Abstract
Pre-incubation of the cumulus-oocyte complex (COCs) may lead to better function of cumulus cells (CCs) and higher oocyte quality by changing the transcriptomic profile of CCs. 140 cumulus cell samples were isolated from 12 participants and divided into two groups based on pre-incubation time. In the T0 group, the COCs were immediately dissected to separate the CCs from around the oocytes. In the T2 group, CCs were prepared after 2 h of incubation. Then, the transcriptomic profile of the CCs of the non pre-incubation group was compared to the 2-h pre-incubation group. Confirmation of RNA sequencing results was done via qRT‑PCR. The CCs transcriptome analysis showed 17 genes were downregulated and 22 genes upregulated in the T2 group compared to the T0 group. Also, the pathways related to ATP production (oxidative phosphorylation, electron transport chain, and Mitochondrial complex I assembly model OXPHOS system), TNF-alpha signaling pathway, and glucocorticoid receptor pathway increased in the T2 group compared to the T0 group. Also, the TGF-β pathway was decreased in the T2 group compared to the T0 group. This study showed that 2 h pre-incubation leads to changes in important pathways in CCs, which positively affects oocyte quality.
Keywords: Pre-incubation, Cumulus cells, RNA-sequencing, Oocyte quality
Subject terms: Cell biology, Developmental biology
Introduction
According to reports, 10–15% of couples worldwide are infertile1. Among the standard techniques for treating infertility is intracytoplasmic sperm injection (ICSI)2. In the ICSI technique, the oocytes collected from the individual are incubated until the ICSI is performed (Pre-incubation), and this incubation time varies depending on the specific conditions of the laboratory3. The evidence is that the complete and final maturation of the nucleus and cytoplasm of the oocyte is necessary for its activation by sperm and also plays a significant role in embryo quality3. Since cytoplasmic maturation of oocytes takes more time than nuclear maturation, oocyte pre-incubation may lead to coordination between them4.
Although there are many differences of opinion regarding the necessity of pre-incubation, and some studies have shown that pre-incubation of oocytes does not affect fertility or pregnancy outcome5, in some studies, it has been stated that the pre-incubation of oocytes before ICSI plays a role in completing the nuclear and cytoplasmic maturation of the oocyte5,6. On the other hand, in pre-incubation, attention should be paid to the passage of time and oocyte aging and its effect on reducing oocyte quality and fertilization rate4. In investigating the impact of different periods of oocyte pre-incubation on nucleus maturation, fertility rate, and embryo quality in IVF and ICSI, 176 IVF and 64 ICSI cycles were studied, and the results showed that at least 2.5 h of pre-incubation is beneficial for IVF and ICSI due to increasing the maturation of oocytes5. In the study comparing zero and 2–4 h of oocyte pre-incubation before ICSI, 2–4 h pre-incubation improved oocyte and embryo quality6. But, pre-incubation for more than 4 h led to a decrease in oocyte quality4. The study results of the effect of different pre-incubation times (from half to 6 h) on the fertilization rate following ICSI showed that the fertilization rate in different time groups before ICSI do not differ significantly from each other7. Therefore, it seems necessary to investigate the appropriate pre-incubation time and its effect on oocyte quality. The issue to be considered is the minimum pre-incubation time required for oocyte maturation and the maximum pre-incubation time that does not lead to oocyte senescence.
In folliculogenesis, the communication between the oocyte and its surrounding granulosa cells, cumulus cells (CC), is critical for oocyte maturation8. The inner row of CC is connected to the zona pellucida of the oocyte, and by establishing a connection with the cytoplasmic membrane, they form the cumulus-oocyte complex (COC), which has three types of connections. One of these communications is the gap junctions that transfer small molecules between the oocyte and the CC. In addition, paracrine signals between these two types of cells are also necessary for the growth and regulation of oocyte meiotic maturation9. Another connection is the production of part of the follicular fluid by the CC, which can also its effects on the oocyte10. Also, the factors secreted from oocytes affect the gene expression and protein pattern of CC. Therefore, CC can reflect the quality of oocytes and a suitable method for selecting or examining oocytes8.
Since there is a difference of opinion about the time of pre-incubation and most of the studies conducted are clinical and little molecular research has been done in this direction, also, because the direct examination of the oocyte is not ethical, in this study, the changes in the transcriptomic profile of cumulus cells under the influence of two time zero and T2 pre-incubation of human COC in patients treated in ART cycles and pathways involved in this process were investigated.
Materials & methods
Patient selection and ovulation stimulation
The study approval was obtained from the Ethics Committee of Iran University of Medical Science (Reference number: IR.IUMS.REC.1396.31837), and all research was performed by relevant guidelines. Written consent was obtained from all participants. 12 healthy oocyte donor women (with at least one natural pregnancy resulting in a live birth) were included in the study. The BMI of all participants was in the normal range (between 18 and 28) with a mean (± SD) age of less than 35 (31.80 ± 3.07). Also, the antagonist protocol was used to stimulate ovulation in all of them.
COC Pre-incubation and cumulus cell isolation
The collection of COCs was done 36–38 h after hCG administration with ultrasound-guided transvaginal puncture. Then, the COCs were washed with GMOPS medium (Vitrolife, Gothenburg, Sweden) to remove the blood contamination. At this stage, the COCs were divided into T0 and T2 groups. In the T0 group, COCs were immediately dissected to separate the cumulus cells from around the oocytes and prepare the oocytes for ICSI. Then, cumulus cells related to mature metaphase II (MII) oocyte were centrifuged at 200 g for 10 min after washing with phosphate-buffered saline (PBS) stored in liquid nitrogen. In the T2 group, dissection and preparation of cumulus cells were done after 2 h of COC incubation. Pre-incubation was performed at 37 °C, 95% humidity, 6% CO2, and 5% O2 condition. All the media were from Vitrolife (Vitrolife, Gothenburg, Sweden). Cumulus cells related to MII oocytes were used in this study.
RNA‑seq and bioinformatics analysis
140 CC samples were isolated from 12 patients and divided into two groups based on pre-incubation time. 70 samples were placed in each group. 40 samples from each group were used for RNA-Seq, and the remaining 30 samples were used to confirm the RNA-Seq results via qRT‑PCR.
Extracted RNA has been sequenced by BGI Hong Kong using an rRNA depletion stranded library preparation kit. More than 13 GB of sequences were generated with Q20 percent more than 97% with 100bp length in PE mode using the DNBseq sequencing platform. We used FastQC11 to double-check the quality of raw data. Since the Phred score of the reads was more than almost 30 for all positions and there were no contaminations, there was no need for trimming and adaptor removal. Next, the Hisat2 algorithm on the GRCh38/hg38 human reference genome was used for alignment12. We also downloaded the RefSeq gtf file for the hg38 reference build from the UCSC database. Next, the aligned outputs from Hisat2 were sorted using Samtools. Then, the set of expressed transcripts and differentially expressed genes were identified using Stringtie and cuffdiff tools13,14. Note that in cuffdiff fragments per kilobase of transcript per million mapped reads (FPKM), library normalization and blind dispersion method were used. It is worth mentioning that this algorithm has been specially developed for cases with no replicates. Next, significantly differentially expressed genes (DEGs) for this study were extracted based on the FDR corrected P value < 0.05 and |log2(FC)|> 1 condition. Then, the statistical power of our test was estimated using the average depth of covering the expressed genes and rnapower function from the RNASeqPower package15. Finally, the gene ontology terms and pathway enrichment analysis of significantly upregulated and down-regulated genes were done using the EnrichR online database16. Integrated Genome Viewer (IGV) tools were used to visualize genes selected for validation using RT-qPCR17.
In the following, the pathways associated with the human cumulus cells in the patients treated in ART cycles influenced by two time zero and T2 pre-incubation were studied. Considering the related enriched pathways to this research, especially in WikiPathway terms and other enrichment analyses such as biological process gene ontology terms and KEGG pathway analysis, four candidates from upregulated and downregulated DEGs genes were selected for qRT-PCR validation.
QRT‑PCR validation
The qRT-PCR method was used to confirm the RNA sequencing data. 4 genes were selected for confirmation: genes CXCL8(C-X-C motif chemokine ligand 8), TNFAIP3 (TNF alpha-induced protein 3), and ND5 (mitochondrially encoded NADH dehydrogenase 5) had the highest expression, and gene EGR2 (early growth response 2) had the lowest expression in the T2 group compared to the T0 group. 30 samples of each group were used for confirmation of RNA-Seq results via qRT‑PCR.
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Germany). Triplicate technical replicates were considered for all samples. The A260/A280 ratio of samples was assessed by a Nanodrop spectrophotometer, and concentration and purity was examined. According to the 'manufacturer's instructions and via the High Capacity cDNA Reverse Transcription Kit (Bio-Rad, 1,725,037) Random Hexamer primers, reverse transcription was done. ABI Prism 7300 Sequence Detector (Applied Biosystems, Foster City, California, USA) was used for qPCR reactions. Standard curves were checked out, and 'Primers' Efficiency was confirmed. The sequence of primers is shown in Table 1. Then, the relative transcript levels (Life Science, 7900HT) were determined using the SYBR Green master mix and Sequence Detection System. Data normalization was done via human GAPDH housekeeping gene. Relative gene expression was quantified using △△ the Ct method and calculated with FC = 2−∆∆CT formula18.
Table 1.
Sequences of the primers used in this study.
| Gene | Primer (5–3) | Tm | Product length | Gene ID |
|---|---|---|---|---|
| CXCL8 | Forward: ATGGCTGCTGAACCAGTAGA | 58.72 | 570 | 3576 |
| Reverse: CTAGTCTTCGTTTTGAACAG | 51.93 | |||
| TNFAIP3 | Forward: AGAGCAACTGAGATCGAGCCA | 61.23 | 145 | 7128 |
| Reverse: CTGGTTGGGATGCTGACACTC | 60.95 | |||
| ND5 | Forward: CAGCAGCCATTCAAGCAATGC | 61.33 | 167 | 4540 |
| Reverse: GGTGGAGACCTAATTGGGCTGATTAG | 62.87 | |||
| EGR2 | Forward: CTTTGACCAGATGAA CGG AGT | 57.95 | 100 | 1959 |
| Reverse: AGCAAAGCTGCTGGGATATG | 58.31 | |||
| GAPDH | Forward: GCAGGGATGATGTTCTGG | 55.07 | 126 | 2597 |
| Reverse: CTTTGGTATCGTGGAAGGAC | 55.86 |
Statistical analysis
One-way ANOVA and ‘Tukey’s multiple comparisons test as post hoc were used for statistical analyses. Also, version 24.0 (IBM) of the SPSS and Prism 8 software were used. P < 0.05 was statistically significant.
Ethical approval
The study was approved by the Ethics Committee of Iran University of Medical Science (Reference number: IR.IUMS.REC.1396.31837).
Results
Bioinformatics analysis
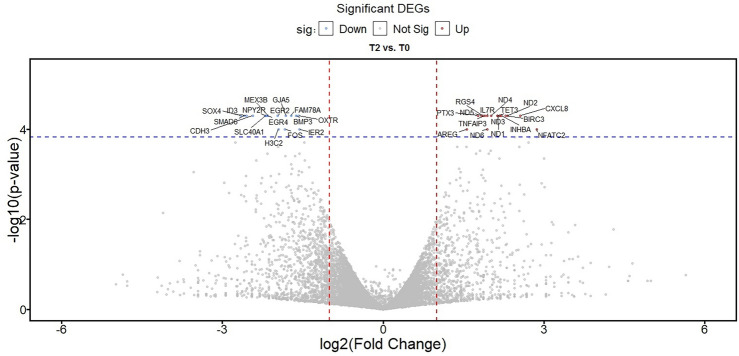
DEGs analysis (q value < 0.05 and |log2(FC)|> 1) resulted in the identification of 39 genes, including 22 upregulated genes and 17 downregulated genes. Tables 2 and 3 shows the upregulated and downregulated genes, respectively. Moreover, in Fig. 1, the volcano plot of expressed genes is depicted. Based on Fig. 1, the dispersion of upregulated and downregulated genes and differentially expressed genes are almost the same. Finally, the statistical power of our test was estimated at 0.8145 > 0.8. So, the sample size for the pooled samples was sufficient to expect statistically meaningful results.
Table 2.
Upregulated DEG in T2 group compared to T0 group.
| Gene symbol | Locus | Expr(0 h) | Expr(2 h) | Log2(FC) | P_value | FDR |
|---|---|---|---|---|---|---|
| CSF3 | chr17:40,015,439–40,017,813 | 0 | 0.782705 | inf | 5.00E−05 | 0.023765 |
| FAM163A | chr1:179,591,612–179,819,418 | 0 | 0.563185 | inf | 5.00E − 05 | 0.023765 |
| SERPINB2 | chr18:63,885,285–63,903,888 | 0 | 1.78659 | inf | 5.00E − 05 | 0.023765 |
| MTATP6P1 | chr1:633,534–634,922 | 18.3315 | 69.3245 | 1.91904 | 5.00E − 05 | 0.023765 |
| HSD3BP3 | chr1:119,538,420–119,546,244 | 0 | 0.909862 | inf | 5.00E − 05 | 0.023765 |
| TET3 | chr2:73,983,630–74,147,912 | 17.3608 | 76.2401 | 2.13472 | 5.00E − 05 | 0.023765 |
| PTX3 | chr3:157,173,699–157,503,605 | 27.6991 | 94.4144 | 1.76917 | 5.00E − 05 | 0.023765 |
| CXCL8 | chr4:73,740,568–73,743,716 | 13.963 | 82.2564 | 2.55852 | 5.00E − 05 | 0.023765 |
| AREG | chr4:74,445,135–74,455,005 | 72.8527 | 215.467 | 1.56441 | 0.0001 | 0.039793 |
| RGS4 | chr1:163,068,870–163,076,802 | 10.896 | 39.5985 | 1.86165 | 5.00E − 05 | 0.023765 |
| IL7R | chr5:35,856,890–35,879,603 | 8.19245 | 31.4328 | 1.93991 | 5.00E − 05 | 0.023765 |
| TNFAIP3 | chr6:137,823,668–137,883,312 | 38.3909 | 142.782 | 1.89498 | 5.00E − 05 | 0.023765 |
| INHBA | chr7:41,685,113–41,779,378 | 5.927 | 27.246 | 2.20067 | 5.00E − 05 | 0.023765 |
| ND1 | chrM:3306–4262 | 107.37 | 472.212 | 2.13684 | 5.00E − 05 | 0.023765 |
| ND2 | chrM:4469–5511 | 64.4693 | 322.528 | 2.32274 | 5.00E − 05 | 0.023765 |
| ND3 | chrM:10,058–10,404 | 125.611 | 568.492 | 2.17818 | 5.00E − 05 | 0.023765 |
| ND4 | chrM:10,469–12,137 | 208.316 | 842.433 | 2.01579 | 5.00E − 05 | 0.023765 |
| ND5 | chrM:12,336–14,148 | 105.397 | 375.965 | 1.83477 | 5.00E − 05 | 0.023765 |
| ND6 | chrM:14,148–14,673 | 25.7703 | 99.2931 | 1.94598 | 0.0001 | 0.039793 |
| BIRC3 | chr11:102,317,483–102,339,403 | 2.65885 | 12.9447 | 2.28349 | 5.00E − 05 | 0.023765 |
| NFATC2 | chr20:51,366,434–51,562,839 | 0.373128 | 2.72307 | 2.86749 | 0.0001 | 0.039793 |
| TNFRSF18 | chr1:1,203,507–1,206,592 | 0 | 0.703968 | inf | 5.00E − 05 | 0.023765 |
Table 3.
Down-regulated DEG in T2 group compared to T0 group.
| Gene symbol | Locus | Expr(0 h) | Expr(2 h) | Log2(FC) | P_value | FDR |
|---|---|---|---|---|---|---|
| IER2 | chr19:13,150,410–13,154,911 | 180.057 | 60.8156 | − 1.56594 | 0.0001 | 0.039793 |
| MIR23A | chr19:13,834,515–13,842,918 | 26.8313 | 7.52007 | − 1.8351 | 5.00E − 05 | 0.0237653 |
| EGR4 | chr2:73,290,928–73,293,548 | 37.651 | 8.33332 | − 2.17572 | 5.00E − 05 | 0.0237653 |
| SLC40A1 | chr2:189,560,589–189,580,786 | 30.8014 | 6.76107 | − 2.18767 | 5.00E − 05 | 0.0237653 |
| GJA5 | chr1:147,700,065–147,789,690 | 86.6778 | 24.6572 | − 1.81366 | 5.00E − 05 | 0.0237653 |
| OXTR | chr3:8,733,801–8,769,613 | 37.7563 | 12.6232 | − 1.58064 | 5.00E − 05 | 0.0237653 |
| BMP3 | chr4:81,030,707–81,057,627 | 45.746 | 14.8048 | − 1.62758 | 5.00E − 05 | 0.0237653 |
| NPY2R | chr4:155,173,722–155,217,076 | 12.8719 | 2.80762 | − 2.1968 | 5.00E − 05 | 0.0237653 |
| SOX4 | chr6:21,593,750–21,598,619 | 70.8297 | 11.2543 | − 2.65388 | 5.00E − 05 | 0.0237653 |
| H3C2 | chr6:26,031,588–26,032,099 | 83.8249 | 21.6336 | − 1.95411 | 0.0001 | 0.039793 |
| FAM78A | chr9:131,258,077–131,281,045 | 26.7168 | 8.1076 | − 1.7204 | 5.00E − 05 | 0.0237653 |
| EGR2 | chr10:62,811,995–62,819,167 | 47.4673 | 12.0846 | − 1.97376 | 5.00E − 05 | 0.0237653 |
| ID3 | chr1:23,539,882–23,573,700 | 69.8879 | 11.9376 | − 2.54953 | 5.00E − 05 | 0.0237653 |
| FOS | chr14:75,278,827–75,282,230 | 939.748 | 263.795 | − 1.83286 | 0.0001 | 0.039793 |
| SMAD6 | chr15:66,702,235–66,782,849 | 21.9378 | 3.62308 | − 2.59813 | 5.00E − 05 | 0.0237653 |
| MEX3B | chr15:82,041,777–82,046,018 | 24.3927 | 5.49967 | − 2.14903 | 5.00E − 05 | 0.0237653 |
| CDH3 | chr16:68,645,309–68,733,771 | 7.28527 | 1.34714 | − 2.43508 | 5.00E − 05 | 0.0237653 |
Figure 1.
Volcano plot for DEGs between the T2 group and T0 group.
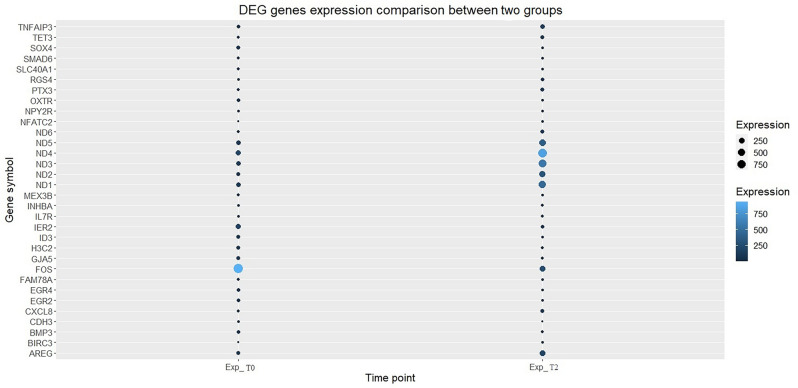
It is worth mentioning that since our samples were pooled and we did not have replicates, the blind dispersion method from the Cuffdiff tool was used to estimate the p-values; as a result, p-value magnitudes were not widely dispersed. Next, a bubble plot was used to demonstrate the expression pattern of differentially expressed gene, as shown in Fig. 2.
Figure 2.
Gene expression pattern between two groups for DEG.
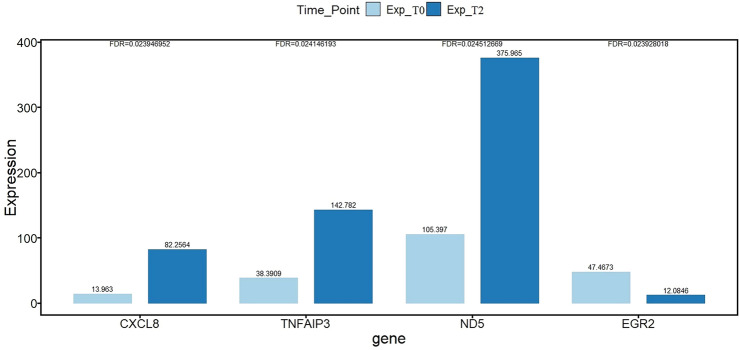
In the next step, enrichment analysis was used to decipher the genes associated with the difference between the two sample groups and shed light on the molecular mechanism of the alteration between the two groups. The WikiPathway enrichment analysis results for upregulated and down-regulated genes are presented in the Tables 4 and 5, respectively. s of less than 0.05 were kept. Only the statistically significant terms with FDR corrected P values of less than 0.05 were kept in this analysis. In Fig. 3, the expression level of the candidate genes in this study is shown. Note that the expression levels have been normalized using FPKM, and the P value corrected using the false discovery method.
Table 4.
Enriched Wiki-Pathway terms for upregulated DEGs in T2 group compared to T0 group.
| Enriched pathway Term (WikiPathway) | q-value | Combined score | Genes |
|---|---|---|---|
| Oxidative phosphorylation WP623 | 3.28E − 09 | 3310.898281 | ND6;ND1;ND3;ND2;ND5;ND4 |
| Electron Transport Chain (OXPHOS system in mitochondria) WP111 | 4.55E − 08 | 1583.835259 | ND6;ND1;ND3;ND2;ND5;ND4 |
| Mitochondrial complex I assembly model OXPHOS system WP4324 | 9.83E − 08 | 2233.049633 | ND6;ND1;ND2;ND5;ND4 |
| Senescence and Autophagy in Cancer WP615 | 0.002335 | 261.8466943 | SERPINB2;CXCL8;INHBA |
| Gastrin signaling pathway WP4659 | 0.002599 | 233.6510707 | SERPINB2;CXCL8;BIRC3 |
| Thymic Stromal LymphoPoietin (TSLP) Signaling Pathway WP2203 | 0.009694 | 297.4766814 | CXCL8;IL7R |
| Hematopoietic Stem Cell Differentiation WP2849 | 0.011176 | 240.7359739 | CSF3;NFATC2 |
| Glucocorticoid Receptor Pathway WP2880 | 0.016611 | 173.5450943 | TNFAIP3;BIRC3 |
| TNF-alpha signaling pathway WP231 | 0.026347 | 119.1462592 | TNFAIP3;BIRC3 |
| Nuclear Receptors Meta-Pathway WP2882 | 0.026697 | 52.16380457 | SERPINB2;TNFAIP3;BIRC3 |
| Cytosine methylation WP3585 | 0.048188 | 549.1072571 | TET3 |
| Regulation of toll-like receptor signaling pathway WP1449 | 0.048188 | 66.53066798 | TNFAIP3 |
Table 5.
Enriched Wiki-Pathway terms for down-regulated DEGs in T2 group compared to T0 group.
| Enriched pathway term (WikiPathway) | q-value | Combined score | Genes |
|---|---|---|---|
| Adipogenesis WP236 | 0.006401258 | 290.6570558 | BMP3;EGR2;ID3 |
| TGF-beta Receptor Signaling WP560 | 0.020494571 | 355.7433228 | FOS;SMAD6 |
| TGF-beta receptor signaling in skeletal dysplasias WP4816 | 0.020494571 | 323.519069 | FOS;SMAD6 |
| Peptide GPCRs WP24 | 0.026577362 | 233.6130223 | OXTR;NPY2R |
| ESC Pluripotency Pathways WP3931 | 0.048946823 | 126.7131058 | FOS;SMAD6 |
| Regulation of toll-like receptor signaling pathway WP1449 | 0.048946823 | 98.52423894 | FOS;SMAD6 |
| Brain-derived neurotrophic factor (BDNF) signaling pathway WP2380 | 0.048946823 | 93.75610919 | EGR2;FOS |
| Myometrial relaxation and contraction pathways WP289 | 0.048946823 | 83.74321389 | OXTR;FOS |
| Iron metabolism in placenta WP2007 | 0.048946823 | 520.8353408 | SLC40A1 |
| Bone morphogenic protein (BMP) signaling and regulation WP1425 | 0.048946823 | 520.8353408 | SMAD6 |
| Neuroinflammation WP4919 | 0.048946823 | 469.1243344 | FOS |
| Transcriptional cascade regulating adipogenesis WP4211 | 0.048946823 | 469.1243344 | EGR2 |
| ncRNAs involved in STAT3 signaling in hepatocellular carcinoma WP4337 | 0.048946823 | 469.1243344 | SOX4 |
| H19 action Rb-E2F1 signaling and CDK-Beta-catenin activity WP3969 | 0.048946823 | 425.939424 | SOX4 |
| ID signaling pathway WP53 | 0.048946823 | 358.0671609 | ID3 |
| MAPK pathway in congenital thyroid cancer WP4928 | 0.048946823 | 358.0671609 | FOS |
Figure 3.
Comparison of normalized gene expression level for the candidate genes for qRT-PCR.
qRT‑PCR
RNA-Seq results were confirmed by using CCs from both groups. The results of qRT-PCR were shown the expression of CXCL8, TNFAIP3, and ND5 genes in the T2 group were significantly higher than in the T0 group (*P < 0.05 / **P < 0.01/ ***P < 0.001/ ****P < 0.0001). EGR2 gene expression in the T2 group was significantly lower than the T0 group (****P < 0.0001) (Fig. 4).
Figure 4.
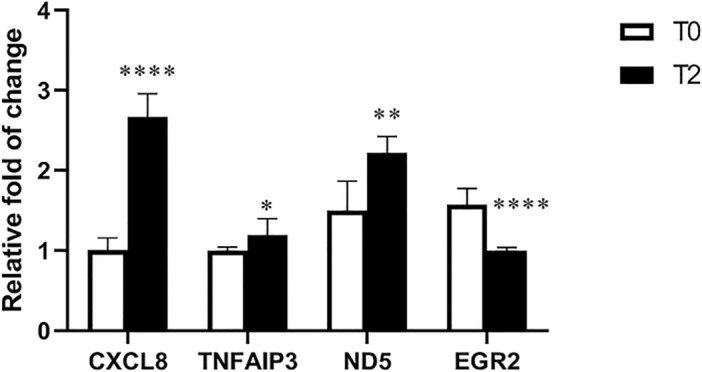
Relative expression of CXCL8, TNFAIP3, ND5, and EGR2 genes in CCs of the T2 group compared with the T0 group. CXCL8: C-X-C motif chemokine ligand 8. TNFAIP3: TNF alpha-induced protein 3. ND5: mitochondrially encoded NADH dehydrogenase 5. EGR2: early growth response 2. (*P < 0.05/ **P < 0.01/ ***P < 0.001/ ****P < 0.0001).
Discussion
In this study, the transcriptomic profile of CCs corresponding to T2 pre-incubated COCs with non-incubated COCs were compared. The results showed that a series of genes and pathways have changed in the CCs of the T2 group compared to the T0 group. Among the most important of them were the paths related to ATP production (oxidative phosphorylation, electron transport chain, and Mitochondrial complex I assembly model OXPHOS system), TNF-alpha signaling pathway, and glucocorticoid receptor pathway, which increased in the T2 group compared to the T0 group. Also, the TGF-β pathway was decreased in the T2 group compared to the T0 group.
In this study, the oxidative phosphorylation, electron transport chain or oxidative phosphorylation system (OXPHOS) in mitochondria, and the complex I system (Mitochondrial complex I assembly model OXPHOS system) pathways increased in the T2 group compared to the T0 group. All the mentioned pathways are involved in ATP production. All cells use ATP as an energy source for survival, proliferation, and differentiation. ATP and other metabolites can pass directly from CCs to oocytes through gap junctions19—these cumulus-oocyte communications attempt to control COC energy metabolism. Energy production in the COC to sustain meiosis during oocyte maturation and subsequent rapid proliferation of cells during embryonic development before implantation is essential20. When these cellular mechanisms are disrupted, molecular damage to the oocyte can induce mitochondrial mutations and reduce ATP production, which is harmful to the oocyte20. Albert et al. showed a statistically significant decrease in the ATP level of CCs from individuals with endometriosis undergoing IVF compared to the control group. Also, their study showed a negative correlation between the ATP of CCs and age21.
On the other hand, suppression of OXPHOS and reduction of ATP production lead to increased apoptosis and suppression of CC proliferation22. By increasing apoptosis in CCs and decreasing the proliferation of these cells, oocytes may not receive proper nutrition from these cells ,, lead to damage to the oocytes. These data suggest that COCs without pre-incubation may have defects in ATP production and mitochondrial function and increased CCs apoptosis. This may lead to reduced oocyte competence and fertilization rate. It is possible that pre-incubating CCs for 2 h with sufficient ATP production would better support the oocyte. Confirmation of these results requires more extensive studies.
The TNF-alpha signaling pathway was among the pathways that increased in the T2 group compared to the T0 group. The TNF signaling pathway involves several processes, including cell proliferation, apoptosis, differentiation, inflammatory and immune response modulation23. The genes were upregulated in this pathway were the TNFAIP3 and BIRC3 genes, both of which are among theapoptosis-inhibiting genes. BIRC3 encodes a member of the inhibitors of apoptosis protein (IAP) family that inhibit apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF224. Also, the protein encoded by TNFAIP3 is a zinc finger protein and ubiquitin-editing enzyme and has been shown to inhibit NF-kappa B activation as well as TNF-mediated apoptosis25. It has been reported that TNFAIP6 and TNFAIP3 are decreased in the CCs of endometriosis patients compared to healthy individuals, which may be related to low embryo quality in these patients23,26. Increased apoptosis in CCs can lead to abnormalities in cleavage divisions of the embryo27,28. Gametes and embryos obtained from COCs without apoptosis or with a minimal amount of apoptosis increase the probability of reaching the blastocyst stage29. With increased apoptosis in CCs, the oocyte may not receive proper nutrition from these cells. Also, increased apoptosis of CCs has been associated with abnormal oocyte morphology30, abnormal embryo cleavage28, reduction of fertilization31, and blastocyst rate29. In addition, the expression level of pro-apoptotic genes in cumulus cells of low-quality embryos was higher than in high-quality embryos32. Based on this, apoptosis in CCs of the T2 group may be less than that of the T0 group. For this reason, the effectiveness of these cells in supporting oocytes is higher in this group.
In the T2 group and the T0 group of our study, one of the pathways that increased was the glucocorticoid receptor pathway. Glucocorticoids (GCs) involve essential processes such as transcription, energy metabolism, and apoptosis. The GC receptor, NR3C1, can bind to DNA and modulate pathways related to these processes. These compounds regulate oxidative phosphorylation gene expression, which is essential in physiological processes that require energy, such as lipid metabolism, stress response, and reproduction. GCs have a unique role in the early stages of reproduction, and NR3C1 has been found to be highly expressed in primordial and antral follicles. ROS imbalance caused by stressful events can alter GC regulation and negatively affect oocyte competence. GC also has an anti-inflammatory effect on ovarian follicular cells and protects the cells from apoptotic signals. The anti-inflammatory effect of GCs is exerted by two complementary mechanisms: induction of apoptosis of inflammatory cells and arrest of apoptosis in resident cells of inflamed tissue. Although the mechanism of GC action on oocyte maturation remains controversial, it has been evaluated positively in horse and mouse species oocytes. Also, in zebrafish, GC receptor knockout led to impaired ovulation, oocyte maturation and defects in fertility.
Furthermore, the knockdown of NR3C1 inhibits the proliferation of cumulus cells33. Proper expansion of CCs is essential for fertility. An expanded cumulus cell is required for ovulation, and it facilitates oocyte extrusion during ovulation, transfer to the fertilization site, and sperm penetration. Cumulus expansion is also related to oocyte competence34. Based on this, the CCs of COCs T2 pre-incubation may have more CCs expansion and less apoptosis, inflammation, leading to the production of competent oocytes with better support from oocytes.
Studies have shown that the exposure of COC's cumulus cells to hydrogen peroxide before fertilization reduced the cleavage rate of the embryo. Also, exposure of the oocyte to hydrogen peroxide leads to the death of the oocyte and defects in the first cleavage35. Lin et al. showed that ROS increases in granulosa cells of endometriosis patients, which causes cellular senescence and contributes to endometriosis-related infertility36. These studies show that CCs protect the oocyte against oxidative stress, and that increased reactive oxygen species in CCs can lead to defects in this support35,36. Transforming growth factor (TGF)-β is one of the factors that leads to an increase in the production of reactive oxygen species (ROS) and a decrease in the concentration of glutathione (GSH) in different types of cells. NAD(P)H oxidase (NOX) is one of the sources of ROS, and TGF-β leads to an increase in ROS through its effect on NOX. This relationship is two-way, and the increase in ROS also leads to a rise in TGF-β gene expression37. TGF receptor is one of the pathways that decreased in our study in the T2 group compared to the T0 group. The upregulation of this pathway in the T0 group may lead to an increase in ROS in the CCs of this group and decrease their ability to support the oocyte.
Many studies have shown that oocyte pre-incubation is effective on oocyte and embryo quality, but the appropriate pre-incubation time is not precisely known. In this study, the effect of 2 h of pre-incubation was investigated and compared with no incubation. The results showed that 2 h leads to changes in important pathways in CCs, which may positively affect oocyte quality. Therefore, T2 pre-incubation may lead to better support of the CCs to the oocyte and thus to a more complete maturation of the oocyte, and more research in this field seems necessary. One of the limitations of this study was the impossibility of direct transcriptomics analysis of oocytes due to ethical problems. Another limitation of this study was the small sample size. It is suggested that more incubation times be compared in future studies and that the examination of the genes and pathways be done in a more specific way. It is also suggested t that the functional validation of essential genes be included in the project.
Acknowledgements
The authors would like to thank Iran University of Medical Sciences (IUMS), Tehran, Iran, for their cooperation throughout study.
Author contributions
M.M.: project administration, designed the study, and reviewed the manuscript. A.G.: performed the experiments and wrote the paper. S.E.: performed the experiments. N.E.G.H. and Z.Z.: The study was designed, and the data was analyzed. M.A.: reviewed the manuscript. R.M.: data collection.
Funding
This work was supported by the Iran University of Medical Sciences (grant no. 31837).
Data availability
The datasets generated and analyzed during the current study are available in the GEO repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE252038"] with GEO accession GSE252038.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Azam Govahi and Sahar Eghbali.
References
- 1.Meng, Q. et al. Incidence of infertility and risk factors of impaired fecundity among newly married couples in a Chinese population. Reprod. Biomed. Online30, 92–100 (2015). 10.1016/j.rbmo.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 2.Devjak, R. et al. Cumulus cells gene expression profiling in terms of oocyte maturity in controlled ovarian hyperstimulation using GnRH agonist or GnRH antagonist. (2012). [DOI] [PMC free article] [PubMed]
- 3.Falcone, P. et al. Correlation between oocyte preincubation time and pregnancy rate after intracytoplasmic sperm injection. Gynecol. Endocrinol.24, 295–299 (2008). 10.1080/09513590802095613 [DOI] [PubMed] [Google Scholar]
- 4.Mohammadi, F. et al. The effect of preincubation time and myo-inositol supplementation on the quality of mouse mii oocytes. J. Reprod. Infertil.21, 259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho, J.Y.-P., Chen, M.-J., Yi, Y.-C., Guu, H.-F. & Ho, E.S.-C. The effect of preincubation period of oocytes on nuclear maturity, fertilization rate, embryo quality, and pregnancy outcome in IVF and ICSI. J. Assist. Reprod. Genet.20, 358–364 (2003). 10.1023/A:1025476910771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isiklar, A. et al. Impact of oocyte pre-incubation time on fertilization, embryo quality and pregnancy rate after intracytoplasmic sperm injection. Reprod. Biomed. Online8, 682–686 (2004). 10.1016/S1472-6483(10)61649-5 [DOI] [PubMed] [Google Scholar]
- 7.Watanabe, H. Correlation between the number of cultured human embryos and embryo development in the wow culture dish system. Fertil. Steril.108, e158 (2017). [Google Scholar]
- 8.Govahi, A. et al. Cutting-edge techniques provide insights regarding repeated implantation failure patients. Reprod. Biomed. Online46, 687–696 (2023). 10.1016/j.rbmo.2022.12.013 [DOI] [PubMed] [Google Scholar]
- 9.Memili, E. et al. Bovine germinal vesicle oocyte and cumulus cell proteomics. Reproduction133, 1107–1120 (2007). 10.1530/REP-06-0149 [DOI] [PubMed] [Google Scholar]
- 10.Revelli, A. et al. Follicular fluid content and oocyte quality: From single biochemical markers to metabolomics. Reprod. Biol. Endocrinol.7, 1–13 (2009). 10.1186/1477-7827-7-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews, S. et al. FastQC. A quality control tool for high throughput sequence data370 (2010).
- 12.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT StringTie, and Ballgown. Nat. Protoc.11, 1650–1667 (2016). 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, S. & Chan, C.-K.K. Analysis of RNA-Seq data using TopHat and Cufflinks 339–361 (Plant Bioinformatics Methods and Protocols, Springer, New York, 2016). [DOI] [PubMed] [Google Scholar]
- 15.Therneau, T., Hart, S. & Kocher, J. P. Calculating samplesSize estimates for RNA Seq studies. R package version 1.10. 0. (2020).
- 16.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform.14, 1–14 (2013). 10.1186/1471-2105-14-S18-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform.14, 178–192 (2013). 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adolfsson, E. & Andershed, A. N. Morphology vs morphokinetics: A retrospective comparison of inter-observer and intra-observer agreement between embryologists on blastocysts with known implantation outcome. JBRA Assist. Reprod.22, 228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunning, K. R., Russell, D. L. & Robker, R. L. Lipids and oocyte developmental competence: the role of fatty acids and b-oxidation. Reproduction148, R15-27 (2014). 10.1530/REP-13-0251 [DOI] [PubMed] [Google Scholar]
- 20.Imanaka, S., Shigetomi, H. & Kobayashi, H. Reprogramming of glucose metabolism of cumulus cells and oocytes and its therapeutic significance. Reprod. Sci.29, 653–667 (2022). 10.1007/s43032-021-00505-6 [DOI] [PubMed] [Google Scholar]
- 21.Hsu, A. L., Townsend, P. M., Oehninger, S. & Castora, F. J. Endometriosis may be associated with mitochondrial dysfunction in cumulus cells from subjects undergoing in vitro fertilization-intracytoplasmic sperm injection, as reflected by decreased adenosine triphosphate production. Fertil. Steril.103, 347–352 (2015). 10.1016/j.fertnstert.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 22.Zhou, X. et al. MiR-126-3p inhibits apoptosis and promotes proliferation by targeting phosphatidylinositol 3-kinase regulatory subunit 2 in porcine ovarian granulosa cells. Asian-Australas. J. Animal Sci.33, 879 (2020). 10.5713/ajas.19.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Da Luz, C. M. et al. Transcriptomic analysis of cumulus cells shows altered pathways in patients with minimal and mild endometriosis. Sci. Rep.12, 5775 (2022). 10.1038/s41598-022-09386-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diop, F. et al. Biological and clinical implications of BIRC3 mutations in chronic lymphocytic leukemia. Haematologica105, 448 (2020). 10.3324/haematol.2019.219550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verstrepen, L. et al. Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem. Pharmacol.80, 2009–2020 (2010). 10.1016/j.bcp.2010.06.044 [DOI] [PubMed] [Google Scholar]
- 26.Allegra, A. et al. The gene expression profile of cumulus cells reveals altered pathways in patients with endometriosis. J. Assist. Reprod. Genet.31, 1277–1285 (2014). 10.1007/s10815-014-0305-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burrows, A. E. et al. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. (2006). [DOI] [PMC free article] [PubMed]
- 28.Faramarzi, A., Khalili, M. A. & Jahromi, M. G. Is there any correlation between apoptotic genes expression in cumulus cells with embryo morphokinetics?. Mol. Biol. Rep.46, 3663–3670 (2019). 10.1007/s11033-019-04781-z [DOI] [PubMed] [Google Scholar]
- 29.Corn, C. M., Hauser-Kronberger, C., Moser, M., Tews, G. & Ebner, T. Predictive value of cumulus cell apoptosis with regard to blastocyst development of corresponding gametes. Fertil. Steril.84, 627–633 (2005). 10.1016/j.fertnstert.2005.03.061 [DOI] [PubMed] [Google Scholar]
- 30.Luo, Y. et al. CTC1 increases the radioresistance of human melanoma cells by inhibiting telomere shortening and apoptosis. Int. J. Mol. Med.33, 1484–1490 (2014). 10.3892/ijmm.2014.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Høst, E., Gabrielsen, A., Lindenberg, S. & Smidt-Jensen, S. Apoptosis in human cumulus cells in relation to zona pellucida thickness variation, maturation stage, and cleavage of the corresponding oocyte after intracytoplasmic sperm injection. Fertil. Steril.77, 511–515 (2002). 10.1016/S0015-0282(01)03006-0 [DOI] [PubMed] [Google Scholar]
- 32.Govahi, A., Amjadi, F., Nasr-Esfahani, M.-H., Raoufi, E. & Mehdizadeh, M. Accompaniment of time-lapse parameters and cumulus cell RNA-sequencing in embryo evaluation. Reprod. Sci.29, 1–15 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Conca, M., Gardela, J., Mogas, T., López-Béjar, M. & Álvarez-Rodríguez, M. Apoptosis and glucocorticoid-related genes mRNA expression is modulated by coenzyme Q10 supplementation during in vitro maturation and vitrification of bovine oocytes and cumulus cells. Theriogenology192, 62–72 (2022). 10.1016/j.theriogenology.2022.08.030 [DOI] [PubMed] [Google Scholar]
- 34.Richani, D., Dunning, K. R., Thompson, J. G. & Gilchrist, R. B. Metabolic co-dependence of the oocyte and cumulus cells: Essential role in determining oocyte developmental competence. Human Reprod. Update27, 27–47 (2021). 10.1093/humupd/dmaa043 [DOI] [PubMed] [Google Scholar]
- 35.Fatehi, A. N. et al. Presence of cumulus cells during in vitro fertilization protects the bovine oocyte against oxidative stress and improves first cleavage but does not affect further development. Zygote13, 177–185 (2005). 10.1017/S0967199405003126 [DOI] [PubMed] [Google Scholar]
- 36.Lin, X. et al. Excessive oxidative stress in cumulus granulosa cells induced cell senescence contributes to endometriosis-associated infertility. Redox Biol.30, 101431 (2020). 10.1016/j.redox.2020.101431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, R.-M. & Pravia, K. G. Oxidative stress and glutathione in TGF-β-mediated fibrogenesis. Free Radical Biol. Med.48, 1–15 (2010). 10.1016/j.freeradbiomed.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the GEO repository, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE252038"] with GEO accession GSE252038.