Abstract
Viral gene products are generally required in widely differing amounts for successful virus growth and assembly. For coronaviruses, regulation of transcription is a major contributor to these differences, but regulation of translation may also be important. Here, we examine the possibility that the 5′ untranslated regions (UTRs), unique for each of the nine species of mRNA in the bovine coronavirus and ranging in length from 70 nucleotides (nt) to 210 nt (inclusive of the common 5′-terminal 65-nt leader), can differentially affect the rate of protein accumulation. When the natural 77-nt 5′ UTR on synthetic transcripts of mRNA 7 (mRNA for N and I proteins) was replaced with the 210-nt 5′ UTR from mRNA 1 (genomic RNA, mRNA for viral polymerase), approximately twofold-less N, or (N) CAT fusion reporter protein, was made in vitro. Twofold less was also made in vivo in uninfected cells when a T7 RNA polymerase-driven transient-transfection system was used. In coronavirus-infected cells, this difference surprisingly became 12-fold as the result of both a stimulated translation from the 77-nt 5′ UTR and a repression of translation from the 210-nt 5′ UTR. These results reveal that a differential 5′ UTR-directed regulation of translation can occur in coronavirus-infected cells and lead us to postulate that the direction and degree of regulation is carried out by viral or virally induced cellular factors acting in trans on cis-acting elements within the 5′ UTR.
Coronaviruses are positive-strand RNA viruses with a genome of approximately 30 kb, the largest known among RNA viruses. Both virus genome replication and transcription take place in the cytoplasm (reviewed in references 23 and 29). Transcription generates a nested set of six to eight subgenomic mRNAs of various lengths that are 3′ coterminal with the genome and translated in most cases from the 5′ terminal open reading frame (ORF). The subgenomic mRNAs are also 5′ coterminal with the genome by virtue of a discontinuous transcription system that places a common leader (making up the 5′-terminal portion of each 5′ untranslated region [UTR]) on the 5′ end of each transcript. A major form of regulation of coronavirus gene expression is documented to be at the level of transcription during which time the shorter mRNAs are produced generally progressively more abundantly than the longer ones. In the bovine coronavirus (BCV), for example, at the time of peak transcription during acute infection (6 h postinfection) there are approximately 1,000 molecules of mRNA 7 to 1 of mRNA 1 (genomic RNA) (20). Regulation of coronavirus gene expression at the level of translation is also documented. (i) It was shown for avian infectious bronchitis virus (IBV) (3), mouse hepatitis virus (MHV) (2), and human coronavirus 229E (HCV 229E) (17), that a −1 ribosomal frameshifting mechanism generates two polypeptides of differing abundance from mRNA 1. (ii) Internal ribosomal entry onto mRNA 3 for IBV (27, 28) and mRNA 5 for MHV (25, 41) are mechanisms used to synthesize a second protein of lesser abundance from a single transcript. (iii) The I protein in BCV and MHV (13, 36), a structural protein of lesser abundance than N but made from an internal ORF on the same mRNA, is synthesized by a leaky scanning mechanism (35). (iv) The coronavirus leader element has been shown to confer a general translational advantage to viral mRNAs in coronavirus-infected cells (39). In addition, mutations arising during persistent infection have been shown to affect translation rates. In the first example, an intraleader ORF in BCV was shown to repress translation of downstream ORFs (19); in the second, an ORF appearing just downstream of the leader in MHV was shown to enhance translation (8).
Here, we examine the possibility that the 5′ UTR, unique for each of the nine species of mRNA in BCV and ranging in length from 70 to 210 nucleotides (nt) (Table 1), affects the rate of translation as evidenced by protein accumulation. The experimental approach was to compare the effects of two 5′ UTRs differing in length by 133 nt on the accumulation of products from mRNA 7 and from an mRNA 7-chloramphenicol acetyltransferase (CAT) fusion transcript. In two different systems of translation in vitro and two in vivo in the absence of coronavirus infection the accumulation from the genomic 5′ UTR of 210 nt was approximately twofold less than from the N mRNA 5′ UTR of 77 nt. In BCV-infected cells this difference became 12-fold as the result of a 0.45-fold stimulation of translation from the 77-nt 5′ UTR and a 3-fold repression of translation from the 210-nt 5′ UTR. Since it was determined that both 5′ UTRs respond in a pattern consistent with a 5′-terminal entry of ribosomes with subsequent scanning, we postulate that the regulation involves the action of viral or virally induced cellular transacting factors on cis-acting elements within the 5′ UTR to affect ribosomal scanning. These results further suggest a possible mechanism, in addition to the regulation of transcription, by which coronaviruses might concurrently maintain high levels of structural protein synthesis and low levels of RNA-dependent RNA polymerase and other products of gene 1 during late infection.
TABLE 1.
5′ untranslated regions on BCV mRNAs
| mRNA species (major protein product) | 5′ UTR sequencea | 5′ UTR lengtha (nt) |
|---|---|---|
| 1 (polymerase) | 5′ (L) UAAACUUUAUAAAAACAUCCACUCCCUGUAUUCUAUGCUUGUGGGCGUAGAUUUUUCAUAGUGGUGUCUAUAUU- | |
| CAUUUCUGCUGUUAACAGCUUUCAGCCAGGGACGUGUUGUAUCCUAGGCAGUGGCCCACCCAUAGGUCACAAUG | 210 | |
| 2 (32 kDa) | 5′ (L) UAAACUUUAAGAAUG | 77 |
| 2-1 (HE) | 5′ (L) UAAACUCAGUGAAAAUG | 79 |
| 3 (S) | 5′ (L) UAAACAUG | 70 |
| 4 (4.8 kDa) | 5′ (L) UAAACCUGUAAUG | 75 |
| 5 (12.7 kDa) | 5′ (L) UAGACCUUAUAACUUUAAGCAUUAUUAAUUGCCAAAGUUUCUAAGGUCACGCCCUAGUAAUG | 124 |
| 5-1 (E) | 5′ (L) CAAACAUUAUGAUAAAUAUUUUGGAGUAAUAACUGGUUUUACAGCAUUCGCUAAUACUGUAGAGGAGGCUGUUA- | |
| ACAAACUGGUUUUCUUAGCUGUUGACUUUAUUACUUGGCGGAGACAGGAGUUAAAUG | 193 | |
| 6 (M) | 5′ (L) CAAACAUUAUG | 73 |
| 7 (N) | 5′ (L) UAAACUUUAAGGAUG | 77 |
Includes the common leader (L) of 65 nt: 5′GAUUGUGAGCGAUUUGCGUGCGUGCAUCCCGCUUCACUGAUCUCUUGUUAGAUCUUUUUAUAAUC. The canonical sequence motif (UC[U/C]AAAC) formed at leader-mRNA junctions and the nonconforming sequence UCUAGAC formed at the leader-mRNA junction for the 12.7 mRNA are underlined. The start and stop codons for an ORF potentially encoding an 8-amino-acid peptide within the 210-nt 5′ UTR of mRNA 1 are identified with bold type in an open box. The start codons thought to initiate translation of the named mRNA species are identified with open boxes. Data come from references 7 and 18.
MATERIALS AND METHODS
Viruses and cells.
Vaccinia virus strains vWR (wt), obtained from J. Weir (University of Tennessee, Knoxville), and vTF7-3 (T7 RNA polymerase expressing), obtained from B. Moss (12), were used at concentrations of 1.8 × 106 and 5.8 × 106 PFU/ml, respectively. BCV was plaque purified and used at a concentration of 3.5 × 106 as described previously (20). Human HRT-18 cells (20) and T7 polymerase-expressing murine OST7-1 cells (12) were grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum (Atlanta Biologicals). Medium for the OST7-1 cells also contained 0.5 mg of Geneticin (G418; Sigma) per ml (12).
Construction of plasmids.
pSP6I, a cDNA clone expressing only the I ORF, was previously described (36). p77N (described previously as pLN [36]), is a cDNA clone of the N mRNA (mRNA 7) complete with the natural leader-containing 77-nt 5′ UTR, a 1,344-nt ORF encoding the N protein, a 291-nt 3′ UTR, and a 3′ poly(A) tail of 21 nt (Fig. 1). p210N, identical to p77N except for the 5′ UTR which comes from genomic RNA, was made by replacing the 77-nt 5′ UTR of p77N with the leader-containing 210-nt genomic 5′ UTR obtained from the cloned DI RNA, pDrep1 (7). For this, the partial gene 1–reporter-containing fragment from pDrep 1 was removed by PCR-based overlap deletion mutagenesis as described elsewhere (34).
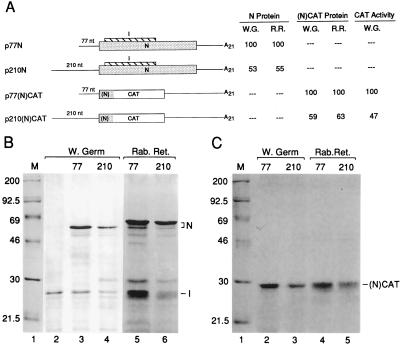
FIG. 1.
Accumulation of protein in vitro from the 210-nt 5′ UTR is twofold less than from the 77-nt 5′ UTR. (A) Schematic representation of constructs and summary of products accumulated in vitro. All constructs contain the promoter for T7 RNA polymerase and the N start codon (not underlined) in the same sequence context AGGAUGU. N coding sequence is indicated by a stippled box, I by a striped box, and CAT by an open box. The ORF for the (N)CAT fusion gene is likewise indicated. Relative amounts of N protein synthesized in wheat germ extract (W.G.) and rabbit reticulocyte lysate (R.R.) were determined by direct scanning of the dried polyacrylamide gel by AMBIS scanning. Relative amounts of (N)CAT fusion protein were determined by densitometry of the autoradiograms. CAT activity was determined by AMBIS scanning of the 14C-labeled product. (B) Autoradiogram of electrophoretically separated radiolabeled in vitro translation products from gene 7. Lane 1, molecular weight markers; lane 2, product from transcripts of pSP6I which encodes only the I protein; lanes 3 and 4, products from transcripts of p77N and p210N in wheat germ extract; lanes 5 and 6, products from transcripts of p77N and p210N in rabbit reticulocyte lysate. (C) Autoradiogram of electrophoretically separated radiolabeled in vitro translation products from the (N)CAT fusion gene. Lane 1, molecular weight markers; lanes 2 and 3, products from transcripts of p77(N)CAT and p210(N)CAT; lanes 4 and 5, products of transcripts from p77(N)CAT and p210(N)CAT.
p77(N)CAT was made by inserting the 654-nt CAT gene into the N ORF of p77N at the XbaI site, 94 nt downstream from the start codon of N. For this, the CAT gene was removed from pCM4 (Pharmacia) with BamHI, blunt ended by fill-in with Klenow enzyme, and ligated into the XbaI-cut, blunt-ended p77N. The N and CAT ORFs were put in frame by digesting them with BamHI (a site recreated at the 5′ end of the CAT insert), blunt ending by fill-in, and religation.
p210(N)CAT was made by replacing the 1,390-nt XmnI fragment of p77(N)CAT with the 1,523-nt XmnI fragment of p210N.
p77(N)CATrz and p210(N)CATrz were made by replacing the 3′-terminal 774-nt SpeI-HindIII fragments in p77(N)CAT and p210(N)CAT, respectively, with the 1,087-nt ribozyme/T7 terminator-containing SpeI-HindIII fragment from pDrep1rz. pDrep1rz was made by ligating the 272-nt StuI-HindIII fragment from the PGEM-based v2.0 vector (described in reference 32; kindly provided by L. A. Ball, University of Alabama at Birmingham) into pDrep1 DNA that had been linearized with MluI, blunt ended with Mung bean nuclease, and digested with HindIII.
pCAT was made by inserting the 654-nt BamHI CAT gene-containing fragment from pCM4 into BamHI-linearized pGEM3Z (Promega Biotech). The 59-nt 5′ UTR on T7 RNA polymerase-generated transcripts of pCAT has the sequence 5′-GGGCGAATTCGAGCTCGGTACCCGGGGATCCGAGATTTTCAGGAGCTAAGGAAGCTAAA-3′.
All constructs were grown in JM-109 strain of Escherichia coli, and all junctions were confirmed by sequencing.
Translation in vitro and analysis of products.
For preparation of capped transcripts, p77N and p210N were linearized with MluI, which cleaves at the 3′ end of the poly(A) tail, and transcribed with T7 RNA polymerase as recommended by the manufacturer (Promega Biotech). RNA quantitation and in vitro translation analyses of N and I were carried out as described previously (19, 35, 36). Briefly, full-length transcripts were quantitated by Northern blot analysis by using an N mRNA-detecting 32P-labeled oligodeoxynucleotide of known specific activity, and 1 μg of each was translated in 50 μl of either wheat germ extract or rabbit reticulocyte lysate (Promega Biotech) in the presence of 35S-labeled methionine; the products were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gels of 10% polyacrylamide. In some cases, as noted, transcripts were quantitated spectrophotometrically. Quantitation of radiolabeled products on the Northern blot (RNA) or in dried polyacrylamide gels (protein) was done with the AMBIS Radioanalytic Imaging System (San Diego, Calif.) or on autoradiograms of the polyacrylamide gels with the Bio-Rad Imaging Spectrophotometer.
CAT assays were done with an enzyme assay kit and 14C-labeled chloramphenicol (ICN), as recommended by the manufacturer (Promega Biotech). Radiolabeled products were spotted onto nitrocellulose and quantitated by scanning with AMBIS.
Transfection with DNA and transient expression in uninfected and virus-infected cells.
Lipofectin (GIBCO-BRL)-mediated transfection was done as recommended by the manufacturer. Briefly, cells in 35-mm dishes at 50 to 80% confluency were transfected with 5 μg of supercoiled plasmid DNA or were infected first at 5 h pretransfection with BCV (at 5 PFU/cell) and/or at 1 h with vaccinia virus (at 5 PFU/cell), as indicated.
CAT assays carried out as described above were done on cell lysates prepared at 24 h posttransfection.
Transfection with RNA and Northern analysis.
Transfection with RNA and quantitation of intracytoplasmic RNA molecules was done as described earlier (6, 20). Here, 1 μg of RNA transcript, quantitated spectrophotometrically, was used per plate, and the probes used for Northern analyses, the CAT gene-detecting probe CAT(+) (5′-GGTGTAACAAGGGTGAACACTATCCC-3′) which binds to nucleotides 253 to 279 of the CAT ORF and the 18S rRNA-detecting probe 18S(+) (6), were 5′ end labeled by the forward reaction. The CAT(+) probe was labeled to a specific activity of 2 × 106 cpm/pmol (AMBIS counts), respectively. Radioactivity quantitation of the Northern blots was done with the AMBIS.
RESULTS
In vitro, and in vivo in uninfected cells, accumulation from the 210-nt genomic 5′ UTR was twofold less than from the 77-nt mRNA 7 5′ UTR.
To compare the effects of the 5′ UTR on the rates of protein accumulation during synthesis in vitro, two 5′ UTRs differing in length by 133 nt and coming from mRNAs encoding proteins of widely differing abundance were tested on reporter mRNAs of two designs. In the first, the 77-nt 5′ UTR of mRNA 7 was replaced by the 210-nt of mRNA 1 (genome) (Table 1 and Fig. 1A). mRNA 7 is the template for the synthesis of the abundant N protein (and the less-abundant I protein from an overlapping internal ORF) (36) and mRNA 1 is the template for the synthesis of polyproteins that are processed into the low-abundance RNA-dependent RNA polymerase, helicase, proteases, and associated proteins (10). Both constructs were flanked at the N start codon by the same sequence (AGGAUGU) identified by Kozak (22) as one of moderate favorability for translation initiation. Full-length synthetic transcripts in subsaturating amounts were translated in wheat germ extract and in rabbit reticulocyte lysate, and radiolabeled N and I proteins were quantitated. Radioactivity in N and I directly reflect molar amounts since each contains eight methionine residues. Figure 1B, lanes 3 through 6, illustrates that the accumulation of N from the 210-nt 5′ UTR is nearly one-half of that from the 77-nt 5′ UTR (summarized in Fig. 1A). Accumulation of I from the 210-nt 5′ UTR was 77% of that from the 77-nt 5′ UTR in wheat germ extract and 76% in rabbit reticulocyte lysate. Accumulation was taken as an indication of efficiency of translation initiation, since transcripts contained the same ORF, 3′ UTR, and poly(A) tail, and the measured intracellular half-lives for similarly constructed BCV mRNAs showed no differences among them (6) (also data shown below and data not shown).
To test for differences with a reporter that could also be used in vivo, the CAT gene was placed in frame within the N gene in p77N and p210N to form p77(N)CAT and p210(N)CAT (Fig. 1A). The fusion protein contains the first 31 amino acids (aa) of N, 11 aa derived from CAT 5′ flanking sequence, and 219 aa of the entire CAT protein. When transcripts of these were quantitated spectrophotometrically and translated in vitro, fusion protein accumulation rates (Fig. 1A; Fig. 1C, lanes 2 to 5) reflected those for N above. Namely, accumulation from the 210-nt 5′ UTR was nearly half of that from the 77-nt 5′ UTR. Nearly twofold-less enzymatic activity was also observed in the in vitro translate, although low activity overall was found (Fig. 1A). These results confirmed the intrinsic differences in translation efficiencies between the two 5′ UTRs and established that CAT is enzymatically active in the context of the N fusion protein.
To determine differences between the two 5′ UTRs in vivo under coronavirus-free conditions, two systems expressing T7 RNA polymerase were used. In the first, HRT cells used previously to characterize the kinetics of BCV replication and gene expression (20) were infected with recombinant T7 RNA polymerase-expressing vaccinia virus 1 h prior to transfection and then either mock transfected or transfected separately with pCAT, p77(N)CAT, or p210(N)CAT DNA; they were then assayed for CAT synthesized from runoff transcripts. The filled bars in Fig. 2A illustrate that CAT expressed in the absence of coronavirus infection, although generally low (ranging from two- to fivefold above the level of mock-transfected cells), was twofold less from the 210-nt 5′ UTR than from the 77-nt 5′ UTR. Expression from the pCAT control was of similar magnitude and demonstrated the feasibility of this experimental system for measuring translation from the various constructs in vivo. Results in vivo for the two 5′ UTR constructs in the absence of a coronavirus infection, therefore, reflected almost exactly those from translations in vitro.
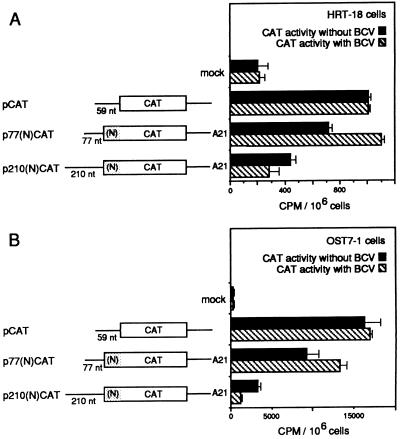
FIG. 2.
Protein accumulation from the 5′ UTRs of 210 and 77 nt in vivo as a function of infection with BCV. (A) HRT-18 cells were infected with T7 RNA polymerase-expressing vaccinia virus at 1 h prior to transfection and then mock transfected or transfected with pCAT, p77(N)CAT, or p210(N)CAT plasmid DNA as schematically depicted; cells were then harvested 24 h later and assayed for CAT activity. Where indicated, cells were also infected with BCV at 5 h prior to transfection. Experiments were done in quadruplicate, and the standard deviations from the mean are indicated. (B) OST7-1 cells were infected with wild-type vaccinia virus at 1 h prior to transfection and transfected as indicated in panel A. Where indicated, cells were also infected with BCV at 5 h prior to transfection. Experiments were done in duplicate, and the ranges are shown.
In the second approach, OST7-1 cells, a stably transfected T7 RNA polymerase-expressing mouse L-cell line (12), were mock transfected or transfected with CAT-containing plasmids to measure differences in translation from transcripts made by the constitutively expressed polymerase. Expression of CAT from the 210-nt 5′ UTR of p210(N)CAT was found to be at nearly basal levels, whereas that from the 59-nt and 77-nt 5′ UTRs of pCAT and p77(N)CAT, respectively, was nearly twofold higher (data not shown). Infection with wild-type vaccinia virus was therefore used to boost T7 RNA polymerase-generated transcript levels in OST7-1 cells (12). The solid bars in Fig. 2B illustrate that while protein accumulation from all three constructs was on average 10-fold higher than that observed in HRT-18 cells (Fig. 2A), translation from the 210-nt 5′ UTR was still nearly half of that from the 77-nt 5′ UTR.
To test whether constructs allowing precise 3′ transcription termination would increase transcript abundance and thus eliminate the need for coinfection with vaccinia virus, plasmids p77(N)CATrz and p210(N)CATrz, which tandemly carry a self-cleaving ribozyme and a T7 RNA polymerase terminator just downstream of the 3′ end of a 68-nt poly(A) tail, were tested. Although in vitro-generated transcripts proved to be abundant and of precise length (data not shown), essentially no advantage over the non-ribozyme-containing constructs was gained. Still, in the presence of vaccinia virus, synthesis from the 210-nt 5′ UTR remained approximately half of that from the 77-nt 5′ UTR (data not shown).
The results of both in vitro and in vivo studies, therefore, suggest that there exist intrinsic translational regulatory properties within the 5′ UTRs of mRNA species 1 and 7.
Intrinsic differences in accumulation rates from the 210 and 77-nt 5′ UTRs are the result of cis-acting influences on a 5′-end-dependent ribosomal entry and scanning mechanism.
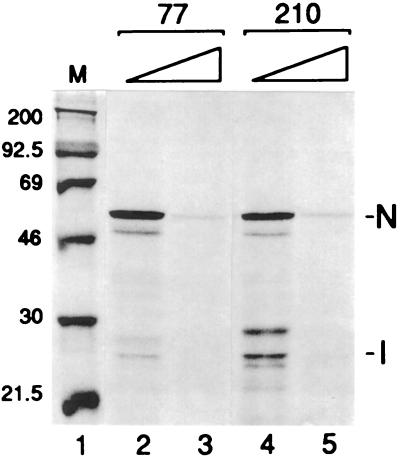
If ribosomes enter both the 210- and 77-nt 5′ UTRs at the 5′ terminus and scan, then a lower translational efficiency from the 210-nt 5′ UTR might be explained by the presence of a greater number of obstacles in the scanning pathway (21, 22). We have demonstrated previously that N and I ORFs on mRNA 7 are accessed by 5′-terminal entry (35). To address whether ORF 1 on mRNA 1 is similarly accessed, two approaches were taken. In the first, capped transcripts of p210N were translated in wheat germ extract in the presence of 0.2 mM 7-methyl GTP to assess the inhibitory potential of this molecule. In the presence of soluble cap, the translation of both N and I was inhibited by 85% (Fig. 3, lanes 4 and 5). This is similar in degree to the inhibition observed for the 77-nt 5′ UTR (95%) (Fig. 3, lanes 2 and 3; see also reference 35) and indicates that a 5′-terminal cap-dependent entry of ribosomes onto the 210-nt 5′ UTR is likely.
FIG. 3.
Inhibition of translation from capped 5′ UTRs by 7-methyl GTP. Capped transcripts of p77N and p210N were made as described in the text and translated in wheat germ extract in the presence of radiolabeled methionine, and the products were analyzed by SDS-PAGE. Lane 1, molecular weight markers; lanes 2 and 4, no 7-methyl GTP; lanes 3 and 5, 0.2 mM 7-methyl GTP.
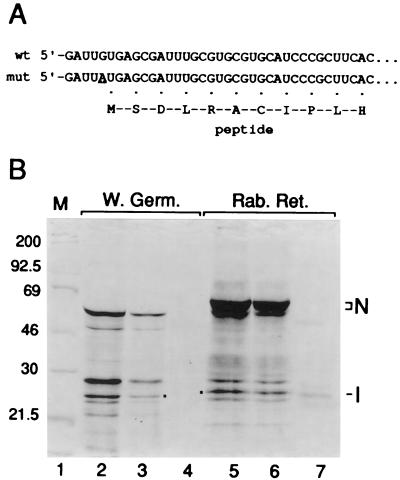
In the second approach, a mutant leader containing an internal ORF of 33 nt (Fig. 4A) and known to cause a diminished translation rate from the 77-nt 5′ UTR (19) was placed within the leader of p210N, and its effect on translation was measured. An intraleader ORF-dependent inhibition of translation of 66% in wheat germ extract (Fig. 4B, lanes 2 and 3) and 30% in rabbit reticulocyte lysate (Fig. 4B, lanes 5 and 6) was found, a finding consistent with a mechanism of ribosomal scanning from the 5′ terminus of the 210-nt genomic 5′ UTR.
FIG. 4.
Repression of translation from the 210-nt 5′ UTR by the intraleader 33-nt ORF. (A) Sequence of the BCV leader and of the peptide generated from the G5→A mutant leader. (B) Repression of translation. Equal amounts of transcript from the p210N (lanes 2 and 5) and p210(intraleaderORF)N (lanes 3 and 6) were translated in wheat germ extract or in rabbit reticulocyte lysate as indicated, and the products were separated by SDS-PAGE. Dried gels were exposed for autoradiography. Lane 1 contains radiolabeled molecular weight marker proteins; lanes 4 and 7 are products of translation reactions in which no RNA was added.
Thus, the differences observed in translation from the 210- and 77-nt 5′UTRs in vitro probably reflect influences of cis-acting elements on ribosomal scanning.
Effect of coronavirus infection on accumulation rates in vivo from mRNAs containing the 210- and 77-nt 5′ UTRs.
In the recent study of Tahara et al. (39), it was demonstrated that in infected cells the common leader element on mouse hepatitis coronavirus mRNAs imparts a translational advantage over cellular mRNAs. The mechanism for this enhancement was not known but was postulated to be the result of an interaction between leader and a viral product. Thus, to determine whether coronavirus infection would alter the translational efficiencies observed in vivo for the transcripts under study here, the two in vivo approaches described above were used except that cells were infected with BCV before infection with vaccinia virus and transfection with plasmid DNA. In the first approach, BCV-infected HRT cells were used. Preliminary experiments had revealed only minimal reciprocal interference of virus growth between BCV and vaccinia virus within the 48-h period used for the experiment. When vaccinia virus at a multiplicity of infection (MOI) of 5 preceded BCV infection at the same MOI by 4 h, no inhibition of BCV was observed as determined by Northern analysis of BCV mRNA throughout infection (data not shown). Upon reversal of the order of infection, a 10% inhibition of vaccinia virus was observed, as determined by titers of progeny virus at 24 and 48 h postinfection (data not shown). When HRT cells were infected with BCV at 5 h and vaccinia virus vTF7-3 at 1 h prior to transfection with reporter plasmids, accumulation of CAT above background levels from runoff transcripts of p77(N)CAT was stimulated by approximately 70% over cells not infected with BCV (880 cpm above background with BCV infection versus 510 cpm without infection; note the hatched bar versus the solid bar in Fig. 2A), suggesting that, in a manner consistent with the findings of Tahara et al. (39), there was a stimulation of translation in trans of this virus leader-containing mRNA molecule as a function of BCV infection. Also consistent with the idea of leader-mediated enhancement was the absence of stimulation from leaderless transcripts of pCAT (Fig. 2A). There was, however, an unexpected >3-fold repression of translation from the 210-nt 5′ UTR-containing transcripts from p210(N)CAT as a function of BCV infection (70 cpm above background with BCV infection versus 230 cpm without infection; note the hatched bar versus the solid bar in Fig. 2A). Together, these changes in BCV-infected HRT cells resulted in a net 12-fold difference between the two 5′ UTRs in the level of CAT expressed (880 versus 70 cpm).
In the second in vivo approach, the effect of BCV infection on translation in OST7-1 cells was measured. Preliminary experiments had determined by Northern analyses at time points throughout a 24-h period and by the titer of progeny virus at 48 h that BCV replicates in OST7-1 cells to nearly the same level as in HRT cells (data not shown). When OST7-1 cells were infected with BCV at 5 h and with wild-type vaccinia virus at 1 h prior to transfection with reporter plasmids, accumulation of CAT at levels above background from p77(N)CAT was stimulated by 46% over cells without BCV infection (12,980 cpm above background versus 8,910 cpm; note the hatched bar versus the solid bar in Fig. 2B), whereas a >3-fold repression of translation was observed from p210(N)CAT (890 cpm above background versus 3,020 cpm; note the hatched bar versus the solid bar in Fig. 2B). This resulted in a net difference in CAT expression of nearly 14-fold (890 versus 12,980 cpm) as a function of BCV infection, a ratio similar to that found in HRT cells. Results with the ribozyme-containing constructs p77(N)CATrz and p210(N)CATrz were similar to those shown in Fig. 2B for p77(N)CAT and p210(N)CAT (data not shown), revealing no advantage to the presence of the T7 RNA polymerase terminator and 3′-terminal ribozyme for boosting overall in vivo expression levels.
Thus, the results of in vivo studies in BCV-infected cells suggest that there are transacting factors resulting from infection differentially influencing the translation of mRNAs 1 and 7 and that these act through the 5′ UTR. Furthermore, the differential influence on translation was not host specific since it was observed in cells from two different host species.
The 210- and 77-nt 5′-UTR-containing transcripts have similar stabilities in virus-infected cells.
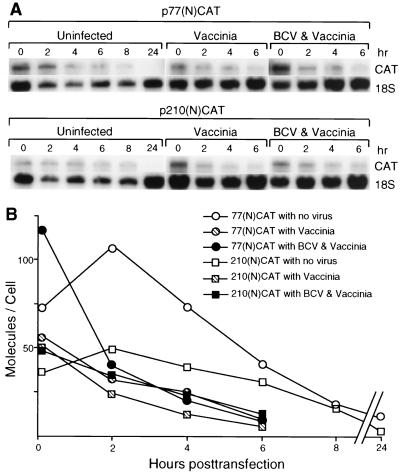
To determine whether conditions in the singly and doubly infected cells would differentially affect transcript stabilities and thus explain the variant protein accumulation patterns noted in Fig. 2, T7 RNA polymerase transcripts generated in vitro were transfected, and amounts surviving over time were quantitated by Northern analysis. As can be observed in Fig. 5, transcripts of both designs entered cells at the rate of approximately 50 to 100 molecules per cell and demonstrated survival half-lives of 2 to 4 h for approximately 90% of the entered population. Curiously, in both infected (data not shown from another experiment) and uninfected cells (Fig. 5 and studies described in reference 6), the remaining molecules appeared to have a prolonged survival with no noticeable decay over a 24-h period. Thus, because of similar decay rates between transcripts harboring the 77- and 210-nt 5′ UTRs, it is unlikely that differing mRNA stabilities can explain the differing protein accumulation patterns noted in Fig. 2.
FIG. 5.
Decay rates of transfected transcripts. RNA transcripts were transfected into cells under the conditions noted, and amounts in the cytoplasm were quantitated by Northern analysis. Quantities of RNA were normalized against the amount of 18S rRNA in the same extract, and the number of molecules per cell was calculated as described in Materials and Methods.
DISCUSSION
We conclude from the results shown here that one of the functions of the unique 5′-UTR structures on mRNA 1 (genome; 210 nt) and mRNA 7 (N mRNA; 77 nt) is to differentially regulate translation during virus replication; effects from the 5′ UTRs of the remaining BCV mRNAs were not examined. Two levels of regulation were revealed. In the first, an intrinsic control was demonstrated by translation in vitro, and in vivo in uninfected cells, wherein ORFs fused to the genomic 5′ UTR were translated with lower efficiency. Since experimental evidence indicated that both 5′ UTRs use a 5′-terminal entry of ribosomes and since the AUG start codons on both were flanked by the same moderately favorable Kozak sequence (AGGAUGU) (22), intrinsic differences are likely to be those affecting ribosomal scanning through the UTR. Based on reported factors known to retard scanning, a number of features in the longer 5′ UTR might be used to explain the difference. (i) Although 5′-UTR distances of up to 100 nt do not intrinsically interfere with translation initiation, distances beyond this might (21). (ii) Two predicted but experimentally unconfirmed stem loops of −13.4 kcal/mol (between nt 85 to 93 and 166 to 175) and −14.2 kcal/mol (between nt 106 to 111 and 193 to 198), components of a postulated stable, higher-order structure (5), potentially impose a structural barrier (21). (iii) An upstream initiator codon for an 8-aa oligopeptide beginning at base 100 (Table 1) could impose a barrier by either utilizing ribosomes for translation that are then dissuaded from further scanning or by initiating synthesis of a transacting repressor oligopeptide (mechanisms reviewed in references 14 and 22). Intriguingly, small ORFs have been found in all sequenced coronavirus genomic 5′ UTRs. In addition to the one described here for BCV, these include ORFs of 24 nt beginning at base 99 in the 209-nt UTR of MHV A59 (26), of 24 nt beginning at base 104 in the 214-nt UTR of MHV JHM (24), of 33 nt beginning at base 81 in the 293-nt UTR of HCV 229E (17), of 9 nt beginning at base 117 in the 313-nt UTR of transmissible gastroenteritis virus (11), and of 33 nt beginning at base 131 in the 527-nt UTR of IBV (4). Alternatively, unique structures within the 210- or 77-nt 5′ UTRs might attract different cellular factors that could affect translation. The cellular La protein, for example, is known to enhance translation after binding to the 5′ internal ribosome entry site element of poliovirus and hepatitis C virus and to the leader sequence of human immunodeficiency virus type 1 (1, 31, 33, 38). The poly(C)-binding protein similarly binds to the 5′ cloverleaf of poliovirus genome and enhances translation (15).
The second level of regulation was observed in BCV-infected cells in which there was found a 0.45-fold stimulation of translation from the 77-nt 5′ UTR and a 3-fold repression of translation from the 210-nt 5′ UTR as a result of infection. What explains the translational stimulation from the 77-nt 5′ UTR? To date we have no data on the character of the effector molecule(s) involved. Our results with the 77-nt 5′ UTR, however, are clearly consistent with the findings of Tahara et al. (39), who showed that MHV infection leads to leader-mediated enhancement of translation. In the study of Tahara et al. the reporter mRNA contained the 73-nt MHV leader (obtained from the 75-nt 5′ UTR of MHV mRNA 6) as the only 5′-UTR sequence, and in a recent study from the same laboratory (40) it was demonstrated that N, with known leader-binding properties (37), mediates a stimulation of translation of leader-containing mRNA in vivo. Thus, N may mediate the stimulation of translation from the 77-nt 5′ UTR in BCV-infected cells. How N stimulates translation in this system will be important to determine since, as noted earlier (40), it is a rare example of a positive regulator of translation in eukaryotic cells. Others that may be mechanistically similar are the 2A protein of poliovirus (16) and NS1 of influenza virus (9), which are also thought to act on the viral 5′ UTRs.
What explains the translational repression from the 210-nt 5′ UTR? Since the 210-nt 5′ UTR also contains the 65-nt leader, the putative element for positive regulation (39, 40), it would be expected that infection-induced repression is one that overrides leader-induced enhancement. One possibility is that there is a specific feedback inhibition by replicase or replicase-associated products of gene 1. In general, replicases are made in relatively low abundance by positive-strand RNA viruses, and for some, as among the procaryotic phages, autoregulation is exquisitely controlled at the level of translation (reviewed in reference 30). The discovery of a translation-replication autoregulatory mechanism involving the poliovirus 5′-terminal cloverleaf structure and viral 3CD protein (15) suggests similar mechanisms might also function in viruses of eukaryotes.
To understand the mechanisms of stimulation and repression observed here, it will be important to determine the nature of the effector molecules and the character of responding cis-acting elements. Are the effector molecules viral or cellular? Are elements within only the 5′ UTR involved, or might sequences elsewhere within the genome be components of the regulatory mechanism? How is translation regulated relative to the time after infection? In this study measurements were made on in vivo products accumulating between 6 and 24 h postinfection, but it will be important to determine the effects during early infection and over shorter time periods. What are the effects of virus infection on translation of the remaining seven unique 5′ UTRs of BCV? Of special interest will be whether repression is also observed from the unusually long 5′ UTRs of mRNAs 5 and 5-1, which are 124 and 195 nt, respectively.
ACKNOWLEDGMENTS
We thank Gwyn Williams and Seulah Ku for the construction of pCAT and Jason Simms for technical help in cloning the 5′ end of mRNA 2.
This work was supported by grants AI 14367 from the National Institutes of Health and 92-37204-8046 from the U.S. Department of Agriculture and with funds from the University of Tennessee, College of Veterinary Medicine, Center of Excellence Program for Livestock Diseases and Human Health.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredenbeek P J, Pachuk C J, Noten A F H, Charite J, Luytjes W, Weiss S R, Spann W J M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59: a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brierly I, Boursnell M E G, Binns M M, Bilimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boursnell M E, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 5.Chang R-Y. Ph.D. dissertation. Knoxville: University of Tennessee; 1994. [Google Scholar]
- 6.Chang R-Y, Brian D A. cis requirement of N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang R-Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Baric R S. Function of a 5′-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J Virol. 1995;69:7529–7540. doi: 10.1128/jvi.69.12.7529-7540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison M, Perlman S. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157:565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer F, Peng D, Hingley S T, Weiss S R, Masters P S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballe A P. Translational control mediated by upstream AUG codons. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 173–197. [Google Scholar]
- 15.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hambidge S, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold J, Raabe T, Schelle-Printz B, Siddell S G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann M A, Chang R-Y, Ku S, Brian D A. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology. 1993;196:163–171. doi: 10.1006/viro.1993.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann M A, Senanayake S D, Brian D A. A translation-attenuating intraleader open reading frame is selected on coronavirus mRNAs during persistent infection. Proc Natl Acad Sci USA. 1993;90:11733–11737. doi: 10.1073/pnas.90.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 22.Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Gagdzhardzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leibowitz J L, Pearlman S, Weinstock G, DeVries J R, Budzilowicz C, Weissemann J M, Weiss S R. Detection of a coronavirus nonstructural protein encoded in a downstream open reading frame. Virology. 1988;164:156–164. doi: 10.1016/0042-6822(88)90631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leparc-Goffart I, Hingley S T, Chua M M, Jiang X, Lavi E, Weiss S R. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239:1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D X, Cavanagh D, Green P, Inglis S C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D X, Inglis S C. Identification of two new polypeptides encoded by mRNAs of the coronavirus infectious bronchitis virus. Virology. 1992;186:342–347. doi: 10.1016/0042-6822(92)90094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luytjes W. Coronavirus gene expression: genome organization and protein synthesis. In: Siddell S G, editor. The coronaviridae. New York, N.Y: Plenum Publishing Corp.; 1995. pp. 33–54. [Google Scholar]
- 30.Mathews M B. Interactions between viruses and the cellular machinery for protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- 31.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 33.Pestova T V, Hellen C U T, Wimmer E. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senanayake S D, Brian D A. Precise large deletions by the PCR-based overlap extension method. Mol Biotechnol. 1995;4:13–15. doi: 10.1007/BF02907467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senanayake S D, Brian D A. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997;48:101–105. doi: 10.1016/S0168-1702(96)01423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senanayake S D, Hofmann M A, Maki J L, Brian D A. The nucleocapsid protein gene of the bovine coronavirus is bicistronic. J Virol. 1992;66:5277–5283. doi: 10.1128/jvi.66.9.5277-5283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stohlman S A, Baric R S, Nelson G N, Soe L H, Welter L M, Deans R J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitkin Y V, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahara S M, Dietlin T A, Bergmann C C, Nelson G W, Kyuwa S, Anthony R P, Stohlman S A. Coronavirus translational regulation: leader affects mRNA efficiency. Virology. 1994;202:621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahara S M, Dietlin T A, Nelson G W, Stohlman S A, Manno D J. Mouse hepatitis virus nucleocapsid protein as a translational effector of viral mRNAs. Adv Exp Med Biol. 1998;440:313–318. doi: 10.1007/978-1-4615-5331-1_41. [DOI] [PubMed] [Google Scholar]
- 41.Thiel V, Siddell S G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J Gen Virol. 1994;75:3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]