Abstract
Analysis of a cat genomic DNA library showed that cats harbor a previously unrecognized endogenous type C retrovirus, whose env gene has homology to the murine Fv-4 resistance gene. This unique retrovirus, designated FcEV (Felis catus endogenous retrovirus), has a type C pol gene, closely related to the primate Papio cynocephalus endogenous virus (PcEV) pol, not overlapping the env gene, unlike in other type C retroviruses, and is presumably present in a higher copy number than RD-114. Phylogenetic analysis of FcEV and RD-114 fragments amplified from cat species and comparison with baboon endogenous virus (BaEV) fragments from monkeys suggested that RD-114 does not represent the cat strain of BaEV but is actually a new recombinant between FcEV type C genes and the env gene of BaEV. Although BaEV did appear to have infected an ancestor of the domestic cat lineage, it was a de novo recombinant that made its way into the cat germ line.
All 38 known cat species, including the big cats, arose during the last 10 to 15 million years (13), with the lineage leading to the domestic cat being formed 7 to 9 million years ago (11). Extant cats belonging to the species Felis probably have a common ancestor much more recently, since they are genetically closely related. An interesting feature of the genus Felis (composed of Felis catus, F. chaus, F. nigripes, F. margarita, F. silvestris, and F. libyca) is the presence of two endogenous retroviruses not found in any other cat species, e.g., endogenous feline leukemia virus (FeLV) and RD-114 (2, 20). RD-114 showed a high level of homology to the monkey virus baboon endogenous virus (BaEV), which was attributed to an ancient cross-species transmission. RD-114 can still be expressed, but there is probably only a single active copy on cat chromosome B3 (19). Analysis of RD-114 proviruses in the domestic cat genome showed that there are approximately 20 related integrations per cell but that most have either lost their env genes or replaced them by a completely unrelated env (20, 23). No complete sequence is available for RD-114; only the env gene and the 3′ end of the pol gene have been sequenced (7), and some long terminal repeat (LTR) sequences are available (22).
BaEV is a complete inducible endogenous retrovirus (1), which is found in the genomic DNA of a subset of African monkeys, e.g., baboons, geladas, mandrills, mangabeys, and African green monkeys (26). Phylogenetic analysis of BaEV sequences from monkeys indicated that two BaEV strains have entered independently into the germ line of the monkey species (26). The last common ancestor of the Cercopithicinae supposedly lived at least 9 million years ago, indicating that exogenous BaEV was spreading in Africa after that time. BaEV is a recombinant retrovirus, containing type C gag and pol genes and a type D env gene, which probably arose by recombination of two primate viruses (simian endogenous retrovirus [SERV] and Papio cynocephalus endogenous virus [PcEV]), whose genomes are also present in monkey genomic DNA (17, 27). RD-114 has the same mosaic genomic structure as BaEV. The env gene of RD-114 is most probably derived from BaEV and not from the primate type D virus SERV, one of the ancestors of BaEV. Although the three env genes are generally very homologous, BaEV and RD-114 have extensive homology at the extreme C-terminal part of this protein, and the 30 nucleotides nt downstream of the env stop codon are highly homologous, in contrast to SERV. Recombination probably took place just downstream of the env gene. The env-LTR intergenic region in type D viruses containing a constitutive transport element, which is absent from type C retroviruses.
To expand our knowledge of the evolution of RD-114 and BaEV, we have analyzed two env genes of presumed RD-114 proviruses isolated from a cat genomic library and have examined RD-114 sequence variation in different species of cats and compared it with BaEV variation in monkey species.
MATERIALS AND METHODS
Cat genomic library.
A cat genomic library in the lambda FIXR II vector was obtained from Stratagene (La Jolla, Calif.). This library was constructed from whole blood of an adult female domestic cat of mixed breed.
Isolation of type C proviral clones.
The cat genomic library was screened by using a 32P-labelled probe homologous to the 3′ end of the RD-114 pol sequence. Lambda DNA was isolated from purified positive plaques by using the Wizard Lambda Preps DNA purification system from Promega (Madison, Wis.).
Cat and primate samples.
DNA samples were obtained from F. catus (domestic cat), F. chaus (jungle cat), and F. silvestris (European wild cat). The F. chaus and F. silvestris DNA samples were kindly donated by Stephen J. O’Brien (National Cancer Institute, Frederick, Md.). Primate samples (peripheral blood mononuclear cells [PBMCs] and serum for Cercocebus aterrimus) were obtained from the following African monkeys: Papio ursinus (chacma baboon), Cercocebus aterrimus (black mangabey), Cercopithecus aethiops aethiops (grivet), Cercopithecus aethiops pygerythrus (vervet), and Cercopithecus aethiops sabaeus (green monkey). The origin of the samples was as published previously (26).
DNA extraction and amplification.
Total DNA was extracted from the samples by a procedure involving silica and guanidine thiocyanate (5). A 590-nt nested fragment of BaEV or RD-114 was amplified by using an outer primer set designated RD. The RD primer set consisted of an upstream primer, 5′ GACCTTAGAGACTGGC 3′ (nt 5655 to 5670 of the BaEV sequence [14]), located in the pol gene and a downstream primer, 5′ GCTGCAATCGCATGG 3′ (nt 6343 to 6357 of BaEV), located in the env gene. A nested set, consisting of primers upstream 5′ GGAGACGCCTCCTATCTC 3′ (nt 5681 to 5698 of BaEV) and downstream 5′ GCGAGGGTCGTCAAACCC 3′ (nt 6289 to 6306 of BaEV), was used in a second amplification reaction, if necessary. A PCR fragment could be obtained from F. chaus only by using the nested primer set directly on the genomic DNA, probably because of mismatches with the outer primer set. For FcEV amplification, the upstream primers were combined with downstream primers based on the env sequence of this virus: 5′ GGGGAAGTTGTCTGGGAGGT 3′ and the nested primer 5′ GAGGGGAACCCACATCAGGT 3′. This primer combination was designated FC. First-round PCR amplifications were done under the following conditions: denaturation for 5 min at 95°C; amplification consisting of 35 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C; followed by an extension of 10 min at 72°C. The protocol for the nested reactions was identical, except that only 25 cycles were performed. The obtained fragments were cloned into the TA vector (Invitrogen, San Diego, Calif.) and sequenced. At least two clones from a single individual were analyzed.
DNA sequencing.
Sequencing of the clones with an ABI 373A or ABI 377 automated sequencer was done in both directions directly from lambda DNA with purified specific primers, as specified by the manufacturer. PCR fragments obtained from genomic DNA were sequenced directly from plasmid DNA with T7 and M13 dye primers.
Sequence analysis.
Alignment of PCR fragments and lambda clone sequences was done with CLUSTAL-W (9) and adjusted by hand. The phylogenetic analyses were done by the neighbor-joining (NJ) method, as implemented in the MEGA package (16). Distances were estimated by Kimura’s two-parameter method (15). One hundred bootstrap replicates were analyzed. The rate constancy in the pol gene was checked by the two-cluster test as described by Takezaki et al. (25), and linearized NJ trees were constructed with a program made available by these authors. The RD-114 sequence used is from reference 7 (GenBank accession no. X87829).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database (accession no. AF155060, AF155061, and AF164903 to AF164923).
RESULTS
Analysis of the Fc21 and Fc41 env genes.
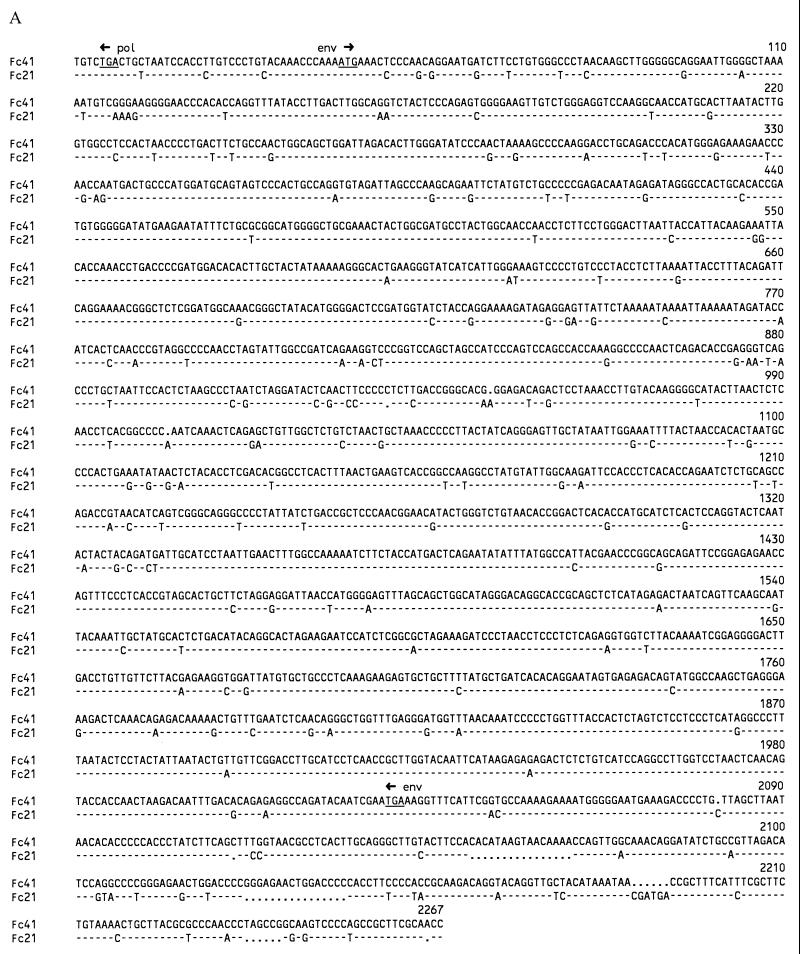
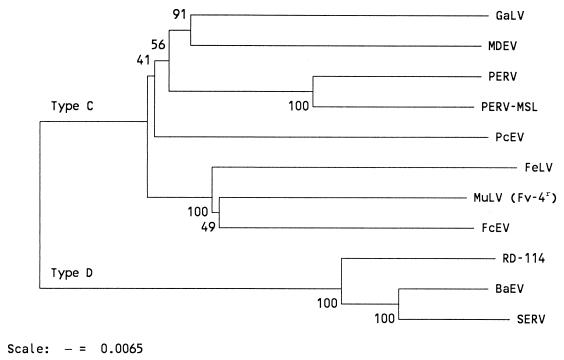
Proviral clones Fc21 and Fc41 were obtained by hybridizing a cat genomic library to a RD-114 pol probe. Sequence analysis showed that both clones contained complete and closely related type C env genes, in contrast to RD-114, which contains a type D env gene. The sequences of part of pol, the pol-env intergenic region, env, and part of the 3′ LTR contained in these clones are shown in Fig. 1A. The env reading frame was completely open in clone Fc21, but in Fc41 it was interrupted three times by the deletion or insertion of single nucleotides. The Env protein sequences encoded by these clones (Fig. 1B) were analyzed together with Env proteins from type C and type D retroviruses (Fig. 2). Fc21 and Fc41 Env proteins were closely related to murine type C viruses, especially to the ecotropic murine leukemia virus (MuLV) group. The endogenous Fv-4r env gene product of Asian mice showed the highest level of amino acid homology to the cat clones, especially in the P15E protein.
FIG. 1.
(A) Alignment of the cat genomic proviral clones Fc21 and Fc41. The stop codons of the pol and env genes and the start codon of the env gene are indicated by arrows and underlined. (B) Alignment of amino acid sequences of Fc21 and Fc41 derived from the sequences in Fig. 1A. The Fc41 reading frame has been modified to allow translation by the removal of 1 nt and the insertion of 2 nt, according to the Fc21 ORF. The putative signal peptide and the gp70 and P15E proteins are indicated. Processing sites were obtained by comparison of Fc21 and Fc41 with the GA-FeLV-B env protein (8). The putative immunosuppressive peptide is shaded.
FIG. 2.
NJ tree based on a p-distance matrix generated from amino acid sequences of retroviral type C env genes. Bootstrap values for 100 replicated trees are indicated. GaLV, gibbon ape leukemia virus (accession no. M26927); MDEV, is Mus dunni endogenous virus (AF053745); PERV (Y17013) and PERV-MSL (AF038599), pig viruses; PcEV, complete type C endogenous monkey virus (16); FeLV (M89997), feline leukemia virus; MuLV Fv-4r, murine leukemia virus env gene present at the Fv-4r locus (M33884). The type D env genes analyzed are from RD-114, BaEV, and SERV (U85506).
At the nucleotide level, an unpublished proviral env sequence from F. silvestris deposited in the GenBank database (accession no. X51929) showed high homology to Fc21 and lower homology to Fc41. Previously, after analysis of a small fragment of X51929, the authors suggested that the clone contained an RD-114/FeLV recombinant virus (4).
Analysis of the Fc21 and Fc41 3′ untranslated region.
Proviral genomes are flanked at both ends by untranslated LTR sequences, which are involved in regulation of gene expression and replication of the viral genome. LTRs are the most variable part of the retroviral genome. Between the stop codon of the env gene and the U3 region of the 3′ LTR, a region containing the polypurine tract, involved in plus-strand synthesis, is found. Analysis of part of the 3′ untranslated regions of both Fc21 and Fc41, containing the unidentified start of the U3 region of the LTR, showed them to be homologous but not identical to each other (Fig. 1A). In a BLAST search of the GenBank database, homology to supposed RD-114 LTRs was found, including the 3′ LTR of the inducible virus, but no homology to any other retrovirus LTRs was found. Of the inducible RD-114, large stretches of homology to BaEV LTRs has also been found, especially at the R and U5 regions of the LTR, which were not sequenced in Fc21 and Fc41.
In the untranslated region located between the stop codon of the env gene and the start of the 3′ LTR, high homology to Fv-4r MuLV and to FeLV was found. For Fc21, the polypurine tract and a stretch of 18 nt downstream showed 100% homology to a rat retroviral transcript. The inducible RD-114 is similar to BaEV in this region and has less homology to Fc21 and Fc41.
Analysis of the Fc21 and Fc41 pol fragment and intergenic region.
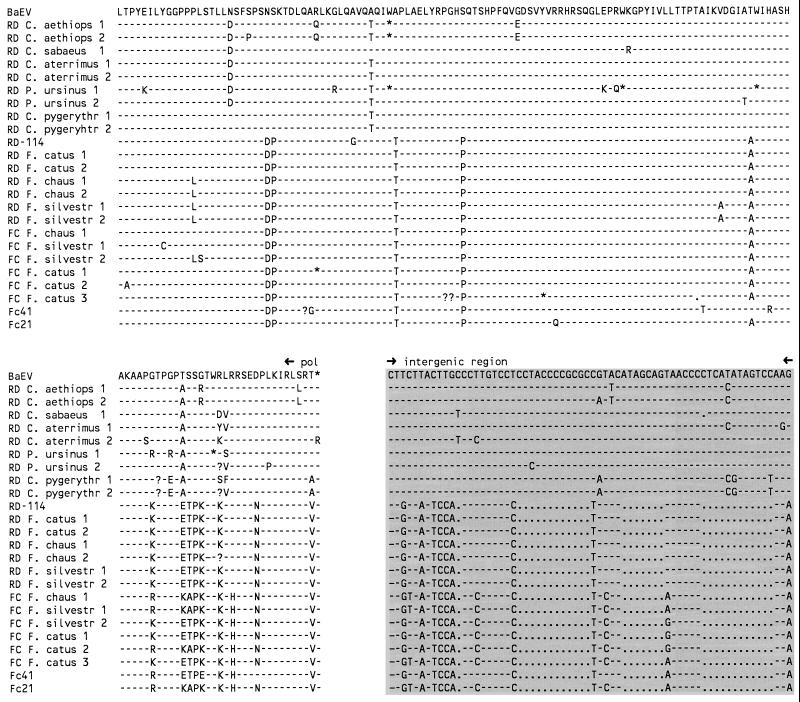
The env gene of Fc21 and Fc41 was found not to overlap the pol open reading frame (ORF), as in many type C viruses, but was separated from it by an untranslated intergenic region. A separate env gene is common in type D retroviruses. From the upstream pol gene of Fc21 and Fc41, approximately 430 nt encoding part of the endonuclease (integrase) was additionally sequenced (see Fig. 4). The pol sequences of Fc21 and Fc41 were then analyzed together with the amplified fragments from cat and monkey genomic DNA (see Fig. 5). The Fc21 and Fc41 pol genes are C type and are closely related to RD-114 pol. The intergenic region of Fc21 and Fc41 is almost identical to that of RD-114 (see Fig. 4). However, based upon the divergent env sequences of the Fc21 and Fc41 genomic clones, we postulated that the proviral genomes present in these clones represent two closely related virus strains distinct from RD-114. This novel endogenous virus we named Felis catus endogenous virus (FcEV).
FIG. 4.
Alignment of a pol-env fragment amplified from monkey and cat species. The pol and env parts of the sequence are shown translated into amino acids, while the intergenic region is shown as nucleotides. The sequence corresponds to nt 5738 to 6288 of the BaEV reference sequence (14). Gaps introduced for optimal alignment are indicated by dots, and identical nucleotides are indicated by dashes. Stop codons are indicated by an asterisk, and incomplete codons (due to single nucleotide deletions) are indicated by a question mark. The primer set (RD or FC) used in generation of the fragments is identified.
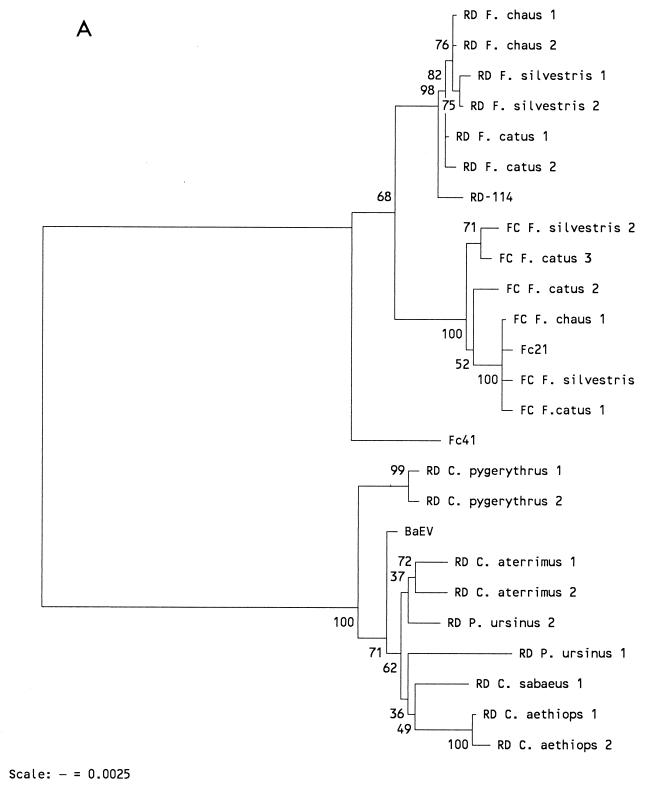
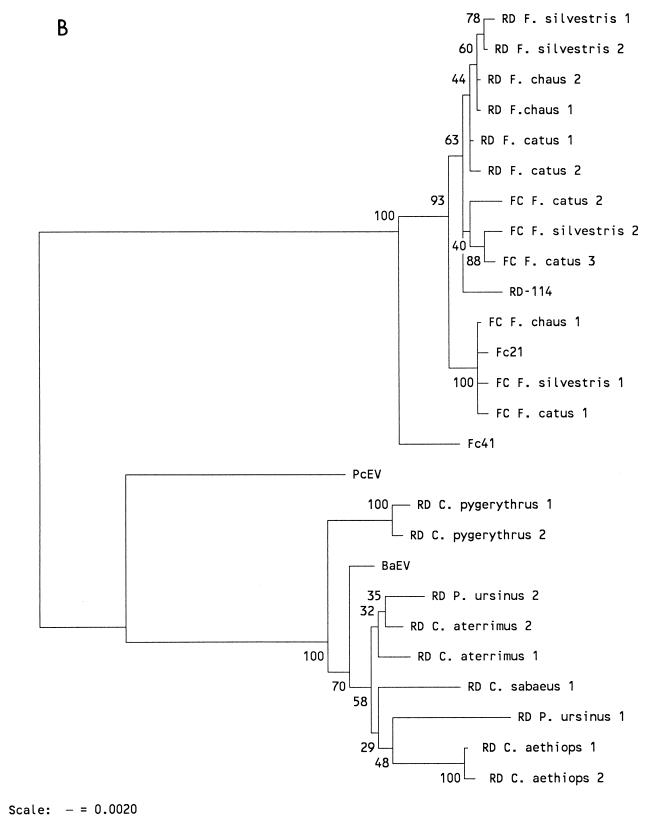
FIG. 5.
(A) NJ tree based on the pol-env nucleotide fragments shown in Fig. 4. Bootstrap values for 100 replicated trees are indicated. Treating gaps introduced for optimal alignment as uninformative, or using them in pairwise comparisons, did not influence the tree. (B) NJ tree based upon only the pol fragment shown in Fig. 4. The corresponding pol fragment of the endogenous primate type C virus PcEV (17) was also included in the analysis. A NJ tree based upon the derived amino acid sequence of the pol fragment showed an identical topology (not shown). The primer set (RD or FC) used in generation of the fragments is identified.
Determination of the recombination breakpoints of RD-114.
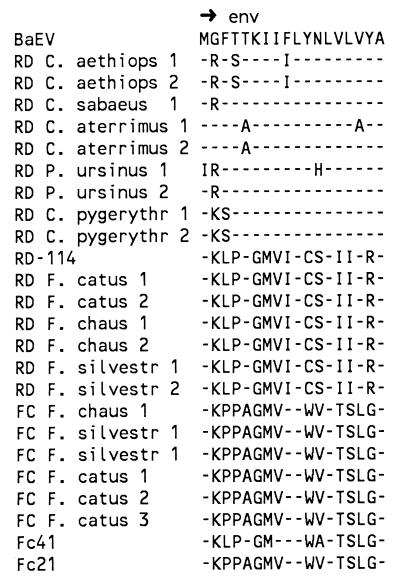
Comparison of the genomes of RD-114, FcEV, and BaEV suggested that RD-114 is a recombinant virus with the following features: the pol gene, pol-env intergenic region, and LTRs were most probably derived from FcEV, while the env gene has high homology to BaEV env, suggesting that it originated from BaEV. Because the pol-env intergenic region of RD-114 was very similar to that of FcEV, it is most likely that recombination occurred somewhere at the 5′ end of the env gene. Alignment of the leader or signal peptide of the gp70 protein region of the env gene suggested that recombination could have occurred in this sequence, since high homology at the nucleotide level was observed (Fig. 3A). Amino acid homology is less obvious in the C-terminal part of the signal peptide, suggesting rapid adaptation to the new host species. The N-terminal part of the signal peptide of all cat viruses was very homologous, suggesting that the point of recombination was located downstream of the first 20 nt of the env reading frame.
FIG. 3.
Analysis of the recombination breakpoints of RD-114. (A) Putative 5′ breakpoint located in the signal peptide sequence of gp70. The start codon of env is underlined. Shading indicates homologous sequences probably involved in recombination. Amino acids derived from the signal peptide sequences are shown. (B) Putative 3′ breakpoint located between the stop codon of env (underlined), and the start of the 3′ LTR (arrow). The possible area of recombination is shaded. P15E C-terminal amino acid sequences are shown.
Homology, at both the nucleotide and amino acid levels, continues throughout env. The amino acid changes observed most probably represent adaptation to the new host. The C-terminal env sequences of RD-114 and BaEV were almost identical, indicating that recombination most probably occurred downstream of the env ORF. However, the RD-114 LTR sequence showed higher homology to FcEV than to BaEV, suggesting that recombination should have occurred between the stop codon of env and the beginning of the 3′ LTR. Indeed, a homologous sequence of approximately 20 nt was found at this position in all three viruses (Fig. 3B). Concluding, the recombination breakpoints of RD-114 are presumably located in the N-terminal part of gp70 (around amino acids 7 to 9) and between the stop codon of env and the boundary of the 3′ LTR sequence. Recombination thus involved almost the complete env gene.
Amplification of BaEV, RD-114, and FcEV fragments.
The derived amino acid sequences of the BaEV, RD-114, and FcEV fragments amplified from monkey and cat genomic DNA are shown in Fig. 4. The fragment included the 3′ end of the pol gene, the pol-env intergenic region, and the 5′ end of the env gene. Translation of the pol ORF showed that many substitutions are silent and do not lead to amino acid replacement compared to BaEV. The RD-114 fragments amplified from the three species of cats are almost identical, suggesting that the time of divergent evolution is short and that probably the same single locus has been amplified. Cat FcEV fragments showed a little more variation than did the RD-114 sequences. BaEV fragments amplified with primer set RD from monkey DNA were even more variable, suggesting that different loci have been amplified or that the total time of divergent evolution is longer. Defective clones, containing premature stop codons or small deletions, were amplified from both monkey and cat DNA.
With the pol-env fragment used, no distinction can be made between the BaEVfor strain infecting mangabeys and mandrills and the BaEVsav strain infecting baboons, geladas, and African green monkeys, since the strains were separated earlier by using LTR sequences and, to a lesser extent, env sequences (26). The few nucleotide differences between the pol-env fragment of the cultured RD-114 isolate (7) and the sequences obtained from cat genomic DNA (Fig. 4) could be due either to mutations acquired during culturing or to sequence artifacts.
Phylogenetic analysis of BaEV, RD-114, and FcEV fragments.
NJ analysis of amplified fragments showed that both endogenous cat viruses clustered together and away from monkey BaEV (Fig. 5A). Surprisingly, the Fc41 fragment was basal to all cat sequences and was most closely related to monkey viruses, which suggested that Fc41 represented the most ancient integration. Analysis of only pol sequences (Fig. 5B) showed more clearly that all cat virus pol fragments had evolved from Fc41 pol. This observation was confirmed in a linearized NJ tree (not shown), where the rate constancy is calculated with respect to an outgroup and subsequently forced upon the sequences. Therefore, RD-114 pol sequences are almost certainly derived from FcEV pol genes. The near absence of divergence in RD-114 from different cat species again suggested that a single locus has been amplified. An NJ analysis of pol-derived amino acid sequences generated a tree identical to the nucleotide pol tree (not shown).
DISCUSSION
To investigate the evolution of the monkey retrovirus BaEV after its presumed transmission to cats, as suggested by sequence comparison, we have sequenced the env genes and adjacent genome parts of two RD-114 proviruses integrated in the genome of the domestic cat (F. catus). Analysis of the two clones (Fc21 and Fc41) revealed that their env genes are of type C, in contrast to the type D env found in RD-114. Earlier hybridization studies had already shown that all putative RD-114 proviruses analyzed possessed env genes unrelated to inducible RD-114 (20, 23). Most of these integrations are full-length proviruses with gag, pol, and env genes flanked by two LTRs. All or most proviruses formerly attributed to RD-114 are more likely to be FcEV integrations, and we assume that the number of 15 to 20 copies per haploid genome is probably the copy number of FcEV. It would be worthwhile to determine the actual FcEV copy number, its gag sequence, and its distribution in feline species. Earlier experiments involving Southern hybridization showed that the RD-114-unrelated env sequences were also unique for the genus Felis and do not occur in primates (20). It is possible that the single inducible RD-114 locus is the only representative of the true RD-114 virus in the cat genome. Domestic cats are polymorphic with respect to the presence or absence of the inducible RD-114 provirus on chromosome B3 (19), suggesting that this integration is recent in evolutionary terms.
The origin of the FcEV env gene is most probably a murine type C virus. Database homology searches and phylogenetic analysis of type C Env proteins showed that FcEV env, and the env-LTR intergenic region, are related to the Fv-4 resistance gene and its downstream sequence of Asian Mus musculus (10). The Fv-4r locus contains the env gene of an ecotropic MuLV, whose expression protects carrier mice from new ecotropic MuLV infections. It would be interesting to see if expression of FcEV env genes occurs and, if so, if it protects cats from related infections. It has been shown previously that FeLV is also derived from rodent viruses. Elder and Mullins (8) noted homologies between the env genes of FeLV-B and a murine mink cell focus-forming virus, while Benveniste et al. (3) observed a larger degree of homology to rat sequences.
The pol gene of Fc41 was found to be closely related to monkey type C pol genes, especially to the recently characterized PcEV pol gene, and less closely related to the BaEV pol gene. PcEV is a full-length type C virus identified in baboons but is present in most Old World monkey genomes (17). PcEV is one of the putative ancestors of BaEV. Phylogenetic analysis showed that all RD-114-like pol genes amplified or isolated from cats, including Fc21 and inducible RD-114, have evolved from the Fc41 pol gene. Thus, the viral integration contained in the Fc41 clone is more ancient than inducible RD-114 but contains a type C env gene. Fc41 thus represents an endogenous type C retrovirus distinct from the prototype RD-114 and should be classified separately as FcEV, together with the closely related Fc21 provirus. Probably, the FcEV provirus present in clone Fc21 represents a less ancient integration of FcEV, integrated much later in the cat genome. Exogenous retroviruses have an increased substitution rate compared to endogenous retroviruses.
RD-114 contains a pol gene distinct from BaEV pol, while cat genomes harbor multiple copies of a type C retrovirus closely related to RD-114 pol. Also, the pol-env intergenic region and the LTRs of RD-114 and FcEV are closely related. The primer binding site of RD-114 and all RD-114-related viruses was found to be complementary to tRNAGly (22), in contrast to the PBS of BaEV, which utilizes tRNAPro to initiate replication (14).
From the phylogenetic analyses, the copy number (15 to 20 per haploid genome, as suggested in reference 20) and the non-inducibility of the FcEV proviruses, it can be assumed that they are older than the RD-114 integration. Although newly integrated proviruses can also be defective, proviruses generally become increasingly suppressed by their host as a function of time. Phylogenetic analysis showed that it is unlikely that the pol gene of RD-114 and the untranslated parts of its genome are derived from BaEV. However, the RD-114 env gene is highly homologous to BaEV env, suggesting that RD-114 is a recombinant virus between the cat virus FcEV and primate BaEV. Recombination could have been facilitated by the two homologous stretches, one located in the signal peptide sequence of the env gene and the other just downstream of the env stop codon. The presence of a separate env gene in FcEV, as in type D viruses, facilitated the exchange of this gene. BaEV thus did infect a cat ancestor, but it was a newly recombined virus that made its way into the cat germ line. Recombination is a common strategy in retroviruses, as is presently seen in the human immunodeficiency virus type 1 pandemic, where approximately 10% of viruses isolated are recombinants (21), either between different subtypes or between different strains. In simian immunodeficiency virus, the virus harbored by sabaeus monkeys is a recombinant between African green monkey and sooty mangabey simian immunodeficiency viruses (12). Also, BaEV itself is a recombinant between C-type (PcEV) and D-type (SERV) primate retroviruses (17, 27).
Cats of the genus Felis thus harbor at least three unique retroviruses in their genome: (i) FcEV, a type C virus, of which presumably 15 to 20 copies exist; (ii) the FcEV-BaEV recombinant virus RD-114, probably present as a single inducible copy; and (iii) multiple endogenous FeLV integrations, which can recombine with exogenous FeLVs (18, 24). Since all three viruses must have infected the common ancestor of the domestic cat lineage, this suggests that in the ancestral cat population, two rodent retroviruses and a primate retrovirus were simultaneously spreading. It should be noted that cats also harbor endogenous virus sequences with homology to the macaque type C virus MAC-1 (6).
REFERENCES
- 1.Benveniste R E, Lieber M M, Livingston D M, Sherr C J, Todaro G J. Infectious C-type virus isolated from a baboon placenta. Nature (London) 1974;248:17–20. doi: 10.1038/248017a0. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste R E, Todaro G J. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature (London) 1974;252:456–459. doi: 10.1038/252456a0. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste R E, Sherr C J, Todaro G J. Evolution of type C viral genes: origin of feline leukemia virus. Science. 1975;190:886–888. doi: 10.1126/science.52892. [DOI] [PubMed] [Google Scholar]
- 4.Beyer W, Möhring R, Drescher B, Nötzel U, Rosenthal S. Molecular cloning of an endogenous cat retroviral element (ECE1)—a recombinant between RD-114 and FeLV-related sequences. Arch Virol. 1987;96:297–301. doi: 10.1007/BF01320971. [DOI] [PubMed] [Google Scholar]
- 5.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for the purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner T I, Todaro G J. Carnivores have sequences in their cellular DNA distantly related to the primate endogenous virus, MAC-1. Virology. 1979;94:224–227. doi: 10.1016/0042-6822(79)90454-9. [DOI] [PubMed] [Google Scholar]
- 7.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins F L. High titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder J H, Mullins J I. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol. 1983;46:871–880. doi: 10.1128/jvi.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Highness D G, Sharp P M. CLUSTAL-W: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H, Laigret F, Martin M A, Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55:768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janczewski D N, Modi W S, Stephens J C, O’Brien S J. Molecular evolution of mitochondrial 12S RNA and cytochrome b sequences in the pantherine lineage of Felidae. Mol Biol Evol. 1995;12:690–707. doi: 10.1093/oxfordjournals.molbev.a040232. [DOI] [PubMed] [Google Scholar]
- 12.Jin M J, Hui H, Robertson D L, Müller M C, Barré-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson W E, O’Brien S J. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J Mol Evol. 1997;44(Suppl.1):S98–S116. doi: 10.1007/pl00000060. [DOI] [PubMed] [Google Scholar]
- 14.Kato S, Matsuo K, Nishimura N, Takahashi N, Takano T. The entire nucleotide sequence of baboon endogenous virus DNA: a chimeric genome structure of murine type C and simian type D retroviruses. Jpn J Genet. 1987;62:127–137. [Google Scholar]
- 15.Kimura M A. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analyses, version 1.02. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 17.Mang R, Goudsmit J, van der Kuyl A C. Novel endogenous type C retrovirus in baboons: complete sequence, providing evidence for baboon endogenous virus gag-pol ancestry. J Virol. 1999;73:7021–7026. doi: 10.1128/jvi.73.8.7021-7026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature (London) 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 19.Reeves R H, Nash W G, O’Brien S J. Genetic mapping of endogenous RD-114 retroviral sequences of domestic cats. J Virol. 1985;56:303–306. doi: 10.1128/jvi.56.1.303-306.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves R H, O’Brien S J. Molecular genetic organization of the RD-114 gene family of endogenous feline retroviral sequences. J Virol. 1984;52:164–171. doi: 10.1128/jvi.52.1.164-171.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 22.Spodick D A, Ghosh A K, Parimoo S, Roy-Burman P. The long terminal repeat of feline endogenous RD-114 retroviral DNAs: analysis of transcription regulatory activity and nucleotide sequence. Virus Res. 1988;9:263–283. doi: 10.1016/0168-1702(88)90035-4. [DOI] [PubMed] [Google Scholar]
- 23.Spodick D A, Soe L H, Roy-Burman P. Genetic analysis of the feline RD-114 retrovirus related endogenous elements. Virus Res. 1984;1:543–555. doi: 10.1016/0168-1702(84)90012-1. [DOI] [PubMed] [Google Scholar]
- 24.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 26.Van der Kuyl A C, Dekker J T, Goudsmit J. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J Virol. 1995;69:7877–7887. doi: 10.1128/jvi.69.12.7877-7887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Kuyl A C, Mang R, Dekker J T, Goudsmit J. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J Virol. 1997;71:3666–3676. doi: 10.1128/jvi.71.5.3666-3676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]