Abstract
The human herpesvirus entry mediator C (HveC/PRR1) is a member of the immunoglobulin family used as a cellular receptor by the alphaherpesviruses herpes simplex virus (HSV), pseudorabies virus, and bovine herpesvirus type 1. We previously demonstrated direct binding of the purified HveC ectodomain to purified HSV type 1 (HSV-1) and HSV-2 glycoprotein D (gD). Here, using a baculovirus expression system, we constructed and purified truncated forms of the receptor containing one [HveC(143t)], two [HveC(245t)], or all three immunoglobulin-like domains [HveC(346t)] of the extracellular region. All three constructs were equally able to compete with HveC(346t) for gD binding. The variable domain bound to virions and blocked HSV infection as well as HveC(346t). Thus, all of the binding to the receptor occurs within the first immunoglobulin-like domain, or V-domain, of HveC. These data confirm and extend those of Cocchi et al. (F. Cocchi, M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume, Proc. Natl. Acad. Sci. USA 95:15700, 1998). Using biosensor analysis, we measured the affinity of binding of gD from HSV strains KOS and rid1 to two forms of HveC. Soluble gDs from the KOS strain of HSV-1 had the same affinity for HveC(346t) and HveC(143t). The mutant gD(rid1t) had an increased affinity for HveC(346t) and HveC(143t) due to a faster rate of complex formation. Interestingly, we found that HveC(346t) was a tetramer in solution, whereas HveC(143t) and HveC(245t) formed dimers, suggesting a role for the third immunoglobulin-like domain of HveC in oligomerization. In addition, the stoichiometry between gD and HveC appeared to be influenced by the level of HveC oligomerization.
Herpes simplex virus (HSV) utilizes several of its 11 membrane glycoproteins during entry into mammalian cells. Glycoprotein C (gC) and/or gB assure the initial attachment to cell surface heparan sulfate proteoglycans but are not sufficient to induce viral entry (23, 60). gD, gB, and the gH-gL complex are required for fusion of the viral envelope with the cell plasma membrane (17, 49). Binding of gD to a cell surface receptor is a key step leading to membrane fusion, which could be inhibited by soluble or membrane-bound gD (6, 17, 26, 27, 45).
Recently, several cellular receptors for HSV have been identified. HveA (41) (previously called HVEM, ATAR [24], or TR2 [31]) can be used as a receptor by most HSV-1 and HSV-2 strains. HveB (PRR2) usage appears to be restricted to HSV-2, some laboratory strains of HSV-1 (rid1 and ANG), and pseudorabies virus (PRV) (15, 55). HveC (PRR1) allows entry of all HSV-1 and HSV-2 strains tested to date, as well as PRV and bovine herpesvirus type 1 (BHV-1) (18, 34). Recently, Cocchi et al. (10) isolated a splice variant of HveC, named HIgR, with an extracellular domain and receptor properties identical to those of HveC. In addition, a monoclonal antibody (MAb) raised against human bladder carcinoma cells 5637 (MAb R1.302) (35), which recognized both HveC and HIgR, could block HSV infection (10).
Unlike HveA, which is a member of the tumor necrosis factor receptor family and a receptor for lymphotoxin alpha and LIGHT (37, 41), HveB and HveC are members of the immunoglobulin (Ig) superfamily (18, 55). They are closely related to the poliovirus receptor (PVR; CD155) (39), which does not function as an HSV receptor but can be used by PRV and BHV-1 for entry into cells (18). CD155, HveB, and HveC are type I membrane glycoproteins harboring three Ig-like domains (V-C2-C2) in their extracellular portion (15, 34, 39). CD155, HveB, and HveC mRNAs are ubiquitously expressed and can be alternately spliced to yield proteins having different transmembrane and intracellular domains (10, 15, 28). The cellular function of CD155 is not known, although HveC and HveB appear to be involved in cell-cell interactions via homophilic binding, both in humans and mice (1, 33, 52).
Cell surface Ig-like molecules are used by a large number of viruses to enter cells. Among them are CD155 (PVR) used by poliovirus (39), CD4 by human immunodeficiency virus (HIV) (12), CAR by coxsackie B virus and adenovirus (4), ICAM-1 by rhinovirus (19, 51), Bgp1a for mouse hepatitis virus (MHV) (57), or NCAM for rabies virus (54). When characterized, the virus-binding site has been localized to the most distal Ig domain of these molecules (14, 16, 32, 38, 42). Evidence for the involvement of the HveC variable domain (V-domain) in HSV infection has also been recently presented (9). Truncated soluble forms of HveA and HveC produced in baculovirus-infected insect cells were shown to interact directly with HSV-gD by enzyme-linked immunosorbent assay (ELISA), in solution, and on viral particles (29, 44, 56). The binding of gD from different strains of HSV to either receptor correlated exactly with the ability of those virus strains to use HveA and/or HveC to enter cells (29, 41, 56). Using a soluble form of HveC, we identified individual residues and antigenic regions of gD that affected receptor binding both in vitro and on viral particles (29, 44). In addition, soluble HveC was an efficient inhibitor of viral infection of neuron-like cell lines in culture such as IMR5 and SY5Y (18).
Recently, Cocchi et al. (9) reported evidence that the most membrane-distal domain (V-domain) of HveC was sufficient to confer susceptibility to HSV infection when expressed at the surface of nonpermissive cells. However, the efficiency of infection appeared to be markedly affected. The same study demonstrated that a soluble form of the HveC V-domain linked to heterologous Ig-domains from the Fc part of an Ig can block HSV infection of cultured cells and interact with a soluble mutant form of gD (9, 45).
In order to extend the characterization of the direct interaction between the receptor and gD, from structure and affinity standpoints, we expressed smaller forms of soluble HveC. Here we show that a protein containing just the single V-domain of HveC and a protein containing the two distal domains of HveC are each able to bind to soluble gD as efficiently as the whole HveC ectodomain. The HveC V-domain protein bound to virus and was also able to block HSV infection of several human neuron-like cell lines as efficiently as the full ectodomain. Using surface plasmon resonance technology, we found that both soluble receptors display similar affinity to several forms of HSV-1 gD. Moreover, both the on and the off rates of gD-HveC complex formation were very similar. Interestingly, the single V-domain protein and the two-domain protein were dimers in solution, whereas the complete ectodomain was a tetramer under the same conditions. This indicated the possibility of several levels of oligomerization of HveC involving at least the first and the third domains.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda Sf9 cells (GIBCO BRL) were maintained in suspension in Sf900II medium or as monolayer cultures in supplemented Grace’s medium (GIBCO BRL) with 10% fetal calf serum (FCS). CHO M3A cells are derived from CHO-IEβ8 cells expressing the β-galactosidase gene under the control of the viral ICP4 promoter (41, 53) and express constitutively the full-length human HveC under the control of a cytomegalovirus promoter. M3A cells were grown in HAM’s F-12 medium supplemented with 10% FCS, 250 μg of G418 per ml, and 150 μg of puromycin per ml. IMR5 and SY5Y (human neuroblastoma cell lines) were grown in Dulbecco modified Eagle medium with 10% FCS. HSV-1 KOS tk12 and HSV-1 rid1 tk12 (41) were purified from infected Vero cells, and titers were obtained on Vero cells (22).
Glycoproteins and antibodies.
Soluble glycoproteins such as gD(306t) and gD(285t) were derived from HSV-1 strain KOS unless otherwise noted; gD-2(306t) was from HSV-2 strain 333 and gD(rid1t) from HSV-1 strain rid1. Construction and purification of these proteins from baculovirus-infected cell supernatant was described elsewhere (43, 45, 47, 48). Rabbit polyclonal serum R7 was raised against HSV-2 gD purified from infected mammalian cells (25). Anti tetra-His MAb was purchased from Qiagen, Inc. Anti-HveC MAb R1.302 (35) was kindly provided by S. McClellan (Beckman Coulter).
Construction of recombinant baculoviruses and purification of soluble receptors.
The strategy to generate soluble HveC(346t) was described previously (29, 58) and was applied here to generate all HveC constructs. Plasmid pBG38 was used as template for PCR amplification by using the upstream primer (C5) 5′-GCGTGATCAGGTGGTCCAGGTGAACGACTCCATGTAT-3′ and the downstream primer 5′-CGGCCCGGGCTAATGATGATGATGATGATGCTGCACGTTGAGAGTGAGGCTTTCC-3′ for HveC(245t) or 5′-CGGCCCGGGCTAATGATGATGATGATGATGCATCACCGTGAGATTGAGCTGGCTTTCT-3′ for HveC(143t). The cloning strategy with the vector pVT-Bac was described earlier (29, 58), and plasmids pCK329 and pCK330, respectively, were generated and used to produce recombinant baculoviruses bac-HveC(245t) and bac-HveC(143t). HveC(245t) contains amino acids 31 to 245 from human HveC with an extra aspartic acid residue at the N terminus and a 6-histidine tail at the C terminus of the protein added during the amplification and cloning process. HveC(143t) contains amino acids 31 to 143 of human HveC with the same C- and N-terminal additions.
All soluble receptors, containing a C-terminal 6-histidine tag, were purified by nickel affinity chromatography (Ni-NTA Superflow; Qiagen, Inc.) as described previously (29, 56, 58). Purified soluble HveC proteins were dialyzed against 100 mM sodium phosphate (pH 8.0)–150 mM NaCl and concentrated.
ELISA.
(i) The standard ELISA with immobilized receptor and soluble gD was as described previously (29). (ii) For the competition ELISA, HveC(346t) at 10 μg/ml in phosphate-buffered saline (PBS) was adsorbed to microtiter plates for 2 h at room temperature. Plates were washed with 0.1% Tween 20 in PBS and blocked with 5% milk–0.2% Tween 20 in PBS (PBST-milk) for 1 h at room temperature. Plates were then incubated overnight at 4°C with PBST-milk containing a constant concentration of gD and variable concentrations of soluble receptors as competitors. The plates were washed, and the bound gD was detected with R7 antiserum (diluted 1:1,000 in PBST-milk) for 1 h, followed by the addition of goat anti-rabbit IgG coupled to horseradish peroxidase (diluted 1:1,000 in PBST-milk) for 30 min. The plate was washed with 20 mM citric acid (pH 4.5) prior to the addition of substrate (ABTS; Moss, Inc.). Absorbance was read at 405 nm.
Blocking assay.
Cells were grown to confluence in 96-well plates in their respective medium and chilled for 15 min at 4°C prior to the addition of virus. HSV-1 KOS tk12 or HSV-1 rid1 tk12 was preincubated with soluble receptors at various concentrations in cold medium containing 30 mM HEPES for 90 min at 4°C. Culture medium was removed, and 100 μl of virus-containing medium was added (multiplicity of infection of 1). Cells were then incubated at 37°C for 5 to 6 h and lysed in NP-40 (0.5% final). Then, 50 μl of cell lysate was mixed with an equal volume of β-galactosidase substrate (chlorophenol red-β-d-galactopyranoside). The level of entry was monitored by reading the absorbance at 595 nm for 50 min to record the enzymatic activity, expressed as the change in optical density per hour (ΔOD/h). Blocking activity of soluble receptors is expressed as the percentage of virus entry into cells under test conditions compared to viral infection in absence of inhibitor (100%).
Gel filtration.
Purified proteins were diluted in PBS and loaded onto a Superdex 200 column (Pharmacia HR 10/30) as described previously (29).
Electrophoresis.
Nondenaturing and nonreducing polyacrylamide gel electrophoresis (PAGE) has been described previously (11). Proteins were separated on precast Tris-glycine gels (Novex) by using 200 mM glycine–25 mM Tris base–0.1% sodium dodecyl sulfate (SDS) as running buffer. Proteins were then visualized by silver staining of the gel (Pharmacia Silver Stain Kit) or transferred to nitrocellulose prior to antibody detection.
Binding of HveC to virus.
Soluble receptor (100 μg) was mixed with 107 PFU of purified HSV-1 KOS (ca. 4 × 108 particles) or 107 PFU of purified HSV-1 rid1 (ca. 3 × 108 particles) in 150 μl of PBS for 90 min at 4°C. The virus was subjected to sedimentation through a sucrose step gradient (10, 30, and 60%) for 4.5 h at 16,000 × g. The viral band was then collected and analyzed by Western blotting as described previously (29, 44) by using anti-tetra-His (Qiagen, Inc.) and anti-VP5 antibodies (21).
Measurement of binding of gD to HveC with an optical biosensor.
Biosensor experiments were carried out on a Biacore X optical biosensor (Biacore AB) at 25°C according to the protocol previously described (47, 59) but with the following modifications. The running buffer was HBS-EP (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20), pH 7.4. Approximately 1,600 response units (RU) of HveC(346t) or 300 RU of HveC(143t) were coupled to flow cell 1 (Fc1) of a CM5 sensor chip via primary amines according to the manufacturer’s specifications. Fc2 was activated and blocked without the addition of protein. Soluble gD was serially diluted in HBS-EP. Each gD sample was injected for 2 min to monitor the association. The sample was then replaced by HBS-EP flow, and the dissociation was monitored for 2 min. During the binding and dissociation phases of gD to HveC, the flow path was set to include both flow cells, the flow rate was 50 μl/min, and the data collection rate was set to high (five measurements/min). To regenerate the HveC surface, brief pulses of 0.2 M Na2CO3 (pH 10) were injected until the response signal returned to baseline. Sensorgrams were corrected for nonspecific binding and refractive index changes by subtracting the control sensorgram (Fc2) from the HveC surface sensorgram (Fc1). Data were analyzed with BIAevaluation software, version 3.0 (5). Model curve fitting was done by using a 1:1 Langmuir interaction with drifting baseline. This models the simple interaction between ligand (L) and receptor (R) as follows: L + R ⇆ LR. The rate of association (kon) was measured from the forward reaction, and koff was measured from the reverse reaction (5).
RESULTS
Production and characterization of baculovirus-expressed HveC receptors.
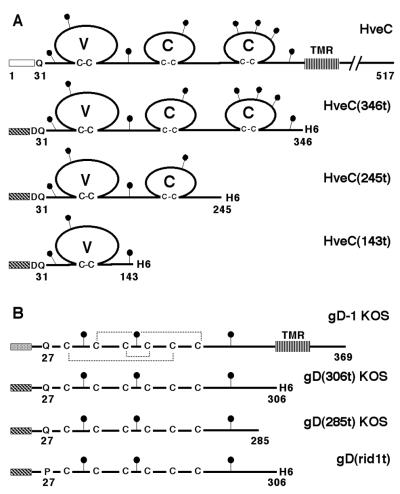
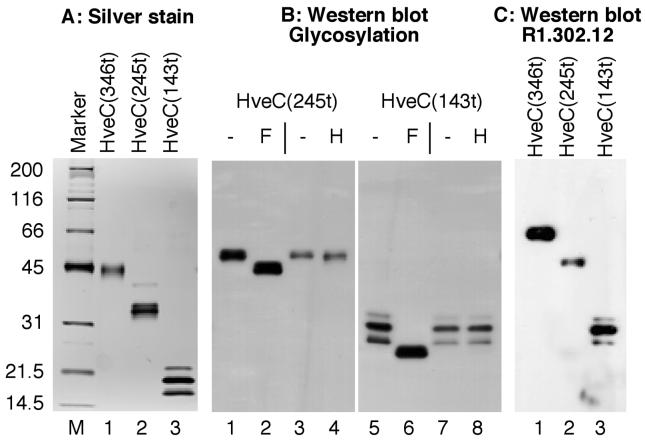
Soluble receptors were purified from supernatant of recombinant baculovirus-infected Sf9 cells. HveC(143t) contains the N-terminal Ig-like V-domain, and HveC(245t) encompasses two N-terminal domains (Fig. 1). Their properties were compared to the previously described HveC(346t) (29), which includes all three Ig-domains of HveC and ends just before the transmembrane region (Fig. 1). As shown previously, HveC(346t) migrated as a 45-kDa glycoprotein as seen by SDS-PAGE, although mass spectrometry revealed a molecular mass of 40 kDa (29). HveC(245t) migrated as a heterogeneous glycoprotein of 33 to 34 kDa (Fig. 2A). Its molecular mass, determined by mass spectrometry, was 28.0 kDa for the main species carrying three N-linked oligosaccharides (N-CHO); minor products with two or four N-CHO were also present (data not shown). By SDS-PAGE, HveC(143t) appeared as three bands, probably representing the protein with one, two, or three N-linked carbohydrates. The apparent sizes were 17, 19, and 22 kDa respectively (Fig. 2A). Mass spectrometric analysis of HveC(143t) detected two major products of 14.5 and 15.6 kDa, as well as a minor product of 16.7 kDa (data not shown). The observed increment of size correlated with the addition of one N-linked carbohydrate chain in insect cells (30, 46). In both cases, treatment of HveC(245t) and HveC(143t) with glycopeptidase F yielded a single, faster-migrating band (Fig. 2B). All N-CHO on HveC(143t) were resistant to endoglycosidase H, whereas N-CHO on HveC(245t) were partially sensitive (Fig. 2B). Thus, the endoglycosidase data suggest that the oligosaccharide on Asn 202 present in HveC(245t) but not HveC(143t) might not be processed from the high-mannose type to the complex type. The presence of three glycosylated forms of HveC(143t) indicated that three N-CHO consensus attachment sites were used. A fourth Asn residue proposed as an N-CHO attachment site in the original sequence of HveC/PRR1 (34) at position 82 in an Asn-Pro-Ser pseudoconsensus site is probably not glycosylated.
FIG. 1.
(A) Schematic representation of HSV receptors. Full-length HveC is shown as a solid line with amino acids numbered from the initial methionine. The open box indicates the HveC natural signal peptide. (B) Schematic representation of gD constructs. gD from HSV-1 KOS is represented with amino acids numbered from the N terminus of the mature gD after cleavage of the gD signal peptide (shaded box). Disulfide bonds are indicated by dotted lines. The black circles represent putative N-linked carbohydrates. The hatched box represents the mellitin signal peptide used in the baculovirus constructs. Baculovirus-expressed proteins are truncated (t) at the indicated amino acid prior to the transmembrane region (TMR). H6, six-histidine tag added at the C terminus.
FIG. 2.
Purified soluble receptors. Receptors purified from recombinant baculovirus-infected Sf9 cell supernatants were separated by SDS-PAGE. (A) Purified proteins were electrophoresed on a 12% polyacrylamide gel under reducing and denaturing conditions and then visualized by silver staining. The molecular mass markers are indicated in kilodaltons. (B) Endoglycosidase digestions. HveC(245t) (lanes 1 to 4) and HveC(143t) (lanes 5 to 8) were subjected to digestion with PNGase F (F) or endoglycosidase H (H) or were mock treated (−) prior to electrophoresis on a 16% acrylamide gel under reducing and denaturing conditions. After the Western blotting, proteins were detected with the anti-tetra-His MAb. (C) Native Western blot. Proteins were run under nondenaturing and nonreducing conditions on a 12% polyacrylamide gel and detected with MAb R1.302 (35).
Detection of HveC truncations with MAbs.
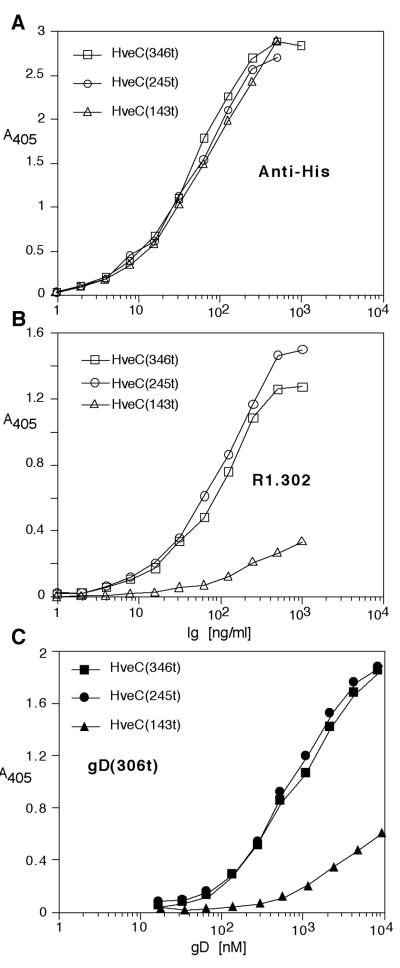
The MAb R1.302 binds to the single V-domain of HveC on cells and blocks virus entry (9, 10). We used this blocking MAb to probe a native Western blot of the three soluble HveC proteins (Fig. 2C). The antibody detected each form of the receptor, including all three glycosylated forms of HveC(143t). Detection of HveC(143t) by MAb R1.302 was abolished when PAGE was performed under reducing and denaturing conditions (data not shown). This indicated that R1.302 recognized a nonlinear epitope on HveC(143t) and that the purified proteins, including the one domain HveC(143t), were correctly folded. We also tested the native conformation of HveC truncations by ELISA (Fig. 3). As expected, all truncated forms of HveC reacted similarly with the anti tetra-His antibody, indicating that comparable amounts of proteins had been immobilized (Fig. 3A). In the same setting, we used MAb R1.302 to detect a conformation-dependent epitope on truncated HveC immobilized on an ELISA plate (Fig. 3B). Both HveC(346t) and HveC(245t) reacted with similar efficiency. In contrast, HveC(143t) was less efficiently recognized by this MAb, suggesting a loss of conformation of this small HveCt.
FIG. 3.
Binding of MAbs and gD to HveC MAbs by ELISA. Detection of immobilized truncated HveC proteins with anti-tetra-His Ig (Qiagen, Inc.) (A) or R1.302 Ig (Beckman Coulter) (B). Bound immunoglobulins were detected with horseradish peroxidase-conjugated anti-mouse IgG secondary antibody and substrate. (C) Plates coated with HveC truncated proteins were incubated with increasing concentrations of gD(306t) from HSV-1 strain KOS. Bound gD was detected with R7 antiserum, followed by peroxidase-conjugated secondary antibody and substrate. Absorbance was read at 405 nm.
Interaction of truncated HveC with gD by ELISA.
Since HveC(346t) binds gD directly in an ELISA (29), we used this method to evaluate gD binding to the three forms of HveC. Various concentrations of soluble gD(306t) or gD(285t) (Fig. 1B) were incubated with each form of HveC which has been immobilized on an ELISA plate. Bound gD was then detected with polyclonal R7 serum. gD(306t) bound to both HveC(346t) and HveC(245t) with similar efficiency, indicating that the most C-terminal Ig domain is dispensable for the interaction of HveC with gD (Fig. 3C). However gD(306t) bound less efficiently to HveC(143t). The shorter gD(285t), which displays an enhanced binding affinity for HveC over that of gD(306t) (see below and reference 47), also bound to HveC(143t) with reduced ability compared to HveC(346t) and HveC(245t) (data not shown).
Binding of HveC truncations to gD in solution.
The ability of each truncated HveC to bind gD by ELISA correlated with its capacity to be recognized by the conformation-dependent blocking MAb R1.302.
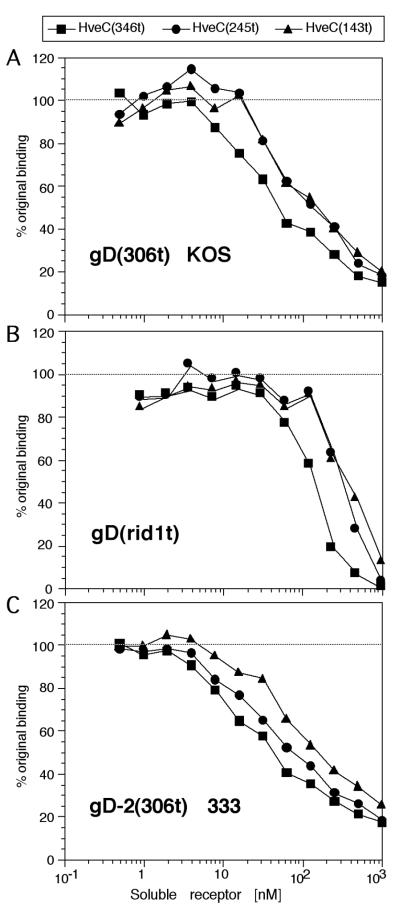
Since heat-denatured HveC(346t) completely failed to bind gD by ELISA (data not shown), the decreased binding ability of HveC(143t) might reflect a partial denaturation that could have occurred during purification or because of immobilization on the ELISA plate. To clarify this point, we performed a competition ELISA where the binding between gD and the three receptor forms occurred in solution. In this experiment HveC(346t) was immobilized on the plate. Then a constant amount of soluble gD(306t), corresponding to the half-saturating concentration for the bound HveC(346t), was added in the absence or presence of one of the soluble receptors which acted as a competitor for binding of gD (Fig. 4A). Soluble HveC(346t), HveC(245t), or HveC(143t) competed the binding of gD(306t) to the immobilized HveC(346t) in a dose-dependent manner. Both soluble HveC(245t) and HveC(143t) were able to compete gD binding nearly as well as soluble HveC(346t) which competes with itself on the plate for gD binding.
FIG. 4.
Competition ELISA. Plates were coated with HveC(346t) and incubated with a constant amount of purified gD, together with increasing concentrations of HveC truncated receptors as competitors. Panels show 1 μM gD-1(306t) from HSV-1 KOS (A), 0.1 μM gD-1(rid1t) (B), and 1 μM gD-2(306t) from HSV-2 strain 333 (C). gD bound to the immobilized receptor was detected with R7 antiserum. Relative binding is indicated as percentage of gD binding in the absence of soluble receptor.
HveC(346t) is also known to bind gD from HSV-2 and gD from the mutant strain HSV-1 rid1 (29, 56). The rid1 form of gD has a point mutation at position 27 (Q27P) (13) which prevents its binding to HveA but enhances its binding to HveC in an ELISA (29, 56). Using the competition assay, we also tested the binding of soluble gD(rid1t) (Fig. 4B) and of gD-2(306t) from HSV-2 strain 333 (Fig. 4C) to truncated HveC forms. For gD-1 rid1 and gD-2, all three truncations competed the binding to HveC(346t) with similar efficiency. Thus, we conclude that soluble gD binds HveC(143t) as well as it does to the other two truncated forms of HveC. Therefore, the poor binding seen by direct ELISA (Fig. 3C) is due to changes in HveC(143t) that occur as a result of immobilization on the ELISA plate. Furthermore, the data in Fig. 4 suggest that the affinity of all three HveC truncations for gD is similar.
Affinity of binding between gD and HveC(346t) or HveC(143t).
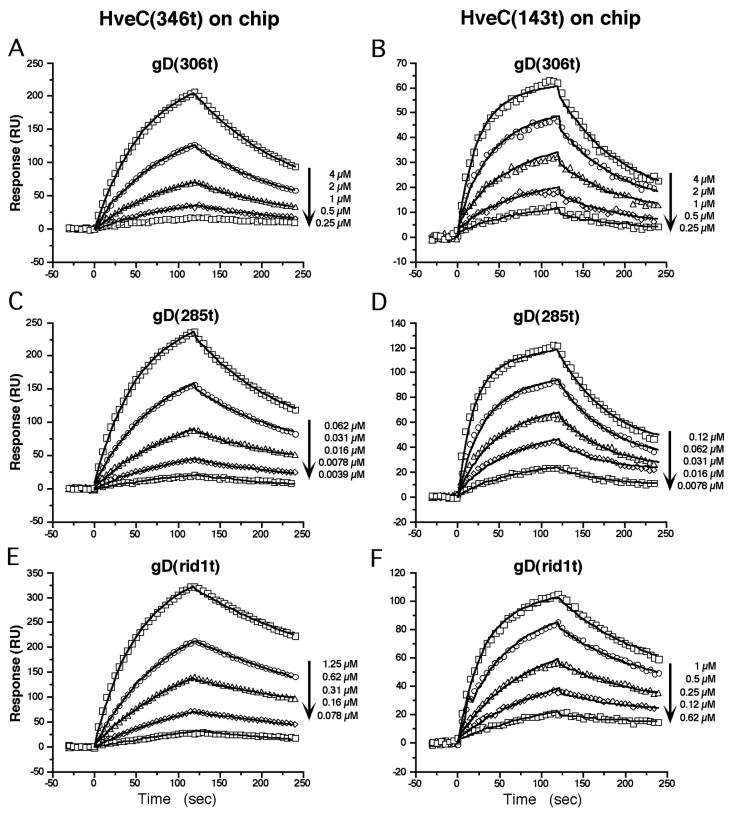
Biosensor analysis was previously used to measure affinity between gD mutant proteins and truncated form of the receptor HveA (47, 59). Here we used the same approach to determine the affinity of gD for HveC(346t) and for the shorter form, HveC(143t). HveC was coupled on the surface of one flow cell (Fc1) of a CM5 chip via primary amines and the second flow cell (Fc2) was left bare. The Fc2 response representing background sticking and bulk change of refractive index was subtracted from the Fc1 response to obtain specific binding data (Fig. 5). Serial dilutions of gD(306t) (Fig. 5A and B), gD(285t) (Fig. 5C and D), and gD(rid1t) (Fig. 5E and F) were flowed over chips carrying HveC(346t) (Fig. 5A, C, and E) or HveC(143t) (Fig. 5B, D, and F). After a baseline was established, the association of gD to immobilized HveC was monitored for 120 s. Buffer was then substituted for gD solution, and the dissociation of the complex was monitored for another 2 min. A global fit of the data was obtained by using the BIAevaluation 3.0 software for a 1:1 Langmuir model (Fig. 5). Kinetics values and affinity constants for each of the pairs tested are summarized in Table 1. In the case of HveC(346t) we calculated a dissociation constant (KD) of 3.2 × 10−6 M for gD(306t). The affinity of gD(285t) for HveC(346t) was enhanced more than 80 times, mainly because of an increase of the on rate (kon). The association between HveC(346t) and gD(rid1t) showed a KD of 1.7 × 10−7 M. The 20-fold increase in affinity compared to gD(306t) was also due to a higher on rate.
FIG. 5.
Analysis of gD binding to HveC in real time. HveC(346t) (A, C, and E) or HveC(143t) (B, D, and F) were immobilized on a CM5 biosensor chip to 1,600 and 300 RU, respectively, in a Biacore X instrument. Various concentrations of gD(306t) (A and B), gD(285t) (C and D), and gD(rid1t) (E and F) were flowed over the chip for 2 min (association) and then replaced by buffer for another 2 min (dissociation). Sensorgrams of corrected data are represented after the subtraction of signal from the control flow cell. Data points were collected at 5 Hz but, for clarity, only one every 25 points is represented here by a symbol. The solid line shows the best fit obtained after global fitting with the BIAevaluation 3.0 software (5).
TABLE 1.
Kinetic and affinity values for HveCt-gDt complex formationa
| HveC type | gD(306t) KOS
|
gD(285t) KOS
|
gD(306t) rid1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M) | kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M) | kon (103 s−1 M−1) | koff (10−2 s−1) | KD (10−6 M) | |
| HveC(346t) | 2.2 | 0.7 | 3.2 | 190 | 0.73 | 0.038 | 27 | 0.47 | 0.17 |
| HveC(143t) | 9.0 | 1.1 | 1.2 | 370 | 1.3 | 0.035 | 39 | 0.8 | 0.2 |
KD = koff/kon. Data represent an average of at least two experiments.
In the case of HveC(143t), the V-domain alone, the affinity for the various forms of gD followed the same trend. gD(306t), with a KD of 1.2 × 10−6 M, had the lowest affinity and gD(285t) (KD = 3.5 × 10−8 M) showed the highest affinity. Again, the variation in affinity was caused primarily by changes in the rate of complex formation. Again, gD(rid1t) showed an intermediate affinity for HveC(143t).
Both HveC(346t) and HveC(143t) displayed similar affinity for gD(285t) or gD(rid1t), with no significant differences in either on or off rates, whereas gD(306t) appeared to have a slightly higher affinity for HveC(143t) than for HveC(346t).
Blocking of HSV infection with soluble truncated forms of HveC.
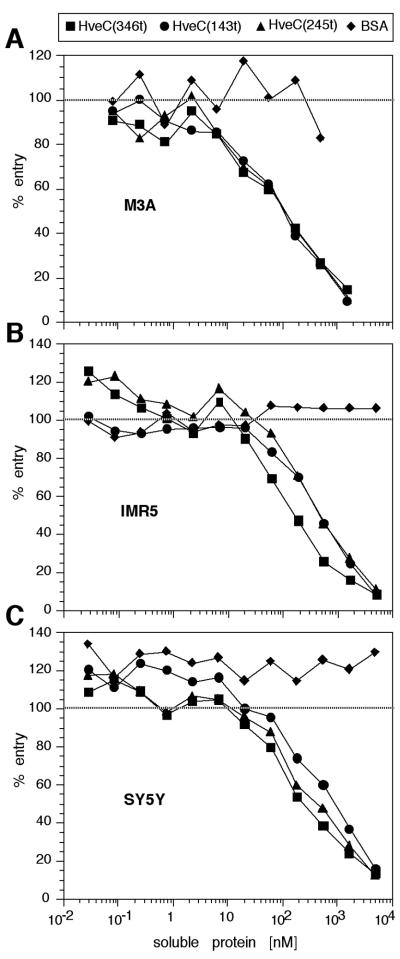
We previously showed that HveC(346t) blocked HSV-1 infection of several human cell lines (18). We therefore examined the ability of HveC(245t) and HveC(143t) to block virus infection. In our standard assay, purified HSV-1 KOS tk12 was preincubated with the three soluble receptors prior to addition to target cells. M3A cells, derived from CHO cells, express full-length human HveC constitutively. Infection of M3A cells with HSV-1 KOS tk12 was efficiently blocked by all three HveC truncations (Fig. 6A). No significant difference was detected in the ability of any of the truncations to inhibit infection.
FIG. 6.
Blocking of HSV infection with soluble HveC truncations. HSV-1 KOS tk12 was preincubated with variable concentrations of soluble HveC truncations or BSA prior to addition to M3A (A), IMR5 (B), or SY5Y (C) cells. Cells were lysed at 5.5 h postinfection, and the β-galactosidase activity was measured. A value of 100% of entry corresponds to the β-galactosidase activity induced after infection with HSV-1 KOS tk12 at a similar multiplicity of infection (0.5 to 1 PFU/cell) in the absence of soluble inhibitor.
We extended this study to include the neuroblastoma cell lines IMR5 and SY5Y. Infection of these cells has also been shown to be inhibited by soluble HveC(346t) (18). Figure 6B and C shows that HveC(245t) and HveC(143t) were able to block HSV-1 infection of these cells as efficiently as HveC(346t). Infection of nondifferentiated NT-2 cells and HeLa cells could also be blocked by all three soluble forms of HveC with similar efficiency (data not shown).
Binding of soluble HveC to virion.
Binding of HveC(346t) to gD on HSV-1 KOS virions was shown previously (29). The ability of the smaller forms of HveC to block infection suggested that these proteins also directly interacted with viral particles. To test this, purified virions were preincubated with the soluble receptors prior to sedimentation through a sucrose gradient. The presence of receptor in the virus band collected from the gradient is indicative of direct binding of receptor to virions. Western blot analysis of the viral band showed that both HveC(346t) and HveC(143t) could be detected and thus had bound to KOS viral particles (Fig. 7). The different glycosylated forms of HveC(143t) could be detected. Similar amounts of each virus were recovered and loaded onto the gel, as reflected by the amount of VP5 protein detected with the NC1 antibody (Fig. 7). Both HveC(346t) and HveC(143t) were also bound to purified HSV rid1 particles in this assay (data not shown).
FIG. 7.

Binding of soluble HveC to HSV particles. Purified HveC(346t) or HveC(143t) (100 μg) were incubated with 107 PFU of purified HSV-1 KOS for 90 min at 4°C. Viral particles were then sedimented through a discontinuous sucrose gradient. The virus-containing fraction was collected, concentrated, and analyzed by Western blot for detection of HveC and virus. Receptors were detected with anti-tetra-His antibody, and VP5 capsid protein was detected with NC-1 polyclonal rabbit serum simultaneously.
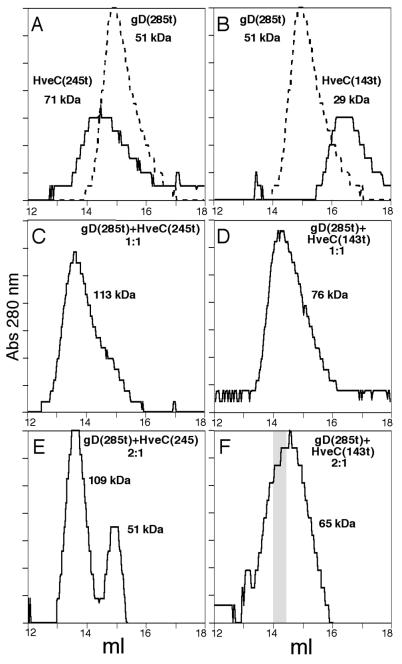
Oligomerization of HveC(143t) or HveC(245t).
We previously showed that HveC(346t) in solution appears as a high-molecular-mass complex of 176 kDa, a size consistent with that of a tetramer (29). Here gel filtration studies were performed with the other two HveC truncations on a Superdex 200 size exclusion column. HveC(245t), with a molecular mass determined by mass spectrometry of 28 kDa, elutes with an apparent size of 71 kDa (Fig. 8A), a finding consistent with the formation of a dimer in solution. Similarly, the 15-kDa HveC(143t) appears to form a 29-kDa dimer in solution (Fig. 8B). The difference in oligomerization between HveC(346t), a tetramer (29), and HveC(245t), a dimer, suggests that the third Ig-domain of HveC can promote a higher level of oligomerization of HveC. gD(285t) eluted from the column with an apparent size of 51 kDa, a result consistent with the presence of a dimeric form of gD (Fig. 8A and B). We previously observed that gD was essentially a dimer in solution and on virus (22, 59).
FIG. 8.
Gel filtration chromatography of HveC(245t) and HveC(143t) alone or in a complex with gD(285t). Elution profiles of HveC(245t) and HveC(143t) loaded at 33 and 66 μM respectively, on a Superdex 200 column are shown in panels A and B as solid lines. An elution profile of gD(285t) (26 μM) is shown as a dotted line in panels A and B. Elution profiles of gD(285t)-HveC(245t) complex (C and E) or gD(285t)-HveC(143t) complex (D and F) at the given ratios are shown. Molecular sizes were determined by calibrating the column with standards of proteins ranging from 13.7 to 669 kDa. The shaded area in panel F indicates the fraction used for quantification of HveC(143t) and gD(285t) in Fig. 9.
Complex formation in solution.
We previously demonstrated direct binding in solution of HveC(346t) to a mutant form of gD from HSV-1 KOS. We found that gD(Δ290-299t) bound to HveC(346t) with a stoichiometry of 2:1 and disrupted the putative HveC(346t) tetramer (29). Here we analyzed the sizes of complexes formed between gD(285t) and HveC(245t) by gel filtration (Fig. 8C and E). When equimolar amounts of HveC(245t) and gD(285t) were mixed, a complex of 113 kDa eluted from the column, a size consistent with a complex containing one dimer of gD and one dimer of HveC(245t) (51 plus 71 kDa), hence with a stoichiometry of 1:1. When twice the amount of gD(285t) is added to HveC(245t) a peak of free gD(285t) dimer could be detected on the profile (Fig. 8E) and by Western blot (data not shown). When initially present at equimolar concentrations, HveC(143t) and gD(285t) formed a complex with an apparent size of 76 kDa (Fig. 8D), a size consistent with a complex containing one dimer of each component (29 plus 51 kDa), also with a stoichiometry of 1:1. No free receptor was detected. When twice the amount of gD(285t) was initially present, the peak shifted to 65 kDa (Fig. 8F). This is due to an excess of free gD in fractions containing minimal amounts of HveC(143t), as detected by Western blot analysis of the fractions (data not shown). Since the 1:1 ratio did not correlate with the stoichiometry obtained earlier with gD(346t) and gD(Δ290-299t), we repeated the study here with HveC(346t) and gD(285t). In the presence of excess gD(285t), the complex eluted with a size of 176 kDa (data not shown), a result consistent with our previously observed complex made of two dimers of gD and one dimer of HveC(346t), a stoichiometry of 2:1.
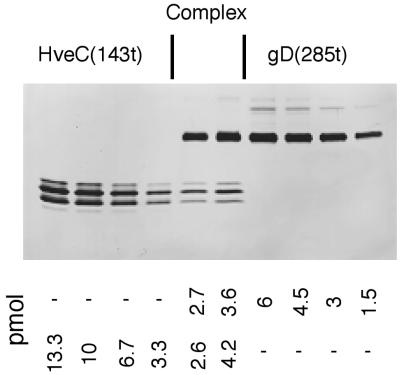
To confirm the stoichiometry of the gD(285t)-HveC(143t) complex, we separated and quantified each component of the complex by PAGE (Fig. 9). A fraction containing the complex formed in the presence of excess gD(285t) but separated from the peak containing free gD was analyzed at two different dilutions (Fig. 9, lanes 5 and 6). By comparing the intensity of the silver-stained proteins in the fraction with known standards (lanes 1 to 4 and 7 to 10), we determined that similar amounts of gD(285t) and HveC(143t) were present in the complex. This indicated a 1:1 ratio of HveC(143t) to gD(285t), supporting the ratio deduced from the size of the complex. This experiment could not be performed with the gD(285t)-HveC(245t) complex because both protein monomers are too close in size to be sufficiently separated on a gel to allow accurate quantification. When the amounts of gD(285t) and HveC(346t) in the complex formed in the presence of excess gD(285t) were quantified by silver-stained gel analysis, the 2:1 ratio was obtained (data not shown), thus confirming our previous observations (29).
FIG. 9.
Quantitation of gD(285t) and HveC(143t) in the complex. Two aliquots of a fraction containing the gD(285t)-HveC(143t) complex separated by gel filtration (Fig. 8F) were loaded onto a 16% polyacrylamide gel (lanes 5 and 6). Standards of known amounts of HveC(143t) and gD(285t) were loaded in lanes 1 to 4 and lanes 7 to 10, respectively, with amounts of proteins as indicated under the gel. After the silver staining of the gel, the intensity of the protein bands was measured by densitometry. The amount of each protein in the complex is indicated and is based on the comparison with the standards.
DISCUSSION
When Ig-like molecules are used as cellular receptors by viruses, the most membrane-distal domain is usually the site of direct contact with the viral ligand (14, 16, 38, 40, 42). The HveC V-domain fused to PVR Ig-domains 2 and 3 or directly anchored on the cell membrane was shown to confer HSV susceptibility to normally resistant cells (9). Cocchi et al. (9) also showed that the most distal Ig-domain of HveC was also the binding site for HSV and for the neutralizing MAb R1.302. However, in that study, the V-domain directly anchored to the cell membrane was poorly used as a receptor, and the soluble construct consisting of the HveC V-domain fused to the Ig Fc portion had a reduced capacity to bind gD. These observations suggest that the second and/or third Ig domains are necessary to ensure proper infectivity by influencing the availability or the affinity to gD. In order to assess the contribution of each Ig-like domain to gD binding, we produced soluble HveC truncations in the baculovirus expression system. We generated three truncations of HveC containing either the full ectodomain [HveC(346t)] (29), the two distal domains [HveC(245t)], or the V-domain alone [HveC(143t)] without the addition of substitutive Ig domains such as the Ig Fc region. The ability of the three truncated forms of HveC to bind virions and gD were compared by using several different assays. We also examined the kinetics and stoichiometry of complex formation with each form of HveC.
The HveC V-domain is sufficient for complete binding to gD.
The HveC V-domain was produced as a soluble 15-kDa glycoprotein and appeared to be correctly folded, since it was recognized by the conformation-dependent MAb R1.302. However, this shortest form of HveC was altered by immobilization on an ELISA plate since it partially lost its ability to bind MAb R1.302. This result is reminiscent of the finding that the single PVR V-domain could not be detected by a specific MAb after immobilization on an ELISA plate (2). Also, stability of the ICAM-1 first Ig domain, where the binding site for rhinovirus was mapped (38, 50), was influenced by mutations in the second Ig domain; however, binding to rhinovirus was not affected (7, 50).
Although the immobilized HveC V-domain showed reduced binding to gD, it could efficiently compete with HveC(346t) for binding with gD in solution. Furthermore, the single V-domain bound HSV particles and blocked infection of HveC expressing CHO cells and neuroblastoma cell lines as well as the full ectodomain. This finding confirms previous observations with the HveC V-domain fused to the Ig Fc region (9) and indicates that additional Ig-like domains are dispensable at least for the binding of gD. In the case of MHV receptor (bgp1a) (57), the binding of viral spike protein to the Ig-like domain 1 is influenced by adjacent Ig-like domains (14). In this instance, the first two Ig-domains of soluble bgp1a have lower affinity for MHV and reduced neutralizing activity. However, when Ig-domain 4 was placed in second position, it affected binding and neutralizing activities differently than the normal Ig-domain 2 (61).
Affinity of the HveC ectodomain for gD.
To study the HveC-gD interaction in real time, we used biosensor technology. First, kinetic and affinity values were calculated for the interaction between the full HveC ectodomain and three truncated forms of HSV-1 gD. The affinity of HveC(346t) for gD(306t) from the KOS strain of HSV-1, which we consider the wild-type form, was in the micromolar range, with a KD comparable to that observed for the interaction between gD(306t) and the structurally unrelated receptor, HveA (47, 59). By ELISA, the C-terminal truncation gD(285t), which lacks functional region IV, and a gD(rid1t) protein, with a point mutation at position 27, showed an enhanced binding to HveC(346t) (29, 47, 59). Biosensor analysis confirmed an 80-fold enhancement of affinity of gD(285t) for HveC and an enhancement of about 20 times of the overall affinity of gD(rid1t) for HveC. In each case the variation in the affinity of these gD proteins for HveC was caused by changes in the rapidity of complex formation (on rates), whereas the off rates were similar. This suggests that, once the complex is formed, its stability is independent of the gD form bound. This observation also held true in the case of HveA binding to most gD mutants studied (47, 59).
It is of note that, unlike the interaction between gD and HveA, the complex formation between gD and HveC did not reach equilibrium even after prolonged contact time, regardless of which form of soluble gD or HveC was used. In addition, when we plotted the concentration-dependent on rate (kobs) versus concentration we obtained a convex curve, indicating that the ligand immobilized on the chip was heterogeneous (data not shown). HveC heterogeneity might result from its direct immobilization on the biosensor chip (5). However, the potential for self-association of HveC could be responsible for heterogeneity and could lead to a more complicated interaction with gD than the simple 1:1 interaction model used to fit biosensor data.
The HveC V-domain affinity for gD compares to the full HveC ectodomain.
We compared the affinity of gD for the HveC V-domain alone with the affinity of gD for the complete HveC ectodomain. In the case of gD(306t), the affinity for HveC(143t) was slightly higher than for HveC(346t), whereas no differences were observed in the affinity of gD(285t) or gD(rid1t) for the two receptor forms. This indicates that the V-domain alone retains all the necessary elements to allow binding to gD variants spanning a broad range of affinity. A biosensor approach was used to study ICAM-1 binding to rhinovirus (7). Truncated ICAM-1 proteins containing the first two domains could be generated without significantly affecting the affinity for rhinovirus. However, an ICAM-1 protein truncated five amino acids upstream, at the bottom of domain 2, had an increased off rate correlated with a decreased ability to block infection and to bind rhinovirus (7, 20, 36). On the contrary, the HveC V-domain alone did not display any variation in the rate of complex dissociation.
Oligomerization of HveC in solution and complex formation with soluble gD.
When run on a size exclusion chromatography column, HveC(346t) had a molecular size of 176 kDa, a result consistent with the formation of a tetramer in solution (29). In contrast, the sizes of HveC(143t) and HveC(245t) in solution were consistent with formation of dimers. By analogy, ICAM-1 domains 1 and 2 also form dimers via the interaction of two domains 1 (3, 8). Our data strongly suggest that the third Ig-like domain (closest to the transmembrane domain) is involved in a higher order of oligomerization of HveC (i.e., a tetramer). In a model proposed by Casasnovas et al. (8) for ICAM-1, the Ig-like domain 5 close to the transmembrane domain is also proposed to be involved in oligomerization. We analyzed the complexes formed in solution between each truncated form of HveC and gD(285t). In the case of HveC(143t) and HveC(245t), the data suggested a complex consisting of one gD dimer and one HveC dimer. The 1:1 ratio obtained in both complexes is in contrast to the 2:1 ratio of gD(Δ290-299t) (29) or gD(285t) bound to HveC(346t). It is possible that the higher degree of oligomerization of HveC(346t) in solution influences the stoichiometry of the complex with gD. It should be noted that in spite of the apparent difference in stoichiometry, gD(285t) binds to HveC(143t) and to HveC(346t) with equal affinity. In addition, all of the biosensor data fit the 1:1 model, which suggests that there is not a second gD binding site on HveC Ig domain 2 or 3. Since the size of the gD-HveC(346t) complex is similar to that of HveC(346t) alone, it is likely that association with gD may disrupt the HveC tetramer into dimers. This distinct property of HveC(346t) suggests that the V-domain is somehow involved with domain 3 in tetramer formation. Further studies need to be done to better correlate the oligomerization of HveC and stoichiometry of the gD-HveC complex. The cell membrane anchored HveC V-domain alone was markedly less efficient as a receptor for HSV entry (9). Since all of the elements necessary for binding gD with full affinity are present in the V-domain of HveC, the decrease in efficiency of the V-domain alone might be due to poor availability, structural instability or incorrect oligomerization due to absence of Ig-domains 2 and/or 3. These observations might become more relevant once the oligomeric status of HveC at the surface of cells is determined.
ACKNOWLEDGMENTS
This investigation was supported by Public Health Service grants AI-18289 to R.J.E. and G.H.C. and AI-36293 to P.G.S. from the National Institute of Allergy and Infectious Diseases (NIAID), grant NS-30606 to R.J.E. and G.H.C. from the National Institute of Neurological Diseases and Stroke, and grant CA-21776 to P. G. S. from the National Cancer Institute. C.K. was supported by a fellowship (823A-053464) of the Swiss National Science Foundation. R.J.G. was supported by a fellowship from the NIAID (F32 AI-09471). We thank the Schools of Dental and Veterinary Medicine of the University of Pennsylvania for supplying funds for the purchase of the Biacore X.
We thank William T. Moore and John D. Lambris of the Protein Chemistry Laboratory of the School of Medicine of the University of Pennsylvania, supported by core grants of the Diabetes and Cancer Centers (DK-19525 and CA-16520), for mass spectrometric analysis. We thank Ruliang Xu for the M3A cells and S. McClellan (Beckman/Coulter) for R1.302 MAb. We are grateful to Nicholas Fasano for technical assistance and to Sharon Willis for critical reading of the manuscript and helpful discussion.
REFERENCES
- 1.Aoki J, Koike S, Asou H, Ise I, Suwa H, Tanaka T, Miyasaka M, Nomoto A. Mouse homolog of poliovirus receptor-related gene 2 product, mPRR2, mediates homophilic cell aggregation. Exp Cell Res. 1997;235:374–384. doi: 10.1006/excr.1997.3685. [DOI] [PubMed] [Google Scholar]
- 2.Arita M, Koike S, Aoki J, Horie H, Nomoto A. Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J Virol. 1998;72:3578–3586. doi: 10.1128/jvi.72.5.3578-3586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bella J, Kolatkar P R, Marlor C W, Greve J M, Rossmann M G. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as LFA-1 integrin ligand. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Biacore Inc. BIAevaluation software handbook, version 3.0. Uppsala, Sweden: Biacore AB; 1997. [Google Scholar]
- 6.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casasnovas J M, Bickford J K, Springer T A. The domain structure of ICAM-1 and the kinetics of binding to rhinovirus. J Virol. 1998;72:6244–6246. doi: 10.1128/jvi.72.7.6244-6246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casasnovas J M, Stehle T, Liu J H, Wang J H, Springer T A. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attribute of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen G H, Isola V J, Kuhns J, Berman P W, Eisenberg R J. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J Virol. 1986;60:157–166. doi: 10.1128/jvi.60.1.157-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalgleish A G, Beverly P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 13.Dean H J, Terhune S S, Shieh M, Susmarski N, Spear P G. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 14.Dveksler G S, Pensiero M N, Dieffenbach C W, Cardellichio C B, Basile A A, Elia P E, Holmes K V. Mouse hepatitis virus strain A59 and blocking antireceptor monoclonal antibody bind to the N-terminal domain of cellular receptor. Proc Natl Acad Sci USA. 1993;90:1716–1720. doi: 10.1073/pnas.90.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberlé F, Dubreuil P, Mattei M-G, Devilard E, Lopez M. The human PRR2 gene, related to the poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 16.Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller A O, Lee W C. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–12. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geraghty R J, Krummenacher C, Eisenberg R J, Cohen G H, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 19.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClellend A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 20.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handler C G, Cohen G H, Eisenberg R J. Crosslinking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handler C G, Eisenberg R J, Cohen G H. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70:6067–6075. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herold B C, Visalli R J, Sumarski N, Brandt C, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 24.Hsu S, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 25.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson R M, Spear P G. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koike S, Horie H, Ise I, Okitsu A, Yoshida M, Iizuka N, Takeuchi K, Takegami T, Nomoto A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9:3217–3224. doi: 10.1002/j.1460-2075.1990.tb07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghry R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda K, Geyer H, Geyer R, Doerfler W, Klenk H-D. The oligosaccharides of influenza virus hemagglutinin expressed in insect cells by a baculovirus vector. Virology. 1990;174:418–429. doi: 10.1016/0042-6822(90)90095-9. [DOI] [PubMed] [Google Scholar]
- 31.Kwon B S, Tan K B, Ni J, Oh K-O, Lee Z H, Kim K K, Kim Y-J, Wang S, Gentz R, Yu G-L, Harrop J, Lyn S D, Silverman C, Porter T G, Truneh A, Young P R. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J Biol Chem. 1997;272:14272–14276. doi: 10.1074/jbc.272.22.14272. [DOI] [PubMed] [Google Scholar]
- 32.Landau N R, Warton M, Littman D R. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 33.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 34.Lopez M, Eberlé F, Mattei M-G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 35.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. CD155 Workshop: identification of a new class of IgG superfamily antigens expressed in hematopoiesis. In: Garland Publishing I, editor. Leukocyte typing VI, white cells differentiation antigens. I. London, England: Garland Publishing; 1997. pp. 1081–1083. [Google Scholar]
- 36.Martin S, Casasnovas J M, Staunton D E, Springer T A. Efficient neutralization and disruption of rhinovirus by chimeric ICAM-1/immunoglobulin molecules. J Virol. 1993;67:3561–3568. doi: 10.1128/jvi.67.6.3561-3568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauri D N, Ebner R, Kochel K D, Montgomery R I, Cheung T C, Yu G-L, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin (LT) a are ligands for herpesvirus entry mediator (HVEM) Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 38.McClelland A, DeBear J, Yost S C, Meyer A M, Marlor C W, Greve J M. Identification of monoclonal antibody epitopes and critical residues for rhinovirus binding in domain 1 of intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 40.Mizukami T, Fuerst T R, Berger E A, Moss B. Binding region for human immunodeficiency virus (HIV) and epitopes for HIV-blocking monoclonal antibodies of the CD4 molecule defined by site-directed mutagenesis. Proc Natl Acad Sci USA. 1988;85:9273–9277. doi: 10.1073/pnas.85.23.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 42.Morrison M E, He Y-J, Wien M W, Hogle J M, Racaniello V. Homolog-scanning mutagenesis reveals poliovirus receptor residues important for virus binding and replication. J Virol. 1994;68:2578–2588. doi: 10.1128/jvi.68.4.2578-2588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicola A V, Peng C, Lou H, Cohen G H, Eisenberg R J. Antigenic structure of soluble herpes simplex virus glycoprotein D correlates with inhibition of HSV infection. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rux A H, Moore W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rux A H, Willis S H, Nicola A V, Hou W, Peng C, Lou H, Cohen G H, Eisenberg R J. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J Virol. 1998;72:7091–7098. doi: 10.1128/jvi.72.9.7091-7098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 50.Staunton D E, Dustin M L, Erickson H P, Springer T A. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 51.Stauton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R, Cohen G H, Eisenberg R J, Spear P G. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thoulouze M I, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. The neural cell adhesion molecule is a receptor for rabies virus. J Virol. 1998;72:7181–7190. doi: 10.1128/jvi.72.9.7181-7190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warner M S, Martinez W, Geraghty R J, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by herpes simplex virus type 2, mutants of herpes simplex virus type 1 and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 56.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the TNFR superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams R K, Jiang G-S, Snyder S W, Frana M F, Holmes K V. Purification of the 110-kilodalton glycoprotein receptor for mouse hepatitis virus (MHV)-A59 from mouse liver and identification of a nonfunctional, homologous protein in MHV-resistant SJL/J mice. J Virol. 1990;64:3817–3823. doi: 10.1128/jvi.64.8.3817-3823.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willis S H, Peng C, Ponce de Leon M, Nicola A V, Rux A H, Cohen G H, Eisenberg R J. Expression and purification of secreted forms of HSV glycoproteins from baculovirus-infected insect cells. In: Brown S M, MacLean A R, editors. Methods in molecular medicine. 10. Herpes simplex virus protocols. Totowa, N.J: Humana Press, Inc.; 1998. pp. 131–156. [DOI] [PubMed] [Google Scholar]
- 59.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W, Salvador L, Cohen G H, Eisenberg R J. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zelus B D, Wessner D R, Williams R K, Pensiero M N, Phibbs F T, DeSouza M, Dveksler G D, Holmes K V. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1b, and Bgp2 murine coronavirus receptors differ in mouse hepatitis binding and neutralizing activities. J Virol. 1998;72:7237–7244. doi: 10.1128/jvi.72.9.7237-7244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]