Abstract
Human herpesvirus 6 variants A and B (HHV-6A and HHV-6B) are closely related viruses that can be readily distinguished by comparison of restriction endonuclease profiles and nucleotide sequences. The viruses are similar with respect to genomic and genetic organization, and their genomes cross-hybridize extensively, but they differ in biological and epidemiologic features. Differences include infectivity of T-cell lines, patterns of reactivity with monoclonal antibodies, and disease associations. Here we report the complete genome sequence of HHV-6B strain Z29 [HHV-6B(Z29)], describe its genetic content, and present an analysis of the relationships between HHV-6A and HHV-6B. As sequenced, the HHV-6B(Z29) genome is 162,114 bp long and is composed of a 144,528-bp unique segment (U) bracketed by 8,793-bp direct repeats (DR). The genomic sequence allows prediction of a total of 119 unique open reading frames (ORFs), 9 of which are present only in HHV-6B. Splicing is predicted in 11 genes, resulting in the 119 ORFs composing 97 unique genes. The overall nucleotide sequence identity between HHV-6A and HHV-6B is 90%. The most divergent regions are DR and the right end of U, spanning ORFs U86 to U100. These regions have 85 and 72% nucleotide sequence identity, respectively. The amino acid sequences of 13 of the 17 ORFs at the right end of U differ by more than 10%, with the notable exception of U94, the adeno-associated virus type 2 rep homolog, which differs by only 2.4%. This region also includes putative cis-acting sequences that are likely to be involved in transcriptional regulation of the major immediate-early locus. The catalog of variant-specific genetic differences resulting from our comparison of the genome sequences adds support to previous data indicating that HHV-6A and HHV-6B are distinct herpesvirus species.
Sequence-based information is an essential precursor to many molecular, biological, and epidemiologic studies. In addition, sequences are important for confirming or clarifying biological and taxonomic classifications, as illustrated for herpesviruses by experiences with Marek’s disease virus, channel catfish virus, and human herpesvirus 6 (HHV-6) (4, 9, 19). While the wealth of information obtained from smaller DNA segments is useful, genetic descriptions of viruses revealed through the determination and analysis of complete genome sequences are uniquely valuable. Thus, the analysis of 17 complete herpesvirus genome sequences has provided detailed information about their coding capacity and genetic architecture, revealing various permutations of conserved gene blocks and clusters of unique genes. This information yielded numerous insights into evolutionary paths within the herpesvirus family.
HHV-6 variants A and B (HHV-6A and HHV-6B) are classified as members of the Betaherpesvirinae subfamily, in the Roseolovirus genus along with human herpesvirus 7 (HHV-7) (47). These viruses share extensive domains of similar genetic organization with other betaherpesviruses, such as human cytomegalovirus (HCMV) (14, 19, 30, 40, 41, 48). The complete genome sequence for HHV-6A strain U1102 [HHV-6A(U1102)] was described by Gompels et al. (19).
HHV-6 was first described by Salahuddin and coworkers as a novel human herpesvirus isolated from the blood of patients with AIDS and other lymphoproliferative diseases (49). Reports describing the isolation of similar viruses soon followed (reviewed in reference 3), including from patients during the acute phase of the common childhood illness roseola or exanthem subitum (60). Characteristics shared by all of these viruses include infection of activated primary CD4+ T cells (33, 34, 54, 55), cross-reactive antigens (6, 43), and similar genomic organizations (30, 32, 36). As the cellular and molecular biologic properties of these viruses were investigated, it became evident that they segregate into two groups that differ with respect to several genetic and biological properties. To recognize the differences between these viruses, a system was established that classified them as either variant A or B. Classification was based on differences in nucleotide sequences, reactivity with panels of monoclonal antibodies, and cell tropism (1). The question of whether the HHV-6 variants should be recognized as distinct viral species was deferred pending the accumulation of additional information.
Although the viruses are closely related, there is no genetic gradient between HHV-6A and HHV6B, and recombinant viruses have never been detected. This situation is in contrast to that seen with the Epstein-Barr virus types, for which concentration of variant-specific changes in a small number of loci does not preclude intervariant recombination (25, 50). To understand the differences that may play a role in segregating the HHV-6 variants into discrete viral species, their genome sequences must be compared. In previous comparisons between subsets of HHV-6A and HHV-6B amino acid sequences, differences ranged from 1 to 5% in the set of genes shared by all herpesviruses (herpesvirus core genes), 19% in the gene encoding a strongly immunoreactive virion protein (U11), and 25% in the IE1 (U89) gene (8, 17, 29, 43, 59). In this report we present the genome sequence of HHV-6B strain Z29 [(HHV-6B(Z29)]. We describe its general organization, protein-coding potential, and relationship with the HHV-6A sequence.
MATERIALS AND METHODS
Sequencing strategy.
A previously described library of clones containing HHV-6B(Z29) restriction endonuclease fragments was used to determine the genome sequence (30). Several regions that were uncloned or cloned as part of a larger insert were amplified by using primers derived from terminal nucleotide sequences of bordering clones or the HHV-6A(U1102) genome sequence (19) (Table 1). The junctions between adjacent genomic restriction endonuclease fragments were confirmed by sequencing junction-spanning PCR amplicons.
TABLE 1.
Oligonucleotides used for amplification of HHV-6B genome segments
| Name | Type | Primer coordinatesa | Sequence | Annealing temp (°C) | Size of amplimer (kb) |
|---|---|---|---|---|---|
| TL | cPCRc | P1, 3–28 | 5′TCGCGTTTCAAAAATTACTTTAAAC | 60 | 1.5 |
| P2, 1456–1480 | 5′TGCGTACACGGCCACCCGCGTCTCC | ||||
| DRL/U | PCRd | P1, 4941–4965 | 5′GTAAGAGAGAGGCCCCGGTCCACAT | 68 | 5.2 |
| P2, 10140–10162 | 5′ATGAGTGGAGTTGACGACGCCCTG | ||||
| U-1 | cPCR | P1, 22633–22653 | 5′GAGATGGACACTGTCATTGAG | 55 | 6.8 |
| P2, 29458–29475 | 5′GTGGTCGGCGGATTAAAT | ||||
| U-2 | cPCR | P1, 34599–34618 | 5′CCGCTTCTCCAATTTCTGTA | 55 | 1.5 |
| P2, 36092–36111 | 5′TGCCTCAATTGCGTCTAAAA | ||||
| U-3 | PCR | P1, 35928–35923 | 5′CACGGCTAGCGTAGAGGTCACCATAG | 60 | 11.5 |
| P2, 47479–47504 | 5′TGTAAATCAAGAGGATGTCCAGATTG | ||||
| U-4 | PCR | P1, 52486–52510 | 5′TTTGTATCTGTAACATTCGACACAC | 60 | 6.2 |
| P2, 58698–58722 | 5′CTCGCTCTCAAAACAACATGTAACT | ||||
| U-5 | PCR | P1, 58624–58641 | 5′AAGACAGGTTACAGAAGC | 50 | 5.5 |
| P2, 64143–64160 | 5′AGGAAGATGAGGTTCAAC | ||||
| U-6 | cPCR | P1, 125916–125934 | 5′CATGTTGAATCATAGGTCGb | 50 | 1.6 |
| P2, 127510–127530 | 5′ATGATAATTTACGCCGCTACGb | ||||
| U-7 | PCR | P1, 127413–127433 | 5′CACCGTCGATGATTGAACACAb | 48 | 2.3 |
| P2, 129765–129785 | 5′ACAGGAAACCTAACTGCAATTb | ||||
| U-8 | PCR | P1, 129765–129785 | 5′AATTGCAGTTAGGTTTCCTGTb | 48 | 2.0 |
| P2, 131811–131829 | 5′GATCCCATATCTTCTCAAGb | ||||
| U-9 | cPCR | P1, 131811–131829 | 5′CTTGAGAAGATATGGCATCb | 50 | 1.5 |
| P2, 133342–133359 | 5′TCAGCAATTTCAACTCCAb |
Coordinates refer to the HHV-6B genome sequence. TL primer sequences are also located at coordinates 153325 to 153349 and 154577 to 154801.
Based on HHV-6A genome sequence (19). Differences between HHV-6A and HHV-6B are in boldface.
cPCR denotes that the PCR product was cloned into a pCR-Blunt vector by using a Zero Blunt PCR cloning kit (Invitrogen, San Diego, Calif.).
PCR conditions differ from those described in Materials and Methods as follows: 94°C for 15 s and 68°C for 9 min (16 cycles); 94°C for 15 s and 68°C for 9 min, with an increase of 20 s every cycle (12 cycles); and 72°C for 10 min (2 cycles).
PCR.
PCR was performed with a proofreading enzyme (Taq Precision Plus; Stratagene, La Jolla, Calif.) on HHV-6B(Z29) nucleocapsid DNA prepared as previously described (32). In most cases, cycles (30) consisted of denaturation (94°C for 45 s), annealing (60 s), and extension (72°C) of 1 min per kb. Primer sequences, genomic coordinates, annealing temperatures, and amplimer sizes are listed in Table 1. PCR amplimers were affinity purified by using standard methods (Qiagen, Santa Clarita, Calif.) prior to direct sequencing. A subset of PCR amplimers was cloned into a pCR-Blunt vector (Invitrogen, San Diego, Calif.) prior to sequencing.
Sequence determination.
Nucleotide sequences were determined with a fourfold or greater redundancy using commercially available primers or by primer walking using custom primers. Double-stranded coverage was not possible in two regions of the direct repeat (DR) because of the presence of repeats or homopolymeric stretches. These regions include a 454-bp segment within the unique region of DR (DRL coordinates 5734 to 6188) and a 694-bp region extending across the (TAACCC)78 repeat array at the right end of DR to the junction of the right end of DR with the left end of the unique segment (U). These regions were sequenced multiple times from different templates, including other PCR amplimers and a lambda phage clone (λH6Z-851) (30) with various primers. In addition, double-stranded information was not obtained for two small regions (totaling 118 bases) at the boundaries of the internal repeat R1 (Fig. 1A).
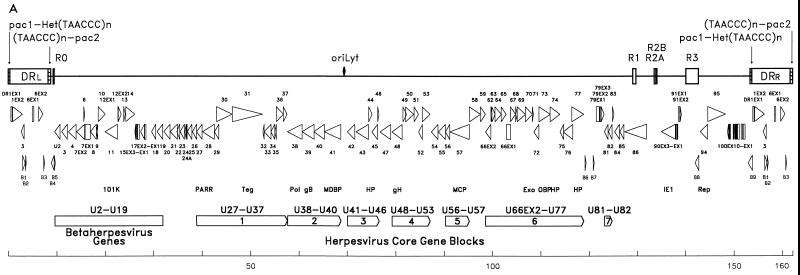
FIG. 1.
(A) HHV-6B genomic and genetic architecture. The upper portion shows the positions and arrangements of the major repeat elements R0, R1, R2A, R2B, and R3, the origin of replication (oriLyt), and the structure of the DR termini. ORF sizes, orientation, and location are indicated. HHV-6B unique ORFs are labeled B1 through B9. The orientation and order of the seven conserved herpesvirus core gene blocks are indicated, as is the block (U2 to U19) found only in betaherpesviruses. 101K, antigenic virion protein 101K; PA, polymerase processivity factor; RR, large ribonucleotide reductase; Teg, large tegument protein; Pol, DNA polymerase; gB, glycoprotein B; MDBP, major DNA binding protein; HP, helicase/primase complex; gH, glycoprotein H; MCP, major capsid protein; Exo, alkaline exonuclease; OBP, origin binding protein; IE1, major immediate-early gene; Rep, parvovirus rep homolog. (B) Nucleotide sequence comparison between HHV-6A and HHV-6B genomes. Genomes were aligned in segments by using GAP with gap and length weights of 50 and 3, respectively, except for the region spanning residues 124000 through 144500 (dashed line), for which weights of 25 and 1, respectively, were used to maximize the alignment. After concatenation of the aligned segments, identities between the aligned sequences were plotted by using PLOTSIMILARITY with a window of 1,000 residues. The horizontal dashed line represents mean identity of 88% across the whole alignment. Several regions with scores less than the mean are labeled; variable intergenic regions are identified by their flanking genes, e.g., U10/U11; the region spanning R2A and R2B is indicated as R2A-R2B. Nucleotide identity (NI) for the indicated regions was determined using GAP for aligned degapped sequences, with gap and length weights of 50 and 3, respectively, except for the segment spanning the right end of U, where weights 250 and 25, respectively, were used in order to omit gaps.
R3 determination.
Repeat R3 is contained within HindIII fragment C, cloned as pH6Z-204 (30). Two complementary sets of nested deletion clones with termini spaced at approximately 200-bp intervals were generated from subclones of pH6Z-204 with exonuclease III (Erase-a-Base; Promega, Wis.). These subclones were then used to sequence both strands of the repeat.
Sequence analysis.
Sequences were assembled and analyzed by using the Wisconsin Package, version 9.0 (Genetics Computer Group, Madison, Wis.). Database searches were done with nonredundant versions of GenBank posted on February 9, 10, and 15, 1999.
Nucleotide sequence accession number.
The sequence reported has been deposited with GenBank under accession no. AF157706.
RESULTS
The HHV-6B genetic content described below is presented in the context of elegant descriptions of HHV-6A and HHV-7 genetic architecture by others (19, 37, 41). Thus, we will not describe coding content in detail but will expand on issues that are unique to HHV-6B. The focus will be on a comparative description between the HHV-6B and HHV-6A genomes. In any such analysis of herpesvirus genomes, it must be remembered that the reported sequences represent a snapshot of a single example from the heterogeneous population of molecules that might be present in an individual or that might have varied on passage in cell culture. Such variation is frequently seen in regions containing repetitive elements (e.g., the het region in DR) (32) but is not necessarily limited to these regions (5).
Sequence assembly.
Over 98% of the sequence was determined on both strands, with an average of fourfold redundancy; the exceptions were three highly repetitive regions described in Materials and Methods. A representative complete genome sequence was compiled by assembling a representative DR element and grafting it to the termini of U. The junctions of U with DRL and DRR were confirmed independently. The DR sequence was assembled by using the following sequences, from left to right: a 1.5-kb cloned PCR amplimer mapping to the 5′ end of DR (TL in Table 1), a 3.8-kb BamHI genomic clone (pH6Z-109, BamHI fragment L) (30), a 5.2-kb PCR amplimer that extends from BamHI L across the DRL-U junction (DRL/U in Table 1), plus clones that span the junctions of DR-DR (from circular or concatemeric genomes) and U-DRR. We previously described three segments that are included in the complete genome sequence (29, 31, 43). One 20-kb segment spans U40 to U57 (GenBank accession no. L16947) and includes the origin of lytic replication (oriLyt). Another 20-kb segment spans U69 to U84 (GenBank accession no. L14772) and includes the homolog of the herpes simplex virus type 1 UL9 origin binding protein. The third is a smaller 3.2-kb segment encoding the antigenic virion protein 101K, which is the product of the U11 gene (GenBank accession no. L13162).
Global features.
The HHV-6B(Z29) genome sequence as assembled is 162,114 bp long. This is in close agreement with values of 159 to 164 kb determined by summation of restriction endonuclease fragment lengths (30, 32). The genome is composed of a 144,528-bp U flanked by 8,793-bp DR segments, DRL and DRR (Fig. 1A).
G+C contents are 40.8 and 59.1% in U and DR, respectively, with an overall G+C content of 42.8%. Nearly identical uneven base distributions between U and DR are also observed in HHV-6A and HHV-7 (19, 37, 41). Similar distributions of low and high G+C content have been described between U segments and long repeats that are present at either genomic termini or termini of long invertible segments of other herpesviruses (2, 7). As previously described, a region with unusually low G+C content (32.2%) is the 1,367-bp region between U41 and U42 that contains the HHV-6B oriLyt (12).
A shared characteristic of betaherpesviruses is CpG suppression with a concomitant increase in TpG in the major immediate-early (IE-A) locus; this feature is in contrast to the global CpG suppression in most gammaherpesvirus genomes and the lack of apparent CpG suppression in alphaherpesviruses (19, 21). Like HHV-6A, HHV-6B is CpG deficient, with a corresponding increase in TpG frequency in the IE-A locus (coordinates 127342 to 139167). CpG deficits in the IE regions of betaherpesviruses have been hypothesized to reflect localized methylation by the host cell during latency (19, 21).
DR structure and genomic termini.
HHV-6B, HHV-6A, and HHV-7 have similarly organized DR segments, which are composed of terminal, unique, and junctional regions. The coding content of the unique region of DR is described in the section on gene content. The assembled HHV-6B DR is 8% longer than that described for HHV-6A as a result of small insertions in the unique region and differences in the copy number of repeats found near the termini (Table 2). The terminal repeats are composed of perfect and imperfect copies of the hexanucleotide TAACCC (telomeric repeat sequence [TRS]). This sequence is also present in repeat arrays at the termini of vertebrate chromosomes (38), near the termini of HHV-6A and HHV-7 (14, 18, 19, 52), and at the junction region between the internal inverted repeats IRS and IRL of Marek’s disease virus (26). Additionally, scattered single copies of TRS are also present in U, distributed with a polarity similar to that of HHV-6A, in which TAACCC is found to the left of oriLyt and the complementary sequence, GGGTTA, to the right of oriLyt (18, 19). This arrangement confers an overall dyad symmetry to the genome, radiating from oriLyt.
TABLE 2.
Coordinates and copy number of repeat arrays in the HHV-6A and HHV-6B genomesa
| Name | Coordinates
|
No. of repeat arrays
|
||
|---|---|---|---|---|
| HHV-6A | HHV-6B | HHV-6A | HHV-6B | |
| DRL | ||||
| htTRS | 56–307 | 59–353 | 42 | 50 |
| TRS | 7655–8008 | 8250–8711 | 59 | 78 |
| DRR | ||||
| htTRS | 151290–151541 | 153380–153674 | 42 | 50 |
| TRS | 158889–159242 | 161571–162032 | 59 | 78 |
| R0 | Not present | 9315–9510 | 13 | |
| R1 | 127497–128181 | 129046–129681 | 51 | 54 |
| R2A | 130797–130950 | 133501–133863 | 2 | 5 |
| R2B | 131022–132254 | 133982–134076 | 1,233 ntb | 8 |
| R3 | 137994–140897 | 140082–142691 | 28 | 26 |
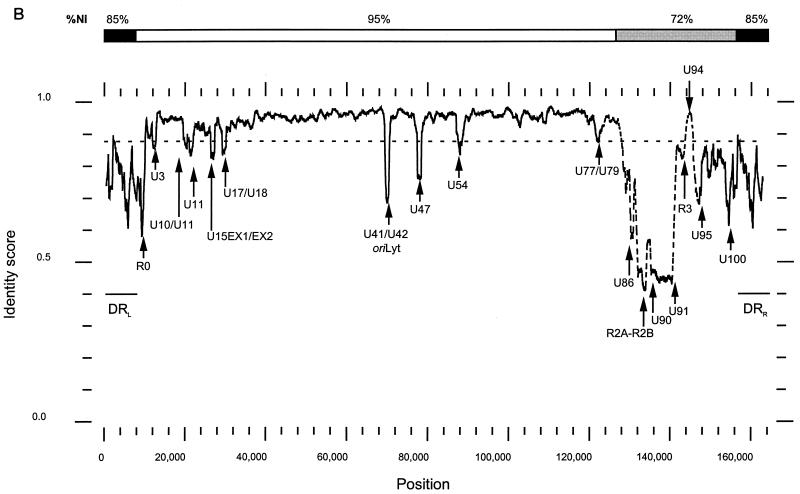
Sequences near the termini of the HHV-6B DR were described previously (57). We confirmed and extended these results by sequencing across the TRS arrays into the adjacent unique DR sequences, by sequencing additional clones that span the junction between the termini of circularized or concatemerized genomes, and by sequencing across the junction between the right end of DR and the left end of U. In summary, DR termini are composed of copies of TRS that are flanked on their left by pac1 at the left terminus and on their right by pac2 at the right terminus (Fig. 2A); pac1 and pac2 are conserved cis-acting herpesvirus packaging signals. At the left terminus of DR, a pac1 cleavage sequence is located 18 nucleotides (nt) from the predicted genomic terminus; adjacent to it are multiple copies of TRS interspersed with the related hexamers TAGGTC and TAGCCC. The right terminus of DR consists of 78 perfectly reiterated copies of TRS, followed by a pac2 signal located 29 nt from the predicted genomic terminus (Fig. 2B and C).
FIG. 2.
(A) DRL and DRR in linear and concatemeric genomes. The locations of terminal sequences from this and other works are indicated relative to the pac1-het(TAACCC)n and (TAACCC)n-pac2 elements at the left and right termini of DR, respectively. The dashed lines inside DR indicate the relative lengths of the TRS arrays. The black bars below the DRs represent the relative location of the sequences aligned in panel B; sequence designations are as shown below the lines. Sources for the sequences are as follows: B2LDR and B2RDR are from this work and were generated from amplimers TL and DRL/U, respectively (Table 1); P3, P10, P15, P4, P9, P1, P7 are DRL-DRR junction-spanning clones derived by PCR using 5′gcggatccTAACCCATCCCCCAACGCGC as the rightward primer (RP) and 5′cggaattcTTAGGGTTAATACCCCCCTTT as the leftward primer (LP) (the BamHI and EcoRI sites used in cloning are in lowercase) on template from HHV- 6B(Z29)-infected Molt-3 cells. The locations of primers RP and LP are indicated in panel B; A1LDR, A1RDR, and A1 (GenBank accession no. X79798, X79799, and X79800, respectively) are from Gompels and Macaulay (18); B1LDR, B1RDR, B1, and B2 (GenBank accession numbers L22337, L23336, and L22335, respectively) are from Thomson et al. (57). (B) Alignment of terminal sequences. Sequences were aligned by using PILEUP with gap creation and extension penalties of 5 and 1, respectively. Sequences A1, A1RDR, and A1LDR are from HHV-6A(U1102); the rest are from HHV-6B(Z29). Differences between the variants are shown in lowercase, and differences between HHV-6B sequences are underlined. Nucleotides found only in DRR-DRL junction-spanning clones are italicized. Putative terminal nucleotides are in bold. (C) Summary diagram of the DR termini. Designations are as for panel A; N indicates any nucleotide.
There were copy number differences in the heterogeneous TRS arrays between the sequence described here and the previously reported left terminal sequence of HHV-6B(Z29) obtained independently by Thomson et al. (57). This copy number variability was also present in different clones obtained from the same PCR amplification (15). Additionally, in electrophoretic analyses, the left terminal BamHI and SalI restriction endonuclease fragments were determined to be 2.7 and 2.0 kb long, respectively (30, 32), compared with 1,389 and 656 bp, respectively, predicted from the sequence. This difference suggests that smaller segments were selectively amplified by PCR from the pool of heterogeneous versions of the region. These results are consistent with the observations of Lindquester and Pellett (32), who found that the length of DR changed from 13 to 10 kb on viral passage in cell culture. The length heterogeneity mapped to the left end of the DR elements. On the basis of the restriction mapping and sequence data described above, it is likely that the variable regions correspond to the heterogeneous TRS arrays. Sequence analysis of uncultured virus will be required to more completely understand the structure of this region in wild-type virus.
The precise genomic termini of HHV-6B have not been directly determined but can be inferred from the sequences of fragments from at or near the genomic termini and that span DR-U boundaries, in the context of motifs conserved at the termini of other herpesvirus genomes. An alignment of our sequences with all previously published HHV-6A and HHV-6B sequences from the terminal region is shown in Fig. 2B. As can be seen in the alignment, terminal and DRR-DRL junction sequences are highly conserved between linear and concatemeric or circular genomes. Additionally, HHV-6B and HHV-6A sequences are highly conserved in this region. Interestingly, of the seven DRR-DRL junction clones that we analyzed, all except P4 had one to eight additional nucleotides at the junction. The mechanism for inserting these nucleotides is not obvious, although the variability is unlikely to be an artifact since it was observed in independently derived clones from different viral stocks and HHV-6A. Similar heterogeneity was also observed in clones derived from plasmid concatemers that had been packaged into extracellular virions or intracellular nucleocapsids (11).
Internal repeats.
In addition to the TRS arrays, five major repeat elements are located in U: R0, R1, R2A, R2B, and R3 (Fig. 1A). Copy number and coordinates for these arrays in both HHV-6A and HHV-6B genomes are given in Table 2. These repeat elements are located in regions of the HHV-6B genome that have lower nucleotide sequence identity with HHV-6A (Fig. 1B). R0 is unique to HHV-6B and is located near the junction of DRL and U and is contained within the putative HHV-6B open reading frame (ORF) B4. R1 is located near the 3′ end of the U86 ORF. Translation of R1 results in a series of serine and arginine (SR) repeats at the carboxy terminus of the U86 protein, the HCMV IE2 (UL122) homolog (19, 42). HHV-6B R1 has greater sequence variation than does HHV-6A R1; the HHV-6B repeat array is assembled from 10 different units, while HHV-6A R1 is assembled from 3 different units. The SR repeats are unique to the HHV-6 version of the protein and reflect divergence from other betaherpesviruses (19).
R2A and R2B are located in the region between U86 and U90 (coordinates 131902 to 138003). This region has only 57.5% nucleotide identity between HHV-6A and HHV-6B. R2A is not unique to HHV-6B; two, rather than five, copies are present in HHV-6A, and they are more divergent than in HHV-6B. R2B is related to HHV-6A R2 but is much shorter. R2B is 94 nt long, compared with the 1.2-kb R2. R2A contains several TATA-like sequences, while R2B has multiple potential binding sites for the transcription factor HNF-5 (TRTTTGY) (16), suggesting a possible role for these sequences in transcription regulation. A point of interest is that a plasmid clone (pH6Z-231) (30) was used as the sequencing template for the region encompassing R2A and R2B; the corresponding region in HHV-6A was refractory to cloning and was sequenced from a PCR-derived template. However, no large deletion is present in pH6Z-231 since the sequenced plasmid had a length similar to the predicted length of the corresponding HHV-6B restriction endonuclease fragment. In addition, R2A and R2B are present in HHV-6B(HST) (27).
R3 is located upstream of the IE-A locus that spans U86 to U91 and is hypothesized to contain cis-acting regulatory sequences that might play a role in transcription regulation of this locus. HHV-6B R3 is composed of 26 copies of 103-, 104-, or 105-bp imperfect repeat units. Individual units from HHV-6B vary considerably in sequence. An alignment of R3 consensus sequences for HHV-6B strains Z29 and HST (27) and HHV-6A (19) is shown in Fig. 3. Sequence variation is scattered throughout individual units of HHV-6B R3, with conserved pockets at positions 32 to 52 and 94 to 96 of the consensus sequence. One of the conserved pockets in all of the individual units of both variants is a potential binding site for the transcription factor PEA3 (AGGAA[A/G]). The PEA3 motif has been found in other viral genomes, including the polyomavirus enhancer and the adenovirus enhancer core element (20, 35). The presence of multiple PEA3 binding sites in R3 is intriguing, as they may represent primary targets of signal transduction in HHV-6A and HHV-6B. Other potential transcription factor binding sites identified in HHV-6A R3 include NF-κB and AP2 (58). In contrast, HHV-6B(Z29) R3 has no NF-κB sites and AP2 sites (CCC[A/C]NG/C[G/C][G/C]) in 11 of the 26 repeat units.
FIG. 3.
Alignment of HHV-6B and HHV-6A consensus R3 sequences. Individual R3 units of HHV-6A (28 copies) (19) and HHV-6B strains HST (24 copies) (27) and Z29 (26 copies) were aligned. Alignments were done by using PILEUP with gap creation and extension penalties of 5 and 1, respectively. Each consensus sequence represents 80% plurality between the aligned units of each virus. Sequences common to all three viruses are in uppercase. Conserved pockets are underlined, and the binding site for the transcription factor PEA3 is in bold. Symbols: Y, C or T; R, A or G; S, C or G; W, A or T; M, A or C; K, G or T; H, A or C or T; V, A or C or G.
HHV-6B gene content.
HHV-6B protein-encoding ORFs were first considered as significant if the translated proteins had sequence counterparts in HHV-6A. Proteins with similarity to HHV-6B-encoded proteins were identified by searching GenBank with the BLAST family of programs. Also, ATG-initiated ORFs as small as 177 nt (59 amino acids) that had no significant overlap with other ORFs and appropriately located polyadenylation signals were also considered as possibly significant. Using these criteria, we identified 127 ORFs; 8 are diploid because they are present in each copy of DR, leaving 119 that are unique (Table 3). These 119 unique ORFs compose 97 genes, based on the predicted splicing patterns for 11 genes. Of the 119 ORFs analyzed, 110 (92%) had their highest similarity score with the set of HHV-6A ORFs initially described by Gompels et al. (19) and modified by Megaw et al. (37). The remaining nine do not have HHV-6A counterparts and are unique to HHV-6B. With the exception of the acceptor site of U91EX2, exon boundaries were in agreement with previously published acceptor and donor splice sites for HHV-6A (37). The nomenclature used is based on that previously employed for HHV-6A and HHV-7 (19, 37, 41). ORFs with HHV-6A counterparts were given the same name; ORFs unique to HHV-6B are designated B1 through B9. Spliced genes are identified by the 5′-proximal exon as was done for HHV-7 (RK) (37). The deduced protein-coding capacity of the HHV-6B genome is listed in Table 3, and the arrangement of the ORFs is shown in Fig. 1A.
TABLE 3.
Locations of HHV-6B ORFs and relative similarities of HHV-6B genes with their HHV-6A and HHV-7 homologs
| ORFa | S | Startb | Endc | % DNA identity with HHV-6A homologd | Codons | %S/%/I with HHV-6A homologe | Substitutionsf
|
%S/%I with HHV-7 homologg | Implied functionh | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ka | Ks | |||||||||
| DR1EX1 | 583 | 841 | 88.4 | 86 | 90.7/87.2 | 0.092 | 0.146 | 50.9/47.6 | HCMV US22 gene family | |
| DR1EX2 | 955 | 2972 | 90.0 | 672 | 84.0/82.4 | 50.7/45.4 | ||||
| DR3 | Ci | 3325 | 2726 | 90.8 | 200 | 87.0/86.4 | 0.072 | 0.174 | ||
| B1 | 3022 | 3498 | 159 | |||||||
| B2 | 3536 | 3772 | 79 | |||||||
| DR6EX1 | 5027 | 5330 | 89.1 | 101 | 86.1/84.1 | 0.112 | 0.223 | 55.4/53.5 | HCMV US22 gene family, transactivator | |
| DR6EX2 | 6329 | 7200 | 88.8 | 290 | 84.3/82.2 | 69.1/62.6 | ||||
| B3 | 7349 | 7525 | 59 | |||||||
| B4 | C | 9492 | 8914 | 193 | Membrane protein? | |||||
| B5 | 9522 | 9758 | 79 | |||||||
| U2 | C | 10715 | 9627 | 92.4 | 363 | 92.1/91.5 | 0.048 | 0.199 | 54.7/47.8 | HCMV US22 gene family |
| U3 | C | 12205 | 11048 | 92.4 | 386 | 89.8/89.3 | 0.064 | 0.211 | 56.1/46.8 | HCMV US22 gene family, transactivator |
| U4 | C | 14040 | 12436 | 96.9 | 535 | 98.9/98.7 | 0.006 | 0.101 | 68.8/58.9 | |
| U7EX1 | C | 16959 | 15890 | 94.9 | 356 | 98.4/97.5 | 0.020 | 0.141 | 74.7/61.6 | HCMV US22 gene family |
| U7EX2 | C | 15800 | 14162 | 94.7 | 546 | 95.2/94.5 | 0.026 | 0.150 | 61.0/51.1 | |
| U6 | 15603 | 15806 | 95.1 | 68 | 97.1/97.1 | 0.017 | 0.113 | |||
| U8 | C | 18198 | 16966 | 95.0 | 411 | 96.4/95.5 | 0.023 | 0.130 | 62.1/50.8 | HCMV US22 gene family |
| U9 | C | 18493 | 18182 | 95.5 | 104 | 94.2/94.2 | 0.026 | 0.083 | ||
| U10 | 18543 | 20051 | 92.9 | 503 | 96.6/94.0 | 0.033 | 0.194 | 61.9/49.4 | ||
| U11 | C | 22534 | 19961 | 89.2 | 858 | 85.6/81.2 | 0.110 | 0.134 | 40.9/29.8 | Antigenic virion protein 101K |
| U12EX1 | 22636 | 22668 | 97.0 | 11 | 100/100 | 0.040 | 0.135 | 72.7/54.5 | G-protein-coupled receptor | |
| U12EX2 | 22746 | 23327 | 93.4 | 194 | 93.8/92.2 | 60.6/46.6 | ||||
| U13 | 23856 | 24176 | 96.2 | 107 | 93.4/92.5 | 0.037 | 0.061 | 44.9/35.7 | ||
| U14 | 24277 | 26106 | 90.5 | 610 | 89.3/85.7 | 0.082 | 0.150 | 56.7/49.9 | ||
| U15EX1 | C | 27048 | 26731 | 94.0 | 106 | 91.4/86.7 | 0.047 | 0.088 | 76.4/67.9 | HCMV UL25/UL35 gene family |
| U15EX2 | C | 26752 | 26500 | 94.5 | 24 | 100/95.8 | 83.3/75.0 | |||
| U15EX3 | C | 26328 | 26146 | 95.1 | 61 | 96.7/91.7 | 82.0/75.4 | |||
| U17EX1 | C | 28421 | 28193 | 95.2 | 76 | 94.7/94.7 | 0.026 | 0.133 | 54.9/43.7 | HCMV US22 gene family, IE-B |
| U17EX2 | C | 28105 | 27333 | 94.6 | 257 | 96.1/94.9 | 66.7/56.5 | |||
| U18 | C | 30485 | 29604 | 93.7 | 294 | 90.8/88.7 | 0.055 | 0.070 | 57.5/47.0 | IE-B |
| U19 | C | 31920 | 30754 | 94.2 | 389 | 92.0/91.5 | 0.043 | 0.098 | 41.5/35.1 | IE-B |
| U20 | C | 33449 | 32148 | 94.2 | 434 | 95.6/95.6 | 0.045 | 0.121 | 31.8/22.2 | |
| U21 | C | 34954 | 33455 | 93.2 | 500 | 91.0/89.8 | 0.050 | 0.121 | 42.8/31.6 | |
| U22 | C | 35459 | 34854 | 95.4 | 202 | 91.2/89.6 | 0.028 | 0.097 | ||
| U23 | C | 36386 | 35490 | 92.6 | 299 | 94.6/94.1 | 0.063 | 0.116 | 26.9/20.9 | |
| U24 | C | 36777 | 36514 | 92.7 | 88 | 88.3/82.7 | 0.080 | 0.054 | 46.2/31.2 | |
| U24A | C | 36969 | 36799 | 93.5 | 57 | 94.7/91.2 | 0.042 | 0.133 | 40.3/28.1 | |
| U25 | C | 37936 | 36989 | 97.4 | 316 | 99.4/98.1 | 0.009 | 0.075 | 45.0/31.2 | HCMV US22 gene family |
| U26 | C | 38931 | 38047 | 95.1 | 295 | 93.8/92.9 | 0.033 | 0.092 | 60.5/47.4 | |
| U27 | C | 40019 | 38922 | 95.5 | 366 | 97.3/95.6 | 0.021 | 0.112 | 45.7/31.1 | Polymerase processivity factor |
| U28 | C | 42549 | 40138 | 95.8 | 804 | 97.1/95.9 | 0.019 | 0.106 | 57.7/46.6 | Large ribonucleotide reductase |
| U29 | C | 43471 | 42575 | 96.0 | 299 | 97.6/96.1 | 0.020 | 0.103 | 66.3/51.9 | Capsid assembly and DNA maturation |
| U30 | 42999 | 46244 | 94.8 | 1082 | 95.2/93.2 | 0.033 | 0.106 | 58.4/46.3 | ||
| U31 | 46265 | 52495 | 94.8 | 2077 | 95.3/93.4 | 0.032 | 0.107 | 57.4/46.5 | Large tegument protein | |
| U32 | C | 52841 | 52575 | 95.8 | 89 | 96.6/96.6 | 0.017 | 0.120 | 71.9/65.2 | |
| U33 | C | 54255 | 52846 | 97.1 | 470 | 98.3/98.1 | 0.009 | 0.082 | 70.6/59.4 | |
| U34 | C | 55036 | 54209 | 95.2 | 276 | 95.3/94.2 | 0.028 | 0.097 | 65.1/57.0 | |
| U35 | C | 55373 | 55056 | 97.8 | 106 | 98.1/98.1 | 0.009 | 0.046 | 68.9/59.2 | |
| U36 | 55372 | 56823 | 96.7 | 484 | 97.9/97.5 | 0.012 | 0.087 | 68.9/58.1 | ||
| U37 | 56830 | 57621 | 97.0 | 264 | 98.1/97.0 | 0.014 | 0.074 | 71.8/61.8 | ||
| U38 | C | 60708 | 57673 | 96.2 | 1012 | 98.7/97.9 | 0.010 | 0.118 | 75.9/67.0 | DNA polymerase |
| U39 | C | 63200 | 60711 | 96.4 | 830 | 97.0/96.1 | 0.019 | 0.109 | 65.8/56.7 | Glycoprotein B |
| U40 | C | 65334 | 63157 | 96.9 | 726 | 99.2/98.8 | 0.008 | 0.093 | 67.8/55.9 | Transport/capsid assembly |
| U41 | C | 68740 | 65345 | 97.3 | 1132 | 99.3/98.8 | 0.006 | 0.070 | 78.7/68.5 | Major DNA binding protein |
| U42 | C | 71653 | 70106 | 95.5 | 516 | 96.1/95.5 | 0.024 | 0.105 | 66.6/56.0 | Transactivator |
| U43 | C | 74460 | 71881 | 97.3 | 860 | 98.8/97.7 | 0.010 | 0.068 | 70.9/61.0 | Helicase/primase complex |
| U44 | 74501 | 75193 | 95.9 | 231 | 98.1/96.7 | 0.014 | 0.121 | 70.4/58.6 | ||
| U45 | C | 76273 | 75146 | 95.8 | 376 | 95.5/94.4 | 0.032 | 0.074 | 59.2/50.5 | |
| U46 | 76346 | 76597 | 95.2 | 84 | 94.1/94.1 | 0.033 | 0.086 | 61.9/51.2 | ||
| U47 | C | 78999 | 76786 | 93.4 | 738 | 89.5/87.2 | 0.065 | 0.080 | 31.8/23.9 | Glycoprotein 0 |
| U48 | C | 81349 | 79268 | 95.3 | 694 | 95.8/94.2 | 0.028 | 0.103 | 52.2/39.0 | Glycoprotein H |
| U49 | 81508 | 82263 | 96.9 | 252 | 97.6/97.2 | 0.015 | 0.059 | 60.7/52.7 | ||
| U50 | 82043 | 83707 | 96.3 | 555 | 98.9/97.3 | 0.015 | 0.090 | 65.4/54.7 | Virion protein | |
| U51 | 83808 | 84710 | 94.9 | 301 | 95.7/94.0 | 0.031 | 0.096 | 48.6/36.1 | G-protein-coupled receptor | |
| U52 | C | 85508 | 84735 | 96.6 | 258 | 98.1/97.3 | 0.012 | 0.081 | 68.1/56.3 | |
| U53 | 85515 | 87098 | 96.2 | 528 | 97.4/96.8 | 0.017 | 0.098 | 59.6/51.1 | Proteinase | |
| U54 | C | 88715 | 87339 | 88.6 | 459 | 84.5/79.7 | 0.120 | 0.118 | 55.3/44.5 | Virion transactivator |
| U55 | C | 90271 | 88796 | 95.1 | 492 | 94.9/93.6 | 0.032 | 0.108 | (A)44.2/32.4 | |
| (B)33.9/21.2 | ||||||||||
| U56 | C | 91162 | 90275 | 98.1 | 296 | 99.0/98.0 | 0.010 | 0.052 | 74.7/65/2 | Capsid protein |
| U57 | C | 95201 | 91167 | 96.6 | 1345 | 98.1/97.8 | 0.011 | 0.087 | 77.9/68.6 | Major capsid protein |
| U58 | 95213 | 97528 | 95.8 | 772 | 97.5/96.9 | 0.015 | 0.109 | 69.8/61.4 | ||
| U59 | 97528 | 98577 | 95.7 | 350 | 96.0/95.1 | 0.022 | 0.100 | 50.9/39.9 | ||
| U62 | 99716 | 99976 | 98.0 | 87 | 96.5/96.5 | 0.019 | 0.062 | 65.8/52.1 | ||
| U63 | 99921 | 100574 | 97.5 | 218 | 99.1/99.1 | 0.004 | 0.090 | 73.9/67.3 | ||
| U64 | 100555 | 101880 | 95.5 | 442 | 94.1/93.7 | 0.031 | 0.093 | 52.6/40.0 | ||
| U65 | 101840 | 102844 | 92.8 | 335 | 89.0/85.7 | 0.073 | 0.074 | 69.1/60.0 | ||
| U66EX1 | C | 103784 | 102912 | 97.5 | 291 | 99.0/99.0 | 0.002 | 0.080 | 72.1/63.8 | Putative terminase |
| U66EX2 | C | 99704 | 98580 | 97.9 | 375 | 100/100 | 86.3/78.3 | |||
| U67 | 103756 | 104814 | 96.7 | 353 | 97.5/96.6 | 0.016 | 0.073 | 63.7/53.2 | ||
| U68 | 104817 | 105158 | 95.9 | 114 | 96.5/94.7 | 0.026 | 0.120 | 62.3/49.1 | ||
| U69 | 105164 | 106852 | 94.8 | 563 | 96.4/94.7 | 0.027 | 0.114 | 63.6/53.7 | Phosphotransferase; ganciclovir kinase | |
| U70 | 106863 | 108326 | 95.9 | 488 | 97.1/96.1 | 0.020 | 0.100 | 64.6/51.8 | Alkaline exonuclease | |
| U71 | 108266 | 108508 | 92.6 | 81 | 90.9/89.6 | 0.060 | 0.136 | 56.2/52.1 | Myristylated virion protein | |
| U72 | C | 109627 | 108596 | 96.6 | 344 | 98.3/97.1 | 0.013 | 0.082 | 72.4/58.7 | Glycoprotein M |
| U73 | 109640 | 111979 | 97.3 | 780 | 98.6/97.6 | 0.014 | 0.067 | 67.3/57.9 | Origin binding protein | |
| U74 | 111951 | 113936 | 96.1 | 662 | 97.6/96.2 | 0.019 | 0.088 | 54.6/41.3 | Helicase/primase complex | |
| U75 | C | 114723 | 113977 | 96.1 | 249 | 97.2/96.0 | 0.021 | 0.103 | 55.8/45.8 | |
| U76 | C | 116620 | 114635 | 97.1 | 661 | 98.9/98.6 | 0.007 | 0.096 | 70.4/58.9 | |
| U77 | 116415 | 118886 | 97.3 | 824 | 99.3/98.9 | 0.005 | 0.083 | 82.7/75.5 | Helicase/primase complex | |
| B6 | 119139 | 119375 | 79 | |||||||
| B7 | 120836 | 121060 | 75 | |||||||
| U79EX1 | 121486 | 122116 | 87.5 | 210 | 84.8/76.7 | 0.094 | 0.103 | 69.3/52.7 | Role in DNA replication | |
| U79EX2 | 122213 | 122412 | 92.4 | 66 | 89.2/84.6 | 57.1/47.6 | ||||
| U79EX3 | 122495 | 123103 | 93.8 | 203 | 88.9/86.9 | 45.4/37.9 | ||||
| U81 | C | 123917 | 123153 | 95.2 | 255 | 95.7/94.5 | 0.030 | 0.109 | 69.2/58.9 | Uracil-DNA glycosylase |
| U82 | C | 124745 | 123996 | 95.5 | 250 | 95.2/94.0 | 0.029 | 0.077 | 49.6/37.4 | Glycoprotein L |
| U83 | 124821 | 125159 | 92.4 | 113 | 87.6/85.6 | 0.071 | 0.098 | Intercrine cytokine | ||
| U84 | C | 126296 | 125271 | 94.2 | 342 | 92.4/90.6 | 0.048 | 0.081 | 51.7/42.4 | |
| U85 | C | 127202 | 126327 | 94.7 | 291 | 93.1/91.7 | 0.044 | 0.083 | 46.9/36.8 | OX-2 homology, glycoprotein |
| U86 | C | 131901 | 127342 | 85.9 | 1520 | 79.2/74.5 | 0.151 | 0.198 | 35.3/29.5 | IE-A |
| U90EX1 | C | 138436 | 138342 | 81.3 | 75 | 73.7/68.4 | 0.171 | 0.127 | 42.8/35.7 | IE-A (IE1), transactivator |
| U90EX2 | C | 138254 | 138043 | 80.9 | 70 | 70.3/67.2 | 67.1/57.1 | |||
| U90EX3 | C | 137932 | 135006 | 85.8 | 975 | 76.7/71.5 | 32.9/25.2 | |||
| U91EX1 | 138591 | 138816 | 76.3 | 75 | 67.8/57.1 | 0.301 | 0.246 | 33.3/25.0 | IE-A | |
| U91EX2 | 138926 | 139167 | 82.0 | 80 | 69.2/67.9 | 45.6/40.0 | ||||
| B8 | C | 142766 | 141972 | 265 | ||||||
| U94 | C | 144565 | 143096 | 96.5 | 490 | 98.4/97.6 | 0.011 | 0.089 | Parvovirus rep homolog | |
| U95 | 144640 | 148275 | 84.0 | 1212 | 72.9/69.0 | 0.188 | 0.150 | 33.4/26.7 | HCMV US22 gene family | |
| U100EX1 | C | 152325 | 151940 | 83.4 | 128 | 78.1/73.4 | 0.115 | 0.178 | 27.2/19.7 | Spliced envelope glycoprotein |
| U100EX2 | C | 151807 | 151526 | 87.6 | 93 | 84.9/81.7 | 53.8/38.7 | |||
| U100EX3 | C | 151121 | 150786 | 88.7 | 111 | 82.9/79.3 | 40.9/32.7 | |||
| U100EX4 | C | 150664 | 150587 | 88.5 | 25 | 96.0/88.0 | 44.0/40.0 | |||
| U100EX5 | C | 150476 | 150369 | 92.6 | 35 | 88.6/80.0 | 34.3/28.6 | |||
| U100EX6 | C | 150272 | 150159 | 95.6 | 37 | 91.9/91.9 | 48.6/37.8 | |||
| U100EX7 | C | 150008 | 149848 | 89.4 | 53 | 88.7/83.0 | 35.3/27.4 | |||
| U100EX8 | C | 149341 | 149332 | 100.0 | 3 | 100/100 | ||||
| U100EX9 | C | 149169 | 149041 | 94.6 | 42 | 92.8/90.5 | 35.7/23.8 | |||
| U100EX10 | C | 148911 | 148668 | 86.5 | 81 | 83.9/76.5 | 24.0/13.3 | |||
| B9 | C | 153252 | 152935 | 106 | ||||||
| DR1EX1 | 153904 | 154162 | 88.4 | 86 | 90.7/87.2 | 0.092 | 0.146 | 50.0/47.6 | HCMV US22 gene family | |
| DR1EX2 | 154276 | 156293 | 90.0 | 672 | 84.0/82.4 | 50.7/45.4 | ||||
| DR3 | C | 156646 | 156047 | 90.8 | 200 | 87.0/86.4 | ||||
| B1 | 156343 | 156819 | 159 | |||||||
| B2 | 156857 | 157093 | 79 | |||||||
| DR6EX1 | 158348 | 158651 | 89.1 | 101 | 86.1/84.1 | 0.112 | 0.223 | 55.4/53.5 | HCMV US22 gene family | |
| DR6EX2 | 159650 | 160521 | 88.8 | 290 | 84.3/82.2 | 69.1/62.6 | ||||
| B3 | 160670 | 160846 | 59 | |||||||
HHV-6B ORFs are named after their HHV-6A counterparts (19), except for spliced genes where the ORF is given the name of the 5′-proximal ORF. Based on the interpretation of the HHV-7 sequence by Megaw et al. (37), some of the ORFs described by Gompels et al. (19) are predicted to be exons: DR2 as DR1EX2; DR7 as DR6EX2; U5 as U7EX2; U16 as U17EX2; U60 as U66EX2; U80 as U79EX3; U89 as U90EX3; and U96, U97, U98, and U99 as U100EX10, EX7, EX3, and EX2, respectively. ORFs B1 to B9 are unique to HHV-6B.
First N-terminal methionine or in spliced genes first nucleotide of predicted exon.
Last nucleotide of the last codon of ORF or exon.
Determined by using GAP with gap and length weights of 50 and 3, respectively.
Percent amino acid similarity (%S) and percent amino acid identity (%I) were determined by using GAP with gap and length weights of 12 and 4, respectively.
Nonsynonymous (Ka) and synonymous (Ks) substitutions between HHV-6B and HHV-6A coding regions were estimated by using DIVERGE. Ka and Ks values given for spliced genes were determined by using the spliced coding sequence and listed in the first exon.
HHV-7 ORFs used in the alignments are from HHV-7(JI) as described by Nicholas (41) plus additional exons described by Megaw et al. (37). Prefixes (A) and (B) denote HHV-7 ORFs U55A and U55B, respectively.
ORFs encoded on the complementary strand are indicated by C.
Nine ORFs (DR4, DR5, DR8, U1, U61, U78, U88, U92, and U93) described by Gompels et al. (19) do not have counterparts in the HHV-6B genome, as a result of either the lack of an initiation codon, truncation, or frameshift mutations. HHV-6B encodes positional counterparts of HHV-6A ORFs LT1, LJ1, and RJ1, but these were excluded from consideration as candidates for being expressed as proteins because they are composed almost entirely of TRS arrays, the large variations in TRS copy number described above, and the lack of amino acid sequence conservation between the variants. Interestingly, DR3, U6, U9, U22, U83, and U94 are present in HHV-6A and HHV-6B but not in HHV-7 and thus are unique to the HHV-6 variants.
The nine putative unique HHV-6B ORFs, B1 through B9, are predicted to encode proteins of 265 amino acids or less (Table 3). B4 and B9 are located near the junctions of U and DR, a region of sequence divergence between the variants (see below). B4 spans R0, while B9 spans a region composed of four copies of an imperfect 62- or 63-bp element. None of the ORFs unique to HHV-6B had significant similarity with any other proteins in GenBank. Experimental data must be obtained to determine if any of the these ORFs encode functional proteins.
Relationships between HHV-6A and HHV-6B amino acid sequences.
Amino acid identity of HHV-6B proteins with their HHV-6A counterparts ranged from 99.5% in the spliced U66 gene to 61.8% for the spliced U91 gene (Table 3). U66 is a highly conserved herpesvirus protein involved in DNA packaging (46). U91 is located in the genomic region that is most divergent between the variants. The function of U91 is not known, but it is likely to be involved in gene regulation since it is part of the IE-A locus (51).
Several HHV-6B ORFs give rise to proteins longer than their HHV-6A counterparts as a result of differences near either their 5′ or 3′ ends. U8, U21, U23, U55, and U83 have 5′ extensions, while U10 and U44 have 3′ extensions. In addition, HHV-6B U47 has a 3′ extension and a 5′ truncation. These ORFs are dispersed in the genome, and none are homologs of conserved herpesvirus genes. In fact, U47 and U55 are located between conserved herpesvirus gene blocks (Fig. 1A). Interestingly, HHV-6B(Z29) U12, which encodes one of the G-protein-coupled receptor homologs, is truncated because of an in-frame termination codon at amino acid 195 of U12EX2. The termination codon lies within transmembrane region 5 of the seven predicted membrane-spanning domains and creates a protein that is 146 amino acids shorter than HHV-6A U12 (19). The HHV-6B(Z29) alteration is not likely to be a cloning or sequencing artifact or error because it was present in plasmid clones derived nearly a decade ago from purified viral DNA and in PCR amplimers derived directly from more recent viral DNA preparations. Nonetheless, the termination codon present in HHV-6B(Z29) is not present in two HHV-6B clinical isolates; translation of the ORF from these viruses should lead to a protein corresponding to the HHV-6A protein (15). Additionally, Isegawa et al. (24) reported that expression of HHV-6B(HST) U12 results in a full-length protein that functions as a β-chemokine receptor. These results indicate that expression of full-length U12 is not required for viral replication in cell culture.
Global variation between HHV-6A and HHV-6B.
The HHV-6A and HHV-6B genomic sequences were aligned in several pieces by using GAP, and the segments were then assembled into complete genomes containing alignment gaps. The resulting alignment was visualized with PLOTSIMILARITY (Fig. 1B). Most of the narrow valleys of low identity, e.g., in the vicinity of U41/U42 and U47, correspond to regions of multiple insertions in one variant relative to the other. The baseline identity level in these and other regions can be easily discerned. Some of the valleys are augmented by differences in copy number of a repeat element between the two genomes, e.g., TRS and R3.
As can be seen, there is high similarity across the middle of the alignment (positions 30000 through 128000), with regions of extensive dissimilarity toward the genomic ends. Highly variable regions are localized to the DRL-U junction, the left end of U, the region spanning U86 to U100, and all of DR. The most variable region is the region between U86 and U90, where there is only 63.2% nucleotide identity between HHV-6B and HHV-6A. Interestingly, the right end of U, spanning ORFs U86 to U100, differs by more than 10%, with the exception of the region encoding U94 (the adeno-associated virus type 2 rep homolog), which has 96.5% nucleotide sequence identity between the variants. In addition, concatemers of HHV-6A and HHV-6B ORFs U75, U76, U77, U79, U81, U82, U83, U84, U85, U86, U90, U91, U94, U95, and U100 that had been aligned codon by codon to correspond to the amino acid sequence alignments used to determine Ks and Ka values (described below) were assembled, and the resulting alignment was visualized as for Fig. 1B. Similar results were observed; i.e., U86, U90, U91, U95, and U100 had the greatest amount of sequence variation (not shown). Three notable areas of low sequence identity located in the region spanning the herpesvirus core genes are the U41-U42 intergenic region (which includes oriLyt), U47, and U54. As noted by others, genes at the junction of conserved herpesvirus gene blocks, e.g., U47 and U54, are frequently more divergent than their conserved neighbors (19); however as discussed later, the basis for variation at these positions in lineages of colinear genomes is not clear.
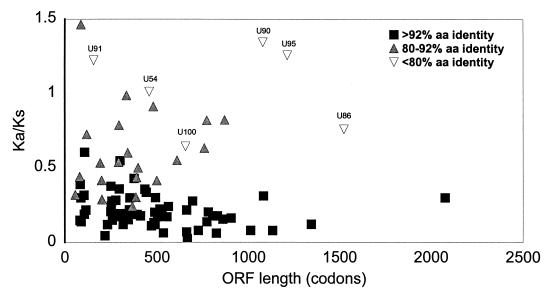
Genetic variation between HHV-6A and HHV-6B was further examined by determining the distribution of nucleotide substitutions within coding sequences. Estimates of the number of nucleotide substitutions per synonymous site (Ks) or per nonsynonymous site (Ka), that is, substitutions that are silent or result in amino acid changes, were computed for the 88 genes present in both HHV-6A and HHV-6B (Table 3). The expected outcome is that Ka will be smaller than Ks, unless positive selective pressure is being exerted on that particular sequence (13, 28). A Ka/Ks ratio of greater than 1 can be an indication that a particular sequence is under a strong selective pressure toward change in the encoded amino acid. Ka/Ks ratios were plotted against the codon length of the aligned pairs, which allows possible stochastic effects of Ka/Ks ratios obtained from short sequences to be visualized (Fig. 4). Of the 88 genes analyzed, U24, U54, U90, U91, and U95 have Ka/Ks ratios of greater than 1. The values for U24 and U91 possibly reflect stochastic effects due to their small size. Of the remaining ORFs with Ka/Ks ratios greater than 1, U90 and U95 appear to be under strong positive selection toward sequence divergence. The products of both genes are hypothesized to have regulatory functions and therefore may have important roles in the establishment of the variant-specific niche in the host. U90 has been shown to be a transcriptional activator of the human immunodeficiency virus type 1 long terminal repeat (61). U95 is a member of the HCMV US22 gene family, and two family members from HCMV have been shown to transactivate gene expression (53). These data add additional support to the hypothesis based on sequence differences that the right-most 24 kb of U of HHV-6A and HHV-6B genomes is not under strong sequence conservation.
FIG. 4.
ORF length versus Ka/Ks ratio. Ka/Ks ratios were calculated by using DIVERGE for 88 aligned HHV-6A and HHV-6B coding sequences. Protein and nucleic acid sequences were aligned by using PILEUP with gap and length weights of 12 and 4, respectively, for proteins and 5 and 1, respectively, for nucleic acids. Ka/Ks ratios for spliced genes were determined for the spliced proteins and are designated by the 5′-proximal exon. ORFs with less than 80% amino acid (aa) identity are labeled.
Relationships of HHV-6B with other herpesviruses.
The relationship of HHV-6B to other herpesviruses is similar to that described for HHV-6A (19). HHV-7 is the next-closest relative of HHV-6B; homologs to 82 HHV-7 genes that are likely to encode proteins are present in HHV-6B (37, 41). Amino acid identities between HHV-6B genes and their HHV-7 homologs range from 75% for U66EX1 and U77 to 22% for U20 (Table 3).
Strain variation.
A 30-kb region encompassing part of U74 through the carboxy terminus of U94 has been reported for HHV-6B(HST) (27). In pairwise comparisons between the two variant B strains, the amino acid identity for complete ORFs ranged from 92% (U90EX3) to 100% (U75). For comparison, amino acid identity between HHV-6A and HHV-6B ORFs in this region range from 57.1% (U91EX1) to 98.9% (U77).
R1 and R3 are regions of greater divergence (Fig. 5); nucleotide identities between R1 and R3 from strains Z29 and HST are 92.4 and 94.4% without gaps, respectively. There are strain differences in the copy number of the repeat elements; R1 has 54 copies in strain Z29 and 53 in strain HST, while R3 has 26 copies in strain Z29 and 24 copies in strain HST. Additionally, Z29 U86 is encoded by a single ORF, while there are two ORFs (U86 and U87) in the HST sequence as a result of a one-base deletion at position 16394 of the HST sequence. The amino-terminal ends of Z29 U86 and HST U87 are related, but the carboxy-terminal end of Z29 U86 is related to HST U86. Overall, the region spanning U75 through the end of R3 has high nucleotide identity between the strains (98.4%). For comparison, the corresponding region in HHV-6A has 79.3% nucleotide identity with HHV-6B (Fig. 5). Most of the intravariant sequence differences are localized in repeat elements R1 and R3; greater interstrain divergence (both insertions and substitutions) has previously been described in regions adjacent to repetitive regions of other herpesviruses (7, 56). The observed sequence differences between the two HHV-6B strains may reflect either geographic or etiologic differences; HST was isolated from a Japanese exanthem subitum patient, whereas Z29 was isolated from an AIDS patient from Zaire.
FIG. 5.
Graphical representation of intra- and intervariant alignments in the region encompassing U75 to the 3′ end of U94 [HHV-6B(Z29) coordinates 113977 to 143096). The HHV-6B(Z29) and HHV-6B(HST) DNA segments were aligned by using GAP with gap and length weights of 50 and 3, respectively, and the graphical representations were generated with GAPSHOW. The corresponding region of HHV-6A(U1102) was aligned with that of HHV-6B(Z29) by using GAP with gap and length weights of 25 and 1, respectively. Nucleotide sequence differences are represented by the longest vertical bars; gaps are depicted by the shorter vertical lines. Regions spanning R1 and R3 are indicated. Locations of R1, R2A, R2B, R3, and ORFs are indicated.
DISCUSSION
In this work we present the general features of the HHV-6B genome sequence, describe its coding potential, and describe its relationships with other herpesviruses. This information will facilitate examination of the biological significance of the genetic differences between HHV-6B and other herpesviruses, in particular between HHV-6A and HHV-6B. The HHV-6B genome sequence allows prediction of 97 unique genes, 88 of which have HHV-6A counterparts, while 82 have counterparts in HHV-7. Thirty-nine of these genes are conserved among all mammalian herpesviruses (herpesvirus core genes), others have been found only in betaherpesviruses, while still others are found only in members of the genus Roseolovirus and some are found only in HHV-6A and HHV-6B. Below we discuss the general aspects of sequence conservation and divergence across the herpesvirus family, as well as the relationship between the HHV-6 variants.
Relationship of HHV-6B and other herpesviruses.
Herpesviruses encode a set of conserved genes, the herpesvirus core genes, that are grouped into seven gene blocks (7). Within gene blocks, the order and transcriptional polarity of the component genes are maintained. The gene blocks have different genomic locations, order, and orientations in the different herpesvirus subfamilies. Products of the core genes include structural components, such as capsid proteins and glycoproteins. Others include enzymes required for DNA replication, such as the major DNA binding protein and the DNA polymerase. The HHV-6B gene organization shares overall similarity with other betaherpesviruses, such as HCMV and HHV-7.
Betaherpesviruses encode several genes that are common to the viruses of this subfamily and that are absent in alpha- and gammaherpesviruses. Many of these genes belong to the US22 family of genes, examples of which are scattered throughout the HCMV genome (7). The function of these genes during infection is unclear, although HCMV TRS1 and IRS1 are transcriptional activators (53). Within the betaherpesviruses, divergence between viruses of the Cytomegalovirus and Roseolovirus genera is reflected by genes that are specific to members of Roseolovirus, including U20, U21, U23, U24, and U26.
It is particularly interesting that the roseoloviruses encode homologs (U73) of both the origin binding protein of alphaherpesviruses and their binding sites in origins of lytic DNA replication (23). As described here and elsewhere (19, 29, 31, 37, 41), the other proteins that are involved in replication at the replication fork are more highly conserved between the roseoloviruses and their HCMV counterparts than with any alphaherpesvirus. This strengthens the previous suggestion (14) that the mechanisms of DNA replication initiation and elongation for roseoloviruses have greater similarities with alphaherpesviruses and cytomegaloviruses, respectively.
Among the roseoloviruses, HHV-7 encodes no genes without counterparts in HHV-6A and HHV-6B, whereas both HHV-6A and HHV-6B encode DR3, U6, U9, U22, U83, and U94. In addition, HHV-6A and HHV-6B each encode variant-specific genes.
Relationship of HHV-6B and HHV-6A.
Comparisons of the HHV-6A and HHV-6B genomes confirmed that while the two genomes are colinear, there are regions of significant variation, including DR, a region spanning the junction of DRL-U and the extreme left end of U, and a 24-kb segment located to the right of U86 (except for U94). The region spanning ORFs U2 through U85, which encompasses approximately 75% of U, is more highly conserved. Of the 89 ORFs in this region, 66 have over 92% amino acid identity. All of the genes belonging to the herpesvirus conserved gene blocks have greater than 94% amino acid identity.
U47 and U54 have less than 90% amino acid identity with their HHV-6A counterparts and Ka/Ks ratios of greater than 0.8, possibly reflecting gene products whose functions do not require specific sequences or that the gene products have different functions in each variant. It is interesting that these genes, which are the most divergent in the segment spanned by the herpesvirus core genes, map at the junctions of blocks of conserved genes. Extensive amino acid sequence variation of genes at these locations has been previously found in comparisons of herpesvirus genomes. This variation is easily rationalized for comparisons between herpesvirus genomes that have been rearranged through these sites, e.g., varicella-zoster virus and Epstein-Barr virus (10), but the basis for the susceptibility of these genes to variation is not obvious in the case of viruses with colinear genomes, e.g., HCMV, HHV-6A, HHV-6B, and HHV-7, for which these sites have not been the location of genomic rearrangements during evolution of the lineage.
The segment from U86 to the right end of U is the most divergent between the variants. This region is likely to be important in defining the biological differences between the variants. Interestingly, this region contains genes that have anomalous sequence compositions due to CpG suppression and complex splicing patterns. As an example, the variants differ in temporal regulation and splicing patterns of U91 transcripts in T-cell lines (39). It will be important to determine whether R3 is involved in transcriptional regulation of the IE-A locus and to ascertain the effect of sequence differences and copy number on R3-mediated transcriptional regulation.
Another source of phenotypic difference between the variants could be the gp82-gp105 complex, which is a major envelope glycoprotein that is composed of a number related polypeptides (44, 45). This glycoprotein is encoded by U100, which encompasses 11 exons (45) and is located in one of the most divergent regions of the genome. Homologs of the gene have been found only in roseoloviruses, with the intron-exon structure being conserved between the HHV-6 variants and HHV-7 (19, 37, 41). Differential splicing accounts for the presence of multiple related protein species, at least in the case of HHV-6A and as postulated for HHV-6B (45). The gp82-105 complex is likely to be important to the biology of HHV-6 since there are variant-specific neutralizing epitopes (44). HHV-6A and HHV-6B U100 share 79.9% amino acid identity, which is much lower than for other glycoproteins, such as glycoproteins B, H, L, and M (Table 3). Because glycoproteins are important determinants of specificity in the initial physical interaction between virus and the host cell, this complex of related proteins may confer different biological properties on the variants.
In addition to the effects of the more dramatically divergent regions, it is likely that the smaller genetic differences elsewhere in the genome reflect subtle adaptations to specific biological niches and cumulatively are likely to have an important effect on the biology of the variants.
Are HHV-6A and HHV-6B distinct herpesvirus species?
The genetic differences found between HHV-6A and HHV-6B are substantially greater than those found between HHV-7 strains RK and JI, even in the conserved region spanning ORFs U2 to U85. Across their lengths, the two HHV-7 genomes differed by a total of only 179 nt, an average of 1 per kb (37). In contrast, the sequence identity between HHV-6A and HHV-6B is 85% in DR, a mean of 95% in the region spanning U2 and U85, and 72% in the region spanning U86 and U100, for an overall identity of 90% (Fig. 1B). Additionally, sequence variation between HHV-6B strains Z29 and HST in the 30-kb region spanning the IE-A locus is substantially less than that between the variants. The accumulated biological, genetic, and epidemiologic data thus converge and make it clear that while HHV-6A and HHV-6B are closely related viruses, they have independent biological niches and meet the criteria for classification into distinct species.
ACKNOWLEDGMENTS
We thank Jodi B. Black, Gary J. Lindquester, and Robert D. Allen for their contributions to this work. We also thank William C. Reeves for his support.
G.D. was a Visiting Fellow at the Centers for Disease Control and Prevention. S.D. was supported by NIH grants KO4 AI01240 and R21 AI34231.
REFERENCES
- 1.Ablashi D, Agut H, Berneman Z, Campadelli-Fiume G, Carrigan D, Ceccerini-Nelli L, Chandran B, Chou S, Collandre H, Cone R, Dambaugh T, Dewhurst S, DiLuca D, Foa-Tomasi L, Fleckenstein B, Frenkel N, Gallo R, Gompels U, Hall C, Jones M, Lawrence G, Martin M, Montagnier L, Neipel F, Nicholas J, Pellett P, Razzaque A, Torrelli G, Thomson B, Salahuddin S, Wyatt L, Yamanishi K. Human herpesvirus-6 strain groups: a nomenclature. Arch Virol. 1993;129:363–366. doi: 10.1007/BF01316913. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun D K, Dominguez G, Pellett P E. Human herpesvirus 6. Clin Microbiol Rev. 1997;10:521–567. doi: 10.1128/cmr.10.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckmaster A E, Scott S D, Sanderson M J, Boursnell M E G, Ross N L J, Binns M M. Gene sequence and mapping data from Marek’s disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J Gen Virol. 1988;69:2033–2042. doi: 10.1099/0022-1317-69-8-2033. [DOI] [PubMed] [Google Scholar]
- 5.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandran B, Tirawatnapong S, Pfeiffer B, Ablashi D V. Antigenic relationships among human herpesvirus-6 isolates. J Med Virol. 1992;37:247–254. doi: 10.1002/jmv.1890370403. [DOI] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Marousek G I. Analysis of interstrain variation in a putative immediate-early region of human herpesvirus 6 DNA and definition of variant-specific sequences. Virology. 1994;198:370–376. doi: 10.1006/viro.1994.1044. [DOI] [PubMed] [Google Scholar]
- 9.Davison A. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J, Taylor P. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J Gen Virol. 1987;68:1067–1079. doi: 10.1099/0022-1317-68-4-1067. [DOI] [PubMed] [Google Scholar]
- 11.Deng H, Dewhurst S. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J Virol. 1998;72:320–329. doi: 10.1128/jvi.72.1.320-329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewhurst S, Dollard S C, Pellett P E, Dambaugh T R. Identification of a lytic-phase origin of DNA replication in human herpesvirus 6B strain Z29. J Virol. 1993;67:7680–7683. doi: 10.1128/jvi.67.12.7680-7683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez G, Black J B, Stamey F R, Inoue N, Pellett P E. Physical and genetic maps of the human herpesvirus 7 strain SB genome. Arch Virol. 1996;141:2387–2408. doi: 10.1007/BF01718639. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez, G., F. R. Stamey, and P. E. Pellett. 1999. Unpublished results.
- 16.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gompels U A, Carrigan D R, Carss A L, Arno J. Two groups of human herpesvirus 6 identified by sequence analyses of laboratory strains and variants from Hodgkin’s lymphoma and bone marrow transplant patients. J Gen Virol. 1993;74:613–622. doi: 10.1099/0022-1317-74-4-613. [DOI] [PubMed] [Google Scholar]
- 18.Gompels U A, Macaulay H A. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J Gen Virol. 1995;76:451–458. doi: 10.1099/0022-1317-76-2-451. [DOI] [PubMed] [Google Scholar]
- 19.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 20.Hearing P, Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983;33:695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- 21.Honess R W, Gompels U A, Barrell B G, Craxton M, Cameron K R, Staden R, Chang Y-N, Hayward G S. Deviations from expected frequencies of CpG dinucleotides in herpesvirus DNAs may be diagnostic of differences in the states of their latent genomes. J Gen Virol. 1989;70:837–855. doi: 10.1099/0022-1317-70-4-837. [DOI] [PubMed] [Google Scholar]
- 22.Huber M T, Compton T. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol. 1998;72:8191–8197. doi: 10.1128/jvi.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue N, Dambaugh T R, Rapp J C, Pellett P E. Alphaherpesvirus origin-binding protein homolog encoded by human herpesvirus 6B, a betaherpesvirus, binds to nucleotide sequences that are similar to ori regions of alphaherpesviruses. J Virol. 1994;68:4126–4136. doi: 10.1128/jvi.68.7.4126-4136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isegawa Y, Ping Z, Nakano K, Sugimoto N, Yamanishi K. Human herpesvirus 6 open reading frame U12 encodes a functional β-chemokine receptor. J Virol. 1998;72:6104–6112. doi: 10.1128/jvi.72.7.6104-6112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1109–1166. [Google Scholar]
- 26.Kishi M, Bradley G, Jessip J, Tanaka A, Nonoyama M. Inverted repeat regions of Marek’s disease virus DNA possess a structure similar to that of the a sequence of herpes simplex virus DNA and contain host cell telomere sequences. J Virol. 1991;65:2791–2797. doi: 10.1128/jvi.65.6.2791-2797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosuge H, Isegawa Y, Yamanishi K. Nucleotide sequence analysis of a 30-kilobase-pair region of human herpesvirus-6B (HHV-6B) genome and strain-specific variations in major immediate-early genes. Virus Res. 1997;52:1–14. doi: 10.1016/s0168-1702(97)00099-3. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Wu C, Luo C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 29.Lindquester G J, Greenamoyer C A, Anton E D, O’Brian J J, Pellett P E, Dambaugh T R. Comparison of a 20 kb region of human herpesvirus 6B with other human betaherpesviruses reveals conserved replication genes and adjacent divergent open reading frames. Arch Virol. 1997;142:193–204. doi: 10.1007/s007050050070. [DOI] [PubMed] [Google Scholar]
- 30.Lindquester G J, Inoue N, Allen R D, Castelli J W, Stamey F R, Dambaugh T R, O’Brian J J, Danovich R M, Frenkel N, Pellett P E. Restriction endonuclease mapping and molecular cloning of the human herpesvirus 6 variant B strain Z29 genome. Arch Virol. 1996;141:367–379. doi: 10.1007/BF01718406. [DOI] [PubMed] [Google Scholar]
- 31.Lindquester G J, O’Brian J J, Anton E D, Greenamoyer C A, Pellett P E, Dambaugh T R. Genetic content of a 20.9 kb segment of human herpesvirus 6B strain Z29 spanning the homologs of human herpesvirus 6A genes U40-57 and containing the origin of replication. Arch Virol. 1997;142:103–123. doi: 10.1007/s007050050062. [DOI] [PubMed] [Google Scholar]
- 32.Lindquester G J, Pellett P E. Properties of the human herpesvirus 6 strain Z29 genome: G + C content, length, and presence of variable-length directly repeated terminal sequence elements. Virology. 1991;182:102–110. doi: 10.1016/0042-6822(91)90653-s. [DOI] [PubMed] [Google Scholar]
- 33.Lopez C, Pellett P, Stewart J, Goldsmith C, Sanderlin K, Black J, Warfield D, Feorino P. Characteristics of human herpesvirus-6. J Infect Dis. 1988;157:1271–1273. doi: 10.1093/infdis/157.6.1271. [DOI] [PubMed] [Google Scholar]
- 34.Lusso P, Markham P D, Tschachler E, di Marzo Veronese F, Salahuddin S Z, Ablashi D V, Pahwa S, Krohn K, Gallo R C. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6) J Exp Med. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin M E, Piette J, Yaniv M, Tang W-J, Folk W R. Activation of the polyomavirus enhancer by a murine activator protein 1 (AP1) homolog and two contiguous proteins. Proc Natl Acad Sci USA. 1988;85:5839–5843. doi: 10.1073/pnas.85.16.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin M E D, Thomson B J, Honess R W, Craxton M A, Gompels U A, Liu M Y, Littler E, Arrand J R, Teo I, Jones M D. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. J Gen Virol. 1991;72:157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- 37.Megaw A G, Rapaport D, Avidor B, Frenkel N, Davison A J. The DNA sequence of the RK strain of human herpesvirus 7. Virology. 1998;244:119–132. doi: 10.1006/viro.1998.9105. [DOI] [PubMed] [Google Scholar]
- 38.Meyne J, Ratliff R L, Moyzis R K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirandola P, Menegazzi P, Merighi S, Ravaioli T, Cassai E, Di Luca D. Temporal mapping of transcripts in herpesvirus 6 variants. J Virol. 1998;72:3837–3844. doi: 10.1128/jvi.72.5.3837-3844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neipel F, Ellinger K, Fleckenstein B. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J Gen Virol. 1991;72:2293–2297. doi: 10.1099/0022-1317-72-9-2293. [DOI] [PubMed] [Google Scholar]
- 41.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholas J. Nucleotide sequence analysis of a 21-kbp region of the genome of human herpesvirus-6 containing homologues of human cytomegalovirus major immediate-early and replication genes. Virology. 1994;204:738–750. doi: 10.1006/viro.1994.1589. [DOI] [PubMed] [Google Scholar]
- 43.Pellett P E, Sanchez-Martinez D, Dominguez G, Black J B, Anton E, Greenamoyer C, Dambaugh T R. A strongly immunoreactive virion protein of human herpesvirus 6 variant B strain Z29: identification and characterization of the gene and mapping of a variant-specific monoclonal antibody reactive epitope. Virology. 1993;195:521–531. doi: 10.1006/viro.1993.1403. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer B, Berneman Z N, Neipel F, Chang C K, Tirwatnapong S, Chandran B. Identification and mapping of the gene encoding the glycoprotein complex gp82-gp105 of human herpesvirus 6 and mapping of the neutralizing epitope recognized by monoclonal antibodies. J Virol. 1993;67:4611–4620. doi: 10.1128/jvi.67.8.4611-4620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeiffer B, Thomson B, Chandran B. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J Virol. 1995;69:3490–3500. doi: 10.1128/jvi.69.6.3490-3500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pringle C R. The universal system of virus taxonomy of the International Committee on Virus Taxonomy (ICTV), including new proposals ratified since publication of the Sixth ICTV Report in 1995. Arch Virol. 1998;143:203–210. doi: 10.1007/s007050050280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruvolo V R, Berneman Z, Secchiero P, Nicholas J. Cloning, restriction endonuclease mapping and partial sequence analysis of the genome of human herpesvirus 7 strain JI. J Gen Virol. 1996;77:1901–1912. doi: 10.1099/0022-1317-77-8-1901. [DOI] [PubMed] [Google Scholar]
- 49.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo R C. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 50.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiewe U, Neipel F, Schreiner D, Fleckenstein B. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J Virol. 1994;68:2978–2985. doi: 10.1128/jvi.68.5.2978-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Secchiero P, Nicholas J, Deng H, Xiaopeng T, van Loon N, Ruvolo V R, Berneman Z N, Reitz M S, Dewhurst S. Identification of human telomeric repeat motifs at the genome termini of human herpesvirus 7: structural analysis and heterogeneity. J Virol. 1995;69:8041–8045. doi: 10.1128/jvi.69.12.8041-8045.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the α gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tedder R S, Briggs M, Cameron C H, Honess R, Robertson D, Whittle H. A novel lymphotropic herpesvirus. Lancet. 1987;ii:390–392. doi: 10.1016/s0140-6736(87)92404-4. [DOI] [PubMed] [Google Scholar]
- 56.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 57.Thomson B J, Dewhurst S D, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68:3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomson B J, Honess R W. The right end of the unique region of the genome of human herpesvirus 6 U1102 contains a candidate immediate early gene enhancer and a homologue of the human cytomegalovirus US22 gene family. J Gen Virol. 1992;73:1649–1660. doi: 10.1099/0022-1317-73-7-1649. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto T, Mukai T, Kondo K, Yamanishi K. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J Clin Microbiol. 1994;32:473–476. doi: 10.1128/jcm.32.2.473-476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;ii:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y, Chang C K, Qian G, Chandran B, Wood C. Trans-activation of the HIV promoter by a cDNA and its genomic clones of human herpesvirus-6. Virology. 1994;199:311–322. doi: 10.1006/viro.1994.1129. [DOI] [PubMed] [Google Scholar]