Abstract
Background
Natural Killer (NK) cells are vital components of the innate immune system, crucial for combating infections and tumor growth, making them pivotal in cancer prognosis and immunotherapy. We sought to understand the diverse characteristics of NK cells within lung adenocarcinoma (LUAD) by conducting single-cell RNA sequencing analyses.
Methods
Using the scRNA-seq dataset for multiple primary lung cancers (MPLCs), we examined two major NK cell groups, NK1 and NK2, comparing the expression profiles of 422 differentially expressed NK signature genes. We identified eight genes (SPON2, PLEKHG3, CAMK2N1, RAB27B, CTBP2, EFHD2, GOLM1, and PLOD1) that distinguish NK1 from NK2 cells. A prognostic signature, the NK gene signature (NKGS) score, was established through LASSO Cox regression. High NKGS scores were linked to poorer overall survival in TCGA-LUAD patients and consistently validated in other datasets (GSE31210 and GSE14814).
Results
Functional analysis revealed an enrichment of genes related to the TGF-β signaling pathway in the high NKGS score group. Moreover, a high NKGS score correlated with an immunosuppressive tumor microenvironment (TME) driven by immune evasion mechanisms. We also observed reduced T-cell receptor (TCR) repertoire diversity in the high-risk NKGS group, indicating a negative association between inflammation and risk score.
Conclusion
This study introduced the innovative NKGS score, differentiating NK1 from NK2 cells. High NKGS scores were associated with the TGF-β pathway and provided insights into LUAD prognosis and immune activities.
Keywords: Single-cell RNA-Sequencing, Lung adenocarcinoma (LUAD), NK gene signature (NKGS), Prognosis, TGF-β signaling, Immunosuppressive
1. Introduction
Non-small cell lung cancer (NSCLC) is a predominant subtype of lung cancer, posing a formidable global health challenge due to its widespread prevalence and high mortality rates [1]. Remarkably, NSCLC constitutes an overwhelming 85 % of all lung cancer cases, underscoring its clinical significance and amplifying the call for advanced therapeutic solutions [2,3]. One of the inherent complexities of NSCLC is its marked genetic and phenotypic heterogeneity. Such diversity not only complicates diagnosis for clinicians but also impedes the creation of universally effective treatment approaches [4,5]. As a result, the spotlight in modern NSCLC research has shifted towards personalized precision therapy [6,7]. In the complex landscape of this disease, biomarkers have become essential tools, holding potential for predicting patient outcomes and refining treatment approaches [[8], [9], [10]].
In recent years, biomarkers such as PD-L1 expression and tumor mutation burden (TMB) have been frequently utilized in the clinical arena, primarily to anticipate the response to immunotherapy, a promising line of treatment that has offered considerable benefits for various malignancies [[11], [12], [13]]. However, these markers do not encapsulate the entire story. The tumor microenvironment (TME) of NSCLC is varied and dynamic, suggesting that relying solely on current biomarkers might not provide a comprehensive understanding of the potential therapeutic outcomes [[14], [15], [16]]. The significance of biomarker research in NSCLC can hardly be overstated. Biomarkers offer insights into the biological behavior of tumors, paving the way for more accurate diagnostic and prognostic tools [16]. Moreover, by identifying specific molecular and cellular features, biomarkers can guide the selection of targeted treatments, enhancing therapeutic efficacy and potentially reducing adverse side effects [[17], [18], [19]]. However, they often successfully predict the therapeutic outcome of immunotherapy only for a subset of LUAD (Lung Adenocarcinoma) patients, indicating a pressing need for expanded biomarker discovery [[20], [21], [22], [23]]. Therefore, a contemporary research hotspot in NSCLC encompasses the development of novel predictive models. This approach is expected to revolutionize therapeutic strategies, by not only improving prognosis accuracy but also by refining treatment modalities, thus aiming to enhance patient outcomes and survival rates [[20], [21], [22], [23]].
Natural Killer (NK) cells, have a profound connection to malignancies, especially lung cancer, a predominant driver of cancer-induced fatalities worldwide [[24], [25], [26]]. Leading this frontier is the adoptive NK cell therapy are administered back to patients [[27], [28], [29]]. These fortified NK cells demonstrate augmented endurance and robust anti-tumor capabilities within the challenging, immunosuppressive landscape of lung cancer's tumor microenvironment (TME) [30,31]. Compounding this challenge, factors such as TGF-β and IL-10, secreted by the tumor cells, serve to dampen NK cell efficacy significantly [[32], [33], [34], [35]]. Add to this the incursion of immunosuppressive cohorts like MDSCs and Tregs, and NK cell activity is further stifled. Disturbingly, preliminary findings indicate that lung tumors may actively induce NK cell apoptosis, depleting their numbers within the TME [[36], [37], [38], [39]]. This potent immunosuppression presents formidable obstacles in treatment efficacy. While frontline modalities like chemotherapy or radiation might successfully target and eliminate cancer cells, an attenuated immune backdrop may falter in maintaining lasting tumor oversight, ushering in potential relapses [40]. In summary, the critical defense role of NK cells against lung cancer is undeniable; however, their compromised cytotoxic prowess in impacted individuals amplifies the urgency for pioneering therapeutic interventions [27].
The advent of single-cell RNA-sequencing (scRNA-seq) technology, coupled with sophisticated data analysis techniques, has unlocked unparalleled potential to decipher the molecular nuances of various immune cell subsets within the TME [[41], [42], [43]]. Previous studies have reported that scRNA-seq datasets can serve as a compelling approach to forecast patient prognosis and their responsiveness to immunotherapy [[44], [45], [46]]. Specifically for NSCLC, scRNA-seq has been adeptly employed to probe alveolar macrophages, an integral pillar of the lung's immunological architecture [[47], [48], [49], [50]]. In this study, we performed an exhaustive analysis of scRNA-seq data from multiple primary lung cancers (MPLCs) [51]. Our aim was to delve into the molecular intricacies of tumor-infiltrating NK cells and pinpoint distinctive marker genes associated with these NK cells. Moreover, we devised a Natural Killer gene signature (NKGS) score as a prognostic tool for LUAD by integrating scRNA-seq and TCGA-LUAD RNA-seq analyses. We further validated the predictive accuracy of the NKGS using two independent cohorts from the Gene Expression Omnibus (GEO) database. Additionally, we delved into the interplay between the NKGS and the immunosuppressive tumor microenvironment (TME) and conducted a potential functional analysis within the LUAD context.
2. Results
2.1. Cellular atlas of multiple primary lung cancers (MPLCs)
To understand the detailed cellular heterogeneity within the tumor microenvironment (TME) of lung cancer, a reanalysis was conducted on the scRNA-seq dataset GSE200972 [51]. This dataset encompasses 19 lung samples (Figure S1A), with 5 tumor samples from the inferior lobe (TI), 4 tumor samples from the middle lobe (TM), and 2 tumor samples from the superior lobe (TS). Additionally, there are 4 normal tissue samples adjacent to TI (NI), 3 normal tissue samples adjacent to TM (NM), and 1 normal tissue sample adjacent to TS (NS). For data processing, a principal component analysis (PCA) with n_comps = 50 was performed, followed by the construction of a neighborhood graph emphasizing the most variable genes and setting n_neighbors at 10. Resultant cell clusters were annotated using CellTypist [52] and canonical cell markers, revealing 84,425 cells categorized into nine primary cell subsets (Fig. 1A). Using the Louvain algorithm (resolution = 0.4) via Scanpy [53], 33 unique cell clusters were discerned (Fig. 1B). Expectedly, these major cell lineages demonstrated consistency across samples based on canonical marker gene expression (Fig. 1C–S1B, and S1C). Cells like T cells, NK cells, B cells, Plasma cells, myeloid cells, mast cells, epithelial cells, fibroblast cells, and endothelial cells displayed prominent characterization through high expressions of CD3D, NKG7, MS4A1, MZB1, S100A9, CAP3, KRT18, DCN, and CLDN5, respectively (Fig. 1C and D). Interestingly, each cell cluster showed enriched expression of its distinguishing canonical marker genes, indicating shared expression patterns within clusters and clear distinctions between cell lineages (Figure S1D). In conclusion, we described the landscapes of cell clusters across 19 MPLCs samples for further analysis.
Fig. 1.
Comprehensive analysis of MPLCs single cell atlas. (A) UMAP plots of nine major cell lineages (T cells, NK cells, B cells, Plasma cells, Myeloid cells, Mast cells, Epithelial cells, Fibroblast cells, and Endothelial cells) from MPLCs patients. (B) UMAP plots of 84,425 cells, colored by 33 cell clusters. (C) Violin plot for expression of one canonical marker gene in each major cell lineage. (D) UMAP plots of nine major cell lineages, colored by corresponding canonical marker gene.
2.2. Characterization of different NK cell clusters
NK cells are crucial components of the innate immune system that play vital roles in tumor immunosurveillance. NK cells are typically categorized based on their surface expression of CD56 and CD16. The two primary subsets are: NK1 (CD56dimCD16hi) and NK2 (CD56brightCD16lo). In lung cancer, the tumor microenvironment can impact NK cell functionality. Tumors often develop mechanisms to suppress NK cell activity, such as producing immunosuppressive cytokines, upregulating inhibitory ligands, or recruiting regulatory cells that inhibit NK cell functions.
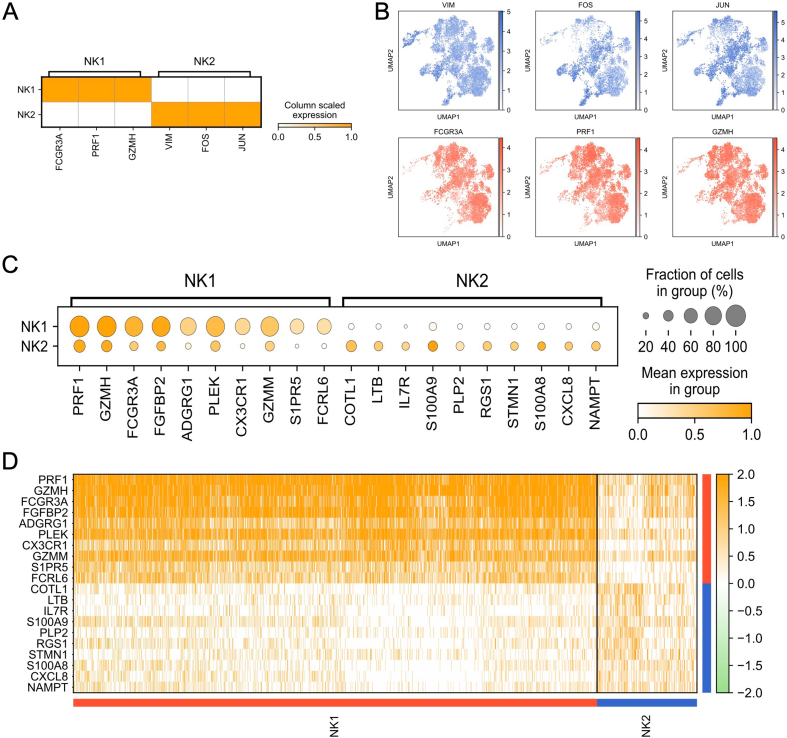
Following stringent quality control measures and filtering, a total of 9099 NK cells were identified and further categorized into two distinct subsets (Fig. 2A). These NK cells were organized into seven unique clusters derived from NI, TI, NM, and TN samples (Fig. 2B). It was noted that NK1 cells exhibited enhanced expression of CD16 (FCGR3A), PRE1, and GZMH, indicated their identity as the CD56dimCD16hi NK cell phenotype (Fig. 2C and D, S2A, and S2B) [54]. In contrast, NK2 cells demonstrated heightened expression levels of VIM, FOS, and JUN (Fig. 2C and D, S2A, and S2B). The data further emphasized that most cellular markers exhibited subset-specific expression patterns (Figure S2C and S2D). Recognized for their potent cytotoxic capabilities, NK1 cells are instrumental in the direct elimination of tumor cells. A reduction in the NK1 cell proportion within tumor samples implies a potential compromise in the anti-tumor defenses in these zones. It's well-observed that tumors can foster an immunosuppressive milieu, thereby limiting the recruitment, activation, and longevity of cytotoxic immune responders, NK1 cells included. A prominent presence of NK1 cells in adjacent normal tissues suggests that the immunosuppressive effects of the tumor might be largely restricted to its immediate surroundings. Collectively, these insights propose that the ratio of NK1 to NK2 cells could offer significant diagnostic or prognostic value.
Fig. 2.
Expression genes of NK cell clusters in MPLCs patients. (A) UMAP plot of 9099 NK cells (NK1, CD56dimCD16high; NK2, CD56brightCD16low), colored by two cell types. (B) UMAP plot of NK cells, colored by seven cell clusters. (C) Dot plot of three represented expressed genes in NK1 and NK2 cell types. (D) Violin plot for expression of three canonical marker genes (NK1, FCGR3A, PRF1, and GZMH; NK2, VIM, FOS, and JUN) in each NK cell type. (E) The proportion of each sample in two NK cell types. (F) The number of cells in two NK cell types.
2.3. Construction of the prognostic signature based on NK cell signature genes
To assess the functionality within NK cell clusters, differentially expressed genes (DEGs) for each cluster were determined by contrasting NK1 and NK2 cell types using the Wilcoxon–Mann–Whitney test. This analysis yielded 218 up-regulated and 203 down-regulated genes (Fig. 3A). Aiming to construct a prognostic signature from these DEGs, the TCGA-LUAD cohort was employed as a training set. Subsequently, a least absolute shrinkage and selection operator (LASSO) Cox proportional hazards regression analysis was executed, culminating in the selection of eight most predictive genes for the risk prognostic model. Risk score = (0.0413 × SPON2 expression) + (0.119 × EFHD2 expression) + (0.139 × PLOD1 expression) + (0.096 × PLEKHG3 expression) + (0.021 × CAMK2N1 expression) + (0.098 × RAB27B expression) + (0.359 × СТВРО expression) + (0.041 × GOLM1 expression) (Fig. 3B). Furthermore, an overview displaying the distribution of risk scores, survival statuses, and gene expression profiles was provided (Fig. 3C). This overview underscored a higher mortality rate in the high-risk group, which also presented elevated gene expression levels. Therefore, we defined a novel prognostic NK gene signature (NKGS) for further analysis.
Fig. 3.
Construction of prognostic model with NK signature genes in the TCGA-LUAD cohort. (A) Volcano plot for expression of differentially expressed genes between NK1 and NK2 cell types. (B) Kaplan–Meier curves of survival analysis compared the overall survival of TCGA-LUAD patients between NKGS-high and NKGS-low groups. (C) ROC curves of the NKGS for predicting the risk of death at 1, 3, and 5 years. (D) The distribution of risk score (top), survival status (middle), and expression (bottom) of the identified eight NK cell marker genes. (E) Validation of the NKGS in GSE31210 (n = 226) cohort. (F) Validation of the NKGS in GSE14814 (n = 114) cohort.
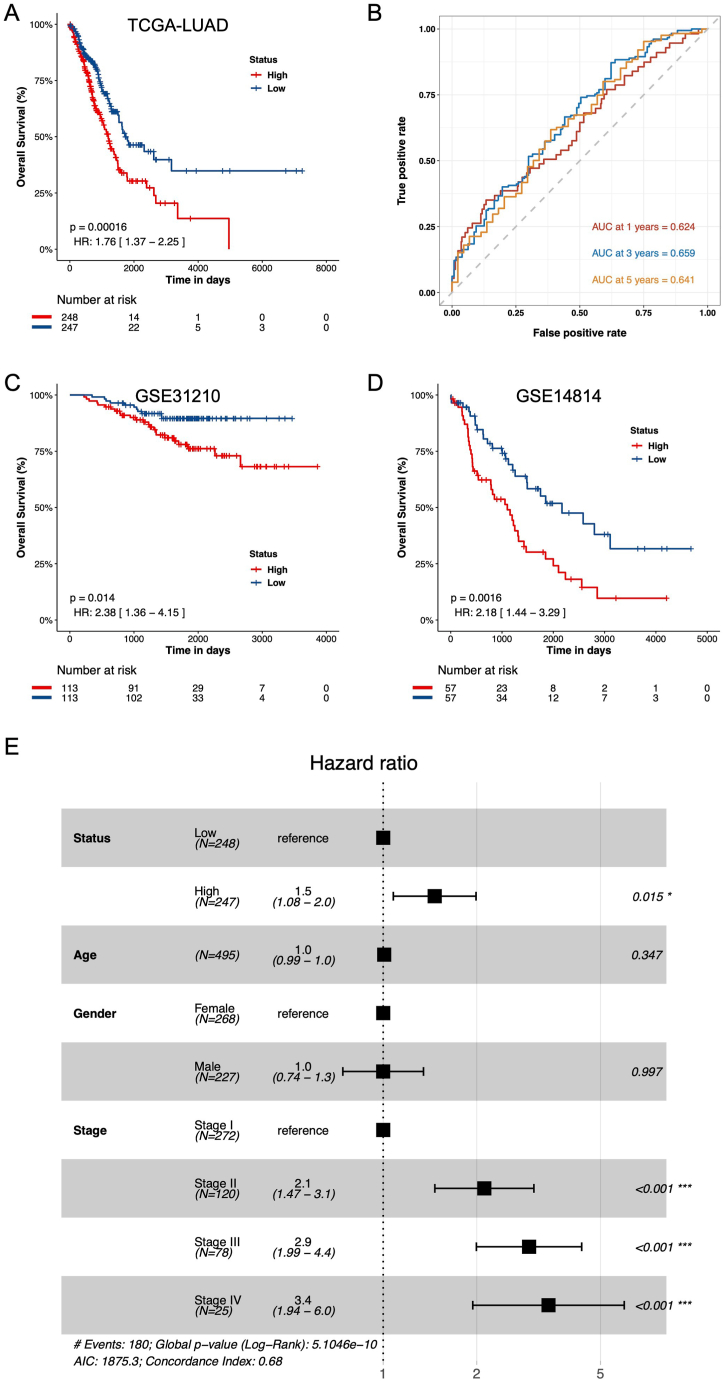
Setting the risk score threshold at 4.67805, achieved by ranking risk scores in descending order, patients were segmented into low-risk (n = 247) and high-risk (n = 248) cohorts. Kaplan–Meier (KM) analysis illustrated that high-risk patients exhibited markedly diminished overall survival (OS) compared to their low-risk counterparts (Fig. 4A, HR = 1.76, P = 0.00016). To gauge the predictive accuracy of this risk prognostic model, time-dependent ROC curves for OS were analyzed, revealing 1-, 3-, and 5-year AUC values of 0.624, 0.659, and 0.641, respectively (Fig. 4B).
Fig. 4.
Cox regression analyses between NKGS-high and NKGS-low groups. (A) Hazard ratios of eight NK signature genes in univariate Cox models that were significantly associated with overall survival. (B) Forest plots show HRs and 95 % confidence intervals (horizontal ranges) derived from cox regression survival analyses for overall survival of eight NK signature genes, including Status, Age, Gender, and Stage.
2.4. Validation of the NKGS in different independent cohorts
In this study, two independent GEO cohorts (GSE31210 and GSE14814) were incorporated to ascertain the reliability of the NKGS score. Initially, using a consistent formula, the risk scores of each patient across the four GEO cohorts were computed. Subsequently, by ranking the risk scores in descending order, the median value was employed to segregate patients into either the high-risk or low-risk category. Kaplan–Meier (KM) analysis was then conducted on both independent GEO cohorts. Results consistently revealed that the high-risk group exhibited a poorer prognosis compared to the low-risk group across both datasets: GSE31210 (Fig. 4C, HR: 2.38, 95 % CI: 1.36–4.15, P = 0.014) and GSE14814 (Fig. 4D, HR: 2.18, 95 % CI: 1.44–3.29, P = 0.0016). Collectively, these outcomes underscore the potential of the NKGS risk score as a predictive tool for the prognosis of LUAD patients. To further validate the application of the NKGS score in external cohorts, we analyzed the 5-year and 10-year survival data from TCGA-LUAD, GSE31210, and GSE14814 using KM curve analysis. The outcomes consistently revealed similar trends for both 5-year and 10-year survival durations, underscoring the efficacy of the NKGS score as a prognostic signature for LUAD (Figure S3A-S3F).
Subsequently, an assessment was undertaken to determine if a high-risk score serves as an independent factor associated with OS. A multivariate Cox proportional hazards analysis, incorporating variables such as age, gender, tumor stage, and risk score, demonstrated that the risk score indeed functioned as an independent predictor of OS in the TCGA-LUAD cohort (P < 0.001, Fig. 4E). In essence, these results suggest that the risk score can prognosticate the outcomes for LUAD patients independently of variables like age and gender, though it remains interdependent with tumor stage.
2.5. Functional enrichment analysis of the NKGS related genes
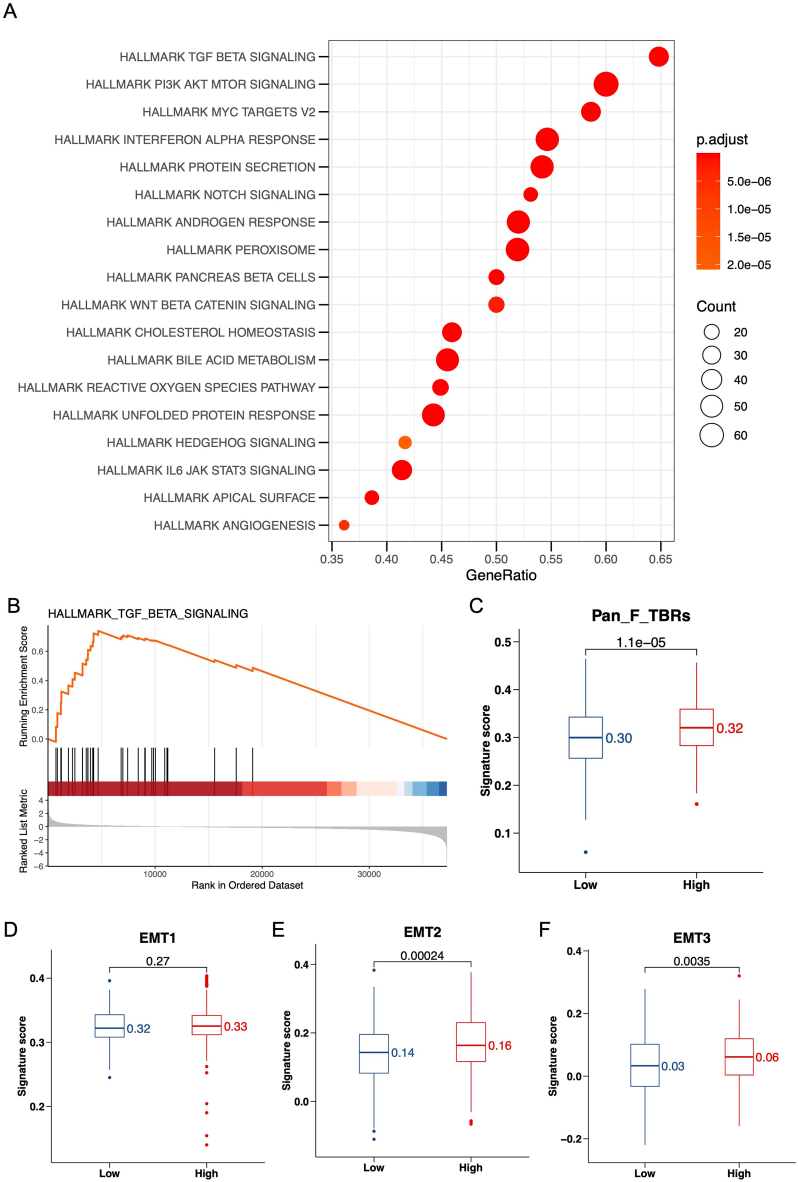
To decipher the biological functions and mechanisms associated with the NKGS risk score, we turned to the Gene Set Enrichment Analysis (GSEA). Our goal was to identify pathways predominantly influenced by genes associated with the NKGS score. Utilizing the hallmark gene sets (c2.cp.v7.4.symbols.gmt) as a reference, we observed a pronounced enrichment of the TGF-β signaling pathway in the high-risk group (Fig. 5A). A more detailed GSEA revealed that the differentially expressed genes (DEGs) related to a heightened NKGS score were predominantly aligned with the TGF-β signaling pathway (Fig. 5B, NES = 2.915, P = 8.77e-11). Concurrently, using the Gene Set Variation Analysis (GSVA), we evaluated the functional activity differences across the NKGS risk score groups. This analysis revealed an increased GSVA pathway activity in the NKGS-high group, notably linked to Pan-F-TBRS (Fig. 5C, P = 1.1e-05) [55]. More interestingly, we found that the NKGS-high group was more enriched in the EMT pathway. While the median value for EMT1 was higher, it was not statistically significant (Fig. 5D, P = 0.27). However, the signature score values for EMT2 (P = 0.00024) and EMT3 (P = 0.0035) were significantly elevated (Fig. 5E and F), indicating a potential for activated EMT pathway. Additionally, when analyzing the expression levels of the signature genes for Pan-F-TBRS and EMT, we found higher expression in the NKGS-high group. Most of the signature gene expressions were more enriched in the NKGS-high group, suggesting that the NKGS score might activate the potential mechanisms of the TGF-β and downstream EMT pathways (Figure S4A-S4D). These observations illuminate the likely attributes of the NKGS-high group, which appears to be defined by enhanced immunosuppression and reduced immune infiltration. Building on this foundation, we aimed to further delineate the disparities in immune infiltration between the NKGS-high and NKGS-low groups.
Fig. 5.
Function analysis between NKGS-high and NKGS-low groups. (A) Function enrichment of NKGS-high and NKGS-low groups in TCGA-LUAD cohort. (B) GSEA analysis of hallmark TGF-β signaling pathway. GSVA activity analysis of Pan-F-TBRs (C), EMT1 (D), EMT2 (E), and EMT3 (F) pathway.
2.6. Analysis of immune infiltration influenced by the NKGS score
To gain a deeper understanding of the attributes and roles of the NKGS-high and NKGS-low groups, we investigated the relationship between NKGS and immune cell infiltration in TCGA-LUAD patients. Leveraging the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm [56], we gauged the potential for immune escape in the TCGA-LUAD cohort. Our analysis revealed that patients in the NKGS-high category had notably increased Cancer Associated Fibroblasts (CAF) scores (Fig. 6A, P = 2.3e-05), heightened exclusion scores (Fig. 6B, P = 0.00068), elevated Myeloid-Derived Suppressor Cells (MDSCs) scores (Fig. 6C, P = 0.00062), and a higher TIDE score (Fig. 6D, P = 0.00068). These metrics derive from gene signatures that signify immune evasion through T-cell exclusion. In sum, the data suggests that the NKGS-high group exhibits elevated values in the aforementioned indicators, implying these patients may be situated in a more immunosuppressive tumor environment. Such an environment could be linked to poorer clinical trajectories and reduced efficacy of immunotherapeutic interventions.
Fig. 6.
Tumor microenvironment analysis between NKGS-high and NKGS-low groups. TIDE analysis in NKGS-high and NKGS-low groups, including CAF score (A), Dysfunction score (B), Exclusion score (C), M2 score (D), MDSC score (E), and TIDE score (F).
3. Prediction of immunotherapy benefits in LUAD patients
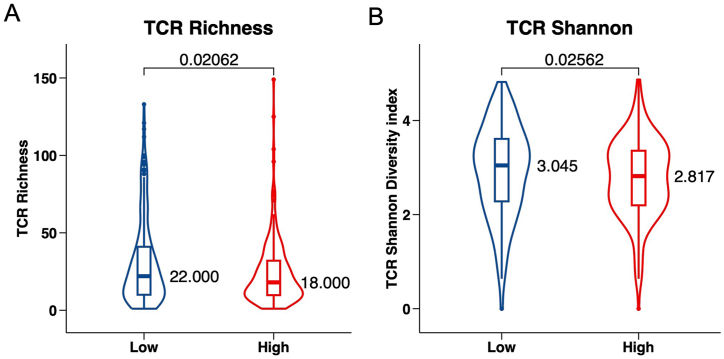
T cells are instrumental in mounting the tumor immune response, making it possible for immunotherapies to accurately identify and target tumor cells. Expanding on previous studies, T cells can be genetically modified to carry T cell receptors (TCRs) tailored to specific cancer antigens, streamlining the detection of antigens presented through the MHC. TCR analysis has emerged as a meaningful biomarker in evaluating anti-tumor immune activities [57]. In our study, we assessed TCR diversity by referencing the TCR repertoire database [45,58] and drew comparisons between the NKGS-high and NKGS-low groups. Intriguingly, we found that the NKGS-high group had considerably diminished TCR diversity (Fig. 7A, P = 0.02062) and a reduced Shannon diversity index (Fig. 7B, P = 0.02562) in contrast to the NKGS-low group. This data indicates that individuals with a lower NKGS risk score could be better positioned to benefit from immunotherapy treatments.
Fig. 7.
The role of NKGS in predicting immunotherapeutic benefit. TCR richness (A) and TCR diversity (B) between NKGS-high and NKGS-low groups.
4. Discussion
Exploring the tumor microenvironment of LUAD has opened the door for us to efficiently decode the molecular characteristics and mechanisms in clinical diagnosis and prognosis research. Single-cell RNA sequencing has further deepened our understanding of the cellular heterogeneity in NSCLC. Some studies even suggest that enhancing NK cell activity might bolster the immune response against cancer. In recent years, particularly the immunotherapy strategies based on NK cells, have emerged and shown new hope in lung cancer treatments. These treatment strategies aim to better utilize the anti-tumor functions of NK cells, offering patients more effective treatment options. Single-cell analysis has also revealed the resistance mechanisms to immunotherapy. The TGF-β signaling pathway, closely related to this, plays a crucial role in the initiation of EMT. Thus, targeting this pathway might further enhance the efficacy of immunotherapy. In conclusion, single-cell analysis techniques provide us with a unique insight into the molecular and cellular heterogeneity of NSCLC and help pinpoint potential therapeutic targets for lung cancer. A deeper understanding of NSCLC heterogeneity and exploration of new treatment strategies may offer a brighter therapeutic outlook for patients afflicted with this severe condition.
In this study, we developed a distinct prognostic prediction model anchored on an NK gene signature tailored for LUAD patients in the TCGA database. We further validated this model rigorously using two separate cohorts from the GEO dataset. The NKGS features nine NK cell marker genes, namely SPON2, PLEKHG3, CAMK2N1, RAB27B, CTBP2, EFHD2, GOLM1, and PLOD1. Most of these genes hold significant associations with LUAD patient prognosis or with NK cell functionality.
Spondin 2 (SPON2) is a protein-coding gene that has been studied in the context of various diseases, including cancer. Identifying potential biomarkers for this condition is crucial. Research reveals that SPON2, while known to be involved in metastasis and cancer progression, plays a vital role in bone metastasis in lung adenocarcinoma (ADC). Elevated SPON2 levels enhance the migration, invasion, and epithelial-to-mesenchymal transition of ADC cells, while its suppression hinders these processes. Additionally, SPON2 levels are higher in metastatic bone tissues than primary ADC tissues, correlating with increased MMP2 and MMP9 expressions. Moreover, in T1a stage NSCLC patients, SPON2 expression is upregulated, facilitated by exosomal HOTAIRM1 interactions in CAFs, promoting NSCLC cell movement [59,60]. Pleckstrin homology and RhoGEF domain-containing G3 (PLEKHG3) is a gene that encodes a protein with a Rho guanine nucleotide exchange factor (RhoGEF) domain, which is involved in activating the Rho family of small GTPases. Research on PLEKHG3 in cancer, especially in lung cancer, is rarely reported. In this study, we found that PLEKHG3 can be used as one of the genes in the prognostic model. This is the first time it has been reported in lung cancer prognosis research. Calcium/Calmodulin Dependent Protein Kinase II Inhibitor 1 (CAMK2N1), in lung cancer, its high expression has been linked to unfavorable outcomes [61]. RAB27B belongs to the RAB family of small GTPases, which are pivotal in vesicle trafficking processes. The heightened RAB27B expression may serve as an adverse prognostic indicator for patients with lung SQCC [62]. C-terminal binding protein 2 (CTBP2) is a well-established transcriptional corepressor with diverse roles in cellular functions such as development, differentiation, and apoptosis. Utilizing the chi-square test and Kaplan-Meier analysis, it was evident that elevated CTBP2 expression was associated with a more aggressive tumor phenotype and adverse prognosis. Furthermore, our findings indicate that CTBP2 knockdown augments NSCLC cell sensitivity to cis-diamminedichloroplatinum (CDDP) by inhibiting the Wnt/β-catenin pathway. Collectively, these insights underscore the pivotal role of CTBP2 in NSCLC progression and CDDP responsiveness, suggesting that targeting CTBP2 could offer a novel therapeutic avenue for NSCLC management [63]. EF-hand domain-containing protein D2 (EFHD2), a protein linked to numerous cellular functions, has been scrutinized in diverse health contexts [64]. Additionally, heightened EFHD2 expression correlates with increased metastasis and epithelial-mesenchymal transition (EMT), making it a promising independent predictor for postsurgical recurrence in stage I lung adenocarcinoma patients [65]. Golgi membrane protein 1 (GOLM1) is a transmembrane glycoprotein found in the Golgi cisternae and has been linked to the carcinogenesis of various cancers. Furthermore, DNA copy number variations and methylation could be key mechanisms driving GOLM1 dysregulation in LUAD [66]. Evidence is mounting that members of the procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD) gene family, namely PLOD1, PLOD2, and PLOD3, play roles in cancer progression and metastasis. Elevated PLOD expression significantly correlates with adverse survival outcomes and immune cell infiltration in LUAD. This suggests that PLOD family genes might serve as emerging biomarkers for adverse prognosis and potential targets for LUAD immunotherapy [67].
The differential distribution of NK1 cells (CD56dimCD16hi) in tissues suggests a significant immunological distinction between tumor and adjacent normal regions. Specifically, higher concentrations of NK1 cells in normal tissues (NI and NM) adjacent to TI and TM hint at a potential suppressive mechanism within the tumor environment, restricting these cytotoxic cells. This may indicate the tumor's ability to modulate its surrounding microenvironment, evading immune surveillance. Conversely, the consistent or diminished presence of NK2 cells (CD56brightCD16lo) across both tumor and surrounding tissues suggests that their primary non-cytotoxic, regulatory functions are not significantly altered by the tumor's presence. Collectively, these patterns underscore the tumor's potential immune-evasive strategies and the intricate interplay between different NK cell subsets in cancer progression. Understanding these distributions could provide insights into therapeutic interventions aimed at harnessing NK cell activity against tumors.
The NKGS prognostic model stands out as a powerful predictor of patient outcomes across both training and validation cohorts. To further understand the underpinnings of the NKGS classifications, we carried out GO pathway and GSEA assessments. Interestingly, the group with a high NKGS score displayed significant involvement in the TGF-β signaling pathway. Through the TIDE algorithms, we discerned diminished immune infiltration and immunosuppression in this group. Additionally, our analysis of the TCR repertoire's complexity and diversity revealed a counter relationship between inflammatory actions and the risk score in the high-risk NKGS cohort.
Furthermore, the elevated exclusion score hints at potential mechanisms that might obstruct effective immune cell infiltration into tumors. Such barriers can be either physical, like a dense extracellular matrix, or biochemical, driven by immunosuppressive cytokines or metabolic alterations. Concurrently, a higher MDSC score reflects an augmented immunosuppressive condition, wherein these cells can suppress T-cell activation, hindering effective anti-tumor immune responses. Lastly, the TIDE score's elevation suggests that tumors in the NKGS-high group might be adept at evading immune responses, possibly rendering them less responsive to conventional immunotherapies. Collectively, these observations intimate that patients in the NKGS-high category may be confronted with more aggressive tumor behavior and could face challenges in leveraging immunotherapies due to a profoundly immunosuppressive microenvironment.
In this study, our mechanistic analyses are largely observational, prompting a need for more in-depth exploration to unravel the link between NKGS gene expression and LUAD prognosis. We have delineated and validated an eight-gene signature rooted in NK cell genes, laying the groundwork for personalized therapeutic strategies for LUAD patients. However, it's vital to approach our results with a clear understanding of their inherent limitations. Future studies should focus on these aspects and further clarify the clinical relevance of the discerned gene signature.
5. Methods and materials
5.1. Collection of single-cell RNA sequencing and GEO datasets
The original scRNA-seq dataset from multiple primary lung cancers (MPLCs) was sourced from GSE200972 [51]. This dataset comprised data from four patients, totaling 19 samples. These included 5 tumor samples from the inferior lobe (TI), 4 tumor samples from the middle lobe (TM), and 2 tumor samples from the superior lobe (TS). Additionally, there were 4 normal tissue samples adjacent to TI (NI), 3 normal tissue samples adjacent to TM (NM), and 1 normal tissue sample adjacent to TS (NS).
Gene expression profiles for lung adenocarcinoma (LUAD) cohorts were procured from the TCGA portal (https://portal.gdc.cancer.gov/). The raw read counts and associated clinical details such as age, gender, stage, overall survival (OS), and vital status for LUAD patients were accessed from the UCSC Xena website (https://xenabrowser.net/datapages/). To further validate the OS status of the proposed gene set, we referenced the microarray data and clinical details from the NCBI GEO database for GSE31210 (n = 226) and GSE14814 (n = 114).
5.2. Single cell analysis and cell clustering
The single-cell dataset matrix was imported into Scanpy (version 1.9.1) [53] for in-depth analysis. Principal component analysis was conducted using ‘sc.tl.pca’ with parameters set to n_comps = 50. The neighborhood graph was clustered using ‘sc.pp.neighbors' with parameters set to n_neighbors = 10. Dimensionality was reduced using highly variable genes, and cell clusters were identified using the Louvain algorithm via the “sc.tl.louvain” module, with the parameter resolution set to 0.4. These cell clusters were subsequently visualized using the uniform manifold approximation and projection (UMAP) module “sc.pl.umap”. The annotations for each cell cluster were determined based on the expression of known marker genes using CellTypist (https://github.com/Teichlab/celltypist) [52].
6. Construction and validation of the prognostic signature
A univariate Cox regression analysis was conducted to determine the prognostic value of the marker genes between NK1 and NK2 cells for OS in TCGA-LUAD patients. Genes with a P < 0.05 were designated as prognostic genes. These genes were then evaluated using the least absolute shrinkage and selection operator (LASSO) Cox proportional hazards regression via the “glmnet” R package. Following this analysis, a risk model was formulated by linearly combining the mRNA expression of the genes with their corresponding risk coefficients. Nine genes were identified as key candidate prognosis-related genes for further investigation. Patients were stratified into NKGS-high risk and NKGS-low risk groups based on the top one-third cutoff value. To verify the prognostic significance of the NKGS score, receiver operating characteristic (ROC) curves [68] were constructed, and the area under the curve (AUC) was computed using the “survivalROC” R package to gauge the accuracy of the NKGS model.
7. Survival analysis
Using the “survival” and “survminer” R packages, Kaplan–Meier (KM) curves were employed to analyze the expression and prognosis of NKGS-related genes in TCGA-LUAD. The KM survival curve analysis indicated a correlation between the NKGS-high group and poorer overall survival (OS). The predictive capability of the NKGS was further corroborated through survival analysis on four independent GEO datasets.
7.1. Differential expressed gene analysis
The expression of individual genes in each cluster was compared to the remaining cells using the “sc.tl.rank_genes_groups” module through the Wilcoxon rank sum test. A gene was defined as up-regulated or down-regulated based on a P < 0.05, with a log (fold change) ≥ 1 or ≤ −1 serving as the cutoff criteria, respectively.
8. Gene set enrichment analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) was conducted to identify significantly enriched pathways using the hallmark gene sets (c2.cp.v7.4.symbols.gmt) from MSigDB (http://software.broadinstitute.org/gsea/msigdb/).
8.1. Gene set variation analysis (GSVA)
Gene Set Variation Analysis (GSVA) was carried out to discern the activity of enriched pathways between the NKGS-high and NKGS-low groups. The enrichment scores for each gene set in the TCGA-LUAD samples were determined using the ssGSEA algorithm via the “GSVA” R package.
9. TIDE analysis
The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm was utilized to predict potential ICB responses [10]. Patients with elevated TIDE scores are more likely to experience antitumor immune evasion, often correlating with reduced response rates to ICB treatment [60]. Recent studies further underscore its effectiveness in predicting or assessing ICB therapy efficacy [43,69].
9.1. TCR analysis
TCR repertoire analysis is recognized as a valuable biomarker for anti-tumor immune responses. In this research, the TCR richness and diversity across each NKGS group were examined in reference to a previous study [58].
9.2. Statistics analysis
Differences of statistical significance were evaluated using a two-tailed Student's t-test on the R platform (R v4.0.3). Multivariate analysis employing the Cox proportional hazards model was executed using the R packages (“survival”, “survminer”, and “forestplot”) to pinpoint independent factors linked to OS in both TCGA-LUAD and GEO cohorts. The P value was corrected using the false discovery rate (FDR), with values or FDR <0.05 deemed significant. An adjusted P < 0.05 served as the threshold criterion.
Funding
Not available.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability statement
The single-cell RNA-seq dataset was sourced from the GEO database using the accession number GSE200972. We obtained the TCGA-LUAD cohort dataset from The Cancer Genome Atlas (TCGA) portal (https://portal.gdc.cancer.gov/). Additionally, we retrieved datasets GSE31210 and GSE14814 from the GEO database.
CRediT authorship contribution statement
Yimin Zhu: Writing – original draft, Visualization, Investigation, Formal analysis, Conceptualization. Xiuhua Wu: Resources, Investigation, Formal analysis. Yunjiao Zhang: Resources, Investigation. Jie Gu: Resources, Formal analysis, Data curation. Rongwei Zhou: Visualization, Investigation. Zhong Guo: Writing – original draft, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank all members in Zhong Guo group for discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33928.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
figs4.
References
- 1.Dela Cruz C.S., Tanoue L.T., Matthay R.A. Lung cancer: epidemiology, etiology, and prevention. Clin. Chest Med. 2011;32(4):605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen J.E., Minna J.D. Molecular biology of lung cancer: clinical implications. Clin. Chest Med. 2011;32(4):703–740. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina J.R., et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodak O., et al. Current landscape of non-small cell lung cancer: epidemiology, histological classification, targeted therapies, and immunotherapy. Cancers. 2021;13(18) doi: 10.3390/cancers13184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z., et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genova C. The long run towards personalized therapy in non-small-cell lung cancer: current state and future directions. Int. J. Mol. Sci. 2023;24(9) doi: 10.3390/ijms24098212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krzyszczyk P., et al. The growing role of precision and personalized medicine for cancer treatment. Technology (Singap World Sci) 2018;6(3–4):79–100. doi: 10.1142/S2339547818300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Restrepo J.C., et al. Advances in genomic data and biomarkers: revolutionizing NSCLC diagnosis and treatment. Cancers. 2023;15(13):3474. doi: 10.3390/cancers15133474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodaghi A., Fattahi N., Ramazani A. Biomarkers: promising and valuable tools towards diagnosis, prognosis and treatment of Covid-19 and other diseases. Heliyon. 2023;9(2) doi: 10.1016/j.heliyon.2023.e13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis K.D., et al. Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: challenges and opportunities. Nat. Rev. Neurol. 2020;16(7):381–400. doi: 10.1038/s41582-020-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klempner S.J., et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncol. 2020;25(1):e147–e159. doi: 10.1634/theoncologist.2019-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarchoan M., et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4(6) doi: 10.1172/jci.insight.126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendriks L.E., Rouleau E., Besse B. Clinical utility of tumor mutational burden in patients with non-small cell lung cancer treated with immunotherapy. Transl. Lung Cancer Res. 2018;7(6):647–660. doi: 10.21037/tlcr.2018.09.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baghban R., et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020;18(1):59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henke E., Nandigama R., Ergun S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 2019;6:160. doi: 10.3389/fmolb.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott A., Salgia R. Biomarkers in lung cancer: from early detection to novel therapeutics and decision making. Biomarkers Med. 2008;2(6):577–586. doi: 10.2217/17520363.2.6.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D.R., Wu X.L., Sun Y.L. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal Transduct. Targeted Ther. 2022;7(1):331. doi: 10.1038/s41392-022-01136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarhadi V.K., Armengol G. Molecular biomarkers in cancer. Biomolecules. 2022;12(8):1021. doi: 10.3390/biom12081021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone E.R., et al. Molecular profiling for precision cancer therapies. Genome Med. 2020;12(1):8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei Y., et al. Progress and challenges of predictive biomarkers for immune checkpoint blockade. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.617335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai R., et al. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 2020;8:34. doi: 10.1186/s40364-020-00209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan T.A., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi M., et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin M., Connolly E., Hussell T. Recruited and tissue-resident natural killer cells in the lung during infection and cancer. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.887503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf N.K., Kissiov D.U., Raulet D.H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 2023;23(2):90–105. doi: 10.1038/s41577-022-00732-1. [DOI] [PubMed] [Google Scholar]
- 26.Levy E.M., Roberti M.P., Mordoh J. Natural killer cells in human cancer: from biological functions to clinical applications. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/676198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Y., Lv X., Du J. Natural killer cell-based immunotherapy for lung cancer: challenges and perspectives. Oncol. Rep. 2021;46(5) doi: 10.3892/or.2021.8183. 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaver K.A., Croom-Perez T.J., Copik A.J. Natural killer cells: the linchpin for successful cancer immunotherapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.679117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bald T., et al. The NK cell-cancer cycle: advances and new challenges in NK cell-based immunotherapies. Nat. Immunol. 2020;21(8):835–847. doi: 10.1038/s41590-020-0728-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altorki N.K., et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen S.K., Gao Y., Basse P.H. NK cells in the tumor microenvironment. Crit. Rev. Oncog. 2014;19(1–2):91–105. doi: 10.1615/critrevoncog.2014011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navin I., Lam M.T., Parihar R. Design and implementation of NK cell-based immunotherapy to overcome the solid tumor microenvironment. Cancers. 2020;12(12) doi: 10.3390/cancers12123871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S.Y., et al. Natural killer cells in cancer biology and therapy. Mol. Cancer. 2020;19(1):120. doi: 10.1186/s12943-020-01238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C., Hu Y., Shi C. Targeting natural killer cells for tumor immunotherapy. Front. Immunol. 2020;11:60. doi: 10.3389/fimmu.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng M., et al. NK cell-based immunotherapy for malignant diseases. Cell. Mol. Immunol. 2013;10(3):230–252. doi: 10.1038/cmi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalfa C., Paust S. Natural killer cell interactions with myeloid derived suppressor cells in the tumor microenvironment and implications for cancer immunotherapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.633205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumino N., et al. Interaction between MDSC and NK cells in solid and hematological malignancies: impact on HSCT. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.638841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melaiu O., et al. Influence of the tumor microenvironment on NK cell function in solid tumors. Front. Immunol. 2019;10:3038. doi: 10.3389/fimmu.2019.03038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindau D., et al. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138(2):105–115. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tie Y., et al. Immunosuppressive cells in cancer: mechanisms and potential therapeutic targets. J. Hematol. Oncol. 2022;15(1):61. doi: 10.1186/s13045-022-01282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang D., et al. Advances in single-cell RNA sequencing and its applications in cancer research. J. Hematol. Oncol. 2023;16(1):98. doi: 10.1186/s13045-023-01494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jovic D., et al. Single-cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med. 2022;12(3):e694. doi: 10.1002/ctm2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G., Ning B., Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen X.Y., et al. Integrating single-cell and bulk RNA sequencing to predict prognosis and immunotherapy response in prostate cancer. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-42858-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song P., et al. Identification and validation of a novel signature based on NK cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma by integrated analysis of single-cell and bulk RNA-sequencing. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.850745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haque A., et al. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bienkowska K.J., Hanley C.J. Improved understanding of NSCLC immunotherapy response mechanisms from single-cell analysis. Transl. Lung Cancer Res. 2023;12(8):1807–1811. doi: 10.21037/tlcr-23-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue Q., et al. Promising immunotherapeutic targets in lung cancer based on single-cell RNA sequencing. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1148061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prazanowska K.H., Lim S.B. An integrated single-cell transcriptomic dataset for non-small cell lung cancer. Sci. Data. 2023;10(1):167. doi: 10.1038/s41597-023-02074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Q., et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med. 2022;14(1):87. doi: 10.1186/s13073-022-01089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., et al. Multidirectional characterization of cellular composition and spatial architecture in human multiple primary lung cancers. Cell Death Dis. 2023;14(7):462. doi: 10.1038/s41419-023-05992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominguez Conde C., et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science. 2022;376(6594):eabl5197. doi: 10.1126/science.abl5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang F., et al. A pan-cancer single-cell panorama of human natural killer cells. Cell. 2023;186(19):4235–4251 e20. doi: 10.1016/j.cell.2023.07.034. [DOI] [PubMed] [Google Scholar]
- 55.Mariathasan S., et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang P., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shafer P., Kelly L.M., Hoyos V. Cancer therapy with TCR-engineered T cells: current strategies, challenges, and prospects. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.835762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830 e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu M., Kong D., Zhang Y. SPON2 promotes the bone metastasis of lung adenocarcinoma via activation of the NF-kappaB signaling pathway. Bone. 2023;167 doi: 10.1016/j.bone.2022.116630. [DOI] [PubMed] [Google Scholar]
- 60.Chen Z., et al. Tumor-derived exosomal HOTAIRM1 regulates SPON2 in CAFs to promote progression of lung adenocarcinoma. Discov Oncol. 2022;13(1):92. doi: 10.1007/s12672-022-00553-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng K., Ren X., Ren Q. NcRNA-mediated upregulation of CAMK2N1 is associated with poor prognosis and tumor immune infiltration of gastric cancer. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.888672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koh H.M., Song D.H. Prognostic role of Rab27A and Rab27B expression in patients with non-small cell lung carcinoma. Thorac Cancer. 2019;10(2):143–149. doi: 10.1111/1759-7714.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang D.P., et al. CtBP2 promotes proliferation and reduces drug sensitivity in non-small cell lung cancer via the Wnt/beta-catenin pathway. Neoplasma. 2018;65(6):888–897. doi: 10.4149/neo_2018_171220N828. [DOI] [PubMed] [Google Scholar]
- 64.Fan C.C., et al. EFHD2 contributes to non-small cell lung cancer cisplatin resistance by the activation of NOX4-ROS-ABCC1 axis. Redox Biol. 2020;34 doi: 10.1016/j.redox.2020.101571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan C.C., et al. EFHD2 promotes epithelial-to-mesenchymal transition and correlates with postsurgical recurrence of stage I lung adenocarcinoma. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-15186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X., Chen L., Zhang T. Increased GOLM1 expression independently predicts unfavorable overall survival and recurrence-free survival in lung adenocarcinoma. Cancer Control. 2018;25(1) doi: 10.1177/1073274818778001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Y., et al. Clinical prognostic value of the PLOD gene family in lung adenocarcinoma. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.770729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Camblor P., Pardo-Fernandez J.C. Parametric estimates for the receiver operating characteristic curve generalization for non-monotone relationships. Stat. Methods Med. Res. 2019;28(7):2032–2048. doi: 10.1177/0962280217747009. [DOI] [PubMed] [Google Scholar]
- 69.Bretz A.C., et al. Domatinostat favors the immunotherapy response by modulating the tumor immune microenvironment (TIME) J Immunother Cancer. 2019;7(1):294. doi: 10.1186/s40425-019-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The single-cell RNA-seq dataset was sourced from the GEO database using the accession number GSE200972. We obtained the TCGA-LUAD cohort dataset from The Cancer Genome Atlas (TCGA) portal (https://portal.gdc.cancer.gov/). Additionally, we retrieved datasets GSE31210 and GSE14814 from the GEO database.