Abstract
A reporter open reading frame (ORF) coding for a fusion of bacterial β-glucuronidase (GUS) with a proteinase domain (Pro) derived from tobacco etch potyvirus was utilized for tagging individual genes of beet yellows closterovirus (BYV). Insertion of this reporter ORF between the first and second codons of the BYV ORFs encoding the HSP70 homolog (HSP70h), a major capsid protein (CP), and a 20-kDa protein (p20) resulted in the expression of the processed GUS-Pro reporter from corresponding subgenomic RNAs. The high sensitivity of GUS assays permitted temporal analysis of reporter accumulation, revealing early expression from the HSP70h promoter, followed by the CP promoter and later the p20 promoter. The kinetics of transcription of the remaining BYV genes encoding a 64-kDa protein (p64), a minor capsid protein (CPm), and a 21-kDa protein (p21) were examined via Northern blot analysis. Taken together, the data indicated that the temporal regulation of BYV gene expression includes early (HSP70h, CPm, CP, and p21 promoters) and late (p64 and p20 promoters) phases. It was also demonstrated that the deletion of six viral genes that are nonessential for RNA amplification resulted in a dramatic increase in the level of transcription from one of the two remaining subgenomic promoters. Comparison with other positive-strand RNA viruses producing multiple subgenomic RNAs showed the uniqueness of the pattern of closterovirus transcriptional regulation.
A variety of evolutionary dissimilar positive-strand RNA viruses employ the formation of subgenomic RNAs (sgRNAs) as a major strategy for gene expression (23). The sgRNAs are normally formed via partial transcription of the genomic minus strand (5, 26, 29). In the viruses producing multiple species of sgRNA, the kinetics and/or levels of sgRNA accumulation are transcriptionally regulated (e.g., see references 5, 9, 20, 25, 30, 35, and 36).
The family Closteroviridae belongs to the Sindbis virus-like supergroup of positive-strand RNA viruses (13). Large closterovirus genomes possess from 9 to 12 open reading frames (ORFs) (37); two of these ORFs (1a and 1b; Fig. 1A) are translated from the genomic RNA, whereas the remaining ORFs are expressed via the formation of a nested set of sgRNAs (10, 16–18, 21). A recent study of the kinetics of accumulation of four citrus tristeza closterovirus (CTV) sgRNAs revealed temporal control of CTV transcription (30). In addition to sgRNAs, closteroviruses possess defective RNAs, some of which were proposed to originate via recombination between genomic RNA and sgRNAs (6).
FIG. 1.
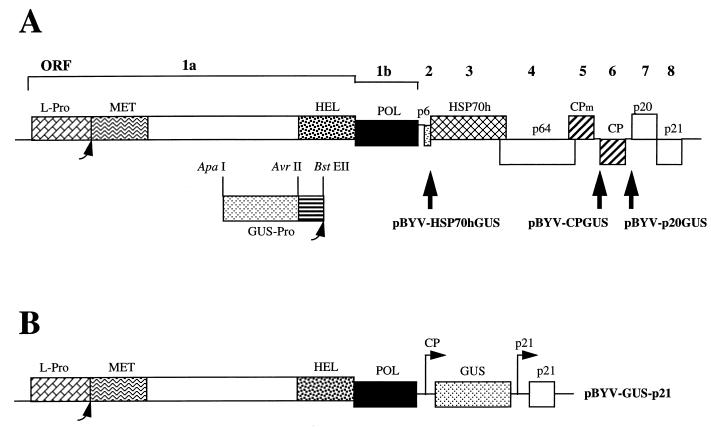
Tagging of BYV by insertion of the self-processing reporter. (A) Diagram of the BYV genome with ORFs 1a to 8 encoding leader proteinase (L-Pro), replication-associated proteins harboring putative methyltransferase (MET), RNA helicase (HEL), and RNA polymerase (POL) domains; p6; HSP70h; p64; CPm; CP; p20; and p21. The bottom part of panel A depicts a reporter GUS-Pro ORF encoding a fusion of GUS with Pro derived from TEV. Large vertical arrows indicate the sites of reporter insertion into BYV cDNA clone and the names of the resulting plasmids. Smaller rounded arrows designate the self-processing sites for the BYV L-Pro and reporter GUS-Pro. (B) Diagram of the deletion variant pBYV-GUS-p21. The designations are the same as those for panel A, except for the arrows marked CP and p21, which show the approximate positions of the 5′ termini of the sgRNAs driven by the CP and p21 promoters.
In this study, we utilized a prototype closterovirus, beet yellows virus (BYV), that possesses a 15.5-kb genomic RNA and at least six 3′-coterminal sgRNAs expressing ORFs 3 to 8 (Fig. 1A) (10, 16, 31). It is not fully established whether the expression of ORF 2 coding for a small hydrophobic 6-kDa protein (p6) involves the formation of an additional, seventh sgRNA species. BYV ORFs 3 through 8 encode an HSP70 homolog (HSP70h), a 64-kDa protein (p64), minor and major capsid proteins (CPm and CP, respectively), a 20-kDa protein (p20), and a 21-kDa protein (p21) (Fig. 1A) (1). Filamentous particles of BYV are composed of a body and a short tail made of the CP and CPm (3). It has been demonstrated that p21 is required for efficient amplification of BYV RNA (31), whereas indirect experiments suggested involvement of the HSP70h in viral cell-to-cell movement (2, 19, 28). The functions of p6, p64, and p20 are obscure.
To further investigate transcriptional regulation in closteroviruses, we used tobacco protoplasts transfected with RNA derived from a cDNA clone of BYV (31). The kinetics of accumulation of genomic RNA and three sgRNAs were examined by using Northern blot analysis. In addition, gene expression involving the most active CP promoter and the least active promoters controlling production of HSP70h and p20 was analyzed by using a self-cleaving reporter protein. Since the reporter possessed β-glucuronidase (GUS) activity, very sensitive GUS assays allowed detection of gene expression at the early phases of viral reproduction. The combination of these two approaches allowed us to reveal the closterovirus gene expression profile and to compare this profile to that of coronaviruses, another family of RNA viruses producing multiple sgRNAs (25).
MATERIALS AND METHODS
Engineering of BYV cDNA clones tagged by insertion of the reporter ORF.
Previously described full-length cDNA clone pBYV-NA and its partial derivatives p65M and p3′-BYV were used throughout this study (31). Recognition sites for restriction endonucleases ApaI and BstEII were introduced between the first and second codons of ORFs 3, 6, and 7 by using site-directed mutagenesis (24). Oligonucleotide 65-ABst (5′-CGGTCGTGTGATGGGGCCCACTGGTAACCAAGTTGTTTTCGGATTAG; the underlined nucleotides here and thereafter represent restriction endonuclease sites) and plasmid p65M were used to mutate ORF 3, whereas oligonucleotides CP-ABst (5′-TTGAGTTTCGTTATGGGGCCCACTGGTAACCTCGAACCTATAAGTGC) and 20-ABst (5′-CACCGGAAAATGACTGGGCCCACTGGTAACCACTCTGTCGAACTAGC) were used to modify ORFs 6 and 7 present in p3′-BYV. The mutations were verified by nucleotide sequencing.
The GUS ORF was PCR amplified with primers 5GUS-Apa (5′-GATGGGCCCATGGTCCGTCCT) and 3GUS-Avr (5′-CTCCTAGGATTTGTTTGCCTCCC), possessing ApaI and AvrII sites, respectively, and pTEV7D-GUS.HC as a template (11). The proteinase domain (Pro) of tobacco etch potyvirus (TEV) HC-Pro (7) specified by TEV nucleotides (nt) 1966 to 2436 (4) was amplified with the same template; the primers were 5PPAvr (5′-CGCCTAGGTAAGGCTCAATATTC) and Pro-Bst (5′-GCGGTTACCTCCAACATTGTAAGT). Restriction endonuclease sites AvrII and BstEII were incorporated into these primers.
To insert DNA fragments encoding two parts of the fusion reporter GUS-Pro into each of ORFs 3, 6, and 7, the three mutant plasmids were digested with ApaI and BstEII, whereas PCR-amplified DNA harboring GUS and Pro ORFs were digested with ApaI plus AvrII and AvrII plus BstEII, respectively. Ligation of the DNA fragments into the corresponding vectors resulted in generation of p65M-GUS-Pro-HSP70h, p3′-BYV-GUS-Pro-CP, and p3′-BYV-GUS-Pro-p20. Each mutant fragment harboring the GUS-Pro ORF fused in frame with ORF 3, 6, or 7 was cloned into the full-length clone as described previously (31). The resulting pBYV-NA derivatives were designated as pBYV-HSP70hGUS, pBYV-CPGUS, and pBYV-p20GUS to indicate that the GUS-Pro reporter is positioned under the control of the BYV promoters directing expression of HSP70h, CP, and p20, respectively (Fig. 1A).
The recombinant variant pBYV-GUS-p21 was engineered by modification of the pBYV-CPGUS. The DNA fragments from the ORF 2 start codon to the BamHI site at nt 13392 and from the AvrII site at the 3′ end of GUS ORF 6 to the HpaI site at nt 14407 were deleted (Fig. 1A and B). As a result, BYV ORFs 2 to 7 were deleted, whereas GUS was expressed under the control of the ORF 6 (CP) promoter (Fig. 1B).
Analyses of RNA accumulation and gene expression.
The in vitro RNA transcription was conducted in 50-μl reaction mixtures with SmaI-linearized plasmids and SP6 RNA polymerase as described previously (31). The reaction mixtures containing ∼50 μg of the RNA transcripts were used to transfect, via electroporation, the protoplasts (∼4 × 106 cells per transfection) obtained from a suspension culture of Nicotiana tabacum cv. Xanthi nc cell line DF (14). Protoplast samples were harvested after 86 h of incubation at room temperature or at the times specified for time course experiments. The RNA samples were isolated by using TRIZOL reagent (Gibco-BRL), and Northern hybridization analysis was performed as previously described (31). The 32P-labeled single-stranded RNA probe of negative polarity was prepared by in vitro transcription by using T7 RNA polymerase and p3′-BYV linearized at the BamHI site (31). The radiolabeled products were detected and quantified using a PhosphorImager (Molecular Dynamics) and ImageQuant, version 5, software package. Four independent protoplast transfections were conducted with each BYV variant in each experiment; means and standard deviations were used to compare the accumulations of the genomic RNA and sgRNAs.
GUS activity in protoplasts was assayed as described before (8). Since the levels of GUS-Pro reporter expression driven by the ORF 3, 6, or 7 promoter were very different (see Results), these levels were expressed as percentages of the maximal expression levels for each variant. This approach allowed us to compare temporal activity patterns of the three promoters with the same scale. Four independent transfections per variant were included in each experiment. Accumulation of the BYV CP in transfected protoplasts was examined by immunoblot analysis with a 1:1,000 dilution of the anti-BYV serum (a gift from Bryce W. Falk, University of California—Davis) as described before (12).
RESULTS
Generation and characterization of the tagged BYV variants.
Although the complex regulation of closterovirus transcription has been revealed via Northern hybridization (16, 17, 30, 31), the low abundance of some RNA species impeded comprehensive analysis of this process. For instance, the time course analysis of the CTV sgRNA encoding HSP70h proved impractical (30), while the BYV sgRNA expressing p20 escaped detection (16). To circumvent these obstacles, we tagged selected subgenomic promoters by insertion of a reporter ORF whose expression permitted sensitive and specific detection of the promoters’ activities.
Reporter protein was engineered by fusing GUS with a potyviral Pro. The chimeric GUS-Pro ORF was inserted between the first and second codons of the ORFs encoding HSP70h, CP, and p20, resulting in plasmids pBYV-HSP70hGUS, pBYV-CPGUS, and pBYV-p20GUS, respectively (Fig. 1A). This design permitted expression of the reporter and original viral product from a single expression unit. Autoproteolytic activity of Pro resulted in cleavage between GUS-Pro and the corresponding viral protein. This cleavage was efficient, since only the free form of GUS-Pro was detected in transfected protoplasts by using anti-GUS serum (data not shown).
In vitro GUS assays conducted at 4 days posttransfection (d.p.t.) demonstrated that tagged BYV variants expressed an enzymatically active reporter. As expected, the highest level of GUS activity was produced by CPGUS, the variant in which reporter expression was driven by the strongest subgenomic promoter. The levels of GUS activity produced by the p20GUS and HSP70hGUS variants were only 5.1% ± 1.3% and 2.3% ± 0.6% of that produced by the CPGUS variant, respectively, demonstrating very low relative activities of the p20 and HSP70h promoters. The background level of GUS activity in protoplasts transfected by the wild-type BYV-NA variant lacking a GUS ORF was less than 5% of that found for the HSP70hGUS variant (data not shown).
Kinetics of gene expression driven by the tagged promoters.
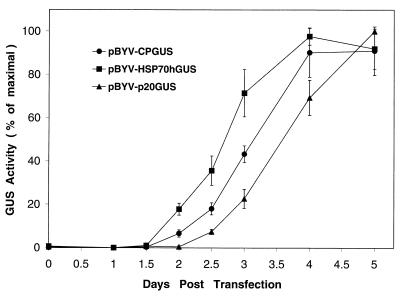
The high sensitivity and specificity of the GUS assays permitted a comparative study of the kinetics of reporter expression in tagged BYV variants. Figure 2 illustrates the results of a typical experiment. The GUS activity induced by the HSP70hGUS and CPGUS variants was first detected at 1.5 d.p.t. In contrast, the p20GUS variant started to produce GUS only at 2 to 2.5 d.p.t. A comparative analysis over the 5-day period revealed that the earliest relative increase in GUS activity was produced by the HSP70h promoter, followed by the CP promoter and the p20 promoter (Fig. 2). These observations allowed us to roughly classify HSP70h and CP promoters as early and the p20 promoter as late.
FIG. 2.
Kinetics of GUS activity accumulation in protoplasts transfected by the three tagged BYV variants. The activity is expressed as a percentage of the absolute maximum for each variant. The means and standard deviations from four independent transfections were used to generate the graph.
Accumulation of the genomic RNA and sgRNAs in wild-type BYV.
The kinetics of accumulation of the genomic RNA and sgRNAs transcribed from the p64, CPm, and p21 promoters were compared by using the wild-type pBYV-NA variant and Northern hybridization analysis. Quantitation of the sgRNAs encoding HSP70h, CP, and p20 was hampered by the high background in corresponding regions of the membrane (see Fig. 6).
FIG. 6.
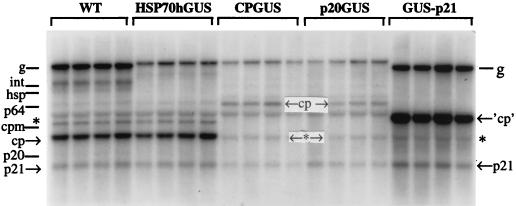
Northern hybridization analysis of the RNAs derived from protoplasts at 86 h posttransfection. The types of the RNA transcripts used are shown on the top. Four lanes for each variant correspond to independent transfections. The RNA probe was the same as that described in the legend for Fig. 3. Positions of the genomic (g) RNAs and sgRNAs encoding HSP70h (hsp), p64, CPm (cpm), CP (cp), p20, and p21 corresponding to the wild-type transfection are shown at the left. WT, wild-type transcripts derived from pBYV-NA clone; int, intermediate-sized, probably defective RNA (6, 31). Asterisks designate the background bands corresponding to plant rRNAs (31). Note that the bands of the CP sgRNAs for the wild-type and HSP70hGUS variants overlap the band of smaller rRNA. The designation ←cp→ shows the position of the sgRNA derived from the CP promoter for the CPGUS and p20GUS variants. The designations on the right indicate positions of the genomic RNA and sgRNAs for the GUS-p21 variant; ‘cp’ corresponds to the GUS-encoding sgRNA derived from the CP promoter. The genomic RNAs and sgRNAs derived from the CP and p21 promoters are quantified in Table 1. The sizes of the genomic RNAs are 15.5 kb for the wild type, 17.8 kb for HSP70hGUS, CPGUS, and p20GUS variants, and 12.6 kb for the GUS-p21 variant. The estimated sizes of the sgRNAs in the wild-type BYV (16) are as follows: 6.1 kb (hsp), 4.4 kb (p64), 2.6 kb (cpm), 1.8 kb (cp), 1.2 kb (p20), and 0.8 kb (p21). The size of the CP sgRNA in CPGUS and p20GUS variants is 4.1 kb, whereas the size of the sgRNA expressing GUS under the control of the CP promoter (‘cp’) is 2.8 kb.
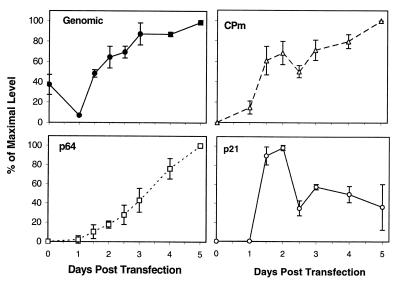
The input RNA transcripts detectable in protoplasts at time zero declined on day 1; then a gradual increase in genomic RNA levels due to replication continued until day 5 (Fig. 3). The accumulation pattern of the p64 sgRNA followed that of the genomic RNA, although with a significant delay (Fig. 3). Dramatically different temporal patterns were seen for the p21 and CPm sgRNAs. The relative level of the p21 sgRNA species approached its maximum at day 2, then declined, and roughly leveled at ∼50% of the maximum (Fig. 3). Similar kinetics were observed for the CPm sgRNA until day 3; at the later times, this RNA species continued to accumulate, approaching its maximum at day 5. The significance of a decline in the levels of p21 and CPm sgRNAs that was reproducibly observed at ∼2.5 d.p.t. awaits further experimentation. Taken together, these data permit us to define p21 and CPm promoters as early and the p64 promoter as late.
FIG. 3.
Kinetics of accumulation of the genomic RNA and three sgRNAs encoding p64, CPm, and p21 in protoplasts transfected by wild-type BYV transcripts. The RNA levels are expressed as percentages of the absolute maximum for each variant. Northern hybridization and a 32P-labeled RNA probe of negative polarity were used to detect and quantify RNA species. The means and standard deviations from four independent transfections were used to generate the graph.
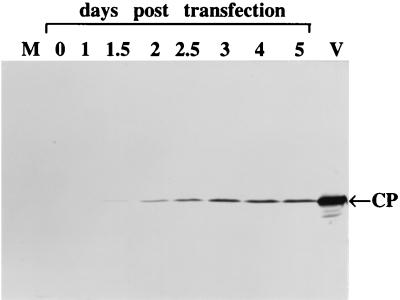
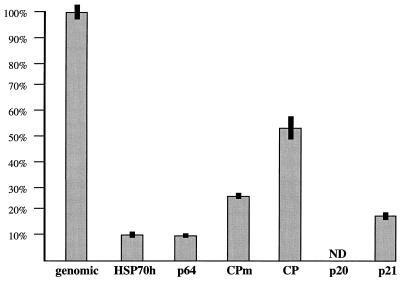
To correlate the data obtained with the tagged and nontagged BYV variants, we analyzed the accumulation of CP directed by wild-type BYV. As illustrated in Fig. 4, CP was detected at 1.5 d.p.t., increased until day 3, and reached a plateau at later times. This pattern parallels that of GUS accumulation driven by the CP promoter in the CPGUS variant for at least 3 d.p.t. Hence, comparison of Fig. 2, 3, and 4 permits a conditional delineation of the two classes of BYV promoters according to the temporal control of their expression. The early promoters include HSP70h, CPm, CP, and p21, while the late promoters are p64 and p20. The maximal relative levels reached by the genomic RNA and sgRNA species are shown in Fig. 5. Its comparison with the genome map (Fig. 1A) indicates no simple correlation between these levels and genome positions of corresponding sgRNA promoters.
FIG. 4.
Immunoblot analysis of the CP accumulation in protoplasts transfected by the wild-type BYV transcripts. M, sample obtained from mock-inoculated protoplasts at 5 d.p.t.; V, capsid protein standard derived from purified and dissociated BYV virions.
FIG. 5.
Relative levels of the genomic RNA and sgRNA species at their respective maxima. The protoplasts were transfected by the wild-type RNA transcripts. The sgRNAs are designated in accord with their coding specificity and presented in order from 5′ to 3′ (see Fig. 1A). The levels of genomic RNA and sgRNAs encoding HSP70h, p64, CPm, and CP were determined at 5 d.p.t., whereas that of p21 sgRNA was measured at 2 d.p.t. Black bars show standard deviations. ND, not determined. Although sgRNA encoding p20 is visible on Northern blots, its quantitation is impractical due to a high background.
Comparative analysis of the four BYV variants.
In order to reveal effects on viral replication and transcription imposed by insertion of the 2.3-kb reporter ORF, we compared the wild-type and tagged BYV variants at ∼4 d.p.t. by using Northern hybridization analysis. The levels of genomic RNAs and of the two sgRNAs derived from the CP and p21 promoters were quantified. Only these three RNA species can be confidently traced among the tagged variants (Fig. 6). It was found that insertion of the reporter ORF resulted in decreased accumulation of the genomic RNAs in the HSP70hGUS, CPGUS, and p20GUS variants (Fig. 6 and Table 1). Surprisingly, accumulation of the sgRNAs was affected differentially. In the HSP70hGUS variant, the levels of the CP and p21 sgRNAs were close to those of the wild type (Table 1). In the CPGUS and p20GUS variants, the levels of the CP sgRNA decreased roughly proportionally to those of the genomic RNAs, whereas accumulation of the p21 sgRNA was affected to a lesser extent (Fig. 6 and Table 1).
TABLE 1.
Accumulation of the genomic RNA and two sgRNA species in BYV variants at 4 d.p.t. of protoplasts
| BYV variant | Accumulation of the following RNA species (% WT ± SD)a
|
||
|---|---|---|---|
| Genomic RNA | CP sgRNAb | p21 sgRNA | |
| Wild type | 100 ± 5 | 100 ± 10 | 100 ± 18 |
| HSP70hGUS | 38 ± 5 | 101 ± 24 | 94 ± 7 |
| CPGUS | 21 ± 5 | 22 ± 5 | 48 ± 16 |
| p20GUS | 22 ± 6 | 10 ± 3 | 71 ± 20 |
| GUS-p21 | 107 ± 14 | 362 ± 39 | 124 ± 25 |
WT, wild type; SD, standard deviation.
Note that this sgRNA species derived from the CP promoter directs expression of the CP in the wild-type, HSP70hGUS, and p20GUS variants; in CPGUS it expresses the CP-GUS-Pro fusion product, whereas in GUS-p21 it expresses GUS.
To assess the impact of genome truncation on RNA amplification and transcription, we used a deletion variant pBYV-GUS-p21 derived from pBYV-CPGUS. This variant possessed only two of six BYV promoters: GUS was expressed by using the CP subgenomic promoter, while p21 ORF remained under the control of its natural promoter (Fig. 1B). ORFs 2 to 7 but not 8 were deleted, since the expression of p6, HSP70h, p64, CPm, CP, and p20 is not essential for replication, whereas p21 is required for efficient accumulation of the RNA (31).
Northern hybridization analysis demonstrated that the level of genomic RNA in protoplasts transfected by using pBYV-GUS-p21 transcripts was five times higher than that for the parental CPGUS variant (Fig. 6 and Table 1). This profound increase in replication can be attributed to the ∼30% decrease in the genome size (17.8 kb in CPGUS versus 12.6 kb in GUS-p21) and/or to the deletion of four sgRNA promoters that could compete for the replicase with the origins for genomic RNA synthesis. Comparison of the GUS-p21 variant to the wild-type virus showed similar accumulation of the genomic RNA and p21 sgRNA, whereas the level of the sgRNA produced by the CP promoter had risen almost fourfold (Table 1). The GUS activity in GUS-p21-transfected protoplasts can be reliably detected at 1 d.p.t. (data not shown), indicating that truncation of the genome resulted in an earlier expression from the CP promoter.
DISCUSSION
Tagging of the BYV ORFs by insertion of the self-processing reporter combined with traditional hybridization analysis allowed us to reveal a complex pattern of transcriptional regulation in a prototype closterovirus. Comparison of the GUS expression under the control of three subgenomic promoters demonstrated earliest activation of the HSP70h and CP promoters and a delayed activation of the p20 promoter. Since the direct quantitation of sgRNAs expressing HSP70h, CP, and p20 was problematic, the utilization of reporter activity offered a more reliable approach for the analysis of these promoters’ regulation. The potential drawback of this system, however, is the high stability of GUS that may result in masking the fine details of regulation.
Hybridization analysis of the remaining three sgRNAs revealed an early and relatively fast accumulation of the p21 and CPm sgRNAs in contrast to a much slower accumulation of the p64 sgRNA. Indeed, the latter RNA species reached 50% of its maximal level after 3 d.p.t., whereas p21 and CPm sgRNAs reached that level before 1.5 d.p.t. A decline in the levels of these two sgRNAs seen at 2.5 d.p.t. adds further complexity to corresponding patterns of temporal regulation. Although direct superposition of the results obtained with tagged and nontagged viruses would be incorrect, the same timing of product appearance driven by the CP promoter in the wild-type virus and in the CPGUS variant allows one to relate these two experimental systems. Hence, we can classify the HSP70h, CPm, CP, and p21 promoters as early and the p64 and p20 promoters as late. It is obvious that the order of promoter activation does not correspond to the order of the ORFs in the BYV genome (Fig. 1A).
The pattern of temporal regulation of multiple transcriptional units demonstrated here for a closterovirus is unique among RNA viruses. Animal coronaviruses that also possess large RNA genomes and produce multiple sgRNAs exhibit no temporal regulation of the transcription (5, 25, 35). It should be remembered, however, that the time scale of coronavirus reproduction is within hours, compared to days for closteroviruses. Among plant viruses producing more than one sgRNA, no temporal regulation was found for two sgRNAs of turnip crinkle carmovirus (36). Strikingly, the plant tobamovirus that also expresses two sgRNAs does regulate the timing of their accumulation (9).
What is common among diverse groups of positive-strand RNA viruses is tight quantitative regulation of subgenomic promoter activities. Several elegant studies have previously demonstrated the roles of the core promoter and additional elements located in adjacent regions of the template (e.g., see references 15, 20, 22, 27, 29, 33, and 36) or even on a distinct RNA molecule (34) in regulating transcription levels. Promoter strength was also shown to depend on the location of the promoter and the presence of additional subgenomic promoters (9, 35). It seems that all these factors modulate closterovirus transcription as well. First, the CP promoter remains the strongest among BYV promoters even though its position varies among four analyzed variants. Specifically, the distance of this promoter from the 3′ end of genomic RNA is 1.8 kb in the wild-type and HSP70hGUS variants, 4.1 kb in the CPGUS and p20GUS variants, and 2.8 kb in the GUS-p21 variant. Second, the activity of the CP promoter decreases in CPGUS and p20GUS but not in HSP70hGUS, likely due to the increased distance from the 3′ end in the first two variants but not in the last variant. Third, the deletion of the adjacent promoters in the truncated GUS-p21 variant results in a dramatic activation of the CP promoter even though it is located farther from the 3′ end than in the wild-type virus.
The ability of BYV to accommodate a 2.3-kb reporter ORF at different positions of the genome demonstrates its utility as a high-capacity gene expression vector. A potential advantage provided by a self-processing reporter is the production of a reporter and a viral product from the same expression unit that minimizes modification of a virus genetic makeup. Preservation of all genes and control regions in a tagged virus appears to be advantageous for studies of viral replication, assembly, and translocation (11, 32).
ACKNOWLEDGMENTS
We thank William Dawson, Theo Dreher, and Bryce Falk for critical reading of the manuscript. The participation of Amit Gal-On in the generation of GUS-tagged BYV variants is greatly appreciated.
This work was supported by grants from the National Institutes of Health (R1GM53190B) and the U.S. Department of Agriculture (NRICGP 97-35303-4515) to V.V.D.
REFERENCES
- 1.Agranovsky A A, Boyko V P, Karasev A V, Lunina N A, Koonin E V, Dolja V V. Nucleotide sequence of the 3′-terminal half of beet yellows closterovirus RNA genome: unique arrangement of eight virus genes. J Gen Virol. 1991;72:15–23. doi: 10.1099/0022-1317-72-1-15. [DOI] [PubMed] [Google Scholar]
- 2.Agranovsky A A, Folimonov A S, Folimonova S Y, Morozov S Y, Schieman J, Lesemann D, Atabekov J G. Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J Gen Virol. 1998;79:889–895. doi: 10.1099/0022-1317-79-4-889. [DOI] [PubMed] [Google Scholar]
- 3.Agranovsky A A, Lesemann D E, Maiss E, Hull R, Atabekov J G. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc Natl Acad Sci USA. 1995;92:2470–2473. doi: 10.1073/pnas.92.7.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison R, Johnston R E, Dougherty W G. The nucleotide sequence of the coding region of tobacco etch virus genomic RNA: evidence for a synthesis of a single polyprotein. Virology. 1986;154:9–20. doi: 10.1016/0042-6822(86)90425-3. [DOI] [PubMed] [Google Scholar]
- 5.An S, Maeda A, Makino S. Coronavirus transcription early in infection. J Virol. 1998;72:8517–8524. doi: 10.1128/jvi.72.11.8517-8524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-Joseph M, Yang G, Gafni R, Mavassi M. Subgenomic RNAs: the possible building blocks for modular recombination of Closteroviridae genomes. Semin Virol. 1997;8:113–119. [Google Scholar]
- 7.Carrington J C, Cary S M, Parks T D, Dougherty W G. A second proteinase encoded by a plant potyvirus genome. EMBO J. 1989;8:365–370. doi: 10.1002/j.1460-2075.1989.tb03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrington J C, Freed D D. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson W O, Lehto K M. Regulation of tobamovirus gene expression. Adv Virus Res. 1990;38:307–342. doi: 10.1016/S0065-3527(08)60865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolja V V, Karasev A V, Agranovsky A A. Organization of beet yellows closterovirus genome. In: Heinz F X, Brinton M A, editors. New aspects of positive-strand RNA viruses. Washington, D.C: American Society for Microbiology; 1990. pp. 31–35. [Google Scholar]
- 11.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase (GUS) into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolja V V, Herndon K L, Pirone T P, Carrington J C. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J Virol. 1993;67:5968–5975. doi: 10.1128/jvi.67.10.5968-5975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolja V V, Karasev A V, Koonin E V. Molecular biology and evolution of closteroviruses: sophisticated build-up of large RNA genomes. Annu Rev Phytopathol. 1994;32:261–285. [Google Scholar]
- 14.Dolja V V, Hong J, Keller K E, Martin R R, Peremyslov V V. Suppression of potyvirus infection by coexpressed closterovirus protein. Virology. 1997;234:243–252. doi: 10.1006/viro.1997.8660. [DOI] [PubMed] [Google Scholar]
- 15.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X-H, Rao A L N, Creamer R. Characterization of beet yellows closterovirus-specific RNAs in infected plants and protoplasts. Phytopathology. 1997;87:347–352. doi: 10.1094/PHYTO.1997.87.3.347. [DOI] [PubMed] [Google Scholar]
- 17.Hilf M E, Karasev A V, Pappu H R, Gumpf D J, Niblett C L, Garnsey S M. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology. 1995;208:576–582. doi: 10.1006/viro.1995.1188. [DOI] [PubMed] [Google Scholar]
- 18.Karasev A V, Hilf M E, Garnsey S M, Dawson W O. Transcriptional strategy of closteroviruses: mapping the 5′ termini of the citrus tristeza virus subgenomic RNAs. J Virol. 1997;71:6233–6236. doi: 10.1128/jvi.71.8.6233-6236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karasev A V, Kashina A S, Gelfand V I, Dolja V V. HSP70-related 65 kDa protein of beet yellows closterovirus is a microtubule-binding protein. FEBS Lett. 1992;304:12–14. doi: 10.1016/0014-5793(92)80578-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim K-H, Hemenway C. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 21.Klaassen V A, Boeshore M, Koonin E V, Tian T, Falk B W. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly transmitted, bipartite closterovirus. Virology. 1995;208:99–110. doi: 10.1006/viro.1995.1133. [DOI] [PubMed] [Google Scholar]
- 22.Koev G, Mohan B R, Miller W A. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel T A, Roberts J D, Zakour R. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 25.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levis R, Schlesinger S, Huang H V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh L E, Dreher T W, Hall T C. Mutational analysis of the core and modulator sequences of the BMV RNA 3 subgenomic promoter. Nucleic Acids Res. 1988;16:981–985. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina V, Peremyslov V V, Hagiwara Y, Dolja V V. Subcellular localization of the HSP70-homolog encoded by beet yellows closterovirus. Virology. 1999;260:173–181. doi: 10.1006/viro.1999.9807. [DOI] [PubMed] [Google Scholar]
- 29.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature (London) 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 30.Navas-Castillo J, Albiach-Marti M R, Gowda S, Hilf M, Garnsey S M, Dawson W O. Kinetics of accumulation of citrus tristeza virus RNAs in host and non-host protoplasts. Virology. 1997;228:92–97. doi: 10.1006/viro.1996.8369. [DOI] [PubMed] [Google Scholar]
- 31.Peremyslov V V, Hagiwara Y, Dolja V V. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J Virol. 1998;72:5870–5876. doi: 10.1128/jvi.72.7.5870-5876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santa Cruz S, Chapman S, Roberts A G, Roberts I M, Prior D A M, Oparka K J. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc Natl Acad Sci USA. 1996;93:6286–6290. doi: 10.1073/pnas.93.13.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sit T L, Vaewhongs A A, Lommel S A. RNA-mediated transactivation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 35.van Marle G, Luytjes W, van der Most R, van der Straaten T, Spaan W J. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Simon A E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology. 1997;232:174–186. doi: 10.1006/viro.1997.8550. [DOI] [PubMed] [Google Scholar]
- 37.Zhu H-Y, Ling K-S, Goszczynski D E, McFerson J R, Gonsalves D. Nucleotide sequence and genome organization of grapevine leafroll-associated virus-2 are similar to beet yellows virus, the closterovirus type member. J Gen Virol. 1998;79:1289–1298. doi: 10.1099/0022-1317-79-5-1289. [DOI] [PubMed] [Google Scholar]