Abstract
Mutagenesis studies have shown that retroviral nucleocapsid (NC) protein Zn2+ fingers (-Cys-X2-Cys-X4-His-X4-Cys- [CCHC]) perform multiple functions in the virus life cycle. Moloney murine leukemia virus mutants His 34→Cys (CCCC) and Cys 39→His (CCHH) were able to package their genomes normally but were replication defective. Thermal dissociation experiments showed that the CCHH mutant was not defective in genomic RNA dimer structure. Primer tRNA placement on the viral genome and the ability of the tRNA to function in reverse transcription initiation in vitro also appear normal. Some “full-length” DNA copies of the viral genome were synthesized in mutant virus-infected cells. The CCCC and CCHH mutants produced these DNA copies at greatly reduced levels. Circle junction fragments, amplified from two-long-terminal-repeat viral DNA (vDNA) by PCR, were cloned and characterized. Remarkably, it was discovered that vDNA isolated from cells infected with mutant virions had a wide variety of abnormalities at the site at which the two ends of the linear precursor had been ligated to form the circle (i.e., the junction between the 5′ end of U3 and the 3′ end of U5). In some molecules, bases were missing from regions corresponding to the U3 and U5 linear vDNA termini; in others, the viral sequences extended either beyond the U5 sequences into the primer-binding site and 5′ leader or beyond the U3 sequences into the polypurine tract into the env coding region. Still other molecules contained nonviral sequences between the linear vDNA termini. Such defective genomes would certainly be unsuitable substrates for integration. Thus, strict conservation of the CCHC structure in NC is required for infection events prior to and possibly including integration.
Retroviral Gag precursors and nucleocapsid (NC) proteins (except those of the spumavirus class) contain Zn2+ fingers. These fingers are involved in a number of processes in the viral life cycle. They are composed of a highly conserved amino acid sequence with invariably spaced Cys and His residues of the form -Cys-X2-Cys-X4-His-X4-Cys- (CCHC). These sequences are found either once or twice depending on the viral species (2, 6).
Over the years, the function of this conserved motif has been investigated by a variety of approaches, including mutational analysis. In one class of mutants, the metal-binding residues of the NC Zn2+ finger are mutated to amino acids other than Cys or His; the resulting finger is no longer able to bind Zn2+ (18). Mutant viruses of this type are defective in their ability to package their RNA genomes; they are, of course, replication defective (for a review, see reference 3).
Another class of mutants in the Moloney murine leukemia virus (Mo-MuLV) system contains mutated NC Zn2+ fingers that retain the ability to coordinate Zn2+ (5). These are the His 34→Cys (CCCC) and Cys 39→His (CCHH) NC Zn2+ finger mutants. These are also replication defective. However, in contrast to mutants in which the conserved Cys or His residues are changed to amino acids other than Cys or His, the CCCC and CCHH mutants package wild-type levels of genomic RNA (16). In these mutants, therefore, the RNA-packaging role performed by NC Zn2+ fingers has successfully been separated from other functions in the viral replication cycle. These mutants provide us with an excellent tool for studying the function of the retroviral NC Zn2+ finger in early infection processes (e.g., reverse transcription and/or integration).
The mutant particles with CCCC and CCHH NC Zn2+ fingers were defective in a number of infectivity assays and were unable to make viral DNA (vDNA), as determined by Southern blot analysis (16). This defect might reflect a requirement for the wild-type Zn2+ finger during reverse transcription or might be due to an abnormality during the assembly of particles in the virus-producing cell. In an effort to identify the function(s) of the Zn2+ fingers as precisely as possible, we have reinvestigated the nature of the functional defect in the mutant particles.
In some of the present experiments, we have analyzed the physical state of the mutant genomic RNA and the presence of primer on the primer-binding sites (PBSs) of these RNAs. No differences from wild-type particles were found in these assays. We have also reexamined the ability of these particles to synthesize vDNA upon infection of new host cells, using a PCR-based approach involving amplification of the circle junction in two-long-terminal-repeat (2-LTR) circles (see Fig. 1). These sensitive assays showed that the mutant particles do synthesize “full-length” DNA, although at a low level. Thus, the defect in reverse transcription, while significant, cannot fully account for the lack of infectivity of these particles. However, when these DNA copies were analyzed in detail, they were found to exhibit a variety of abnormalities at their ends: in some molecules, bases were missing from the normal junction site, while in others, foreign sequences were inserted at the junction site or the viral sequences extended beyond their normal termini. None of these aberrant DNA molecules can serve as a substrate for the viral integrase (IN); this property and the reduction in vDNA synthesis presumably are responsible for the detrimental effects on infectivity these mutant virions. These results shed new light on the function(s) of the NC Zn2+ fingers during the infectious process and underscore the potential usefulness of these structures as targets for antiviral therapy.
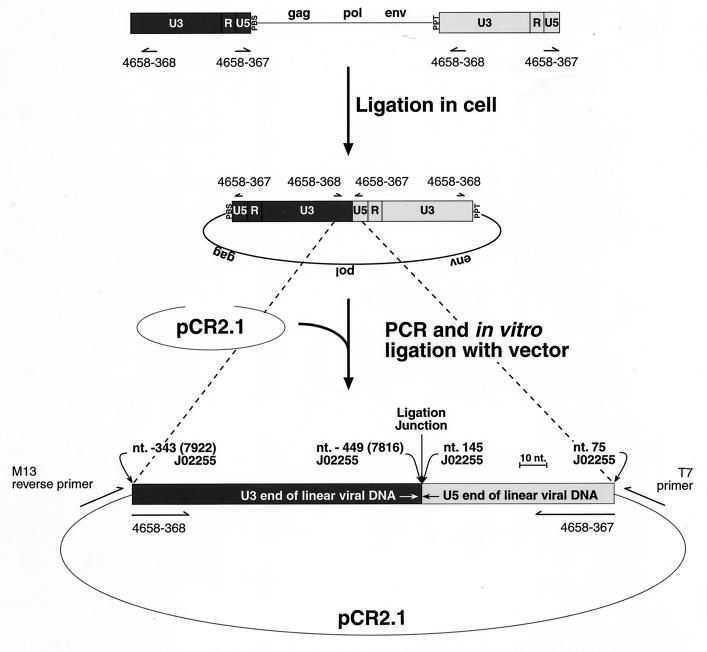
FIG. 1.
Ligation of the linear vDNA to form the 2-LTR circularized species. The linear form of the vDNA is diagrammed with the 5′-LTR (position and orientations described are with respect to plus-strand sequences) in dark gray and the 3′-LTR in light gray. The 5′ end shows the U3, R, U5 region of the LTR as well as the neighboring PBS. The 3′ end shows the ppt region and the subsequent 3′ LTR. The positions of the PCR primers used to amplify the 2-LTR circularized vDNA species are indicated. The 2-LTR species in the middle portion of the figure shows the expected vDNA species that results after ligation of the linear vDNA form in the cell. PCR amplification with primers 4658-367 and 4658-368 with subsequent cloning into the TA cloning site of the pRC2.1 vector gives the expected form shown at the bottom of the panel. Points of reference from the Mo-MuLV genome (GenBank accession no. J02255 [30]) are indicated in the lower portion of the diagram. The diagram shows one of two possible orientations in the pCR2.1 vector. The positions of the M13-reverse and T7 primers, which were used for sequence analysis of the 2-LTR circularized, vDNA, PCR product inserts, are indicated.
MATERIALS AND METHODS
Cell lines and transfections.
293 cells (adenovirus-transformed human embryonal kidney cells) which express the large T antigen (293T) were obtained and cultured as described previously (16). Mutant and wild-type proviral clones were transfected with the calcium phosphate mammalian cell transfection kit from 5′→3′, Inc. (Boulder, Colo.). Log-phase 293T cells, grown in 150-cm2 flasks, were transfected and virus was harvested as described previously (16).
Plasmid constructs.
The mutant and wild-type Mo-MuLV proviral clones used in this study have been described previously (15–17). Another group of viruses used in this study are designated Mo(10A1), and proviral clones of this type were constructed as described by Ott et al. (25). Mutant and wild-type Mo(10A1) viruses contained the 10A1 Env in place of the Mo-MuLV Env, enabling them to infect 293T cells.
Exogenous template RT assays.
For virus characterization, reverse transcriptase (RT) assays were performed on clarified supernatants as described previously (16).
Viral RNA analysis.
Viral RNA (vRNA) was isolated as described previously (15). Samples were ethanol precipitated and stored at −20°C until they were used for the analyses described below.
The melting temperatures of the vRNA dimers were determined as described previously (15). The occupancy of the PBS by the tRNAPro primer on vRNAs was determined by nondenaturing Northern blot analysis and primer-tagging procedures as described previously, using Mo-MuLV RT from Life Technologies, Inc. (14, 36).
Viral DNA isolation.
To monitor reverse transcription processes, vDNA was obtained from cells infected with mutant and wild-type Mo(10A1)-MuLV virions. Viruses were obtained by transfecting 293T cells, collecting cell-free supernatants at 24-h intervals, and storing the supernatants at −70°C. Before 150-cm2 flasks of 293T cells (∼90% confluent) were infected with the mutant and wild-type Mo(10A1) viruses, monolayers were incubated for 20 min at 37°C with 15 ml of Dulbecco’s modified Eagle’s medium containing 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 10 U of penicillin G, 10 U of streptomycin sulfate, and 20 μg of DEAE-dextran HCl (Sigma Chemical Co., St. Louis, Mo.) per ml. The monolayers were washed with 20 ml of Hanks’ balanced salt solution (Life Technologies, Inc.). For each mutant, 100 ml of supernatant was thawed and used to infect a 150-cm2 flask of 293T cells at 80 to 90% confluency. In some experiments, wild-type virus was diluted with Dulbecco’s modified Eagle’s medium containing 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, 10 U of penicillin G, and 10 U of streptomycin sulfate before being applied to the monolayers.
A reverse transcription-negative control was prepared as follows. At 18 h prior to infection, a flask containing 293T cells was incubated with 20 μM 3′-azido-3′-deoxythymidine (AZT; Sigma Chemical Co.) and 20 μM 2′,3′-dideoxycytidine (ddC; Sigma Chemical Co.) in 30 ml of medium. The next day, drug-containing medium was removed and the flask was DEAE-dextran treated as described above (in the presence of 20 μM each AZT and ddC). Then 100 ml of undiluted supernatant containing wild-type Mo(10A1)-MuLV and the RT inhibitors (20 μM each AZT and ddC) was applied to the drug-treated 293T monolayer. In addition Hirt supernatant DNA was isolated from a flask of 293T cells that was incubated with culture fluids harvested from monolayers transfected with sheared salmon sperm DNA; this served as a negative control.
DNA samples were harvested after the monolayers were incubated with the various culture fluids for 24 h. The infected 293T cells were removed from the flask surface by incubation in phosphate-buffered saline (without Ca2+ or Mg2+) (Life Technologies, Inc.) containing 1 mM EDTA. The cells were pelleted at 750 × g for 5 min, and vDNA was isolated by the procedure described by Hirt (20). Cell pellets were lysed in a final volume of 0.63 ml. Ethanol-precipitated samples were resuspended in 50 μl of 10 mM Tris HCl–1 mM EDTA (pH 8.0) (TE buffer) for PCR analysis.
Viral DNA analysis.
PCR analysis of 2-LTR circularized vDNA was performed by using the following primers and protocols. The primers used to detect 2-LTR circularized DNA are 4658-367 (5′-TTG TGG TCT CGC TGT TCC TTG-3′) (the 5′ end corresponds to nucleotide (nt) 75 of the Mo-MuLV genome in U5; Genbank accession no. J02255 [30]) and 4658-368 (5′-CTG ACC TTG ATC TGA ACT TCT CTA TTC TC-3′) (the 5′ end corresponds to the complement of nt 7922 of the Mo-MuLV genome in U3; Genbank accession no. J02255). Figure 1 shows the locations of the primers and the orientation of the ends of the linear vDNA before and after ligation. Reactions were performed in 50-μl volumes with AmpliTaq core reagents supplemented with 4 mM MgCl2 and AmpliTaq Gold polymerase (Perkin-Elmer, Roche Molecular Systems, Inc., Branchburg, N.J.) as specified by the manufacturer. The PCR cycling parameters were one cycle at 94°C for 9 min; 35 cycles of 94°C for 10 s, 56°C for 15 s, and 61°C for 30 s; and one cycle at 72°C for 8 min. Then 20 μl of the PCR products from the vDNA analyses were fractionated on a 2% (wt/vol) MetaPhor agarose (FMC BioProducts, Rockland, Maine) gel in 100 mM Tris–90 mM boric acid–1 mM EDTA (pH 8.3) (TBE) buffer. The PCR products were visualized by staining with ethidium bromide.
The PCR products were also cloned into the pCR2.1 vector by using the TA cloning kit (Invitrogen Corp., Carlsbad, Calif.). To reduce the levels of primer-dimer incorporation into the cloning vector, PCR products were washed with TE buffer by using a Microcon 30 centrifugal filter (Millipore Corp., Bedford, Mass.). The retentates were then ligated with the pCR2.1 vector. Clones were screened for 2-LTR inserts by using the same primers and PCR conditions described above (Fig. 1).
The sizes of the PCR products present in individual clones were determined. The PCR products were fractionated on 2% (wt/vol) MetaPhor agarose gels in TBE buffer along with φX174-HaeIII DNA markers (Life Technologies, Inc.), and the gels were stained with ethidium bromide. The migration distances from the sample wells of the φX174-HaeIII DNA marker bands plotted against the logarithm of the sizes (in base pairs) of these bands were used to generate standard curves. The Microsoft Corp. (Redmond, Wash.) EXCEL 97 GROWTH function was used to calculate the sizes of the PCR products, based on their migration distances, by using the data generated from the φX174-HaeIII DNA marker bands. The GROWTH function returns the y values (size) from x values (PCR product migration distances) that are specified by the existing x and y values from the φX174-HaeIII DNA marker bands.
The pCR2.1 clones containing 2-LTR circularized vDNA PCR products were sequenced with the T7 (5′-AAT ACG ACT CAC TAT AG-3′) and M13-reverse (5′-AAC AGC TAT GAC CAT G-3′) primers that flank the TA cloning site of pCR2.1 (Invitrogen Corp.) (Fig. 1). A sampling of clones of ∼180 bp or smaller were chosen at random. Clones larger than ∼180 bp were also sequenced to obtain information on the structure of the inserts. Sequencing was performed with the ABI Dye Terminator cycle-sequencing ready reaction kit with DNA AmpliTaq DNA polymerase FS (PE Biosystems, Foster City, Calif.) on an ABI model 373 automated sequencer. Sequence alignments and comparisons were performed with the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.).
RESULTS
It is known that retroviral NC proteins (and the parental Gag polyproteins containing NC as a domain) facilitate RNA-RNA interactions during virus assembly and maturation. Thus, it appears that the Gag polyprotein, acting through its NC domain, catalyzes the annealing of primer tRNA to the PBS before or during virus assembly (11, 21, 22). The polyprotein may also catalyze the dimerization of genomic RNAs. In addition, the NC protein in the mature retroviral particle induces a conformational change in the RNA dimer, termed “maturation,” after the particle is released from the cell and Gag is cleaved (12, 13, 15). It seemed possible that replacement of the normal CCHC Zn2+ finger with CCHH or CCCC fingers would affect these functions of NC and the Gag polyprotein.
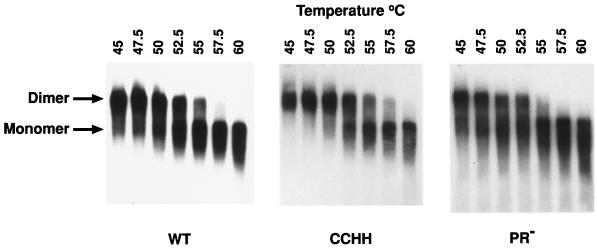
“Immature” retroviral dimeric RNAs (e.g., those present in PR− virions [13, 15]) are less thermostable than dimers isolated from mature, wild-type particles. Therefore, we compared the physical state of the genomic RNA in the mutant particles with that of RNA from wild-type and PR− particles by measuring the temperature required to dissociate the dimers into monomers. As shown in Fig. 2, the thermostability of the dimers from the CCHH particles was indistinguishable from that of the wild-type particles and clearly greater than that of dimers from PR− particles (e.g., the wild-type and CCHH RNAs were both ∼50% dimeric after exposure to 52.5°C and some dimers are still present after treatment at 55°C, whereas less than half of the PR− RNA dimers survived exposure to 50°C and none were detectable in the 55°C-treated sample). These results suggest that the CCHH NC protein is able to induce the same conformational changes in the initially packaged, “immature” dimeric RNA as the wild-type protein.
FIG. 2.
Melting analyses of Mo-MuLV dimeric RNAs isolated from wild-type and CCHH and PR− mutant virions. RNAs extracted from the respective virions were resuspended and heated for 10 min at the temperatures indicated in the figure. The positions of the monomer and dimer RNA species are indicated at the left. A comparison of the melting temperatures of the wild type (WT), the CCHH mutant, and the PR− mutant RNA dimers is presented. Wild-type virus contains a “mature” RNA dimer, and PR− virus contains an “immature” RNA dimer (15). The melting analysis of the CCHH mutant RNA was compared with that of the wild-type RNA and the PR− RNA in separate experiments, but in the interest of saving space, only one CCHH analysis is presented. The melting analyses were identical for the CCHH mutant in both experiments.
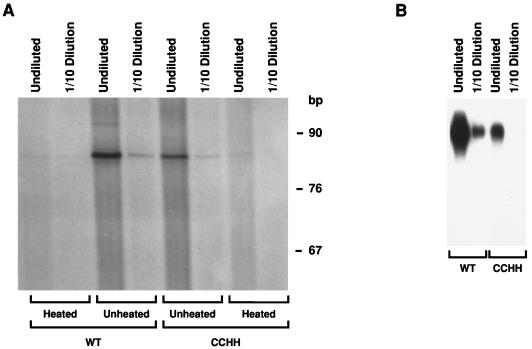
We also tested the genomic RNA isolated from CCHH mutant particles for the presence of primer tRNA at its PBS. These assays were performed by incubating the RNA isolated from virus with Mo-MuLV RT in the presence of [α-32P]dATP; tRNA annealed to the PBS is radiolabeled by the addition of one or two radioactive dAMP residues under these conditions (14, 36). As seen in Fig. 3A, primer was present on the CCHH RNA. The level of radioactivity in the mutant band was somewhat lower than in the wild-type band; however, when the two RNA preparations were compared with respect to the amount of vRNA, the mutant sample was found to have a correspondingly lower vRNA concentration (Fig. 3B). The quantitative analysis of these results is presented in Table 1 and shows that the efficiency of primer tagging per molecule of genomic RNA was virtually the same in wild-type and mutant RNAs. Thus, the change from CCHC to CCHH in the Zn2+ finger does not appear to affect the placement of primer tRNA on the PBS of vRNAs.
FIG. 3.
PBS occupancy determination in genomic RNAs isolated from wild-type (WT) and CCHH mutant virions. (A) Primer tagging was performed to determine the levels of primer tRNA at the PBS. Undiluted and 1:10-diluted vRNAs were examined as indicated. Heated (samples heated to 100°C for 5 min and immediately chilled on ice prior to primer tagging) and unheated samples were tested, as indicated. (B) Nondenaturing Northern blot analysis of CCHH mutant and wild-type vRNA were tested undiluted and at a 1:10 dilution (RNA from 7.5 and 0.75 ml of culture supernatant, respectively) is indicated.
TABLE 1.
Occupancy of the PBS in Mo-MuLV CCHH mutant and wild-type vRNA
| Sample | Genomic RNA content (% of wild type)a | tRNA content of PBS (% of wild type)b | tRNA/genomic RNA (% of wild type)c |
|---|---|---|---|
| Wild type undiluted | 100 | 100 | 100 |
| Wild type 1:10 Dilution | 12.7 | 11.6 | 91 |
| CCHH undiluted | 30.8 | 41.4 | 134 |
| CCHH 1:10 Dilution | 1.3 | 5.2 | 400 |
vRNA band intensity from nondenaturing Northern blot analysis (Fig. 3B), measured by phosphorimaging.
[α-32P]dAMP-radiolabeled tRNA band intensity from primer tagging (Fig. 3A), measured by phosphorimaging.
Ratio of tRNA intensity to genomic RNA intensity expressed as a percentage of the wild-type value.
The results presented above suggest that the RNA in the CCHH particles should be a suitable primer/template for reverse transcription. Therefore, reverse transcription steps were examined by PCR techniques. PCR is much more sensitive than the Southern blot procedure we previously used to analyze reverse transcription products in the CCCC and CCHH mutant particles (16). Along with the CCHH mutant, we examined the CCCC mutant and other mutants that have been characterized previously. The SSHC mutant contains a defective NC Zn2+ finger since two of the ligand-binding residues (Cys) were replaced with non-ligand-binding residues (Ser). Two other mutants with aromatic residue mutations, Y28S and W35S, were examined; these mutants contain CCHC NC Zn2+ fingers (17).
We examined 2-LTR circularized vDNA by the PCR procedures. Presumably, this product arises from the ligation of linear forms of vDNA in the nucleus of the infected cell (Fig. 1) (29, 33). The 2-LTR circles are formed only during infection and can easily be isolated (20). Specific primers, similar to those used by Stevenson et al. (31) and Engelman et al. (10) for human immunodeficiency virus type 1 (HIV-1), were used to specifically amplify sequences flanking the junction formed upon joining of the ends of the linear vDNA.
Mutant and wild-type viruses were incubated with cells, and the vDNA species were isolated. Figure 4 shows that correct-sized PCR products can be detected in vDNAs isolated from the infected cells, although they are diffuse for the CCCC, CCHH, and SSHC mutants. These results are in contrast to the Southern blot analysis of the CCCC and CCHH mutants, reflecting the greater sensitivity of PCR. The sizes of the bands correspond to the expected size of 178 bp. The Hirt supernatant DNA from cells infected with the wild-type virus could be diluted 1,000-fold and a positive PCR signal was still observed. The CCCC, CCHH, and SSHC mutants were barely visible, only in undiluted samples (Fig. 4); PCR products were detected in the Y28S mutant at a 1:100 (not at a 1:1,000) dilution; and bands were visible in W35S between the 1:10 and 1:100 dilutions.
FIG. 4.
Agarose gel analysis of 2-LTR circularized vDNA PCR products. Hirt supernatants, isolated from 293T cells infected with mutant and wild-type viruses, were amplified with primers specific for 2-LTR circularized vDNA (see Fig. 1). The RT activities (in cpm per milliliter) of the undiluted inocula (corrected for background) are as follows: (−)-Control, 0; CCCC, 451,760; CCHH, 748,370; SSHC, 637,170; Y28S, 630,670; W35S, 564,340; and wild type, 551,430. The location of the expected 178-bp band is shown on the right. Viruses incubated with the cells are indicated at the top, and the dilutions tested by PCR are indicated at the bottom. The lane labeled (−)-Control contains PCR products from the Hirt supernatant from cells incubated with transfected cell culture supernatant that did not contain any virus. The AZT/ddC lane contains PCR products from the Hirt supernatant from cells incubated with wild-type virus in the presence of the RT-inhibiting compounds AZT and ddC. The No DNA lane contains PCR products generated in the absence of added Hirt supernatant.
In this analysis, the wild-type inoculum used to infect 293T monolayers could be diluted 10- and 100-fold and the PCR signal still remained strong; correspondingly lower levels of PCR products were detected, indicating that the amount of virus used to infect the 293T cells was not saturating with respect to the levels of available receptors. The level of the AZT- and ddC-treated wild-type control was reduced by 100-fold, indicating that reverse transcription in the infected cell was required for formation of the PCR product. The sample labeled “(−)-Control” was from cells incubated with virus-free supernatants (supernatants from monolayers transfected with sheared salmon sperm DNA), and the negative result here again shows that the product arises only as a result of infection by MuLV in the inoculum.
The presence of 2-LTR circularized DNA (Fig. 4) indicates that the primer/template complex observed in the CCHH mutant (Fig. 3 and Table 1) and the other mutants can be extended by reverse transcription processes. Thus, a block in reverse transcription cannot fully account for the ∼105-fold reduction in infectivity in this mutant (16). Note, however, that the PCR product bands for the CCCC, CCHH, and SSHC mutants in Fig. 4 appeared quite smeared. This suggests that the products may be heterogeneous in size. To further investigate the diffuse character of the bands, we cloned the PCR products into the pCR2.1 vector (Fig. 1) and examined the clones in detail.
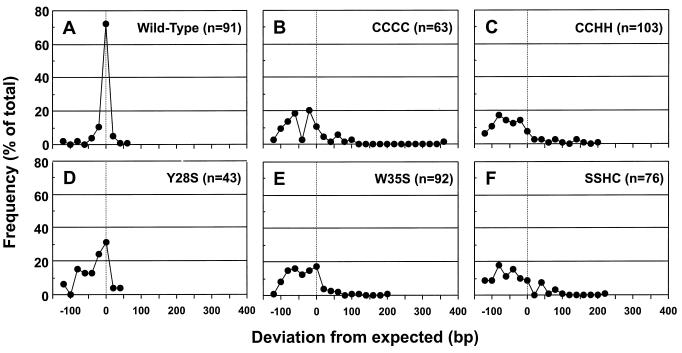
The sizes of the inserts cloned into the pCR2.1 vector were measured, and the frequencies of the various insert sizes are presented in Fig. 5. A majority of the inserts (∼70%) from the wild-type virus were close to the expected size (178 bp) (Fig. 5A). The deviation from the expected size was relatively small, and those that were different from the expected size were generally shorter. In contrast, the sizes of the inserts from the mutants were widely distributed (Fig. 5B to F). Lower levels of inserts of the correct size were observed, ranging from ∼30% for the Y28S mutant to ∼7% for the CCHH mutant. The deviation from the expected insert size in the mutants is much greater than that observed from the wild-type virus, with most of the inserts being shorter and some being longer. This explains why some of the PCR products observed in Fig. 4 appear smeared.
FIG. 5.
Frequency distribution of the sizes of cloned 2-LTR circularized vDNA PCR products from cells incubated with mutant and wild-type viruses. Individual clones of the PCR products obtained in the experiment in Fig. 4 were analyzed for the sizes of the inserts. Results are reported as the deviation from the expected size of 178 bp (Fig. 1). The frequencies were compiled in 20-nt windows from the expected size. Frequencies are reported as a percentage of the total number of samples examined. Distributions are shown for PCR product inserts obtained from cells incubated with the wild-type (A), CCCC (B), CCHH (C), Y28S (D), W35S (E), and SSHC (F) viruses.
Results from Fig. 5 indicate that there are deletions, insertions, or rearrangements in the PCR inserts from the mutants. This suggests that the full-length linear vDNA in the mutants was defective prior to ligation. Such alterations could account for the 105-fold reduction in infectivity in these mutants. What accounts for the various defects in the inserts from the mutants? To determine how the vDNA that flanks the junction formed by the ligation of linear vDNA is defective, the PCR inserts, examined in Fig. 5, were sequenced.
Sequences obtained for the mutant and wild-type PCR inserts were compared with what would be expected during the normal course of reverse transcription. The inserts in the pCR2.1 clones were sequenced with the T7 and/or M13 reverse primer, and the expected nucleotide sequence locations are denoted in Fig. 1. The insert sequences could be aligned and arranged into various groups; the results are presented in Fig. 6.
FIG. 6.
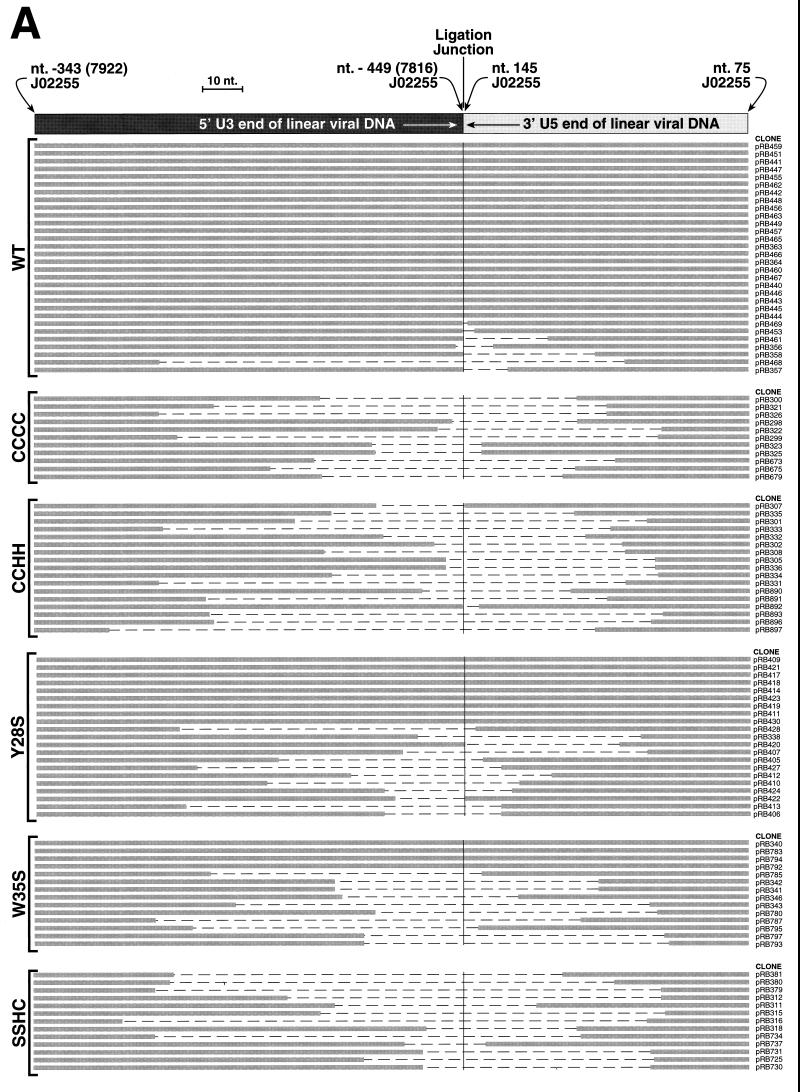
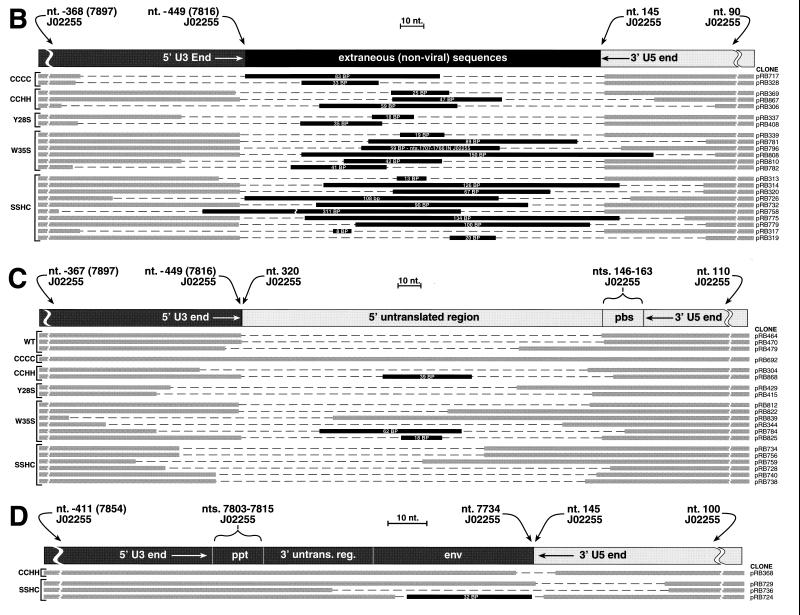
Schematic of alignments of PCR product inserts. The cloned inserts of products from the PCR of Hirt supernatants isolated from cells incubated with mutant and wild-type (WT) viruses were sequenced. The individual clone designations are indicated on the right, and the mutant examined is indicated on the left. The thick gray lines in the lower portion of each panel represent nucleotides that are present in each of the clones. The dashed regions depict the nucleotides that are missing from the vDNA product insert. The black regions depict insertions of extraneous (nonviral or noncontiguous viral) DNA sequences with the size of the fragment indicated inside the black region. (A) Alignment of full-length or truncated inserts that are isogenic with the expected sequence for authentic 2-LTR circularized vDNA. The diagram at the top is the same as that shown in Fig. 1. (B) Alignment of clones containing extraneous DNA inserts between the ends of the linear vDNA termini. The diagram at the top shows the arrangement of the termini and fragments. (C and D) Alignments of clones that continue through the PBS (C) or the ppt (D) are depicted. The diagrams at the top of each panel show the arrangement of the consensus viral sequences.
Most of the inserts obtained from cells infected with the wild-type virus had the sequence expected from a ligated, full-length vDNA (Fig. 6A). That is, the vDNA contained the terminal dinucleotides on each of the ends (i.e., the ends have not been processed by the viral IN protein [4, 28]). A few of the inserts were missing nucleotides in regions that correspond to the ends of the “full-length” vDNA (Fig. 1 and 6A). However, in contrast to the wild type, none of the inserts from the CCCC, CCHH, and SSHC mutants were full length: all clones lacked bases from both the U3 and U5 ends at the junction point. Some inserts from the Y28S and W35S mutants had sequences that correspond to full-length linear vDNAs, identical to those seen in the wild-type inserts. However, the Y28S and W35S mutants had ∼50% and only ∼25% of full-length inserts, respectively; the remaining inserts from the Y28S and W35S mutants had the same types of truncations observed in the CCCC, CCHH, SSHC, and wild-type samples.
Additional forms of 2-LTR circularized vDNAs were also obtained, and the alignments are presented in Fig. 6B to D. In Fig. 6B, most of the clones appear to have one correct end (either U3 or U5) lacking the terminal dinucleotide, as if this end had been processed by the viral IN protein. The other vDNA end was usually truncated (>2 bp removed). Nonviral (or, for clone pRB796, noncontiguous viral) sequences of different lengths were attached between the termini.
Another set of PCR inserts (Fig. 6C) had complete 3′ U5 ends (referring to viral plus-strand sequences) lacking the terminal dinucleotide. However, there were additional contiguous viral sequences after the normal 3′ end of U5 that continued into and in some cases through the PBS into the untranslated region prior to the gag open reading frame (ORF). The region that corresponds to the other end of the linear vDNA was either full length or truncated to different degrees. Wild-type as well as mutant viruses generated some clones of this type. Three clones (pRB784, pRB825, and pRB868) contained sequences of nonviral origin inserted between the vDNA termini.
Another set of inserts was also obtained, resembling the mirror image of those in Fig. 6C. As shown in Fig. 6D, these contained complete 5′ U3 ends (referring to the plus strand) with additional viral sequences extending through the ppt into the env ORF or the untranslated region between the env ORF and the ppt. No wild-type inserts were observed in these alignments. One clone (pRB724) contained nonviral sequences that were inserted between vDNA termini.
DISCUSSION
Early mutagenesis studies of retroviral NC Zn2+ fingers indicated that they performed some function(s) in infection processes, in addition to their role in RNA packaging. Thus, some mutants were able to package detectable levels of vRNA but were found to be profoundly defective in sensitive infectivity assays that can measure even single integration events (17, 26). More recently, we have designed and characterized noninfectious Mo-MuLV mutants that are able to package wild-type levels of vRNA (16). These mutants have provided us with valuable reagents that can now be used to determine the function of the NC Zn2+ finger in early infection processes in the absence of RNA packaging defects.
We found that the initial step in reverse transcription (addition of the first nucleotide to the tRNA primer) was not impaired in CCHH mutant particles (Fig. 3; Table 1). While all of the mutant virions tested were able to reverse transcribe their genomes to some extent, the CCCC, CCHH, and SSHC mutants produced approximately full-length products at greatly reduced levels (Fig. 4). In cells infected with wild-type virus, the 2-LTR circle junction fragments obtained by PCR were nearly all of the expected length (178 bp). In contrast, the sizes of products from mutant-infected cells were far more heterogeneous (Fig. 5), which was also reflected in the apparent smearing of the band representing the total population of PCR products (Fig. 4).
Sequence analysis revealed that the great majority of clones obtained from the PCR of wild-type-infected cells were apparently generated by direct, end-to-end joining of the termini of full-length, unprocessed vDNA molecules (Fig. 6A). In contrast, the mutants produced a remarkable variety of forms of vDNA, and only a minority of the Y28S and W35S products (and none of the products from the other mutants) were the normal junction fragment (Fig. 6). It should be remembered that full-length 2-LTR circles are formed far less frequently by the CCCC, CCHH, and SSHC mutants than by the wild type; thus, it is possible that the aberrant forms seen with the mutants are also present in the wild-type sample but represent a small minority of the circles in the wild-type case and are therefore difficult to detect in this analysis.
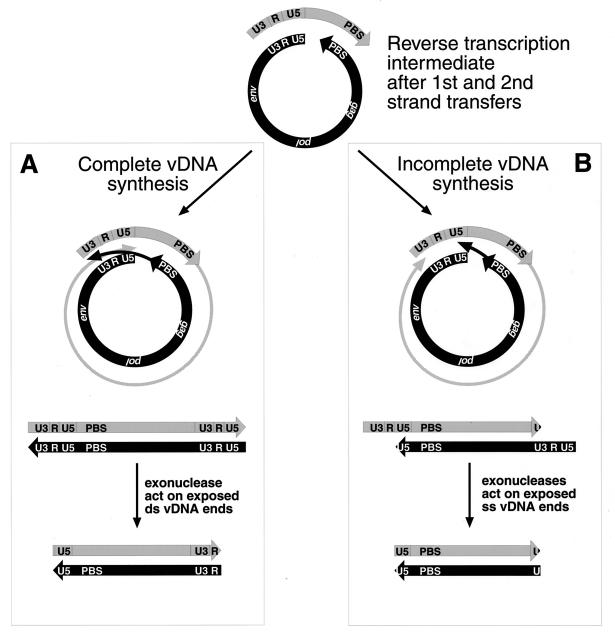
In the majority of the products obtained from the mutant-infected cells, bases were missing from both U3 and U5 partners in the junction fragment (Fig. 6A). As diagrammed in Fig. 7, these truncated forms could arise either by exonucleolytic digestion of the ends of a full-length vDNA molecule before ligation in vivo (Fig. 7A) or by a failure of RT to complete DNA synthesis (followed by exonucleolytic removal of overhanging 5′ ends) before ligation in vivo (Fig. 7B). We cannot distinguish between these two possibilities from the current data.
FIG. 7.
Mechanisms for vDNA terminal truncations. (A) The path on the left shows how the termini become truncated after complete synthesis of the vDNA by unabated reverse transcription. Exonuclease degradation of the double-stranded vDNA (ds vDNA) would account for the missing nucleotides at the vDNA termini. (B) The scheme on the right shows how incomplete synthesis of vDNA could occur, due to the inability of NC to completely melt the double-stranded vDNA complementary regions, U3|R|U5 and PBS, thus blocking RT from completing synthesis to the proper ends. Nucleases would then degrade single-stranded vDNA overhangs (ss vDNA).
Other products cloned from the mutant-infected cells are presented in Fig. 6B. The formation of these products appeared symmetrical, since either U3 or U5 was processed (and the other end truncated) with roughly similar frequencies. It seems possible that these products arise by a “half-integration” reaction, in which IN processes one end and joins it to cellular DNA but fails to insert the other end; the integrated end might then be excised together with a fragment of host DNA, which could then be joined to a truncated version of the other end. Species similar to these were previously described by Dunn et al. in experiments with wild-type Rous sarcoma virus (9).
Initiation of minus-strand DNA synthesis within the PBS or leader, rather than at the normal position (the U5-PBS boundary), appears to be the most plausible explanation for the formation of the products in Fig. 6C. These species contained additional contiguous 5′ viral sequences in the 2-LTR junction fragments. We do not know why this should happen at an appreciable frequency, since the mutants appeared to have the normal level of primer tRNA, presumably at the normal position on the PBS. For clones pRB464, pRB470, pRB728, pRB784, pRB825, and pRB868, it is possible that the tRNA was not completely removed by RNase H and reverse transcription continued into the attached tRNA primer.
Finally, a handful of products were found (Fig. 6D) which appear to be the mirror image of those in Fig. 6C: the sequences from the 3′ end of the vRNA extend beyond the U3-ppt boundary to include ppt, the 3′ untranslated region, and the env coding sequence. In all four of these clones, the U5 portion of the junction fragment was truncated; one of them also contained an insert of nonviral sequence. It seems possible that these products were generated by abnormal priming of plus-strand DNA synthesis at a site 5′ of the usual position (the ppt-U3 boundary); alternatively, they might arise if the acceptor template in the second-strand transfer event were a molecule of genomic RNA rather than plus-strand strong stop DNA; such a template would, of course, possess the additional ppt, 3′ untranslated region, and env sequences found in these junction fragments.
The primary goal of the present experiments was to identify a functional defect(s) underlying the near-absolute lack of infectivity of MuLV Zn2+ finger mutants. In a sense, there appear to be several defects, each of which can partially explain the biological defectiveness of these viruses. In three of the mutants (17), but not in CCCC or CCHH mutants (16), packaging of vRNA is very inefficient during particle assembly. Further, the production of approximately full-length DNA products is reduced ∼1,000-fold (based on the limiting-dilution analysis [Fig. 4]) when CCCC or CCHH particles infect new host cells (16). The nature of this reverse transcription defect will obviously be addressed in future studies. Finally, we now report that the approximately full-length products, i.e., those containing two LTRs which can be ligated in the infected cell, show a dramatic new phenotype: the ends of the LTRs have a variety of aberrations, including truncations, additional viral sequences, and inserts of nonviral sequences. These ends are not suitable substrates for the viral IN protein, and thus these DNA molecules cannot carry out a normal infectious cycle.
A substantial amount of knowledge, showing that the NC protein performs a variety of functions during the retroviral life cycle, has accumulated in recent years (reviewed in references 3, 8, and 27). Thus, as a domain of the Gag precursor, NC plays an essential role in the selection of vRNA for packaging during virus assembly. This domain also appears to anneal the primer tRNA to the PBS (7, 11, 14, 23, 32) and may promote the initial interaction of genomic RNA molecules to form the “immature dimer” (11, 13, 15). The mature NC protein (i.e., after cleavage from the Gag precursor) also appears to facilitate reverse transcription, both by assisting RT through sites of secondary structure in the template (35, 37) and by promoting efficient strand transfer during reverse transcription (1, 19) (this function may be particularly important in the case of minus-strand transfer in HIV-1 reverse transcription, where the initial DNA product has a very stable secondary structure). The mature protein also condenses the dimeric RNA into a more stable dimeric structure during maturation of the particle (12, 13, 15, 24).
The mutants studied here are clearly not defective with respect to several of these functions: CCCC and CCHH mutants package vRNA normally, and primer tRNA has been efficiently annealed to vRNA in CCHH particles (Fig. 3). The dimeric RNA also appears to have the normal, “mature” conformation in these particles (Fig. 2). In addition, in vitro assays with mutant NC proteins suggest that the Zn2+ fingers are probably not crucial in the maturation of the dimeric RNA (12) or in the facilitation of reverse transcription at sites of secondary structure in the template (37). Thus, it is difficult to explain why infection with a particle with an altered Zn2+ finger in NC leads to the production of “full-length” DNA products with aberrant ends.
It is intriguing to suggest that a single functional defect may be responsible for the two phenotypes observed in reverse transcription by CCCC or CCHH particles, i.e., inefficient synthesis of “full-length” DNA molecules and aberrations at the ends of those that are made. In other words, perhaps there are aberrations or missteps at all stages of reverse transcription in these mutants, so that many or all of the particles initiate reverse transcription but most do not make products which can be detected as full-length by Southern blotting and do not form circles from which “2-LTR junction fragments” can be amplified. In any case, the results show that the Zn2+ finger plays a crucial accessory role in reverse transcription (or in protection of the full-length DNA molecule from exonucleolytic attack) (Fig. 7) in vivo. The fact that some of the aberrant ends have undergone partial processing and partial integration (Fig. 6B) raises the possibility that the NC Zn2+ finger participates in normal integration as well. We are beginning experiments to test these possibilities directly.
A recent report (34) described experiments with an analogous mutant of HIV-1, in which the N-terminal finger was changed to CCCC. However, the results of these investigations diverged from those presented here with MuLV in a number of respects: the HIV-1 CCCC mutant was reported to package vRNA inefficiently, and no circular vDNAs or 5′ LTRs were detected in cells infected with the mutant particles. Perhaps the difference in experimental systems is responsible for the differences in the results obtained.
ACKNOWLEDGMENTS
We acknowledge the Frederick Biomedical Super Computing Laboratory for allocation of computer time and staff support. We also thank Stephen H. Hughes of ABL and David E. Ott of SAIC Frederick for their helpful suggestions and discussions.
Research was sponsored in part by the National Cancer Institute, Department of Health and Human Services (DHHS), under contract NO1-CO-56000 with SAIC-Frederick and contract NO1-CO-46000 with ABL-Basic Research Program.
REFERENCES
- 1.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg J M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casas-Finet J R, Urbaneja M A, Nower P N, Gorelick R J, Bosche W J, Kane B P, Johnson D, Henderson L E. Functional properties of point-mutated MoMuLV nucleocapsid (NC) protein p10. FASEB J. 1997;11:A981. [Google Scholar]
- 6.Covey S N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the Gag and Pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlix J-L, Lapadat-Tapolsky M, De Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 9.Dunn M M, Olsen J C, Swanstrom R. Characterization of unintegrated retroviral DNA with long terminal repeat-associated cell-derived inserts. J Virol. 1992;66:5735–5743. doi: 10.1128/jvi.66.10.5735-5743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The HIV-1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green L M, Berg J M. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: spectroscopic studies and a proposed three-dimensional structure. Proc Natl Acad Sci USA. 1989;86:175–178. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix J L, Wainberg M A, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA(3Lys) genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Khorchid A, Wang J, Parniak M A, Darlix J L, Wainberg M A, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160(gag-pol) and tRNA(Lys) incorporation into human immunodeficiency virus type 1. J Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg M A, Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muriaux D, De Rocquigny H, Roques B P, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 25.Ott D E, Keller J, Sill K, Rein A. Phenotypes of murine leukemia virus-induced tumors: influence of 3′ viral coding sequences. J Virol. 1992;66:6107–6116. doi: 10.1128/jvi.66.10.6107-6116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982;120:251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 27.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 28.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 29.Shank P R, Varmus H E. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978;25:104–114. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature (London) 1981;201:187–199. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 31.Stevenson M, Haggerty S, Lamonica C A, Meier C M, Welch S K, Wasiak A J. Integration is not necessary for expression of human immunodeficiency virus type 1 protein products. J Virol. 1990;64:2421–2425. doi: 10.1128/jvi.64.5.2421-2425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanstrom R, DeLorbe W J, Bishop J M, Varmus H E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci USA. 1981;78:124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanchou V, Gabus C, Rogemond V, Darlix J L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 36.Whitcomb J M, Ortiz-Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]