Abstract
Infectious molecular clones have been isolated from two maedi-visna virus (MVV) strains, one of which (KV1772kv72/67) is an antigenic escape mutant of the other (LV1-1KS1). To map the type-specific neutralization epitope, we constructed viruses containing chimeric envelope genes by using KV1772kv72/67 as a backbone and replacing various parts of the envelope gene with equivalent sequences from LV1-1KS1. The neutralization phenotype was found to map to a region in the envelope gene containing two deletions and four amino acid changes within 39 amino acids (positions 559 to 597 of Env). Serum obtained from a lamb infected with a chimeric virus, VR1, containing only the 39 amino acids from LV1-1KS1 in the KV1772kv72/67 backbone neutralized LV1-1KS1 but not KV1772kv72/67. The region in the envelope gene that we had thus shown to be involved in escape from neutralization was cloned into pGEX-3X expression vectors, and the resulting fusion peptides from both molecular clones were tested in immunoblots for reactivity with the KV1772kv72/67 and VR1 type-specific antisera. The type-specific KV1772kv72/67 antiserum reacted only with the fusion peptide from KV1772kv72/67 and not with that from LV1-1KS1, and the type-specific VR1 antiserum reacted only with the fusion peptide from LV1-1KS1 and not with that from KV1772kv72/67. Pepscan analysis showed that the region contained two linear epitopes, one of which was specific to each of the molecularly cloned viruses. This linear epitope was not bound by all type-specific neutralizing antisera, however, which indicates that it is not by itself the neutralization epitope but may be a part of it. These findings show that mutations within amino acids 559 to 597 in the envelope gene of MVV virus result in escape from neutralization. Furthermore, the region contains one or more parts of a discontinuous neutralization epitope.
Maedi-visna virus (MVV) is a member of the lentivirus subgroup of retroviruses (40, 41). This virus causes slowly progressive diseases that affect mainly the central nervous system and the lungs of sheep (reviewed in reference 14). The primary target cells of MVV infection are cells of the monocyte/macrophage lineage, and virus gene expression is activated upon macrophage maturation (12, 28). In MVV infection, type-specific neutralizing antibodies appear after 1 to 6 months, and in most sheep other more broadly neutralizing antibodies appear up to 3 years later. A similar pattern has been found in human immunodeficiency virus (HIV) infection. Domains in the envelope glycoprotein that induce virus-neutralizing antibodies in HIV-1 infection have been well characterized (16, 20, 25, 45, 48). The type-specific antibodies which arise shortly after infection are directed mainly against the V3 loop (29, 33).
The ability of viruses to escape neutralization has been described for various lentivirus systems. There have been several reports on the emergence of HIV-1 neutralization escape mutants, both in vitro and in vivo (1, 10, 23). Most of these escape mutants have mutations that map in the V3 loop (10, 26, 51). The major neutralizing epitopes in simian immunodeficiency virus (SIV) are thought to be conformational or discontinuous, and it has been shown that the V3-corresponding region of SIVmac does not serve as a target of neutralizing antibodies (18). Neutralization-resistant variants of SIV have been mapped in the V4 region of the SIV envelope glycoprotein (7, 19). In feline immunodeficiency virus (FIV), a single amino acid substitution in V5 of the envelope protein allows escape from virus neutralization (38).
Many studies have demonstrated the occurrence of neutralization-resistant mutants during long-term MVV infection of sheep (17, 21, 27, 46). The characterization of these mutants was, however, hampered by the lack of molecular clones. In this study we have used two molecular clones of MVV which elicit a different neutralization response. The two molecular clones, KV1772kv72/67 and LV1-1KS1, differ by 22 amino acids, and there are two deletions in the envelope gene in strain KV1772kv72/67 (3, 44).
To identify the type-specific neutralization epitope, we constructed recombinant viruses in which env sequences of strain KV1772kv72/67 were replaced by those of strain LV1-1KS1. The neutralization phenotype was transferred with a fragment from the envelope gene containing two deletions and four amino acid changes within 39 amino acids (aa 559 to 597).
MATERIALS AND METHODS
Virus, cells, and sera.
The molecularly cloned virus KV1772kv72/67 is derived from visna virus KV1772, which was selected for neurovirulence by serial passage of strain K1514 in sheep (13, 22). The molecularly cloned virus LV1-1KS1 was reported to be derived from K1514 (44), but sequencing data from a number of virus isolates indicates that it is most closely related to K796, a virus appearing early in the lineage of visna virus strains (see Fig. 2).
FIG. 2.
Lineage of MVV strains following passage through sheep and tissue culture (T.C.). The origin of the molecular clones KV1772kv72/67 and LV1-1KS1 is given in this diagram.
Sheep choroid plexus (SCP) cells established as described previously (31, 42) were grown at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco’s minimal essential medium (Gibco) supplemented with 2 mM glutamine, 100 IU of penicillin per ml, 100 IU of streptomycin per ml and either 10% normal lamb serum (growth medium) or 1% lamb serum (maintenance medium). Macrophage cultures were established as follows. Heparinized blood (100 ml) was collected from normal sheep, and peripheral blood mononuclear cells obtained by sedimentation on Histopaque-1077 (Sigma) were washed repeatedly in phosphate-buffered saline and resuspended at 12 × 106 cells/ml in growth medium supplemented with 5 × 10−5 M mercaptoethanol. They were then seeded into plastic four-chamber (1-ml) tissue culture slides (180 mm2; Permanox, Nunc Inc.) or into 24-well plates. After incubation at 37°C in a humidified atmosphere of 5% CO2 for 24 h, supernatant and unattached cells were removed, the slide was washed twice with phosphate-buffered saline, and 1 ml of new growth medium was added to each chamber. Adherent cells were further incubated for at least 7 days before they were infected.
Transfections were performed by using subconfluent monolayers of ovine fetal synovial (FOS) cells. DNA was transfected with Lipofectamine as specified by the manufacturer (Life Sciences, Inc.). Transfected FOS cells were passaged (1:3 split) and incubated in maintenance medium until syncytia appeared (5 to 8 days). Supernatants from transfected cells were also tested for the presence of reverse transcriptase (RT) activity before passage into SCP cells.
The sera used for immunoblots and pepscan were the following: serum 21372 from sheep 1923 after 26 weeks of infection with KV1772kv72/67, serum 21160 from sheep 1923 preinfection, serum 20494 from sheep 1896 after 8 weeks of infection with KV1772kv72/67 (this serum neutralized KV1772kv72/67 at a titer of 2,048 to 4,096 but did not neutralize LV1-1KS1), serum 20464 from sheep 1896 preinfection, serum 22156 from sheep 1998 after 16 weeks of infection with VR1, and serum 22077 from sheep 1998 preinfection. The KV1772kv72/67 type-specific antiserum used for neutralization was serum 20494 from sheep 1896.
RT assay.
Viral particles from 0.5 ml of cell-free supernatants from infected cells were pelleted at 70,000 rpm for 10 min in a Beckman TLA 100.4 rotor. RT activity was determined as described previously (49).
Virus neutralization test.
Virus (100 50% tissue culture infective doses [TCID50]) was mixed with serial twofold dilutions of serum in DMEM with 2% lamb serum. The samples were incubated at room temperature for 24 h and then inoculated in quadruplicate onto monolayers of SCP cells in 96-well tissue culture plates (Nunc) and kept at 37°C in a humidified atmosphere of 5% CO2. Cytopathic effect was monitored after 7, 14, 21, and 28 days. The neutralization titer was calculated as the reciprocal of the serum dilution which caused complete neutralization in 50% of inoculated cultures. In macrophages, neutralization was assayed by infecting 24-well plates containing blood-derived macrophages with 100 TCID50 of virus mixed with twofold dilutions of serum, four wells for each dilution. Growth was monitored by measuring RT activity in parallel cultures without antiserum; at two time points at maximum RT activity, all the samples were assayed for RT.
Generation of a clone containing chimeric env gene.
The construction of chimeric molecular clone VR1 is schematically presented in Fig. 1. The 5′ sector of KV1772kv72/67 in Bluescript II (Stratagene), called p8XSp5, was used in the first subcloning step. This clone contains an XbaI fragment of the KV1772kv72/67 visna virus clone, including 1.5 kb of cellular flanking sequences and the 5′ long terminal repeat through nucleotide (nt) 7768 (4) (Fig. 1A). This plasmid contains a unique StuI site in the cellular flanking sequence about 700 nt upstream from the 5′ long terminal repeat. The StuI- and XbaI-generated fragment was subcloned into HincII- and XbaI-digested BluescriptII to eliminate the HincII site. This clone was named p8XSp5-RK1 (Fig. 1B), and from it a HincII (nt 6393)-XbaI (nt 7768) fragment was generated and subcloned into HincII- and XbaI-digested pUC19 (pRK2). PCR was performed to amplify the segment from the LV1-1KS1 clone containing a StyI (nt 7614) site and an XbaI (nt 7768) site. The amplified fragment was digested with StyI and XbaI, and the 154-bp fragment was isolated on 4% MetaPhor agarose (FMC BioProducts) and subcloned into StyI- and XbaI-digested pUC19 clone pRK2 to generate subclone pRK3. From this clone, the HincII-XbaI fragment was excised and subcloned into HincII- and XbaI-digested p8XSp5-RK1, to construct a chimeric kv72 clone with an LV1-1KS1 StyI-XbaI fragment (pRK4) (Fig. 1C). This clone was mixed in equimolar quantities with the 3′ clone of KV1772kv72/67 in BluescriptII, called p67r (4), digested with XbaI, and ligated. The full-length viral DNA with a chimeric env gene, named VR1, was transfected into FOS cells as described above. When RT activity could be measured (5 to 8 days), culture supernatant was transferred to SCP cells, virus progeny were collected, and proviral DNA was PCR amplified with env-specific primers, which were biotinylated to allow direct solid-phase sequencing (Dynal) to confirm the origin of the virus. Intermediate clone constructs were also confirmed by sequence analysis. The clones VB1, VB5, and VR2 were constructed in a similar way by using the restriction sites indicated in Fig. 4.
FIG. 1.
Schematic presentation of the construction of chimeric MVV clone VR1 (see the text for details). The sheep flanking sequence is indicated by a bold zigzag line, and the vector is indicated by a thin zigzag line. Restriction enzyme cleavage sites are indicated as follows: X, XbaI; St, StuI; H, HincII; S, StyI; B, BamHI.
FIG. 4.
Diagrammatic representation of recombinant maedi-visna viruses derived from the molecular clones KV1772kv72/67 and LV1-1KS1. The restriction sites used to construct the viruses are shown. The neutralization titer of KV1772kv72/67 type-specific antiserum for each virus is shown on the right.
Construction of pGEX-3X–GST fusion peptide expression vectors.
The env gene fragment between nt 7499 and 7793 from clones LV1-1KS1 and KV1772kv72/67 was amplified by 30 cycles of PCR with the following thermal profile: denaturing at 94°C for 30 s, annealing at 55°C for 15 s, and extension at 72°C for 90 s. The forward primer was 5′GCGGGATCCAAAATAGCACCATAACAGGAA3′, and the reverse primer was 5′CCGGAATTCTGGTATCGYTGCACYAACAT3′. The primers provide BamHI and EcoRI cloning sites (underlined) for cloning the KV1772kv72/67 and LV1-1KS1 PCR products in frame with glutathione S-transferase (GST) of the pGEX-3X expression vector. The presence of correct inserts was confirmed by sequence analysis. Expression of GST fusion peptides followed standard procedures (4).
Immunoblotting.
Immunoblotting was performed with the Mini-proteanII system (Bio-Rad) as specified by the manufacturer. Dilutions of fusion peptides were boiled, diluted 1:1 in Laemmli sample buffer (125 mM Tris buffer supplemented with 5% mercaptoethanol and 2% sodium dodecyl sulfate [SDS] [pH 6.8]) and separated by SDS-polyacrylamide gel electrophoresis. Half of the gels were used for silver staining (Bio-Rad) to determine the sizes and amounts of peptides, and half were transferred to a nitrocellulose membrane in 25 mM Tris glycine buffer (pH 8.8) containing 20% methanol. Transfer of the fusion peptides was carried out for 60 min at 200 V and 1.2 mA/cm2 in a Milliblot graphite electroblotter I (Millipore). After the transfer, the nitrocellulose membranes were blocked for 1 h at room temperature with 0.1 M Tris-HCl-buffered saline (pH 7.8) containing 0.5% Tween 20. The sera and conjugate were diluted in 0.1 M Tris-HCl-buffered saline (pH 7.8) containing 0.1% Tween 20 (TBS-T). After the blocking step, the nitrocellulose membranes were washed once with TBS-T and incubated overnight at 4°C on a roller with serum samples at a dilution 1/500. Rabbit anti-goat immunoglobulin G conjugated to horseradish peroxidase (Sigma A4174) at a dilution of 1/10,000 was added for 1 h at room temperature. The blots were washed extensively in TBS-T between each step. The enhanced chemiluminescence system from Amersham was used for developing the blots.
Pepscan analysis.
Peptides were synthesized on polyethylene rods and tested for their reactivity with antisera in an enzyme-linked immunosorbent assay by established procedures (15). Twelve-mer peptides were synthesized and tested for reactivity to the sera at a 1:250 dilution.
RESULTS
Generation of chimeric virus.
The two molecular clones used in this study both have their origin in a transmission experiment where virus was passaged serially through tissue culture and sheep (Fig. 2). The LV1-1KS1 clone was derived from strain K796, but the KV1772kv72/67 molecular clone is a derivative of strain K1772, a descendant of strain K1010, which is a neutralization escape mutant of strain K796 (17). The molecularly cloned viruses retained the neutralization phenotypes of their parental strains. The envelope proteins derived from the two molecular clones differ by 22 amino acids and two deletions of 5 and 1 amino acids (Fig. 3). For mapping the type-specific neutralization epitope, four recombinants were constructed by using restriction fragments from the env gene of LV1-1KS1 with KV1772kv72/67 as a backbone. All recombinant viruses were tested for neutralization by KV1772kv72/67 type-specific antiserum (Fig. 4). The KV1772kv72/67 type-specific antiserum neutralized the KV1772kv72/67 virus very efficiently at a dilution of 1:2,048, whereas it did not neutralize LV1-1KS1 at a dilution of 1:8. The XbaI-SacI region of the envelope gene comprising the carboxyl end of SU and the TM protein did not affect neutralization (recombinant VB5), and although sequences in front of the StyI site at nt 7613 influenced the neutralization (recombinant VR2), mutations in the region between StyI at nt 7613 and XbaI at nt 7768 were necessary and sufficient to abolish neutralization (recombinants VB1 and VR1).
FIG. 3.
Comparison of the amino acid sequences of the envelope genes of the two molecularly cloned MVV strains, KV1772kv72/67 and LV1-1KS1. Dots represent identical amino acids, and dashes represent deletions. The proteolytic cleavage site between SU and TM is indicated by an arrow.
Infection of lambs with the chimeric virus.
Two lambs (animals 1997 and 1998) were infected intracerebrally with the recombinant virus VR1. Both lambs became infected, as evidenced by frequent virus isolations and strong antibody responses to the virus. Both lambs seroconverted within 6 weeks, and one of the lambs (animal 1998) acquired a high titer of neutralizing antibodies to the inoculated virus. Serum was collected biweekly for the first 8 weeks, and every 4 weeks thereafter until sacrifice at 28 weeks postinfection. The two parental virus strains and the recombinant virus were tested for neutralization by antiserum obtained from lamb 1998. Neutralizing antibodies against the infecting virus, VR1, appeared after 6 weeks, and its level stayed high till the end of the experiment. LV1-1KS1 was neutralized almost to the same extent as VR1, whereas KV1772kv72/67 was not neutralized (Fig. 5). We have thus shown that the specific neutralization phenotype resides in one or more of the four substitutions and two deletions that were transferred.
FIG. 5.
Neutralization titers of serial antisera from sheep 1998 (infected with the recombinant virus VR1). The neutralization titer was determined as described in Materials and Methods.
Growth of chimeric virus in macrophages.
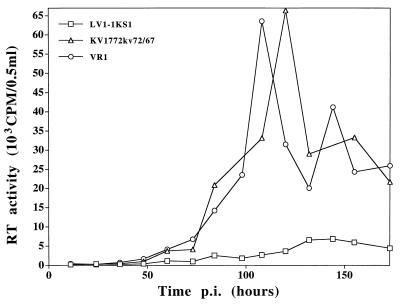
The progeny of the molecular clone LV1-1KS1 grow less efficiently in macrophages than do the progeny of the molecular clone KV1772kv72/67 (47). We tested the growth of the chimeric virus in macrophages to test whether there was a linkage between macrophage tropism and neutralization specificity. Figure 6 shows the growth curves of the two parent strains and the chimeric virus in macrophages. The impaired replication of strain LV1-1KS1 in macrophages was not transferred with the 39-amino-acid fragment.
FIG. 6.
Growth curves of the two parental strains KV1772kv72/67 and LV1-1KS1 and the chimeric virus VR1 in sheep monocyte-derived macrophages as measured by RT activity. p.i., postinfection.
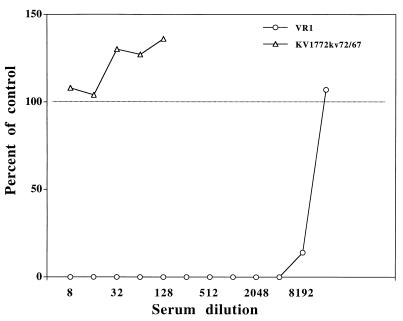
Neutralization of macrophage-grown virus with type-specific antiserum.
The neutralization specificity of the antisera was routinely determined by using SCP cells as a host system. However, macrophages are considered to be the natural target cells for MVV. We therefore tested the specificity of the antisera on viruses grown in macrophages. The virus strains KV1772kv72/67 and VR1 were passed twice in macrophages, and neutralization assays were carried out with serial twofold dilutions of antiserum obtained from sheep infected with VR1. The VR1 antiserum neutralized the infecting strain up to a dilution of 4,000 to 8,000 in macrophages as well as in SCP cells. Strain KV1772kv72/67 was not neutralized (Fig. 7).
FIG. 7.
Neutralization of macrophage-grown virus with twofold serial dilutions of VR1 type-specific antiserum.
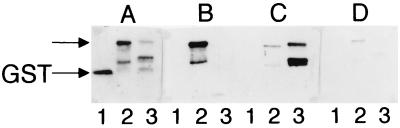
Reactivity of MVV-specific antisera to GST fusion peptides derived from the two molecular clones, KV1772kv72/67 and LV1-1KS1.
A 293-bp stretch from both parental strains, covering the region where the neutralization epitope mapped, was cloned in a pGEX3X expression vector (the region is indicated in Fig. 8). The production of fusion peptides was examined by silver staining of SDS-polyacrylamide gels and by using rabbit antiserum to GST on Western blots. Low levels of fusion peptides were obtained, and these proved to be quite unstable. Expression of a 38.5-kDa peptide, consistent with the predicted size, was observed, as was a series of partial proteolytic fragments. The KV1772kv72/67 and LV1-1KS1 fusion peptides can be distinguished by their size, since the KV1772kv72/67 fusion peptide is 6 amino acids (approximately 5%) smaller (Fig. 9A). Antiserum from a sheep infected with KV1772kv72/67 reacted with the fusion peptide from KV1772kv72/67 in immunoblots, whereas it did not bind to the LV1-1KS1 fusion peptide. With a VR1 type-specific antiserum, which neutralized LV1-1KS1 and VR1 but not KV1772kv72/67, the reverse was true. This antiserum bound to the LV1-1KS1 fusion peptide but only weakly to KV1772kv72/67 (Fig. 9B and C). No reaction was seen with preinfection sera (data not shown). The region in the Env glycoproteins defined by these peptides thus contains B-cell epitopes that are differentially recognized by type-specific antisera.
FIG. 8.
Amino acid sequences from the two strains KV1772kv72/67 and LV1-1KS1 of the region cloned into GEX vectors. Dots indicate identical amino acids; dashes indicate deletions. The region that was exchanged between the two clones is indicated by boldface letters.
FIG. 9.
Western blots of the GST fusion peptides with different sera. Lanes: 1, GST; 2, KV1772kv72/67-GST fusion peptide; 3, LV1-1KS1-GST fusion peptide. (A) Rabbit antiserum to GST; (B) KV1772kv72/67 type-specific serum (sheep 1998, 23 weeks postinfection); (C) VR1 type-specific serum; (D) KV1772kv72/67 serum with a high neutralization titer. The size of the fusion peptides (approximately 40 kDa) is indicated by the arrow.
The fusion peptides were also reacted with antiserum from a sheep that was infected with strain KV1772kv72/67 and had developed a strong neutralization antibody response by 8 weeks after infection. This antiserum showed only a very faint reaction to the KV1772kv72/67 fusion peptide (Fig. 9D). It therefore appears that the species-specific linear epitope(s) in this region is not identical to the neutralization epitope.
A pepscan analysis clarified these results further. Twelve-mer peptides with an overlapping sequence of 11 amino acids were synthesized. The amino acid sequences of the peptides corresponded to the region covered by the two fusion peptides, starting with peptides TTMWNIYQNCSK and TTMWNIYQNCSR for LV1-1KS1 and KV1772kv72/67, respectively (starting at aa 529 [Fig. 8]). The 12-mer peptides were analyzed for reactivity with the sera used for the Western blots. Serum from the sheep infected with the chimeric virus VR1 revealed two linear epitopes on the LV1-1KS1 peptide, with the core sequences TVNDLK and KGSRR (Fig. 10). The corresponding sequences of the KV1772kv72/67 peptide were TVNNLK and KSQR, respectively. The latter of these epitopes was not recognized on the KV1772kv72/67 peptide; it differs from that of the other strain by one amino acid substitution and a deletion of one amino acid. Type-specific antiserum against KV1772kv72/67 recognized the former epitope on the KV1772kv72/67 peptide but not on the LV1-1KS1 peptide. The sheep had not developed antibodies against the latter epitope, however. When the 8-week antiserum with the high neutralization titer against KV1772kv72/67 was tested, there was no reaction to either of the epitopes (Fig. 10). These results indicate that although there are at least two linear epitopes in the region, none of them is by itself the neutralization epitope but may be a part of it.
FIG. 10.
Pepscan analysis of three sheep sera by using 12-mer peptides spanning the region of the neutralizing epitope in the two parental strains (amino acid positions are shown). (A and B) VR1 serum 16 weeks postinfection; (C and D) KV1772kv72/67 serum (sheep 1998) 23 weeks postinfection; (E and F) KV1772kv72 serum (sheep 1896) with a high neutralization titer 8 weeks postinfection.
DISCUSSION
We have demonstrated that mutations in the region between amino acids 559 and 597 in the outer envelope glycoprotein gene of MVV result in escape from neutralization and creation of a new type of neutralization specificity. This is a variable region in the MVV SU glycoprotein (2, 35) in an analogous position to V4-V5 in SIV and in FIV (30). Western blots with GST fusion peptides of this region from two virus strains with different neutralization specificities and pepscan analysis revealed two linear epitopes in this region, which were differentially recognized by type-specific antisera. The absence of antibodies to these epitopes in a sheep that mounted a very strong type-specific neutralizing response indicates, however, that neither of the linear epitopes is by itself the neutralization epitope. It is more likely that one or both of the linear epitopes are part of a discontinuous neutralization epitope. This conclusion is supported by the fact that when this region was sequenced in 10 viral isolates that were antigenic variants, 7 of them had mutations in either of the linear epitopes, in addition to a few that were mutated in a potential glycosylation site (2a). We propose that a disulfide bond between two conserved cysteines is formed. This would bring the two linear epitopes into proximity (Fig. 11). We have found two antigenic variants that have one of the cysteines mutated; one of these strains shows impaired growth in macrophages, and the growth rate of the other has not been tested (2a). This is further evidence for the importance of the cysteine in the spatial structure of the protein. These cysteines are widely conserved in maedi-visna virus strains (3, 6, 32, 35, 43, 44) and also in caprine arthritis-encephalitis virus CAEV (34).
FIG. 11.
The region between amino acids 553 and 586 of LV1-1KS1 (numbering as in reference 34). The proposed disulfide cross bridge between the two conserved cysteines is shown. The two linear epitopes are indicated by boxes.
This cysteine loop, which is in the fourth variable region in the envelope gene of MVV, V4 (34, 35), may have an analogous function to V3 in HIV-1. The V3 loop of HIV-1 gp120 is the major target for neutralizing antibodies (33), and it is also a recognition site for coreceptors and a determinant of viral cell tropism (9, 11, 24). In FIV, the neutralization domain also seems to determine cell tropism (50). Virus derived from one of the molecular clones used in this study, LV1-1KS1, grow poorly in macrophages but very efficiently in SCP cells (47). For this particular clone of MVV, however, the change in cell tropism is not related to neutralization, since the poor growth in macrophages was not transferred with the fragment that contained the part of the discontinuous neutralization epitope we have mapped in this study, nor was it transferred with any part of the MVV gp135 (16a).
Several studies have shown that the neutralization sensitivity of lentivirus antisera often is dependent on the cell type used to propagate the virus (5, 8, 36, 52). The mechanism for this is unclear; in some cases several passages in the cell type are needed for adaptation of the virus, and mutations are probably involved, whereas in other cases, cell-type-specific neutralization characteristics are established within a few passages and are reversible (36). We routinely use SCP cells to propagate the virus and for neutralization tests. These cells are fibroblast-like cells from the choroid plexus of sheep and are very efficient for propagating the virus. Macrophages are, however, the target cells for MVV in vivo. Our finding that the neutralization specificity was retained when we used macrophages as host cells indicates that SCP cells are valid for assessing the neutralization specificity.
Our results can best be explained by a discontinuous neutralization epitope being located, at least in part, in the V4 region of the MVV env gene. Other regions of the env gene may also contribute to the neutralization phenotype. Although the changes in the StyI-XbaI fragment (nt 7613 to 7768) were necessary and sufficient to abolish neutralization and create a new neutralization phenotype, amino acid changes in front of this region also affected neutralization (Fig. 4). Furthermore, when antiserum to the recombinant virus containing this StyI-XbaI fragment from LV1-1KS1 in the KV1772kv72/67 backbone was tested, the titer against the input recombinant virus was consistently 1 to 3 dilutions higher than that for LV1-1KS1 (Fig. 5), indicating that the neutralization epitope on the recombinant virus was not completely identical to the epitope on LV1-1KS1. This is further evidence for the discontinuous nature of the epitope. Discontinuous neutralization epitopes appear to be common in other lentiviruses. Although neutralizing activity can be induced by synthetic linear peptides from the V3 loop in HIV-1 (29), the initial type-specific neutralizing antibodies in HIV-1 infection are directed mainly against a discontinuous epitope in the V3 loop (25, 37). Also, in SIV and FIV, antigenic variants are found to map in discontinuous epitopes in the V4-V5 region (7, 18, 19, 39). The cellular receptor(s) for MVV has not been determined, but by analogy to HIV-1, it is reasonable to suggest that the V4 region defines a binding site for a cellular receptor or a coreceptor for MVV.
It is clear from the results presented in this study that the V4 region of MVV is an important determinant for infection. Further analysis of this region should lead to an elucidation of its role in virus entry.
ACKNOWLEDGMENTS
This study was supported by The Icelandic Research Council, The University of Iceland Research Fund, and The Icelandic Research Fund for Graduate Students.
We thank Karl Skírnisson for helping with photography. We are also indebted to Svava Högnadóttir, Steinunn Árnadóttir, Sigurdur Torfi Gudmundsson and Gudmundur Einarsson for expert technical help.
REFERENCES
- 1.Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nyström G, Fenyo E M. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Andrésdóttir V, Tang X, Agnarsdóttir G, Andrésson Ó, Georgsson G, Skraban R, Torsteinsdóttir S, Rafnar B, Benediktsdóttir E, Matthíasdóttir S, Árnadóttir S, Högnadóttir S, Pálsson P A, Pétursson G. Biological and genetic differences between lung- and brain-derived isolates of maedi-visna virus. Virus Genes. 1998;16:281–293. doi: 10.1023/a:1008030706308. [DOI] [PubMed] [Google Scholar]
- 2a.Andrésdóttir, V., et al. Unpublished results.
- 3.Andrésson Ó, Elser J E, Tobin G J, Greenwood J D, Gonda M A, Georgsson G, Andrésdóttir V, Benediktsdóttir E, Carlsdóttir H M, Mäntylä E O, Rafnar B, Pálsson P A, Casey J W, Pétursson G. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology. 1993;193:89–105. doi: 10.1006/viro.1993.1106. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, More D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988. [Google Scholar]
- 5.Baldinotti F, Matteucci D, Mazzetti P, Gianelli C, Bandecchi P, Tozzini F, Bendinelli M. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J Virol. 1994;68:4572–4579. doi: 10.1128/jvi.68.7.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun M J, Clements J E, Gonda M A. The visna virus genome: evidence for a hypervariable site in the env gene and sequence homology among lentivirus envelope proteins. J Virol. 1987;61:4046–4054. doi: 10.1128/jvi.61.12.4046-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W S, Collignon C, Thiriart C, Burns D P W, Stott E J, Kent K A, Desrosiers R C. Effects of natural sequence variation on recognition by monoclonal antibodies that neutralize simian immunodeficiency virus infectivity. J Virol. 1994;68:5395–5402. doi: 10.1128/jvi.68.9.5395-5402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook R F, Berger S L, Rushlow K E, McManus J M, Cook S J, Harrold S, Raabe M L, Montelaro R C, Issel C J. Enhanced sensitivity to neutralizing antibodies in a variant of equine infectious anemia virus is linked to amino acid substitutions in the surface unit envelope glycoprotein. J Virol. 1995;69:1493–1499. doi: 10.1128/jvi.69.3.1493-1499.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.di Marzo Verones F, Reitz M S, Gupta G, Robert-Guroff M, Boyer-Thompson C, Louie A, Gallo R C, Lusso P. Loss of a neutralizing epitope by a spontaneous point mutation in the V3 loop of HIV-1 isolated from an infected laboratory worker. J Biol Chem. 1993;268:25894–25901. [PubMed] [Google Scholar]
- 11.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 12.Gendelman H E, Narayan O, Kennedy-Stoskopf S, Kennedy P G E, Ghotbi Z, Clements J E, Stanley J, Pezeshkpour G. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J Virol. 1986;58:67–74. doi: 10.1128/jvi.58.1.67-74.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgsson G, Houwers D J, Pálsson P A, Pétursson G. Expression of viral antigens in the central nervous system of visna-infected sheep: an immunohistochemical study on experimental visna induced by virus strains of increased neurovirulence. Acta Neuropathol. 1989;77:299–306. doi: 10.1007/BF00687582. [DOI] [PubMed] [Google Scholar]
- 14.Georgsson G. Maedi-visna: pathology and pathogenesis. In: Pétursson G, Hoff-Jörgensen R, editors. Maedi-visna and related diseases. Boston, Mass: Kluwer Academic Publishers; 1990. pp. 19–54. [Google Scholar]
- 15.Geysen H M, Meloen R H, Barteling S J. Use of synthetic peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci USA. 1984;81:3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goudsmit J, Boucher C A B, Meloen R H, Epstein L G, Smit L, van der Hoek L, Bakker M. Human antibody response to a strain-specific HIV-1 gp120 epitope associated with cell fusion inhibition. AIDS. 1988;2:157–164. [PubMed] [Google Scholar]
- 16a.Gudmundsson, B., et al. Unpublished results.
- 17.Gudnadóttir M. Visna-maedi in sheep. Prog Med Virol. 1974;18:336–349. [PubMed] [Google Scholar]
- 18.Javaherian K, Langlois A J, Schmidt S, Kaufmann M, Cates N, Langedijk J P M, Meloen R H, Desrosiers R C, Burns D P W, Bolognesi D P, LaRosa G J, Putney S D. The principal neutralization determinant of simian immunodeficiency virus differs from that of human immunodeficiency virus. Proc Natl Acad Sci USA. 1992;89:1418–1422. doi: 10.1073/pnas.89.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinsey N E, Anderson M G, Unangst T J, Joag S V, Narayan O, Zink M C, Clements J E. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology. 1996;221:14–21. doi: 10.1006/viro.1996.0348. [DOI] [PubMed] [Google Scholar]
- 20.Lasky L A, Nakamura G, Smith D J. Delineation of a region of a human immunodeficiency virus type 1 gp 120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 21.Lutley R, Pétursson G, Pálsson P A, Georgsson G, Klein J, Nathanson N. Antigenic drift in visna: virus variation during long-term infection of Icelandic sheep. J Gen Virol. 1983;64:1433–1440. doi: 10.1099/0022-1317-64-7-1433. [DOI] [PubMed] [Google Scholar]
- 22.Lutley R E, Pétursson G, Georgsson G, Pálsson P A, Nathanson N. Strains of visna virus with increased neurovirulence. In: Sharp J M, Hoff-Jørgensen R, editors. Slow viruses in sheep, goats and cattle. Luxembourg: Directorate-General for Agriculture and Coordination of Agricultural Research, Commission of the European Communities; 1985. pp. 45–49. [Google Scholar]
- 23.McKeating J A, Gow J, Goudsmit J, Pearl L H, Mulder C, Weiss R A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 24.McKnight A, Weiss R A, Shotton C, Takeuchi Y, Hoshino H, Clapham P R. Change in tropism upon immune escape by human immunodeficiency virus. J Virol. 1995;69:3167–3170. doi: 10.1128/jvi.69.5.3167-3170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore J P, Ho D D. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nara P, Smit L, Dunlop N, Hatch W, Merges M, Waters D, Kelliher J, Gallo R C, Fischinger P J, Goudsmit J. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J Virol. 1990;64:3779–3791. doi: 10.1128/jvi.64.8.3779-3791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayan O, Griffin D E, Chase J. Antigenic shift of visna virus in persistently infected sheep. Science. 1977;197:376–378. doi: 10.1126/science.195339. [DOI] [PubMed] [Google Scholar]
- 28.Narayan O, Wolinsky J S, Clements J E, Strandberg J D, Griffin D E, Cork L C. Slow virus replication: the role of macrophages in the persistence and expression of visna virus in sheep and goats. J Gen Virol. 1982;9:345–356. doi: 10.1099/0022-1317-59-2-345. [DOI] [PubMed] [Google Scholar]
- 29.Palker T J, Clark M E, Langlois A J, Matthews T J, Weinhold K J, Randall R R, Bolognesi D P, Haynes B F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancino G, Ellerbrok H, Sitbon M, Sonigo P. Conserved framework of envelope glycoproteins among lentiviruses. Curr Top Microbiol Immun. 1993;188:77–100. doi: 10.1007/978-3-642-78536-8_5. [DOI] [PubMed] [Google Scholar]
- 31.Pétursson G, Nathanson N, Georgsson G, Panitch H, Pálsson P A. Pathogenesis of visna. I. Sequential virologic, serologic, and pathologic studies. Lab Investig. 1976;35:402–412. [PubMed] [Google Scholar]
- 32.Querat G, Audoly G, Sonigo P, Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1989;175:434–447. doi: 10.1016/0042-6822(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 33.Rusche J R, Javaherian K, McDanal C, Petro J, Lynn D L, Grimaila A, Langlois A, Gallo R C, Arthur L O, Fishinger P J, Bolognesi D P, Putney S D, Matthews T J. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino-acid sequence of the viral envelope gp120. Proc Natl Acad Sci USA. 1988;85:3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saltarelli M, Querat G, Konings D A M, Vigne R, Clements J E. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology. 1990;179:347–364. doi: 10.1016/0042-6822(90)90303-9. [DOI] [PubMed] [Google Scholar]
- 35.Sargan D R, Bennet I D, Cousens C, Roy D J, Blacklaws B A, Dalziel R G, Watt N J, McConnell I. Nucleotide sequence of EV1, a British isolate of maedi-visna virus. J Gen Virol. 1991;72:1893–1903. doi: 10.1099/0022-1317-72-8-1893. [DOI] [PubMed] [Google Scholar]
- 36.Sawyer L S W, Wrin M T, Crawford-Miksza L, Potts B, Wu Y, Weber P A, Alfonso R D, Hanson C V. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schreiber M, Wachsmuth C, Muller H, Odemuyiwa S, Schmitz H, Meyer S, Meyer B, Schneider-Mergener J. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J Virol. 1997;71:9198–9205. doi: 10.1128/jvi.71.12.9198-9205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siebelink K H J, Rimmelzwaan G F, Bosch M L, Meloen R H, Osterhaus A D M E. A single amino acid substitution in hypervariable region 5 of the envelope protein of feline immunodeficiency virus allows escape from virus neutralization. J Virol. 1993;67:2202–2208. doi: 10.1128/jvi.67.4.2202-2208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siebelink K H J, Huisman W, Karlas J A, Rimmelzwaan G F, Bosch M L, Osterhaus A D M E. Neutralization of feline immunodeficiency virus by polyclonal feline antibody: simultaneous involvement of hypervariable regions 4 and 5 of the surface glycoprotein. J Virol. 1995;69:5124–5127. doi: 10.1128/jvi.69.8.5124-5127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigurdsson B, Grimsson H, Palsson P A. Maedi, a chronic, progressive infection of sheep’s lungs. J Infect Dis. 1952;90:233–241. doi: 10.1093/infdis/90.3.233. [DOI] [PubMed] [Google Scholar]
- 41.Sigurdsson B, Palsson P A. Visna of sheep. A slow demyelinating infection. Br J Exp Pathol. 1958;39:519–528. [PMC free article] [PubMed] [Google Scholar]
- 42.Sigurdsson B, Thormar H, Pálsson P A. Cultivation of visna virus in tissue culture. Arch Gesamte Virusforsch. 1960;10:368–380. doi: 10.1007/BF01241886. [DOI] [PubMed] [Google Scholar]
- 43.Sonigo P, Alizon M, Staskus K, Klatzmann D, Cole S, Danos O, Retzel E, Tiollais P, Haase A, Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985;42:369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- 44.Staskus K A, Retzel E F, Lewis E D, Silsby J L, St. Cyr S, Rank J M, Wietgrefe S W, Haase A T, Cook R, Fast D, Gieser P, Harty J T, Kong S H, Lahti C J, Neufeld T P, Porter T E, Shoop E, Zachow K R. Isolation of replication-competent molecular clones of visna virus. Virology. 1991;181:228–240. doi: 10.1016/0042-6822(91)90488-w. [DOI] [PubMed] [Google Scholar]
- 45.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp 120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thormar H, Barshatzky M R, Arnesen K, Kozlowski P B. The emergence of antigenic variants is a rare event in long-term visna virus infection in vivo. J Gen Virol. 1983;64:1427–1432. doi: 10.1099/0022-1317-64-7-1427. [DOI] [PubMed] [Google Scholar]
- 47.Torsteinsdottir S, Agnarsdottir G, Matthiasdottir S, Rafnar B, Andresdottir V, Andresson O S, Staskus K, Petursson G, Palsson P A, Georgsson G. In vivo and in vitro infection with two different molecular clones of visna virus. Virology. 1997;229:370–380. doi: 10.1006/viro.1996.8428. [DOI] [PubMed] [Google Scholar]
- 48.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp 120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turelli P, Pétursson G, Guiguen F, Mornex J-F, Vigne R, Querat G. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J Virol. 1996;70:1213–1217. doi: 10.1128/jvi.70.2.1213-1217.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verschoor E J, Boven L A, Blaak H, Van Vliet A I W, Horzinek M C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J Virol. 1995;69:4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfs T F W, Zwart G, Bakker M, Valk M, Kuiken C L, Goudsmit J. Naturally occurring mutations within HIV-1 V3 genomic RNA lead to antigenic variation dependent on a single amino acid substitution. Virology. 1991;185:195–205. doi: 10.1016/0042-6822(91)90767-6. [DOI] [PubMed] [Google Scholar]
- 52.Zhuge W, Jia F, Adany I, Narayan O, Stephens E B. Plasmas from lymphocyte- and macrophage-tropic SIVmac-infected macaques have antibodies with a broader spectrum of virus neutralization activity in macrophage versus lymphocyte cultures. Virology. 1997;227:24–33. doi: 10.1006/viro.1996.8300. [DOI] [PubMed] [Google Scholar]