Summary
Here, we present a protocol describing the quantification of oxygen consumption rate (OCR) and maximal respiration rate (MRR) in living induced pluripotent stem cell (iPSC)-derived neurons using the Seahorse analyzer. We guide you through the whole process: culture amplification and seeding of neural progenitor cells (NPCs), their differentiation into neurons, and normalization of the results to cell number in the analytical phase. The assessment of cellular mitochondrial function, by analyzing mitochondrial respiration, could be useful in various diseases as well as in drug screening.
For complete details on the use and execution of this protocol, please refer to Aleo et al.1
Subject areas: Cell Biology, Metabolism, Stem Cells
Graphical abstract

Highlights
-
•
Differentiation of NPCs into neurons in XFe96 cell culture plate
-
•
Assessment of oxygen consumption and maximal respiration in iPSC-derived neurons
-
•
Normalization of the results to cell number in the analytical phase
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol describing the quantification of oxygen consumption rate (OCR) and maximal respiration rate (MRR) in living induced pluripotent stem cell (iPSC)-derived neurons using the Seahorse analyzer. We guide you through the whole process: culture amplification and seeding of neural progenitor cells (NPCs), their differentiation into neurons, and normalization of the results to cell number in the analytical phase. The assessment of cellular mitochondrial function, by analyzing mitochondrial respiration, could be useful in various diseases as well as in drug screening.
Before you begin
Neurons are highly dependent on oxidative metabolism to fulfil their function, both during development and after maturation. In this scenario, mitochondria play an important role as they are involved in the production of adenosine triphosphate (ATP) and in the maintenance of metabolism and calcium balance. Impairment of the electron transport chain leads to membrane leakage, electrolyte imbalance, activation of pro-apoptotic signaling pathways and mitophagy.2 These processes are associated with the pathogenesis of many neurodegenerative diseases. Here, we propose a well-established protocol3 adapted for the study of metabolic changes in neurons to analyze the consequences of mutations, affecting mitochondrial or nuclear DNA, and impairing mitochondrial functionality. Our approach is broadly applicable, not only for researchers working exclusively on mitochondrial diseases, but also for those working in a variety of fields, including metabolic studies.
Note: Given the delicate nature of neurons, conducting experiments with these cells is challenging. Therefore, we have previously shown how mitochondrial respiration can be assessed using the Seahorse XF Analyzer in iPSC-derived neural progenitor cells (NPCs).1
Note: Our protocol is optimized for a Seahorse XFe96 analyzer. Adjust the number of cells and the volume of buffer according to the manufacturing instructions of the XFe24 analyzer. Please check the Agilent website before starting your experiments (https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| DMEM/F-12 | Life Technologies | Cat#31330095 |
| 100× N-2 supplement | Life Technologies | Cat#17502001 |
| 50× B-27 serum-free Supplement | Life Technologies | Cat#17504001 |
| MEM non-essential amino acids solution (NEAA) 100× | Euroclone | Cat#ECB3054D |
| FGF human recombinant | Tebubio | Cat#100-18B-1mg |
| EGF recombinant | Tebubio | Cat#AF-100-15-500ug |
| BDNF protein, human | D.B.A. | Cat#HY-P7116A-100ug |
| GDNF protein, human | D.B.A. | Cat#HY-P7182-100ug |
| N-[(3,5-difluorophenyl) acetyl-]-L-alanyl-2-phenyl] glycine-1,1-dimethylethyl ester (DAPT) γ-secretase inhibitor | Merck Life Science | Cat#D5942-5MG |
| Penicillin-Streptomycin 100× | Euroclone | Cat#ECB3001D |
| L-glutamine 100× | Euroclone | Cat#ECB3000D |
| Rho-associated protein kinase inhibitor (ROCKi, Y-27632) | D.B.A. | Cat#SC-281642A |
| Phosphate-buffered saline (PBS), pH 7.4 | Euroclone | Cat#ECB4004L |
| Poly-L-ornithine | Merck Life Science | Cat#P3655-100MG |
| Laminin 1 mg | Merck Life Science | Cat#L2020 |
| Trypsin-EDTA 1× in PBS | Euroclone | Cat#ECB3052D |
| XF calibrant solution | Agilent Technologies | Cat#100840-000 |
| Agilent Seahorse XF DMEM medium pH 7.4 | Seahorse Bioscience | Cat#103575-100 |
| Sodium pyruvate 100 mM | Euroclone | Cat#ECM0542D |
| 2-Deoxy-D-glucose | Merck Life Science | Cat#D8375-5G |
| Oligomycin from Streptomyces diastatochromogenes | Merck Life Science | Cat#O4876-25MG |
| FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) | Merck Life Science | Cat#C2920 |
| Rotenone | Merck Life Science | Cat#R8875 |
| Antimycin A from Streptomyces sp. | Sigma | Cat#A8674-50MG |
| Critical commercial assays | ||
| Seahorse XFe96 FluxPak mini | Seahorse Bioscience | Cat#102601-100 |
| CyQuant Direct Cell Proliferation Assay Kit | Life Technologies | Cat#C35011 |
| Experimental models: Cell lines | ||
| iPSCs | This paper and Peron et al.4 | FINCBi001-A (https://hpscreg.eu/cell-line/FINCBi001-A) |
| Neural progenitor cells (control, patients carrying the 3460/ND1 or the 11778/ND4 mutation) | This paper and Peron et al.2 | N/A |
| Software and algorithms | ||
| Seahorse Wave | This paper; Agilent Technologies | https://www.agilent.com/zh-cn/product/cell-analysis/real-time-cell-metabolicanalysis/xf-software/seahorse-wave-desktopsoftware-740897 |
| GraphPad Prism | GraphPad | https://www.graphpad.com |
| Microsoft Excel | Microsoft | https://www.microsoft.com/it-it/microsoft-365/excel?market=it |
Materials and equipment
Medium recipes
| NPC Expansion medium (NEM) | ||
|---|---|---|
| Components | Final conc. | 50 mL |
| DMEM/F12 | – | 46.7 mL |
| N2 100× | 1× | 500 μL |
| B27 50× | 1× | 1 mL |
| FGF (10 μg/mL) | 30 ng/mL | 150 μL |
| EGF (10 μg/mL) | 30 ng/mL | 150 μL |
| Penicillin-Streptomycin 100× | 1× | 500 μL |
| L-Glutamine 100× | 1× | 500 μL |
| NEAA 100× | 1× | 500 μL |
The medium should be stored at 4°C for up to 1 week.
| Neuronal differentiation medium (NDM) | ||
|---|---|---|
| Components | Final conc. | 50 mL |
| DMEM/F12 | – | 46.8 mL |
| N2 100× | 1× | 500 μL |
| B27 50× | 1× | 1 mL |
| BDNF (200 μg/mL) | 30 ng/mL | 7.5 μL |
| GDNF (200 μg/mL) | 30 ng/mL | 7.5 μL |
| DAPT (10 mM) | 1 μM | 5 μL |
| Penicillin-Streptomycin 100× | 1× | 500 μL |
| L-Glutamine 100× | 1× | 500 μL |
| NEAA 100× | 1× | 500 μL |
The medium should be stored at 4°C for up to 1 week.
| Seahorse assay medium | ||
|---|---|---|
| Components | Final conc. | 50 mL |
| Agilent Seahorse XF DMEM medium pH 7.4 | – | 48.5 mL |
| Sodium Pyruvate 100 mM | 1 mM | 500 μL |
| L-Glutamine 100× | 2 mM | 500 μL |
| D-Glucose (1 M) | 10 mM | 500 μL |
The medium should be stored at 4°C for up to 1 week.
| Other solutions | |
|---|---|
| Poly-L-Ornithine | Dissolve in PBS at a concentration of 0.1 mg/mL and store at 4°C for up to 3 months. |
| Laminin | Make aliquots and store at −20°C for up to 3 months. Dilute 1:500 in PBS freshly on demand. |
| Trypsin | Dilute the Trypsin 1:10 to obtain a 0,1× concentration and store at 4°C for up to 1 month. |
| ROCK inhibitor (Y-27632) | Dissolve in DMSO at a concentration of 10 mM. Make aliquots and store at −20°C for up to 6 months. Protect from light. |
| FGF Human Recombinant | Reconstitute in sterile, distilled water at a concentration of 10 μg/mL. Make aliquots and store at −20°C for up to 6 months. |
| EGF Recombinant | Reconstitute in sterile, distilled water at a concentration of 10 μg/mL. Make aliquots and store at −20°C for up to 6 months. |
| BDNF Protein, Human | Reconstitute in sterile, distilled water a concentration of 200 μg/mL and store at −20°C for up to 6 months. |
| GDNF Protein, Human | Reconstitute in sterile, distilled water at a concentration of 200 μg/mL and store at −20°C for up to 6 months. |
| DAPT | Dissolve in DMSO at a concentration of 10 mM. Make aliquots and store at −20°C for up to 6 months. |
| 2-Deoxy-D-Glucose | Reconstitute in sterile, distilled water at a concentration of 1 M and store at 4°C for up to 1 year. |
| Oligomycin | Resuspend in DMSO to have a final concentration of 1 mM. Make aliquots and store at −20°C for up to 6 months. |
| FCCP | Resuspend in DMSO to have a final concentration of 1 mM. Make aliquots and store at −20°C for up to 6 months. |
| Rotenone | Resuspend in EtOH to have a final concentration of 1 mM. Store at −20°C for up to 6 months. |
| Antimycin A | Resuspend in EtOH to have a final concentration of 1 mM. Store at −20°C for up to 6 months. |
Step-by-step method details
Coating T25 flasks (or XFe96 cell culture plates) with poly-L-ornithine -laminin (POL)
Timing: 5 h
Neural progenitor cells (NPCs) and neurons are grown on Poly-L-ornithine/laminin (POL) coated supports to favor proliferation and differentiation. Poly-L-ornithine promotes preferential differentiation of neural progenitor cells via ERK signaling pathway.5 Furthermore, stem cells in vivo are located in niches. A common feature of niches is the presence of extracellular matrix molecules. Therefore, laminin is used to mimic the microenvironment in vivo.6
Alternatives: Fibronectin (FN) could replace POL as a coating material. However, the efficiency of differentiation is lower because FN does not activate ERK signaling pathway.5
CRITICAL: Work under sterile conditions.
-
1.
Coat T25 flasks (or XFe96 cell culture plates) with 2 mL (20 μL/w) of cold Poly-L-ornithine (0.1 mg/mL) solution.
-
2.
Incubate for 1 h at 37°C.
-
3.
Remove Poly-L-Ornithine and wash three times with cold PBS.
-
4.
Add 2 mL/T25 (50 μL/w) of cold laminin (2 μg/mL) and incubate for 3 h at 37°C.
Note: Laminin solution should be prepared fresh every time.
-
5.
Aspirate laminin solution and proceed to seed the cells.
Alternatives: Flasks or plates can be stored at 4°C for 2–3 days. Remember to add 2 mL/T25 (20 μL/w) of PBS and seal the plate with Parafilm to prevent the plate from drying.
NPCs culture amplification
Timing: 1–2 weeks (this is variable and is dependent on the cell lines)
Neural progenitor cells are obtained through embryoid body formation. Briefly, embryoid body suspensions are cultured for 5 days and then plated onto Matrigel-coated plates to generate neuro-ectodermal rosette structures. After 5–7 days, rosettes are picked and plated on Poly-L-Ornithine/Laminin (POL) to generate and maintain NPC cultures. NPCs can be frozen for later utilization. See Brafman, 20157 for more details.
Expand NPCs in POL-coated T25 flasks to have enough cells for seeding.
Note: Adjust the splitting schedule according to the growth rate of each cell line to ensure that sufficient cells are available to begin neuronal differentiation on the designated seeding day for each line. A minimum number of cells (500.000) are required.
-
6.
Warm NEM medium and trypsin 0.1× at 20–25°C for approximately 1 h.
-
7.Detach NPCs:
-
a.Aspirate NEM from NPC cultures and wash with 2 mL of PBS.
-
b.Add 2 mL of trypsin 0.1× solution to each flask and incubate 5 min in the CO2 incubator at 37°C.
-
c.Observe under the microscope to check if cells are detached. If not, gently tap the sides of the flask and observe again to determine if additional incubation and tapping is required.
-
d.Add 4 mL/flask of PBS and using a 5 mL serological pipette gently wash off the remaining attached cells.
-
e.With a 2 mL serological pipette mildly triturate the cell suspension to break up any visible cell clumps and transfer the cell suspension to a 15 mL conical tube.Optional: It is possible to wash the flask again with another 4 mL of PBS to collect the cells remaining in the flask.
-
f.Centrifuge tubes at 1200 rcf for 3 min at 20–25°C.
-
g.Aspirate the supernatant and re-suspend cells in the appropriate amount of NEM medium supplemented with Rock inhibitor (1:1000). Carefully pipette the cell suspension up and down 2–3 times with a 2 mL serological pipette until all cell aggregates are dissociated.
-
h.Aspirate laminin solution or PBS from each POL-coated flask.
-
i.Add the appropriate volume of cell suspension to each flask.
-
j.Place the flask into the CO2 incubator. Gently move the flask in several rapid horizontal and vertical motions to distribute the cells evenly over the culture surface.
 CRITICAL: Check for expression of neural progenitors’ markers (Sox1, Pax6, Nestin) and absence of pluripotent markers (Oct4 and Nanog) before starting the differentiation.
CRITICAL: Check for expression of neural progenitors’ markers (Sox1, Pax6, Nestin) and absence of pluripotent markers (Oct4 and Nanog) before starting the differentiation.
-
a.
Seeding of NPCs in XFe96 cell culture plate
Timing: variable (2–4 h depending on the number of cell lines)
To obtain idNeurons, NPCs are seeded at 2–3 × 104 cells/well on POL-coated XFe96 cell culture plate and differentiated for 30 days in the plate.
CRITICAL: For differentiation into neurons, it is preferable to use NPCs after passage 3, but no later than passage 7.
-
8.
Detach NPCs as described in 6–7f.
-
9.Cells counting:
-
a.Aspirate the supernatant and re-suspend cells in 1 mL of NEM medium.
-
b.Use 10 μL of the cell suspension to count the cells.
-
a.
Note: Use Trypan Blue staining to distinguish between live and dead cells.
-
10.Seed NPCs in the POL-coated XFe96 cell culture plate:
-
a.Re-suspend cells in the appropriate amount of NEM medium supplemented with rock inhibitor (1:1000) to reach a final concentration of 2–3 × 104 cells/well.
-
b.Aspirate laminin solution or PBS from each well of the XFe96 cell culture plate.
-
c.Gently pipette the cell suspension up and down to homogenize it.
-
d.Pour the homogenized suspension on a reagent reservoir.
-
e.Use a multichannel pipette to distribute the right number of cells in each well.Note: It is important to retain 4 cell-free wells to be used as blanks during the Seahorse Cell Mito Stress Test. These wells, usually the 4 corner wells, must be handled in the same manner as the cell loaded wells.
-
f.Keep the plate for 30 min under the hood to allow cells to attach uniformly to the plate (see problem 1).
 CRITICAL: This step is important to obtain a uniform concentration of cells between the wells.
CRITICAL: This step is important to obtain a uniform concentration of cells between the wells. -
g.Place the plate into the CO2 incubator. Carefully agitate the plate with swift horizontal and vertical movements to ensure uniform distribution of cells across the culture surface.Note: The number of wells per cell line depends on the experimental set-up. Since idNeurons are very sensitive cells it is possible that differentiation problems may occur in some wells. For this reason, we strongly recommend using at least 12 wells per condition.
-
a.
Differentiation of NPCs into neurons
Timing: 30 days
We follow the 2015 Brafman protocol7 with some modifications to differentiate NPCs into neurons. The day after seeding, check under the microscope that all NPCs are attached. Check the uniformity of the cells' number between the samples and between the wells. If all the checks are passed proceed with the first medium change to start the differentiation experiment (see problem 2).
Note: Warm NDM medium at 20–25°C before use for approximately 1 h.
-
11.
Aspirate the NEM medium from each well of the plate.
-
12.
Add 100 μL /well of NDM medium using a multichannel pipette.
-
13.
Place the plate into the CO2 incubator.
Medium is changed every other day for 30 days. After 4 weeks in NDM medium, the cells should acquire a neuronal morphology and can be used for the experiments.
CRITICAL: After a few weeks, the idNeurons become delicate, and it is better not to stress them. Change only ¾ of the medium every other day.
Prepare for Agilent Seahorse Cell Mito Stress Test
Timing: 1 h
One essential element of the XF assay is the sensor cartridge. Each probe tip within the sensor cartridge is layered with a solid-state sensing material, capable of detecting variations in pH and oxygen concentration over time to determine rates. Proper hydration is crucial for ensuring optimal functionality of these sensors.
Note: The following steps are performed the day before the experiment.
-
14.Hydrate the Seahorse Cartridge.
-
a.Place the Sensor Cartridge upside down next to the utility plate.
-
b.Add 200 μL of sterile water to each well of the utility plate using multi-channel pipette.
-
c.Place the XF hydrobooster (the pink mask provided with the cartridge) on the top of the plate.
-
d.Close the cartridge above the hydrobooster and verify that the water level is high enough to keep the sensors submerged. Avoid bubbles formation.
-
e.Incubate overnight in a non-CO2 incubator set at 37°C. Ensure adequate humidification inside the incubator or wrap the cartridge with aluminum foil to create a humidified environment to prevent water evaporation.
-
a.
Alternatives: If the hydration step is performed the same day of the experiment, use directly calibrant instead of water. It is recommended to do at least 4 h of hydration.
-
15.Design experiment in Wave software.
-
a.Create the assay template file to run on the Seahorse Analyzer with Wave Desktop software.
-
b.Save the template to use the next day.
-
c.Create the Assay protocol as in Table 1 and save for the next day.
-
a.
Note: See the Instrument User Manual (https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay) for additional details.
-
16.
Prepare the Seahorse assay medium according to the recipe (see the section Medium preparation in materials and equipment).
-
17.
Turn on the Agilent Seahorse XF Analyzer, and let it warm up to 37°C overnight (minimum of 5 h).
Alternatives: Rotenone and Antimycin A can be loaded together into the same port.
Table 1.
Seahorse assay protocol
| Seahorse assay protocol | ||
|---|---|---|
| Time (min.) | Perform | |
| Calibrate | ||
| Equilibrate | ||
| Mix | 4 | 3× |
| Measure | 5 | |
| Inject port A (Oligomycin) | ||
| Mix | 4 | 3× |
| Measure | 5 | |
| Inject port B (FCCP) | ||
| Mix | 4 | 3× |
| Measure | 5 | |
| Inject port C (Rotenone) | ||
| Mix | 4 | 3× |
| Measure | 5 | |
| Inject port D (Antimycin A) | ||
| Mix | 4 | 3× |
| Measure | 5 | |
| End | ||
Agilent Seahorse Cell Mito Stress Test
Timing: 4–5 h
The Seahorse XF Analyzer performs metabolic measurements on living cells in real time. The Seahorse Mitochondrial Stress Test evaluates mitochondrial function by measuring oxygen consumption rate (OCR), an indicator of mitochondrial respiration. During the assay, respiratory modulators are added to the cells via injection ports, providing a detailed understanding of cellular energetics.
Note: Before starting, examine the cells under a light microscope to confirm the health, morphology (see problem 3), uniformity, and purity (no contamination) of the cells and to ensure cell attachment with a uniform monolayer. Make sure that there are no cells in the background correction wells.
-
18.
Verify that the pH of the Seahorse assay medium is still at 7.4 and warm at 37°C for 15 min.
-
19.Prepare the Seahorse cartridge 60 min before running assay.
-
a.Place the sensor cartridge upside down next to the utility plate.
-
b.Remove the XF hydrobooster from the plate.Note: Do not replace the XF hydrobooster on the plate for the rest of the experiment.
-
c.Remove the water from the utility plate.
-
d.Fill each well of the utility plate with 200 μL of the pre-warmed (30 min) XF Calibrant using multi-channel pipette.
-
e.Lower the sensor cartridge onto the utility plate and verify that the sensors are immersed in the calibrant.
-
f.Place assembled sensor cartridge with utility plate in 37°C non-CO2 incubator for 60 min.
-
a.
-
20.Change the cell medium to Seahorse Assay Medium.
-
a.Gently aspirate NDM medium from each well.
 CRITICAL: Do not remove all the medium. Since idNeurons are very sensitive cells, it is better not to let them dry out.
CRITICAL: Do not remove all the medium. Since idNeurons are very sensitive cells, it is better not to let them dry out. -
b.Add 20 μL of PBS per well using multi-channel pipette.
-
c.Carefully aspirate the PBS.
-
d.Add 180 μL of the pre-warmed Seahorse assay medium using multi-channel pipette.
-
e.Place the XFe cell culture plate in 37°C non-CO2 incubator for 60 min.
-
a.
-
21.
Prepare working solutions of compounds to be loaded into the sensor cartridge according to Table 2.
Alternatives: Piericidin A (AR-054), a natural mitochondrial Complex I inhibitor,8 can be used as an alternative to Rotenone. Piericidin A and Rotenone share a common binding domain in Complex I.9,10
-
22.Load Sensor Cartridge with compounds.
-
a.Orient the Agilent Seahorse XF Assay Cartridge. Align the row labels (labeled A-H) to the left. The triangular notch will be in the lower left corner.
-
b.Place the A/D loading guide flat on top of the assay cartridge. Align the loading guide so that the letter “A” is in the upper left corner.Note: Use your fingertips to hold the outer edges of the loading guide to stabilize it during loading so that the pipette tips do not come off the loading guide.
-
c.Carefully dispense the correct volume of the appropriate injection solution into the ports.
 CRITICAL: Avoid creating air bubbles.Note: Angle the tips very slightly, but do not force the tips completely into the holes.
CRITICAL: Avoid creating air bubbles.Note: Angle the tips very slightly, but do not force the tips completely into the holes. -
d.Remove the A/D loader, switch to the B/C loading guide and align it and align it with the letter “B” in the upper left corner.
-
e.Repeat for each port.Note: Visually check injection ports for uniform load.
-
a.
-
23.Run the assay.
-
a.Open the assay template file (see 15a) and verify that everything is correct.
-
b.Check the parameters of the Cell Mito Stress Test.
-
c.Press the “Start run” button to start the sensor cartridge calibration (20 min).
-
d.Load the cartridge with the utility plate on the tray.Note: Remove the lid from the sensor cartridge.
 CRITICAL: Position the plate according to the correct direction. Visit the Agilent Web site for more details. (https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay).
CRITICAL: Position the plate according to the correct direction. Visit the Agilent Web site for more details. (https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay). -
e.When calibration is complete (about 20 min), the sensor cartridge remains inside the device and the utility plate is ejected.
-
f.Remove the lid from the cell plate and place it on the tray, making sure the plate is in the correct orientation.
-
g.Click “Load Cell Plate” and the assay will automatically begin after equilibration is complete.
-
h.After approximately 2 h, the assay is complete. Remove the sensor cartridge with the cell plate and save the results.
-
i.Discard the sensor cartridge and replace the lid on the cell plate.
-
a.
-
24.
Insert a USB drive and export the results (see problem 4 and problem 5).
Table 2.
Preparation of working solutions of compounds
| Working solutions of compounds | |||||
|---|---|---|---|---|---|
| Port | Compound | Stock Conc. (mM) | Intermediate conc. in the port (μM) | Intermediate volume to add to port (μL) | Final Conc. in the well (μM) |
| A | Oligomycin | 1 mM | 10 μM | 20 μL /well | 1 μM |
| B | FCCP | 1 mM | 21 μM | 22 μL /well | 2.1 μM |
| C | Rotenone | 1 mM | 10 μM | 25 μL /well | 1 μM |
| D | Antimycin A | 1 mM | 10 μM | 28 μL /well | 1 μM |
CyQUANT assay
Timing: 1.30 h
Various methods are used to normalize the results. However, since the plate is POL-coated, normalization using total protein is not applicable.11,12 One of the cheapest, least time-consuming and simple method is cell counting using the CyQUANT Direct Cell Proliferation Assay, a DNA-based assay. A cell-permeant DNA-binding dye is combined with a masking dye reagent. The masking dye blocks the staining of dead cells, so that only healthy cells are stained. No washing, cell lysis or radioactivity is required. It also has high sensitivity and dynamic range.
Note: Refer to the Assay Protocol on the supplier website for additional details (https://www.thermofisher.com/it/en/home/references/protocols/cell-and-tissue-analysis/protocols/cyquant-direct-microplate-reagent-for-cell-viability-protocol.html).
CRITICAL: It is necessary to generate a cell number standard curve with known numbers of idNeurons.
Note: Although we have counted cells prior to plating, the number of cells before OCR measurement may vary due to different death rates during the 30 days of differentiation.
-
25.
Warm reagents at 20–25°C for 1 h.
-
26.
Mix the stain with Background Suppressor I and PBS in a tube (see Table 3).
-
27.
Use a multichannel pipette to remove 172 μL of Seahorse Medium from each well of the cell culture plate (see problem 6).
-
28.
Add 100 μL/w of the CyQUANT mix using a multichannel pipette.
Note: The ratio should be 1:1.
-
29.
Wrap the plate in aluminum foil to keep it in the dark.
-
30.
Place the plate in a 37°C non-CO2 incubator for 60 min.
-
31.
Read the signal using a microplate reader.
-
32.
Save the measurement.
Table 3.
Preparation of CyQUANT mix
| CyQUANT mix | |
|---|---|
| Volume (100 wells – 100 μL /well) | |
| CyQUANT™ Direct nucleic acid stain (component A) | 40 μL |
| CyQUANT™ Direct background suppressor I (component B) | 200 μL |
| PBS | 9.70 mL |
Expected outcomes
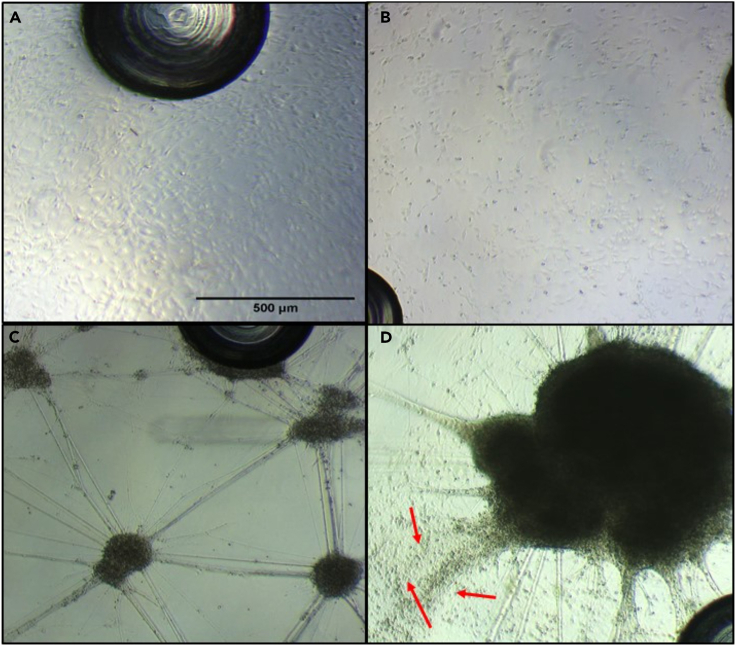
NPCs should begin to form neurite outgrowths after 10 days of differentiation and begin to cluster at approximately 15 days. The number and size of clusters may vary as they are cell lines dependent. There are usually many neurite extensions between clusters and they are located at different levels. Prior to the evaluation of mitochondrial respiration, the plate should be free of undifferentiated cells (see Figure 1).
Figure 1.
Examples of confluence of NPC (one day after seeding) and of derived neurons
(A) shows the correct confluence to start differentiation, (B) an example of insufficient cell number. After 30 days of differentiation, idNeurons should appear as in (C), with many small clusters connected by neurite extensions. The situation shown in (D) is undesirable because there are still many undifferentiated cells (red arrows) and a large cluster where the compound may not penetrate. Magnification 4×.
We expect an OCR trend as shown in Figure 2.
Figure 2.
Oxygen consumption rate graph
The basic parameters of mitochondrial function are highlighted in boxes: basal respiration in green, ATP-linked respiration in pink, proton leak in light green, maximal respiration in blue and spare respiratory capacity with a bracket. OCR is measured in pmol/min and time in minutes. The red line is the background line. The blue line shows the graph of the cell line.
To determine basal oxygen consumption, three measurements of OCR are taken before the addition of any drugs. A reduction in oxygen consumption follows the addition of oligomycin because it blocks the catalytic activity of ATP synthase by binding to OSCP (oligomycin sensitivity conferral protein), a 23 kDa polypeptide of the F0 subunit. Thus, the proton gradient between the mitochondrial matrix and the intermembrane space is no longer dissipated, resulting in a slowing of electron transport. To obtain the maximum rate of oxygen consumption, the uncoupler FCCP is added. It dissipates the proton gradient of the inner mitochondrial membrane, causing an increase in the proton pump demand, so that the ETC (electron transport chain) works at its maximum, consuming a large amount of oxygen. Finally, a reduction in the OCR level is due to the addition of the inhibitor of C (complex) I, rotenone, and the inhibitor of CIII, antimycin A. A large reduction in the OCR level indicates that most of the oxygen consumption is dependent on electrons generated by CI-mediated NADH (nicotinamide adenine dinucleotide) oxidation. By inhibiting CIII, we can measure non-mitochondrial oxygen consumption.
Variations in the OCR profile are an indication of ETC function impairment.
Quantification and statistical analysis
To obtain reliable and reproducible results, it is important to normalize the OCR measurement versus the cell number.
It is recommended to perform at least three biological replicates for each experiment.
Before proceeding with calculations, check that the blanks are similar and that the measurements in each well are correct: if the trend is not as shown in Figure 1, the evaluation of this well should be discarded.
Note: Open the exported data with Excel software.
-
1.Normalization of the CyQUANT Assay luminescence measurement.
-
a.Calculate the average of the 4 blank wells.
-
b.Divide the measured value by it.
-
c.Normalize each well using the calibration curve.
-
d.The final result is the normalized cell count per well.
-
a.
-
2.Normalization of OCR measurement.
-
a.Discard wells whose measurements failed.
-
b.Normalize each OCR measurement to the number of cells per well.
-
c.Calculates, for each cell line, the average of each OCR measurement.
-
d.Do this for each condition to obtain the measures for basal, oligomycin, FCCP, rotenone and antimycin conditions.
-
e.Subtract the average oligomycin measurements from the average FCCP measurements to obtain the maximum respiratory rate (MRR).
-
f.Calculate the standard deviation for each measurement.
-
a.
-
3.
To better compare different experiments, data are expressed as a percentage of the baseline average of the control line.
Statistical analyses are performed using GraphPad Prism v.8.
Data are expressed as mean ± standard deviation (SD). Statistical analysis of OCR measurements is performed using a t-test between the measurement of control and mutated line for each time point of the time course.
Statistical significance is defined as p value < 0.05, ∗∗p value < 0.01, and ∗∗∗p value < 0.001.
Limitations
The efficiency of differentiation depends on several factors and may be cell line dependent. However, the cell population must be enriched in neurons for a reliable assay. The presence of nuclear or mitochondrial gene mutations may affect the ability of NPCs to differentiate into neurons, as they rely heavily on oxidative metabolism to perform their function. In addition, switching from glycolysis to oxidative phosphorylation is essential for the generation of neurons.
If even adjusting the timing and factor concentrations depending on the cell line is not sufficient to obtain a pure population, this protocol cannot be used.
Troubleshooting
Problem 1
Wells at the edge of the plate have a lower cell concentration than wells in the center of the plate (related to step 10f).
Potential solution
After seeding, it is important to keep the plate for 30 min under the hood to allow cells to attach uniformly to the plate.
Problem 2
Differentiation efficiency is not very high (related to step 11).
Potential solution
A high initial number of seeded NPCs may affect differentiation efficiency. Reduce the number of neural precursor cells seeded on the culture plate.
Problem 3
Uncertainty about the purity of the neural cell culture (related to step 18).
Potential solution
We recommend seeding NPCs in parallel into chamber slides and T25 or 6-multiwell cell plates on the same day that NPCs are seeded into XFe96 cell culture plates. At the end of differentiation, the chamber slides can be used for immunofluorescence characterization of specific neuronal markers (Map2; β-tub III). In addition, idNeurons in T25 or 6Mw are used to obtain pellets to perform quantitative PCR for molecular characterization.
Problem 4
Poor basal signal (related to step 24).
Potential solution
-
•
If it is caused by insufficient cell number, increase the number of neural precursor cells seeded on the culture plate.
-
•
Another cause may be over-incubation of the XFe cell culture plates in the non-CO2 incubator, which may reduce cell viability. Make sure that the incubation time is at least 30 min but not more than 1 h.
Problem 5
The OCR measurement does not increase after the injection of FCCP (related to step 24).
Potential solution
-
•
Verify that the compound has been added properly by looking at the cartridge after the assay is complete. If the wells are empty, FCCP has been added; if not, the compound remains in the well.
-
•
If the compound is still in the cartridge, the experiment must be repeated paying attention to the way the cartridge is loaded. Be careful to load compounds with the pipette tips oriented at a very slight angle, less than 5°, to avoid bubbles. Do not enter the pipette tips too profound inside the hole which, as this may result in a leak of compounds in the utility plate.
-
•
If the compound has been added properly, the problem can be the concentration of FCCP. We recommend making a titration curve with FCCP for each cell line as the correct concentration may be cell line dependent.
Problem 6
Cells detach or suffer after the Seahorse Assay.
In this case, be careful not to aspirate cells when removing the Seahorse Medium and adding the CyQUANT Mix (related to step 27).
Potential solution
-
•
Try to be gentle when changing the Seahorse Assay Medium.
-
•
Incubate XFe cell culture plates in the non-CO2 incubator for less than 1 h.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Camille Peron (camille.peron@istituto-besta.it).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be fulfilled by the technical contact, Chiara Fasano (chiara.fasano@istituto-besta.it) and Camille Peron (camille.peron@istituto-besta.it).
Materials availability
iPSCs were generated from a patient carrying the homoplasmic m.3460G > A/MT-ND1 mutation using the Sendai virus non-integrating virus.4 iPSCs FINCBi001-A are available at https://hpscreg.eu/cell-line/FINCBi001-A.
Data and code availability
This study did not generate datasets or code.
Acknowledgments
This work was completely or partially supported by Italy grant 2018-01, the Italian Ministry of Health as part of the REORION project (RF-2018-12366703) and the Italian Ministry of Health (RRC). A.C. is a recipient of SG-2021-12374454 from the Italian Ministry of Health. We would like to thank the project POS, Linea Azione 4.1; T4-AN-09 of the Italian Ministry of Health. All the authors are part of the Center for the Study of Mitochondrial Pediatric Diseases funded by the Mariani Foundation. C.F. is supported by Associazione Luigi Comini, Onlus.
Author contributions
Conceptualization, C.P. and V.T.; investigation, C.P.; writing – original draft, C.F.; writing – review and editing, C.F., C.P., A.C., and V.T.; supervision, V.T.; funding acquisition, V.T. and A.C.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Valeria Tiranti, Email: valeria.tiranti@istituto-besta.it.
Camille Peron, Email: camille.peron@istituto-besta.it.
References
- 1.Aleo S.J., Del Dotto V., Romagnoli M., Fiorini C., Capirossi G., Peron C., Maresca A., Caporali L., Capristo M., Tropeano C.V., et al. Genetic variants affecting NQO1 protein levels impact the efficacy of idebenone treatment in Leber hereditary optic neuropathy. Cell Rep. Med. 2024;5 doi: 10.1016/j.xcrm.2023.101383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danese A., Patergnani S., Maresca A., Peron C., Raimondi A., Caporali L., Marchi S., La Morgia C., Del Dotto V., Zanna C., et al. Pathological mitophagy disrupts mitochondrial homeostasis in Leber’s hereditary optic neuropathy. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu X., Ma Y., Liu Y., Wan Q. Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2020.100245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peron C., Mauceri R., Iannielli A., Cavaliere A., Legati A., Rizzo A., Sciacca F.L., Broccoli V., Tiranti V. Generation of two human iPSC lines, FINCBi002-A and FINCBi003-A, carrying heteroplasmic macrodeletion of mitochondrial DNA causing Pearson’s syndrome. Stem Cell Res. 2021;50 doi: 10.1016/j.scr.2020.102151. [DOI] [PubMed] [Google Scholar]
- 5.Ge H., Tan L., Wu P., Yin Y., Liu X., Meng H., Cui G., Wu N., Lin J., Hu R., Feng H. Poly-L-ornithine promotes preferred differentiation of neural stem/progenitor cells via ERK signalling pathway. Sci. Rep. 2015;5 doi: 10.1038/srep15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall P.E., Lathia J.D., Caldwell M.A., ffrench-Constant C. Laminin enhances the growth of human neural stem cells in defined culture media. BMC Neurosci. 2008;9:71. doi: 10.1186/1471-2202-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brafman D.A. In: Stem Cell Renewal and Cell-Cell Communication: Methods and Protocols Methods in Molecular Biology. Turksen K., editor. Springer; 2015. Generation, Expansion, and Differentiation of Human Pluripotent Stem Cell (hPSC) Derived Neural Progenitor Cells (NPCs) pp. 87–102. [DOI] [PubMed] [Google Scholar]
- 8.Bridges H.R., Fedor J.G., Blaza J.N., Di Luca A., Jussupow A., Jarman O.D., Wright J.J., Agip A.-N.A., Gamiz-Hernandez A.P., Roessler M.M., et al. Structure of inhibitor-bound mammalian complex I. Nat. Commun. 2020;11:5261. doi: 10.1038/s41467-020-18950-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okun J.G., Lümmen P., Brandt U. Three Classes of Inhibitors Share a Common Binding Domain in Mitochondrial Complex I (NADH:Ubiquinone Oxidoreductase) J. Biol. Chem. 1999;274:2625–2630. doi: 10.1074/jbc.274.5.2625. [DOI] [PubMed] [Google Scholar]
- 10.Gutman M., Singer T.P., Casida J.E. Studies on the respiratory chain-linked reduced nicotinamide adenine dinucleotide dehydrogenase. XVII. Reaction sites of piericidin A and rotenone. J. Biol. Chem. 1970;245:1992–1997. doi: 10.1016/S0021-9258(18)63196-5. [DOI] [PubMed] [Google Scholar]
- 11.He F. Bradford Protein Assay. Bio-101. 2011:e45. https://bio-protocol.org/exchange/protocoldetail?id=45&type=1. [Google Scholar]
- 12.Bradford M.M. A rapid and sensitive for the quantitation of microgram quantitites of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate datasets or code.