Abstract
Mason-Pfizer monkey virus (M-PMV), the prototypical type D retrovirus, assembles immature capsids within the cytoplasm of the cell prior to plasma membrane interaction. Several mutants of M-PMV Gag have been described which display altered transport, assembly, or both. In this report, we describe the use of an in vitro synthesis and assembly system to distinguish between defects in intracellular transport and the process of assembly itself for two previously described gag gene mutants. Matrix domain mutant R55W converts the type D morphogenesis of M-PMV particles into type C and has been hypothesized to alter the transport of Gag, redirecting it to the plasma membrane where assembly subsequently occurs. We show here that R55W can assemble in both the in vitro translation-assembly system and within inclusion bodies in bacteria and thus has retained the capacity to assemble in the cytoplasm. This supports the concept that R55 is located within a domain responsible for the transport of Gag to an intracellular site for assembly. In contrast, deletions within the p12 domain of M-PMV Gag had previously been shown to affect the efficiency of particle formation such that under low-level expression conditions, Gag would fail to assemble. We demonstrate here that the efficiency of assembly in the in vitro system mirrors that seen in cells under expression conditions similar to that of an infection. These results argue that the p12 domain of this D-type retrovirus plays a critical role in the membrane-independent assembly of immature capsids.
Retrovirus assembly can proceed by either of two morphogenic pathways. D-type retroviruses such as Mason-Pfizer monkey virus (M-PMV) preassemble their capsids within the cytoplasm. These capsids then migrate to the plasma membrane, where they are enveloped and released. In contrast, retroviruses that follow C-type morphogenesis, such as human immunodeficiency virus (HIV) and Rous sarcoma virus, assemble their capsids at the underside of the plasma membrane in a process combined with budding (27). Both assembly processes are driven by the Gag polyprotein precursor. During or shortly after virions are released from the cell, capsids are proteolytically matured by the viral proteinase (14). Cleavage of Gag results in the formation of a compact nucleoprotein core that varies in shape among retroviruses (29). For either morphogenic pathway, viral glycoproteins are translocated into the endoplasmic reticulum, transported to the plasma membrane through the secretory pathway, and incorporated into the virus during capsid envelopment. The RNA genome is presumed to be packaged into the virion at the site of capsid assembly.
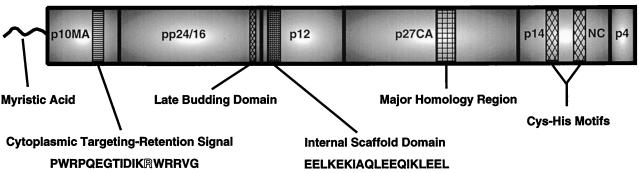
All retroviral Gag proteins contain three major domains—matrix (MA), capsid (CA), and nucleocapsid (NC)—as inferred from the matured species following proteolytic maturation (18). Processing of M-PMV Gag results in the production of matured Gag proteins MA, pp24/16, p12, CA, NC, and p4 (Fig. 1). As described for other retroviruses (28) MA is assumed to be associated with the membrane within matured virions, and CA is responsible for the formation of the virion core which contains the genomic RNA complexed with NC. In addition to the canonical retroviral Gag domains, M-PMV contains three additional major domains. The potential functions of these additional mature proteins, pp24/16, p12, and p4 in the mature virion are unknown. However, functional elements within the different domains have been defined that operate in the context of the Gag precursor. The known functional elements, beginning with the N terminus of the precursor protein, are as follows. (i) The N-terminal myristic acid moiety is dispensable for assembly of capsids, but is necessary for the transport of completed capsids to the plasma membrane (20). (ii) The putative cytoplasmic targeting-retention signal (CTRS) within MA appears to be responsible for the transport of Gag precursors to an intracellular site for assembly. This sequence in M-PMV is homologous to a similarly located sequence in the B-type retrovirus mouse mammary tumor virus (MMTV) (21). (iii) A short four-amino-acid motif (PPPY) located within the pp24/16 region functions as the “late budding domain” that is responsible for release of enveloped capsids from the plasma membrane (34). (iv) An acidic stretch of amino acids within the p12 region appears to be a putative internal scaffold domain (ISD) required for the efficient intracellular assembly of immature capsids (24). (v) The major homology region is the most conserved stretch of amino acids within the Gag proteins of retroviruses. Mutations within this sequence can have diverse affects, including the abrogation of particle assembly (26). (vi) The Cys-His motifs within NC, although they have not been directly investigated in M-PMV Gag, have been shown to be critical for the packaging of the viral RNA genome in other retroviruses (reviewed in reference 2).
FIG. 1.
Identified and potential morphological determinants within the M-PMV Gag precursor protein. The structure as inferred from the major proteolytic products of processing by the viral protease is shown. These domains are indicated from the N terminus to the C terminus as follows: p10MA, matrix; pp24/16, phosphoprotein; p12; p27CA, capsid; p14NC, nucleocapsid; and p4. Within these domains are depicted discrete amino acid sequences for which specific morphologic functions have been predicted or defined. The N-terminal myristic acid modification is also shown. The specific amino acid sequences that comprise the two domains of interest in this report are shown below. For the cytoplasmic targeting-retention signal, a previously defined arginine residue critical for function is displayed in outline.
Evidence for the existence of the CTRS and ISD domains comes from mutational studies in which altered or partially deleted gag genes are expressed in cultured cells. For the CTRS domain, a single amino acid substitution in MA (R55W) was found to redirect the morphogenic pathway of M-PMV Gag from D type to C type (21). While the designation of the region containing R55 as a transport determinant appears logical, it is still possible that the effect of the mutation is due instead to a defect in assembly such that the additional interaction with the plasma membrane becomes necessary for Gag multimerization to occur. For the ISD, it was found that deletions within this region result in an inability of Gag to assemble into immature capsids when the precursor is expressed under the control of the M-PMV promoter. However, when these species of Gag are overexpressed by the vaccinia virus/T7 system, all are able to assemble, a result which led to the hypothesis that this region is not required for assembly but instead influences its efficiency (24). Alternatively, the failure of ISD deletion mutants to assemble could be due to a defect in transport. In this case, the ability to overcome this defect by overexpression could be explained by the intracytoplasmic concentration of Gag reaching a level sufficient for multimerization to occur.
We have extended the analysis of the putative CTRS and ISD regions by examination of Gag precursors containing mutations in these domains in an in vitro assembly system. The rationale for this analysis is to examine the assembly of Gag in a system that lacks the potentially confounding effects of intracellular transport in order to determine unambiguously the nature of the two functional domains. We have previously described a system in which wild-type M-PMV Gag precursors are synthesized by in vitro translation and assemble into structures that, by gradient analysis and electron microscopy, are indistinguishable from immature capsids produced in transfected cells in culture (23). Because of the very low levels of expression possible in vitro, we also conducted our analysis of the CTRS and the ISD by utilizing the very-high-level bacterial T7 expression system for comparison. We report here that the R55W mutation, which lies within the putative CTRS, did not affect the efficiency with which Gag precursors assemble into immature capsid-like particles in either bacteria or in vitro. In contrast, deletions within the p12 domain that removed the putative ISD region rendered Gag incapable of assembly in vitro. Consistent with previous results obtained with an overexpression system in cultured cells (24), all Gag variants containing deletions of sequence within the ISD could efficiently assemble in bacteria.
MATERIALS AND METHODS
DNA constructs.
Plasmid pTFCG.M100A is a derivative of plasmid pTFCG, which contains the encephalomyocarditis virus cap-independent translation enhancer element and M-PMV gag, pro, and pol flanked by a T7 promoter and a T7 terminator (23). Plasmid pTFCG was modified to create pTFCG.M100A by replacing the methionine codon at position 100 with that for alanine. This was accomplished by substitution of the PacI to SacI fragment of pTFCG with the same fragment containing the modification from pGAG78, an infectious molecular clone of M-PMV based upon pSHRM15 (20). The phenotype of virus produced after transfection of pGAG78 into cells in culture was previously found to be indistinguishable from that of the wild type (22).
Plasmid pTFCG.R55W.M100A was constructed in the following manner: plasmid pTFCG was partially digested by Psp406I. Linear vector was isolated and then further digested with BsgI, and the desired 8.6-kb fragment was isolated. This fragment was then ligated with the 1.9-kb Psp1406I to BsgI fragment of pSHRM15.R55W (21) containing the arginine-to-tryptophan codon substitution. The resulting plasmid, pTFCG.R55W, was then also modified, as described above for pTFCG, to introduce the M100A substitution. The presence of the two mutations was confirmed by sequencing.
Plasmids pTFCGΔ8-58, pTFCGΔ1-83, pTFCGΔ1-25, pTFCGΔ26-53, and pTFCGΔ54-83 were constructed by moving the ∼1.5- to 2.3-kb ScaI to BsgI fragment containing the p12 domain deletions from pSHRM15Δ8-58, pSHRM15Δ1-83, pSHRM15Δ1-25, pSHRM15Δ26-53, and pSHRM15Δ54-83 (24), respectively, into the corresponding position of pTFCG.M100A that had been digested with BsgI and partially digested with ScaI. The M100A substitution was retained by this strategy.
Plasmids pETΔ8-58, pETΔ1-83, pETΔ1-25, pETΔ26-53, and pETΔ54-83 were constructed by replacing the PacI-NdeI fragment of pETGagHis6 with the corresponding fragments of pTFCGΔ8-58, pTFCGΔ1-83, pTFCGΔ1-25, pTFCGΔ26-53, and pTFCGΔ54-83, respectively. Plasmid pETGagHis6 (a generous gift of Robert A. Weldon, Jr.) was constructed by PCR amplification and subcloning steps. First, the p12 and CA coding regions were amplified by PCR with the primers PR1141 (5′-GGCGGTTGTTAATCC), which was designed to anneal to gag sequences located just upstream of the p12 coding sequence, and p4XhoI (5′-CAGCTCGAGATACTTGTGTGG), which was designed to insert an XhoI site at the 3′ end of gag such that six histidine codons would be placed directly adjacent to the last codon of gag. After PCR amplification of pSHRM15 with these primers, the SacI-XhoI fragment of the PCR product was cloned into corresponding sites of pET-21d (Novagen, Inc.). To subclone the 5′ end of gag, the NcoI-SacI fragment of pSITGAGPP a derivative of pSIT (1) containing gag, pro, and pol of M-PMV, was inserted into the homologous sites to finally construct pETGagHis6. After each subcloning step, the plasmid DNAs were sequenced to ensure that unwanted mutations were not inadvertently created.
Plasmid pET.R55W.M100A was constructed by transfer of the 1.5-kb BssHI to PacI fragment of pET.ΔNC.R55W, containing the 5′ sequence of gag and including the R55W substitution, into the 5.8-kb PacI to BssHI fragment of pET.M100A. Plasmid pET.M100A was constructed by first PCR amplifying the MA-p12 coding regions of pGAG78 by using the primers Nco485 (5′-GATATACCATGGGGCAA), which contains an NcoI site, and RP1179 (5′-TCCTCTAATTGAGCAA). After digestion of the PCR product with NcoI and SacI, the fragment was used to replace the NcoI-SacI fragment of pETGagHis6.
In vitro analysis of immature capsid assembly.
Transcription and translation were performed simultaneously with the pTFCG series plasmids and the TNT Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine. Products of these synthesis reactions were analyzed on sucrose gradients. Reactions were diluted to a total volume of 200 μl with 30% (wt/wt) sucrose in gradient buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100. Diluted samples were then loaded onto 2.2-ml continuous 30 to 55% (wt/wt) sucrose gradients in gradient buffer. Gradients were centrifuged in a TLS-55 rotor (Beckman Instruments) for 2 h at 55,000 rpm. Approximately 200-μl fractions were taken by hand with a Pipetman (Gilson) from the top of the gradient. The pellet was resuspended in 200 μl of 55% (wt/wt) sucrose in gradient buffer. Aliquots (5 μl) of each fraction were dissolved in sodium dodecyl sulfate (SDS) sample buffer and then loaded onto an SDS–10% polyacrylamide gel. After electrophoresis, radioactive bands were visualized by fluorography of sodium salicylate-impregnated gels.
Expression of Gag species in bacteria.
Bacterial expression plasmids of the pET series were transfected into Escherichia coli strain BL21(DE3) which had already been transformed with plasmid pBB131 which contains the Saccharomyces cerevisiae gene for protein N-myristoyltransferase (plasmid pBB131 was the kind gift of J. I. Gordon, Department of Molecular Biology and Pharmacology, Washington University School of Medicine, St. Louis, Mo.). Cells were grown in Luria broth containing 500 μM myristic acid. Production of both Gag and myristoyltransferase was induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested after 4 h of induction by centrifugation at 14,000 rpm in an Eppendorf microcentrifuge (Brinkman).
Electron microscopy.
For analysis of in vitro synthesis products by electron microscopy, reaction mixtures were diluted to a total of 1 ml in gradient buffer without detergent and centrifuged at 70,000 rpm for 1 h in a TLA-100.3 rotor with microcentrifuge inserts (Beckman Instruments). For analysis of Gag species produced in bacteria, 1-ml culture samples were centrifuged at 14,000 rpm in an Eppendorf microcentrifuge for 10 min. The resulting pellets, derived from either translation reactions or bacterial cultures, were then fixed overnight in 1% glutaraldehyde in phosphate-buffered saline (pH 7.0) at 4°C. After a rinse in phosphate-buffered saline, pellets were postfixed in 1% buffered osmium tetroxide for 1 h. These pellets were rinsed once more and then dehydrated in a graded series of ethanol solutions beginning with 50% and ending with 100%. After dehydration, pellets were rinsed three times with propylene oxide and then embedded in Polybed. Ultrathin sections were acquired by using a Rechert-Jung Ultra Cut E ultramicrotome. After staining with uranyl acetate and lead citrate, sections were examined and photographed by using a Hitachi-7000 transmission electron microscope.
RESULTS
Morphogenesis mutant R55W forms particulate structures as efficiently as the wild type in vitro.
As discussed above, the matrix domain mutant R55W alters the morphogenic pathway of M-PMV Gag such that immature capsids assemble under the plasma membrane rather than within the cytoplasm. The altered site of assembly for the R55W variant of M-PMV Gag has been hypothesized to be the result of the disruption of a specific signal within the matrix domain which is responsible for the transport of precursors to the intracellular assembly site (21). Indeed, examination of the nuclear magnetic resonance (NMR) structure of M-PMV MA has revealed that residue 55 is located within the base of an external loop not found in any other MA structure solved to date (8). Such an exposed loop might provide an interaction site for the cellular transport machinery responsible for the intracellular targeting of M-PMV Gag. However, it could be argued that the R55W mutation has no effect upon transport, but, instead, imposes a necessary requirement for membrane interaction in order for Gag multimerization to occur. To distinguish between these two possibilities—transport defect versus assembly defect—we have analyzed the potential assembly behavior of the R55W variant of Gag in an in vitro translation system.
As reported previously, the Gag and Gag-Pro precursor proteins of M-PMV assemble into immature capsid structures (23) in an ATP-dependent manner (32) upon synthesis in an in vitro system. These assembled structures can be detected by sucrose gradient analysis of translation reactions as well as by electron microscopy. Before analysis of R55W was undertaken, however, we modified our expression constructs to alter the second methionine codon of gag at residue 100 to that of alanine. The purpose of this modification is to prevent initiation of translation at this internal position, which produces an N-terminally-truncated Gag protein. In experiments previously reported, these truncated species represented a significant proportion of the total synthesized protein (23). The M100A mutation was introduced into the M-PMV gag, pro, and pol T7 expression plasmid pTFCG (23) to make plasmid pTFCG.M100A. After DNA sequencing to confirm the presence of the alteration, this new expression construct was tested for its ability to express Gag precursor proteins and was found, by both sucrose gradient and electron microscopic analyses, to produce assembled structures with an efficiency equivalent to that of the wild-type species (not shown). Since it had been shown previously that an infectious proviral clone of M-PMV containing this M100A alteration behaved essentially like the wild type in tissue culture infection assays (22), the M100A species of Gag will be considered equivalent to the wild type in the analyses presented here.
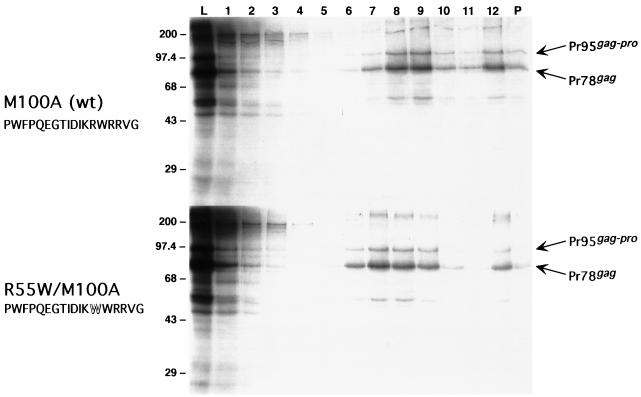
After introduction of R55W into the pTFCG.M100A background to make plasmid pTFCG.R55W.M100A and confirmation by sequencing, the R55W form of Gag was examined in parallel with M100A in a coupled transcription and translation reaction. The resulting products were then subjected to sucrose gradient centrifugation and analysis by SDS-polyacrylamide gel electrophoresis (PAGE). Protein synthesis reactions and gradient analyses were performed as previously described (23). The profiles of Gag protein species across the gradient for R55W and M100A are essentially identical (Fig. 2). For both species, a significant proportion of Gag and Gag-Pro migrates into the gradient to a position indicative of immature capsid structures (Fig. 2, fractions 7 to 9, density equal to ∼1.19 to 1.21 g/ml). Thus, by the biochemical analysis described above, the C-type morphogenesis mutant of M-PMV Gag is capable of efficient in vitro assembly.
FIG. 2.
Comparison of in vitro-synthesized and -assembled mutant matrix domain Gag species by sucrose gradient analysis. Aliquots of gradient fractions were electrophoresed on an SDS–10% polyacrylamide gel. Lane numbers indicate gradient fractions, beginning with the top fraction at no. 1. Lanes L contain an equivalent aliquot of the translation reactions before gradient fractionation. Lanes P contain material remaining as a pellet in the tube after removal of the gradient. Lanes 7 to 9, containing material indicative of immature capsids, are of a density of ∼1.19 to 1.21 g/ml. The numbers to the left indicate the positions of prestained molecular size standards in kilodaltons. Pr78gag and Pr95gag-pro indicate the positions of the Gag and Gag-Pro precursor polypeptides of M-PMV, respectively. The upper and lower panels depict the fractionation of translation reactions programmed with the M100A and R55W/M100A gag genes, respectively. The amino acid sequence of the region homologous with one in MMTV and containing residue 55 is also shown. The residue altered to tryptophan in the R55W mutant is depicted by the outlined W.
An amino-terminal sequence within the p12 domain is necessary for assembly in vitro.
Previous characterization of p12 domain deletion mutants had revealed an expression-level-dependent requirement for this domain in assembly (24). When expressed in HeLa cells under control of the long terminal repeat (LTR) promoter, several of these mutants failed to assemble particles; however, all Gag species containing deleted regions of p12 were able to assemble particles, when expressed by the vaccinia virus/T7 system. Thus, overexpression compensates for the defect created by the deletion. The interpretation of these results was that p12 functions to promote Gag multimerization in the process of assembly (24). However, as with R55W, the phenotype of these mutants might be explained by a defect in transport. Loss of specific transport to the site of assembly could lead to a loss of particle formation. This defect could then be overcome by overexpression by providing a sufficiently increased concentration of Gag within the cytoplasm for assembly to occur.
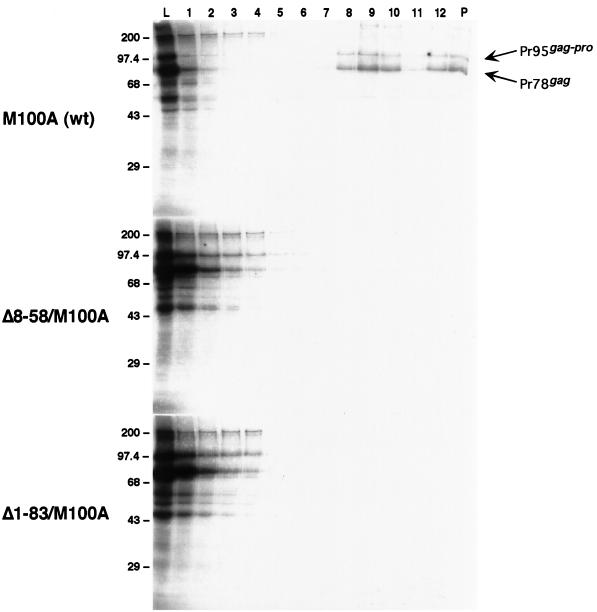
In order to distinguish between a role for p12 in assembly versus a role in transport, we examined the behavior of these mutants in our in vitro assembly system. The rational, as it was for R55W, is to utilize a system that lacks the elements of cytoplasmic transport in order to make the distinction. First we examined two mutants in which either a large proportion or the entire p12 domain within Gag was deleted. Gradient analysis of these two mutants, Δ8-58 and Δ1-83, showed that neither is capable of producing particulate material similar to that formed by M100A when synthesized in vitro (Fig. 3).
FIG. 3.
Comparison of in vitro-synthesized p12 domain deletion mutant Gag species by sucrose gradient analysis. The experiment was performed and lane designations and molecular size standards are as described for Fig. 2. Lanes 8 to 10 containing material indicative of immature capsids are of a density of ∼1.19 to 1.21 g/ml. M100A, Δ8-58/M100A, and Δ1-83/M100A indicate analyses of translation reactions programmed with the corresponding gag genes.
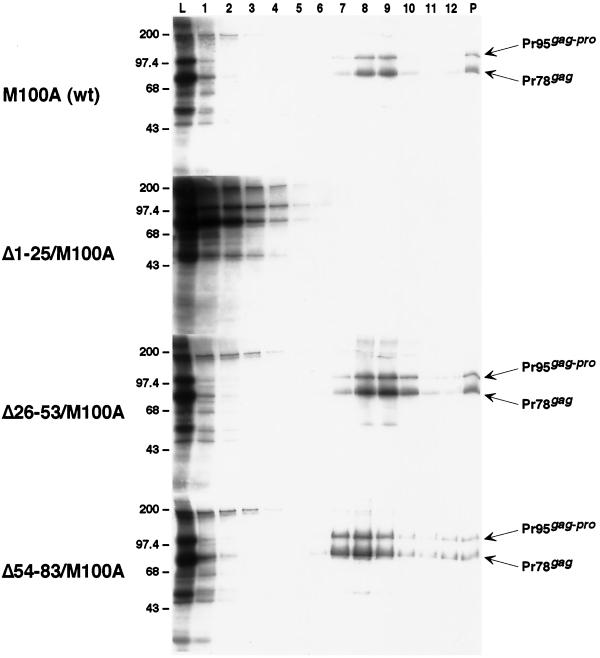
To further delineate the region within p12 responsible for this assembly-defective phenotype, we examined a series of smaller deletion mutants, Δ1-25, Δ25-53, and Δ54-83, in which approximately one-third of the p12 domain is deleted. Two of these mutants assembled particulate material which sedimented, in a manner similar to that encoded by the M100A construct, into the gradient (Fig. 4). In contrast, the third mutant (Δ1-25) failed to assemble material of sufficient size and density to migrate into the denser region of the gradient (Fig. 4) and produced a profile like that of the larger p12 domain deletions (Fig. 3).
FIG. 4.
Further comparative analysis of in vitro-synthesized p12 domain deletion mutant Gag species by sucrose gradient fractionation. The experiment was performed and lane designations and molecular size standards are as described for Fig. 2. Lanes 7 to 9 containing material indicative of immature capsids are of a density of ∼1.19 to 1.21 g/ml. M100A, Δ1-25/M100A, Δ26-53/M100A, and Δ54-83/M100A indicate analyses of translation reactions programmed with the corresponding gag genes.
Comparison among all the assembly defective mutants reveals that the function within p12 critical for assembly in this in vitro system requires, in whole or in part, residues 8 through 25 of this domain. Furthermore, the inability of mutants lacking this domain to assemble in the in vitro system confirms the hypothesis that p12 functions in the process of assembly itself rather than in transport of Gag to the site of assembly.
R55W and two partial p12 deletion mutants produce immature capsid-like structures in vitro.
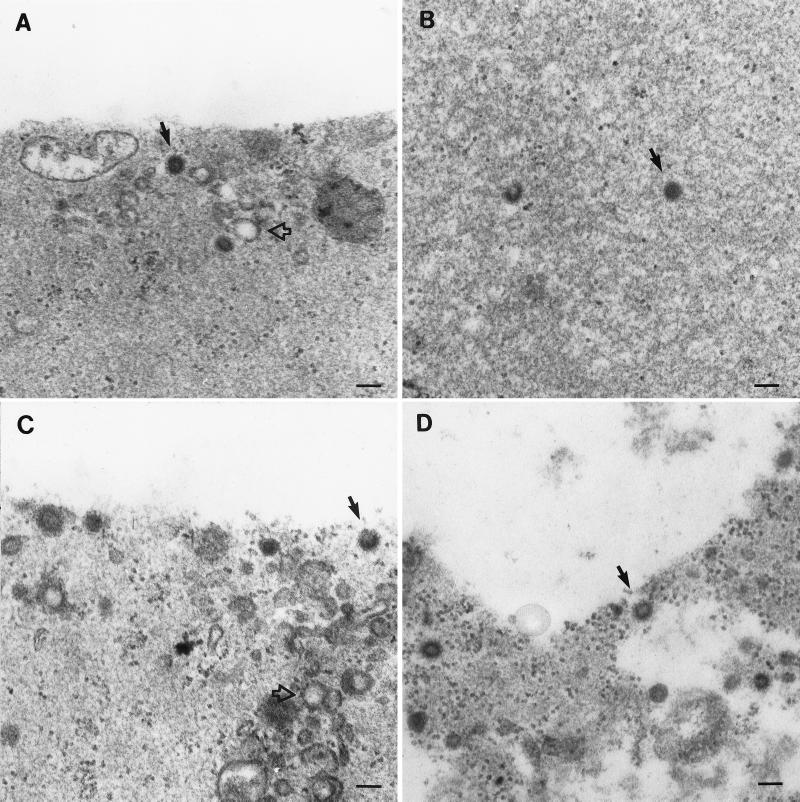
Although the above gradient analysis strongly suggests that the several Gag species which sediment into gradients are assembled capsids, we sought to confirm this by electron microscopy. Analysis by electron microscopy was performed as previously described (23). Briefly, whole transcription-translation reaction mixtures were diluted into buffer and then centrifuged at high speed. The resulting pellets were then processed for thin-section electron microscopy. Immature capsid-like structures were observed, along or near the base of the pellet in micrographs, for all mutants in which particulate material sediments into sucrose gradients (Fig. 5). The structures assembled by the R55W Gag precursors were indistinguishable from those composed of M100A, providing further confirmation that R55W is not defective in assembly (Fig. 5A and D). Two p12 deletion mutants, Δ26-53 and Δ54-83, also produced structures similar to those found in the M100A samples (Fig. 5B and C). In contrast, capsid-like structures were not observed for Gag species that were unable to assemble as indicated by gradient analysis (not shown).
FIG. 5.
Analysis of coupled transcription and translation reactions by thin-section electron microscopy. M-PMV gag genes were expressed in vitro, the reaction mixtures were diluted and centrifuged at high speed, and the resulting pellets were fixed and processed for microscopy. (A to D) Pellets derived from lysates expressing M100A (wild type) Δ26-53, Δ54-83, and R55W, respectively. Solid arrows indicate apparently completed immature capsid structures. Open arrows indicate vesicle-like structures in these non-detergent-treated samples.
Mutant Gag species assemble in bacteria.
Since the in vitro system provides only a low level of expressed protein—less than can be detected by silver stain of gels (<10 ng [data not shown])—it was of interest to compare the above results with those obtained by high-level expression. It had already been established that M-PMV Gag can assemble into immature particle-like structures when expressed at a high level in bacteria—amounts easily detected by Coomassie stain of gels (≫10 μg [data not shown]) (15). Although E. coli possesses transport machinery, it would not be expected that this machinery would specifically interact with M-PMV Gag. Thus, expression in bacteria would serve as a comparison to the in vitro system in the same way that the vaccinia virus/T7 overexpression system had served as a comparison to LTR-driven expression in cultured cells (24).
To achieve high-level bacterial expression of the mutant Gag species in bacteria, the regions of gag containing the mutated MA and p12 coding sequences were moved into plasmid pETGagHis6, a T7 bacterial expression vector containing the M-PMV gag gene. All the pET-based gag expression vectors also contained the M100A substitution to prevent, as in the case of in vitro expression, the production of N-terminally-truncated Gag species by internal initiation of translation. In addition to the gag expression vectors, the BL21(DE3) bacteria used in these experiments also harbored a plasmid, pBB131, maintained by kanamycin selection, that expresses the yeast myristyl transferase (16). Thus, when these cells are induced in the presence of 500 μM myristic acid, a significant proportion of the resulting Gag protein is N-terminally myristylated (31).
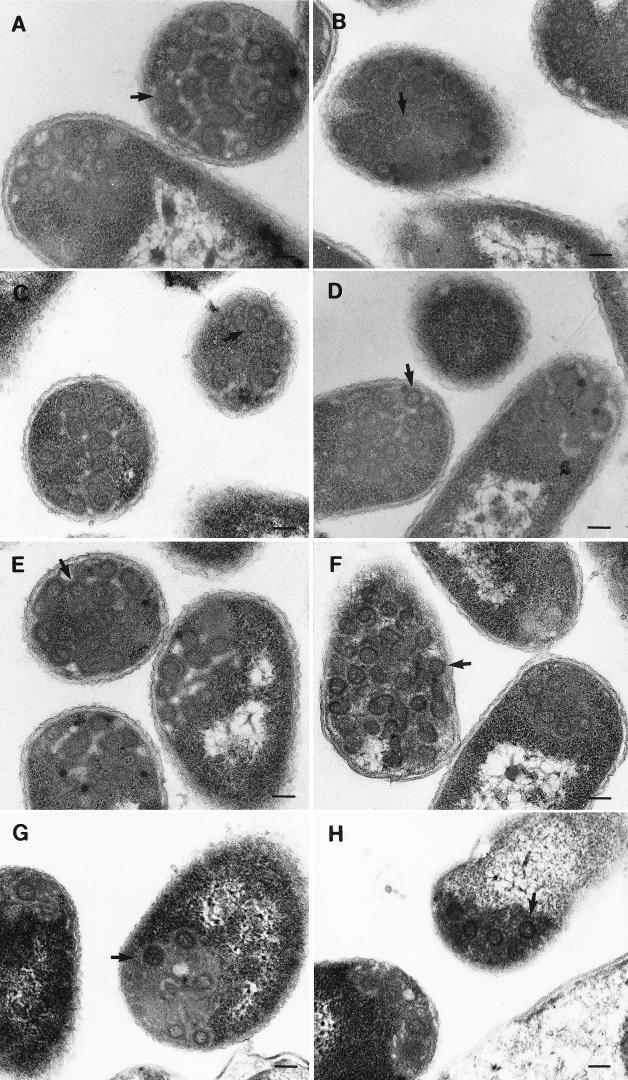
After induction of expression, the bacterial cells were centrifuged at low speed and the resulting pellets were processed for thin-section electron microscopy. Before centrifugation, small aliquots of these cells were lysed and examined by SDS-PAGE for comparison with cells containing a control gag-minus vector to confirm the production of Gag (not shown). For all examined Gag species, immature capsid-like structures were found within or associated with inclusion bodies in bacteria (Fig. 6). Aberrant or incomplete structures were also observed for all species, as has been reported previously for wild-type gag expression in either bacteria (15) or Cos-1 cells (23). Thus, this bacterial expression system provides further evidence that R55W is not defective in assembly and also confirms the previous results that overexpression of p12 deletion mutants can overcome a defect in the ability to multimerize into assembled structures. The fact that overexpression can compensate for the assembly defect indicates that p12 is not absolutely required for, but influences the efficiency of, capsid assembly.
FIG. 6.
Analysis of M-PMV Gag species expressed in bacteria by thin-section electron microscopy. After 4 h of induction with IPTG, BL21(DE3) cells expressing the various M-PMV Gag species were centrifuged at low speed, and the resulting pellets were fixed and further processed for microscopy. (A to F) Pelleted bacteria expressing M100A, Δ8-53, Δ1-83, Δ1-25, Δ26-53, and Δ54-83, respectively. (G and H) Pelleted bacteria expressing R55W. Arrows indicate apparently completed immature capsid structures.
DISCUSSION
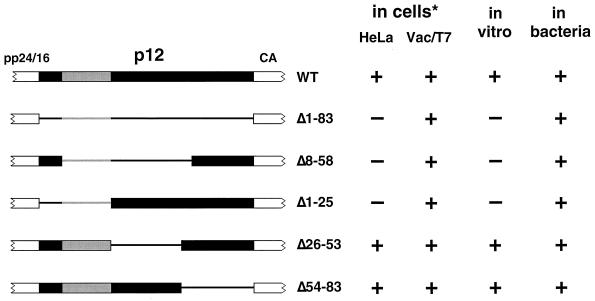
By the utilization of two different expression systems in which retrovirus particle assembly can be observed in the absence of the complicating action of eukaryotic cytoplasmic transport, we have been able to unambiguously define the functions of two critical domains within the M-PMV Gag precursor protein. The region within matrix containing the R55 residue has now been definitively identified as a transport domain, since alteration of its sequence has no discernible effect upon Gag assembly in vitro and in bacteria. Likewise, the acidic stretch of residues within p12 constitutes an auxiliary assembly domain, since its deletion results in an inability of Gag to assemble under low expression conditions, even in the absence of a eukaryotic transport mechanism. Furthermore, these two systems, because of their contrasting levels of protein expression, varying from minute quantities in vitro to gross overexpression in bacterial cells, have allowed us to make direct comparisons with results from similarly varied levels of expression in tissue culture. The ability of the various p12 deletions to assemble in vitro mirrors their ability to assemble in HeLa cells, where expression levels mimic those of an infection (Fig. 7). Similarly, overexpression in bacteria allows for the assembly of all of the deletion mutants, just as overexpression with the vaccinia virus/T7 system compensates for the defect observed in cultured cells.
FIG. 7.
Summary of assembly by p12 deletion Gag mutants. The ability of various p12 deletion species to assemble in vitro and in bacteria is compared to previously reported data from the results of expression in tissue culture cell lines (* [see reference 24]). Schematics of the p12 domain of Gag are shown to the left, with regions of the domain present in each species depicted by the wide bar and regions absent depicted by the thin line. A region apparently critical to assembly under conditions of lower expression is depicted in gray. HeLa refers to results of transfected provirus expression in HeLa cells. Vac/T7 refers to overexpression by the vaccinia virus/T7 polymerase system in CV-1 cells. Plus and minus signs indicate the presence and absence, respectively, of assembled immature capsid structures.
Numerous studies have identified the matrix domain of retroviral Gag precursors as containing a signal or signals important for proper intracellular targeting (9, 17, 35). The R55W mutation lies within a region that is highly conserved between M-PMV and MMTV MA proteins. Since both of these viruses assemble within the cytoplasm, it would be expected for both Gag proteins to contain a similar targeting signal for the intracellular assembly site. In the NMR structure of the M-PMV matrix protein, R55 lies at the base of an external, solvent-exposed loop not found in the structures of any other matrix protein solved to date (8). One could envision this loop as functioning as a docking structure for the cellular transport machinery. Further evidence for the transport function of the R55 loop region of M-PMV matrix has been provided by Choi et al. (7), who have engineered this region into the matrix domain of murine leukemia virus (MuLV) Gag resulting in the production of intracytoplasmic particles. Similarly, the homologous region of MMTV can also induce the assembly of intracytoplasmic immature particles when engineered into the MuLV matrix domain (7). These results led the authors of that study to term this region a cytoplasmic transport-retention signal (CTRS).
Just as the CTRS is found in both M-PMV and MMTV, so too does the auxilliary assembly domain of M-PMV have an apparent counterpart in MMTV. In both cases, the sequence is very highly negatively charged, with seven glutamic acid residues within the M-PMV sequence and nine aspartic acid plus four glutamic acid residues in the homologous p3 region of MMTV (24). How might this region function to increase the efficiency of Gag multimerization and assembly? One possibility is that it functions as an internal scaffold that assists the remainder of the Gag precursor to assemble into the immature particle. Once this particle has formed and budded out of the cell, proteolytic maturation would then excise the p12 domain and leave the matured CA and NC proteins free to reassort into a cylindrical core. Precedence for an internal scaffold domain (ISD) within a virus capsid protein comes from studies of bacteriophage HK97, which unlike most phage, does not have a separate scaffold protein. Instead, the amino-terminal delta domain of the major shell protein is postulated to provide this function (13).
If the ISD indeed functions as an internal scaffold for B- and D-type retroviruses, what then provides the scaffold function for C-type viruses? Campbell and Vogt, in studies of in vitro assembly of Rous sarcoma virus Gag proteins, demonstrated a requirement for RNA for the formation of virus-like particles and hypothesized that this RNA serves as a scaffold upon which Gag proteins assemble (5, 6). However, it seems unlikely that RNA alone can serve as a required scaffold for immature particle formation, since HIV Gag or fragments of Gag can assemble into immature-like particles in vitro in the absence of added RNA (12, 30). Furthermore, since B- and D-type viruses must each incorporate a genomic RNA, the ISD region must be required in addition to RNA for efficient assembly of an infectious virus.
Do all retroviruses have a requirement for a scaffold-like function for efficient assembly? In the in vitro systems using purified Gag or Gag fragments referred to above, assembly is achieved at very high protein concentrations—several milligrams per milliliter. Similarly, fragments of HIV Gag, including those lacking the MA domain, can assemble intracellularly when expressed at extremely high levels by recombinant baculoviruses (reviewed in reference 3). This concentration effect is, perhaps, the same phenomenon we observed with p12-deleted M-PMV Gag, in which overexpression compensates for the deletion and allows assembly to occur. Thus, C-type and, indeed, ISD-deleted D-type Gag precursors are competent to assemble without the assistance of a putative scaffold, provided they are sufficiently concentrated. We would propose, therefore, that the ISD functions by increasing the effective concentration of M-PMV Gag to facilitate assembly, independent of it binding to RNA.
If, as postulated, the ISD provides the concentrating factor for B- and D-type retroviruses, what provides this function for the C-type retroviruses? The obvious candidate for this role is the plasma membrane. Several studies have demonstrated that even Gag proteins with very large deletions—precursors that should be severely handicapped in their ability to assemble—can form particulate structures that bud from the cell provided they reach the plasma membrane. A Rous sarcoma virus Gag fragment, p25M1, consisting of only MA plus the spacer peptides p2A and p2B between MA and p10 can still produce budded particles, although these are of a lower density than those produced by full-length Gag, due to the absence of the interaction (I) domain in NC (33). Similarly, the matrix protein alone of simian immunodeficiency virus is capable of producing membrane-enveloped particles (10). Thus, membranes may serve as the concentrating factor for C-type viruses and could even be considered to function as a scaffold upon which Gag proteins can multimerize. Consistent with this idea, C-type Gag proteins that are not myristylated fail to assemble, presumably because the precursors are not transported to and/or do not interact with membranes (4, 11, 19, 25). In contrast, myristylation-minus D-type Gag is capable of assembly, although the resulting immature capsids fail to be transported to the cell surface (20). Indeed, in the present study, we have tested the hypothesis that an interaction with the plasma membrane might be necessary to overcome a putative assembly defect in the R55W mutant of M-PMV, but have shown that this variant of Gag, which contains the ISD, is competent to assemble in the absence of such an interaction.
The studies presented here have extended the analysis of two critical morphogenic domains within the Gag precursor of M-PMV and have served to more precisely define their functions. Because it provides the ability to examine assembly in the absence of a functioning cellular transport machinery, the technique of in vitro synthesis and assembly has emerged as a powerful tool for the dissection of transport and assembly functions within the retrovirus Gag precursor.
ACKNOWLEDGMENTS
We are grateful to Eugene Arms at the UAB Comprehensive Cancer Center Electron Microscopy Core Facility for excellent assistance with electron microscopy. We also thank Christina Ochsenbauer, Scott Parker, and Sabine Piller for critical reading of the manuscript.
This work was supported by Public Health Service grants CA-27834 to E.H. and AI-09301 to M.S.
REFERENCES
- 1.Andreansky M, Hunter E. Phagemid pSIT permits efficient in vitro mutagenesis and tightly controlled expression in E. coli. BioTechniques. 1994;15:626–633. [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Boulanger P, Jones I. Use of heterologous expression systems to study retroviral morphogenesis. Curr Top Microbiol. 1996;214:237–260. doi: 10.1007/978-3-642-80145-7_8. [DOI] [PubMed] [Google Scholar]
- 4.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi G, Park S, Choi B, Hong S, Lee J, Hunter E, Rhee S S. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J Virol. 1999;73:5431–5437. doi: 10.1128/jvi.73.7.5431-5437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fäcke M, Janetzko A, Shoeman R L, Kräusslich H-G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González S A, Affranchino J L, Gelderblom H R, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 11.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type I. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrix R W, Duda R L. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–283. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoll L J, Gordon J I. Use of Escherichia coli strains containing fad mutations plus a triple plasmid expression system to study the import of myristate, its activation by Saccharomyces cerevisiae acyl-CoA synthetase, and its utilization by S. cerevisiae myristoly-CoA:protein N-myristoyltransferase. J Biol Chem. 1993;268:4281–4290. [PubMed] [Google Scholar]
- 17.Kräusslich H-G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol. 1996;214:25–64. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 18.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 22.Rhee, S. S., and E. Hunter. Unpublished results.
- 23.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommerfelt M A, Rhee S S, Hunter E. Importance of p12 protein in Mason-Pfizer monkey virus assembly and infectivity. J Virol. 1992;66:7005–7011. doi: 10.1128/jvi.66.12.7005-7011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spearman P, Wang J-J, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 28.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 29.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 30.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weldon, R. A., Jr., and E. Hunter. Unpublished results.
- 32.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]