two F2 crosses (C57BL/6 Ifng−/− × 129/Sv) F2 Ifng−/− and (129/Sv Ifngr−/− × C57BL/6) F2 Ifngr−/−, for level of viral persistence and appearance of symptoms. (For the sake of simplicity, these crosses will be designated F2 Ifng−/− and F2 Ifngr−/−, respectively, in the rest of this article). If the difference in susceptibility were due to the functional differences between interferon gamma and its receptor, the F2 Ifng−/− and C57BL/6 Ifng−/− mice should have the same phenotype and the F2 Ifngr−/− and 129/Sv Ifngr−/− mice should also have the same phenotype. If, on the other hand, the difference in susceptibility were due to the difference in genetic background, both crosses should have similar phenotypes, with large variations between animals.
One hundred and seventeen F2 Ifng−/− and 151 F2 Ifngr−/− animals were inoculated and monitored for clinical symptoms. After 45 days, the level of viral persistence was measured for each mouse. As shown in Fig. 3, both groups of F2 mice were infected at a significantly higher level than the C57BL/6 Ifng−/− strain (P = 0.0405 and P = 0.0014, respectively, for F2 Ifng−/− and F2 Ifngr−/−) and at a significantly lower level than the 129/Sv Ifngr−/− strain (P < 0.0001 and P = 0.0077, respectively, for F2 Ifng−/− and F2 Ifngr−/−). In contrast to the C57BL/6 Ifng−/− mice, which remained asymptomatic, 14.5 and 26% of the F2 Ifng−/− and F2 Ifngr−/− mice, respectively, showed paralysis. Mortality was 2.5 and 4%, respectively (Table 1). The incidence of clinical disease was lower for both F2 crosses than for the 129/Sv Ifngr−/− strain (78%). Taken together, these results imply that the difference in susceptibility between the C57BL/6 Ifng−/− and the 129/Sv Ifngr−/− mice is due mainly to the difference in genetic background. We cannot rule out a contribution of the difference in mutation, since the F2 Ifngr−/− mice were infected at a slightly, but significantly, higher level than F2 Ifng−/− mice (P = 0.0039) and showed more clinical symptoms (P = 0.0241). Only males contributed to the difference in susceptibility between the F2 crosses (mF2Ifngr−/−male = 2.7 ± 0.1 [n = 81]; mF2Ifng−/−male = 2.0 ± 0.20 [n = 57]; P = 0.0008; mF2Ifngr−/−female = 2.2 ± 0.2 [n = 53]; mF2Ifng−/−female = 2.1 ± 0.2 [n = 53]; NS).
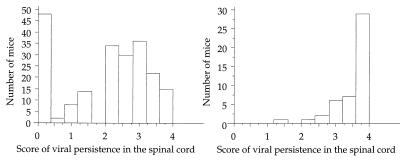
Clinical symptoms appeared at the same time in the two groups of F2 mice (mean time of appearance, 37 days p.i.). Interestingly, in both groups, clinical signs were associated with a high level of persistent infection: 42 of 46 mice with paralysis had a persistence score higher than 3 (Fig. 4).
FIG. 4.
Amount of viral RNA at 45 days p.i. in the spinal cords of asymptomatic (left) and symptomatic (right) mice.
Localization of loci controlling viral persistence.
The 145 mice of the F2 Ifngr−/− cross and the 111 mice of the F2 Ifng−/− cross were genotyped in an attempt to map loci controlling viral persistence. The entire genome was screened with 99 and 108 microsatellite markers, respectively. Two to eight loci were analyzed for each chromosome, depending on the size of the chromosome and on the presence or absence of a putative linkage. The distance between adjacent loci varied from 1 centi-Morgan (cM) to 27 cM. No marker was linked with viral persistence, even at the suggestive-linkage level, in either cross. However, in the F2 Ifng−/− cross, weak linkage with viral persistence was observed on chromosome 19, close to D19Mit123 (P = 0.0028), on chromosome 15 between D15Mit154 (P = 0.0059) and D15Mit156 (P = 0.0043), and on chromosome 12 between D12Mit218 (P = 0.0029) and D12Mit106 (P = 0.0094). These results were not confirmed with the F2 Ifngr−/− cross.
Localization of loci controlling clinical disease.
The number of clinically affected mice (n = 39) was large enough in the F2 Ifngr−/− cross to warrant looking for linkage with polymorphic markers. This was not the case for the F2 Ifng−/− cross (n = 17). The F2 Ifngr−/− mice were separated into two groups, affected and nonaffected, regardless of the time of appearance of the symptoms. Linkage was tested at each locus by comparing the numbers of affected and nonaffected mice as a function of genotype with a χ2 test. There was significant linkage with clinical disease on chromosome 11 for marker D11Mit179 (P < 0.0001; LOD score = 4.8) and suggestive linkage with the two flanking markers, D11Mit4 and D11Mit99 (P = 0.0008, LOD score = 3.1, and P = 0.0012, LOD score = 2.9, respectively) (Table 2). In the nonaffected group, alleles segregated randomly. Susceptibility to clinical disease was conferred by the C57BL/6 allele at the D11Mit179 locus. Indeed, among the 39 affected mice, 26 were homozygous for the C57BL/6 allelic form, 5 were homozygous for the 129/Sv allelic form, and 8 were heterozygous. This result was confirmed with the Logrank test of the nonparametric Kaplan-Meier survival analysis (χ2 = 24.468 [2 df]; P < 0.0001).
TABLE 2.
Association of chromosome 11 markers with clinical disease
| Locus
|
No. of affected miceb
|
No. of nonaffected miceb
|
P value | LOD scorec | |||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Location (cM)a | B6 | B6/129 | 129 | B6 | B6/129 | 129 | ||
| D11Mit62 | 1 | 11 | 21 | 7 | 19 | 56 | 27 | 0.7300 | |
| D11Mit21 | 21.5 | 15 | 21 | 3 | 27 | 62 | 23 | 0.0852 | |
| D11Mit4 | 33.4 | 23 | 10 | 6 | 29 | 57 | 26 | 0.0008 | 3.1 |
| D11Mit179 | 41.1 | 26 | 8 | 5 | 28 | 58 | 26 | <0.0001 | 4.8 |
| D11Mit99 | 47.7 | 22 | 13 | 4 | 28 | 55 | 29 | 0.0012 | 2.9 |
| D11Mit258 | 52.6 | 20 | 15 | 4 | 29 | 47 | 36 | 0.0040 | |
Distance from the D11Mit74 marker.
B6, homozygous C57BL/6 allele; B6/129, heterozygous allele; 129, homozygous 129/Sv allele.
The LOD score is indicated only for suggestive or significant linkage.
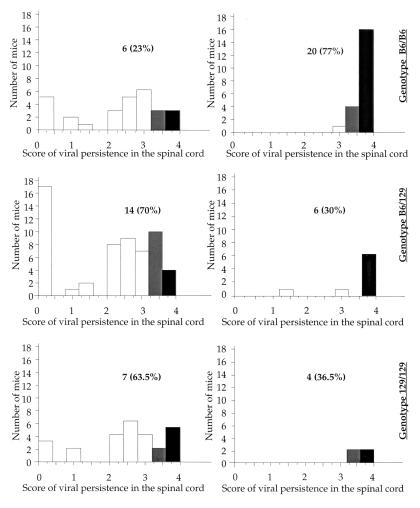
The results reported in a previous section of this paper suggested the existence of a relationship between clinical symptoms and a high level of viral persistence. To examine the relationship between clinical signs, viral persistence, and genotype at the D11Mit179 locus, the amounts of viral RNA in the nonaffected and affected mice were compared as a function of genotype (Fig. 5). Clinical signs were associated with a high level of persistent infection regardless of the genotype. However, when one compares mice with high viral loads (with scores equal or superior to 3.5), 77% of the C57BL/6 homozygous mice were affected as opposed to 30% of the heterozygous mice and 36.5% of the 129/Sv homozygous mice (Fig. 5). Interestingly, there was only weak linkage between viral persistence and the D11Mit179 marker (P = 0.0167). In conclusion, our results strongly suggest the presence on chromosome 11 of a locus, which we named Tmevd5, which controls the severity of clinical disease but not the viral load.
FIG. 5.
Amount of viral RNA 45 days p.i. in the spinal cords of asymptomatic (left-hand graphs) and symptomatic (right-hand graphs) mice as a function of genotype at the D11Mit179 locus. For each group of animals, the absolute number and the percentage of mice with a score of viral persistence equal or superior to 3.5 are given. These animals are indicated by hatched and solid boxes, respectively. Six asymptomatic mice died before 45 days p.i. (five were C57BL/6 homozygous, and one was 129/Sv homozygous for the D11Mit179 marker).
In the F2 Ifngr−/− cross, males had 2.2 times more viral RNA in their spinal cords than females. Therefore, we decided to study susceptibility to clinical signs in males and females separately. When doing so, two markers, D19Mit65 and D19Mit119, had suggestive linkage with disease only in females (P = 0.0011 and P = 0.0013, respectively) (Table 3). The segregation of alleles in males and in nonaffected females was random, but in affected females there was a deficit in heterozygous animals. Therefore, there might be another locus, on chromosome 19, which controls clinical symptoms.
TABLE 3.
Association of chromosome 19 markers with clinical disease
| Sex of mouse | Locus
|
No. of affected miceb
|
No. of nonaffected miceb
|
P value | LOD scorec | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Location (cMa) | B6 | B6/129 | 129 | B6 | B6/129 | 129 | |||
| Female | D19Mit16 | 15 | 5 | 6 | 4 | 5 | 31 | 15 | 0.0758 | |
| D19Mit65 | 35.9 | 10 | 1 | 4 | 10 | 24 | 17 | 0.0011 | 2.9 | |
| D19Mit119 | 38.2 | 10 | 1 | 4 | 10 | 22 | 19 | 0.0013 | 2.9 | |
| D19Mit83 | 42.8 | 9 | 1 | 5 | 9 | 22 | 20 | 0.0023 | 2.6 | |
| D19Mit123 | 46.8 | 9 | 2 | 4 | 10 | 23 | 18 | 0.0070 | ||
| D19Mit1 | 59.8 | 9 | 2 | 4 | 12 | 22 | 17 | 0.0199 | ||
| D19Mit137 | 56.6 | 8 | 3 | 4 | 12 | 24 | 15 | 0.0638 | ||
| Male | D19Mit16 | 15 | 7 | 11 | 6 | 15 | 25 | 21 | 0.6988 | |
| D19Mit65 | 35.9 | 7 | 9 | 8 | 14 | 28 | 19 | 0.7503 | ||
| D19Mit119 | 38.2 | 8 | 8 | 8 | 14 | 28 | 19 | 0.5002 | ||
| D19Mit83 | 42.8 | 8 | 8 | 8 | 15 | 28 | 18 | 0.5457 | ||
| D19Mit123 | 46.8 | 7 | 12 | 5 | 13 | 30 | 18 | 0.6272 | ||
| D19Mit1 | 49.8 | 7 | 13 | 4 | 12 | 31 | 18 | 0.4007 | ||
| D19Mit137 | 56.6 | 6 | 14 | 4 | 11 | 36 | 14 | 0.6905 | ||
Distance from the D19Mit32 marker.
B6, homozygous C57BL/6 allele; B6/129, heterozygous allele; 129, homozygous 129/Sv allele.
The LOD score is indicated only for suggestive or significant linkage.
DISCUSSION
In a previous work, we used a cross between the SJL/J and B10.S strains to identify a locus, close to Ifng, which controls the persistence of Theiler’s virus in the spinal cord (5). Because of the antiviral and immunoregulatory properties of interferon gamma, the Ifng gene was a good candidate for the control of viral persistence. Studies with 129/Sv mice in which the gene coding for the interferon gamma receptor had been inactivated demonstrated the role of the interferon gamma pathway in resistance to persistent infection (16, 35). The recent development of a C57BL/6 strain whose gene coding for interferon gamma has been inactivated allowed us to test the role of the cytokine directly. Unexpectedly, these mice were less susceptible to viral persistence and to clinical signs than the 129/Sv Ifngr−/− mice. The difference in susceptibility between the 129/Sv Ifngr−/− and C57BL/6 Ifng−/− strains could have been due to the functional differences existing between the cytokine and its receptor or to the difference in genetic background between the two mouse strains. By comparing the phenotypes of the F2 Ifng−/− and the F2 Ifngr−/− crosses (see Results for the definition of these crosses), we showed that 80% of the difference in susceptibility to viral persistence was due to the difference in genetic background. Since there is a small, but statistically significant, difference in viral load between the F2 Ifng−/− and F2 Ifngr−/− lines (Fig. 3), part of the difference in susceptibility between the parental strains might be due to the fact that the inactivated gene was different for each of them. However, we cannot rule out the possibility that some 129/Sv susceptibility loci were still present in the C57BL/6 Ifng−/− strain, since it was obtained from a 129/Sv ES cell line after only 10 backcrosses. 129/Sv Ifng−/− mice have been described by Cantin et al. since this article was submitted for publication (7). They will be invaluable to compare the roles of gamma interferon and its receptor in the pathogenesis of this model disease.
The comparison of the phenotypes of the parental strains and of the two F2 crosses showed that the level of viral persistence was genetically controlled (Fig. 3). In spite of this, we did not find a locus linked to viral persistence when a complete genome scan of both crosses was performed. This is most likely because the number of loci involved is too high to allow the detection of linkages with a cross of approximately 100 animals. It has been reported that the C57BL/6 strain is more resistant to demyelination than other H-2Db strains (32). The existence of a large number of non-H-2 resistance genes in the C57BL/6 strain might put limits on the use of null-mutant mice with this background to study the pathogenesis of Theiler’s virus infection. The phenotypes of the mutant mice might be less pronounced than on another resistant background, and also more difficult to interpret.
The main conclusion of the present work is that the appearance of symptoms in this model disease depends on at least two independent factors: high viral load and a locus on chromosome 11 which we named Tmevd5. In previous publications, we noted that the mouse strains with high persistent viral load, as measured with a dot blot assay, were usually the strains identified by other groups as susceptible to clinical disease (6, 27). In the present study we confirmed the association between high viral load and clinical disease among mice from two different F2 crosses (Fig. 4 and 5). However, others who measured viral load with an infectivity assay did not find a correlation between viral persistence and clinical disease (9, 23, 31). The reason for this discrepancy is not clear at present. It could be due in part to trivial technical difficulties with the infectivity assay (the DA virus remains extremely membrane associated in tissue extracts).
The Tmevd5 locus is significantly linked to clinical disease but not, or only weakly, to viral persistence. This locus could act in two different ways: (i) it could change the threshold of viral load required for the appearance of symptoms, and (ii) it could change the likelihood of appearance of symptoms for mice with a given viral load. The data shown in Fig. 5 clearly favor the second possibility: all symptomatic mice have a large viral load, regardless of genotype. However, the fraction of animals with a large viral load which are symptomatic is much higher when the animals are C57BL/6 homozygous at the Tmevd5 locus. The characterization of the Tmevd5 locus is now an important step in the study of the pathogenesis of this model disease. Interestingly, Idd4, a locus involved in the insulin-dependent diabetes of the nonobese diabetic mouse, is located in the same region (39). The Tmevd5 C57BL/6 allele confers susceptibility, although the C57BL/6 strain is resistant. Although this may seem surprising at first, similar results have been reported with other polygenic diseases (13, 17, 29). It is likely that, in the C57BL/6 strain, the effect of a susceptible allele at one locus is overcome by the effects of resistant alleles at other loci. As shown by our work, the C57BL/6 allele at Tmevd5 affects clinical disease only in mice with a high viral burden. The level of viral infection was probably too low to observe the effect of this locus in the C57BL/6 Ifng−/− mice.
Our findings may have implications for the study of diseases of humans. Persistent viral infections may be asymptomatic or associated with severe diseases in humans. Infection by human T-cell leukemia virus type I is a case in point. With other viruses, such as human immunodeficiency virus, the duration of the asymptomatic phase can be highly variable from individual to individual. It is usually assumed that, in these situations, the main determinant of clinical disease is high viral burden. Our work with Theiler’s virus shows that the situation might be more complex and that host genetic factors can influence the occurrence of clinical disease in individuals with the same high viral load.
Multiple sclerosis is a multifactorial disease in which environmental, presumably infectious, and genetic factors (14, 15, 18, 36) interact to cause white matter damage. In mice, several common viruses, including Theiler’s virus and strains of coronaviruses, cause diseases which resemble multiple sclerosis in genetically susceptible inbred strains. By analogy with the murine models, one has to consider the possibility that multiple sclerosis results from infections by common human viruses in individuals with a specific genetic background which renders them susceptible to demyelination by a particular virus. If such were the case, the epidemiological studies performed so far to test the viral hypothesis of multiple sclerosis would be unable to establish a causal relationship between a given virus and the disease.
In conclusion, high viral load is necessary but not sufficient to cause clinical disease in mice persistently infected with Theiler’s virus. The Tmevd5 locus on chromosome 11 modifies the risk of disease in mice with a high viral burden. These results might have important implications for the study of multiple sclerosis and other multifactorial human diseases.
ACKNOWLEDGMENTS
We thank F. Bihl for helpful discussion, J.-L. Guénet, G. Millon, X. Montagutelli, and S. Wain-Hobson for critical reading of the manuscript, and M. Gau for secretarial aid.
This study was supported by grants from the Institut Pasteur Fondation, the Centre National de la Recherche Scientifique, the National Multiple Sclerosis Society USA, the European Community Human Capital and Mobility Program, and the Association pour la Recherche sur la Sclérose en Plaques. S.A. is the recipient of a scholarship from the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Aubert C, Chamorro M, Brahic M. Identification of Theiler’s virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb Pathog. 1987;3:319–326. doi: 10.1016/0882-4010(87)90002-7. [DOI] [PubMed] [Google Scholar]
- 2.Azoulay A, Brahic M, Bureau J-F. FVB mice transgenic for the H-2Db gene become resistant to persistent infection by Theiler’s virus. J Virol. 1994;68:4049–4052. doi: 10.1128/jvi.68.6.4049-4052.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahic M, Bureau J-F. Genetics of susceptibility to Theiler’s virus infection. Bioessays. 1998;20:627–633. doi: 10.1002/(SICI)1521-1878(199808)20:8<627::AID-BIES5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Bureau J-F, Drescher K M, Pease L R, Vikoren T, Delcroix M, Zoecklein L, Brahic M, Rodriguez M. Chromosome 14 contains determinants that regulate susceptibility to Theiler’s virus-induced demyelination in the mouse. Genetics. 1998;148:1941–1949. doi: 10.1093/genetics/148.4.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bureau J-F, Montagutelli X, Bihl F, Lefebvre S, Guénet J-L, Brahic M. Mapping loci influencing the persistence of Theiler’s virus in the murine central nervous system. Nat Genet. 1993;5:87–91. doi: 10.1038/ng0993-87. [DOI] [PubMed] [Google Scholar]
- 6.Bureau J-F, Montagutelli X, Lefebvre S, Guénet J-L, Pla M, Brahic M. The interaction of two groups of murine genes determines the persistence of Theiler’s virus in the central nervous system. J Virol. 1992;66:4698–4704. doi: 10.1128/jvi.66.8.4698-4704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantin E, Tanamachi B, Openshaw H, Mann J, Clarke K. Gamma interferon (IFN-γ) receptor null-mutant mice are more susceptible to herpes simplex virus type 1 infection than IFN-γ ligand null-mutant mice. J Virol. 1999;73:5196–5200. doi: 10.1128/jvi.73.6.5196-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Clatch R J, Lipton H L, Miller S D. Class II-restricted T cell responses in Theiler’s murine encephalomyelitis virus (TMEV)-induced demyelinating disease. II. Survey of host immune responses and central nervous system virus titers in inbred mouse strains. Microb Pathog. 1987;3:327–337. doi: 10.1016/0882-4010(87)90003-9. [DOI] [PubMed] [Google Scholar]
- 10.Clatch R J, Miller S D, Metzner R, Dal Canto M C, Lipton H L. Monocytes/macrophages isolated from the mouse central nervous system contain infectious Theiler’s murine encephalomyelitis virus (TMEV) Virology. 1990;176:244–254. doi: 10.1016/0042-6822(90)90249-q. [DOI] [PubMed] [Google Scholar]
- 11.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Multiple defects of immune cell function in mice with disrupted interferon-γ genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 12.Daniels J B, Pappenheimer A M, Richardson S. Observations on encephalomyelitis of mice (DA strain) J Exp Med. 1952;96:22–24. doi: 10.1084/jem.96.6.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Gouyon B, Melanitou E, Richard M F, Requarth M, Hahn I H, Guénet J L, Demenais F, Julier C, Lathrop G M, Boitard C, Avner P. Genetic analysis of diabetes and insulitis in an interspecific cross of the nonobese diabetic mouse with Mus spretus. Proc Natl Acad Sci USA. 1993;90:1877–1881. doi: 10.1073/pnas.90.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Diaz, J. S. 16 May 1999, revision date. Primer sequences. [Online.] http: //www-genome.wi.mit.edu/cgi-bin/mouse/index. [26 July 1999, last date accessed.]
- 14.Ebers G C, Bulman D E, Sadovnick A D, Paty D W, Warren S, Hader W, Murray T J, Seland T P, Duquette P, Grey T, Nelson R, Nicolle M, Brunet D. A population-based study of multiple sclerosis in twins. N Engl J Med. 1986;315:1638–1642. doi: 10.1056/NEJM198612253152603. [DOI] [PubMed] [Google Scholar]
- 15.Ebers G C, Kukay K, Bulman D E, Sadovnick A D, Rice G, Anderson C, Armstrong H, Cousin K, Bell R B, Hader W, Paty D W, Hashimoto S, Oger J, Duquette P, Warren S, Gray T, O’Connor P, Nath A, Auty A, Metz L, Francis G, Paulseth J E, Murray T J, Pryse-Phillips W, Nelson R, Freedman M, Brunet D, Bouchard J-P, Hinds D, Risch N. A full genome search in multiple sclerosis. Nat Genet. 1996;13:472–476. doi: 10.1038/ng0896-472. [DOI] [PubMed] [Google Scholar]
- 16.Fiette L, Aubert C, Müller U, Huang S, Aguet M, Brahic M, Bureau J-F. Theiler’s virus infection of 129Sv mice that lack the interferon α/β or interferon γ receptors. J Exp Med. 1995;181:2069–2076. doi: 10.1084/jem.181.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fijneman R J, Jansen R C, van der Valk M A, Demant P. High frequency of interactions between lung cancer susceptibility genes in the mouse: mapping of Sluc5 to Sluc14. Cancer Res. 1998;58:4794–4798. [PubMed] [Google Scholar]
- 18.Haines J L, Ter-Minassian M, Bazyk A, Gusella J F, Kim D J, Terwedow H, Pericak-Vance M A, Rimmler J B, Haynes C S, Roses A D, Lee A, Shaner B, Menold M, Seboun E, Fitoussi R-P, Gartouix C, Reyes C, Ribierre F, Gyapay G, Weissenbach J, Hauser S L, Goodkin D E, Lincoln R, Usuku K, Garcia-Merino A, Gatto N, Young S, Oksenberg J R. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatibility complex. Nat Genet. 1996;13:469–471. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Immune response in mice that lack the interferon-γ receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 20.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 21.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 22.Lipton H L. Theiler’s virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipton H L, Dal Canto M C. Susceptibility of inbred mice to chronic central nervous system infection by Theiler’s murine encephalomyelitis virus. Infect Immun. 1979;26:369–374. doi: 10.1128/iai.26.1.369-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipton H L, Melvold R, Miller S D, Dal Canto M C. Mutation of a major histocompatibility class I locus, H-2D, leads to an increased virus burden and disease susceptibility in Theiler’s virus-induced demyelinating disease. J Neurovirol. 1995;1:138–144. doi: 10.3109/13550289509113960. [DOI] [PubMed] [Google Scholar]
- 25.Manly K F. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- 26.McAllister A, Tangy F, Aubert C, Brahic M. Molecular cloning of the complete genome of Theiler’s virus, strain DA, and production of infectious transcripts. Microb Pathog. 1989;7:381–388. doi: 10.1016/0882-4010(89)90041-7. [DOI] [PubMed] [Google Scholar]
- 27.Miller S, Gerety S. Immunologic aspects of Theiler’s murine encephalomyelitis virus (TMEV)-induced demyelinating disease. Semin Virol. 1990;1:263–272. [Google Scholar]
- 28.Montagutelli X, Serikawa T, Guénet J L. PCR-analyzed microsatellites: data concerning laboratory and wild-derived mouse inbred strains. Mamm Genome. 1991;1:255–259. doi: 10.1007/BF00352333. [DOI] [PubMed] [Google Scholar]
- 29.Morel L, Mohan C, Yu Y, Schiffenbauer J, Rudofsky U H, Tian N, Longmate J A, Wakeland E K. Multiplex inheritance of component phenotypes in a murine model of lupus. Mamm Genome. 1999;10:176–181. doi: 10.1007/s003359900964. [DOI] [PubMed] [Google Scholar]
- 30.Njenga M K, Asakura K, Hunter S F, Wettstein P, Pease L R, Rodriguez M. The immune system preferentially clears Theiler’s virus from the gray matter of the central nervous system. J Virol. 1997;71:8592–8601. doi: 10.1128/jvi.71.11.8592-8601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patick A K, Pease L R, David C S, Rodriguez M. Major histocompatibility complex-conferred resistance to Theiler’s virus-induced demyelinating disease is inherited as a dominant trait in B10 congenic mice. J Virol. 1990;64:5570–5576. doi: 10.1128/jvi.64.11.5570-5576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelka A, Olsberg C, Miller S, Waltenbaugh C, Creighton T M, Dal Canto M C, Melvold R. Effects of irradiation on development of Theiler’s murine encephalomyelitis virus (TMEV)-induced demyelinating disease in genetically resistant mice. Cell Immunol. 1993;152:440–455. doi: 10.1006/cimm.1993.1303. [DOI] [PubMed] [Google Scholar]
- 33.Pena Rossi C, Delcroix M, Huitinga I, McAllister A, van Rooijen N, Claassen E, Brahic M. Role of macrophages during Theiler’s virus infection. J Virol. 1997;71:3336–3340. doi: 10.1128/jvi.71.4.3336-3340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez M, David C S. H-2Dd transgene suppresses Theiler’s virus-induced demyelination in susceptible strains of mice. J Neurovirol. 1995;1:111–117. doi: 10.3109/13550289509111015. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez M, Pavelko K, Coffman R L. Gamma interferon is critical for resistance to Theiler’s virus-induced demyelination. J Virol. 1995;69:7286–7290. doi: 10.1128/jvi.69.11.7286-7290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawcer S, Jones H B, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow P N, Compston A. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet. 1996;13:464–468. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- 37.Teuscher C, Rhein D M, Livingstone K D, Paynter R A, Doerge R W, Nicholson S M, Melvold R W. Evidence that Tmevd2 and eae3 may represent either a common locus or members of a gene complex controlling susceptibility to immunologically mediated demyelination in mice. J Immunol. 1997;159:4930–4934. [PubMed] [Google Scholar]
- 38.Theiler M, Gard S. Encephalomyelitis of mice. I. Characteristics and pathogenesis of the virus. J Exp Med. 1940;72:49–67. doi: 10.1084/jem.72.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yui M A, Muralidharan K, Moreno-Altamirano B, Perrin G, Chestnut K, Wakeland E K. Production of congenic mouse strains carrying NOD-derived diabetogenic genetic intervals: an approach for the genetic dissection of complex traits. Mamm Genome. 1996;7:331–334. doi: 10.1007/s003359900097. [DOI] [PubMed] [Google Scholar]