Visual Abstract
Key Words: azidonorleucine (ANL), mass spectrometry, methionyl-tRNA synthetase (MetRS), myocardial infarction, newly synthesized protein
Highlights
-
•
A mouse model was designed for identifying and visualizing newly synthesized proteins in cardiomyocytes in vivo.
-
•
Proteins associated with sarcomeres and mitochondria are synthesized at a high level and exhibit high dynamics in cardiomyocytes.
-
•
Myocardial infarction decreases the protein synthesis in cardiomyocytes in the infarcted zone.
-
•
Cardiomyocytes in the infarcted zone but are still perfused display increased mRNA expression but decreased protein synthesis, suggesting a decoupling of transcription and translation 24 hours after myocardial infarction.
Summary
Gene expression involves transcription, translation, and mRNA and protein degradation. Advanced RNA sequencing measures mRNA levels for cell state assessment, but mRNA level does not fully reflect protein level. Identifying heart cell proteomes and their stress response is crucial. Using a cardiomyocyte-specific mouse model, we tracked protein synthesis after myocardial infarction. Our results showed that myocardial infarction suppresses protein synthesis and unveils a decoupling of translation and transcription regulation in cardiomyocytes.
The leading cause of global mortality is heart failure, primarily from myocardial infarction (MI).1,2 After MI, the heart undergoes extensive myocardial remodeling characterized by the accumulation of fibrosis, cardiac inflammation, and hypertrophic growth, which distorts the heart’s structure, increases heart stiffness, and leads to ventricular dysfunction.3 Cardiomyocytes (CMs) are the cells that provide contractile function in the heart and respond to the altered environment after MI by initiating a complex series of transcriptional, signaling, structural, electrophysiologic, and functional alterations. The CM response to the pathophysiologic stress of MI regulates CM death,4 hypertrophic growth,5 metabolic shifts,6,7 electrophysiologic remodeling,8,9 and sarcomere reorganization.10, 11, 12 Understanding the underlying mechanisms of the CM stress response and developing therapeutic strategies to intervene in this process may beneficially reverse remodeling after MI and improve cardiac function.
Gene expression comprises several steps, including transcription, translation, and degradation of messenger RNAs and proteins. The emergence of advanced RNA-sequencing technologies has enabled accurate measurement of mRNA expression and the use of mRNA profiles to evaluate cell conditions.13 Yet it is important to note that mRNA levels do not entirely mirror protein levels.14,15 It is critical to identify the proteomes of specific cell types of the heart and understand how they change in the stress response. To this end, bio-orthogonal and synthetic biology strategies based on metabolic precursors have been developed to label proteins or introduce measurable protein modifications.16, 17, 18 For example, methionine tRNA can be charged with the methionine surrogate azidonorleucine (ANL) by expressing a methionyl-tRNA synthetase (MetRS) with an expanded amino acid binding site (MetRS L274G, hereafter referred to as MetRS∗).19 By controlling the transcription of a gene encoding MetRS∗ with cell type–specific promoters, one can direct the cell-type–specific incorporation of ANL into nascent polypeptides, which can then be tagged by means of a Click chemistry reaction and imaged or isolated with the use of affinity purification.20
Immediately after MI, millions of CMs die and are not replenished. The dead CMs trigger the inflammatory response, which recruits immune cells and facilitates fibrosis. To respond to the changing microenvironment, CMs undergo a remodeling process, including limited cell cycle entry,21,22 excitation-contraction coupling alternation,23,24 metabolic shift from oxidative phosphorylation to glycolysis,25 and hypertrophic growth.26 Here, we engineered a mouse line in which the conditional expression of MetRS∗ is controlled by the cardiac-specific promoter αMHC-MerCreMer (MCM) Cre recombinase. This approach enables the metabolic labeling of newly synthesized proteins in CMs in vivo. In mice that underwent a sham procedure or MI, we characterized the proteome to determine the molecular factors that are up-regulated or down-regulated in response to MI. Our results showed that, at homeostasis, proteins associated with sarcomeres and metabolism are synthesized at a high level in CMs. After MI, the levels of newly synthesized proteins related to mitochondrial metabolism are substantially decreased in CMs, while proteins associated with mitogen-activated protein kinase (MAP kinase) activity increase. Furthermore, we observed that MI-induced genes are concentrated in CMs localized in the infarct area, and these CMs showed decreased protein synthesis, indicating a separation between the control of transcription and translation after MI.
Methods
Mice
Mouse studies were performed following the Institutional Animal Care and Use Committee at Baylor College of Medicine (Houston, Texas). Fluorescent noncanonical amino acid tagging (FUNCAT) mice (floxed-STOP-MetRS∗; cat. 028071) were purchased from the Jackson Laboratory. Cardiac-specific MetRS∗ mice were generated by crossing FUNCAT mice with MCM Cre mice. To activate Cre recombinase in CMs, we intraperitoneally injected MCM/+;MetRS∗/+ mice with 3 doses of tamoxifen (50 mg/kg body weight per injection). For labeling newly synthesized proteins for mass spectrometry (MS) and immunofluorescence staining, ANL (200 mmol/L, 5 mL/kg body weight) was administered intraperitoneally immediately after the mice underwent sham surgery or MI. The same dose of ANL was injected 12 hours later. Twenty-four hours after the first ANL injection, mice were killed and their hearts harvested and snap-frozen in liquid nitrogen or fixed with 10% formalin overnight until future use.
Left anterior descending coronary artery occlusion
MI was induced on 8–12-week-old MCM/+;MetRS∗ mice by left anterior descending coronary artery (LAD) occlusion as previously described.27 Briefly, Nylon sutures (8-0 nonabsorbable) were used to occlude the LAD. Proper occlusion was marked by blanching of the myocardium seen on visual inspection during tissue collection. Vicryl sutures (6-0 absorbable) were used to close the thoracic cavity. The entire procedure required approximately 12 minutes from the onset of hypothermia to recovery. Sham procedures excluded placement of a suture around the LAD.
Bio-orthogonal noncanonical amino acid tagging enrichment
Frozen heart samples were homogenized for 20 seconds in 1% sodium dodecyl sulfate with phosphate-buffered saline solution (PBS). Lysates were heated at 75 °C for 15 minutes and then centrifuged at 4 °C for 10 minutes. The supernates were separated, and the protein concentrations were determined with the use of Ionic Detergent Compatibility Reagent for Pierce 660nm Protein Assay Reagent. For Click chemistry labeling, 50 μL (∼200 μg) protein was incubated with 100 μL PBS, 4 μL alkyne-biotin (1 mg/mL; TA105-5, Click Chemistry Tools), 10 μL tris-hydroxypropyltriazolylmethylamine (THPTA) (100 mmol/L; 1010-100, Click Chemistry Tools), 10 μL CuSO4 (20 mmol/L), and 10 μL sodium ascorbate (300 mmol/L; A4034, Sigma-Aldrich) at 4 °C overnight. The biotin-labeled protein could be used directly for Western blotting or further purified with the use of methanol-chloroform. To enrich the biotin-labeled proteins, the purified protein samples were incubated with NeutrAvidin beads (29200, ThermoFisher) for immunoprecipitation (IP). After IP, proteins were eluted for Western blotting or MS. Antibodies used for Western blotting were as follows: mouse anti–troponin T (MS-295, ThermoFisher), rabbit anti–α-actinin (ab68167, Abcam), mouse anti–α-actin (A2172, Sigma-Aldrich), rabbit anti-GAPDH (2118S, Cell Signaling), and mouse anti-biotin (SAB4200680, Sigma-Aldrich).
Fluorescent noncanonical amino acid tagging
After fixation, hearts were dehydrated in alcohol gradients, treated with xylene, and embedded in paraffin for sectioning. Slides were sectioned at 7-μm intervals for immunofluorescence staining. For FUNCAT labeling, we used the Click-it EdU Imaging Kit (C10646, Thermo Fisher Scientific) for the Click chemistry reactions by replacing the Alexa Fluor picolyl azide dye with TAMRA-alkyne (900932, Sigma-Aldrich). Then, the slides were co-stained with other antibodies for immunofluorescence staining. The following antibodies were used: rabbit anti-Rab8b (ab222017, Abcam) and rabbit anti-Zc3hav1 (ab154680, Abcam).
Liquid chromatography and MS analysis
The affinity-purified samples were resolved on NuPAGE 10% Bis-Tris Gel (Life Technologies), and the gel pieces were processed for in-gel digestion with the use of Trypsin enzyme. Each lane was excised into 5 equal pieces and combined to make 3 peptide pools. The tryptic peptides were dissolved in 12 μL 5% methanol containing 0.1% formic acid buffer and analyzed with the use of a nanoflow Easy-nLC-1000 system coupled to an Orbitrap Fusion (Thermo Fisher Scientific) MS. The peptides were loaded on a pre-column (2 cm × 100 μm inner diameter) and separated on an in-line 20 cm × 75 μm inner diameter column (ReproSil-Pur 120 C18-AQ, Dr Maisch HPLC). The heated column was maintained at 60 °C. Peptide separation was done at a flow rate of 200 nL/min over a 110-minute gradient time with different concentrations of solvent B (2%-30% for 86 minutes, 30%-60% for 6 minutes, 60%-90% for 8 minutes, and hold at 50% for 10 minutes). The MS was operated in a top 30 data-dependent mode. The MS1 scan was collected in Orbitrap (120,000 resolution, 300-1,400 m/z, 50 ms injection time), followed by the MS2 scan in IonTrap (higher-energy collisional dissociation 30%, rapid scan, 30 ms injection time). The dynamic exclusion was set to 5 seconds. The raw data were searched by with the use of Proteome Discoverer 2.0 (Thermo Scientific) with Mascot 2.4 (Matrix Science) against the National Center for Biotechnology Information RefSeq mus musculus database (updated March 24, 2020). The precursor mass tolerance was confined to 20 ppm, and the fragment mass tolerance to 0.5 daltons. A maximum of 2 missed cleavages with the enzyme Trypsin/P was allowed. The dynamic modification of oxidation on methionine, protein N-terminal acetylation, and destreak on cysteine was allowed. The peptide spectrum match output file was used for grouping at the gene level, and the area quantification was performed as described previously.28
SoRa super-resolution imaging
Confocal imaging was carried out on a Zeiss LSM 880 with an Airysacn FAST Confocal Microscope at the Optical Imaging and Vital Microscopy Core Laboratory of the Baylor College of Medicine.
SoRa super-resolution imaging was performed on a Yokagowa SoRa super-resolution spinning disk microscope equipped with a ×63/1.4 oil immersion objective lens in the Confocal Imaging Core Laboratory at Cincinnati Children’s Hospital Medical Center. Excitation wavelengths were 405 nm, 488 nm, 561 nm, and 642 nm with exposure time 50-200 ms depending on the signal strength. Fluorescence images were collected with the use of appropriate filters.
Single-nucleus and spatial RNA sequencing analysis
Published snRNA-seq29 and spatial transcriptomics (ST) data29,30 were downloaded and the original cell or spot-level meta data, such as cell-type annotations, were directly used for analyses without additional modification. For the ST data, we performed the integration of 5 sections across 2 data sources. We filtered the spots based on the same criteria and log-normalized all unique molecular identifier (UMI) counts using the same parameters to ensure comparability across sections. We labeled remote, border, and ischemic zones within the tissue section based on the expression of zone-specific gene sets reported in the original paper by Calcagno et al.29 These include 57 genes for the border zone (eg, Nppa, Myh7), 50 genes for the remote zone (eg, Ckm, Cox7a1), and 50 genes for the ischemic zone (eg, Spp1, Apoe). All gene set expression z-scores were calculated with the use of the AddModuleScore function provided by the Seurat V4 package using default parameters.
Statistical analysis
The gpGrouper algorithm was used to calculate peptide area based on MS1 expression estimates, also called intensity-based absolute quantification (iBAQ). The “median” normalized iBAQ values were used for data analysis. The differential expression of proteins was calculated using the moderated t-test to determine P values and log2 fold changes in the R package limma. The false discovery rate–corrected P value was calculated by means of the Benjamini-Hochberg procedure. Reproducibility among replicate proteome data is tested using 1-way random intraclass correlation. Gene set enrichment analysis was performed with the use of the ClusterProfiler package in R. Canonic pathway gene sets derived from Kyoto Encyclopedia of Genes and Genomes and Gene Ontology terms were used for the enrichment analyses. Quantitative data are presented as mean ± SEM with individual data points plotted and compared using 1-way or 2-way repeated-measures analysis of variance to consider that multiple samples were analyzed from a given mouse. Quantitative data for the 2 groups were statistically evaluated by means of the Wilcoxon signed-rank test.
R (version 4.3.2) and several packages were used for data processing and visualization. Specifically, Seurat (v5.0.1) was used to normalize the single cell and ST data. DEseq2 (version 1.42.0) was used for principal component analysis of the protein expression data. And ClusterProfiler (version 4.10.0) was used for gene set and gene ontology enrichment analyses.
Results
Label the newly synthesized proteins in vivo
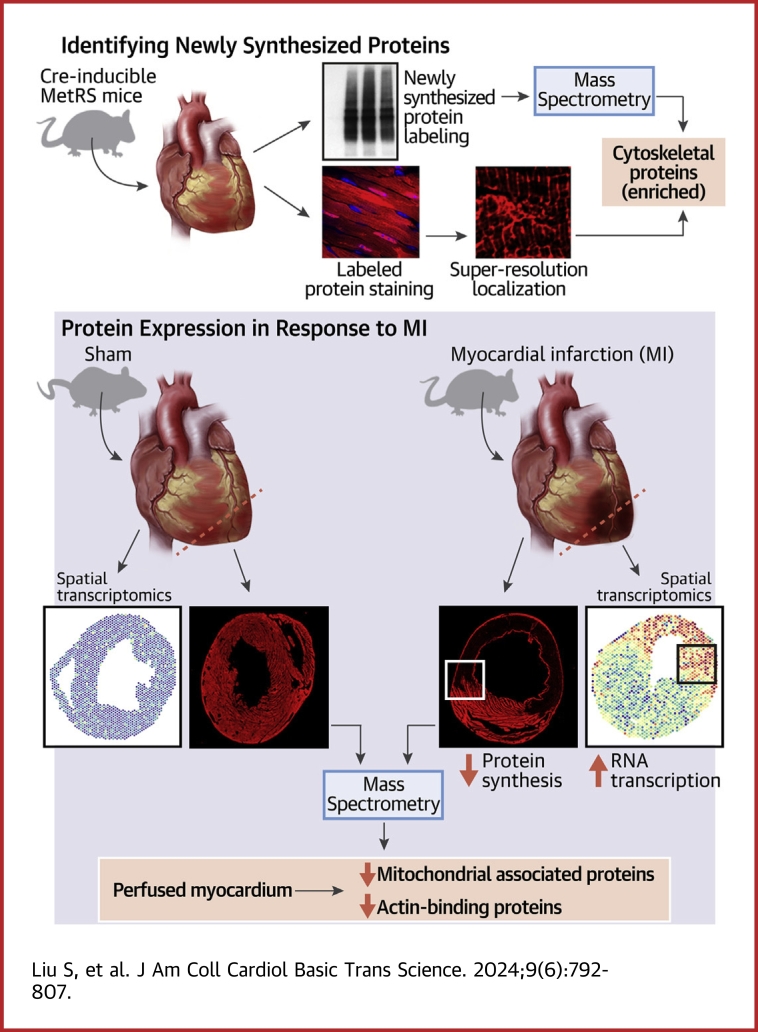
To label newly synthesized proteins in CMs, we used transgenic mice expressing the floxed-STOP-MetRS∗ transgene and crossed them with MCM Cre mice to enable the CM-specific expression of MetRS∗ in CMs. In the ribosome of these mice, MetRS∗ incorporated the methionine surrogate ANL, a noncanonical amino acid, into newly synthesized peptides (Figure 1A). ANL has a structure that is slightly different from methionine and is not recognized by endogenous MetRS. Therefore, labeling occurs only in cells with MetRS∗ expression. Importantly, ANL carries a reactive azide group, which enables alkyne to be covalently attached to ANL with the use of copper-catalyzed azide-alkyne ligation (bio-orthogonal, noncanonical amino acid tagging [BONCAT]) for downstream applications (Figure 1A).
Figure 1.
CM-Specific Nascent Protein Labeling in Mice
(A) Schematic showing how the translational machinery labels newly synthesized proteins. (B) Western Blotting shows the newly synthesized proteins labeled by alkyne biotin, which could also be pull-down by the biotin beads for enrichment. Samples were harvested 24 hours after ANL injection. (C) TAMRA-alkyne labels ANL in the newly synthesized proteins. ANL indicates the signal of newly synthesized protein, arrowheads indicate the ANL signal in the nuclei, and arrows indicate the ANL signal in the intercalated discs. Note that there is no ANL signal in non-CMs (star). Scale bar = 10 μm. (D) Experimental design for testing the half-life of newly synthesized proteins in vivo. To activate Cre recombinase in CMs, we intraperitoneally injected MCM/+;MetRS∗/+ mice with 3 doses of tamoxifen (TAM). For the labeling of newly synthesized proteins, ANL was administered intraperitoneally. Mice were killed and their hearts removed on days 1, 2, 3, or 5 after injection. (E) Newly synthesized proteins detected by Western blotting, with quantification of protein levels for (F) input and (G) biotin-IP. Data are presented as mean ± SEM and were compared by means of 1-way repeated-measures analysis of variance (ANOVA), n = 3 per time point. (H) ANL-labeled proteins in CMs from hearts 1 day and 4 days after a single ANL injection. White arrowheads indicate the nucleus ANL signal, and yellow arrowheads indicate the sarcomeric ANL signal. Scale bar = 5 μm. ANL intensity was quantified in (I). Data are presented as the mean ± SEM and were compared by means of Wilcoxon signed-rank test; the ANL intensity was measured in a total of 90 CMs from each group (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. ANL = azidonorleucine; CM = cardiomyocyte; DAPI = 6-diamino-2-phenylindole; WGA = wheat germ agglutinin.
We initiated the induction of MetRS∗ expression in CMs of 3–4-month-old MCM/+;MetRS∗/+ mice by administering 3 doses of tamoxifen. To mitigate potential side-effects associated with tamoxifen injection, we allowed a 1-week interval before introducing ANL via IP injection. Previous studies have indicated that maximal ANL labeling occurs approximately 16 hours after ANL injection in hippocampal neurons.31 Accordingly, we removed the hearts 24 hours after ANL injection to reach the maximal labeling and conducted protein labeling assessments in CMs. Through Western blotting analysis, we successfully detected biotin-labeling proteins in the heart lysates obtained from MCM/+;MetRS∗/+ mice that were injected with ANL (Figure 1B). In addition, we further enriched the labeled proteins via biotin-immunoprecipitation (Figure 1B). To visualize the localization of newly synthesized proteins, we used TAMRA-alkyne, a fluorescent dye that selectively binds to the ANL by Click chemistry. Twenty-four hours after the ANL injection, we observed a TAMRA fluorescent signal in CMs, particularly concentrated at the intercalated disc and nuclei of CMs. The ANL intensity varies in each CMs but is not dependent on its distance to the vessels (Supplemental Figure 1A). However, no fluorescence signal could be detected in non-CMs of MCM/+;MetRS∗/+ mice (Figure 1C).
The half-life of ANL-labeled newly synthesized proteins
As a proof of concept, we first used this model to test the half-lives of newly synthesized proteins in CMs during homeostasis. One week before injection with ANL, we injected MCM/+;MetRS∗/+ mice with 3 doses of tamoxifen to induce MetRS∗ expression in CMs. Then, a single dose of ANL was injected for the pulse labeling of newly synthesized proteins. After ANL injection, we removed mouse hearts 1, 2, 3, and 5 days later and measured the levels of labeled proteins in CMs with the use of Western blotting (Figures 1D and 1E). We detected the highest level of protein labeling 1 day after ANL injection, when most of the proteins were labeled. The levels of labeled protein gradually decreased with time owing to endogenous protein turnover. Five days after ANL injection, only a small portion of the long-lived proteins could be detected, and other proteins showed a half-life of the turnover rate of about 2 days (Figures 1E to 1G). Consistently, immunofluorescence staining revealed a 3-fold decrease in ANL-labeled proteins 4 days after ANL injection compared with the samples 1 day after ANL injection (Figures 1H and 1I; Supplemental Figure 1B). The most decreased ANL-labeled proteins primarily localized in the nucleus, while the cytoplasmic became more diffused, resulting in the loss of the sarcomere pattern observed in CMs 1 day after ANL injection (Figure 1H).
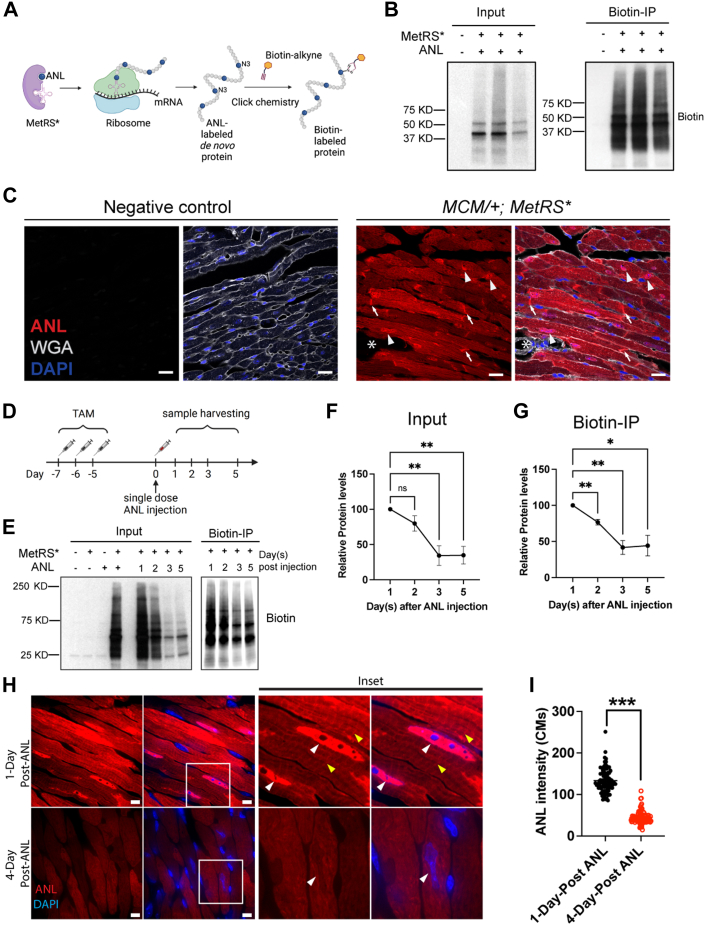
Determining the ANL-labeled newly synthesized proteins by mass spectrometry
To examine the proteome and identify newly synthesized proteins in CMs in vivo, we performed biotin-IP MS analysis. Various ANL labeling periods, such as 10 or 21 days, have been previously tested.32, 33, 34 However, given our hypothesis that many CM proteins are short-lived, we decided to focus on a short-term labeling protocol. To examine nascent protein synthesis without bias, we removed mouse hearts 1 day after ANL injection (Figure 2A). Using MS, we identified an approximate average of 3,000 proteins from each sample and 1,500 proteins with high confidence. After background normalization, 1,029 newly synthesized proteins were identified in CMs from sham-treated hearts (Figure 2B, Supplemental Table 1). The distribution pattern of protein size was similar to the publicly available entire mouse proteome (Supplemental Figure 2). Gene Ontology analysis showed that most of the proteins were associated with the molecular functions of actin binding, adenosine triphosphate hydrolysis activity, oxidoreduction-driven transmembrane transporter activity, RNA binding, and nicotinamide adenine dinucleotide (NADH) dehydrogenase activity (Figure 2C). These proteins are components of the mitochondrial protein-containing complex, myofibril, sarcomere, and respiratory chain complex (Figure 2D). The most abundant proteins in CMs were components of the sarcomere structure, such as ACTC1, ACTA1, MYH6, TPM1, and DES, and components of the tricarboxylic acid cycle and respiratory electron transport, such as ATP5A1, ATP5B, MDH2, ACO2, and IDH2. We also identified some ion channel proteins with increased expression, such as Vdac1, Vdac-2, and Vdac-3, which localize at the outer mitochondrial membrane and are essential for mitochondrial function (Figures 2B and 2E).
Figure 2.
Identifying Newly Synthesized Proteins in CMs With Mass Spectrometry
(A) Schematic showing the workflow for labeling, detecting, and identifying newly synthesized proteins specifically in CMs with the use of mass spectrometry. (B) Proteins enriched in CMs from MCM/+;MetRS∗ mice compared with CMs from negative control mice. (C, D) Results of Gene Ontology analysis of the newly synthesized proteins in CMs at homeostasis. (E) Heat map showing protein levels in the representative categories. (F, G) Validation with biotin-immunoprecipitation (IP) showing the enrichment of cardiac troponin T (cTNT) and cardiac α-actinin protein expression in newly synthesized proteins. (H) Time-course ANL pulse labeling of newly synthesized sarcomeric α-actin and GAPDH at different time points after ANL injection. (I) Superresolution images show the fluorescent noncanonical amino acid tagging labeling of newly synthesized proteins in situ. Arrowheads indicate the nucleus. Scale bars = 5 μm. ANL = azidonorleucine; CM = cardiomyocyte; DAPI = 6-diamino-2-phenylindole; WGA = wheat germ agglutinin.
For validation, we enriched the newly synthesized proteins by performing biotin-IP and determined the expression of the full length of 2 cardiac-specific proteins, cardiac troponin T and sarcomeric α-actinin, as well as α-actin and GAPDH with the use of Western blotting (Figures 2F to 2H). Time-course pulse labeling revealed the turnover rate of sarcomere α-actin and GAPDH, both half-lives being around 2 days (Figure 2H). Using TAMRA-alkyne, we labeled the newly synthesized proteins in situ. In agreement with the MS results, the newly synthesized proteins localized primarily in the intercalated discs, sarcomeres, and nuclei (Figure 2I, Supplemental Figure 3). We inferred that these proteins are highly dynamic in CMs during homeostasis, suggesting the fast turnover rate of these proteins in the sarcomere and metabolic processes.
Decreased protein synthesis in CMs in the border zone
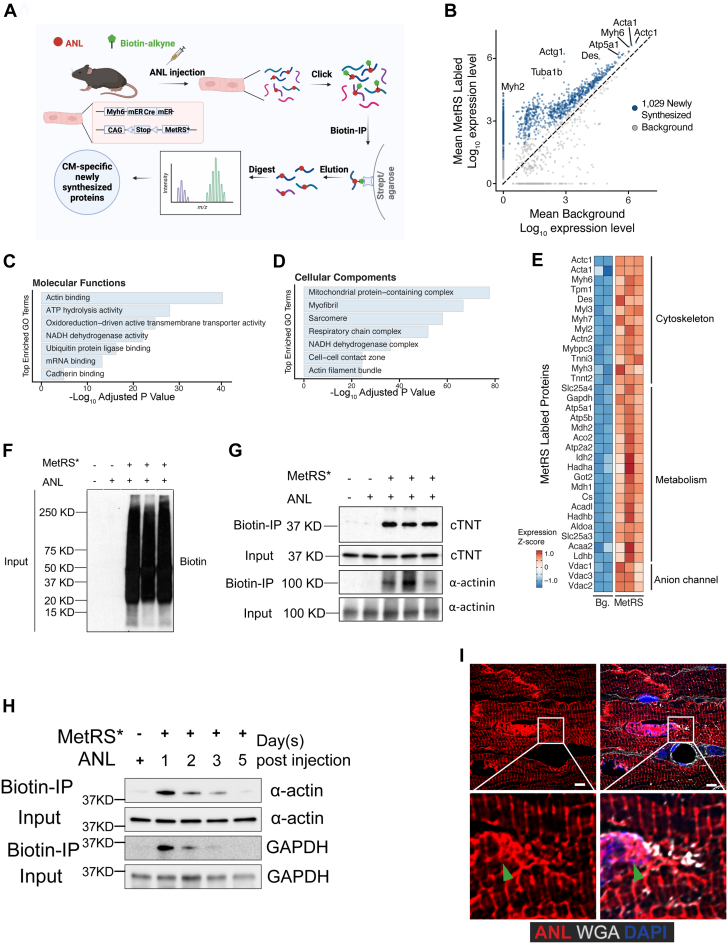
Subsequently, we proceeded to investigate protein synthesis in CMs following MI. Using immunofluorescence staining, we were able to visualize protein synthesis in CMs at the different regions (Figures 3A and 3B). In mice that underwent a sham procedure, the ANL signal appeared evenly distributed among CMs, suggesting that most CMs produced proteins at a similar level. Interestingly, 24 hours after MI, immunofluorescence staining revealed a substantial area of CMs lacking the ANL signal (Figures 3A and 3B). Despite exhibiting normal cellular morphology, CMs within this area were either not adequately perfused or experienced inhibition in the translation of new peptides due to the challenging microenvironment after MI. Consequently, we defined this region as the infarct area (Figure 3B). Furthermore, we observed that a few layers of CMs situated at the subepicardium and subendocardium of both the left and right ventricles, in close proximity to the infarct areas, exhibited the expression of newly synthesized proteins. Notably, these CMs were distinctly separated from the CMs within the infarct area (Figures 3A and 3B). By combining the immunofluorescence staining, we observed the CMs localized in the infarct area also lack the expression of the highly synthesized proteins, such as cardiac troponin T and α-actin, indicating that the protein synthesis is disrupted in those CMs (Figures 3C and 3D). In addition, we identified some CMs adjacent to the infarct area that displayed a decreased ANL signal, indicating decreased protein synthesis in those CMs (Figures 3E and 3F). Similarly, in a model where we injected ANL 6 hours before the MI, with all CMs receiving ANL, the CMs localized in the infarcted area still showed decreased ANL intensity (Supplemental Figures 4A and 4B). Together, these results suggest that MI alters protein synthesis in CMs in a regional-dependent manner.
Figure 3.
Visualization of Newly Synthesized Proteins in the Hearts of Mice That Underwent a Sham Procedure or MI
(A, B) Immunofluorescence staining at (B) increasing magnifications showing the ANL-labeled newly synthesized proteins in CMs of mice that underwent a sham procedure or MI. Dark areas in the sections are the ischemic zones after MI. Scale bar = 500 μm in A, 100 μm in B. (C, D) Immunofluorescence staining show the expression of cardiac troponin T (cTNT), sarcomeric α-actin, and ANL in border zone CMs. Scale bar = 10 μm. (E) ANL staining in border zone CMs. Arrowheads indicate CMs with decreased ANL signal. Scale bar = 10 μm. (F) Quantification of ANL intensity in CMs of sham and MI hearts as shown in E (80 CMs in each group from 4 biological mouse hearts). Data are presented as mean ± SEM and were compared by means of the Wilcoxon signed-rank test. ∗∗∗P < 0.001. ANL = azidonorleucine; CM = cardiomyocyte; DAPI = 6-diamino-2-phenylindole; LV = left ventricle; MI = myocardial infarction; NC = negative control (heart sample from mice without ANL injection); RV = right ventricle WGA = wheat germ agglutinin.
Identifying the differentially expressed proteins after MI
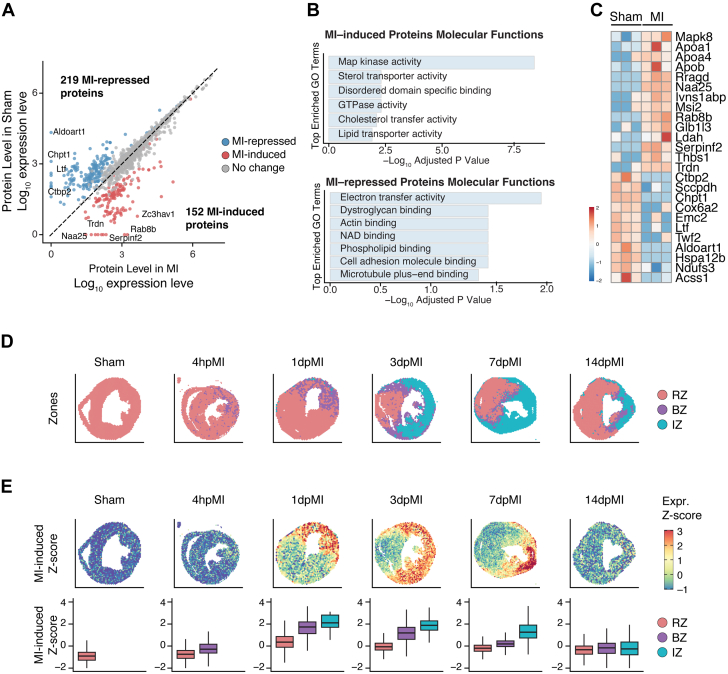
We were interested in how the survived CMs near the infarct area respond to MI through protein synthesis. To identify the proteome of the CMs after MI, we extracted the proteins from hearts of MCM/+;MetRS∗/+ mice treated with sham or MI and performed biotin-IP followed by MS analysis. Our results revealed similar enrichments of the newly synthesized proteins from hearts treated with sham or MI (Supplemental Figure 5A). Principal-component analysis of the proteomics data indicated that MI disease status had the greatest influence on protein expression variance (Supplemental Figure 5B) and that the sex of the mice did not significantly contribute to the variance (Supplemental Figure 5C). Pairwise Pearson correlation between the sham and MI group replicates demonstrated strong reproducibility across the proteome (Supplemental Figure 5D). We conducted a comparative analysis of proteome data from MCM/+;MetRS∗/+ mice that underwent MI compared with the corresponding MCM/+;MetRS∗/+ mice that underwent a sham procedure. Our results identified 152 proteins that had increased synthesis (termed MI-induced) and 219 proteins that had decreased synthesis (termed MI-repressed) in the post-MI CMs compared with the sham (Figure 4A, Supplemental Table 2). The differentially expressed proteins that had increased levels in CMs after MI were associated with MAP kinase activity, sterol transporter activity, GTPase activity, and lipid metabolism (Figures 4A and 4B). The proteins that had decreased levels in CMs after MI were associated with mitochondrial oxidative phosphorylation and NADH dehydrogenase activity, such as NDUFA3 and COX6A2. In addition, the expression of some protein kinase–binding proteins, such as CTBP2 and TWF2, was reduced in CMs after MI (Figures 4A to 4C). The protein that decreased the most in CMs after MI was ALDOART1, which encodes an enzyme named aldolase 1A, retrogene 1. This enzyme is related to glycolysis, suggesting that MI may decrease glycolysis in a subset of CMs (Figures 4A and 4C). The most significantly increased proteins in CMs after MI were related to lipid remodeling, such as APOA1 and APOA4; and GTPase activity, such as RAB8B, and RRAGD (Figures 4A and 4C).
Figure 4.
Mass Spectrometry Analysis of Newly Synthesized Proteins in the Hearts of Mice That Underwent a Sham Procedure or MI
(A) Differentially expressed newly synthesized proteins in CMs 24 hours after mice underwent MI vs a sham procedure. The proteins with the most significant differential expression are specified. (B) Gene Ontology (GO) analysis of the differentially expressed proteins in the CMs of mice that underwent MI vs a sham procedure (GO terms shown for decreased proteins or increased proteins, as indicated). (C) Heat map showing differentially expressed proteins in the CMs of mice that underwent MI vs a sham procedure. (D) Spatial transcriptomics analysis of spatial feature plots for the remote zone (RZ), border zone (BZ), and ischemic zone (IZ) in adult hearts at 4 hours, 1 days, 3 days, 7 days, and 14 days after MI (pMI). (E) Z-score shows the spatial pattern of MI-induced protein genes. CM = cardiomyocyte; MI = myocardial infarction.
Transcriptional and translational decoupling in CMs after MI
Next, we asked whether the protein synthesis revealed by proteomics data can be recapitulated at the transcriptional level. We examined published single-nucleus RNA-sequencing data29 and observed that the protein genes induced and repressed by MI were predominantly enriched in CMs, which aligned with our proteomic results derived from a CM-specific mouse model (Supplemental Figure 6). Using published ST data,29,30 we further obtained the spatial information of MI-regulated protein genes. We analyzed a series of 5 ST data sets generated in adult mouse hearts at 4-hour, 1-day, 3-day, 7-day, and 14-day post-MI time points, as well as a sham-operated heart as control.29,30 Using published marker genes, we were able to annotate the remote zone, border zone, and ischemic zone on all sections based on their transcription signatures (Figure 4D).29 Consistently with our proteomics data, the MI-induced genes displayed a robust expression starting at 1 day after MI, with particularly high levels in the border and ischemic zones (Figure 4E). This expression pattern persisted at 3 days after MI and peaked in the ischemic zone at 7 days after MI. After 2 weeks, the expression of MI-induced protein genes returned to a near baseline level across the zones (Figure 4E). These results showed increased mRNA transcription but decreased overall protein synthesis in CMs within the infarct area, suggesting a decoupling of transcriptional and translational regulation in these CMs after MI.
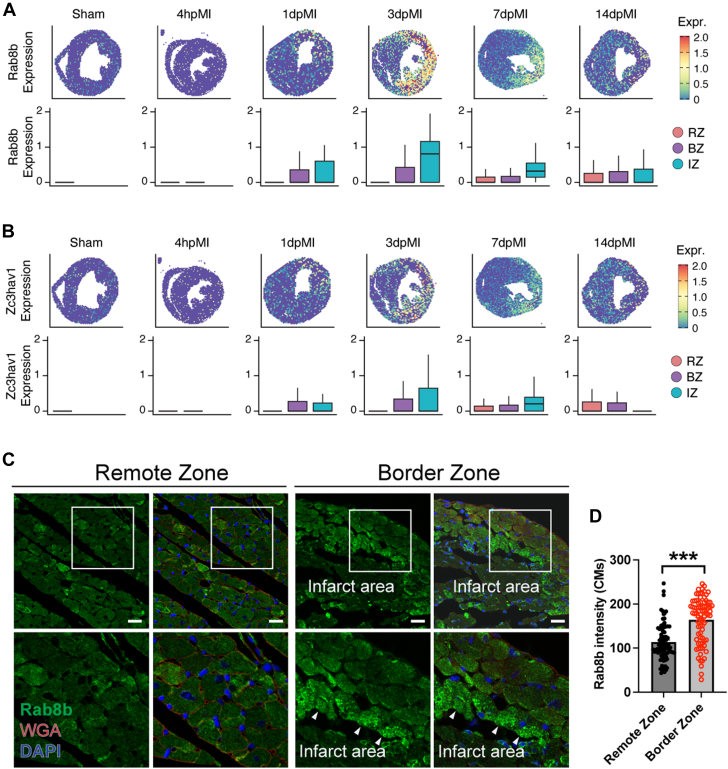
To further validate the expression of the newly synthesized proteins in CMs, we performed immunofluorescence co-staining for ANL with RAB8A and ZC3HAV1, the 2 proteins with the most increased expression in CMs 24 hours after MI (Supplemental Figure 7). RAB8B is a small GTPase that regulates intracellular vesicle transportation,35 and ZC3HAV1 is a CCCH-type zinc finger protein that plays an essential role in the innate immune response against multiple DNA and RNA viruses.36 Immunofluorescence staining showed increased expression of RAB8B and ZC3HAV1 and their co-localization with ANL staining in border zone CMs (Supplemental Figure 7). In accordance with the result that MI-induced genes are significantly concentrated in the infarct area, the mRNA levels of both Rab8b and Zc3hav1 were observed to be higher in the ischemic zone (Figures 5A and 5B). In contrast, we observed elevated RAB8B protein in border zone CMs but not in the remote zone CMs, suggesting a separation of transcriptional regulation and translational regulation in CMs within the infarct area (Figures 5C and 5D). Interestingly, the border zone CMs exhibited more prominent punctate-like RAB8B signals than the CMs in the remote zone, indicating a higher activity level of intracellular vesicle transportation (Figure 5C). Taken together, these findings suggest that the MCM/+;MetRS∗ mouse model is valuable for identifying newly synthesized proteins in CMs under various disease conditions.
Figure 5.
Validation of RAB8B Expression in the Heart After the Treatment of MI
(A, B) Spatial transcriptomics analysis of Rab8b and Zc3hav1 expression at different time points as indicated, both are MI-induced genes. (C) Immunofluorescence staining of RAB8B in hearts after MI. Arrowheads indicate CMs with high RAB8B expression in the border zone. Scale bar = 25 μm. (D) Quantification of RAB8B expression in CMs localize at border zone or remote zone. 80 CMs in each group from 4 biological mouse hearts. Data are presented as mean ± SEM and were compared by means of the Wilcoxon signed-rank test. ∗∗∗P < 0.001. BZ = border zone; CM = cardiomyocyte; DAPI = 6-diamino-2-phenylindole; IZ = ischemic zone; MI = myocardial infarction; RZ = remote zone; WGA = wheat germ agglutinin.
Discussion
In response to physiologic or pathologic stress, cells change the proteome and adapt to the environment, either by modifying protein structure via posttranslational modification37,38 or by altering protein composition, which is regulated by protein synthesis and degradation. With the development of advanced RNA-sequencing technologies, the amount of mRNA can be precisely measured, and the mRNA profiles can be used to assess cell states.13 However, mRNA levels may not fully reflect protein levels.14,15 In the present research, we observed an overall diminished protein synthesis in CMs within the infarct area. However, our ST data showed that mRNAs induced by MI are highly enriched in this specific area, suggesting a decoupling of transcriptional and translational regulation after MI. Moreover, a recent study has highlighted the significance of translational regulation in controlling protein activity during the early stage of injury response and regeneration.39 Therefore, to accurately evaluate cell conditions and understand how cells respond to the environment, examining the proteome is essential.
By coupling BONCAT with MS proteomics analysis,16,33,40 we labeled the newly synthesized proteins in CMs in vivo. Under homeostatic conditions, we determined that the most highly expressed proteins, such as ACTC1, ACTA1, MYH6, TPM1, and DES, are associated with sarcomeres, which is consistent with the conclusion that the sarcomere is dynamic and requires constant replenishment for functional maintenance. This phenomenon is consistent with recent studies showing that the assembly and turnover of sarcomere proteins are essential for the contractile function of CMs.11,41, 42, 43, 44 The proteins associated with the tricarboxylic acid cycle and electron transport chain are also highly expressed in CMs. These proteins are required for energy production to supply the constant beating of CMs.
Our ANL pulse labeling experiment indicated that the general approximate protein half-life in CMs is 2-3 days, a slightly shorter duration compared with those reported using other methods, such as heavy water and heavy amino acids labeling.45, 46, 47, 48, 49, 50 Specifically, reports suggest that the half-life of myosin heavy chain is about 5-6 days,46,50 cardiac troponins range from 3.2 days to 5.3 days,47 and titin has a half-life of about 70 hours in the skeletal muscle cells.51 Notably, using deuterium oxide labeling combined with MS, Lau et al found that the half-life of cardiac proteins is about 7.7 days,49 and the protein turnover rates were variable, ranging from <1 day to >3 weeks. Moreover, cytosolic proteins exhibit a half-life of ∼6.5 days, whereas mitochondrial proteins have a half-life of ∼15 days.48 The discrepancies between these studies and ours could stem from different labeling periods (long-term labeling vs short-term labeling) and the isotopes used for the protein labeling (radioactive vs metabolic). Long-term labeling requires mathematical calculation of the radioactivity to determine the protein turnover rate, which is potentially confounded by the radioactivity from the intracellular free isotope pools. It has been reported that calculation on various labeling isotope pools resulted in different turnover rates for myosin, ranging from 2.7 days to 5-6 days.46
With the animal model used in this study, we measured only newly synthesized protein, which contributes to the total proteome that determines the CM state. This allowed us to differentiate the nascent proteomic profile from the total proteome, which is also affected by protein degradation. These newly synthesized proteins represent the actively gained functions of the CMs in response to the altered environment in a real-time manner, enabling a direct evaluation of CM state changes in different contexts. After MI, the most decreased newly synthesized proteins were associated with oxidative phosphorylation in the mitochondria. The activity of NADH dehydrogenase was reduced, which may be the reason for the decreased level of NAD+ in CMs after MI.52, 53, 54 This observation suggests that CMs actively respond to oxygen deficiency after MI by reducing the synthesis of oxygen consumption proteins. This is a good example of how CMs adjust the translational machinery to adapt to the harsh ischemic microenvironment.
Study limitations
The present study has certain limitations. The ANL-immunofluorescence staining showed that newly synthesized proteins localize in the sarcomere and nucleus of CMs, but most of the highly expressed proteins identified with the use of MS are not nuclear proteins. The reason for this discrepancy could be either that our protocol failed to enrich proteins from the nuclear fraction or that there was a lower expression of nuclear proteins. This study specifically focused on determining the newly synthesized proteins in CMs. Owing to technologic limitations, we could only enrich the newly synthesized proteins in which methionine had been replaced with ANL, constituting only a fraction of the total proteome. Conducting MS on the entire proteome would have helped us to evaluate the ratio of newly synthesized proteins more comprehensively. In addition, conducting MS on various fractions, including the nuclear, cytosolic, and mitochondrial fractions, would provide valuable insights into how CMs use translational machinery in response to pathophysiologic stimuli. Based on our immunofluorescence staining data, we inferred that most enriched proteins were from the CMs with high ANL signaling. Given the regional differences that we observed in the myocardium after MI, the possibility remains that the CMs near the ischemic areas were overrepresented, which means we may not have detected the proteomes in the CMs at risk but only the proteomes in the viable CMs. This may be why we did not see proteins associated with necrosis and apoptosis, which are the primary mechanisms for CM death.55,56 We chose the time point of 24 hours after MI, which allowed us to obtain information on how CMs directly respond to injury. In the future, collecting samples at later time points may help us to understand how CMs adapt to the harsh environment after MI. Because cardiac remodeling is a dynamic process, determining the spatial and temporal molecular mechanisms of CM remodeling may aid in developing improved strategies for treating patients with MI-induced heart failure.
Conclusions
In this study, we developed an animal model to identify and visualize the newly synthesized proteins in CMs. Our findings revealed that, under normal conditions, proteins associated with sarcomeres and metabolism are synthesized at a high rate in CMs. After MI, the synthesis of newly synthesized proteins decreases in the CMs in the infarcted area. Proteins associated with mitochondrial metabolism are significantly decreased in CMs, whereas proteins related to MAP kinase activity increase. In addition, we observed that MI-induced genes are concentrated in CMs localized in the infarct area, but these CMs showed decreased protein synthesis, indicating a separation between the control of transcription and translation after MI.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: After myocardial infarction, cardiomyocytes undergo a series of remodeling processes, resulting in structural changes in the heart and deteriorating heart function. In this study, we developed an animal model that enables us to identify and visualize the newly synthesized proteins in cardiomyocytes. We examined the proteomic profile in cardiomyocytes and evaluated how cardiomyocytes respond to myocardial injury in the early stage.
TRANSLATIONAL OUTLOOK: Understanding the molecular mechanisms that drive cardiac remodeling may lead to medical interventions that could potentially delay or prevent adverse cardiac remodeling. We identified proteins induced and repressed in cardiomyocytes in response to myocardial infarction, which could serve as potential targets for drug or gene therapy treatment for heart failure.
Funding Support and Author Disclosures
This study received funding from the National Institutes of Health (NIH; HL 127717, HL 130804, and HL 118761 to Dr Martin) and the Vivian L. Smith Foundation (to Dr Martin). Dr Martin was supported by the LeDucq Foundation’s Transatlantic Networks of Excellence in Cardiovascular Research (14CVD01: “Defining the Genomic Topology of Atrial Fibrillation”), the MacDonald Research Fund Award (16RDM001), and a grant from the Saving Tiny Hearts Society (to Dr Martin). This study was also supported in part by the American Heart Association Postdoctoral Fellowship (18POST34060186) and Career Development Award (849706) to Dr Liu and the BCM Mass Spectrometry Proteomics Core, which is supported by the Dan L. Duncan Comprehensive Cancer Center NIH award (P30 CA125123) and the CPRIT Core Facility Award (RP210227). Dr Martin is a co-founder of and owns shares in Yap Therapeutics. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank Thuy Tien Tran and Ann Bromley for their technical support in the Martin laboratory and Sara Marshall in the Liu laboratory at Cincinnati Children’s Hospital Medical Center. Nicole Stancel, PhD, ELS(D), of the Department of Scientific Publications at the Texas Heart Institute in Houston provided editorial support.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures and a table, please see the online version of this paper.
Contributor Information
Shijie Liu, Email: shijie.liu@cchmc.org.
James F. Martin, Email: jfmartin@bcm.edu.
Appendix
References
- 1.Ambrosy A.P., Fonarow G.C., Butler J., et al. The global health and economic burden of hospitalizations for heart failure: lessons learned from hospitalized heart failure registries. J Am Coll Cardiol. 2014;63(12):1123–1133. doi: 10.1016/j.jacc.2013.11.053. [DOI] [PubMed] [Google Scholar]
- 2.Braunwald E. The war against heart failure: the Lancet lecture. Lancet. 2015;385(9970):812–824. doi: 10.1016/S0140-6736(14)61889-4. [DOI] [PubMed] [Google Scholar]
- 3.Burchfield J.S., Xie M., Hill J.A. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation. 2013;128(4):388–400. doi: 10.1161/CIRCULATIONAHA.113.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyata S., Takemura G., Kawase Y., et al. Autophagic cardiomyocyte death in cardiomyopathic hamsters and its prevention by granulocyte colony-stimulating factor. Am J Pathol. 2006;168(2):386–397. doi: 10.2353/ajpath.2006.050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott P.M., Anastasakis A., Anastasakis A., et al. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35(39):2733–2779. doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 6.Kolwicz S.C., Jr., Purohit S., Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res. 2013;113(5):603–616. doi: 10.1161/CIRCRESAHA.113.302095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuurbier C.J., Bertrand L., Beauloye C.R., et al. Cardiac metabolism as a driver and therapeutic target of myocardial infarction. J Cell Mol Med. 2020;24(11):5937–5954. doi: 10.1111/jcmm.15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuculich P.S., Zhang J., Wang Y., et al. The electrophysiological cardiac ventricular substrate in patients after myocardial infarction: noninvasive characterization with electrocardiographic imaging. J Am Coll Cardiol. 2011;58(18):1893–1902. doi: 10.1016/j.jacc.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendonca Costa C., Plank G., Rinaldi C.A., Niederer S.A., Bishop M.J. Modeling the electrophysiological properties of the infarct border zone. Front Physiol. 2018;9:356. doi: 10.3389/fphys.2018.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kötter S., Kazmierowska M., Andresen C., et al. Titin-based cardiac myocyte stiffening contributes to early adaptive ventricular remodeling after myocardial infarction. Circ Res. 2016;119(9):1017–1029. doi: 10.1161/CIRCRESAHA.116.309685. [DOI] [PubMed] [Google Scholar]
- 11.Martin T.G., Myers V.D., Dubey P., et al. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun. 2021;12(1):2942. doi: 10.1038/s41467-021-23272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avner B.S., Shioura K.M., Scruggs S.B., et al. Myocardial infarction in mice alters sarcomeric function via post-translational protein modification. Mol Cell Biochem. 2012;363(1-2):203–215. doi: 10.1007/s11010-011-1172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwanhäusser B., Busse D., Li N., et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 15.Tebaldi T., Re A., Viero G., et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012;13:220. doi: 10.1186/1471-2164-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieterich D.C., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103(25):9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsässer S.J., Ernst R.J., Walker O.S., Chin J.W. Genetic code expansion in stable cell lines enables encoded chromatin modification. Nat Methods. 2016;13(2):158–164. doi: 10.1038/nmeth.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laughlin S.T., Baskin J.M., Amacher S.L., Bertozzi C.R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320(5876):664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahdavi A., Hamblin G.D., Jindal G.A., et al. Engineered aminoacyl-tRNA synthetase for cell-selective analysis of mammalian protein synthesis. J Am Chem Soc. 2016;138(13):4278–4281. doi: 10.1021/jacs.5b08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngo J.T., Champion J.A., Mahdavi A., et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5(10):715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W.E., Li L., Xia X., et al. Dedifferentiation, proliferation, and redifferentiation of adult mammalian cardiomyocytes after ischemic injury. Circulation. 2017;136(9):834–848. doi: 10.1161/CIRCULATIONAHA.116.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisner D.A., Caldwell J.L., Kistamás K., Trafford A.W. Calcium and excitation-contraction coupling in the heart. Circ Res. 2017;121(2):181–195. doi: 10.1161/CIRCRESAHA.117.310230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez A.M., Guatimosim S., Dilly K.W., Vassort G., Lederer W.J. Heart failure after myocardial infarction: altered excitation-contraction coupling. Circulation. 2001;104(6):688–693. doi: 10.1161/hc3201.092285. [DOI] [PubMed] [Google Scholar]
- 25.Tran D.H., Wang Z.V. Glucose metabolism in cardiac hypertrophy and heart failure. J Am Heart Assoc. 2019;8(12) doi: 10.1161/JAHA.119.012673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehat I., Molkentin J.D. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. 2010;122(25):2727–2735. doi: 10.1161/CIRCULATIONAHA.110.942268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heallen T., Morikawa Y., Leach J., et al. Hippo signaling impedes adult heart regeneration. Development. 2013;140(23):4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saltzman A.B., Leng M., Bhatt B., et al. gpGrouper: a peptide grouping algorithm for gene-centric inference and quantitation of bottom-up proteomics data. Mol Cell Proteomics. 2018;17(11):2270–2283. doi: 10.1074/mcp.TIR118.000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calcagno D.M., Taghdiri N., Ninh V.K., et al. Single-cell and spatial transcriptomics of the infarcted heart define the dynamic onset of the border zone in response to mechanical destabilization. Nat Cardovasc Res. 2022;1(11):1039–1055. [Google Scholar]
- 30.Yamada S., Ko T., Hatsuse S., et al. Spatiotemporal transcriptome analysis reveals critical roles for mechano-sensing genes at the border zone in remodeling after myocardial infarction. Nat Cardovasc Res. 2022;1(11):1072–1083. [Google Scholar]
- 31.Evans H.T., Bodea L.G., Götz J. Cell-specific noncanonical amino acid labelling identifies changes in the de novo proteome during memory formation. Elife. 2020;9 doi: 10.7554/eLife.52990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez-Castelao B., Schanzenbächer C.T., Hanus C., et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat Biotechnol. 2017;35(12):1196–1201. doi: 10.1038/nbt.4016. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Castelao B., Schanzenbächer C.T., Langer J.D., Schuman E.M. Cell-type-specific metabolic labeling, detection and identification of nascent proteomes in vivo. Nat Protoc. 2019;14(2):556–575. doi: 10.1038/s41596-018-0106-6. [DOI] [PubMed] [Google Scholar]
- 34.Azizian N.G., Sullivan D.K., Nie L., et al. Selective labeling and identification of the tumor cell proteome of pancreatic cancer in vivo. J Proteome Res. 2021;20(1):858–866. doi: 10.1021/acs.jproteome.0c00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattula K., Furuhjelm J., Arffman A., Peränen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13(9):3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao G., Guo X., Goff S.P. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 37.Yan K., Wang K., Li P. The role of post-translational modifications in cardiac hypertrophy. J Cell Mol Med. 2019;23(6):3795–3807. doi: 10.1111/jcmm.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aggarwal S., Banerjee S.K., Talukdar N.C., Yadav A.K. Post-translational modification crosstalk and hotspots in sirtuin interactors implicated in cardiovascular diseases. Front Genet. 2020;11:356. doi: 10.3389/fgene.2020.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhulyn O., Rosenblatt H.D., Shokat L., et al. Evolutionarily divergent mTOR remodels translatome for tissue regeneration. Nature. 2023;620(7972):163–171. doi: 10.1038/s41586-023-06365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKay C.S., Finn M.G. Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem Biol. 2014;21(9):1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis Y.E., Moskovitz A., Mutlak M., Heineke J., Caspi L.H., Kehat I. Localization of transcripts, translation, and degradation for spatiotemporal sarcomere maintenance. J Mol Cell Cardiol. 2018;116:16–28. doi: 10.1016/j.yjmcc.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken) 2010;67(11):677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh S.R., Hope I.A. Determination of the mobility of novel and established Caenorhabditis elegans sarcomeric proteins in vivo. Eur J Cell Biol. 2010;89(6):437–448. doi: 10.1016/j.ejcb.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph F., Hüttemeister J., da Silva Lopes K., et al. Resolving titin’s lifecycle and the spatial organization of protein turnover in mouse cardiomyocytes. Proc Natl Acad Sci U S A. 2019;116(50):25126–25136. doi: 10.1073/pnas.1904385116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morkin E., Kimata S., Skillman J.J. Myosin synthesis and degradation during development of cardiac hypertrophy in the rabbit. Circ Res. 1972;30(6):690–702. doi: 10.1161/01.res.30.6.690. [DOI] [PubMed] [Google Scholar]
- 46.Martin A.F., Rabinowitz M., Blough R., Prior G., Zak R. Measurements of half-life of rat cardiac myosin heavy chain with leucyl-tRNA used as precursor pool. J Biol Chem. 1977;252(10):3422–3429. [PubMed] [Google Scholar]
- 47.Martin A.F. Turnover of cardiac troponin subunits. Kinetic evidence for a precursor pool of troponin-I. J Biol Chem. 1981;256(2):964–968. [PubMed] [Google Scholar]
- 48.Lam M.P., Wang D., Lau E., et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J Clin Invest. 2014;124(4):1734–1744. doi: 10.1172/JCI73787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau E., Cao Q., Lam M.P.Y., et al. Integrated omics dissection of proteome dynamics during cardiac remodeling. Nat Commun. 2018;9(1):120. doi: 10.1038/s41467-017-02467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Everett A.W., Prior G., Zak R. Equilibration of leucine between the plasma compartment and leucyl-tRNA in the heart, and turnover of cardiac myosin heavy chain. Biochem J. 1981;194(1):365–368. doi: 10.1042/bj1940365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacs W.B., Kim I.S., Struve A., Fulton A.B. Biosynthesis of titin in cultured skeletal muscle cells. J Cell Biol. 1989;109(5):2189–2195. doi: 10.1083/jcb.109.5.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diguet N., Trammell S.A.J., Tannous C., et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation. 2018;137(21):2256–2273. doi: 10.1161/CIRCULATIONAHA.116.026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horton J.L., Martin O.J., Lai L., et al. Mitochondrial protein hyperacetylation in the failing heart. JCI Insight. 2016;2(1) doi: 10.1172/jci.insight.84897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin A.S., Abraham D.M., Hershberger K.A., et al. Nicotinamide mononucleotide requires SIRT3 to improve cardiac function and bioenergetics in a Friedreich’s ataxia cardiomyopathy model. JCI Insight. 2017;2(14) doi: 10.1172/jci.insight.93885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Chen Y., Doan J., Murray J., Molkentin J.D., Liu Q. Transforming growth factor beta-activated kinase 1 signaling pathway critically regulates myocardial survival and remodeling. Circulation. 2014;130(24):2162–2172. doi: 10.1161/CIRCULATIONAHA.114.011195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thygesen K., Alpert J.S., Jaffe A.S., et al. Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth universal definition of myocardial infarction (2018) J Am Coll Cardiol. 2018;72(18):2231–2264. doi: 10.1016/j.jacc.2018.08.1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.