Abstract
Background and Aim:
Staphylococci, which inhabit skin and mucous membranes in humans and animals, are opportunistic pathogens. Coagulase-positive and coagulase-negative staphylococci (CoNS) are the two main groups. Clinical abscesses in rabbits often harbor Staphylococcus aureus and CoNS. This study estimated S. aureus and CoNS prevalence, resistance profiles, antimicrobial-resistant genes, and the accessory gene regulator (agr) group in rabbit clinical abscesses.
Materials and Methods:
Sixty-seven abscesses were gathered from 67 rabbits who visited Prasu-Arthorn Animal Hospital in Nakornpathom, Thailand, from January 2014 to October 2015. Thirty-four subcutaneous, 29 dental, 2 ocular, 1 mammary gland, and 1 uterine abscess were present. Conventional methods, including Gram staining, mannitol fermentation, hemolysis on blood agar, catalase testing, and coagulase production, identified and isolated S. aureus and CoNS from all abscesses. All S. aureus and CoNS isolates underwent antimicrobial susceptibility testing using the disk diffusion method. Polymerase chain reaction was used to detect the presence of blaZ, aacA-aphD, msrA, tetK, gyrA, grlA, dfrG, and cfr antimicrobial-resistant genes. Methicillin resistance was identified through the detection of a cefoxitin-resistant phenotype and the presence of mecA gene. Further investigation was conducted on the agr group of S. aureus isolates.
Results:
In 67 abscesses, we found 19 S. aureus isolates in 9 abscesses (13.43%) and 37 CoNS isolates in 18 abscesses (26.87%), both majorly located at subcutaneous sites. About 59.46% of CoNS isolates were methicillin-resistant compared to 5.26% of S. aureus isolates. Methicillin-resistant S. aureus (MRSA) and methicillin-resistant CoNS (MRCoNS) both displayed multidrug resistance (MDR). Both MRSA and MRCoNS expressed multiple antimicrobial resistance genes, including blaZ, aacA-aphD, gyrA, grlA, msrA, tetK, and dfrG. Approximately 73.68% of the agr groups were agr I, 15.79% were agr III, and 10.53% were agr II.
Conclusion:
This study found a high prevalence of MRCoNS with antimicrobial resistance and multiple antimicrobial-resistant genes in rabbits with clinical abscesses. The effectiveness of antibiotics against infections caused by such strains is a matter of concern. Owners should be educated about the importance of good hygiene practices and judicious antibiotic use to prevent widespread antimicrobial resistance.
Keywords: antimicrobial resistance, antimicrobial resistance genes, coagulase-negative staphylococci, rabbit, Staphylococcus aureus
Introduction
Staphylococci inhabit the upper respiratory tract of humans and animals as normal flora on the skin and mucous membranes. These bacteria can adapt and thrive in the environment under adverse conditions, originating from it. Coagulase-positive staphylococci (CoPS) and coagulase-negative staphylococci (CoNS) are the two main groups [1, 2]. Staphylococcus aureus, a globally present zoonotic pathogen under CoPS, notoriously known for its broad virulence factors and antibiotic resistance, causes infections of high morbidity and mortality. In humans, S. aureus can cause a spectrum of diseases, from minor skin infections to severe conditions such as endovascular infections, pneumonia, septic arthritis, endocarditis, osteomyelitis, and sepsis [3]. Animals serve as significant reservoirs for human infections with S. aureus [4]. CoNS are commensal bacteria in humans and several animal species. These opportunistic pathogens pose substantial risks in health-care facilities, particularly through the application of medical devices. In small animal medicine, research on CoNS is less prevalent than that of S. aureus. Although isolated from different infection sites like the skin, ear canal, and respiratory tract, the complete virulence potential of CoNS remains uncertain due to their ability to rapidly acquire multiple resistance traits leading to multidrug resistance (MDR) against various antimicrobials [5-7]. Staphylococcus epidermidis and other species can make infections related to implanted medical devices particularly difficult to treat due to their ability to form biofilms. Despite extensive research on multidrug-resistant CoNS in humans, there is a paucity of data on this topic within veterinary medicine [8]. Staphylococci predominantly infect small dermal lesions in rabbits, making them a significant host for these bacteria. These infections have the potential to penetrate the subcutaneous tissue, leading to conditions such as pododermatitis, subcutaneous abscesses, and mastitis. Internal organ abscesses, including those of the lungs, liver, and uterus, can occasionally form. In severe cases, rabbit infections can lead to infertility, adverse production outcomes, and mortality [3, 9]. Staphylococci infections present a major concern due to the high prevalence of antibiotic resistance in these microbes. Methicillin-resistant staphylococci hold significant clinical importance. Staphylococci are also widely resistant to other antibiotics. They exhibit near-universal resistance to β-lactam antibiotics, such as penicillin (P) and its derivatives, which can be inactivated by β-lactamases. They can exhibit resistance to almost all antibiotics in a combined form [10]. Studies have consistently reported high levels of antimicrobial resistance in staphylococci obtained from companion animals [11–14]. Limited information is available on the antimicrobial resistance of staphylococci found in rabbits [9, 11].
The pathogenicity of S. aureus depends on the emergence of diverse accessory genes for cell wall-related and extracellular proteins. Genes responsible for virulence factors, secretory exotoxins, and hemolysins significantly impact S. aureus’s virulence and antimicrobial resistance. The accessory gene regulator (agr) locus, consisting of the genes hld, agrB, agrD, agrC, and agrA, manages the gene expression. The grouping of agr I, II, III, and IV is determined by distinct polymorphisms in agrD and agrC [15]. Agr groups have lately been linked to specific clinical conditions [16–22]. For instance, community-acquired methicillin-resistant S. aureus (MRSA) and isolates producing S. aureus toxic shock syndrome toxin-1 typically belonged to agr III. Meanwhile, glycopeptide–intermediately resistant S. aureus (GISA) isolates and exfoliatin-producing strains are more prevalent among isolates in agr II and IV, respectively [16]. The preponderance of clinical isolates is found to be comprised of agr I strains [15, 17–24].
This study estimated the prevalence of S. aureus and CoNS in rabbits, determined the antimicrobial resistance of these isolates, identified resistant genes, and classified S. aureus strains according to their agr group.
Materials and Methods
Ethical approval
Sample collection was conducted by an experienced veterinarian, following the guidelines of “Guide for the Care and Use of Laboratory Animals” [25]. All experimental procedures involving animals were approved by The Faculty of Veterinary Science-Animal Care and Use Committee, Mahidol University (protocol number MUVS-2013-35 and MUVS-2023-07-42).
Study period and location
The study was conducted from January 2014 to October 2015. Samples were collected at Prasu-Arthorn Animal Hospital, Faculty of Veterinary Science, Mahidol University, Nakhonpathom Province.
Specimen collection and bacterial isolation
Samples were obtained from 67 rabbits presenting with clinical abscesses at Prasu-Arthorn Animal Hospital in Nakornpathom Province. The owners granted permission for sample collection. Sixty-seven specimens were gathered, including 34 subcutaneous abscesses, 29 dental abscesses, 2 ocular abscesses, 1 mammary gland abscess, and 1 uterine abscess. Within 8 h of collection, all specimens were transferred to the Department of Veterinary Science, Mahidol University, in an Amies transport medium (Oxoid, UK). Three yellow colonies per specimen were grown on mannitol salt agar (Oxoid) at 37°C for 24–48 h and subsequently transferred to blood agar (Oxoid). The colonies were identified based on their hemolysis patterns using classical biochemical techniques, including Gram staining (Merck, Germany), catalase production (Merck), coagulation (Oxoid), and latex agglutination (Dryspot Staphytect plus, Oxoid) for protein A detection.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing and interpretation were conducted using the disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines [26]. A total of 13 antimicrobial drugs were tested, including amikacin (30 μg), azithromycin (AZM) (15 μg), cefazolin (30 μg), cefoxitin (FOX) (30 μg), ceftriaxone (30 μg), chloramphenicol (C) (30 μg), ciprofloxacin (5 μg), doxycycline (DO) (30 μg), gentamicin (10 μg), moxifloxacin (MXF) (5 μg), norfloxacin (NOR) (10 μg), P (10 units), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). S. aureus ATTC® 25923 (Manassas, USA) served as the control strain. Inhibition zones were measured and categorized according to the CLSI guidelines as susceptible (S), intermediate (I), or resistant (R) [26]. Methicillin-resistant strains were confirmed by the FOX-resistant phenotype.
Detection of antimicrobial resistance and agr genes
The DNA extraction kit (Geneaid, Taiwan) was used to extract bacterial genomic DNA. By adding lysis buffer, the bacterial cells were broken down. The membrane retained the DNA in the extraction column, which was then eluted with an elution buffer. The amount of extracted DNA was assessed by spectrophotometric measurement at 260 nm (A260). The antimicrobial resistance genes (blaZ, mecA, aacA-aphD, msrA, tetK, gyrA, grlA, dfrG, and cfr) [27] and agr [21] were identified through polymerase chain reaction (PCR). The specific primers for each gene are given in Table-1 [21, 27]. This PCR mixture, with a total volume of 25 μL, included 1 μM each antimicrobial resistance gene primer, 2 μM agr primer, 2.5 μL 10 U Taq PCR buffer, 0.2 mM deoxyribonucleotide triphosphates (dNTPs), 2 mM MgCl2, and 1 U Taq DNA polymerase (Thermo-scientific, Germany). The PCR mixture underwent a thermal cycling process on the Flexcycler2 (Analytik Jena, Germany) consisting of an initial denaturation at 95°C for 5 min, 30 cycles with denaturation at 95°C for 30 s (Table-1), annealing at gene-specific temperatures for 30 s, extension at 72°C for 60 s, and a final extension at 72°C for 10 min. About 1.5% agarose gel electrophoresis was used to analyze the amplified products, which were then stained with SYBR Safe (Invitrogen, USA). The DNA bands were observed under ultraviolet light using the Invitrogen UVP Bioimaging system (USA).
Table-1.
PCR primer for amplification of antimicrobial resistant genes and agr group.
| Target gene | Primer (5′–3′) | PCR amplicon (bp) | Reference |
|---|---|---|---|
| Beta-lactams blaZ | CAGTTCACATGCCAAAGAG TACACTCTTGGCGGTTTC | 772 | [27] |
| mecA | AAAATCGATGGTAAAGGTTGGC AGTTCTGCAGTACCGGATTTGC | 533 | [27] |
| Aminoglycosides aacA-aphD | CAAGAGCAATAAGGGCATAC CAATAGTTTCAATAGGATAA | 936 | [27] |
| Fluoroquinolones gyrA | AATGAACAAGGTATGACACC TACGCGCTTCAGTATAACGC | 223 | [27] |
| grlA | ACTTGAAGATGTTTTAGGTGAT TTAGGAAATCTTGATGGCAA | 459 | [27] |
| Macrolides msrA | GGCACAATAAGAGTGTTTAAAGG AAGTTATATCATGAATAGATTGTCCTGTT | 940 | [27] |
| Tetracyclines tetK | TTATGGTGGTTGTAGCTAGAAA AAAGGGTTAGAAACTCTTGAAA | 348 | [27] |
| Folate pathway inhibitors dfrG | CCCAAGGACTGGGAATATG TCCTCATAATTCACTTCTGG | 326 | [27] |
| Phenicols cfr | TGAAGTATAAAGCAGGTTGGGAGTCA ACCATATAATTGACCACAAGCAGC | 746 | [27] |
| agr group | |||
| pan | ATGCACATGGTGCACATGC | 439 | [21] |
| agr I | GTCACAAGTACTATAAGCTGCGAT | 572 | |
| agr II | TATTACTAATTGAAAAGTGCCATAGC | 320 | |
| agr III | GTAATGTAATAGCTTGTATAATAATACCCA | 657 | |
| agr IV | GCGATAATGCCGTAATACCCG |
PCR=Polymerase chain reaction, agr=Accessory gene regulator
Statistical analysis
The data were compiled and analyzed using GraphPad Prism version 5 (La Jolla, CA, USA). The antimicrobial resistance phenotypes of S. aureus and CoNS are presented as percentages with 95% confidence intervals.
Results
Rabbit abscess
Figure-1 depicts the various types of abscesses, such as the subcutaneous abscess (1A), dental abscess (1B), ocular abscess (1C), and mammary gland abscess (1D).
Figure-1.
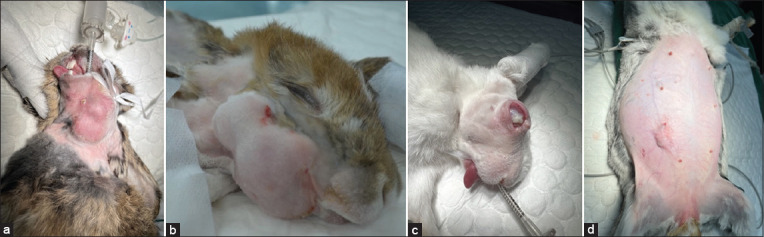
Illustrates abscesses in rabbits, encompassing (a) subcutaneous, (b) dental, (c) ocular, and (d) mammary gland manifestations.
Bacterial isolation and antimicrobial susceptibility testing
Among 67 samples collected from 67 rabbits, 19 isolates of S. aureus were identified from 9 abscesses (13.43%), whereas 37 isolates of CoNS were obtained from 18 abscesses (26.87%). Both S. aureus and CoNS were predominantly found in subcutaneous abscesses (Table-2). These isolates included 1 strain (5.26%) of MRSA and 22 strains (59.46%) of methicillin-resistant CoNS (MRCoNS).
Table-2.
Staphylococcus aureus and coagulase-negative staphylococci (CoNS) isolated from clinical abscess.
| Type of abscess specimen | No. of specimen collection (%) | S. aureus | CoNS | ||
|---|---|---|---|---|---|
|
| |||||
| No. of specimen (%) | No. of isolated | No. of specimen (%) | No. of isolated | ||
| Subcutaneous | 34 (50.75) | 4 (5.97) | 11 | 10 (14.93) | 20 |
| Dental | 29 (43.28) | 3 (4.48) | 4 | 7 (10.45) | 16 |
| Ocular | 2 (2.99) | 1 (1.49) | 3 | 0 (0) | 0 |
| Mammary gland | 1 (1.49) | 1 (1.49) | 1 | 0 (0) | 0 |
| Uterine | 1 (1.49) | 0 (0) | 0 | 1 (1.49) | 1 |
| Total | 67 (100) | 9 (13.43) | 19 | 18 (26.67) | 37 |
The susceptibility of S. aureus and CoNS isolates to antimicrobials was determined. About 42.11% of S. aureus and 78.38% of CoNS were identified as P-resitant (Figure-2). However, S. aureus isolates exhibited greater resistance to DO (15.79%) and C (10.53%), whereas CoNS had greater resistance to AZM (59.46%), MXF (54.05%), and NOR (54.05%). CoNS exhibited a higher resistance rate to a broader array of antimicrobial drugs compared to S. aureus. Fifty-nine-point four six percent of the CoNS isolates displayed MDR. The antimicrobial resistance of S. aureus and CoNS is presented in Tables-3 and 4.
Figure-2.
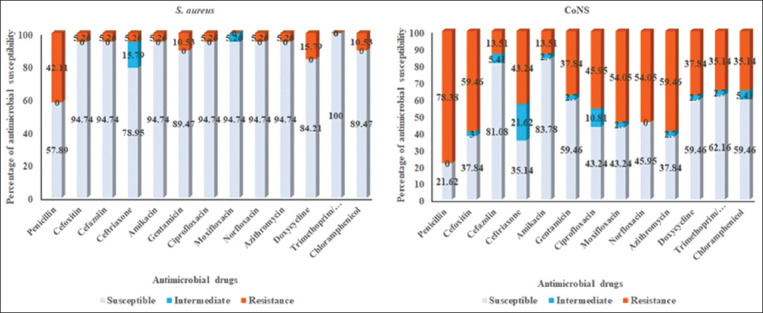
Antibiogram profile of Staphylococcus aureus and coagulase-negative staphylococci isolated from rabbits with clinical abscesses.
Table-3.
Pattern of antimicrobial resistance of Staphylococcus aureus.
| Pattern of antimicrobials resistance of Staphylococcus aureus | Resistant gene | No. of resistance isolates (%) | agr group (No.; %) |
|---|---|---|---|
| Susceptible to all drugs | - | 8 (42.11) | I (8; 42.11) |
| P | blaZ | 5 (26.32) | I (3; 15.79), II (2; 10.53) |
| C | cfr | 2 (10.53) | I (2; 10.53) |
| AK, CN | aacA-aphD | 1 (5.26) | III (1; 5.26) |
| P, DO | blaZ, tetK | 2 (10.53) | III (2; 10.53) |
| P, FOX, KZ, CRO, CN, CIP, MXF, NOR, AZM, DO | blaZ, mecA, gyrA, grlA, msrA | 1 (5.26) | I (1; 5.26) |
AK=Amikacin, AZM=Azithromycin, C=Chloramphenicol, CIP=Ciprofloxacin, CN=Gentamicin, CRO=Ceftriaxone, DO=Doxycycline, FOX=Cefoxitin, KZ=Cephazolin, MXF=Moxifloxacin, NOR=Norfloxacin, P=Penicillin
Table-4.
Pattern of antimicrobial resistance of coagulase-negative staphylococci.
| Pattern of antimicrobials resistance of coagulase-negative staphylococci | Resistant gene | No. of isolate |
|---|---|---|
| Susceptible to all drugs | - | 4 |
| P | - | 1 |
| P | blaZ | 4 |
| C | - | 2 |
| C | cfr | 1 |
| P, FOX | mecA | 1 |
| AK, AZM | msrA | 1 |
| P, FOX, MXF | blaZ, mecA | 1 |
| P, FOX, CRO, MXF | blaZ, mecA | 1 |
| P, FOX, MXF, NOR, AZM | blaZ, mecA, gyrA, grlA, msrA | 1 |
| P, AK, CN, CIP, NOR, AZM | blaZ, aacA-aphD, gyrA, grlA, msrA | 1 |
| P, AK, CN, CIP, AZM, SXT | blaZ, aacA-aphD, gyrA, grlA, msrA, dfrG | 1 |
| P, FOX, CRO, CN, NOR, AZM, C | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA | 1 |
| P, FOX, CN, CIP, MXF, NOR, AZM | blaZ, mecA, gyrA, grlA, msrA | 2 |
| P, FOX, CRO, CN, CIP, MXF, NOR, AZM | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA | 1 |
| P, FOX, CN, CIP, MXF, NOR, AZM, DO, C | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA | 1 |
| P, FOX, CRO, CN, CIP, MXF, NOR, AZM, DO, SXT | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA, tetK, dfrG | 1 |
| P, FOX, CRO, CN, CIP, MXF, NOR, AZM, DO, C | blaZ, mecA, gyrA, grlA, msrA, tetK | 1 |
| P, FOX, CRO, CIP, MXF, NOR, AZM, DO, SXT, C | blaZ, mecA, gyrA, grlA, msrA, tetK, dfrG | 3 |
| P, FOX, CRO, CIP, MXF, NOR, AZM, DO, SXT, C | blaZ, mecA, gyrA, grlA, msrA, tetK, dfrG, cfr | 3 |
| P, FOX, KZ, CRO, CN, MXF, NOR, AZM, DO, SXT | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA, tetK, dfrG | 1 |
| P, FOX, KZ, CRO, AK, CN, MXF, NOR, AZM, DO, SXT | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA, tetK, dfrG | 1 |
| P, FOX, KZ, CRO, CN, CIP, MXF, NOR, AZM, DO, SXT | blaZ, mecA, aacA-aphD, gyrA, grlA, mrsA, tetK, dfrG | 1 |
| P, FOX, KZ, CRO, AK, CN, CIP, MXF, NOR, AZM, DO, SXT | blaZ, mecA, aacA-aphD, gyrA, grlA, msrA, tetK, dfrG | 1 |
| P, FOX, KZ, CRO, CN, CIP, MXF, NOR, AZM, DO, SXT, C | blaZ, mecA, aacA-aphD, gyrA, grlA, dfrG | 1 |
AK=Amikacin, AZM=Azithromycin, C=Chloramphenicol, CIP=Ciprofloxacin, CN=Gentamicin, CRO=Ceftriaxone, DO=Doxycycline, FOX=Cefoxitin, KZ=Cephazolin, MXF=Moxifloxacin, NOR=Norfloxacin, P=Penicillin, SXT=Trimethoprim/sulfamethoxazole
Detection of antimicrobial resistance genes and agr
Antimicrobial resistance genes were detected in the S. aureus isolates. CoNS bore more antimicrobial resistance genes, among which blaZ was the predominant one linked to P resistance in S. aureus. CoNS showed a higher frequency of antimicrobial resistance genes such as gyrA, grlA, and msrA (Figure-3), while carrying more combinations of such genes than S. aureus (Table-4). Nineteen S. aureus isolates were analyzed, determining that 73.68% belonged to agr group I, whereas 15.79% and 10.53% belonged to agr groups III and II, respectively. (Table-3) In this study, agr group IV was not detected.
Figure-3.
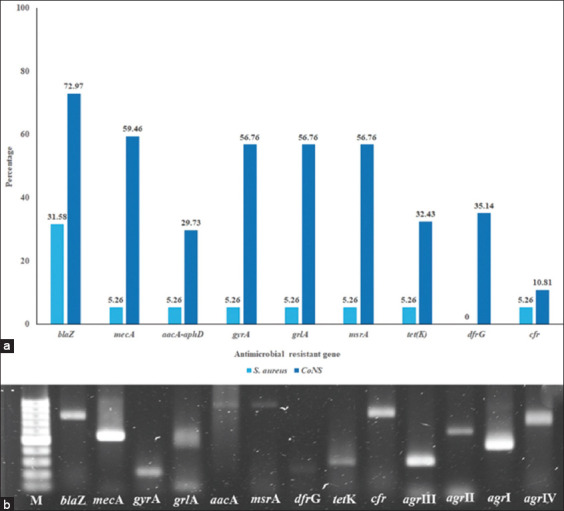
Antimicrobial resistance genes carried by Staphylococcus aureus and coagulase-negative staphylococci isolated from rabbits with clinical abscesses (a) and antimicrobial resistance genes product amplification by PCR (b). Lane M represents 100 bp ladder, lane 2 represents blaZ amplicon, lane 3 represents mecA amplicon, lane 4 represents gyrA amplicon, lane 5 represents grlA amplicon, lane 6 represents aacA-aphD amplicon, lane 7 represents msrA amplicon, lane 8 represents dfrG amplicon, lane 9 represents tetK amplicon, lane 10 represents cfr amplicon, lane 11 represents agr III amplicon, lane 12 represents agr II amplicon, lane 13 represents agr I amplicon, and lane 14 represents agr IV amplicon. PCR=Polymerase chain reaction, agr=Accessory gene regulator.
Discussion
Clinical abscesses commonly harbor pathogenic staphylococci bacteria. Studies on staphylococci in companion animals have mostly focused on other species, with little data available on rabbits [9, 11–14]. About 13.43% of S. aureus and 26.87% of CoNS were diagnosed from clinical abscesses in rabbits (19 and 37 isolates, respectively) in our study. Subcutaneous abscesses were common in rabbits and were mostly caused by both S. aureus and CoNS [28]. Our study recorded a smaller proportion of S. aureus compared to 76% [28], 71% [29], and 19.1% [3]. The discrepancy seen in our study compared to previous research is due to the difference in rabbit populations, with ours consisting of individually raised animals and those in earlier studies being part of farm populations [3, 28, 29]. Although opportunistic, CoNS was frequently isolated from infected lesions in our study. The prevalence of CoNS in our study on rabbits falls within the reported range of 17.5%–63% in cats and dogs studies [5, 30]. CoNS attention is drawn due to their presence in sites of infection, high drug resistance rates and role as resistance gene reservoirs [1, 8, 31].
Antimicrobial resistance has gradually increased due to the overuse of antibiotics. Although less attention is given to it, the regulation of antibiotic use in companion animals is markedly different from that of food animal production systems [32]. Since 2004, numerous studies, including those by Goni et al. [9] and Vancraeynest et al. [11] have reported antimicrobial resistance in S. aureus isolated from rabbits, which varied according to the research’s time and location. While Vancraeynest et al. [9], Wang et al. [4], and Chai et al. [3] found low resistance rates (<25%). Attili et al. [29] identified high resistance rates with S. aureus strains being resistant to tetracycline (95.8%), clindamycin (93.8%), and erythromycin (93.8%).
About 5.26% of MRSA isolates in this study showed MDR. Antimicrobial resistance genes such as tetK, msr, erm, and aac(6’)-aph(2’) have earlier been detected in MRSA strains derived from rabbits [9, 28]. Tetracycline- and macrolide-resistant S. aureus isolates from our study carried the tetK and msrA genes, respectively. However, the isolates exhibiting aminoglycoside resistance in this study carried the aacA-aphD gene, whereas isolates from other studies carried the aac(6’)-aph(2’) gene [28]. Additionally, we also identified other resistance genes such as blaZ (P resistance), gyrA and grlA (quinolone resistance), and cfr (phenicol resistance).
The role of CoNS in causing disease in animals is still under debate in veterinary medicine. Previous research by Suepaul et al. [1], Elnageh et al. [30], and Michels et al. [33] indicates that animals with CoNS bacteria exhibit a wide range of antimicrobial resistance, making them potential repositories for resistance genes. The prevalence of CoNS in this study (26.87%) aligns with the range observed in companion animals (6%–63%) [5, 7, 30, 32] and is double that of S. aureus (13.43%). About 78.38% of CoNS in this study showed resistance to pencillin, 59.46% to AZM, 54.05% to MXF, and 54.05% to NOR, all due to the presence of antimicrobial resistance genes (blaZ, msrA, gyrA, and grlA). Surprisingly, the occurrence of MRCoNS was more frequent than S. aureus, even outnumbering reports in companion animals [5, 34]. Methicillin-resistant strains are known to exhibit heightened resistance across various antibiotic classes and pose a potential risk for transferring antimicrobial resistance genes between staphylococci, particularly those with higher pathogenicity, such as S. aureus [2, 34]. Our study overlooked the significance of the mecC gene, a critical factor in methicillin resistance in Staphylococcus, potentially limiting our understanding of antibiotic resistance mechanisms. Further study of this gene may lead to a more comprehensive understanding of resistance mechanisms and improved treatment outcomes. MDR strains are disproportionately represented among MRSA and CoNS isolates. The data in Table-4, along with the findings of Conner et al. [32], confirm the higher antimicrobial resistance prevalence in MRCoNS.
The agr is responsible for managing the expression of staphylococcal virulence factors and additional accessory genes. It significantly boosts the secretion of virulence factors at the expense of reducing those attached to the cell membrane. The correlation between clinical conditions, including enterotoxins diseases, infective endocarditis, toxic shock syndrome, and exfoliative diseases, and the four agr groups (agr I, II, III, and IV), has been emphasized by numerous studies [15]. US nosocomial MRSA isolates have been primarily agr II, while community-acquired strains predominantly carry the agr III genotype [18, 24, 35, 36]. Cafiso et al. [37] linked GISA and biofilm production to the agr II genotype.
Isolates with a positive agr system harbor more virulence genes than those with a negative agr system [38]. Isolates having the agr system were more vulnerable to antibiotics than those without it, with agr III strains being more susceptible than agr I strains to each tested antibiotic [23, 38]. In agreement with previous studies by Sakoulas et al. [15], Yoon et al. [17], Bibalan et al. [18], Javdan et al. [19], and Cheraghi et al. [20], agr I was the most common agr group found in our isolates. No agr IV isolates were found, indicating regional differences in agr group occurrence. Agr I was identified predominantly in samples from Brazil, Portugal, Hungary, and Berlin; agr II in isolates from Japan and North America; and agr III in European samples [36]. The distribution pattern of agricultural groups across diverse geographical regions reveals distinct populations of S. aureus. Monitoring drug resistance trends over time is essential for preserving antibacterial product efficacy. Veterinarians rely on antimicrobial susceptibility data to determine the most effective antibiotics for treating infections. Effective implementation of preventive measures and antimicrobial stewardship in veterinary and animal medicine are crucial for thwarting antimicrobial resistance [30, 34, 39].
Conclusion
In rabbits, S. aureus, a Staphylococcus species, stands out for its heightened pathogenicity, causing clinical abscesses. CoNS is most commonly isolated from abscesses in studies. These bacteria carry multiple antimicrobial resistance genes, such as blaZ, mecA, aacA-aphD, msrA, tetK, gyrA, grlA, dfrG, and cfr, rendering them resistant to various antimicrobial drugs. The effectiveness of drugs against infections caused by these bacteria is restricted. Effective use of antibiotics in rabbits is necessary to prevent the propagation of antibiotic resistance. Educating rabbit owners to implement appropriate antimicrobial programs is essential in addressing this concern.
Authors’ Contributions
SB and NI: Conceptualized and designed the study. NS: Carried out sample collection and animal care. NS, AJ, SS, TK, WT, NI, and SB: Conducted the study. NS, AJ, NI, and SB: Analyzed the data. SB: Provided reagents, materials, and analysis tools. NS, SS, NI, and SB: Wrote and edited the manuscript. All authors have read, reviewed, and approved the final manuscript.
Acknowledgments
We express our gratitude to Prasu-Arthorn Animal Hospital for their assistance in specimen collection. Special thanks to the Faculty of Veterinary Science for providing laboratory space and equipment. The research received support from the Annual Research Fund of the Faculty of Veterinary Science, Mahidol University, in 2014.
Footnotes
The research received support from the Annual Research Fund of the Faculty of Veterinary Science, Mahidol University, in 2014.
Competing Interests
The authors declare that they do not have any competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Suepaul S, Georges K, Unakal C, Boyen F, Sookhoo J, Ashraph K, Yusuf A, Butay P. Determination of the frequency, species distribution and antimicrobial resistance of staphylococci isolated from dogs and their owners in Trinidad. PLoS One. 2021;16(7):e0254048. doi: 10.1371/journal.pone.0254048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez-Sanz E, Ceballos S, Ruiz-Ripa L, Zarazaga M, Torres C. Clonally diverse methicillin and multidrug-resistant coagulase-negative staphylococci are ubiquitous and pose transfer ability between pets and their owners. Front. Microbiol. 2019;10:485. doi: 10.3389/fmicb.2019.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai M.H, Sukiman M.Z, Najib N.M, Mohabbar N.A, Azizan N, Mohamad N.M, Ariffin S.M.Z, Ghazal M.F. Molecular detection and antibiogram of Staphylococcus aureus in rabbits, rabbit handlers, and rabbitry in Terengganu, Malaysia. J. Adv. Vet. Anim. Res. 2021;8(3):388–395. doi: 10.5455/javar.2021.h527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Sang L, Sun S, Chen Y, Chen D, Xi X. Characterisation of Staphylococcus aureus isolated from rabbits in Fujian, China. Epidemiol. Infect. 2019;147:e256. doi: 10.1017/S0950268819001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phumthanakorn N, Prapasarakul N, Yindee J, Gronsan D. Frequency, distribution, and antimicrobial resistance of coagulase-negative staphylococci isolated from clinical samples in dogs and cats. Microb. Drug Resist. 2022;28(2):236–243. doi: 10.1089/mdr.2020.0586. [DOI] [PubMed] [Google Scholar]
- 6.Heilmann C, Ziebuhr W, Becke K. Are coagulase-negative staphylococci virulent? Clin Microbiol Infect 2019. 2022;25(9):1071–1080. doi: 10.1016/j.cmi.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Lane M.J, Roy A.F, Kearney M.T, Pucheu-Hasto C.M. Characterization, distribution, antimicrobial resistance and resistance risk factors in staphylococci isolated from cats from 2001 to 2014. Vet. Med. Sci. 2018;4(4):315–325. doi: 10.1002/vms3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marincola G, Liong O, Schoen C, Abouelfetouh A, Hamdy A, Wencker F.D.R, Marciniak T, Becker K, Köck R, Ziebuh W. Antimicrobial resistance profiles of coagulase-negative staphylococci in community-based healthy individuals in Germany. Front Public Health. 2021;9:684456. doi: 10.3389/fpubh.2021.684456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vancraeynest D, Hermans K, Martel A, Vaneechoutte M, Devriese L.A, Haesebrouc F. Antimicrobial resistance and resistance genes in Staphylococcus aureus strains from rabbits. Vet Microbiol. 2004;101(4):245–251. doi: 10.1016/j.vetmic.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Cheung G.Y.C, Bae J.S, Ott M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goni P, Vergara Y, Ruiz J, Albizu I, Vila J, Gomez-Lu R. Antibiotic resistance and epidemiological typing of Staphylococcus aureus strains from ovine and rabbit mastitis. Int. J. Antimicrob. Agents. 2004;23(3):268–272. doi: 10.1016/j.ijantimicag.2003.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Loncaric I, Künzel F, Licka T, Simhofer H, Spergser J, Rosengarte R. Identification and characterization of methicillin-resistant Staphylococcus aureus (MRSA) from Austrian companion animals and horses. Vet. Microbiol. 2014;168(2–4):381–387. doi: 10.1016/j.vetmic.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Youn J.H, Park Y.H, Hang'ombe B, Sugimot C. Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia, Africa. Comp. Immunol. Microbiol. Infect. Dis. 2014;37(2):123–130. doi: 10.1016/j.cimid.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Drougka E, Foka A, Koutinas C.K, Jelastopulu E, Giormezis N, Farmaki O, Sarrou S, Anastassiou E.D, Petinaki E, Spiliopoulo I. Interspecies spread of Staphylococcus aureus clones among companion animals and human close contacts in a veterinary teaching hospital. A cross-sectional study in Greece. Prev. Vet. Med. 2016;126:190–198. doi: 10.1016/j.prevetmed.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Sakoulas G, Eliopoulos G.M, Moellering R.C, Jr, Wennersten C, Venkataraman L, Novick R.P, Gol H.S. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46(5):1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moise-Broder P.A, Sakoulas G, Eliopoulos G.M, Schentag J.J, Forrest A, Moellerin R.C., Jr Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. 2004;38(12):1700–1705. doi: 10.1086/421092. [DOI] [PubMed] [Google Scholar]
- 17.Yoon H.J, Choi J.Y, Lee K, Yong D, Kim J.M, Song YG. Accessory gene regulator group polymorphisms in methicillin-resistant Staphylococcus aureus:An association with clinical significance. Yonsei Med. J. 2007;48(2):176–183. doi: 10.3349/ymj.2007.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibalan M.H, Shakeri F, Javid N, Ghaemi A, Ghaem E.A. Accessory gene regulator types of Staphylococcus aureus isolated in Gorgan, North of Iran. J. Clin. Diagn. Res. 2014;8(4):DC07–DC09. doi: 10.7860/JCDR/2014/6971.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheraghi S, Pourgholi L, Shafaati M, Fesharaki S.H, Jalali A, Nosrati R, Borouman M.A. Analysis of virulence genes and accessory gene regulator (agr) types among methicillin-resistant Staphylococcus aureus strains in Iran. J. Glob. Antimicrob. Resist. 2017;10:315–320. doi: 10.1016/j.jgar.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Robinson D.A, Monk A.B, Cooper J.E, Feil E.J, Enrigh M.C. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J. Bacteriol. 2005;187(24):8312–8321. doi: 10.1128/JB.187.24.8312-8321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saedi S, Derakhshan S, Ghader E. Antibiotic resistance and typing of agr locus in Staphylococcus aureus isolated from clinical samples in Sanandaj, Western Iran. Iran J. Basic. Med. Sci. 2020;23(10):1307–1314. doi: 10.22038/ijbms.2020.46064.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azimian A, Najar-Pirayeh S, Mirab-Samiee S, Nader M. Occurrence of methicillin-resistant Staphylococcus aureus (MRSA) among clinical samples in Tehran-Iran and its correlation with polymorphism of specific accessory gene regulator (AGR) groups. Braz. J. Microbiol. 2012;43(2):779–785. doi: 10.1590/S1517-83822012000200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javdan S, Narimani T, Abadi M.S.S, Gholipou A. Agr typing of Staphylococcus aureus species isolated from clinical samples in training hospitals of Isfahan and Shahrekord. BMC Res. Notes. 2019;12(1):363. doi: 10.1186/s13104-019-4396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Indrawattana N, Sungkhachat O, Sookrung N, Chongsa-Nguan M, Tungtrongchitr A, Voravuthikunchai S.P, Kong-Ngoen T, Kurazono H, Chaicump W. Staphylococcus aureus clinical isolates:Antibiotic susceptibility, molecular characteristics, and ability to form biofilm. Biomed. Res. Int. 2013;2013:314654. doi: 10.1155/2013/314654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guide for the Care and Use of Laboratory Animals. The National Academies Collection:Reports funded by National Institutes of Health. 8th ed. Washington, DC: 2011. [Google Scholar]
- 26.CLSI. Performance Standards for Antimicrobial Susceptibility Testing;Twenty Informational Supplement. CLSI Document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. 2012 [Google Scholar]
- 27.Jangsangthong A, Suriyakhun N, Tunyong W, Kong-Ngoen T, Santajit S, Indrawattana N, Indrawattana N, Buranasinsu S. Occurrence of antimicrobial resistance and antimicrobial resistance genes in methicillin-resistant Staphylococcus aureus isolated from healthy rabbits. Vet. World. 2022;15(11):2699–2704. doi: 10.14202/vetworld.2022.2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva V, De Sousa T, Gomez P, Sabenca C, Vieira-Pinto M, Capita R, Alonso-Calleja C, Torres C, Capelo J.L, Igrejas G, Poet P. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) in purulent subcutaneous lesions of farm rabbits. Foods. 2020;9(4):439. doi: 10.3390/foods9040439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attili A.R, Nebbia P, Bellato A, Galosi L, Papeschi C, Rossi G, Linardi M, Fileni E, Cuteri V, Chiesa F, Robin P. The effect of age and sampling site on the outcome of Staphylococcus aureus infection in a rabbit (Oryctolagus cuniculus) farm in Italy. Animals (Basel) 2020;10(5):774. doi: 10.3390/ani10050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elnageh H.R, Hiblu M.A, Abbassi M.S, Abouzeed Y.M, Ahme M.O. Prevalence and antimicrobial resistance of Staphylococcus species isolated from cats and dogs. Open Vet. J. 2021;10(4):452–456. doi: 10.4314/ovj.v10i4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bora P, Datta P, Gupta V, Singhal L, Chande J. Characterization and antimicrobial susceptibility of coagulase-negative staphylococci isolated from clinical samples. J. Lab Physicians. 2018;10(4):414–419. doi: 10.4103/JLP.JLP_55_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conner J.G, Smith J, Erol E, Locke S, Phillips E, Carter C.N, Odo A. Temporal trends and predictors of antimicrobial resistance among Staphylococcus spp. Isolated from canine specimens submitted to a diagnostic laboratory. PLoS One. 2018;13(8):e0200719. doi: 10.1371/journal.pone.0200719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michels R, Last K, Becker S.L, Papa C. Update on coagulase-negative staphylococci what the clinician should know. Microorganisms. 2021;9(4):830. doi: 10.3390/microorganisms9040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Jong A, Youala M, El Garch F, Simjee S, Rose M, Morrissey I, Moyaert H. Antimicrobial susceptibility monitoring of canine and feline skin and ear pathogens isolated from European veterinary clinics:results of the ComPath Surveillance programme. Vet. Dermatol. 2020;31(6):431–e114. doi: 10.1111/vde.12886. [DOI] [PubMed] [Google Scholar]
- 35.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesc F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002;70(2):631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohsenzadeh M, Ghazvini K, Azimia A. Frequency of specific agr groups and antibiotic resistance in Staphylococcus aureus isolated from bovine mastitis in the Northeast of Iran. Vet. Res. Forum. 2015;6(4):295–299. [PMC free article] [PubMed] [Google Scholar]
- 37.Cafiso V, Bertuccio T, Santagati M, Demelio V, Spina D, Nicoletti G, Stefan S. Agr-genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2007;51(1):220–227. doi: 10.1111/j.1574-695X.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- 38.Derakhshan S, Navidinia M, Hagh F. Antibiotic susceptibility of human-associated Staphylococcus aureus and its relation to agr typing, virulence genes, and biofilm formation. BMC Infect. Dis. 2021;21(1):627. doi: 10.1186/s12879-021-06307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung W.K, Shin S, Park Y., Lim S.K, Moon D.C, Par K.T. Distribution and antimicrobial resistance profiles of bacterial species in stray cats, hospital-admitted cats, and veterinary staff in South Korea. BMC Vet. Res. 2020;16(1):109. doi: 10.1186/s12917-020-02326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


