Abstract
Background:
American Indian and Alaska Native people in the United States experience high rates of stomach cancer. Helicobacter pylori infection is a significant risk factor for stomach cancer, and H. pylori strains that carry the cagA gene are linked to greater gastrointestinal disease severity. Yet, little is known about H. pylori and cagA infections in American Indian and Alaska Native people, particularly at the tribal level. We assessed the prevalence and risk factors of H. pylori infection and cagA gene carriage in tribal members from the Navajo Nation.
Materials and Methods:
We conducted a cross-sectional study with adults from the Navajo Nation. Stool samples collected from participants were analyzed with droplet digital PCR for H. pylori 16S ribosomal and cagA virulence genes. Self-administered health and food questionnaires were mailed to participants to collect information on sociodemographic, health, lifestyle, and environmental risk factors for H. pylori infection. Logistic regression assessed the association between risk factors and H. pylori infection and cagA gene carriage.
Results:
Among 99 adults, the median age was 45 (age range: 18 to 79 years), and 73.7% were female. About 56.6% (95% CI: 46.2-66.5) of participants were infected with H. pylori. Of H. pylori-infected participants, 78.6% (95% CI: 65.6-88.4) were cagA-gene positive. No significant associations of relevant risk factors with H. pylori and cagA-gene positive infections were noted.
Conclusions:
In a community-based study population, a substantial proportion of adult tribal members had H. pylori and cagA-gene positive infections. Given these high proportions, culturally appropriate prevention strategies and interventions addressing H. pylori infections present an avenue for additional research and stomach cancer prevention in the Navajo Nation.
Keywords: Indigenous, stomach cancer, cancer disparities, risk factors, Helicobacter pylori, cagA
1. INTRODUCTION
A substantial burden of stomach cancer incidence continues to be observed in American Indian and Alaska Native populations in the United States (US)1,2. The stomach cancer incidence rate was 1.98 times higher in American Indian and Alaska Native people than in White people (10.4 per 100,000 vs. 5.3 per 100,000)2. For the largest American Indian tribe in the US, the Navajo Nation is experiencing higher incidence and mortality rates for stomach cancer than the surrounding White population (Incidence: 15.0 per 100,000 vs. 4.3 per 100,000; Mortality: 9.8 per 100,000 vs. 2.2 per 100,000)3. A contributing factor to this elevated burden of stomach cancer in the Navajo people may be related to a high burden of Helicobacter pylori (H. pylori) infection, an infectious pathogen that is an important risk factor for stomach cancer4. Therefore, identifying the prevalence and risk factors of H. pylori and its most important virulence factor, cagA, in American Indian people can inform prevention strategies and interventions to reduce the burden of stomach cancer.
H. pylori is a spiral-shaped bacterium that colonizes and infects the stomach lining and causes gastrointestinal diseases, such as gastric ulcers and stomach cancer5,6. H. pylori infections are predominately acquired during childhood, although transmission can occur at any time during a person’s life through person-to-person contact, oral-to-oral or fecal-to-oral routes, consuming contaminated food and/or water7. Other known risk factors associated with H. pylori infection include older age, lower socioeconomic status, household crowding, and living with someone infected with H. pylori7–14.
The virulence factors of the H. pylori strain further determines the development of severe gastrointestinal disease. H. pylori strains vary in their production of virulence factors (e.g., CagA, VacA, BabA), which form the causal link between H. pylori and stomach cancer4. In particular, H. pylori strains that contain the cytotoxin-associated antigen gene pathogenicity island (cag PAI) encode more than 20 proteins that produced a specialized secretion system that delivers the CagA protein effector and other bacterial metabolites into host cells15,16. CagA interrupts normal gastric epithelial cell activity, promotes inflammation, and increases gastrointestinal disease severity, including stomach cancer risk17,18. Further, the cagA gene has two allele variations: classified as East Asian or Western. The cagA gene with the East Asian allele encodes an EPIYA-D motif and is associated with a greater risk of stomach cancer than the cagA gene with the Western allele with an EPIYA-C motif19.
Given the long-standing stomach cancer disparity among American Indian people in the US, gaining insight into the epidemiology of H. pylori among American Indian people can provide valuable information for prevention efforts. Therefore, we assessed the prevalence of H. pylori and cagA-positive infections in adults from the Navajo Nation. We also explored sociodemographic, health, lifestyle, and environmental risk factors associated with H. pylori and cagA-positive infections6–9,20. We hypothesized that adults from the Navajo Nation would have a disproportionately higher prevalence of H. pylori and cagA-positive infections than White people in the US.
2. MATERIALS AND METHODS
Study methods and participants
A cross-sectional community-based study was conducted from January to November 2021 in two regions of the Navajo Nation: the central and northeast regions. Participants learned about the Navajo ABID (Assessing the gut microbiota and Individual Diet) Study through online and offline recruitment platforms, such as a study website, social media pages (i.e., Facebook and Instagram), newspaper ads, flyers/postcards posted in the community, and in-person community events. Participants opted into the study by contacting the study team by phone or email or completing a participant intake form on the study website. Due to COVID-19 pandemic, online recruitment platforms were used throughout the study, and offline recruitment platforms were used in the last four months of the study when COVID-19 vaccines were available, and the tribal community lifted physical distancing mandates.
Eligible participants had to identify as a Navajo tribal member, be at least 18 years old, reside in the study regions, not be pregnant, not have used oral or intravenous antibiotics in the past 3 months, not be using proton pump inhibitors, and not be undergoing any cancer treatment. Each eligible participant received a copy of a consent form in either English or Navajo. Those who participated gave verbal consent over the phone and were mailed a study packet containing questionnaires, a stool sample kit, instructions for completing the study packet, and prepaid envelopes to mail back the questionnaires and stool samples to the study group in Seattle, WA.
This study was approved by the Navajo Nation Human Research Review Board (NNR-20.384T) and the University of Washington Human Subjects Division (00011217). Verbal consent from each eligible participant was obtained and recorded by the study group.
Measurement of Helicobacter pylori and cagA genotypes with droplet digital PCR (ddPCR)
The presence of H. pylori (positive or negative) and cagA genotypes (positive or negative and EPIYA-C or EPIY-D allele type) were determined from stool samples collected by the participants in their homes. Self-sampling stool collection instructions were provided to participants with the following modifications21. Participants placed stool samples (approximately 1 teaspoon) in a vial with 5 mL of 95% ethanol preservative (Fisher Scientific), rather than RNAlater, and stored at room temperature before mailing to the Salama Lab at the Fred Hutchinson Cancer Center (FHCC) in Seattle, WA. Pilot studies showed similar ddPCR assay performance between the two nucleic acid preservatives.
Bacterial DNA was extracted from stool samples using the QIAamp Stool DNA Mini Kit (Qiagen) and analyzed in duplicates using droplet digital Polymerase Chain Reaction (ddPCR) assays according to the manufacturer instructions for the QX200 ddPCR System (BioRad), which have been validated in adult stool in other studies in the Salama Lab22. Briefly, 0.5 ml of stool-ethanol slurry were extracted for each subject yielding concentrations from 5-200 ng/μl stool DNA. We utilized 10 μl stool DNA for duplicate reactions for each sample and assay. H. pylori gene detection was not correlated with stool DNA concentration. Separate sample reactions contained probes and primers for H. pylori 16S and cagA, or for cagA EPIYA-C and EPIYA-D genotyping using primers and probes described previously22. Droplets were generated using the QX200 Droplet Generator (BioRad). Droplets were then analyzed for fluorescent amplitude using the QX200 Droplet Reader (BioRad). Data were analyzed using the QuantaSoft software version 1.6.6 (BioRad). For quality control, a positive control (stool DNA from a confirmed H. pylori-positive volunteer) and negative control (molecular grade water) were included in each batch of samples analyzed. Additional controls included 0.2 pg/μl genomic DNA from strains EM17-1 (cagA+, EPIYA-C), EM41-2 (cagA+, EPIYA-D), EM16-4 (cagA-) as appropriate23. A sample with greater than 5 droplets observed above recommended fluorescence intensity thresholds for H. pylori, cagA, and cagA EPIYA typing was categorized as positive. The fluorescence intensity threshold values were set to 4500 for H. pylori 16S assay, 2000 for the cagA assay, and 2000 for the EPIYA-C and EPIYA-D assays22. In addition, the thresholds for each assay were visually evaluated by inspecting the amplitude plot of the fluorescence intensity threshold between a positive control sample and the cluster of positive droplets and the cluster of negative droplets, as well as ensuring the threshold was greater than two standard deviations from the negative droplets.
Participant data collection
Data on study variables were collected from self-administered health and food questionnaires that were mailed to consented participants. Self-reported sociodemographic characteristics (age, sex, educational attainment); health conditions (i.e., diabetes, family history of stomach cancer, body mass index); medication use (daily aspirin use, monthly over the counter stomach medicine use, monthly vitamin use); lifestyle factors (smoking, alcohol use, physical activity); and environmental exposure (drinking water source) were adapted by prior items developed by Harris et al. and the Centers for Disease Control and Prevention, Behavioral Risk Factor Surveillance Survey24,25.
The Food Frequency Questionnaire (FFQ), developed by the Nutrition Assessment Shared Resource (NASR) of Fred Hutchinson Cancer Center, Seattle, WA, assessed dietary intake. Participants were mailed an FFQ to report the frequency of consumption and portion size of 181-food items over the last year. Nutrient calculations were performed with the Nutrient Data System for Research software version v2020, developed by the Nutrition Coordinating Center, University of Minnesota, Minneapolis, MN. The annual intake of each food item was calculated using the frequency of consumption (never or less than once per month, 1 per month, 2-3 per month, 1 per week, 2 per week, 3-4 per week, 5-6 per week, 1 per day, 2+ per day) and portion size (small, medium, large). Three diet variables derived from these data were included in the present analysis: daily sodium intake, daily vegetable intake (including green salad, tomatoes, carrots, peppers, broccoli, cauliflower, cabbages, green beans, peas, corn and hominy, squash, zucchini, yams, sweet potatoes, cooked greens potatoes, coleslaw, tomato juice and other vegetable juice), and daily fruit intake (including apples, applesauce, pears, bananas, peaches, plums, apricots, dried fruits, citrus fruits, berries, melons, orange juice, grapefruit juice, and other 100% fruit juice). Dietary cutoffs were established with the US Department of Agriculture and US Department of Health and Human Services Dietary Guidelines for Americans for daily intakes of vegetables (3.0 cups/day) and fruit (2.0 cups/day)26. Sodium daily intake was categorized into a tertile (low, medium, high) based on the sodium daily intake distribution in H. pylori-negative participants (≤2,028 mg/day, 2,209-3,765 mg/day, >3,765 mg/day).
Statistical analyses
The sample size for this study was determined based on practical considerations (cost and feasibility) and formal statistical considerations. We assumed the H. pylori prevalence in Navajo adults to be 58%27. Therefore, in a sample of 150 participants, the H. pylori prevalence in this study was estimated with an absolute precision of 8% with 95% confidence.
For descriptive and univariate analyses of study variables, we calculated the frequency and percentages for categorical measures for the overall study population and by H. pylori and cagA-gene positive infection status. We then analyzed the distribution of study variables by H. pylori and cagA-gene positive infection status (positive vs. negative) using Pearsons chi-squared (χ2) test. Study variables included birth year cohort (years; 1940-1965, 1966-1975, 1976-1985, 1985+), age quartiles (18-35, 36-45, 46-55, 56+), sex (male, female), education (≤high school, >high school), aspirin daily use (yes, no), monthly medication use (over the counter stomach medication, vitamins), family history of stomach cancer (yes, no), self-reported diabetes health condition (yes, no), body mass index (<25.0, 25.0-30.0, >30.0 kg/m2), smoking (never, ever smoked, current smoker), alcohol use (never, drank in the past, current use), sodium daily intake (low, medium, high), vegetable daily intake (≤3.0 cups/day, >3.0 cups/day), fruit daily intake (≤2.0 cups/day, >2.0 cups/day), and type of drinking water consumed (filtered water or unfiltered tap water, bottled water).
Multivariate logistic regression analyses were used to calculate odds ratios (OR) and 95% CI to test associations of sociodemographic, family history, lifestyle, and environmental variables with H. pylori infection and cagA-gene positive infection. All models were adjusted for birth year cohort and sex, and the reference group was H. pylori-negative people. All analyses were performed using R Studio version 4.2.2 (R Core Team, Vienna, Austria). A value of p<0.05 was considered statistically significant.
3. RESULTS
From January to November 2021, 260 potential participants expressed interest in participating in the study (Figure 1). After attempting to contact all 260 individuals regarding the study, 115 were ineligible, and 145 were eligible. Of the 145 consented participants, 99 (68%) participants with complete questionnaires and H. pylori and cagA ddPCR results were included in these analyses. The remaining 41 individuals never completed the study procedures or decided not to participate, and five individuals were excluded due to calculated extreme energy intake values. Participants with extreme calorie intake were removed because these extreme values would have influenced the calculated nutrient variables analyzed in this study. It is likely that these 5 participants were extremely malnourished (<500 kcal/day), overnourished (>5,500 kcal/day), or did not understand the food frequency questionnaire.
FIGURE 1.
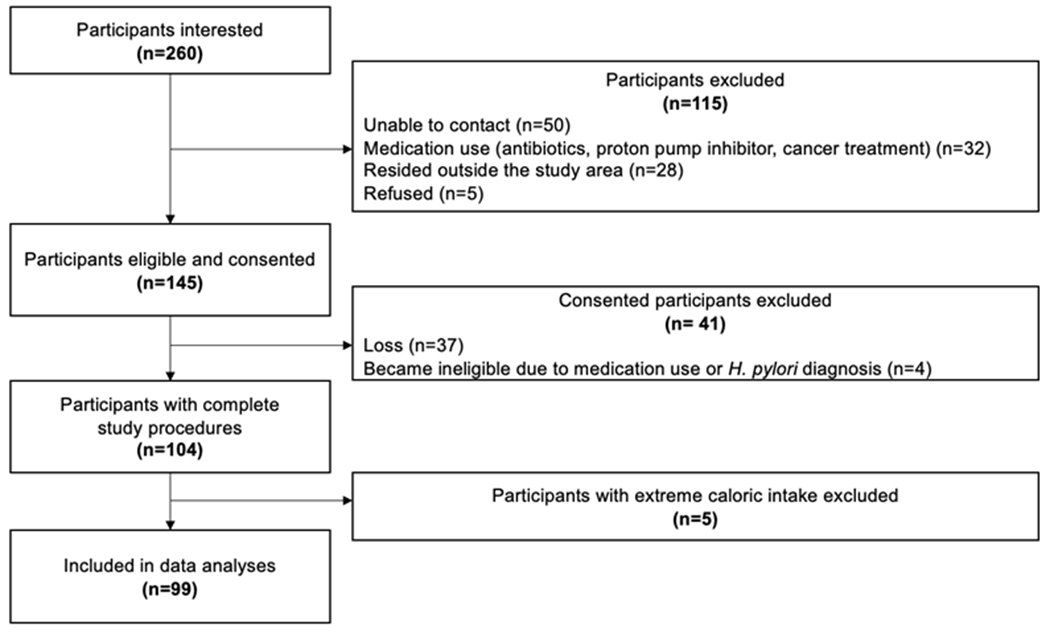
Flowchart of participant recruitment to the Navajo ABID study: January to November 2021.
Participant characteristics
Of the 99 participants, the median age was 45 years old (age range: 18 to 79 years), 25.3% were in the 1940-1965 birth year, 21.2% in the 1966-1975 birth year, 29.3% in the 1976-1985 birth year, 24.2% in the 1985+ birth year, 73.7% were female, 77.8% had greater than high school education, 16% had a family history of stomach cancer, 21.2% reported they had diabetes, 49.5% had a body mass index greater than or equal to 30 kg/m2, 36.4% ever smoked, 29.3% consumed alcohol in the past month, 40.4% reported high sodium daily intake, and 53.5% consumed filtered or unfiltered tap water (Table 1).
TABLE 1.
Percent distribution of overall participant characteristics and by Helicobacter pylori (H. pylori) status.
| Characteristics | Overall (N=99) | H. pylori-negative (n=43) | H. pylori-positive (n=56) | p-valuea |
|---|---|---|---|---|
| Age group, n (%) | ||||
| 18-35 | 24 (24.2) | 13 (30.2) | 11 (19.6) | 0.190 |
| 36-45 | 29 (29.3) | 8 (18.6) | 21 (37.5) | |
| 46-55 | 21 (21.2) | 11 (25.6) | 10 (17.9) | |
| 55+ | 25 (25.3) | 11 (25.6) | 14 (25.0) | |
| Birth year, n (%) | ||||
| 1940-1965 | 25 (25.3) | 11 (25.6) | 14 (25.0) | 0.190 |
| 1966-1975 | 21 (21.2) | 11 (25.6) | 10 (17.9) | |
| 1976-1985 | 29 (29.3) | 8 (18.6) | 21 (37.5) | |
| 1985+ | 24 (24.2) | 13 (30.2) | 11 (19.6) | |
| Sex, n (%) | ||||
| Female | 73 (73.7) | 32 (74.4) | 41 (73.2) | 1.000 |
| Male | 26 (26.3) | 11 (25.6) | 15 (26.8) | |
| Education, n (%) | ||||
| ≤High school | 21 (21.2) | 12 (27.9) | 9 (16.1) | 0.257 |
| >High school | 77 (77.8) | 31 (72.1) | 46 (82.1) | |
| Aspirin daily use, n (%) | 18 (18.2) | 9 (20.9) | 9 (16.1) | 0.752 |
| OTC stomach medicine, monthly use, n (%) | 27 (27.3) | 8 (18.6) | 19 (33.9) | 0.142 |
| Vitamins, monthly use, n (%) | 60 (60.6) | 22 (51.2) | 38 (67.9) | 0.140 |
| Family health history of stomach cancer, n (%) | 16 (16.2) | 8 (18.6) | 8 (14.3) | 0.762 |
| Diabetes, n (%) | 21 (21.2) | 10 (23.3) | 11 (19.6) | 0.851 |
| BMI (kg/m2), n (%) | ||||
| <25.0 | 16 (16.2) | 5 (11.6) | 11 (19.6) | 0.449 |
| 25.0-30.0 | 31 (31.3) | 13 (30.2) | 18 (32.1) | |
| >30.0 | 49 (49.5) | 24 (55.8) | 25 (44.6) | |
| Physical activity, n (%) | 0.147 | |||
| Low (0-149 mins / week) | 47 (47.5) | 17 (39.5) | 30 (53.6) | |
| Moderate (150-300 mins / week) | 24 (24.2) | 12 (27.9) | 12 (21.4) | |
| High (>300mins / week) | 14 (14.1) | 9 (20.9) | 5 (8.9) | |
| Smoking, n (%) | ||||
| Never | 61 (61.6) | 29 (67.4) | 32 (57.1) | 0.376 |
| Ever smoked | 36 (36.4) | 13 (30.2) | 23 (41.1) | |
| Alcohol use, n (%) | ||||
| Never | 17 (17.2) | 9 (20.9) | 8 (14.3) | 0.132 |
| Drank in the past | 50 (50.5) | 17 (39.5) | 33 (58.9) | |
| Current use | 29 (29.3) | 16 (37.2) | 13 (23.2) | |
| Sodium daily intake (mg/day), n (%) | ||||
| Low (≤2,028) | 21 (26.3) | 14 (32.6) | 12 (21.4) | 0.307 |
| Medium (2,209-3,765) | 33 (33.3) | 15 (34.9) | 18 (32.1) | |
| High (>3,765) | 40 (40.4) | 14 (32.6) | 26 (46.4) | |
| Vegetable daily intake (cups/day), n (%) | ||||
| ≤3.0 | 63 (63.6) | 28 (65.1) | 35 (62.5) | 0.954 |
| >3.0 | 36 (36.4) | 15 (34.9) | 21 (37.5) | |
| Fruit daily intake (cups/day), n (%) | ||||
| ≤2.0 | 58 (58.6) | 27 (62.8) | 31 (55.4) | 0.590 |
| >2.0 | 41 (41.1) | 16 (37.2) | 25 (44.6) | |
| Type of water consumed, n (%) | ||||
| Filtered or Unfiltered Tap water | 53 (53.5) | 24 (55.8) | 29 (51.8) | 0.998 |
| Bottled water | 44 (44.4) | 19 (44.2) | 25 (44.6) |
Abbreviations: SD=standard deviation; BMI=Body mass index, OTC=Over the Counter.
Pearson’s chi-squared test
Prevalence of H. pylori infection
Based on analysis of ddPCR results of collected stool samples, active H. pylori infection was found in 56 participants, with an overall prevalence of 56.6% (95% CI: 46.2-66.5). Overall, the distribution of sociodemographic, health, lifestyle, and environmental factors was similar across H. pylori status (Table 1).
Prevalence of cagA gene carriage
The cagA genotyping assay detected the cagA gene in 78.6% (44/56, 95% CI: 65.6%-88.4%) of H. pylori-positive participants. Of these samples, 95.5% (42/44, 95% CI: 84.5%-99.4%) were the Western allele type (EPIYA-C motif). The distribution of sociodemographic, health, lifestyle, and environmental factors was similar between H. pylori-negative and cagA-positive participants (Table 2).
TABLE 2.
Percent distribution of demographic characteristics by cagA status.
| Characteristics | H. pylori-negative (n=43) | cagA-positive (44) | p-valuea |
|---|---|---|---|
| Age group, n (%) | |||
| 18-35 | 13 (30.2) | 8 (18.2) | 0.340 |
| 36-45 | 8 (18.6) | 15 (34.1) | |
| 46-55 | 11 (25.6) | 10 (22.7) | |
| 55+ | 11 (25.6) | 11 (25.0) | |
| Birth year, n (%) | |||
| 1940-1965 | 11 (25.6) | 11 (25.0) | 0.340 |
| 1966-1975 | 11 (25.6) | 10 (22.7) | |
| 1976-1985 | 8 (18.6) | 15 (34.1) | |
| 1985+ | 13 (30.2) | 8 (18.2) | |
| Sex, n (%) | |||
| Female | 32 (74.4) | 33 (75.0) | 1.000 |
| Male | 11 (25.6) | 11 (25.0) | |
| Education, n (%) | |||
| ≤High school | 12 (27.9) | 7 (77.8) | 0.299 |
| >High school | 31 (72.1) | 36 (78.3) | |
| Aspirin daily use, n (%) | 9 (20.9) | 7 (15.9) | 0.782 |
| OTC stomach medicine, monthly use, n (%) | 8 (18.6) | 16 (36.4) | 0.107 |
| Vitamins, monthly use, n (%) | 22 (51.2) | 31 (70.5) | 0.104 |
| Family history of stomach cancer, n (%) | 8 (18.6) | 7 (15.9) | 0.961 |
| Diabetes, n(%) | 10 (23.3) | 10 (22.7) | 1.000 |
| BMI (kg/m2), n (%) | |||
| <25.0 | 5 (11.6) | 9 (20.5) | 0.514 |
| 25.0 – 30.0 | 13 (30.2) | 13 (29.5) | |
| >30.0 | 24 (55.8) | 21 (47.7) | |
| Physical activity, n (%) | |||
| Low (0-149 mins / week) | 17 (39.5) | 22 (50.0) | 0.260 |
| Moderate (150-300 mins / week) | 12 (27.9) | 10 (22.7) | |
| High (>300mins / week) | 9 (20.9) | 4 (9.1) | |
| Smoking, n (%) | |||
| Never | 29 (67.4) | 25 (56.8) | 0.413 |
| Ever smoked | 13 (30.2) | 18 (40.9) | |
| Alcohol use, n (%) | |||
| Never | 9 (20.9) | 5 (11.4) | 0.074 |
| Drank in the past | 17 (39.5) | 28 (63.6) | |
| Current use | 16 (37.2) | 10 (22.7) | |
| Sodium daily intake (mg/day), n (%), | |||
| Low (≤2,028) | 14 (32.6) | 11 (25.0) | 0.654 |
| Medium (2,209-3,765) | 15 (34.9) | 15 (34.1) | |
| High (>3,765) | 14 (32.6) | 18 (40.9) | |
| Vegetable daily intake (cups/day), n (%) | |||
| ≤3.0 | 28 (65.1) | 29 (65.9) | 1.000 |
| >3.0 | 15 (34.9) | 15 (34.1) | |
| Fruit daily intake (cups/day), n (%) | |||
| ≤2.0 | 27 (62.8) | 25 (56.8) | 0.727 |
| >2.0 | 16 (37.2) | 19 (43.2) | |
| Type of water consumed, n (%) | |||
| Filtered or Unfiltered Tap water | 24 (55.8) | 24 (54.5) | 1.000 |
| Bottled water | 19 (44.2) | 19 (43.2) |
Abbreviations: BMI=Body mass index, OTC=Over the Counter.
Pearson’s chi-squared test
Association between risk factors and H. pylori infection and cagA gene carriage
Table 3 shows the crude and adjusted odds ratios (ORs) for H. pylori infection and cagA gene carriage associated with selected risk factors. Older birth year cohort, being male, lower education, family history of stomach cancer, ever smoking, current alcohol use, high daily sodium intake, and drinking bottled water were not statistically significantly associated with H. pylori infection or cagA gene carriage.
TABLE 3.
Association between risk factors and Helicobacter pylori (H. pylori)-positive infection and cagA-positive genotype.
| H. pylori-positive | cagA-positive | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Characteristics | n (%) | Univariate OR (95% CI) | Adjusteda AOR (95% CI) | n (%) | Univariate OR (95% CI) | Adjusteda AOR (95% CI) |
| Birth year | ||||||
| 1940-1965 | 14/25 (56.0) | Ref. | Ref. | 11/22 (50.0) | Ref. | Ref. |
| 1966-1975 | 10/21 (47.6) | 0.71 (0.22-2.29) | 0.71 (0.22-2.30) | 10/21 (47.6) | 0.91 (0.27-3.03) | 0.90 (0.27-3.02) |
| 1976-1985 | 21/29 (72.4) | 2.06 (0.67-6.60) | 2.06 (0.67-6.61) | 15/23 (65.2) | 1.88 (0.57-6.40) | 1.88 (0.57-6.42) |
| 1985+ | 11/24 (45.8) | 0.66 (0.21-2.04) | 0.66 (0.21-2.04) | 8/21 (38.1) | 0.62 (0.18-2.06) | 0.62 (0.18-2.06) |
| Sex | ||||||
| Female | 41/73 (56.2) | Ref. | Ref. | 33/65 (50.8) | Ref. | Ref. |
| Male | 15/26 (57.7) | 1.06 (0.43-2.68) | 0.98 (0.38-2.55) | 11/22 (50.0) | 0.96 (0.37-2.57) | 0.93 (0.34-2.54) |
| Education | ||||||
| ≤High school | 9/21 (42.9) | Ref. | Ref. | 7/19 (36.8) | Ref. | Ref. |
| >High school | 46/77 (59.7) | 1.98 (0.75-5.39) | 1.76 (0.63-5.06) | 36/67 (53.7) | 1.99 (0.71-5.94) | 1.83 (0.61-5.80) |
| Family history of stomach cancer | ||||||
| No | 48/83 (57.8) | Ref. | Ref. | 37/72 (84.1) | Ref. | Ref. |
| Yes | 8/16 (50.0) | 0.73 (0.25-2.16) | 0.74 (0.24-2.30) | 7/15 (15.9) | 0.87 (0.26-2.54) | 0.80 (0.25-2.57) |
| Smoking | ||||||
| Never | 32/61 (52.5) | Ref. | Ref. | 25/54 (46.3) | Ref. | Ref. |
| Ever smoked | 23/36 (63.9) | 1.60 (0.69-3.80) | 1.52 (0.62-3.79) | 18/31 (58.1) | 1.61 (0.66-3.98) | 1.54 (0.60-4.03) |
| Alcohol use | ||||||
| Never | 8/17 (47.1) | Ref. | Ref. | 5/14 (35.7) | Ref. | Ref. |
| Drank in the past | 33/50 (66.0) | 2.18 (0.71-6.84) | 2.04 (0.56-7.67) | 28/45 (62.2) | 2.96 (0.88-11.07) | 3.06 (0.76-13.64) |
| Current use | 13/29 (44.8) | 0.91 (0.27-3.08) | 0.58 (0.14-2.41) | 10/26 (38.5) | 1.13 (0.30-4.56) | 0.87 (0.18-4.16) |
| Sodium daily intake (mg/day) | ||||||
| Low (≤2,028) | 12/26 (46.2) | Ref. | Ref. | 11/25 (44.0) | Ref. | Ref. |
| Medium (2,209-3,765) | 18/33 (54.5) | 1.40 (0.50-3.98) | 1.27 (0.43-3.80) | 15/30 (50.0) | 1.27 (0.44-3.75) | 1.25 (0.41-3.90) |
| High (>3,765) | 26/40 (65.0) | 2.17 (0.80-6.05) | 2.12 (0.74-6.15) | 18/32 (56.3) | 1.64 (0.57-4.78) | 1.70 (0.57-5.19) |
| Type of water consumed | ||||||
| Filtered or Unfiltered Tap water | 29/53 (54.7) | Ref. | Ref. | 24/48 (50.0) | Ref. | Ref. |
| Bottled water | 25/44 (56.8) | 1.09 (0.49-2.45) | 0.96 (0.41-2.25) | 19/38 (50.0) | 1.00 (0.43-2.35) | 0.84 (0.34-2.07) |
Abbreviations: OR=Odds ratio; AOR=Adjusted odds ratio; CI=Confidence Interval
Adjusted for sex and birth year cohort.
4. DISCUSSION
This is the first study to investigate the prevalence and correlates of H. pylori infection and cagA status from stool samples in the Navajo Nation. We observed an H. pylori prevalence of 56.6% (95% CI: 46.2%-66.5%) in Navajo adults and a cagA prevalence of 78.6% (95% CI: 65.6%-88.4%) in H. pylori-infected Navajo adults. The H. pylori prevalence in our study was 2.7 times higher than the prevalence reported in non-Hispanic White adults from the 1999-2000 National Health and Nutrition Examination Survey (21% H. pylori seroprevalence)28,29, and the cagA prevalence among H. pylori-positive adults was 1.3 times higher than the prevalence reported in H. pylori-positive White adults (non-Hispanic and Hispanic) from five prospective cohorts in the US (59% cagA seroprevalence)29,30. We further found no statistically significant associations between birth year cohort, sex, education, family history of stomach cancer, smoking, alcohol use, sodium daily intake, and drinking water source with H. pylori infection.
Participants in our study experienced a high burden of H. pylori infections (56.6%). This finding is comparable with a study of H. pylori infections in a community-based random sampling of Navajo adults residing in the western region of the Navajo Nation (56.4% (Urea Breath Test (UBT))27. Because evidence has shown H. pylori infection increases the risk of peptic ulcers and stomach cancer and successful eradication of H. pylori infection is associated with reduced risk of these conditions, these findings may explain the stomach cancer disparity in the Navajo Nation and the need to address H. pylori with prevention and eradication strategies6,31. Also, the H. pylori prevalence among the birth year cohort was as follows: 1940-1965, 56.0% H. pylori (95% CI, 34.9-75.6); 1966-1975, 47.6% (95% CI, 25.7-70.2); 1976-1985, 72.4% (95% CI, 52.8-87.3); and 1985+, 45.8% (95% CI, 25.6-67.2) (Table 3). Statistical analyses show no significant difference or decrease over the generations, as seen in other studies32,33. A relative stable H. pylori prevalence across the birth cohort may indicate continued challenges with socioeconomic factors such as living in a crowded household and access to clean drinking water across the birth cohorts, as was observed during the COVID-19 pandemic34,35. Therefore, culturally appropriate H. pylori prevention strategies and education for all generations in the Navajo Nation are needed.
Among H. pylori-positive participants, we found a cagA prevalence of 78.6% (95% CI: 65.6-88.4), which was consistent with a reported 77.0% cagA prevalence from a study of biopsy samples of Navajo H. pylori patients with gastric disease36. Additional genes encoded in the cag PAI assemble into a specialized secretion system to deliver CagA protein and bacterial metabolites into host cells that promote inflammation and increase gastrointestinal disease severity by activating or disrupting normal cellular pathways17,18. Further, the cagA gene can possess a C-terminal region with a motif of five amino acid residues, which is known as the EPIYA motif, that play an important role in the relationship of CagA proteins with cell-to-cell interaction and tyrosine phosphorylation-dependent interactions37. There are four EPIYA segments, EPIYA-A, -B, -C, and -D defined by the surrounding amino acid sequence. The EPIYA-A, -B, and -C segments are characteristic of CagA of H. pylori in non-Asian countries, known as the cagA Western allele type, while the EPIYA-A, -B, and D segment is specific to CagA of H. pylori in Asian countries, known as the cagA East Asian allele type; some strains contain neither EPIYA-C or EPIYA-D motifs37. Both EPIYA-C and EPIYA-D motifs bind to SHP2 (SH2-containing protein tyrosine phosphatase), a known oncogene. While EPIYA-D motifs have higher affinity for SHP2 and the cagA East Asian allele type is associated with a greater risk of stomach cancer than cagA Western allele type, recent studies have shown that duplication of the EPIYA-C motif leads to substantial increases in SHP2 activation38 and presence of two more EPIYA-C motifs is associated with increased cancer risk39,40. In our study, we found that 95.5% of H. pylori-cagA-positive participants showed presence of an EPIYA-C motif or cagA Western allele type. This finding is consistent with another study in Navajo H. pylori-infected patients, where the majority carried a cagA Western allele type36. However, the ddPCR method used in our study does not indicate the number of EPIYA-C motifs present. Future studies that isolate Navajo H. pylori strains and characterize the full sequence and cellular activities of their cagA alleles should be prioritized to further understand factors contributing to stomach cancer risk in this population. Because stomach cancer is associated with the cagA gene, there may be a large segment of the Navajo Nation at much higher risk for stomach cancer. To address stomach cancer in the Navajo Nation, local cancer prevention strategies may require the consideration of the greater burden of the cagA gene in the Navajo people.
Our study used stool-based ddPCR to detect H. pylori shed into the stool. Compared to serum-based analysis methods, which measure the antibody response to H. pylori and determine asymptomatic or previously exposed individuals, the ddPCR approach determines active H. pylori infection41. Therefore, the use of stool-based ddPCR for the detection of active H. pylori infection in our study was appropriate, and the disparity between the prevalence of stool-based H. pylori prevalence in our study (56.6%) and the seroprevalence in non-Hispanic Whites in the US (21.0%) is much greater because seroprevalence includes inactive and active infections. Moreover, a study by Talarico et al. reported that the stool-based ddPCR assay method had a sensitivity of 84% and 100%, and specificity of 100% and 71% compared to serology and stool antigen tests, respectively22.
This study was subject to several limitations. First, our study is cross-sectional and not able to determine causality. It is uncertain when H. pylori infection was acquired, which could be during childhood or earlier compared to when the risk factors were assessed in the study. Thus, it cannot be determined if the risk factors preceded H. pylori infection. Second, our study population was a non-random sample of the Navajo adult population. Those motivated to participate in the study may have been influenced to participate because of a history of gastrointestinal conditions and/or symptoms they were experiencing at the time of recruitment or having a family history of stomach cancer. In addition, the demographic distribution of our study population does not fully reflect the demographic profile of the Navajo adult population. Based on the 2021 American Community Survey (ACS) 5-year Census estimates, our study did not adequately capture the male population (23.1% vs. 48.1%, respectively)42. Therefore, the findings may not generalize to the general Navajo adult population. In addition, our H. pylori prevalence is likely an underestimate because of the low participation rate of males because H. pylori is more common in males. Third, while we made efforts to recruit participants through offline approaches (i.e., flyers, word of mouth, and in-person community events), this study primarily used online approaches (i.e., website, social media) to recruit participants due to the COVID-19 pandemic, which may have unintentionally excluded a large segment of the Navajo population who do not have internet access43,44. Fourth, our study has limited power to detect statistical significance in several analyses, particularly among the adjusted models. Thus, some of our analyses may have imprecise estimates and should be interpreted with caution. Fifth, participants may not have accurately recalled dietary information such as specific foods they consumed, frequency, or portion sizes, thus limiting our ability to obtain accurate estimates, even though we accounted for energy intake to reduce potential measurement error. Sixth, the FFQ used in our study was not designed for the Navajo people and may not have fully captured nutrient intake. Therefore, developing a culturally appropriate FFQ with a comprehensive food list and food/beverage portion sizes with the tribe can improve the assessment of dietary intake in the Navajo people. Moreover, food sources were limited in these Navajo communities due to the COVID-19 pandemic, which impacted the study area throughout the data collection period, and thus the foods consumed may have differed from the usual foods participants ate. Seventh, because the ddPCR method used to analyze stool samples is not a clinically approved test, we notified all participants of their H. pylori results with recommendations for further testing; however, we did not confirm our results with any follow-up clinical tests participants received. Eighth, we did not assess additional antigens of H. pylori because of the problem of multiple testing, and cagA was the focus of this study because it has a strong association with stomach cancer. Lastly, the H. pylori and cagA prevalence comparisons between the study population in the Navajo Nation and White adults in the US are not direct statistical conclusions. These studies have distinct age- and sex- distribution and have been conducted at different times.
Our study has several strengths. This study is among a few studies that investigated H. pylori infection and cagA status in an Indigenous tribal population with disproportionate stomach cancer rates. Secondly, our study focuses on a community-based population and not a hospital-based population, which adds to the evidence of H. pylori infection in a “healthy” population. Thirdly, tribal leaders and community members of the two study regions supported the study, despite the logistic challenges and stress of the COVID-19 pandemic. Lastly, the testing of H. pylori infection was non-invasive, and the ddPCR methods used to detect H. pylori have a sensitivity and specificity greater than 84%22.
5. CONCLUSION
In conclusion, our study showed a high prevalence of H. pylori and cagA infections in Navajo adults living in the Navajo Nation. Therefore, prevention strategies and interventions are warranted to reduce H. pylori infections in the Navajo Nation. These strategies should be tailored to the needs of Navajo people and individuals at higher risk for stomach cancer. Further larger studies are needed to elucidate the risk factors of H. pylori infection and stomach cancer in American Indian populations with high stomach cancer incidence and mortality.
Significance Statement.
This original study provides an understanding of the Helicobacter pylori disease burden in adults from the Navajo Nation, a tribal nation with higher rates of stomach cancer than White populations in the Southwest. These tribal-specific results inform tribal cancer prevention strategies to reduce the Helicobacter pylori burden to ensure the health of the Navajo Nation.
Acknowledgments
We thank the study participants for their participation, including the support from members of the community and the Navajo Nation Human Research Review Board. We further give thanks to Frank Morgan and Martha Garrison for assisting with Navajo translation; to Note Louis and Joycelyn Becenti for designing social media and website content; to Rebecca Becenti, Marilyn Becenti, and Karla Chavez for conducting community outreach, and to Salama lab members particular to Ali Meyer and Eli Le for receiving and analyzing specimens.
Funding
This study was supported by the F99 National Institutes of Health/National Cancer Institute Fellowship Award (F99CA253685), the Tribal Researchers’ Cancer Control Research Awards (S06GM123543), and the National Institute of Allergy and Infectious Diseases R01 AI054423. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement
No conflict of interest to declare.
Informed Consent
All participants gave verbal informed consent for participation in this study.
Ethics Statement
This study was approved by the Navajo Nation Human Research Review Board (NNR-20.384T) and the University of Washington Human Subjects Division (00011217).
Data Availability
The Navajo Nation oversees the data, and requests for data can be made through the Navajo Nation Human Research Review Board (https://nnhrrb.navajo-nsn.gov/index.html).
REFERENCES
- 1.Wiggins CL, Perdue DG, Henderson JA, Bruce MG, Lanier AP, Kelley JJ, Seals BF, Espey DK. Gastric cancer among American Indians and Alaska Natives in the United States, 1999–2004. Cancer. 2008; 113(S5): 1225–1233. [DOI] [PubMed] [Google Scholar]
- 2.Melkonian SC, Jim MA, Haverkamp D, Wiggins CL, McCollum J, White MC, Kaur JS, Espey DK. Disparities in Cancer Incidence and Trends among American Indians and Alaska Natives in the United States, 2010–2015. Cancer Epidemiology Biomarkers & Prevention. 2019; 28(10): 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Among the Navajo 2005-2013 FINAL.pdf. http://www.nec.navajo-nsn.gov/Portals/0/Reports/Cancer-Among-the-Navajo-2005-2013-FINAL.pdf. Accessed February 11, 2018.
- 4.Ahn HJ, Lee DS. Helicobacter pylori in gastric carcinogenesis. World J Gastrointest Oncol. 2015; 7(12): 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench. 2015; 8(Suppl1): S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 6.Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin Microbiol Rev. 2010; 23(4): 713–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayali S, Manfredi M, Gaiani F, Bianchi L, Bizzarri B, Leandro G, Di Mario F, De’Angelis G. Helicobacter pylori, transmission routes and recurrence of infection: state of the art. Acta Biomed. 2018; 89(Suppl 8): 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter pylori Infection. Helicobacter. 2014; 19(s1): 1–5. [DOI] [PubMed] [Google Scholar]
- 9.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000; 22(2): 283–297. [DOI] [PubMed] [Google Scholar]
- 10.Krueger WS, Hilborn ED, Converse RR, Wade TJ. Environmental risk factors associated with Helicobacter pylori seroprevalence in the United States: a cross-sectional analysis of NHANES data. Epidemiology and Infection. 2015; 143(12): 2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanakala VV, Thomas J, Vijayaraghavan S. Alcohol Consumption and Active Helicobacter Pylori Infection. Clinical Gastroenterology and Hepatology. 2017; 15(1): e18. [Google Scholar]
- 12.Park JY, Forman D, Waskito LA, Yamaoka Y, Crabtree JE. Epidemiology of Helicobacter pylori and CagA-Positive Infections and Global Variations in Gastric Cancer. Toxins (Basel). 2018; 10(4): E163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cover TL, Peek RM Jr. Diet, microbial virulence, and Helicobacter pylori-induced gastric cancer. Gut Microbes. 2013; 4(6): 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: Epidemiological and biological evidence. World J Gastroenterol. 2009; 15(18): 2204–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miernyk K, Morris J, Bruden D, McMahon B, Hurlburt D, Sacco F, Parkinson A, Hennessy T, Bruce M. Characterization of Helicobacter pylori cagA and vacA Genotypes among Alaskans and Their Correlation with Clinical Disease. Journal of Clinical Microbiology. 2011; 49(9): 3114–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayehmiri F, Kiani F, Sayehmiri K, Soroush S, Asadollahi K, Alikhani MY, Delpisheh A, Emaneini M, Bogdanovic L, Varzi AM, Zarrilli R, Taherikalani M. Prevalence of cagA and vacA among Helicobacter pylori-infected patients in Iran: a systematic review and meta-analysis. J Infect Dev Ctries. 2015; 9(7): 686–696. [DOI] [PubMed] [Google Scholar]
- 17.Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Reviews Cancer. 2002; 2(1): 28–37. [DOI] [PubMed] [Google Scholar]
- 18.Jones KR, Whitmire JM, Merrell DS. A Tale of Two Toxins: Helicobacter pylori CagA and VacA Modulate Host Pathways that Impact Disease. Front Microbiol. 2010; 1: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talarico S, Leverich CK, Wei B, Ma J, Cao XG, Guo YJ, Han GS, Yao L, Self S, Zhao Y, Salama NR. Increased H. pylori stool shedding and EPIYA-D cagA alleles are associated with gastric cancer in an East Asian hospital. PLoS One. 2018; 13(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mhaskar RS, Ricardo I, Azliyati A, Laxminarayan R, Amol B, Santosh W, Boo K. Assessment of Risk Factors of Helicobacter Pylori Infection and Peptic Ulcer Disease. J Glob Infect Dis. 2013; 5(2): 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hullar MAJ, Jenkins IC, Randolph TW, Curtis KR, Monroe KR, Ernst T, Shepherd JA, Stram DO, Cheng I, Kristal BS, Wilkens LR, Franke A, Le Marchand L, Lim U, Lampe JW. Associations of the gut microbiome with hepatic adiposity in the Multiethnic Cohort Adiposity Phenotype Study. Gut Microbes. 2021; 13(1): 1965463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talarico S, Safaeian M, Gonzalez P, Hildesheim A, Herrero R, Porras C, Cortes B, Larson A, Fang FC, Salama NR. Quantitative detection and genotyping of Helicobacter pylori from stool using droplet digital PCR reveals variation in bacterial loads that correlates with cagA virulence gene carriage. Helicobacter. 2016; 21(4): 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talarico S, Gold BD, Fero J, Thompson DT, Guarner J, Czinn S, Salama NR. Pediatric Helicobacter pylori isolates display distinct gene coding capacities and virulence gene marker profiles. J Clin Microbiol. 2009; 47(6): 1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behavioral Risk Factor Surveillance System Survey Questionnaire (2019, 2020). Atlanta, Georgia: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. [Google Scholar]
- 25.Harris RB, Brown HE, Begay RL, Sanderson PR, Chief C, Monroy FP, Oren E. Helicobacter pylori Prevalence and Risk Factors in Three Rural Indigenous Communities of Northern Arizona. Int J Environ Res Public Health. 2022; 19(2): 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietary Guidelines for Americans, 2020-2025. U.S. Department of Agriculture and U.S. Department of Health and Human Services; 2020. Available at DietaryGuidelines.gov. Accessed May 24, 2023. [Google Scholar]
- 27.Harris RB, Sanderson PR, Chief C, Monroy FP, Brown HE, Oren E, Harris R. Helicobacter pylori infections in Navajo communities of Northern Arizona. Poster Presentation presented at the: Annual Meeting; The American Soceity of Preventative Oncology. https://aspo.org/wp-content/uploads/Robin-Harris-Helicobacter-pylori-in-Northern-Arizona_Harris_ASPO2020.pdf. Accessed January 10, 2021. [Google Scholar]
- 28.Grad YH, Lipsitch M, Aiello AE. Secular Trends in Helicobacter pylori Seroprevalence in Adults in the United States: Evidence for Sustained Race/Ethnic Disparities. Am J Epidemiol. 2012; 175(1): 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown H, Cantrell S, Tang H, Epplein M, Garman KS. Racial Differences in Helicobacter pylori Prevalence in the US: A Systematic Review. Gastro Hep Adv. 2022; 1(5): 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga MG, Butt J, Blot WJ, Le Marchand L, Haiman CA, Chen Y, Wassertheil-Smoller S, Tinker LF, Peer RM Jr, Potter JD, Cover TL, Hyslop T, Zeleniuch-Jacquotte A, Berndt SI, Hildesheim A, Waterboer T, Pawlita M, Epplein M. Racial Differences in Helicobacter pylori CagA Sero-prevalence in a Consortium of Adult Cohorts in the United States. Cancer Epidemiol Biomarkers Prev. 2020; 29(10): 2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Jiang SF, Lei NY, Shah SC, Corley DA. Effect of Helicobacter pylori Eradication Therapy on the Incidence of Noncardia Gastric Adenocarcinoma in a Large Diverse Population in the United States. Gastroenterology. 2023; 165(2): 391–401.e2. [DOI] [PubMed] [Google Scholar]
- 32.Shah SC, Halvorson AE, Lee D, Bustamante R, McBay B, Gupta R, Denton J, Dorn C, Wilson O, Peek RM Jr, Gupta S, Liu L, Hung A, Greevy R, Roumie CL. Helicobacter pylori Burden in the United States According to Individual Demographics and Geography: A Nationwide Analysis of the Veterans Healthcare System. Clin Gastroenterol Hepatol. 2024; 22(1): 42–50.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Nishiyama T, Kikuchi S, Inoue M, Sawada N, Tsugane S, Lin Y. Changing trends in the prevalence of H. pylori infection in Japan (1908-2003): a systematic review and meta-regression analysis of 170,752 individuals. Sci Rep. 2017; 7(1): 15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Lonebear D, Barceló NE, Akee R, Carroll SR. Research Full Report: American Indian Reservations and COVID-19: Correlates of Early Infection Rates in the Pandemic. Journal of Public Health Management and Practice. 2020; 26(4): 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emerson MA, Montoya T. Confronting Legacies of Structural Racism and Settler Colonialism to Understand COVID-19 Impacts on the Navajo Nation. Am J Public Health. 2021; 111(8): 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monroy FP, Brown HE, Sanderson PR, Jarrin G, Mbegbu M, Kyman S, Harris RB. Helicobacter pylori in Native Americans in Northern Arizona. Diseases. 2022; 10(2): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatakeyama M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2017; 93(4): 196–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagase L, Hayashi T, Senda T, Hatakeyama M. Dramatic increase in SHP2 binding activity of Helicobacter pylori Western CagA by EPIYA-C duplication: its implications in gastric carcinogenesis. Sci Rep. 2015; 5: 15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tserentogtokh T, Gantuya B, Subsomwong P, Oyuntsetseg K, Bolor D, Erdene-Ochir Y, Azzaya D, Davaadorj D, Uchida T, Matsuhisa T, Yamaoka Y. Western-Type Helicobacter pylori CagA are the Most Frequent Type in Mongolian Patients. Cancers (Basel). 2019; 11(5): 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freire de Melo F, Marques HS, Rocha Pinheiro SL, Bueno Lemos FF, Luz MS, Teixeira KN, Souza CL, Oliveira MV. Influence of Helicobacter pylori oncoprotein CagA in gastric cancer: A critical-reflective analysis. World J Clin Oncol. 2022; 13(11): 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang YK, Kuo FC, Liu CJ, Wu MC, Shih HY, Wang SSW, Wu JY, Kuo CH, Huang YK, Wu DC. Diagnosis of Helicobacter pylori infection: Current options and developments. World J Gastroenterol. 2015; 21(40): 11221–11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Census Bureau (2017-2021). Sex by Age American Community Survey 5-year estimates. Retrieved from <https://censusreporter.org. https://censusreporter.org/data/table/?table=B01001&geo_ids=25200US2430R,01000US&primary_geo_id=25200US2430R#valueType|estimate. Accessed July 11, 2023.
- 43.Navajo Nation Looks to Build Broadband Network – MeriTalk State & Local. https://www.meritalkslg.com/articles/navajo-nation-looks-to-build-broadband-network/. Accessed March 31, 2022. [Google Scholar]
- 44.Apr 6 NT|, Updates | 2020 | Coronavirus. COVID-19 Across the Navajo Nation. Navajo Times. https://navajotimes.com/coronavirus-updates/covid-19-across-the-navajo-nation/. Published April 6, 2020. Accessed May 15, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Navajo Nation oversees the data, and requests for data can be made through the Navajo Nation Human Research Review Board (https://nnhrrb.navajo-nsn.gov/index.html).


