Abstract
Background
Rotor syndrome (RS, OMIM#237450) is an extremely rare autosomal digenic recessive disorder characterized by mild non-hemolytic hereditary conjugated hyperbilirubinemia, caused by biallelic variation of SLCO1B1 and SLCO1B3 genes that resulted in OATP1B1/B3 dysfunction in the sinusoidal membrane leading to impaired bilirubin reuptake ability of hepatocytes.
Methods
One RS pedigree was recruited and clinical features were documented. Whole genome second-generation sequencing was used to screen candidate genes and mutations, Sanger sequencing confirmed predicted mutations.
Results
This study detected a homozygous nonsense variant c.1738C > T (p.R580*) in the coding region of the SLCO1B1 (NM006446) gene in a family with RS and hepatitis B virus infection by Variants analysis and Sanger sequencing, and confirmed by Copy Number Variation (CNV) analysis and Long Range PCR that there was a homozygous insertion of intron 5 of the SLCO1B3 gene into intron 5 of long-interspersed element 1 (LINE1). A few cases of such haplotypes have been reported in East Asian populations. A hepatitis B virus infection with fatty liver disease was indicated by pathology, which revealed mild-moderate lobular inflammation, moderate lobular inflammation, moderate hepatocellular steatosis, and fibrosis stage 1–2 (NAS score: 4 points/S1-2) alterations. Heterozygotes carrying p.R580* and LINE1 insertions were also detected in family members (I1, I2, III2, III3), but they did not develop conjugated hyperbilirubinemia.
Conclusion
The mutations may be the molecular genetic foundation for the presence of SLCO1B1 c.1738C > T(p.R580*) and SLCO1B3 (LINE1) in this RS pedigree.
Keywords: Rotor syndrome, SLCOB1, SLCOB3, OATP1B1/B3, Autosomal digenic recessive disorder, Hereditary conjugated hyperbilirubinemia
1. Introduction
Rotor syndrome (RS) is a rare autosomal digenic recessive disorder characterized clinically by mild and non-hemolytic conjugated hyperbilirubinemia and increased urinary coproporphyrin excretion [[1], [2], [3], [4]], often occurring shortly after birth or in childhood and adolescence, with the most common symptom being mild jaundice. RS was first reported in 1948 and has long been noticed to be familial [4], and animal experiments by the van de Steeg team in 2012 and case series based on genome analysis showed that organic anion transporting polypeptide-related gene (OATP) defects are necessary for the pathogenesis of RS [5]. In 2014, Kagawa 's study of genotypes in RS patients also corroborated van de Steeg' s work [6]. The molecular mechanism of RS is that homozygous or compound heterozygous nonfunctional variants of SLCOB1 and SLCOB3 genes lead to dysfunction of OATP1B1 and OATP1B3 proteins in the sinusoidal/basolateral surface of hepatocytes and weaken the uptake of conjugated bilirubin, conjugated bile acids, coproporrin, drugs and other substances by hepatocytes.
RS and Dubin-Johnson syndrome (DJS) have a fair prognosis, do not influence life, pregnancy, liver fibrosis, or treatment, and the primary harm is increased drug sensitivity [7]. After ruling out other hepatobiliary disorders, an inherited hyperconjugated bilirubinaemia may be considered in patients with recurrent mild jaundice that began in childhood or adolescence and is caused by medications, other disorders, or pregnancy [8,9]. In addition to elevated serum bilirubin, liver injury tests such as alkaline phosphatase and transaminase activity, serum albumin concentration, and prothrombin time are usually normal and show no signs of hemolysis. Bile acids are usually elevated in RS [3]. Gene sequencing is now commonly used to identify RS and DJS [10,11].In the past, liver biopsy [12], Hepatobiliary scintigraphy with 99mTc radiotracers [13], dynamic observation of Bromosulphthaleine (BSP) clearance were usually used to identify RS and DJS, which are now no longer used because of side effects.
2. Methods
2.1. Study subjects
A 42-year-old male Han Chinese patient presented to our hospital with complaints of recurrent yellow eyes, yellow skin, and yellow urine for more than 40 years. He was diagnosed with congenital jaundice for 40 years and found to have a history of chronic hepatitis B for 16 years. He usually did not receive antiviral therapy and had no adverse special hobbies including smoking, drinking, and drugs. In addition, he did not use intravenous or oral medications. The father had a history of bowel cancer, the mother had a history of breast cancer, and there was no other family history or genetic history. There was no history of similar jaundice in the family.
2.2. Clinical features and laboratory findings
Patients' clinical characteristics and biochemical parameters were gathered by making an appointment at the hospital, and peripheral venous blood was drawn for tests on serum antibodies, coagulation, blood routine, and biochemistry. Urine routine tests were conducted, along with liver biopsies and immunohistochemistry analyses including Masson, D-PAS, reticulin, and HE stainings.
2.3. Genomic DNA extraction
2–3 ml of peripheral venous blood was collected from the proband with an Ethylene Diamine Tetraacetic Acid (EDTA) anticoagulant tube; genomic DNA was extracted with a blood genomic DNA extraction kit (TIANamp Blood DNA Kit, DP348-03, Beijing, China). After approval by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University, the investigated family members or their guardians signed informed consent forms.
2.4. Library construction and whole exome sequencing
Next Generation Sequencing Library construction, whole genome library construction (Hieff NGS OnePot DNA Library Prep Kit for Illumina, YEASEN, Shanghai, China) was performed, and whole exome library capture was performed with xGen Exome Research Panelv1.0 (Integrated DNA Technologies, Inc., USA) to construct the whole exome library of the proband. Paired -end sequencing was performed on the Illumina (San Diego, CA) sequencing platform using the PE150 mode.
2.5. Sequencing data analysis
Sequencing data analysis first removed Reads that did not meet quality control requirements from the sequencing raw data and then used BWA (Burrows – Wheeler Aligner) software with UCSC (Fig. https://genome.ucsc.edu/), the provided hg19 version of the human genome reference sequence was aligned, and finally variants were identified by GATKv3.70. Only variants in 1552 genes included in the Endocrine and Metabolic Genetic Disease Panel were analyzed [14]. Variant pathogenicity classification refers to American College of Medical Genetics (ACMG) and Genomics genetic variation classification criteria and guidelines, and variants were classified as pathogenic variants, possible pathogenic variants, clinically unknown variants, possible benign variants, and benign variants.
2.6. PCR
Using Primer3 Plus (Fig. http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi), against the SLCO1B1 gene c.Primers were designed for intron insertion positions in the 1738C > T and SLCO1B3 genes and used In-Silico PCR (Fig. http://genome.ucsc.edu/cgi-bin/hgPcr), to verify primer specificity.C. of SLCO1B1 (NM006446)The amplified fragment length of the sequence in which 1738C > T (p.R580*) resides was 581 bp and the primers were F: TGGGGCCATTCAACTGTGAG; R: GCCCTTCACTGCCTACTG.A forward primer LINEck-F: TCAATGGAACATCACCTGAGA was designed at exon 5 where the SLCO1B3 mutation was located, a pair of reverse primers LINEck-R2: GTGGTGGGTTTCTCCTTCTG and a pair of reverse primers LINEck-R1: CCGTTTCTTAAGCCGGTCTG were designed at intron 5, and a 277-bp fragment could be amplified by the primer pair LINEck-F: LINEck-R1 when there was no insertion; no fragment could be amplified by the primer pair LINEck-F/LINEck-R2 when there was homozygous insertion, and a 349-bp fragment could be amplified by the primer pair LINEck-F/LINEck-R1 [6]. PCR amplification of the target variants of SLCO1B1 gene and SLCO1B3 gene was completed on an ABI PCR (Veriti 96-Well Thermal Cycler) amplification instrument at an annealing temperature of 60 °C. After PCR amplification, 2 % agarose gel electrophoresis was used to detect the amplification effect.
2.7. Sanger sequencing and result analysis
PCR amplification products from family members were sequenced using an automated DNA sequence analyzer (ABI3730 DNA Analyzer), and sequencing results were analyzed and aligned using CodonCode Aligner software (CodonCode Corporation, USA).
3. Results
3.1. Clinical phenotyping
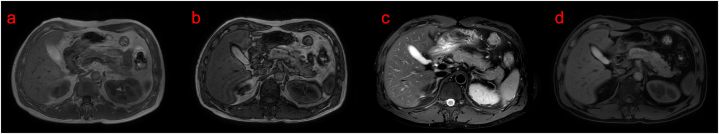
The proband had recurrent jaundice for more than 40 years, and repeated blood biochemical parameters before previous hospitalization revealed elevated total bilirubin (TBIL), direct bilirubin (DBIL), and indirect bilirubin (IBIL), mainly elevated DBIL. Examination after admission showed TBIL 119.1 μmol/L, DBIL 91.1 μmol/L, IBIL 28.0 μmol/L, total cholesterol 5.80 mmol/L, cystatin C 1.18 mg/L (Table 1). Urinalysis showed bilirubin 17 (2 +) umol/L. FibroScan showed controlled attenuation parameter (CAP) 277 dB/m and liver Stiffness Measurement (LSM) 5.9 kPa, which were considered as moderate fatty liver and mild fibrosis. Viral hepatitis type B showed that HBsAg, HBeAb and HBcAb IgG were positive, HBsAb and HBeAg were negative, and hepatitis B virus copy number was <5.00E + 02 IU/mL. Serum IgG4:41.18 g/L (normal value, 8–151 mg/L). Upper abdominal color ultrasound showed mild fatty liver, gallbladder bulge-like lesions, liver and gallbladder MRI showed mild fatty liver, small nodular lesions in segment IV of the liver, considered benign (Fig. 1).
Table 1.
Biochemical markers in probands suffering from Rotor syndrome (II4).
| Test item | Value | Normal value | Test item | Value | Normal value |
|---|---|---|---|---|---|
| Blood test | Coagulation function | ||||
| WBC ( × 109/L) | 6.7 | 4.0–10.0 | TT (sec) | 15.3 | 12.3–22.0 |
| RBC ( × 1012/L) | 4.9 | 4.3–5.8 | APTT (sec) | 31.2 | 20.0–45.9 |
| Hb (g/L) | 145 | 120∼140 | PT (sec) | 11.6 | 9.7–15.7 |
| PLT ( × 109/L) | 204 | 125∼350 | Fg (g/L) | 3.3 | 2.0–4.0 |
| MCV (fl) | 30 | 27–34 | INR | 0.9 | 0.8–1.2 |
| MCH (pg) | 331 | 316∼354 | D-D(mg/L) | 0.23 | 0.8–1.2 |
| RET ( × 109/L) | 53 | 24–84 | hemolysis test | ||
| SF (ng/mL) | 126 | 15∼200 | Haptoglobin (g/L) | 0.74 | 0.16–2.0 |
| TSAT(%) | 33 | 33∼35 | DAT | (−) | (−) |
| Biochemical indexes | Ham test | (−) | (−) | ||
| TP (g/L) | 69.6 | 60∼80 | Serum test | ||
| ALB (g/L) | 44.7 | 35∼55 | HbsAg (IU/ml) | 5.56 | 0∼0.05 |
| GLO (g/L) | 24.9 | 20∼30 | HbsAb (mIU/ml) | 2.30 | 0∼10.0 |
| A/G | 1.8 | 1.2–2.4 | HbeAg (S/CO) | 0.35 | 0∼1.0 |
| TBIL (umol/L) | 119.1 | 5.1–19 | HBeAb (S/CO) | 0.01 | >1.0 |
| DBIL (umol/L) | 91.1 | 1.7–6.8 | HBcAb (S/CO) | 11.98 | 0∼1.0 |
| IBIL (umol/L) | 28.0 | 0∼12 | HAVAb (S/CO) | 0.02 | 0∼1.0 |
| ALT (U/L) | 51.0 | 5∼40 | HCVAb (S/CO) | 0.12 | 0∼1.0 |
| AST (U/L) | 25.0 | 8∼40 | HDVAb (S/CO) | 0.22 | 0∼1.0 |
| GGT (U/L) | 65 | 10∼60 | HIVAb (S/CO) | 0.01 | 0∼1.0 |
| BUN (mmol/L) | 4.92 | 2.86–8.2 | AMA | (−) | (−) |
| CREA(μmol/L) | 58.9 | 62∼116 | ANA | (−) | (−) |
| UA (umol/L) | 36.1 | 208∼428 | SMA | (−) | (−) |
| ALP (U/L) | 96.0 | 45∼125 | Cu (μmol/L) | 15.3 | 11.0–22.0 |
| TG (mmol/L) | 2.69 | <1.7 | CER(mg/L) | 301.2 | 210∼450 |
| HDL-C (mmol/L) | 0.87 | 1.16–1.5 | AAT(g/L) | 0.12 | 0.78–2.0 |
| LDL-C (mmol/L) | 4.65 | 1.56–3.37 | ESR (mm/h) | 7.3 | 0∼15 |
| TCHO (mmol/L) | 5.80 | 0∼5.17 | Tumor markers | ||
| Urine test | AFP (ng/mL) | 3.2 | 0∼25.0 | ||
| uCu(μmol/24h) | 0.33 | 0.24–0.94 | CEA (ng/mL) | 10.7 | 0∼5.17 |
Note: WBC, White blood cell; Hb, Hemoglobin. PLT, Platelet; RBC, Red blood cell; MCV, Mean corpuscular volume; MCH, Mean corpuscular hemolobin; MCHC, Mean corpuscular hemolobin concentration; RET, Reticulocyte count; SF, Serum Ferritin; TSAT, Transferrin Saturation; TP, Total protein; ALB, Albumin; GLO, Globulin; A/G, Ratio of albumin to globulin; TBIL, Total bilirubin; DBIL, Direct bilirubin; IBIL, Indirect bilirubin; ALT, Alanine aminotransferase; AST, Aspartate transferase; BUN, Blood urea nitrogen; CREA, Creatinine; UA, Uric acid; TG, Triglyceride; TCHO, Total cholesterol; HDL, High-density lipoprotein; LDL, Low-density lipoprotein; APTT, Activated partial thromboplastin time; PT, Prothrombin time; TT, Thrombin time; Fg, Fibrinogen; INR, International normalized ratio; DD, D-Dimer; DAT, Direct antiglobulin test; HBsAg, Hepatitis B surface antigen; HBsAb, Antibody to hepatitis surface antigen; HBcAg, Hepatitis B core antigen; HBcAb, Antibody to hepatitis B core antigen; HBeAg, Hepatitis B e-antigen; HBeAb, Antibody to hepatitis B e-antigen; HAVAb, Hepatitis A virus antibody; HCVAb, Hepatitis C virus antibody; HDVAb, Hepatitis D virus antibody; HIVAb. Human immunodeficiency virus antibody; AMA, Anti-mitochondrial antibody; ANA, Antinuclear antibody; SMA, Anti-smooth muscle antibody; Cu, Serum copper; uCu, Urine copper; CER, Serum ceruloplasmin; AAT, Alpha 1 antitrypsin; ESR, Erythrocyte sedimentation rate; AFP, Alpha-fetoprotein; CEA, Carcinoembryonic antigen.
Fig. 1.
Magnetic resonance imaging (MRI) of the liver in probands with Rotor syndrome. a, b are T1WI forward and reverse phases; c is fat-compression T2WI; d is fat-compression T1WI. The clinical impression is mild fatty liver and benign micronodular lesions in segment IV of the liver.
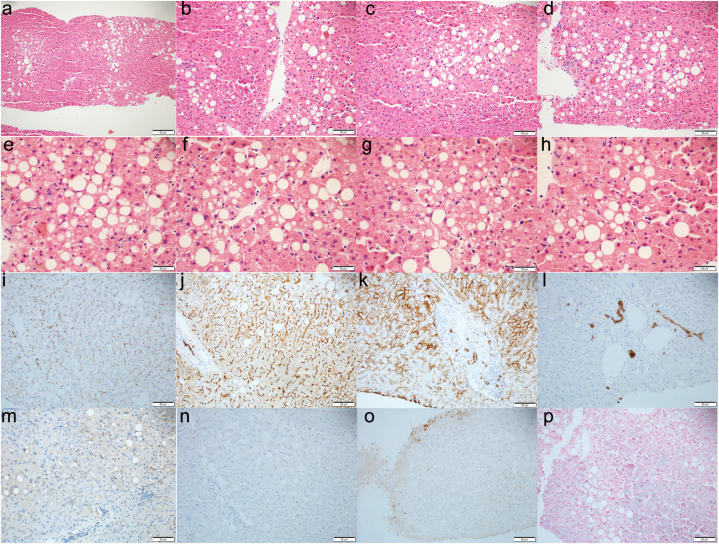
Liver puncture (Fig. 2) results were as follows: 1. Microscopically, it was described as: (HE and reticulin staining) (lesion range was defined as + ∼ + + + +, 25 % as +): hepatic lobular structures were present, central veins about (11), ballooning degeneration (−), ground-glass degeneration (−), mixed vesicular steatosis (+, 40 %), eosinophilic necrosis (−), punctate necrosis (++), debris-like necrosis (−), portal-portal/portal-moderate bridging necrosis (−), Kupffer cell hyperplasia (+), hepatocyte cholestasis (−), cholangiolithiasis (−), glycogen (+), and mild perisinusoidal fibrosis.Portal areas were about (16), enlarged (++), fibrous hyperplasia (−), arcuate fibrogenesis (−), pseudohour formation (−), lymphoid and mononuclear cell infiltration (scattered - +), small bile duct cholestasis (−), and mild atrophy of some bile duct epithelium.2, immunohistochemical staining (positive range defined as + to + + + +, 25 % as +), HBsAg positive range of hepatocytes (+), including cytoplasmic staining (+); HBcAg positive range (−); CK7: biliary epithelial cells (+), progenitor cells (+); CD10: bile capillaries (+).3, in situ hybridization staining: EBER (−).4. Special staining: Masson staining showed a small amount of fibrous hyperplasia and mild perisinusoidal fibrosis around the portal area; reticular fiber staining showed normal liver plate structure; D-PAS staining confirmed the above results, no α1-antitrypsin bodies were observed; iron staining showed no hemosiderin deposition; aldehyde fuchsin staining showed lipofuscin deposition in some hepatocytes; Rodanyu staining showed no copper particle deposition. Pathological diagnosis: mild-moderate lobulitis with moderate lobulitis with moderate hepatocyte steatosis and liver fibrosis stage 1–2, combined with clinical history and immunohistochemical results supporting hepatitis B virus infection with fatty liver disease (NAS score: 4 points/S1-2) changes.
Fig. 2.
Pathology of liver biopsy in patients with Rotor syndrome. a-h was HE staining (e-f, × 400-fold; a, × 100-fold; b-d, × 200-fold). Immunohistochemical staining: i was ABCB11 (+) in bile capillaries, J was CD10 (+) in bile capillaries, k was CD138 (+) in hepatocytes, l was CK7 (+) in bile duct epithelial cells, m was EBER (−), n was HBcAg (−), o was HbsAg (+), and p was iron staining (−). The clinical impression was mild-moderate lobulitis with moderate hepatocellular steatosis and fibrosis stage 1–2 consistent with hepatitis B virus infection with fatty liver disease (NAS score: 4 points/S1-2) changes.
3.2. Screening for mutations in Rotor syndrome
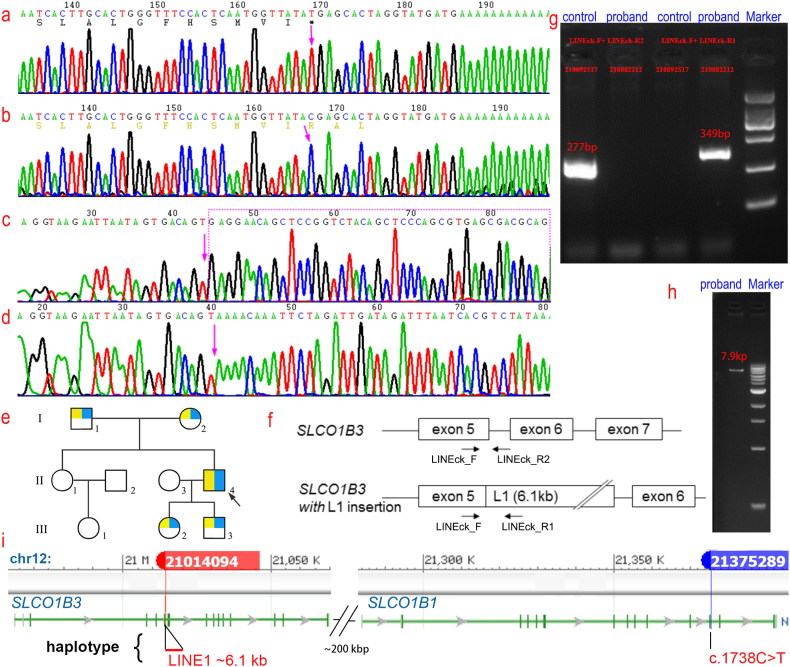
Whole exome sequencing and clinical Gene Panel analysis of the proband revealed a homozygous nonsense variant c.1738C > T (p.R580*) in the coding region of the SLCO1B1 (NM006446) gene in the subject by Single Nucleotide Variants analysis, which changed from coding arginine to a stop codon. Copy Number Variation analysis with Sanger sequencing suggested that there may be a homozygous insertion of the retrotransposon long-interspersed element 1 in intron 5 of the SLCO1B3 gene (Fig. 3c–d), considering this insertion variant as a causative variant. Abnormal SLCO1B1 gene and abnormal SLCO1B3 gene lead to the digenic recessive disorder Rotor hyperbilirubinemia (OMIM#237450), which occurs when homozygous or compound heterozygous variants occur in both SLCO1B1 gene and SLCO1B3 gene (Fig. 3i).
Fig. 3.
a–d, Sanger sequencing plot. a, A homozygous nonsense mutation c.1738C > T (p.R580*) was found in the coding region of the SLCO1B1 gene in patients with Rotor syndrome; b, the corresponding wild-type SLCO1B1 gene; c, a homozygous insertion of the retrotransposon long-interspersed element 1 (LINE1) was found in the coding region of the SLCO1B3 gene in patients with Rotor syndrome; d, the corresponding wild-type SLCO1B3 gene. e, Pedigree mapping of families with Rotor type hyperbilirubinemia. Arrows indicate the proband, yellow represents carriers of the SLCO1B1 mutation and blue represents carriers of the SLCO1B3 mutation. f, Pattern plot of insertion LINE1 variants in the SLCO1B3 gene. g, Long Range PCR electropherogram of Line1 insertion mutation. Control lanes failed to amplify any fragment in LINEck-F/LINEck-R1, 277 bp in LINEck-F/LINEck-R2, 349 bp in LINEck-F/LINEck-R1, and none in LINEckF/LINEckR2.h, a 7.9 kb band was amplified from the proband lane in the Long Range PCR electropherogram of the LINE1 insertion mutation. i, A schematic comparison of the haploid versions of the SLCO1B1 and SLCO1B3 genes.
3.3. Gene mutation validation of Rotor syndrome
Sanger sequencing confirmed a homozygous nonsense variant c.1738C > T (p.R580*) in SLCO1B1 in the proband (Fig. 3a–b), which was not detected in control samples, II1 and III1, while p.R580* heterozygotes were detected in I1, I2, III2, and III3. PCR was performed by designing primers on exon 5 and intron 5 of the SLCO1B3 gene and the homozygous retrotransposon LINE1 (Fig. 3f, g, h), which showed that intron 5 was inserted into LINE1. Sanger sequencing and Long Range PCR showed that there was a homozygous insertion of the retrotransposon LINE1 at intron 5 of the SLCO1B3 gene in the proband (II4), and no LINE1 insertion was detected in the control samples, II1 and III1, while heterozygotes for LINE1 insertion were detected in I1, I2, III2, and III3 (Fig. 3e).
3.4. Tertiary structure prediction of SLCO1B1 R580X variants
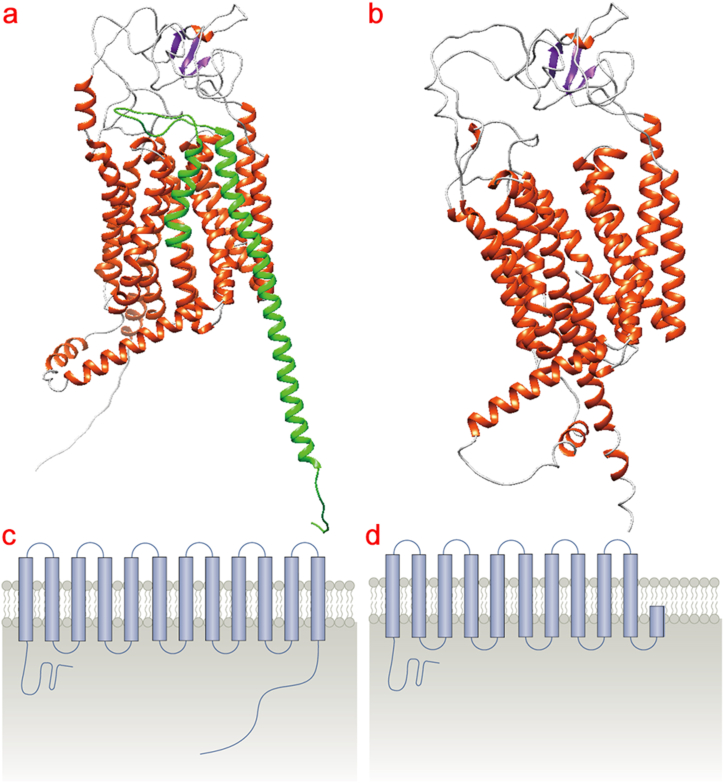
Changes in SLCO1B1 tertiary structure by the R580X mutation were predicted by AlphaFold protein homology structure simulation (Fig. https://www.bkunyun.com/), the R580X mutation resulted in a truncated protein causing the loss of an amino acid sequence encoding a transmembrane protein, resulting in the lack of a helical structure in a three-dimensional structure. This impacts the integrity of the SLCO1B1 transmembrane domain and may affect signaling of the receptor (Fig. 4).
Fig. 4.
Predicted effect of the R580X mutation on SLCO1B1 tertiary structure. a, b, AlphaFold protein homology structure simulations predicted changes in SLCO1B1 tertiary structure by the R580X mutation (Fig. https://www.bkunyun.com/). a, wild-type; b, R580X mutant. Green indicates missing amino acid sequence. Schematic representation of the effects of the c-d, R580X mutation on the SLCO1B1 transmembrane domain.
4. Discussions
The clinical manifestations and positive results of auxiliary examinations of RS can be explained by OATP1B1 and OATP1B3 dysfunction caused by their defective genes. ATP is mainly responsible for the transport of organic anions into hepatocytes, and its transport substrates include conjugated bilirubin, conjugated bile acids, coproporphyrin, and some drugs. OATP1B1/B3 is involved in a mechanism in hepatocytes that prevents damage caused by toxin saturation in upstream hepatocytes, while downstream hepatocytes work too little. In the case of bilirubin metabolism, unconjugated bilirubin enters hepatocytes from the perisinusoidal space and becomes conjugated bilirubin after glucuronic acid modification, some of which is excreted into bile through the canalicular/apical surface of hepatocytes, and the MRP3 protein (also known as ABCC3) of the sinusoidal/basolateral space of hepatocytes also reexcretes partially conjugated bilirubin into the perisinusoidal space, which is retaken up by downstream (located near the central vein) hepatocytes through OATP1B1/B3 and excreted into bile through downstream hepatocytes [15]. Since OATP1B does not dominate the uptake of unconjugated and conjugated bilirubin, and only conjugated bilirubin, which is used to balance upstream and downstream hepatocellular loads, enters the blood upon inactivation of OATP1B alone, serum bilirubin elevations in RS are mainly conjugated and only slightly elevated, usually between 34.2 and 85.5 μmol/L. RS occurs only when OATP1B1 or OATP1B3 are simultaneously inactivated [3], since the two transporters can compensate for each other in conjugated bilirubin transport [7,16]. It is worth noting that serum UCB is sometimes mildly elevated [17,18], cross-sectional studies have found an association between SLCO1B3 variants and serum UCB levels [19], and plasma UCB is also significantly increased nearly 2-fold in Slco1a/1b (a gene similar to human SLCO1B1/B3 function) knockout mice. This is partially reversed by the expression of human OATP1B1/B3 [5], although whether human OATP1B1/B3 protein can transport unconjugated bilirubin needs to be demonstrated by cellular experiments, there is a link between the two, and UCB is sometimes elevated in the probands of this study.
OATP1B is responsible for the transport of substrates between upstream and downstream hepatocytes with glucuronidated BAs at C-3 (BA-3G), mainly transported by OATP1B1 [10]. Radiotracers are not or rarely taken up by the liver during hepatobiliary scintigraphy in RS patients, and the liver is not or poorly imaged, due to inactivation of OATP1B1/B3 and decreased ability of the liver to take anionic radiotracers [4,5]. In contrast, due to the different genes of the lesions, the liver can be clearly imaged during 99Tc-HIDA biliary imaging in DJS, and gallbladder imaging is delayed compared to RS [1,13], and the liver tissue of DJS patients is mostly black, dark green, or grayish black [11,12], and the liver tissue of RS patients is mostly normal color.
BSP is a substrate of OATP1B1/B3, so the uptake and storage capacity of BSP is also significantly impaired in RS patients [16], and urinary excretion of coproporphyrins is increased in RS patients because OATP1B1 interacts with several porphyrins [20]. In addition to OATP1B1/B3, it has also been explored whether RS has other causative genes. ABCC2, an important protein excreted by CB into bile, which is the causative protein of DJS, has been shown to function normally in RS patients, and its ABCC2 gene sequence is normal [21]. If there is a problem with CB processing, elevated levels should be unconjugated bilirubin. Therefore, the currently recognized causative gene of RS is OATP1B1/B3.
OATP1B1/B3 plays an important role in drug detoxification. Prior to the discovery of the association between OATP1B1/B3 and RS, studies of OATP transporters and associated drug toxicity were abundant and could provide a reference for contraindications to the use of drugs in RS patients [22]. Even if no drug accumulation or toxicity has been reported in RS patients to date, caution is needed from a safety point of view, particularly in RS patients with jaundice. Drugs transported by OATP1B1, or drugs inhibiting OATP1B1 activity include: cyclosporin A, atorvastatin, pravastatin, simvastatin, gemfibrozil; drugs transported by OATP1B3, or drugs inhibiting OATP1B3 activity include cyclosporin A, pravastatin, rifampicin, taurocholate, troglitazone sulfate [7].
In 2012, van de Steeg 's study also found three haplotypes of RS [5], called R1, R2, R3, R1 was found in Central Europeans, R2 was found in Central Europeans and Saudi Arabians, R3 was found in Filipinos, haplotype R1 included 7.2-kbp deleted in SLCO1B3 (resulting in exon 12 removal) and c.1738C > T (p.R580X) nonsense mutation in SLCO1B1; haplotype R2 was defined as a ∼405-kbp deletion of homozygous, including exon 3–15 of SLCO1B3 and the entire SLCO1B1; haplotype R3 was c.1747 + 1G > a splice site mutation in SLCO1B3 and c.757C > T (p.R253X) mutation in SLCO1B1. In 2014, Kagawa team [6] discovered the presence of another RS haplotype in Japanese characterized by intron 5 of SLCO1B3 (ClinVar ID: 977762, corresponding to intron 6 23 in Kim 's study in 2021 [23]) inserted a 6-kbp Line1 retrotransposon, a common transposon in the human genome that was confirmed to be the cause of some genetic diseases, while mutations in SLCO1B1 are p.R580X retroposition-active in R1 [6]. This mutation type was also detected in a 12-year-old child with jaundice in China in 2021 [24]. This is consistent with our findings, thus considering that this haploid is more common in East Asian populations. Insertion of Line1 resulted in skipping of exon 5 or exon 5–7 and introduction of a premature stop codonin the SLCO1B3 transcript, resulting in complete inactivation of the OATP1B3 protein, while Kagawa observed one homozygote (0.18 %) and 58 heterozygotes (10.4 %) in 554 healthy Japanese and found that Line1 insertion was not rare in the Japanese population (5.4 %), but the c.1738C > T (p.R580X) mutation in SLCO1B1 was not screened [6]. The incidence of RS is below 1:1,000,000 [7], which may be mainly limited by the low carrier frequency of SLCO1B1, such as 0.0039 in the East Asian population in the genomeAD database. In 2021, among 725 people in Korea, Kim 's team confirmed 91 (12.55 %) carrying the LINE-1 insertion site of SLCO1B3 intron 6 by long-PCR (4 (0.55 %) of them homozygous and 87 (12 %); 5 (0.69 %) were heterozygous carriers of SLCO1B1 p.R580* type, while 2 (0.28 %) of them also carried heterozygotes of SLCO1B3 LINE-1 insertion [23]. Interestingly, Kim also searched the 1000genomes public database for the LINE-1 insertion variant with the highest carrier frequency (18.5 %) in southern Han Chinese, suggesting that the insertion has a founder effect in this population, compared with 10.1 % in East Asian mean carrier frequency [23]. This suggests that SLCO1B3 Line1 insertion variants predominate in southern Han RS patients. Because a significant number of causative variants are structural variants, such as deletions, inversions, and insertions of large genetic elements, which cannot be detected by conventional high-throughput sequencing methods, unknown inactivating mutations of these types of SLCOB1/B3 cannot be obtained by census, so RS patient populations remain to be further studied to determine whether other haplotypes exist.
The proband of this family had onset in infancy and was mainly characterized by yellowish discoloration and non-hemolytic conjugated hyperbilirubinemia, combined with hepatitis B virus infection, and pathology showed mild-moderate lobular inflammation with moderate lobular inflammation with moderate hepatocyte steatosis and liver fibrosis stage 1–2, with normal liver biopsy. The haplotype found by Kagawa and Kim was detected in this RS patient [6,23]. RS is extremely rare clinically, often occurring shortly after birth or in childhood and adolescence, and serum total bilirubin levels are usually between 34 and 85 μmol/L, but may be higher [25]. The clinical manifestations of RS and DJS are similar, mostly increased DBIL, and thus easily misdiagnosed [26]. Molecular genetic testing is now used for differential diagnosis [27], offering the possibility of being non-invasive and allowing diagnosis confirmation and avoidance of misguided therapeutic approaches.
5. Conclusions
In this study, a heterozygous mutation of c.1738C > T (p.R580*) in SLCO1B1 in the proband and that intron 5 of the SLCO1B3 gene inserted into LINE1 was identified as a pathogenic mutation that caused OATP1B1/B3 dysfunction in the sinusoidal membrane leading to impaired bilirubin reuptake ability of hepatocytes. The mutations may be the molecular genetic foundation for the presence of SLCO1B1 c.1738C > T (p.R580*) and SLCO1B3 (LINE1) in this RS pedigree.
Ethics approval and consent to participate
All procedures were performed in accordance with the tenets of the Declaration of Helsinki and the study was approved by the Branch for Medical Research and Clinical Technology Application, Ethics Committee of First Affiliated Hospital of Fujian Medical University (APPROVAL NUMBER: MTCA, ECFAH of FMU [2015]084-2). All participants and legal guardians of the minors involved in the present study provided written informed consent.
Availability of data and materials
The data that underlie and support the findings of this study can be made available from the corresponding author upon reasonable request.
Funding
This work was supported by the Fujian Province Natural Science Fund Project (2021J02053, 2022J01996, 2020J011064, 2021J01704), Fujian Traditional Chinese Medicine Scientific Research Project (2021zyjc09), Fujian Provincial Youth Scientific Program on health (2021QNB001), Startup Fund for scientific research of Fujian Medical University (2021QH1272), the Special Research Foundation of Fujian Provincial Department of Finance (2020-822, 2021-848, 2021-917, 2022-840), National famous and old Chinese medicine experts (Xuemei Zhang, Xiaohua Yan) inheritance studio construction project, and the Fujian Province Medical Innovation Foundation (2021CXB001, 2022CXB002).
CRediT authorship contribution statement
Li-zhen Lin: Writing – original draft. Qiu-yan Wu: Writing – original draft. Jian-hui Zhang: Data curation. Shi-jie Li: Data curation. Wei-zhen Wu: Data curation. Dan-dan Ruan: Data curation. Min Wu: Formal analysis. Qian Chen: Formal analysis. Li-sheng Liao: Software. Zhu-Ting Fang: Methodology. Jie-wei Luo: Supervision. Zuo-an Li: Supervision. Zhou Li: Supervision. Hong Li: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all participants for their valuable contributions to this article.
Contributor Information
Jie-wei Luo, Email: docluo0421@aliyun.com.
Zuo-an Li, Email: 61395776@qq.com.
Zhou Li, Email: 616768863@qq.com.
Hong Li, Email: 913836396@qq.com.
References
- 1.Bar-Meir S., Baron J., Seligson U., Gottesfeld F., Levy R., Gilat T. 99mTc-HIDA cholescintigraphy in Dubin-Johnson and Rotor syndromes. Radiology. 1982;142(3):743–746. doi: 10.1148/radiology.142.3.7063695. [DOI] [PubMed] [Google Scholar]
- 2.Wolkoff A.W., Wolpert E., Pascasio F.N., Arias I.M. Rotor's syndrome. A distinct inheritable pathophysiologic entity. Am. J. Med. 1976;60(2):173–179. doi: 10.1016/0002-9343(76)90426-5. [DOI] [PubMed] [Google Scholar]
- 3.The familial conjugated hyperbilirubinemias. Semin. Liver Dis. 1994;14(4):386–394. doi: 10.1055/s-2007-1007329. [DOI] [PubMed] [Google Scholar]
- 4.Jirsa M., Knisely A.S., Schinkel A., Kmoch S. In: GeneReviews((R)) Adam M.P., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. 1993. Rotor syndrome. Seattle (WA) [Google Scholar]
- 5.van de Steeg E., Stranecky V., Hartmannova H., Noskova L., Hrebicek M., Wagenaar E., van Esch A., de Waart D.R., Oude Elferink R.P., Kenworthy K.E., et al. Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J. Clin. Invest. 2012;122(2):519–528. doi: 10.1172/JCI59526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagawa T., Oka A., Kobayashi Y., Hiasa Y., Kitamura T., Sakugawa H., Adachi Y., Anzai K., Tsuruya K., Arase Y., et al. Recessive inheritance of population-specific intronic LINE-1 insertion causes a rotor syndrome phenotype. Hum. Mutat. 2015;36(3):327–332. doi: 10.1002/humu.22745. [DOI] [PubMed] [Google Scholar]
- 7.Morais M.B., Machado M.V. Benign inheritable disorders of bilirubin metabolism manifested by conjugated hyperbilirubinemia-A narrative review. United European Gastroenterol J. 2022;10(7):745–753. doi: 10.1002/ueg2.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendez-Sanchez N., Qi X., Vitek L., Arrese M. Evaluating an outpatient with an elevated bilirubin. Am. J. Gastroenterol. 2019;114(8):1185–1188. doi: 10.14309/ajg.0000000000000336. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan J.I., Rockey D.C. Diagnosis and evaluation of hyperbilirubinemia. Curr. Opin. Gastroenterol. 2017;33(3):164–170. doi: 10.1097/MOG.0000000000000354. [DOI] [PubMed] [Google Scholar]
- 10.Kimura A., Kagawa T., Takei H., Maruo Y., Sakugawa H., Sasaki T., Murai T., Naritaka N., Takikawa H., Nittono H. Rotor syndrome: glucuronidated bile acidemia from defective reuptake by hepatocytes. Hepatol Commun. 2021;5(4):629–633. doi: 10.1002/hep4.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao C., Guan J.X., Zhong B., Shao C.K., Tang L.Y., Chen J.N. [Clinical and pathological features of Dubin-Johnson syndrome] Zhonghua Bing Li Xue Za Zhi. 2021;50(8):929–933. doi: 10.3760/cma.j.cn112151-20201122-00859. [DOI] [PubMed] [Google Scholar]
- 12.Robinson J.T., Thorvaldsdottir H., Wenger A.M., Zehir A., Mesirov J.P. Variant review with the integrative genomics viewer. Cancer Res. 2017;77(21):e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimopoulou D., Lyra V., Dimopoulou A., Papaevangelou V., Fessatou S. Is hepatobiliary scintigraphy sufficient to diagnose rotor syndrome in a 3-year-old boy? J. Nucl. Med. Technol. 2021;49(2):193–194. doi: 10.2967/jnmt.120.257618. [DOI] [PubMed] [Google Scholar]
- 14.Chen H.L., Li H.Y., Wu J.F., Wu S.H., Chen H.L., Yang Y.H., Hsu Y.H., Liou B.Y., Chang M.H., Ni Y.H. Panel-based next-generation sequencing for the diagnosis of cholestatic genetic liver diseases: clinical utility and challenges. J. Pediatr. 2019;205:153–159 e156. doi: 10.1016/j.jpeds.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Chen H.L., Wu S.H., Hsu S.H., Liou B.Y., Chen H.L., Chang M.H. Jaundice revisited: recent advances in the diagnosis and treatment of inherited cholestatic liver diseases. J. Biomed. Sci. 2018;25(1):75. doi: 10.1186/s12929-018-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erlinger S., Arias I.M., Dhumeaux D. Inherited disorders of bilirubin transport and conjugation: new insights into molecular mechanisms and consequences. Gastroenterology. 2014;146(7):1625–1638. doi: 10.1053/j.gastro.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 17.Fedeli G., Rapaccini G.L., Anti M., Miggiano G., Sabelli C., De Vitis I., Focacci C., Galli G. Impaired clearance of cholephilic anions in Rotor syndrome. Z. Gastroenterol. 1983;21(5):228–233. [PubMed] [Google Scholar]
- 18.Kawasaki H., Kimura N., Irisa T., Hirayama C. Dye clearance studies in Rotor's syndrome. Am. J. Gastroenterol. 1979;71(4):380–388. [PubMed] [Google Scholar]
- 19.Sanna S., Busonero F., Maschio A., McArdle P.F., Usala G., Dei M., Lai S., Mulas A., Piras M.G., Perseu L., et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 2009;18(14):2711–2718. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell S.D., Lau W.F., Xu J.J. Interaction of porphyrins with human organic anion transporting polypeptide 1B1. Chem. Biol. Interact. 2009;182(1):45–51. doi: 10.1016/j.cbi.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Hrebicek M., Jirasek T., Hartmannova H., Noskova L., Stranecky V., Ivanek R., Kmoch S., Cebecauerova D., Vitek L., Mikulecky M., et al. Rotor-type hyperbilirubinaemia has no defect in the canalicular bilirubin export pump. Liver Int. 2007;27(4):485–491. doi: 10.1111/j.1478-3231.2007.01446.x. [DOI] [PubMed] [Google Scholar]
- 22.Konig J. Uptake transporters of the human OATP family: molecular characteristics, substrates, their role in drug-drug interactions, and functional consequences of polymorphisms. Handb. Exp. Pharmacol. 2011;201:1–28. doi: 10.1007/978-3-642-14541-4_1. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y.G., Sung H., Shin H.S., Kim M.J., Lee J.S., Park S.S., Seong M.W. Intronic LINE-1 insertion in SLCO1B3 as a highly prevalent cause of rotor syndrome in East Asian population. J. Hum. Genet. 2022;67(2):71–77. doi: 10.1038/s10038-021-00967-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang D., Li X., Lai P., Zheng L. [Analysis of genetic variants in a case with Rotor syndrome] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2021;38(4):359–362. doi: 10.3760/cma.j.cn511374-20200131-00051. [DOI] [PubMed] [Google Scholar]
- 25.Memon N., Weinberger B.I., Hegyi T., Aleksunes L.M. Inherited disorders of bilirubin clearance. Pediatr. Res. 2016;79(3):378–386. doi: 10.1038/pr.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morais M., Couvert P., Jeru I., Machado M.V. Rotor syndrome presenting as dubin-johnson syndrome. Case Rep Gastroenterol. 2022;16(2):452–455. doi: 10.1159/000525517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gumus E., Karaca M., Deveci U., Jirsa M. The first Turkish family with Rotor syndrome diagnosed at the molecular level. Turk Pediatri Ars. 2020;55(4):430–433. doi: 10.14744/TurkPediatriArs.2019.55798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that underlie and support the findings of this study can be made available from the corresponding author upon reasonable request.