Abstract
Analysis of flavivirus polyprotein processing has revealed the presence of a substrate for the virus-encoded NS2B-NS3 protease at the carboxy-terminal end of the C (capsid or core) protein. Cleavage at this site has been implicated in the efficient generation of the amino terminus of prM via signal peptidase cleavage. Yellow fever virus has four basic residues (Arg-Lys-Arg-Arg) in the P1 through P4 positions of this cleavage site. Multiple alanine substitutions were made for these residues in order to investigate the substrate specificity and biological significance of this cleavage. Mutants were analyzed by several methods: (i) a cell-free trans processing assay for direct analysis of NS2B-NS3-mediated cleavage; (ii) a trans processing assay in BHK-21 cells, using a C-prM polyprotein, for analysis of prM production; (iii) an infectivity assay of full-length transcripts to determine plaque-forming ability; and (iv) analysis of proteins expressed from full-length transcripts to assess processing in the context of the complete genome. Mutants that exhibited severe defects in processing in vitro and in vivo were incapable of forming plaques. Mutants that contained two adjacent basic residues within the P1 through P4 region were processed more efficiently in vitro and in vivo, and transcripts bearing these mutations were fully infectious. Furthermore, two naturally occurring plaque-forming revertants were analyzed and shown to have restored protein processing phenotypes in vivo. Finally, the efficient production of prM was shown to be dependent on the proteolytic activity of NS3. These data support a model of two coordinated cleavages, one that generates the carboxy terminus of C and another that generates the amino terminus of prM. A block in the viral protease-mediated cleavage inhibits the production of prM by the signal peptidase, inhibits particle release, and eliminates plaque formation.
Yellow fever virus (YF) is a member of the Flavivirus genus of the Flaviviridae, a family of small enveloped positive-strand RNA viruses which also includes the Pestivirus and hepatitis C virus genera. The flavivirus genome encodes a single long open reading frame which yields a long polyprotein. This polyprotein is processed by a combination of host and viral proteases to generate the mature viral proteins (for a review, see reference 32). The three structural proteins are located within the N-terminal one-quarter of the polyprotein, in the order C-prM-E, where C is the capsid protein, prM is a glycosylated precursor of the viral envelope-bound M protein, and E is the envelope protein. The nonstructural proteins (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) are located in the remaining portion of the polyprotein. The amino termini of prM, E, NS1, and NS4B are generated by signal peptidase cleavage within the lumen of the endoplasmic reticulum (ER). A trypsin-like serine protease residues within the N-terminal one-third of NS3; the activity of this protease requires the presence of NS2B as a cofactor. Several cleavages within the nonstructural region are mediated by this protease complex, namely the 2A/2B, 2B/3, 3/4A, and 4B/5 cleavages. In addition, the NS2B-NS3 protease is also responsible for mediating cleavages at the C terminus of the C protein and the C terminus of NS4A, as well as alternative cleavages within NS2A and NS3.
Several lines of evidence support a significant role for the NS2B-NS3-mediated cleavage at the C terminus of the C protein. The preferred substrate for the viral protease is a pair of basic amino acids in the P2 and P1 positions, followed usually by a serine or glycine (sometimes alanine or threonine) in the P1′ position (nomenclature as per reference 4). For YF, such a motif is located 20 amino acids N terminal to the eventual amino terminus of prM. This motif is a conserved feature among the flaviviruses, suggesting a functional role. Cleavage at this location has been demonstrated for YF with a cell-free trans-processing assay in a manner dependent on NS2B and the protease domain of NS3 (2). A similar assay for a related flavivirus, West Nile virus, suggests cleavage at the analogous location (44). The C proteins from two flaviviruses, Kunjin virus (40) and West Nile virus (31), have been shown to contain pairs of basic amino acids at their C termini. Interestingly, efficient generation of the prM protein is dependent on the presence of the viral protease; in the absence of the protease, a C-prM polyprotein is commonly detected and release of structural proteins is impaired (2, 22, 37, 45). This has led to a model of two coordinated cleavages, one on either side of the ER membrane (22). A hydrophobic domain between these two sites serves to direct translocation of prM into the lumen, where efficient signal peptidase cleavage occurs only after cleavage at the upstream dibasic site. More direct evidence for this model comes from the demonstration of posttranslational signal peptidase cleavage following trypsin digestion of the cytoplasmic portion of a C-prM polyprotein (41).
Despite the evidence that NS2B-NS3-mediated cleavage of the C protein plays an important role in the processing of the structural protein, it remains to be demonstrated that this processing plays an essential role in the viral replication cycle. To investigate the substrate specificity of the capsid dibasic-site cleavage and to examine the importance of this cleavage for the replication of the virus, a number of site-directed mutations were made around the NS2B-NS3-mediated cleavage site. An assay developed previously allowed the direct analysis of NS2B-NS3-dependent cleavage at the dibasic site (2), while the YF infectious clone was used to investigate the plaque-forming ability of mutant transcripts (5, 33). In addition, a transient-expression assay was used to express mutant C-prM polyproteins in the presence or absence of the viral protease. It was also possible to analyze protein processing directly in cells transfected with full-length transcripts, since protein expression levels for both plaque-forming and non-plaque-forming mutants were similar. Viral protease-mediated processing at the conserved dibasic site was found to correlate with proper processing of the C-prM polyprotein and with the ability to generate plaques on two cell lines.
MATERIALS AND METHODS
Cell lines, virus, and antisera.
Growth of YF stocks and BHK-21 and SW-13 monolayers has been summarized previously (9). A BHK-21 derivative (BHK-21/J) was kindly provided by Paul Olivo (Washington University, St. Louis, Mo.) and was used for experiments involving full-length YF transcripts. Antisera specific for prM, E, NS2B, and NS3 have been described previously (8), and a rabbit antiserum raised against a fusion protein containing residues 1 to 94 of the YF C protein was kindly provided by M. Bouloy (Institute Pasteur, Paris, France).
Plasmid constructions.
Standard recombinant DNA techniques (3) were used for plasmid constructions. The construction of the pTM3-NS2B, pTM3-NS3181, and pTM3-NS2B-3181 plasmids has been described previously (10). pTM3-NS3181(S→A) has also been described previously (17). pTM3-C-prM was constructed through a three-step process: (i) the NcoI site of pTM3 (28) was removed by digestion with NcoI, treatment with mung bean nuclease to generate blunt ends, and religation; (ii) the C-prM cassette from pBS/C-prM (2) was cloned into this pTM3 derivative by using the EcoRI and SstI sites of each; and (iii) a 41-bp region preceding the translation initiation site for the C-prM polyprotein was removed by digestion with EcoRI and BstEII, generation of blunt ends with mung bean nuclease, and religation. The regions around the deleted NcoI site and the 41-bp deletion were verified by sequencing.
Mutagenesis.
Site-directed mutagenesis was performed by a modification of the Kunkel method (15, 18) with either pBS-anchC (2) or pBS-YFS (which contains a cassette encoding the YF structural proteins [14a]) as a template. Restriction sites were introduced by using silent mutations for easy identification. The 78-bp MscI-BsmI fragment from each mutant was substituted into pBS-anchC.3 (2) for use in the in vitro assay; the identity of the subcloned fragment was verified by sequencing. Mutants were initially subcloned into pYF5′3′IV (33) by a three-piece ligation: a 3,868-bp SalI-MscI fragment from pYF5′3′IV plus a 2,960-bp SalI-partial BsmI fragment from pYF5′3′IV plus the 78-bp MscI-BsmI fragment from each pBS-anchC or pBS-YFS mutant. Mutant derivatives of a full-length YF cDNA clone, pACNR-FLYF (5), were then constructed by substitution of a 1,903-bp EagI-NsiI fragment from the appropriate pYF5′3′IV mutant. The pTM3-C-prM mutant derivatives were constructed by substitution of the 394-bp NcoI-MscI fragments from the pYF5′3′IV mutants into pTM3-C-prM.
Cell-free trans-processing assay.
The protease assay has been described previously (2). The small peptides produced in this reaction were resolved by using the Tricine system of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (38) with 16.5% acrylamide in the separating portion of the gel and an acrylamide/bisacrylamide ratio of 21:1. Tricine gels were fixed for 30 min in 10 volumes of 25% ethanol–10% acetic acid, washed twice with distilled H2O (15 min per wash), and dried for 4 h under a vacuum at 60°C, followed by visualization with a Bio-Rad Molecular Imager System (model GS-363). Quantitation of protein bands was performed with Bio-Rad software. The proportion of cleavage was defined as the ratio of radioactivity in the two product bands to the total radioactivity present in all three bands (substrate and two products).
Transient expression.
The vaccinia virus-T7 system was used to transiently express transfected plasmids (14). BHK-21 monolayers in 35-mm-diameter dishes were infected with vTF7-3 (a vaccinia virus recombinant expressing T7 RNA polymerase) at a multiplicity of infection of 10 in a total volume of 0.2 ml of phosphate-buffered saline plus 1% fetal bovine serum (FBS). The inoculum was kept on the monolayers for 0.5 h at 37°C and then replaced with 0.5 ml of minimal essential medium (MEM; Life Technologies, Inc.) containing 12 μg of Lipofectamine (Gibco-BRL) and 1 μg of each of the relevant plasmids. Following a 2.5-h transfection period at 37°C, cells were labeled for 4.5 h at 37°C with 30 μCi of [35S]methionine (ICN Translabel)/ml in MEM containing 1/40th of the normal concentration of methionine and 2% FBS. After being labeled, monolayers were washed twice with MEM and lysed with 0.3 ml of a lysis buffer containing 0.5% SDS, 50 mM Tris-Cl (pH 7.5), 150 mM NaCl, and 1 mM EDTA. One-sixth of each SDS lysate was immunoprecipitated with the appropriate antiserum as described previously (9) and separated by conventional SDS-PAGE using 13% acrylamide (3). Gels were treated for fluorography as described elsewhere (2) and exposed to X-ray film (Kodak X-Omat) at −80°C.
For the transient-expression experiment (see Fig. 5), the above procedure was modified as follows. Only 10 ng of pTM3-NS2B-3181 was used for each transfection, although pTM3-C-prM derivatives were added at 1 μg per transfection. Following transfection, the transfection mix was replaced with MEM plus 2% FBS. After 50 min at 37°C, cells were labeled for 20 min with methionine-free MEM plus 100 μCi of [35S]methionine/ml and then lysed as described above.
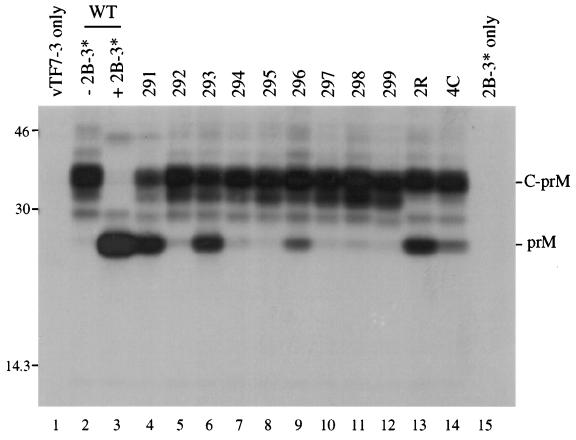
FIG. 5.
Coexpression of C-prM mutants and NS2B-3181 following transfection of plasmids at a 100:1 ratio. One microgram of pTM3-C-prM derivative and 10 ng of pTM3-NS2B-3181 were transfected for each 35-mm-diameter dish (lanes 3 to 14). SDS lysates were immunoprecipitated with prM antiserum and separated by SDS–13% PAGE. Lanes: 1, vTF7-3 alone; 2 and 3, wild-type (WT) pTM3-C-prM with (lane 3) or without (lane 2) pTM3-NS2B-3181; 15, pTM3-NS2B-3181 in the absence of pTM3-C-prM plasmid. The positions of protein standards (in kilodaltons) are shown on the left, and the positions of prM and the C-prM polyprotein are indicated on the right.
Specific infectivity measurements.
YF transcripts were synthesized with SP6 RNA polymerase (33), using XhoI-linearized pACNR-FLYF (5) as a template. Incorporation of trace [5,6-3H]UTP was assayed by adsorption to DE81 (Whatman) filter paper as a means of quantitating RNA yields (35), and transcript integrity was confirmed by electrophoresis in agarose gels followed by ethidium bromide staining (1 μg/ml for 20 min). For electroporation, 3 μg of each transcript was mixed with 8 × 106 (BHK-21) or 4 × 106 (SW-13) cells in a phosphate-buffered solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and pulsed in 2-mm-gap electroporation cuvettes (BTX) with an electroporator (BTX Electro Square Porator model T820) set for 5 pulses at 960 V with a pulse length of 99 μs (BHK-21) or for 3 pulses at 800 V with a pulse length of 60 μs (SW-13). After a 10-min recovery phase at room temperature, 35-mm-diameter dishes were seeded with serial 10-fold dilutions of transfected cells in the presence of untransfected cells (6 × 105 BHK-21 cells or 8 × 105 SW-13 cells for each 35-mm-diameter dish). Cell medium was replaced with an agarose overlay (MEM with 0.6% agarose and 2% FBS) after allowing 4 to 6 h for attachment. Cells were incubated for 4 (BHK-21) or 5 (SW-13) days at 37°C in an atmosphere of 5% CO2, fixed with 7% formaldehyde, and stained with 1% crystal violet in 5% ethanol. Data presented in Tables 1 and 2 are the averages of values from two or three experiments, except for the SW-13 results in Table 1, which are from a single experiment. However, this particular experiment was performed several times via Lipofectin-mediated transfection with comparable results, although with lower overall efficiency.
TABLE 1.
Multiple alanine substitutions introduced at the capsid dibasic cleavage site and the resultant phenotypes
| Transcript | Amino acid sequence around the capsid dibasic cleavage site | % of substrate cleaved in vitroa | Specific infectivity of transcript (PFU/μg) in cell line:
|
|
|---|---|---|---|---|
| BHK-21 | SW-13 | |||
| ↓ | ||||
| WTb | S S R K R R S H D | 38 | 1.0 × 106 | 1.6 × 105 |
| 291 | . . A A . . . . . | 24 | 1.0 × 106 | 1.2 × 105 |
| 292 | . . A . A . . . . | <3 | <10 | <10 |
| 293 | . . A . . A . . . | 10 | 5.6 × 105 | 1.8 × 105 |
| 294 | . . . A A . . . . | <3 | <10 | 14 |
| 295 | . . . A . A . . . | <3 | <10 | <10 |
| 296 | . . . . A A . . . | 16 | 2.7 × 105 | 1.5 × 105 |
| 297 | . . A A A . . . . | <3 | <10 | <10 |
| 298 | . . A . A A . . . | <3 | <10 | <10 |
| 299 | . . A A A A . . . | <3 | <10 | <10 |
As calculated from Fig. 2.
WT, wild type.
TABLE 2.
Comparison of infectivities of naturally occurring revertants and their parents
| Transcript | Amino acid sequence around the capsid dibasic cleavage site | Specific infectivity of transcript (PFU/μg) in cell line:
|
|
|---|---|---|---|
| BHK-21 | SW-13 | ||
| ↓ | |||
| WTa | S R K R R S H D | 5.4 × 105 | 8.2 × 104 |
| 292 | S A K A R S H D | <10 | <10 |
| 2R | S A K V R S H D | 3.5 × 102 | 1.0 × 105 |
| 294 | S R A A R S H D | <10 | 14 |
| 4C | S R A V R S H D | 6.9 × 102 | 9.2 × 104 |
WT, wild type.
Protein analysis of cells transfected with full-length YF transcripts.
BHK-21 cells were transfected as described above, diluted in MEM plus 10% FBS, and used to seed 35-mm-diameter dishes (106 cells per dish). Cells were labeled beginning at 18 h (see Fig. 8) or 19 h (see Fig. 7) postelectroporation, using 120 (see Fig. 8) or 150 (see Fig. 7) μCi of [35S]methionine (ICN Translabel)/ml in MEM containing 1/40th of the normal concentration of methionine and 2% FBS. At 24 h postelectroporation, cells were lysed and subjected to immunoprecipitation as described above, using 1/6 (see Fig. 7) or 1/15 (see Fig. 8) of the total lysate from each 35-mm-diameter dish. For one of the experiments (see Fig. 8), the labeling medium was harvested and clarified (16,000 × g for 2 min) immediately prior to cell lysis. One-sixth of the total harvested medium was denatured by adding SDS to a final concentration of 0.5% and heating at 75°C for 5 min; immunoprecipitation was performed as described above.
FIG. 8.
Analysis of envelope protein distribution in cells transfected with full-length mutant transcripts of YF cDNA. (A) Transfected BHK-21 cells were labeled with [35S]methionine for 6 h (18 to 24 h postelectroporation); media was harvested after the labeling period, solubilized with SDS, and immunoprecipitated with antiserum to the E protein. (B) SDS lysates of the cell monolayers from the above experiment were prepared and immunoprecipitated with E antiserum. (C) SDS lysates of the cell monolayers from the above experiment were prepared and immunoprecipitated with NS3 antiserum. Immunoprecipitation of medium or of lysate of cells transfected with no RNA is shown in the first lane of each panel. The positions of E and NS3 are indicated on the right side of their respective panels, and the positions of protein standards (in kilodaltons) are shown on the left. WT, wild type.
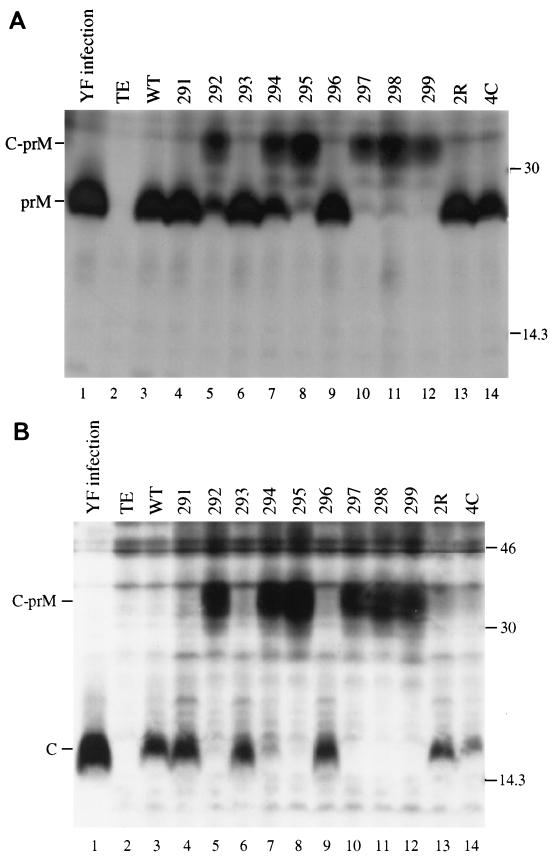
FIG. 7.
Analysis of C-prM processing in cells transfected with full-length mutant transcripts of YF cDNA. Transfected BHK-21 cells were labeled with [35S]methionine for 5 h (19 to 24 h postelectroporation), and SDS lysates were immunoprecipitated with prM (A) or C (B) antiserum. As a control, cells were infected with YF at a multiplicity of infection of 10 and labeled under the same conditions (16 to 21 h postinfection). Immunoprecipitation of YF-infected cell lysates was performed with only one-half the volume used for transfected-cell lysates. Lane 1, YF control; lane 2, cells electroporated with no RNA (TE only); lane 3, electroporation of wild-type (WT) transcript; lanes 4 to 14, mutant transcripts (as denoted). The positions of C, prM, and the C-prM polyprotein are indicated on the left, and those of the molecular mass standards (in kilodaltons) are shown on the right.
RT-PCR.
Viral stocks of mutant or potentially revertant populations were precipitated with one-fourth volume of 40% polyethylene glycol 8000 in TNE (100 mM NaCl, 10 mM Tris-Cl [pH 8], 1 mM EDTA) for 2 h on ice, pelleted (30 min at 4°C and 16,000 × g), and resuspended in 40 μl of 2% SDS containing 0.1 mg of proteinase K/ml. This mixture was incubated at 37°C for 30 min, extracted twice with phenol and twice with chloroform, precipitated, and resuspended in TE (10 mM Tris-Cl [pH 8.0], 0.1 mM EDTA). The resulting RNA preparation served as a template for cDNA synthesis with SuperScript II reverse transcriptase (Gibco-BRL) and a YF genome-specific primer (CMR#45, which is the minus-strand complement to YF nucleotides 1050 to 1065 [5′-ACACACTTGTCTTGCT-3′]). Amplification of cDNA by PCR was done with two additional YF-specific primers (PCL#8108, which corresponds to YF nucleotides 1 to 18 [5′-AGTAAATCCTGTGTGCTA-3′] and CMR#44, which is the minus-strand complement to YF nucleotides 836 to 851 [5′-ATCTCTCAATCTTTTG-3′]) and KlenTaq LA DNA polymerase (supplied by Wayne Barnes, Washington University, St. Louis). Reverse transcription (RT)-PCR fragments were sequenced with a Thermo-Sequenase [33P]ddNTP Cycle Sequencing Kit (Amersham). A 78-bp MscI-BsmI fragment derived from the RT-PCR fragments for populations 2R and 4C was cloned into pBS-anchC.3 (2), verified by sequencing, and further subcloned into pYF5′3′IV, pACNR-FLYF, and pTM3-C-prM as described above.
RESULTS
Mutagenesis of the capsid dibasic cleavage site.
Mutagenesis around the dibasic cleavage sites of the YF nonstructural region has demonstrated little substrate specificity beyond the P2, P1, and P1′ residues (11, 18, 30). However, it is likely that there are important structural determinants beyond this motif, since several regions of the polyprotein contain a similar sequence and are not known to be cleaved. In addition, some tolerance to nonconservative substitution has been observed in the P1 position of the 3/4A and 4B/5 sites (18), the P1′ position of the 2B/3 site (11), and the P2 position of the 2B/3 and 4B/5 sites (11, 18). The predicted dibasic cleavage site of the C protein is especially basic, with typically four or five basic residues located within the P1 through P6 positions; cleavage sites in the nonstructural region of the polyprotein are generally less basic (7). For YF, the site of the capsid cleavage has been experimentally determined in vitro (2). The amino acids in the P6 through P1 positions are Ser-Ser-Arg-Lys-Arg-Arg (Fig. 1). The presence of multiple basic residues has several implications for mutagenesis. First, it is possible that this plays a role in substrate determination. For example, a minimum of three or four basic amino acids may be a requirement for trans cleavage, in contrast to the probable cis cleavages of the nonstructural portion of the polyprotein. Second, this feature may be involved in some other aspect of viral replication, such as genome packaging. Finally, amino acid substitutions in this region might ablate the original cleavage site while permitting cleavage at a secondary site. One example of this possibility is the introduction of a serine in the P1 position, which results in a sequence predicted to be a good substrate for the viral protease (Arg-Lys-Arg-Ser-Ser). This consideration made it necessary to focus on all four basic residues preceding the NS2B-NS3-mediated cleavage site.
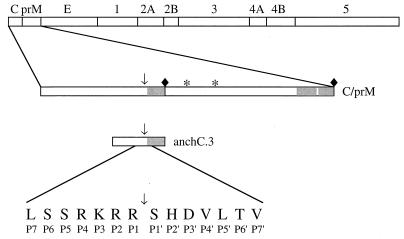
FIG. 1.
Schematic diagram of the substrate used in the cell-free trans-processing assay. The YF polyprotein is shown at the top; the locations of the structural proteins (C, prM, and E) and the nonstructural proteins (NS1 through NS5) are indicated. Below the YF polyprotein is an expanded view of the C-prM cassette; the glycosylation sites (∗), the signal peptidase cleavage site (⧫), and the capsid dibasic cleavage site (↓) are noted, and the hydrophobic regions of the C-prM polyprotein are shaded. The substrate used for the protease assay, denoted anchC.3, is 51 amino acids in length. Its size and position relative to the C-prM polyprotein are shown. The sequence around the cleavage site is indicated, using the one-letter amino acid code.
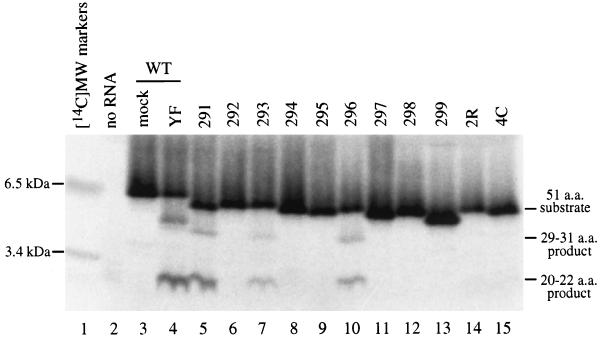
Figure 1 depicts the substrate used for the cell-free assay of capsid cleavage. Multiple alanine substitutions were introduced in the P1 through P4 positions of the capsid cleavage site (Table 1), and these mutants were subcloned into pBS-anchC.3 (2). These constructs were linearized with XbaI, transcribed with T7 RNA polymerase, and translated with rabbit reticulocyte lysate; the in vitro translation products were incubated with Triton X-100 lysates of YF-infected SW-13 cells and separated by SDS-PAGE in the presence of Tricine. Figure 2 is a phosphorimage of such a gel. Cleavage products are detectable for only three of the nine mutants tested: 291, 293, and 296 (Fig. 2 and Table 1). Each of these three mutants contain two adjacent basic residues, and these are the only three mutants with this feature. Based on the substrate specificity of the NS2B-NS3 protease (32), this suggests that the cleavage site can be shifted by one (mutant 293) or two (mutant 296) residues. It should be pointed out that with this gel system, the migration of small peptides is quite sensitive to amino acid composition. The overall charge of the peptide seems to have the most dramatic effect on migration, since substrates with identical charges migrate similarly (the presence of fewer basic residues is associated with faster migration).
FIG. 2.
Cell-free protease assay. Transcripts generated in vitro from mutant derivatives of pBS-anchC.3 (Tables 1 and 2) were translated in a cell-free system and radiolabeled by the inclusion of [35S]methionine. The resultant protein mixture was incubated with a Triton X-100 lysate from YF-infected SW-13 cells (lanes 4 to 15) or from mock-infected SW-13 cells (lane 3) and then separated by Tricine-SDS-PAGE. Visualization of the image was performed with a Bio-Rad Molecular Imager System. RNA was omitted from the translation mixture in lane 2. Lanes 3 and 4 show the wild-type (WT) substrate, while the substrates used in the other lanes are indicated. A set of 14C-labeled molecular mass standards was run in lane 1 (indicated on the left). The positions of the substrate and products are shown on the right. The wild-type substrate generates products of 31 and 20 amino acids (a.a.). However, the products seen for mutants 293 (lane 7) and 296 (lane 10) likely result from cleavage on the carboxy-terminal side of the basic pair of residues that generate products of 30 and 21 amino acids (293) or 29 and 22 amino acids (296).
Processing of the C-prM polyprotein.
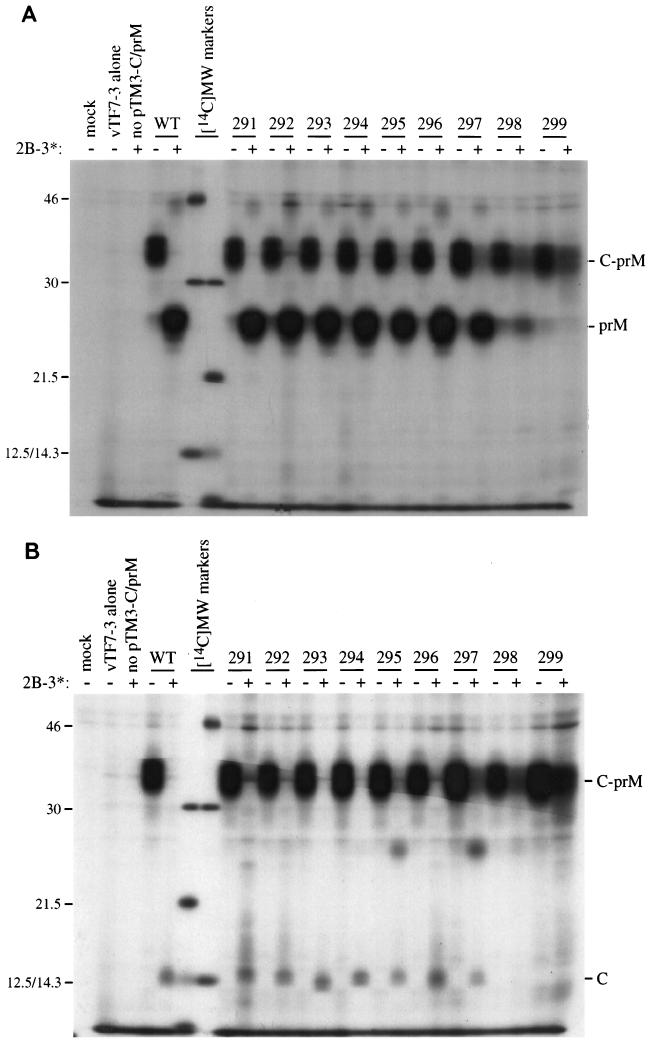
The efficient production of prM is thought to be dependent on prior cleavage at the dibasic cleavage site (22). However, this hypothesis is not universally accepted since it has been reported that efficient prM production can occur with mutants that do not cleave at the dibasic site (43). The production of prM was examined in the context of the dibasic cleavage site mutations by subcloning the 291 to 299 panel into a vector expressing the two structural proteins C and prM (pTM3-C-prM). Transient expression in the absence of the viral NS2B-NS3 protease yielded primarily an unprocessed C-prM molecule, although small amounts of prM were also visible. This was true for the wild-type construct as well as for each of the mutants (Fig. 3). Pulse-chase experiments demonstrate a very slow accumulation of prM over time (data not shown), indicating that the viral protease is not essential for the signal peptidase cleavage that generates prM. Expression of C-prM in the presence of the NS2B-NS3 protease was initially performed by transfecting an equimolar ratio of plasmids encoding C-prM and NS2B-NS3. Surprisingly, prM (Fig. 3A) and C (Fig. 3B) were produced quite efficiently for nearly all of the mutants, with the exception of 298 (AKAA) and 299 (AAAA). A similar result was obtained when the entire YF structural region (C-prM-E) was expressed (data not shown).
FIG. 3.
Coexpression of C-prM mutants and NS2B-3181. Expression of the transfected plasmids was driven by a vaccinia virus recombinant that expresses T7 RNA polymerase (vTF7-3). The C-prM cassettes contained the mutations at the dibasic capsid site that are listed in Table 1. SDS lysates were immunoprecipitated with antiserum to either prM (A) or C (B). Mock infection, vTF7-3 alone, and pTM3-NS2B-3181 without pTM3-C-prM are shown in the first three lanes. Wild-type (WT) and mutant C-prM are indicated, either without (−) or with (+) cotransfection of pTM3-NS2B-3181. Positions of molecular mass standards ([14C]MW markers) are shown, with sizes in kilodaltons noted on the left (the 12.5- and 14.3-kDa markers comigrate on SDS–13% PAGE). The positions of C, prM, and the C-prM polyprotein are indicated on the right.
Efficient C-prM processing is dependent on a proteolytically active NS3.
Although prM production is inefficient in the absence of NS2B-NS3, the above result, as well as data provided by others (43), might suggest that the mechanism of upregulation is independent of proteolytic cleavage at the dibasic site. It has been demonstrated previously that C-prM processing is dependent on the active-site serine of NS3 (22); however, this experiment was done in the context of a long nonstructural polyprotein. When the protease is inactivated by site-directed mutagenesis, polyprotein processing does not occur. Thus, it can still be argued that prM production is independent of the viral protease per se but is dependent on the proper mature nonstructural domain (43, 46). The strategy employed to examine prM production, therefore, was to express the proteolytic domain of NS3 (residues 1 to 181) and the NS2B cofactor via separate plasmids. An alanine substitution for Ser138 renders the protease catalytically inactive (12).
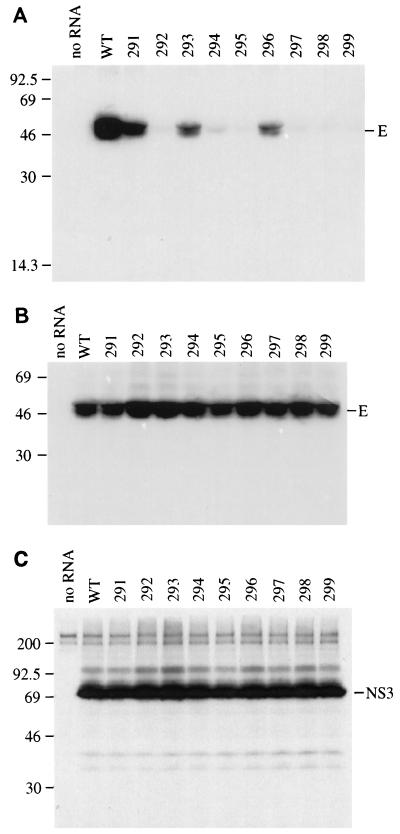
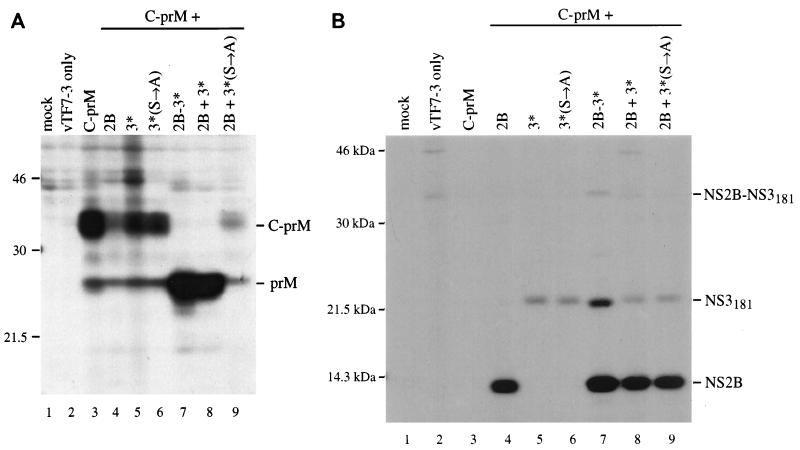
Production of prM was examined by transient expression, using different plasmid combinations. When C-prM was expressed alone, the primary product was an uncleaved polyprotein that ran as a smear of about 34 to 37 kDa (Fig. 4A, lane 3). Also seen was a minor band of about 26 kDa, which is the size of prM alone. Expression of C-prM with NS2B had a dramatic effect on the level of detectable C-prM polyprotein, although there was no enhancement of the prM band (compare lanes 3 and 4 of Fig. 4A). This effect could also be seen in lane 9 (NS2B plus inactive NS3181) and was found to be reproducible. Expression of C-prM with either form of NS3181, wild type or the Ser138→Ala variant, had no obvious effect on processing (Fig. 4A, lanes 5 and 6). However, expression of C-prM with the NS2B-3181 polyprotein is correlated with efficient production of prM and very little unprocessed C-prM polyprotein (compare lanes 3 and 7 of Fig. 4A). A similar result was seen when NS2B and NS3181 were expressed from separate plasmids (Fig. 4A, lane 8), but not when NS2B was expressed in combination with the inactive NS3 (lane 9). Figure 4B demonstrates that the components of the protease were being expressed appropriately. These results are consistent with a role for the active viral protease in upregulating prM production.
FIG. 4.
Coexpression of wild-type C-prM and NS2B-3181 components. Following infection with vTF7-3, BHK-21 cells were transfected with pTM3-C-prM alone (lane 3) or in combination with pTM3-NS2B (lane 4), pTM3-NS3181 (lane 5), pTM3-NS3181(S→A) (lane 6), pTM3-NS2B-3181 (lane 7), pTM3-NS2B plus pTM3-NS3181 (lane 8), or pTM3-NS2B plus pTM3-NS3181(S→A) (lane 9). Following immunoprecipitation with antiserum to prM (A) or a combination of antisera to NS2B and NS3 (B), proteins were separated by SDS–13% PAGE. Mock-infected cell lysate was immunoprecipitated in lane 1, and untransfected vTF7-3-infected cell lysate was immunoprecipitated in lane 2. The identities of the proteins are noted on the right side of each panel, and the positions of standards (in kilodaltons) are indicated on the left. A small amount of unprocessed NS2B-3181 can be seen in lane 7 of panel B. NS3181 has only a single internal methionine residue, while NS2B has four, contributing to the difference in band intensity.
Efficiency of C-prM processing correlates with cleavage efficiency at the capsid dibasic site.
Both the experiment described in the previous section and the transient expression of the C-prM mutants (Fig. 3) demonstrate the requirement for an active NS2B-NS3 protease in the efficient production of prM. However, most of the C-prM mutants were efficiently processed in the presence of NS2B-NS3, while the in vitro data suggest that only three of the nine mutants are adequate substrates for the viral proteolytic activity (Fig. 2). To reconcile the results of these experiments, we attempted to demonstrate more subtle differences in cleavage efficiencies between the mutant constructs, using the transient expression assay. Variations in efficiency could be demonstrated by transfecting a substrate-encoding plasmid (pTM3-C-prM) at a 100-fold excess relative to an enzyme-encoding plasmid (pTM3-NS2B-3181). Presumably this had the effect of increasing the substrate-to-enzyme ratio, although the relative levels of these proteins in the transfected cells are not known. As seen in Fig. 5 (compare lanes 2 and 3), the wild type C-prM polyprotein was completely processed to prM in the presence of the viral protease under these conditions. None of the mutants were completely processed (lanes 4 to 14), and in fact only three of the mutants generated appreciable amounts of prM (lanes 4, 6, and 9). With this experiment, then, we were able to establish that the three mutants that displayed the most efficient prM production under conditions of limiting NS2B-NS3 (mutants 291, 293, and 296) were the same three mutants that were cleaved most efficiently at the dibasic site in vitro.
Infectivity of mutant transcripts correlates with capsid dibasic-site cleavage efficiency.
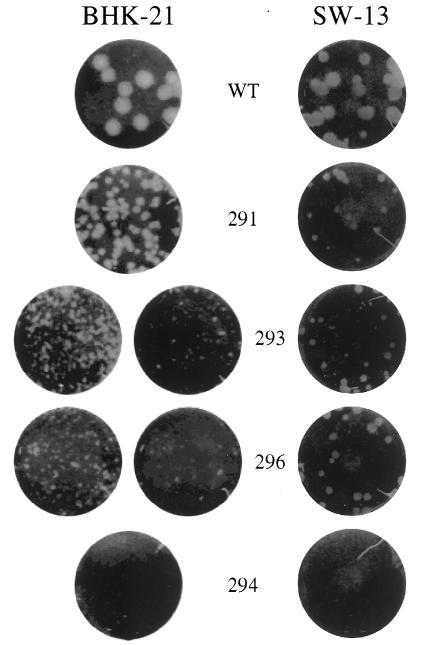
To test the phenotype of the cleavage site mutants in the context of the virus, the mutant panel was subcloned into a full-length infectious YF cDNA clone (5). The specific infectivity of transcripts generated from the full-length cDNA template was tested on two cell types, BHK-21 and SW-13 (Table 1). Mutants 291, 293, and 296 were fully infectious compared to the wild type, although mutant plaques were smaller than wild-type plaques (Fig. 6). Higher specific infectivities were seen in general with BHK-21 than with SW-13 due to the difference in their electroporation efficiencies (data not shown). Among the mutants forming plaques on BHK-21, mutant 296 displayed the smallest plaque phenotype as well as a slightly lower specific infectivity, mutant 291 plaques were the largest, and mutant 293 plaques were intermediate in size. This may reflect the effect of shifting the probable cleavage site by one (293) or two (296) positions. However, on SW-13 cells these phenotypes did not hold. Mutant 296 transcripts generated plaques of the same size as or even larger size than those of the other two mutants and did not display a reduction in specific infectivity. In general, the remaining mutants were unable to form plaques, and attempts to visualize plaques from these mutants over longer time periods were unsuccessful (data not shown). However, isolated plaques occasionally appeared for the lowest cell dilutions, equivalent to a specific infectivity of about 10−4 to 10−5 of that of the wild type. These were the most common for mutant 294 on SW-13 cells.
FIG. 6.
Plaque assays of YF containing mutations at the capsid dibasic cleavage site. BHK-21 and SW-13 cells were transfected with the mutant transcripts shown, and serial 10-fold dilutions were seeded on 35-mm-diameter dishes in the presence of untransfected cells. Monolayers were seeded with 0.2 ml of the following dilutions: for BHK-21, 10−5 (wild type [WT]), 10−4 (291), 10−3 (left) and 10−4 (right) (293 and 296), and 10−1 (294); and for SW-13, 10−4 except for 294, which was a 10−1 dilution.
C-prM processing is inefficient for non-plaque-forming transcripts.
Given that the cleavage site mutations were introduced into the structural region, it might be expected that RNA replication and translation of the viral polyprotein would be unaffected. The processing phenotypes of the mutants were examined by exploiting this possibility. Although viral proteins were undetectable at early time points, it was possible to examine viral proteins directly in transfected cells by radiolabeling cells between 18 and 24 h posttransfection. Presumably this reflects amplification of the viral genome and the subsequent elevation of protein expression levels. This labeling period precedes the peak of wild-type virus release (20). Figure 7 shows the results of such an experiment, involving immunoprecipitation with antiserum to either prM (Fig. 7A) or C protein (Fig. 7B). In several lanes, a significant amount of protein migrates in the range of an unprocessed C-prM polyprotein. These lanes represent the non-plaque-forming mutants, and among them, 292 (lane 5) and 294 (lane 7) exhibited the most robust prM production. The viable, plaque-forming mutants appeared to demonstrate complete processing to C and prM (lanes 4, 6, and 9). Thus, poor substrates at the dibasic cleavage site are associated with incomplete C-prM processing and with the inability to form plaques.
Release of structural protein is impaired for non-plaque-forming mutants.
Defects in any one of a number of processes, including RNA replication, particle assembly, virus release, particle infectivity, and viral cytopathogenicity, can eliminate the plaque-forming phenotype. It would seem to be a strong possibility that assembly and/or release is defective in the non-plaque-forming mutants, given the observed C-prM processing phenotype. Confirmation that the release of structural proteins is impaired in the non-plaque-forming mutants was obtained by immunoprecipitation of the E protein from the medium of transfected, labeled cells (Fig. 8A). The observed doublet is the result of incompletely reduced E protein (data not shown). No significant differences in the level of intracellular E protein were observed in SDS lysates prepared from the cell monolayers (Fig. 8B). Immunoprecipitation with an antiserum to a nonstructural protein, NS3, also shows equivalent intracellular expression levels across the mutant panel (Fig. 8C). Lysates were tested to confirm that the amount of protein was not saturating (data not shown). Although a defect in RNA replication cannot be formally ruled out, the fact that equivalent levels of radiolabeled viral proteins were detected between 18 and 24 h postinfection makes this unlikely.
Mutants 291, 293, and 296 secrete E protein, although at levels well below what is seen for the wild-type transcript. Although the proportion of E that is in the form of infectious particles is unknown, these results suggest that even the plaque-forming mutants are impaired with respect to particle release. This would be consistent with the reduced processing efficiency at the capsid dibasic site (Fig. 2 and Table 1), less-efficient C-prM processing (Fig. 5), and the smaller-plaque phenotype (Fig. 6). In fact, the level of E detected in Fig. 8A correlates nicely with plaque size on BHK-21 monolayers (291 > 293 > 296).
Characterization of revertants.
The three plaque-forming mutants could be passaged on either SW-13 or BHK-21 cells, with titers approaching wild-type levels, while maintaining the original cleavage site mutation (data not shown). Although the specific infectivities of the non-plaque-forming mutant transcripts were very low, it was sometimes possible to isolate stocks of plaque-forming virus from cells transfected with these transcripts. This was accomplished through a combination of extended incubation of the transfected cells and passaging of the virus by using the medium of transfected cells as an inoculum for untransfected cells. RT-PCR fragments derived from these resulting virus populations were sequenced across the region coding for the capsid dibasic cleavage site. In one case, a virus stock derived from the 292 parent (P4 to P1 = Ala-Lys-Ala-Arg) contained a single nucleotide change (GCC→GTC) resulting in a valine substitution in the P2 position (Table 2). The same substitution was observed in a population recovered from transfection with the 294 parent mutation. In addition, several independently derived virus populations that did not contain reversions or additional mutations in the structural-region coding sequences were obtained from the 294 transcripts. Presumably these populations contain an additional mutation(s) in either the noncoding sequences or the nonstructural protein coding sequence which allows the virus to overcome the original defect. These populations displayed one of at least three distinct plaque phenotypes, suggesting that they are not genetically identical.
To verify that the substitutions detected in the revertant populations of 292 and 294 (denoted 2R and 4C, respectively) were alone sufficient for the plaque phenotypes exhibited, a full-length cDNA clone containing this region within a wild-type genetic background was constructed. When tested on the cell line from which the revertants were derived (SW-13), the specific infectivities of the 2R and 4C transcripts were similar to that of the wild type (Table 2). However, on BHK-21 cells, the infectivities of the revertant transcripts were dramatically reduced and plaques were barely discernible. The nature of this host-specific difference is unknown.
No cleavage products were detected when the 2R and 4C sequences were tested with the in vitro cleavage assay (Fig. 2, lanes 14 and 15). However, 2R and 4C displayed enhanced production of prM in comparison to the parent sequences during transient expression of C-prM (Fig. 5; compare lanes 5 and 7 with lanes 13 and 14). This enhancement was dependent on the expression of NS2B-3181 (data not shown). Enhanced processing was also apparent in cells transfected with the full-length transcripts (Fig. 7; compare lanes 5 and 7 with lanes 13 and 14).
DISCUSSION
This report is the first demonstration that the NS2B-NS3-mediated cleavage at the C terminus of the C protein is an essential component of the flavivirus life cycle. A panel of nine mutants that contained two, three, or four alanine substitutions within the region of four basic amino acids that comprise the P1 through P4 residues, based on the previously identified cleavage site (2), was constructed. Processing of the C-prM polyprotein, release of structural proteins into the medium, and plaque formation were all found to correlate with the presence of a pair of adjacent basic residues and with in vitro cleavage efficiency at this site.
Two naturally arising revertants were isolated, presenting another opportunity to test the relationship between the cleavage phenotype and the viability of the virus, as measured by plaque formation. The substitution of valine for alanine in the P2 position is a fairly conservative change and thus does not create an obviously superior substrate for a dibasic-residue-directed protease. Substitution of valine at the P2 position of the 2B/3 cleavage site reduces processing efficiency only slightly (11), although the substitution of alanine in the P2 position of the 4B/5 site also results in residual processing activity (18). Using the anchC.3 in vitro assay, no cleavage activity on the revertant substrates could be detected (Fig. 2). It should be pointed out that this assay is inherently inefficient, since even the wild-type substrate is only partially processed during a 3-h incubation. In fact, C-prM processing was shown to be more complete for 2R and 4C than for 292 and 294 (Fig. 5 and 7). Since C-prM processing of 2R and 4C remains dependent on the NS2B-NS3 region, it seems most likely that these pseudorevertant sequences were selected from the virus population as more efficient substrates for the viral protease.
The amino terminus of prM is consistent with a signal peptidase cleavage (6, 23, 41), and translocation of prM into the ER occurs independently of the nonstructural region; however, several reports have shown that the production of prM is inefficient in the absence of the virus-encoded NS2B-NS3 protease (for a discussion, see references 2 and 22). The inefficiency of signal peptidase cleavage is proposed to be due to a lack of carboxy-terminal polar residues within the signal sequence, as well as some determinants within C and prM (42). A model of coordinated cleavages in which the viral protease mediates upstream cleavage at a conserved cluster of basic residues at the carboxy terminus of the C protein, followed by a secondary cleavage mediated by signal peptidase, has thus evolved. The first cleavage allows the second cleavage to occur more efficiently. Studies with Murray Valley encephalitis virus have demonstrated that prM can be generated posttranslationally in the absence of NS2B-NS3 by digesting the cytoplasmic portion of a structural polyprotein with trypsin (41). The amino terminus of the prM species produced in this way was determined to be consistent with signal peptidase cleavage. Presumably the NS2B-NS3 protease functions in a similar manner. By removing the capsid protein from the cytoplasmic side of the C-prM polyprotein, the signal peptidase site is somehow made more accessible to cleavage on the lumenal side of the ER membrane.
An alternative model is that the signal peptidase cleavage, while still occurring posttranslationally, is upregulated by interaction with the nonstructural region, specifically some portion of NS2B (43, 45, 46). In this model, cleavage at the dibasic site is not necessary for efficient signal peptidase cleavage. Evidence for this model comes from the analysis of a mutation at the dibasic site which appears to block cleavage using an in vitro system; contrary to the prediction of the first model, this mutation had no effect on secretion of a prM-E heterodimer (43). However, these experiments were done in the presence of the proteolytic domain of NS2B-NS3, and no evidence was presented to show either that cleavage at the dibasic site did not occur or that the prM-E secretion was protease independent. Also, the results obtained with 2R and 4C, as well as the results of transient expression of C-prM (Fig. 3), demonstrate that care should be taken when extrapolating the results of an in vitro cleavage assay to in vivo processing. The experiment shown in Fig. 4 was designed to address the possibility that upregulation of prM production is independent of the proteolytic activity of NS3. This experiment demonstrates that NS2B alone is insufficient to upregulate prM production, although it appears to have an impact on the amount of unprocessed C-prM that is detectable. The nature of this effect is unknown, although possibly NS2B has a destabilizing influence on the structural region in the absence of proper processing. It is difficult to completely disprove the alternative, NS2B-mediated model, since NS2B is a necessary component of and is generated by the active protease. However, no prM upregulation was detected when NS2B was expressed with a mutant NS3181 containing an alanine at the active-site serine. The upregulation of prM was restored by using a wild-type NS3181. We cannot rule out the possibility that NS2B has a function other than its role as a cofactor for the viral protease. For example, NS2B is likely to be involved in the membrane association of the active protease, and it may in fact be involved in direct interactions with the structural region. The observation that expression of NS2B alone had a significant effect on the observable level of C-prM polyprotein may reflect such an interaction. However, our data demonstrate that the efficient production of prM requires an enzymatically active viral protease.
It is interesting that processing of C-prM can occur in mutants with drastically altered dibasic substrates (Fig. 3). This probably indicates that important structural determinants beyond the primary amino acid sequence are preserved in these mutants. Despite the efficiency of C-prM processing seen with this experiment, the processing is much less efficient in the same mutants when the entire genome is expressed (Fig. 7). This could be a consequence of the high levels of expression that are induced by T7 RNA polymerase in the transient-expression assay. Alternatively, it may reflect different specific activities of two forms of the protease. Only the N-terminal one-third of NS3 is expressed from pTM3-NS2B-3181, whereas within infected cells, the full-length NS3 protein would be expected to be a component of the replication complex.
The partial processing of C-prM observed for some of the non-plaque-forming mutants (Fig. 7) suggests that some low level of particle assembly and release is possible. Certainly there must be a limit to the rate of infectious-particle release below which it becomes very difficult to resolve plaques by the standard assay. It is noteworthy that the two mutants from which revertants were derived, 292 and 294, showed higher levels of C-prM processing than the other non-plaque-forming mutants (Fig. 7A, lanes 5 and 7). Mutant 294 in particular was the easiest mutant from which to derive revertant populations. This may reflect a less-severe defect to overcome genetically, or it may be a consequence of low levels of cell-to-cell spread and, hence, more replication cycles in which mutations could arise.
The correlation seen among cleavage at the dibasic site, C-prM processing, release of structural protein, and plaque formation indicates a critical role for the mechanism of coordinated cleavages. In addition, substitutions which optimize the prM signal sequence and allow efficient signal peptidase cleavage independent of NS2B-NS3 also eliminate plaque formation (16). One conspicuous explanation for this mechanism of coordinated cleavages is that it functions as a regulatory device. Processing intermediates may have functions distinct from their terminal products, such that the local concentration of active NS2B-NS3 protease plays an important regulatory role. For example, perhaps the capsid dibasic-site cleavage is delayed until the levels of NS2B-NS3 reach a certain threshold, thereby preventing or delaying nucleocapsid formation until sufficient levels of viral genome are available for encapsidation. Another possibility is that the uncleaved C-prM blocks premature transport out of the ER.
By any of these scenarios, the envelopment of assembling or assembled nucleocapsids might be coordinated by the two cleavages and thus might account for the inability to detect a nucleocapsid intermediate in flavivirus-infected cells (29). The local concentration of viral RNA might also be involved in this regulatory scheme. Interaction of the capsid protein with RNA might make the dibasic site more accessible to cleavage; thus, the capsid molecules most likely to be cleaved by NS2B-NS3 would be complexed with RNA. This could be in addition to or instead of regulation by the concentration of NS2B-NS3. It has been demonstrated that prM and E can form empty, subviral particles in the absence of C (1, 26, 39); perhaps the tethering of the core protein to the prM-E complex is a mechanism to ensure the envelopment of genome-containing core particles, particularly given that prM and E are predicted to contain very few residues on the cytoplasmic side of the ER membrane with which to interact with C.
Another possible regulator of cleavage at the dibasic site is the specific activity of the protease, rather than its concentration. The protease is likely to be a part of the replication complex, and it is conceivable that rearrangements of the complex result in altered specific activity for cleavage at the capsid dibasic site. Thus, a signal for cleavage of the capsid protein might be a modification of the replication complex preceding a hypothetical shift from synthesis of minus strands to synthesis of plus strands.
A situation that parallels the two coordinated cleavages in the structural region of the polyprotein appears to exist in the nonstructural region (17). NS2B-NS3-mediated cleavage at the carboxy terminus of NS4A, denoted the 4A/2K site, appears to be a necessary precursor to the signal peptidase cleavage that generates the amino terminus of NS4B (at the 2K/4B site). This raises the question of why there might be two such coordinated cleavages in the same polyprotein. Sequence alignments indicate that these cleavages are completely conserved across the Flavivirus genus. Perhaps there is some interaction between the two transmembrane signal sequences. Possibly this is a means of synchronizing two distinct aspects of replication; for example, the simultaneous upregulation of signal peptidase cleavage within the structural and nonstructural regions would be a way of coordinating the initiation of the assembly process and a shift from the synthesis of minus strands to plus strands. Finally, either or both of the cleaved signal sequences may have some function independent of regulation. The fate of these peptides, 20 and 23 amino acids in length for YF, is unknown. If they remain integrated in the membrane, either or both could conceivably be incorporated into virions. The prM signal sequence in particular would likely be localized at the site of viral assembly. Although there has never been a report of these peptides being found in virions or infected cells, they could easily go undetected due to their small size. Among the flaviviruses, the distance between the capsid dibasic site and the prM signal peptidase site ranges from 14 to 22 amino acids, while the 2K fragment preceding the amino terminus of NS4B invariably contains 23 amino acids. Reports on the fate of eukaryotic signal peptide fragments have recently become available (24); they can be detected in the cytosol, can bind to major histocompatability complex class I molecules in the ER lumen, can be further processed by an unidentified signal peptide peptidase, and can bind to calmodulin. The signal sequence of human immunodeficiency virus type 1 p-gp160 has been shown to be released into the cytoplasm following cleavage and to interact with calmodulin, although no function for this interaction is known (25).
The flaviviruses have evolved a coordinated processing scheme that exploits the host cell signal peptidase. Analysis of structural-protein processing has been complicated by the presence of two neighboring cleavages. With the use of a cell-free trans-processing assay, we were able to directly observe cleavage at the dibasic site. These data were correlated with plaque formation and prM production to demonstrate that this processing is a critical step in the viral life cycle. This particular mechanism, in which the viral protease plays a central role, appears to be unique to the Flavivirus genus. However, the C protein of the mature hepatitis C virus particle is also truncated, in a delayed fashion, with respect to the amino terminus of E1 by an unknown enzyme, possibly a signal peptidase (21, 36, 47). In addition, delayed signal peptidase cleavages also appear to play a role in the processing of the E2-p7-NS2 region of hepatitis C virus (13, 19, 27) and in cleavage of the E0-E1 polyprotein of pestiviruses (34). It remains to be seen whether the delayed cleavages observed in this family reflect any common function. Such a determination will require a better understanding of the role of these processing cascades.
ACKNOWLEDGMENTS
We thank Brett Lindenbach and Keril Blight for careful reviews of the manuscript.
This work was supported in part by a grant from the Public Health Service (AI31501). Much of this work was conducted while S.M.A. was a predoctoral candidate supported by the Division of Biology and Biomedical Sciences at Washington University and by NRSA grants 5T32 GM 07067 and 5T32 AI 07172.
REFERENCES
- 1.Allison S L, Stadler K, Mandl C W, Kunz C, Heinz F X. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg S M, Nestorowicz A, McCourt D W, Rice C M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol. 1994;68:3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1998. [Google Scholar]
- 4.Barrett A J. Classification of peptidases. Methods Enzymol. 1994;244:1–15. doi: 10.1016/0076-6879(94)44003-4. [DOI] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., E. Kooi, B. D. Lindenbach, M. Lucassen, N. Huijkman, W. J. M. Spaan, and C. M. Rice. Unpublished data.
- 6.Castle E, Nowak T, Leidner U, Wengler G, Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985;145:227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 7.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 8.Chambers T J, McCourt D W, Rice C M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990;177:159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- 9.Chambers T J, McCourt D W, Rice C M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 10.Chambers T J, Nestorowicz A, Amberg S M, Rice C M. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers T J, Nestorowicz A, Rice C M. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J Virol. 1995;69:1600–1605. doi: 10.1128/jvi.69.3.1600-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Grakoui, A., and C. M. Rice. Unpublished data.
- 15.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, E., C. E. Stocks, S. M. Amberg, C. M. Rice, and M. Lobigs. Submitted for publication.
- 17.Lin C, Amberg S M, Chambers T J, Rice C M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993;67:2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin C, Chambers T J, Rice C M. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993;192:596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, Lindenbach B D, Prágai B, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach B D, Rice C M. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Tackney C, Bhat R A, Prince A M, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657–662. doi: 10.1128/jvi.71.1.657-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion requires the function of the viral proteinase NS3. Proc Natl Acad Sci USA. 1993;90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989;63:3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 25.Martoglio B, Graf R, Dobberstein B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 1997;16:6636–6645. doi: 10.1093/emboj/16.22.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason P W, Pincus S, Fournier M J, Mason T L, Shope R E, Paoletti E. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology. 1991;180:294–305. doi: 10.1016/0042-6822(91)90034-9. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima H, Hijikata H, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 29.Murphy F A. Togavirus morphology and morphogenesis. In: Schlesinger R W, editor. The togaviruses: biology, structure, replication. New York, N.Y: Academic Press; 1980. pp. 241–316. [Google Scholar]
- 30.Nestorowicz A, Chambers T J, Rice C M. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication and evidence for alternative processing of the NS2A protein. Virology. 1994;199:114–123. doi: 10.1006/viro.1994.1103. [DOI] [PubMed] [Google Scholar]
- 31.Nowak T, Färber P M, Wengler G, Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- 32.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–960. [Google Scholar]
- 33.Rice C M, Grakoui A, Galler R, Chambers T J. Transcription of infectious yellow fever virus RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989;1:285–296. [PubMed] [Google Scholar]
- 34.Rümenapf T, Unger G, Strauss J H, Thiel H-J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67:3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato T, Takamura C, Yasuda A, Miyamoto M, Kamogawa K, Yasui K. High-level expression of the Japanese encephalitis virus E protein by recombinant vaccinia virus and enhancement of its extracellular release by the NS3 gene product. Virology. 1993;192:483–490. doi: 10.1006/viro.1993.1064. [DOI] [PubMed] [Google Scholar]
- 38.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 39.Schalich J, Allison S L, Stiasny K, Mandl C W, Kunz C, Heinz F X. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speight G, Westaway E G. Carboxy-terminal analysis of nine proteins specified by the flavivirus Kunjin: evidence that only the intracellular core protein is truncated. J Gen Virol. 1989;70:2209–2214. doi: 10.1099/0022-1317-70-8-2209. [DOI] [PubMed] [Google Scholar]
- 41.Stocks C E, Lobigs M. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J Virol. 1995;69:8123–8126. doi: 10.1128/jvi.69.12.8123-8126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stocks C E, Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J Virol. 1998;72:2141–2149. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamshchikov V F, Compans R W. Formation of the flavivirus envelope: role of the viral NS2B-NS3 protease. J Virol. 1995;69:1995–2003. doi: 10.1128/jvi.69.4.1995-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamshchikov V F, Compans R W. Processing of the intracellular form of the West Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J Virol. 1994;68:5765–5771. doi: 10.1128/jvi.68.9.5765-5771.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamshchikov V F, Compans R W. Regulation of the late events in flavivirus protein processing and maturation. Virology. 1993;192:38–51. doi: 10.1006/viro.1993.1006. [DOI] [PubMed] [Google Scholar]
- 46.Yamshchikov V F, Trent D W, Compans R W. Upregulation of signalase processing and induction of prM-E secretion by the flavivirus NS2B-NS3 protease: roles of protease components. J Virol. 1997;71:4364–4371. doi: 10.1128/jvi.71.6.4364-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi S-I, Ichikawa M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]