Abstract
Cell-mediated immunity plays an essential role in the control of infection with the human cytomegalovirus (HCMV). However, only a few CD8+-T-cell epitopes are known, with the majority being contained in the pp65 phosphoprotein, which is believed to dominate the CD8+-T-cell response to HCMV. Here, we have readdressed the issue of CD8+ T cells specific for the 72-kDa major immediate-early protein (IE-1), which is nonstructural but is found very early and throughout the replicative cycle. Using a novel flow-cytometric assay, we were able to identify CD8+-T-cell epitopes (by IE-1 peptide-specific induction of cytokine synthesis) and simultaneously measure the frequency of cells directed against them. For this purpose, 81 pentadecamer peptides covering the complete 491-amino-acid sequence of IE-1 were tested on peripheral blood mononuclear cells of anti-HCMV immunoglobulin G-seropositive donors. At least 10 new epitopes were identified, and the fine specificity and presenting HLA molecule of the first of them was determined. The frequencies of CD8+ T cells directed against IE-1 were similar to those directed against pp65 in donors tested with known pp65-derived peptides. Importantly, additional testing of a corresponding set of peptides covering the complete sequence of pp65 on 10 of these donors identified individuals whose CD8+ T cells recognized IE-1 but not pp65 and vice versa, clearly illustrating that either protein may be a major target. In summary, our results suggest that IE-1 is far more important as a CD8+-T-cell target than current opinion suggests.
Primary infection with or reactivation of the human cytomegalovirus (HCMV) is a major complication after bone marrow transplantation and solid-organ transplantation, and in persons infected with human immunodeficiency virus (11, 19–21, 24). Cell-mediated immunity plays an essential role in the control of persistent infection, induction of latency, and recovery from acute disease (11, 14, 15, 19–21, 24). Adoptive transfer of HCMV-specific CD8+-T-cell clones in bone marrow transplant recipients has successfully prevented viremia and disease (11, 20, 21, 24), illustrating the importance of a sufficient CD8+-T-cell response in particular. Boosting the CD8+-T-cell response with HCMV-derived peptides could potentially be a useful alternative to adoptive T-cell transfer in these patients. In this regard, a previously defined HLA-2.1-presented cytotoxic T-lymphocyte (CTL) epitope from the HCMV pp65 protein is being evaluated as a candidate for vaccination therapy (5).
Unfortunately, to date, our knowledge of HCMV CD8+-T-cell epitopes is limited to only a few, most of which originate from the pp65 lower matrix phosphoprotein (UL83) (25) and some of which originate from the 72-kDa major immediate-early protein (IE-1) (1, 9). The presenting HLA molecules, as far as they are known, are HLA-A2, HLA-B7, HLA-B8, HLA-B18, and HLA-B35 (1, 9, 25), so that only approximately 50 to 60% of a Caucasian population could theoretically benefit from vaccinations or immune therapy based on these known epitopes. Despite the existence of several CTL epitopes in IE-1 (1, 9) and the observation of IE-1-directed CTL activity in early studies on the anti-HCMV T-cell response (2), the pp65 phosphoprotein is presently believed to largely dominate the anti-HCMV CTL response (13, 25). This is partly because structural proteins, such as pp65, which is presented by infected cells prior to virus protein synthesis, are thought to be more effective targets per se than are nonstructural proteins (such as IE-1) (13). That major histocompatibility complex class I-presented peptides are typically 9 amino acids long (16–18) was first revealed in 1991 (8) and later reflected in the use of at least nine overlaps in the design of peptides used for epitope mapping. The only major study attempting to map CTL epitopes in IE-1 was in fact published in 1991 and used only six overlaps between adjacent peptides (1). Because pp65 had only recently been studied with regard to CTL epitopes by using a modern peptide design (25), we chose IE-1 as the first HCMV protein to readdress the issue of CD8+-T-cell epitopes by using a novel and very efficient flow cytometry-based method (12). This new method examines rapid peptide-specific induction of effector cytokine synthesis at the single-cell level (12). For this purpose, 81 15-amino-acid peptides (nine overlaps between adjacent peptides) derived from the 491-amino-acid protein sequence of IE-1 (SwissProt accession no. P13202) were tested on peripheral blood mononuclear cells (PBMC) from 20 HCMV-seropositive healthy blood donors. At least 10 15-amino-acid peptides that induced gamma interferon (IFN-γ) synthesis in subsets of donors were identified. For the first one of these, we have now determined the minimal epitope and the presenting HLA molecule. Interestingly, the measured CD8+-T-cell frequencies were of the same order of magnitude as those elicited by known pp65-derived peptides in HLA-A2- and HLA-B7-positive donors (12). More importantly even, additional testing with a corresponding set of 15-amino-acid peptides covering the complete sequence of pp65 in some of the donors identified several subjects with a strong CD8+-T-cell response against IE-1 but not pp65. Conversely, we identified several donors who had no CD8+-T-cell response to IE-1 but did show a response to pp65-derived peptides. These results may have important implications with regard to the putative role of IE-1 as a factor in HCMV-associated immune system pathology. In summary, our results suggest that in some individuals IE-1 may be of the same importance as pp65, and in a subset who do not have pp65-specific CD8+ T cells, IE-1 may even dominate the CD8+ immune response (possibly together with yet unidentified epitopes from other proteins).
MATERIALS AND METHODS
Citrated blood was obtained from HLA-typed HCMV-seropositive (immunoglobulin G IgG) blood donors. Following standard Ficoll-Paque (Pharmacia, Uppsala, Sweden) density centrifugation, cells were washed with sterile phosphate-buffered saline (PBS; Gibco-BRL), resuspended in RPMI 1640 (Biochrom, Berlin, Germany) containing 0.1% (wt/vol) bovine serum albumin (Biochrom) and 50 mM glutamine (Biochrom), and adjusted to 107 cells/ml. Then 200 μl each of cell suspensions and peptide solutions (10 μg/ml in RPMI 1640 containing 0.1% bovine serum albumin) were placed in Cellstar polystyrene tubes (Greiner, Frickenhausen, Germany) and placed in an incubator (5° slant) at 37°C under a humidified 5% CO2 atmosphere. After 1 h, 1,600 μl of RPMI 1640 containing 12.5% (vol/vol) fetal calf serum (Biochrom), 50 mM glutamine, and 12.5 μg of brefeldin A (Sigma, Munich, Germany) per ml was added. After an additional 5 h, the cells were washed with cold PBS, resuspended in PBS–1 mM EDTA (Merck, Darmstadt, Germany), incubated for 10 min at 37°C, and washed again with cold PBS. After surface staining with monoclonal antibodies (for 30 min at 4°C in the dark), the cells were fixed for 5 min at 37°C in PBS containing 4% (wt/vol) paraformaldehyde (Merck) and washed in PBS prior to permeabilization (permeabilizing solution; Becton Dickinson, Heidelberg, Germany) as specified by the manufacturer. Following intracellular staining, the cells were washed in PBS and analyzed on a FACScalibur flow cytometer (Becton Dickinson) by using the CellQuestTM software package. Unstimulated samples were analyzed to verify the effect of stimulation. Data files were analyzed with CellQuest or Paint-a-Gate software (Becton Dickinson).
On day 1, candidate peptides were selected according to the results obtained with pooled peptides. These candidate peptides were tested individually on day 2. For that purpose, PBMC from day 1 were kept in a standard incubator. Stimulation was performed with individual peptides by the same method as on day 1, i.e., with the same concentration of individual peptides as within the peptide pool. Parallel stimulation with (noncandidate) control peptides and unstimulated samples was always performed to rule out unspecific stimulation.
Antibodies and peptides.
Fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ, phycoerythrin (PE)-conjugated anti-CD69, Peridinin-chlorophyl-protein (PerCP)-conjugated anti-CD8, allophyocyanin (APC)-conjugated anti-CD3, and the corresponding isotype- and fluorescent conjugate-matched control reagents were purchased from Becton Dickinson. Peptides (synthesized by the standard Fmoc method) were purchased from NMI (Reutlingen, Germany) or produced at our own facility. Peptides were stored freeze-dried or in dimethyl sulfoxide (DMSO) at 4 mg/ml. Peptide pools were generated from peptides dissolved in DMSO. DMSO concentrations in all assay mixtures were kept below 0.1% (vol/vol).
RESULTS AND DISCUSSION
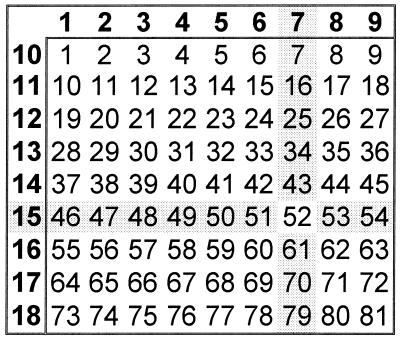
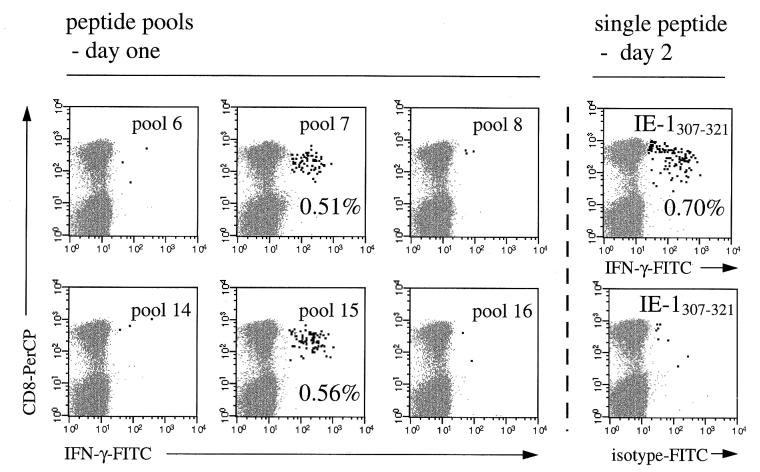
Measurement of peptide-specific induction of IFN-γ in CD8+ T cells is a rapid and efficient way to identify CD8+-T-cell epitopes (12). By testing 81 overlapping peptides, the complete amino acid sequence of the IE-1 protein was covered. Peptide pools were set up in such a way that every peptide was contained in exactly two pools. Thus, it was possible to identify candidate peptides by testing not more than 18 peptide pools on day 1 (Fig. 1). The following day, selected candidate peptides were tested individually. For this purpose, PBMC from the same donors had been kept in complete medium in a standard incubator overnight. Interestingly, stimulation with previously identified pp65-derived peptides (12) on days 1 and 2 revealed that this preincubation slightly increased the frequency of responding CD8+ T cells without increasing nonspecific stimulation as tested with an irrelevant peptide (Fig. 1, top right). We believe that this effect was due to preactivation of antigen-presenting cells and/or a reduction of inhibitory monocytic activity. The shading in Fig. 1 illustrates how candidate peptides were chosen. The combination of the two positive pools, 7 and 15, clearly identifies the candidate peptide 52 (i.e., IE-1307–321, the only one contained in both pools). Figure 2 corresponds to Fig. 1 and shows the CD8+-T-cell IFN-γ responses obtained with six different pools, the two positive pools and the four “neighboring” negative pools, 6, 8, 14, and 16. Table 1 summarizes the results from 15 experiments with samples from healthy HCMV IgG-seropositive donors, which led to the identification of several CD8+-T-cell-inducing IE-1-derived peptides. Table 1 also shows the results obtained by testing a corresponding set of 15-amino-acid peptides covering the complete sequence of the pp65 protein (SwissProt accession no. P06725) in 10 of these donors.
FIG. 1.
Design of peptide pools. The numbers of the pools (left column and top row) are shown in bold. Individual peptides (n = 81) in these 18 pools correspond to the numbers in the respective columns and rows, with peptide 1 = IE-11–15, peptide 2 = IE-17–21, etc., according to the complete sequence of the 72-kDa major IE protein.
FIG. 2.
Identification of a CD8+-T-cell inducing peptide from peptide pools. Stimulation of PBMC from an HCMV IgG-seropositive donor with overlapping 15-amino-acid peptides originating from the IE-1 protein. The figure shows results obtained with six different peptide pools on day 1, two of which (pool 7 and pool 15) gave positive results, and stimulation with the candidate peptide IE-1307–321 (EFCRVLCCYVLEETS), the only peptide contained in both positive pools, on day 2. IFN-γ-positive events are highlighted. Results for 50,000 CD3 T cells are displayed in each diagram. PBMC were stained with anti-IFN-γ–FITC, anti-CD69–PE, anti-CD8–PerCP, and anti-CD3–APC. Axes show log fluorescence intensity.
TABLE 1.
Listing of identified CD8+ T cell epitopes in IE-1 and pp65a
| Donor | HLA type | IE-1
|
pp65
|
||
|---|---|---|---|---|---|
| Peptide | %b | Peptide | % | ||
| D1 | A2, A66, Bw6, B7, Cw7, B41 | IE-1307–321 | 0.67 | Not tested | |
| IE-173–87 | 0.50 | ||||
| D2 | A2, A3, Bw6, B7, Cw7 | IE-1307–321 | 0.47 | pp65413–427c | 0.20 |
| IE-1181–195 | 0.15 | pp65417–431c | 0.33 | ||
| D3 | A3, Bw4, Bw6, B7, B51, Cw7 | IE-1307–321 | 0.78 | Not tested | |
| D4 | A1, A3, Bw6, B7, B8, Cw7 | IE-1307–321 | 5.46 | Not tested | |
| IE-1199–213 | 1.36 | ||||
| D5 | A1, A2, Bw6, B8, B60, Cw3, Cw7 | IE-137–51 | 2.72 | pp65489–503d | 0.13 |
| IE-1193–207 | 0.18 | pp65493–507d | 0.10 | ||
| IE-1199–213 | 0.17 | ||||
| D6 | A1, A29, Bw4, Bw6, B8, B57, Cw6 | IE-185–99 | 1.59 | Nonee | None |
| D7 | A2, B35, B37, Cw2, Cw9 | IE-1421–435 | 0.13 | pp65489–503 | 0.19 |
| pp65493–507 | 0.55 | ||||
| D8 | A1, A3, Bw4, B37, Cw6 | IE-1289–303 | 1.44 | None | None |
| D9 | A3, A23, Bw4, Bw6, B37, B62, Cw3, Cw6 | IE-1181–195 | 0.56 | Not tested | |
| D10 | A1, Bw6, B8, Cw7 | IE-185–99 | 1.73 | None | None |
| IE-1193–207 | 0.22 | ||||
| D11 | A1, B8, B44, Cw4, Cw7 | IE-1193–207 | 2.17 | Not tested | |
| IE-1199–213 | 1.46 | ||||
| D12 | A1, A2, Bw4, Bw6, B52, B61, Cw3 | IE-1379–393 | 0.46 | pp65289–303 | 1.03 |
| pp65293–307 | 0.90 | ||||
| pp65489–503 | NDf | ||||
| pp65493–507 | 0.16 | ||||
| pp65517–531 | 0.15 | ||||
| D13 | A30, A32, Bw4, B49, B51, Cw7 | None | None | pp65109–123 | 0.16 |
| D14 | A2, A28, Bw4, B27, B37, Cw3, Cw6 | None | None | pp65205–219 | 0.78 |
| pp65489–503 | 0.02 | ||||
| pp65493–507 | 0.02 | ||||
| D15 | A2, A30/31, Bw6, B8, B60, Cw3, Cw7 | None | None | pp65489–503 | 0.33 |
| pp65493–507 | 0.30 | ||||
| D16–D18 | B7… | IE-1307–321g | None | Not tested | Not tested |
All peptides giving positive responses are listed for each donor. The complete sets of overlapping 15-mer peptides covering the complete sequences of IE-1 and pp65 were tested.
Percentage of total CD8+ T cells expressing IFN-γ (background from control samples subtracted).
Contains the known HLA-B7-presented epitopes pp65417–425 and pp65418–426 (12).
Contains the known HLA-A2 presented epitope pp65495–503 (12).
Negative responses were defined as absence of a population of positive events (not found in control samples, i.e., unstimulated samples or samples with irrelevant peptide) with at least 50,000 CD3+ events gated. Positive events in control samples (‘background’) ranged from 0.01 to 0.06% of CD8+ T cells.
pp65493–503 was not tested individually in this donor for lack of material; however, a response would be expected (like pp65493–507, it contains the known HLA-A2 presented epitope pp65495–503).
By way of exclusion, this peptide was found to be HLA-B7 presented (see HLA types in bold type in the table) and was the only peptide tested in these HCMV IgG-seronegative, HLA-B7-positive controls. Responses did not exceed those from unstimulated samples (0.01 to 0.06% of CD8+ T cells).
Among the IE-1-derived peptides, IE-1307–321 (EFCRVLCCYVLEETS), IE-1193–207 (ARAKKDELRRKMMYM), and IE-1199–213 (ELRRKMMYMCYRNIE) were the most frequently identified. All donors reactive to IE-1307–321 were HLA-B7, HLA-Bw6, and HLA-Cw7 positive (D1, D2, D3, and D4); however, donors who were HLA-Cw7 or HLA-Bw6 but not HLA-B7 positive did not react to this peptide (for example D5, D6, D9, D10, and D11), identifying HLA-B7 as the presenting allomorph.
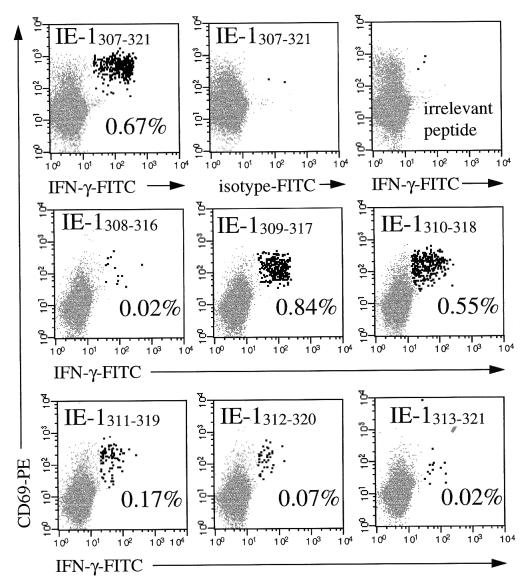
To define the epitope contained in IE-1307–321 more precisely, all seven consecutive 9-amino-acid peptides derived from its sequence were synthesized (i.e., IE-1307–315, IE-1308–316, IE-1309–317, through IE-1313–321) and tested in two donors. Several of these nonamer peptides were able to stimulate IFN-γ induction. In both donors, IE-1309–317 induced the highest frequency of CD8+ T cells (0.84 and 0.64%), IE-1310–318 gave a somewhat smaller response (0.50 and 0.42%), and two additional consecutive peptides, IE-1309–317 and IE-1310–318, gave increasingly weak responses (Fig. 3). These different frequencies of responding CD8+ T cells obtained in response to stimulation with largely overlapping peptides suggested that T cells of overlapping or different specificities were addressed or that, owing to the relatively high concentrations of peptides we used (on the order of 109 mol/liter), the incomplete epitope also stimulated T-cell cytokine induction in much the same way as incomplete epitopes can induce target cell lysis in CTL assays if their concentrations are high enough (7). This will have to be examined in additional experiments.
FIG. 3.
Fine mapping of a 15-amino-acid peptide. According to the amino acid sequence of the HLA-B7-presented peptide IE-1307–321 (EFCRVLCCYVLEETS), seven additional nonamer peptides were synthesized and tested on PBMC of an HLA-B7-positive donor whose CD8+ T cells were reactive to the original peptide (top). Several of these nonamer peptides led to IFN-γ induction in CD8+ T cells, with IE-1309–317 giving the strongest response (middle and bottom). IFN-γ-positive events are highlighted. The activation marker CD69 is used to increase the specificity of the analysis. Results for 50,000 CD8+ T cells are displayed. PBMC were stained with anti-IFN-γ–FITC, anti-CD69–PE, anti-CD8–PerCP, and anti-CD3–APC. Axes show log fluorescence intensity.
Finally, to show that previous infection with CMV was necessary for cytokine induction by IE-1307–321, we tested this peptide on PBMC from three HCMV-seronegative, HLA-B7-positive donors under identical conditions but observed no response (Table 1, bottom). Phorbol myristate acetate/ionomycin stimulation performed in parallel showed that the CD8+ T cells from all individuals were able to produce cytokines upon stimulation. We assumed, therefore, that reactivity to this peptide in our HLA-B7-positive donors was indeed dependent on immunization with HCMV and not likely to result from cross-reactivity with peptides of different origin.
Unlike IE-1307–321, the other identified epitopes did not allow for easy determination of their presenting HLA molecule. Notably, the peptides IE-1193–207 and IE-1199–213 (Table 1) share 9 amino acids, suggesting that in donors who were reactive to both peptides (D5 and D11 [Table 1]) this overlap may define the epitope. Interestingly, computerized binding motif analysis carried out by SYFPEITHI (16), a computer program based on listings of MHC ligands and binding motifs (17, 18) predicted HLA-B8 as the most likely presenting allomorph for this candidate epitope. An even higher score, however, was obtained for HLA-A1 and the decapeptide, IE-1198–207 (DELRRKMMYM [contained only in IE-1193–207]). Nevertheless, at this stage the data is inconclusive, because although HLA-A1 and HLA-B8 are found in all donors reactive to one or both of these peptides (D4, D5, D10, and D11), other donors with just HLA-A1 (D8 and D12), just HLA-B8 (D15), or both HLA-A1 and HLA-B8 (D6) did not respond to either. Fine mapping of the epitope(s) with smaller peptides, however, is likely to give additional clues.
It is important to note that despite the identification of many new epitopes and high frequencies of CD8+ T cells directed against them in some donors, samples from other individuals tested with the complete set of IE-1 peptides showed no response to any of them. Testing some of our donors with a corresponding set of overlapping peptides covering the complete 561-amino-acid sequence of pp65 identified several individuals who, despite a strong response to one or several IE-1-derived peptides, did not respond to any of the pp65-derived peptides and vice versa. By testing the pp65-derived peptides, several new epitopes were identified, and some of these are also shown in Table 1 (the presenting HLA allomorphs are currently being determined). In some donors, the responses to certain peptides were clearly positive (a distinct population of IFN-γ-positive events not observed in the control) yet extremely small. For example, in D15, the frequency of CD8+ T cells responsive to pp65489–503 and pp65493–507 (both containing the known epitope pp65495–503) was only 0.02%, while 0.78% responded to the newly identified epitope pp65205–219. This probably illustrates that when several different epitopes can be presented, individual peptides may become dominant epitopes. The same phenomenon may also be observed in other donors, where single peptides give much stronger responses than others (Table 1).
In donors reactive to both pp65 and IE-1, the frequencies of responsive CD8+ T cells may be compared; however, it should be noted that the responses obtained with overlapping peptides cannot simply be added, since complete or partial epitopes may be contained in the overlap. More important than the difference in the frequencies of CD8+ T cells responding to one or the other protein in donors responsive to both seems to be the fact that some donors responded to only one of the two proteins. So far we have tested 12 donors with both complete sets of peptides. In total, 7 donors (58%) had IE-1-specific CD8+ T cells, 9 donors (75%) had pp65-specific T cells, and 4 donors (33%) had CD8+ T cells responsive to both proteins. Three donors (25%) responded only to IE-1, and 5 (42%) responded only to pp65. The finding that IE-1-specific CD8+ T cells exist in some but not all individuals may be very important with regard to IE-1-associated immune system pathology (3, 4). IE-1 is known to induce adhesion molecule upregulation in infected cells, which may lead to nonspecific activation of other cells (3, 4). This process is not disrupted by therapy with ganciclovir or foscarnet (4). The role of IE-1-recognizing CD8+ T cells in such a situation will be an interesting subject to study.
Notably, all donors nonresponsive to the pp65 peptides were HLA-A2 or HLA-B7 negative. Conversely, most individuals not responding to IE-1 were HLA-A2 positive and HLA-B7 negative. The data, though preliminary, suggests that preferences for IE-1 over pp65 and vice versa are directly related to the HLA type, and additional studies are in progress to corroborate these results. Dominant peptides may be derived from pp65 or IE-1, and clearly there is no obvious hierarchy of presenting allomorphs at this stage.
Interestingly, Gilbert et al. reported in 1996 that IE-1-specific CTL were able to lyse IE-1-transfected but not HCMV-infected autologous fibroblasts (10), and they found that pp65 apparently abrogated IE-1 peptide presentation by restricting its access to the antigen-processing machinery or diverting it to a different degradation pathway. This in contrast to earlier data obtained by Borysiewicz et al., who found that in two subjects, 18 to 58% of the CTL clones able to lyse HCMV-infected autologous fibroblasts were IE-1 specific (2). In their study, only a small number of IE-1-specific CTL clones were unable to lyse HCMV-infected targets. The phenomenon described by Gilbert et al. may thus apply to only some IE-1-specific clones. This could be explained if some but not all IE-1 epitopes escaped presentation under the influence of pp65. Nevertheless, the high frequencies of IE-1-specific CD8+ T cells in our donors clearly argue that sufficient presentation of IE-1-derived peptides takes place for these cells to be generated. It is important to note in this regard that in HCMV reactivation, IE proteins are expressed prior to the early protein pp65 (whose expression is regulated by IE-1- and IE-2-dependent promoters) (22). Recent studies indicate that HCMV reactivation is a frequent event even in healthy donors (23), seems to be stress related, and is probably mediated by tumor necrosis factor alpha (TNF-α) (6, 23).
To compile a selection of epitopes which may be useful for immunotherapy in a majority of the population, additional work must be done. Since the protein-coding content of HCMV is enormous, it will hardly be possible to scan all proteins for epitopes. However, the need to identify T-cell targets in HCMV proteins will increase if vaccination trials using, for example, the A2-presented pp65495–503 are successful (5). Our results should encourage the search for T-cell epitopes in other nonstructural HCMV proteins to complete the repertoire of peptides which may be potential candidates for vaccine development.
REFERENCES
- 1.Alp N J, Allport T D, Van Zanten J, Rodgers B, Sissons J G, Borysiewicz L K. Fine specificity of cellular immune responses in humans to human cytomegalovirus immediate-early 1 protein. J Virol. 1991;65:4812–4820. doi: 10.1128/jvi.65.9.4812-4820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borysiewicz L K, Hickling J K, Graham S, Sinclair J, Cranage M P, Smith G L, Sissons J G. Human cytomegalovirus-specific cytotoxic T cells. Relative frequency of stage-specific CTL recognizing the 72-kD immediate early protein and glycoprotein B expressed by recombinant vaccinia viruses. J Exp Med. 1988;168:919–931. doi: 10.1084/jem.168.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns L J, Pooley J C, Walsh D J, Vercellotti G M, Weber M L, Kovacs A. Intercellular adhesion molecule-1 expression in endothelial cells is activated by cytomegalovirus immediate early proteins. Transplantation. 1999;67:137–144. doi: 10.1097/00007890-199901150-00023. [DOI] [PubMed] [Google Scholar]
- 4.Craigen J L, Grundy J E. Cytomegalovirus induced up-regulation of LFA-3 (CD58) and ICAM-1 (CD54) is a direct viral effect that is not prevented by ganciclovir or foscarnet treatment. Transplantation. 1996;62:1102–1108. doi: 10.1097/00007890-199610270-00014. [DOI] [PubMed] [Google Scholar]
- 5.Diamond D J, York J, Sun J Y, Wright C L, Forman S J. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- 6.Docke W D, Prosch S, Fietze E, Kimel V, Zuckermann H, Klug C, Syrbe U, Kruger H D, von Baehr R, Volk H D. Cytomegalovirus reactivation and tumour necrosis factor. Lancet. 1994;343:268–269. doi: 10.1016/s0140-6736(94)91116-9. [DOI] [PubMed] [Google Scholar]
- 7.Falk K, Rotzschke O, Stevanovic S, Gnau V, Sparbier K, Jung G, Rammensee H-G, Walden P. Analysis of a naturally occurring HLA class I-restricted viral epitope. Immunology. 1994;82:337–342. [PMC free article] [PubMed] [Google Scholar]
- 8.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 9.Gavin M A, Gilbert M J, Riddell S R, Greenberg P D, Bevan M J. Alkali hydrolysis of recombinant proteins allows for the rapid identification of class I MHC-restricted CTL epitopes. J Immunol. 1993;151:3971–3980. [PubMed] [Google Scholar]
- 10.Gilbert M J, Riddell S R, Plachter B, Greenberg P D. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature. 1996;383:720–722. doi: 10.1038/383720a0. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg P D, Reusser P, Goodrich J M, Riddell S R. Development of a treatment regimen for human cytomegalovirus (CMV) infection in bone marrow transplantation recipients by adoptive transfer of donor-derived CMV-specific T cell clones expanded in vitro. Ann N Y Acad Sci. 1991;636:184–195. doi: 10.1111/j.1749-6632.1991.tb33450.x. [DOI] [PubMed] [Google Scholar]
- 12.Kern F, Surel I P, Brock C, Freistedt B, Radtke H, Scheffold A, Blasczyk R, Reinke P, Schneider-Mergener J, Radbruch A, Walden P, Volk H D. T-cell epitope mapping by flow cytometry. Nat Med. 1998;4:975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 14.Quinnan G V, Jr, Burns W H, Kirmani N, Rook A H, Manischewitz J, Jackson L, Santos G W, Saral R. HLA-restricted cytotoxic T lymphocytes are an early immune response and important defense mechanism in cytomegalovirus infections. Rev Infect Dis. 1984;6:156–163. doi: 10.1093/clinids/6.2.156. [DOI] [PubMed] [Google Scholar]
- 15.Quinnan G V, Jr, Kirmani N, Rook A H, Manischewitz J F, Jackson L, Moreschi G, Santos G W, Saral R, Burns W H. Cytotoxic T cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N Engl J Med. 1982;307:7–13. doi: 10.1056/NEJM198207013070102. [DOI] [PubMed] [Google Scholar]
- 16.Rammensee, H. G., J. Bachmann, N. Emmerich, and S. Stevanovic. 15 January 1999, posting date. SYFPEITHI—an Internet database for MHC ligands and peptide motifs. [Online] http://134.2.96.221/scripts/hlaserver.dll/home.htm. [20 April 1999, last date accessed.] [DOI] [PubMed]
- 17.Rammensee H G, Bachmann J, Stevanovic S. MHC ligands and peptide motifs. Georgetown, Tex: Landes Bioscience; 1997. [Google Scholar]
- 18.Rammensee H G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 19.Reusser P, Riddell S R, Meyers J D, Greenberg P D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 20.Riddell S R, Reusser P, Greenberg P D. Cytotoxic T cells specific for cytomegalovirus: a potential therapy for immunocompromised patients. Rev Infect Dis. 1991;11:966–973. doi: 10.1093/clind/13.supplement_11.s966. [DOI] [PubMed] [Google Scholar]
- 21.Riddell S R, Walter B A, Gilbert M J, Greenberg P D. Selective reconstitution of CD8+ cytotoxic T lymphocyte responses in immunodeficient bone marrow transplant recipients by the adoptive transfer of T cell clones. Bone Marrow Transplant. 1994;4:78–84. [PubMed] [Google Scholar]
- 22.Stenberg R M. Immediate-early genes of human cytomegalovirus: organization and function. In: Becker Y, Darai G, Huang E S, editors. Molecular aspects of human cytomegalovirus diseases. Berlin, Germany: Springer-Verlag KG; 1993. pp. 330–359. [Google Scholar]
- 23.Toro A I, Ossa J. PCR activity of CMV in healthy CMV-seropositive individuals: does latency need redefinition? Res Virol. 1996;147:233–238. doi: 10.1016/0923-2516(96)89654-3. [DOI] [PubMed] [Google Scholar]
- 24.Walter E A, Greenberg P D, Gilbert M J, Finch R J, Watanabe K S, Thomas E D, Riddell S R. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 25.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]