Abstract
In polarized epithelial cells, the assembly and release of human immunodeficiency virus type 1 (HIV-1) occur at the basolateral side of the plasma membrane, and the site of assembly is determined by the site of expression of the Env protein. In order to investigate whether the expression of the Env proteins exclusively in the endoplasmic reticulum (ER) can alter the site of virus assembly, we coexpressed the simian immunodeficiency virus (SIV) Gag protein and mutant SIV Env proteins having an ER retrieval signal (KKXX motif). In cells expressing the wild-type (wt) Env protein or coexpressing Env and Gag proteins, the Env protein was processed into the surface (SU) and transmembrane (TM) proteins. In contrast, in cells expressing the mutant Env proteins alone or in combination with Gag, the Env proteins were retrieved to the ER and were not proteolytically processed. Coexpression of the Gag and ER-retained mutant Env proteins resulted in a transient decrease in the release of the Gag protein into the medium, suggesting an interaction between the Gag and ER-retrieved Env proteins. Using saponin-permeabilized cells coexpressing Gag and Env proteins, we obtained further evidence for Env-Gag interaction. A monoclonal antibody specific to the SIV Gag protein was found to coimmunoprecipitate both the Gag and Env proteins. The interaction was specific, as coexpressed SIV Env proteins without the cytoplasmic tail or a chimeric HIV-1 Env proteins with the CD4 cytoplasmic tail were not coimmunoprecipitated by the Gag-specific antibody. Electron microscopic analyses indicated that assembly of virus particles occurred only at the surfaces of cells in which the Gag protein was coexpressed with either the wt or ER-retrieved mutant Env protein. These data indicate that although the Env and Gag proteins interact intracellularly, the site of assembly of SIV is not redirected to an intracellular organelle by the retrieval of the Env protein to the ER.
The Env proteins of retroviruses are synthesized on ribosomes in the endoplasmic reticulum (ER), undergo oligosaccharide processing and proteolytic cleavage in the Golgi complex, and are transported to the cell surface. In contrast, the Gag and Pol proteins are synthesized on cytoplasmic polysomes and are thought to be directly transported to the plasma membrane. A unique property of the retroviruses is the ability of the Gag protein to assemble into noninfectious virus-like particles (VLPs) which are released by budding at the cell surface, without a requirement for the Env protein (7, 15, 18, 31). The assembly of enveloped viruses may occur at the plasma membrane or at intracellular organelles, and the site of assembly varies for different virus families (28, 36). Although human and simian immunodeficiency viruses (HIV and SIV, respectively) are usually found to assemble by budding at the plasma membrane, certain cell types are reported to support virus assembly at intracellular compartments. For example, electron microscopic studies have indicated that HIV can assemble and accumulate within cytoplasmic vesicles in macrophages (25). SIVmac239 is a T-lymphocyte-tropic virus which was reported to be unable to assemble intracellularly or at the cell surface in macrophages, whereas a variant macrophage-tropic chimeric virus (SIVmac239/17E) was found to produce virus particles which accumulated in cytoplasmic vacuoles of macrophages (37). Retroviruses of another genus, the foamy viruses, characteristically assemble at intracellular compartments, and this phenomenon was attributed to the localization of the Env protein in the ER by virtue of a KK motif (16, 17).
Previous studies in our laboratory indicated that when expressed alone, the HIV type 1 (HIV-1) envelope protein is expressed preferentially at the basolateral surface (26), whereas the Gag protein is transported to both the apical and basolateral sides of polarized epithelial cells (27). When both the Env and Gag proteins were coexpressed, the expression of the Env protein at the basolateral surface resulted in the preferential release of virus particles containing Gag and Env at this plasma membrane domain (27). Similarly, in neurons, the Gag protein is expressed both in the somatic and axonal regions, whereas the Env protein is preferentially expressed only at the somatic region, and coexpression of Env and Gag proteins resulted in the exclusion of the Gag protein from the axons (41). Thus, the Env protein plays a major role in determining the cellular site of virus assembly.
We have recently characterized a series of env mutants which encode SIVmac239 Env proteins with COOH-terminal sequences comprising a KK motif with lysines at the −3 and −4 positions. Some of these mutant proteins, when expressed in HeLa T4 cells, were found to be effectively retained in the ER and hence do not undergo proteolytic processing and have defective cell fusion activity (39). In order to investigate whether the ER-retained Env proteins interact with the Gag protein or result in the alteration of the site of virus assembly, we have analyzed the localization of viral proteins and virus assembly in cells expressing SIVmac239 Env mutants having ER retention signals together with the SIV Gag protein.
MATERIALS AND METHODS
Cells, virus, and reagents.
HeLa T4 and HeLa cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum. The recombinant vaccinia virus vTF7-3, which expresses bacteriophage T7 RNA polymerase in infected cells (14), was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health). Plasmids Env164RS, Env37RS, and Env18RS encode proteins with 164, 37, and 18 amino acids in the cytoplasmic tail, respectively, and the amino acids at the −3 and −4 positions of the C-terminal end encode a KKXX motif (ER retention signal). EnvΔ164aa encodes a protein in which the entire cytoplasmic tail is deleted. For expression of the SIVmac239 Gag protein, a plasmid pGEM-11Z Gag described by Vzorov and Compans (40) was used. Since the gene encodes a truncated version of the Gag protein and does not code for the protease, the resulting protein had a molecular mass of 46 kDa and no proteolytic products were observed (40). This construct was used because the resulting protein has the necessary and sufficient domains for interaction with the Env protein and the lack of Gag processing simplified further analyses. The rhesus antiserum to SIV were kindly provided by P. Marx and Shawn O’Neil, and monoclonal anti-p28 serum was purchased from Intracel.
Transfection, radioimmunoprecipitation, and protein analyses.
Transfection and protein analyses were done as described previously (39). Briefly, HeLa T4 or HeLa cells (5 × 105) were seeded onto 35-mm-diameter dishes 1 day prior to transfection. The cells were infected for 1 h with vaccinia virus vTF7-3 (multiplicity of infection, 10) which expresses the T7 RNA polymerase. DNA (5 μg each of Gag- and Env-expressing plasmids) was mixed with Lipofectin (10 μg) and added to the cells after removal of the virus inoculum. At 15 h posttransfection, the cells were starved in medium lacking methionine and cysteine and then labeled for 30 min with 100 μCi of 35S labeling mix containing methionine and cysteine (NEN Life Science Products). At the end of the labeling period, the label was removed and Dulbecco’s modified Eagle medium with fetal calf serum was added and chased for different times up to 12 h. Cells lysed in radioimmunoprecipitation buffer (0.15 M NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 20 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride) were clarified, and proteins were immunoprecipitated with polyclonal SIV-specific antiserum. The immunoprecipitated proteins were extensively washed and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. The media were also collected at different chase times, immunoprecipitated, and analyzed similarly.
Quantification of Gag proteins released into the media.
The cells expressing the Gag protein alone or coexpressing either wild-type (wt) Env or Env164RS were labeled with 35S and the Gag proteins released into the media were collected at the times indicated in the figures. The cell debris was removed by centrifugation at 12,000 rpm (Eppendorf centrifuge) for 10 min, and the supernatants were immunoprecipitated with SIV antiserum and analyzed by SDS-PAGE and autoradiography. The proteins were quantified with a PhosphorImager, and the relative units are plotted in a graph.
Assay for intracellular interaction of SIV Gag and Env proteins.
HeLa T4 cells coexpressing Gag and various SIV Env mutants were labeled with 100 μCi of [35S]methionine/cysteine for 3 h. At the end of the labeling period, the cells were washed with phosphate-buffered saline (PBS) and permeabilized with saponin (50 μg/ml) (30, 39). After permeabilization, the cells were incubated with anti-Gag antibody for 60 min at 4°C. The cells were then washed with PBS to remove the unbound antibody and lysed in 3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propanesulfonate (CHAPS) buffer (4). The cell extract was centrifuged at 14,000 rpm for 10 min at 4°C. Protein A (5 mg) was added to the supernatant to detect the Env-Gag protein complexes. After the protein A beads were washed with CHAPS buffer, the proteins were analyzed by SDS-PAGE and visualized by autoradiography.
Electron microscopy.
HeLa T4 cells coexpressing Gag and the different forms of the Env proteins were fixed with 1% glutaraldehyde in PBS, and postfixation and staining were done with osmium tetroxide and tannic acid, respectively (1). After dehydration with ethanol, the samples were embedded in Epon (Fluka). Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Philips CM 10 electron microscope at 80 kV.
RESULTS
Coexpression of Gag with ER-retained SIVmac239 Env proteins.
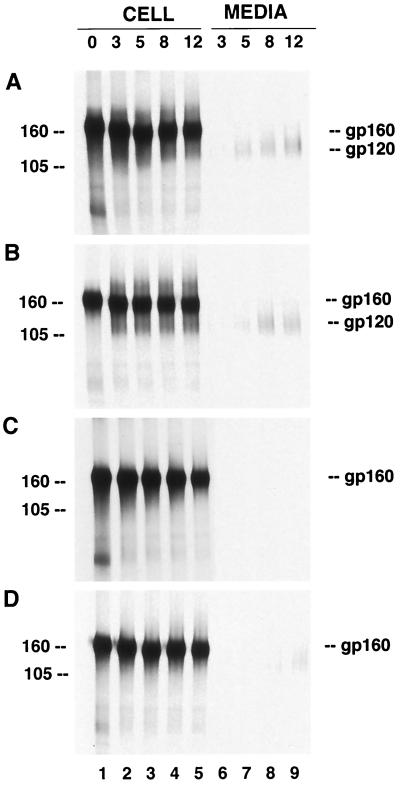
We recently described the characterization of SIV Env protein mutants having an ER retrieval signal (39). In order to investigate the processing kinetics of the Env proteins and their release in cells coexpressing Gag with such ER-retrieved Env proteins, we transfected HeLa T4 cells with plasmids encoding the SIV Gag protein alone or in combination with SIV wt Env or SIV Env164RS. Using indirect immunofluorescence, we found that 70 to 75% of the transfected cells expressed both Env and Gag proteins. When cells coexpressing the wt SIV Env proteins were analyzed by immunoprecipitation, the gp160 precursor was observed during the 30-min pulse and was processed, as evidenced by the presence of gp120 during the chase periods (Fig. 1A, lanes 1 to 5). Secretion of gp120 into the culture medium was readily detected from wt-Env-expressing cells after a 3-h chase period and progressively increased during further chase periods (lanes 6 to 9). When wt Env was coexpressed with the Gag protein, similar intracellular processing (Fig. 1B, lanes 1 to 5) and gp120 secretion kinetics were observed (lanes 6 to 9). In contrast, in cells transfected with plasmid Env164RS, the precursor protein remained a 160-kDa protein without detectable processing into gp120 (Fig. 1C, lanes 1 to 5) and gp120 was not secreted from the cells (lanes 6 to 9). The lack of proteolytic processing was expected, based on our previous observations, and was due to the retrieval of the proteins to the ER before the proteins reach the cellular compartment where the processing occurs (39). In the case of cells coexpressing SIV Env164RS and Gag, intracellular processing of Env was also not observed (Fig. 1D, lanes 6 to 9). Interestingly, however, coexpression of Gag with SIV Env164RS resulted in the release of low levels of the mutant Env proteins into the media (lanes 6 to 9). Similar results were obtained with HeLa cells which do not express the CD4 receptor.
FIG. 1.
Coexpression of the SIVmac239 Gag and Env proteins. HeLa T4 cells were infected with vTF7-3 and transfected with plasmids encoding the Gag and wt Env proteins or SIV Env164RS. For better resolution of gp160 and gp120, an aliquot of the immunoprecipitated proteins was analyzed by SDS–7% PAGE. Fifteen hours posttransfection, the cells were labeled with [35S]methionine/cysteine for 30 min and chased for 3, 5, 8, and 12 h, as indicated above the gels. Samples of cell lysates or media were immunoprecipitated, analyzed by SDS-PAGE, and visualized by autoradiography. Expression of wt SIV Env proteins (A), Env proteins coexpressed with the Gag proteins (B), SIV Env164RS protein (C), and mutant Env164RS coexpressed with Gag (D). gp160 and gp120 denote the Env precursor and surface proteins, respectively. The positions of molecular mass markers (in kilodaltons) used are indicated to the left of the gels.
Release of Gag protein in cells coexpressing wt and ER-retained Env proteins.
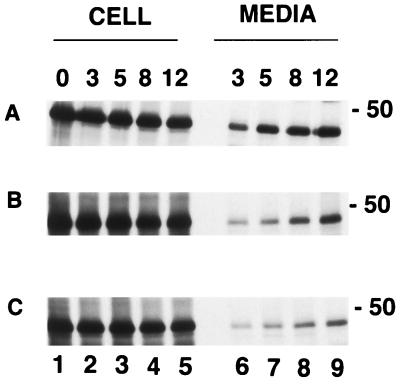
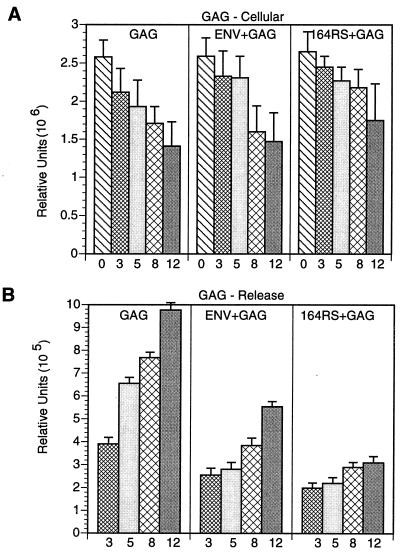
In order to examine the effect of ER-retrieved Env on the Gag protein, the patterns of Gag protein expression and release were analyzed from cells coexpressing Gag with either the wt or ER-retrieved Env proteins. The Gag construct used had a deletion of the coding sequences for the protease (40); therefore, the synthesized protein had a molecular mass of 46 kDa and remained a precursor without processing into p17 and p27. The intracellular level of the protein was found to be gradually reduced during the chase period (Fig. 2A, lanes 1 to 5; Fig. 3A). The Gag protein was detected in the media in the 3-h samples, and the amount progressively increased during the chase periods. The level of Gag protein synthesized when coexpressed with the wt Env protein was similar to that observed when Gag was expressed alone (Fig. 2B, lanes 1 to 5). However, the amount of Gag protein released from cells coexpressing both Gag and Env proteins was found to be reduced (lanes 6 to 9). Interestingly, the amount of Gag protein released from cells coexpressing Gag and Env164RS was further reduced, especially during the 12-h chase period, compared to that from cells coexpressing Gag and wt Env (Fig. 2C, lanes 6 to 9), although the intracellular levels were similar. Quantitation of proteins with a PhosphorImager indicated that the reduction in Gag release from cells expressing Gag and Env164RS compared to that of Gag and Env, was at least 50 to 60% (Fig. 3B). These experiments were repeated three times in CD4-positive and -negative cell lines, and the decrease in the release of Gag was found to be reproducible. Thus, these data suggested that the mutant Env protein might interact with the Gag protein, resulting in slowing the kinetics of Gag release into the media.
FIG. 2.
Expression and release of Gag when coexpressed with wt Env or Env164RS. Cells expressing the Gag proteins were radiolabeled, and aliquots of cell lysate or medium were immunoprecipitated by SIV antiserum. For the detection of Gag from cells coexpressed with wt Env or 164RS, an aliquot from the samples of panels B and D in Fig. 1 was subjected to SDS–8% PAGE. The cells were labeled in a 30-min pulse and chased for 3, 5, 8, and 12 h, as indicated over the gels. Intracellular levels of Gag protein alone (A), Gag protein when expressed with wt Env protein (B), and Gag protein when expressed with Env164RS (C) and the Gag proteins released into the media after the chase periods (in hours) indicated over the gels.
FIG. 3.
Quantitation of Gag proteins. The intracellular levels of the Gag proteins during the times indicated (in hours) under the bars in the figures were quantified by exposing the gel to a PhosphorImager, and the relative units are plotted in panel A. Similar quantification was performed for the Gag proteins released into the media (B). Shown are Gag protein levels in cells expressing Gag (GAG), Gag plus Env, or Env164RS (64RS). The average values for the triplicate assays are plotted in the graph.
Physical association between Gag and Env proteins.
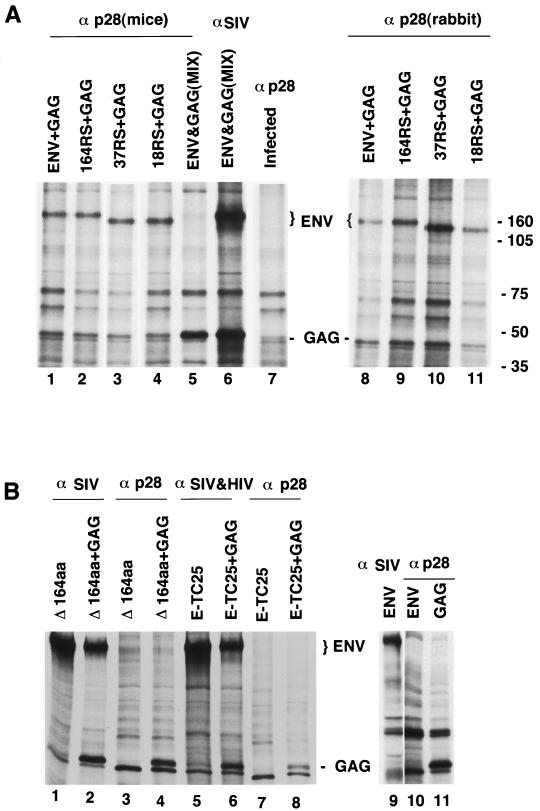
The results described above suggested a possible intracellular interaction between the SIV Env and Gag proteins. To further investigate such interactions, we cotransfected HeLa T4 cells with plasmids encoding the Gag protein together with wt- or mutant-Env-expressing plasmids. Fifteen hours posttransfection, the cells were labeled with [35S]methionine and cysteine for 3 h, permeabilized with saponin (30, 39), and then incubated with anti-Gag antibody. After lysis and immunoprecipitation as described in Materials and Methods, the radiolabeled polypeptides were analyzed by SDS-PAGE (Fig. 4). The results indicate that a fraction of the wt Env proteins was coprecipitated with the Gag protein when a Gag-specific antibody was used (Fig. 4A, lane 1). Interestingly, the mutant Env proteins which are effectively retrieved to the ER were also coprecipitated with the Gag protein, similar to the wt Env protein (lanes 2 to 4). For a control, we mixed the lysates from cells separately expressing the wt Env and the Gag proteins and carried out similar experiments. The presence of both proteins in the lysates was confirmed by immunoprecipitation with anti-SIV antiserum (lane 6). However, the Gag antibodies did not coprecipitate the SIV Env proteins in the lysate (lane 5). These results provide evidence for an intracellular interaction between the Gag and Env proteins of SIV. A similar Env-Gag interaction was also observed with another Gag-specific antibody (rabbit anti-p28 antibody) (Fig. 4, lanes 8 to 11).
FIG. 4.
Intracellular interaction of Gag with SIV Env proteins having an ER retention signal. HeLa T4 cells coexpressing the Gag protein with wt Env, Env164RS, Env37RS, or Env18RS were labeled with [35S]methionine/cysteine for 3 h and permeabilized with saponin for 2 min. The cells were incubated with anti-Gag antibody (α p28) for 60 min, lysed in CHAPS buffer, and processed as described in Materials and Methods. The protein complexes were resolved by SDS-PAGE. (A) Lane 1 shows coimmunoprecipitation of wt Env (ENV) and Gag (GAG) with Gag antibody; lanes 2 to 4 show the coimmunoprecipitation of Gag with Env164RS (164RS), Env37RS (37RS), or Env18RS (18RS) with Gag antibodies. For a control, lysates from cells separately expressing wt Env and Gag proteins were mixed and one portion was immunoprecipitated with Gag antibodies (lane 5) and the other portion was immunoprecipitated with SIV antiserum (lane 6). Lane 7 is a control showing the proteins from vaccinia virus-infected but untransfected cells immunoprecipitated with Gag antiserum. Lanes 8 to 11 show Env-Gag complexes detected by a rabbit antibody to the Gag protein. (B) Cells expressing EnvΔ164aa (Δ164aa) alone (lanes 1 and 3) or coexpressed with Gag (lanes 2 to 4) and cells expressing a fusion protein (E-TC25) having the extracellular domain of HIV-1 gp160 and the anchor and 25 amino acids in the cytoplasmic domains of CD4 alone (lanes 5 and 7) or in combination with Gag (lanes 6 and 8). Rhesus SIV antiserum was used in lanes 1 and 2, and mouse p28 antibody was used in lanes 3 and 4. Lanes 5 and 6 used a combination of SIV and HIV-1 antisera, and lanes 7 and 8 used anti-Gag (mouse p28) antiserum. For another set of controls, we transfected HeLa T4 cells with Env-expressing plasmids, treated with anti-p28 antibody, and lysed in CHAPS buffer. To one portion of the lysate, SIV antiserum was added to detect the Env protein (lane 9) and the other portion (lane 10) was processed as described in above for panel A. Lane 11 shows that the Gag protein reacted with anti-p28 antibody after saponin permeabilization.
The relative levels of Gag and Env proteins precipitated by p28 antibody were found to be reduced compared to the total proteins detected in the cell lysates. We observed that a large portion of the Gag protein leaked out during the saponin permeabilization and antibody treatment, whereas Env proteins were barely detected extracellularly. This is probably due to the membrane-anchored property of the Env protein. We also observed similar leakage of β-galactosidase under these permeabilization conditions, which suggests that proteins with cytoplasmic localization are more prone to leakage during the procedure. Because of this factor, we could not quantitatively compare the Env-Gag interaction in the wt and mutant Env proteins. The p28 antibody did not detect Env-Gag complexes after lysis of coexpressing cells in CHAPS buffer (data not shown), suggesting possible dissociation of the proteins during lysis in this buffer.
In order to determine whether the intracellular interaction involves specific sequences in the Env cytoplasmic tail, we constructed a plasmid (EnvΔ164aa) which encodes an Env protein lacking the cytoplasmic tail and analyzed its ability to interact with the Gag protein. The Env protein without the cytoplasmic tail was expressed at similar levels in the absence or presence of the Gag protein (Fig. 4B, lanes 1 and 2). Interestingly, unlike the full-length Env protein, this protein was not coimmunoprecipitated by the p28 antibody (lanes 3 and 4). These results indicate a requirement for the cytoplasmic tail of the Env protein to interact specifically with the Gag protein. To confirm the specificity of this interaction with the Gag protein, we also analyzed a chimeric protein (E-TC25) having the extracellular domain of HIV-1 gp160 and the anchor and 25 amino acids of the cytoplasmic domain of CD4 (38). Again, this protein was found to be expressed at similar levels alone or with the Gag protein (lanes 5 and 6) but failed to interact with the Gag protein, as evidenced by the lack of detectable coimmunoprecipitation by the p28 antibody (lanes 7 and 8). These results demonstrate a requirement for the Env cytoplasmic domain sequences in order to interact specifically with the Gag protein, as observed by coprecipitation. In order to exclude the possibility that the p28 antibody by itself can react with the Env protein in the absence of the Gag protein, we subjected the Env-expressing cells to saponin permeabilization and treatment with p28 antibody. The results clearly demonstrate that the p28 antibody did not react with Env protein in the absence of the coexpressed Gag protein (lanes 9 to 11).
Electron microscopy of VLP assembly.
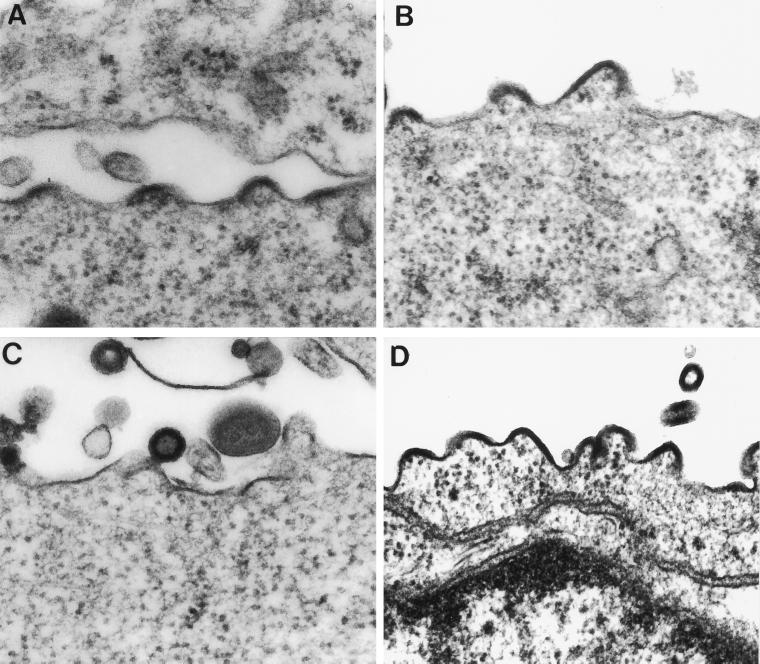
Since we observed an interaction between the Env proteins having an ER retrieval signal and Gag proteins, we investigated whether such an interaction could result in an alteration of the site of virus assembly. If the intracellular accumulation of the Env protein could redirect the site of virus assembly, virus particles might be observed in association with the ER. This was analyzed by cotransfecting HeLa T4 cells with plasmids encoding the wt or mutant Env proteins and the Gag protein. In cells cotransfected with plasmids encoding the wt Env and Gag proteins, VLP assembly and release were observed at the plasma membrane (Fig. 5A). In cells transfected with plasmids encoding Gag and Env164RS, we did not observe a change in the assembly site, as budding VLPs were observed only at the cell surface (Fig. 5B). We also analyzed the site of virus assembly by coexpression of Gag with Env truncation mutants in which the C terminus is modified to contain a KK motif (Env37RS or Env18RS). These proteins also were not found to redirect virus assembly to intracellular compartments, as budding virus particles were found only at the cell surface (Fig. 5C and D). In cells expressing Gag and Env18RS, numerous budding particles can be visualized at the cell surface, confirming the site of assembly at the plasma membrane. We also analyzed whether mutant Env proteins could be identified near or at budding sites at the plasma membrane by employing immunogold labeling (40). The results (not shown) indicated that the wt Env protein was present in large amounts on the surfaces of cells coexpressing wt Env and Gag proteins. However, no gold labeling specific for Env proteins was observed on the surfaces of cells coexpressing Gag and Env164RS, Env37RS, or Env18RS.
FIG. 5.
Analysis of the site of SIV assembly by electron microscopy. HeLa T4 cells were transfected, fixed, and embedded, and ultrathin sections were stained with tannic acid and examined by electron microscopy. VLPs budding from cells coexpressing wt Env and Gag proteins (A), Gag and Env164RS (B), Gag and Env37RS (C), and Gag and Env18RS (D). Magnification, ×80,000.
DISCUSSION
The assembly of HIV and SIV involves an interaction between the matrix protein and the cytoplasmic tail of the Env protein (11–13, 18, 19). Recently, Pfeiffer et al. (29) and Salzwedel et al. (34) studied the effects of ER retrieval of the HIV-1 Env protein and reported that its retrieval in the ER did not result in intracellular virus assembly. In addition, such Env proteins were not incorporated into the released virus (29, 34). However, it is uncertain whether the inability of the ER-retrieved Env proteins to redirect the site of assembly was due at least in part to the lack of interaction between the ER-retrieved Env and Gag proteins. Recently we demonstrated that the SIV Env protein can be retrieved to the ER by modifying the cytoplasmic tail sequences to contain a KK motif at the −3 and −4 positions (39). In the present study, we investigated the effects of retention of the SIV Env protein in the ER on the interaction with the Gag protein and site of SIV assembly. Our data provide evidence that there is indeed an interaction between the ER-retained Env and Gag proteins. The finding that anti-Gag antibody precipitates the Env precursor protein indicates that such interactions occur before the Env proteins reach the processing compartment. It is possible that such an early interaction directs the Gag proteins to the basolateral domain of polarized epithelial cells when expressed with Env proteins. Interestingly, we also demonstrated an intracellular interaction between the Gag protein and a truncated Env protein with 18 amino acids including the retrieval signal. A number of studies have documented that serial passage of SIV in human cells resulted in viruses which contain a truncation after 18 amino acids of the cytoplasmic tail (3, 21, 23). The Env proteins with truncations were found to be more fusogenic (32, 35, 43), and such truncated Env proteins are incorporated more efficiently into virus particles (22, 40, 42) and retain the ability to be preferentially expressed on basolateral membranes (2). These results also indicate that the domains or protein structures required for SIV Gag interaction and Env incorporation lie within the 18-amino-acid cytoplasmic tail. Although SIV and HIV-1 share similarities, truncated Env proteins are not efficiently incorporated into HIV-1 virions (8, 12), whereas truncated Env proteins are more efficiently incorporated by SIV (40, 42, 43). Using glutathione S-transferase–HIV-1 Env fusion proteins, Cosson (4) reported that the C-terminal 67 amino acids were essential for the interaction of HIV-1 Env and matrix protein. We have provided evidence that SIV Env proteins having 164, 37, or 18 amino acids and containing a KK motif at the −3 and −4 positions of the C terminus still associate intracellularly with the Gag protein. Since SIV with a premature stop codon resulting in an Env protein with 18 amino acids is selected during propagation in human cell lines (23), the 18-amino-acid tail is likely to be sufficient for Gag interactions involved in virus assembly.
We also observed that the release of the Gag protein into the media was transiently retarded when it was coexpressed with SIV Env protein constructs that were retained in the ER. When we coexpressed Gag with the ER-retained SIV Env164RS, we observed that a fraction of the Env proteins was released into the media. Subsequent gradient analysis indicated that those proteins were released in membrane vesicles (not shown). We previously found that the TM protein was not generated from Env164RS when coexpressed with the Gag protein. It is possible that a fraction of transport vesicles containing these proteins (8 to 10% compared to the amount of gp120 shed from wt-Env-expressing cells) is not retrieved due to the Gag protein masking the retention signal. The retrieval and retention of proteins containing the KKXX motif are mediated by the coatomer complex (5, 6, 10, 20). Recently, we have also presented evidence that the retrieval of SIV Env proteins with a KKXX motif is mediated by their association with βCOP, a subunit of the coatomer complex (39). In cells coexpressing the ER-retained Env and the Gag proteins, the coatomers and the Gag proteins might compete for the cytoplasmic tail of the Env protein. Under these circumstances, a fraction of the mutant Env proteins may bind to the Gag protein, and this may sterically mask the ER retrieval signal located in the cytoplasmic tail. Similar masking of a retrieval signal has been found to exist during the assembly of the immunoglobulin E receptor. The cytoplasmic tail of the α chain of the human high-affinity receptor for immunoglobulin E contains a KKXX motif. This signal was demonstrated to be masked when the α chain assembled with the γ chain, which enabled the cell surface transport of the assembled receptor (24). In an analogous phenomenon, the interaction between the Gag and Env proteins of HIV-1 resulted in the lack of recognition of the Tyr-based endocytosis signal in the cytoplasmic tail (9, 33).
In summary, we conclude that an interaction of the Gag protein with SIV Env proteins can occur even when the Env proteins are preferentially retained in the ER. The interaction can occur with Env proteins having a cytoplasmic tail containing as few as 18 amino acids. An interaction was not detected, however, when the entire cytoplasmic tail was deleted. Despite evidence for such protein interactions, the site of virus assembly was not redirected from the plasma membrane to the ER. For efficient assembly and Env protein incorporation, exposure of large numbers of the Env protein cytoplasmic tails may be required at the cytoplasmic surfaces of membranes, and this may not occur in the ER retention mutants because of their interaction with βCOP proteins or because of a higher density of cellular proteins at such sites. However, this does not appear to impede the assembly of foamy viruses, which are also retroviruses belonging to the Spumavirus genus. The interesting difference is that foamy virus glycoproteins are targeted to the ER by a KKXX motif in the cytoplasmic tail, and ER localization promotes intracellular virus assembly and budding (16, 17). In the case of HIV-1 and SIV, it is possible that certain factors which may mediate the interaction between the Env and Gag proteins to promote virus assembly may be available only at the cell surface. In the absence of such factors, Gag and Env proteins may interact intracellularly but may not be able to assemble into virus particles. It is also possible that differences in lateral mobility of Env proteins in the ER and on the plasma membrane could play a role in determining the ability of such proteins to participate in virus assembly.
ACKNOWLEDGMENTS
This study was supported in part by NIH grants AI 34242, AI 00165, and AI 38501 from the National Institute of Allergy and Infectious Diseases.
The following reagents were obtained through the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HeLa T4 from R. Axel and vaccinia virus (vTF7-3) from B. Moss. We thank P. Marx and S. O’Neil for providing the rhesus anti-SIV antiserum, M. Jabbar for providing the plasmid expressing the HIV-1 Env-CD4 fusion protein, A. Vzorov and D. Taylor for their help and suggestions, and T. Cassingham for assistance in the preparation of the manuscript.
REFERENCES
- 1.Allan J S, Whitehead E M, Strout K, Short M, Kanda P, Hart T K, Bugelski P J. Strong association of SIVagm envelope glycoprotein heterodimers: possible role in receptor-mediated activation. AIDS Res Hum Retroviruses. 1992;8:2011–2020. doi: 10.1089/aid.1992.8.2011. [DOI] [PubMed] [Google Scholar]
- 2.Ball J M, Mulligan M, Compans R W. Basolateral sorting of the HIV type 2 and SIV envelope glycoproteins in polarized epithelial cells: role of the cytoplasmic domain. AIDS Res Hum Retroviruses. 1997;13:665–675. doi: 10.1089/aid.1997.13.665. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 5.Cosson P, Demolliere C, Hennecke S, Duden R, Letourneur F. δ- and ζ-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- 6.Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;262:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 7.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The Gag precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faulstich D, Auerbach S, Orci L, Ravazzola M, Wegehingel S, Lottspeich F, Stenbeck G, Harter C, Wieland F T, Tschohner H. Architecture of coatomer: molecular characterization of δ-COP and protein interactions within the complex. J Biol Chem. 1996;135:53–61. doi: 10.1083/jcb.135.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 16.Goepfert P A, Shaw K L, Ritter G D, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez S A, Affranchino J L, Gelderblom H R, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez S A, Burny A, Affranchino J L. Identification of domains in the simian immunodeficiency virus matrix protein essential for assembly and envelope glycoprotein incorporation. J Virol. 1996;70:6384–6389. doi: 10.1128/jvi.70.9.6384-6389.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harter C, Pavel J, Coccia F, Draken E, Wegehingel S, Tschochner H, Wieland F. Nonclathrin coat protein γ, a subunit of coatomer, binds to the cytoplasmic dilysine motif of membrane proteins of the early secretory pathway. Proc Natl Acad Sci USA. 1996;93:1902–1906. doi: 10.1073/pnas.93.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J I. SIV adaptation to human cells. Nature (London) 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 22.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodama T, Wooley D P, Naidu Y M, Kestler III H W, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letourneur F, Hennecke S, Demolliere C, Cosson P. Steric masking of a dilysine endoplasmic retention signal motif during assembly of the human high affinity receptor for immunoglobulin E. J Cell Biol. 1995;129:971–978. doi: 10.1083/jcb.129.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orenstein J M, Meltzer M S, Phipps T, Gendelman H E. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J Virol. 1988;62:2578–2586. doi: 10.1128/jvi.62.8.2578-2586.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens R J, Compans R W. Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J Virol. 1989;63:978–982. doi: 10.1128/jvi.63.2.978-982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens R J, Dubay J, Hunter E, Compans R W. The human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 30.Rasenick M M, Lazarevic M, Watanabe M, Hamm H E. Permeable cell systems as models for studying disruption, by site-specific synthetic peptides, of receptor-G protein-effector coupling. Methods Companion Methods Enzymol. 1993;5:252–257. [Google Scholar]
- 31.Rhee S S, Hui H X, Hunter E. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J Virol. 1990;64:3844–3852. doi: 10.1128/jvi.64.8.3844-3852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritter G D, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 33.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:538–544. [PubMed] [Google Scholar]
- 34.Salzwedel K, West J T, Jr, Mulligan M, Hunter E. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J Virol. 1998;72:7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens E B, Compans R W. Assembly of animal viruses at cellular membranes. Annu Rev Microbiol. 1988;42:489–516. doi: 10.1146/annurev.mi.42.100188.002421. [DOI] [PubMed] [Google Scholar]
- 37.Stephens E B, McClure H M, Narayan O. The proteins of lymphocyte- and macrophage-tropic strains of SIV are processed differently in macrophages. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 38.Vincent M J, Raja N U, Jabbar M A. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J Virol. 1993;67:5538–5549. doi: 10.1128/jvi.67.9.5538-5549.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent M J, Martin A S, Compans R W. Function of the KKXX motif in endoplasmic reticulum retrieval of a transmembrane protein depends on the length and structure of the cytoplasmic domain. J Biol Chem. 1998;273:950–956. doi: 10.1074/jbc.273.2.950. [DOI] [PubMed] [Google Scholar]
- 40.Vzorov A N, Compans R W. Assembly and release of SIV Env proteins with full-length or truncated cytoplasmic domains. Virology. 1996;221:22–33. doi: 10.1006/viro.1996.0349. [DOI] [PubMed] [Google Scholar]
- 41.Weclewicz K, Ekstrom M, Kristensson K, Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamshchikov G V, Ritter G D, Vey M, Compans R W. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214:50–58. doi: 10.1006/viro.1995.9955. [DOI] [PubMed] [Google Scholar]
- 43.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]