Abstract
Rift Valley fever virus (RVFV), a phlebovirus of the Bunyaviridae family, is an arthropod-borne virus which emerges periodically throughout Africa, emphasizing that it poses a major threat for animal and human populations. To assess the genetic variability of RVFV, several isolates from diverse localities of Africa were investigated by means of reverse transcription-PCR followed by direct sequencing of a region of the small (S), medium (M), and large (L) genomic segments. Phylogenetic analysis showed the existence of three major lineages corresponding to geographic variants from West Africa, Egypt, and Central-East Africa. However, incongruences detected between the L, M, and S phylogenies suggested that genetic exchange via reassortment occurred between strains from different lineages. This hypothesis, depicted by parallel phylogenies, was further confirmed by statistical tests. Our findings, which strongly suggest exchanges between strains from areas of endemicity in West and East Africa, strengthen the potential existence of a sylvatic cycle in the tropical rain forest. This also emphasizes the risk of generating uncontrolled chimeric viruses by using live attenuated vaccines in areas of endemicity.
Rift Valley fever is a serious emerging arthropod-borne viral anthropozoonosis caused by a phlebovirus (Rift Valley fever virus [RVFV], family Bunyaviridae), which was reported primarily to infect domestic cattle and more recently to cause massive epidemics in human populations across Africa. Modifications in the ecological and/or environmental conditions appeared to be responsible for the emergence of the virus (16, 24). Disease in humans exhibits clinical manifestations ranging from acute febrile illness to severe complications, including hepatitis, encephalitis, hemorragic fever, and ocular sequelae (13). Periodic large-scale epidemics, such as the ones in Mauritania in 1987 and 1998 (7, 43), Madagascar in 1990–1991 (17–19), and Egypt in 1977 and 1993 (2, 16), as well as in East Africa (Kenya, Somalia, and Tanzania) in 1997–1998, reiterate the potential of this virus as a considerable threat to human health, the latter epidemic affecting some 89,000 people and causing 500 deaths (1). Analysis of one strain isolated from a fatal human case during this epidemic showed a close relatedness with a strain isolated in Madagascar during the 1990–1991 outbreak (28), revealing that the virus could spread across considerable distances, possibly beyond Africa. This threat becomes more disquieting when we consider that numerous mosquito species around the world are competent laboratory vectors for RVFV (9, 35–37). Therefore, understanding the mechanism underlying its dispersal and evolution is of paramount importance to the control of this disease.
The RVFV genome consists of three negative-sense single-stranded RNA segments designated L (large), M (medium), and S (small) (for reviews, see references 10 and 31). The L segment codes for the L viral polymerase. The M segment codes for the precursor to the envelope glycoproteins, G1 and G2, which, after cleavage, generate two additional nonstructural proteins of 78 and 14 kDa. The S segment codes for the nucleocapsid protein N and the nonstructural protein NSs by using an ambisense strategy. Because of its segmented nature, the genome of members of the Bunyaviridae family allows RNA segment reassortment (exchange of a whole segment) when cells are coinfected by two closely related viruses of the same genus or serogroup (reviewed in reference 26). Reassortment between strains of RVFV has been demonstrated experimentally in tissue cultures (30) and in mosquitoes that were dually infected (38). Naturally occurring reassortants have been suggested or demonstrated for bunyavirus (8, 12, 40) and hantavirus (11, 14, 27). However, to date, despite some clues derived from unexpected groupings in a phylogenetic tree based on the NSs coding region (29), reassortment among natural isolates of RVFV has not been investigated. In this report, we address the question of RVFV genetic reassortment under natural conditions and its consequences on the evolution and epidemiology of the disease.
MATERIALS AND METHODS
Virus propagation and RNA extraction.
The origins and years of isolation of RVFV isolates are shown in Table 1. Propagation of viruses and cytoplasmic RNA extraction from infected cells were done as previously described (29).
TABLE 1.
Origins and years of isolation of RVFV isolates
| Code | Strain | Yr of isolation | Origin | Source | Genotypea (S/M/L) |
|---|---|---|---|---|---|
| SNSb | Smithburn | 1944 | Uganda | Entebbe strain | C/C/C |
| ArUGA-55 | Lunyo | 1955 | Uganda | Mosquito | C/C/C |
| ArCAr-69 | Ar B 1976 | 1969 | CAR | Mosquito | C/C/C |
| C13 | Clone 13 | 1974 | CAR | Human | */C/C |
| HEGY-77 | ZH 548 | 1977 | Egypt | Human | E/E/E |
| MP12b | MP12 | 1977 | Egypt | ZH548 strain | E/E/E |
| ArMAD-79 | Ar Mg 811 | 1979 | Madagascar | Mosquito | E/?/E |
| ArSEN-84 | Ar D 38661 | 1984 | Senegal | Mosquito | E/W/W |
| AnGUI-84 | An K 6087 | 1984 | Guinea | Bat | W/C/C |
| ArBUF-84 | Ar D 38457 | 1984 | Burkina Faso | Mosquito | W/W/W |
| H1MAU-87 | H D 47502 | 1987 | Mauritania | Human | C/W/W |
| H2MAU-87 | H D 47311 | 1987 | Mauritania | Human | W/W/W |
| H3MAU-87 | H D 47408 | 1987 | Mauritania | Human | W/W/W |
| H4MAU-87 | H D 48255 | 1987 | Mauritania | Human | W/?/W |
| AnMAD-91 | An Mg 990 | 1991 | Madagascar | Bovine | C/C/C |
| ArSEN-93 | Ar D 104769 | 1993 | Senegal | Mosquito | C/C/W |
| AnSEN-K-93 | An D 106417 | 1993 | Senegal | Zebu | W/W/C |
| BEGY-93 | B EGY 93 | 1993 | Egypt | Buffalo | E/E/E |
| HEGY-93 | H EGY 93 | 1993 | Egypt | Human | E/E/E |
| HKEN-97 | 384-97.1 | 1997 | Kenya | Human | C/C/C |
E, W, and C, Egyptian, Western African, and Central-East African lineage, respectively; ∗, sequence too short to be assigned a lineage; ?, sequence not determined.
Attenuated strain.
Reverse transcription (RT)-PCR and sequencing procedures.
Three different sets of primers, NS3a-NS2g, MRV1a-MRV2g, and Wag-Xg, were used to amplify portions of the NSs, G2, and L coding regions, respectively. The NSs coding region is located in the S segment, whereas the regions coding for G2 and L are in the M and L segments, respectively. The antisense primers NS3a, MRV1a, and Wag were used to synthesize the first-strand complementary DNA. Primer sequences, protocols for cDNA synthesis, PCR amplification, and direct sequencing of PCR products were described previously (20, 21, 29, 34).
Phylogenetic reconstruction.
The phylogenetic analyses of each segment of the RVFV genome were performed with data sets for the NSs, G2, or L protein coding region by using maximum likelihood and maximum parsimony methods in PAUP (Phylogenetic Analysis Using Parsimony, beta version 4.0; kindly provided by David L. Swofford, Smithsonian Institution, Washington, D.C.). For a more realistic tree reconstruction and branch length estimates for each data set, the optimal values for the transition probabilities among different nucleotides and the value for the shape parameter (alpha) for the gamma distributed variable rates among sites were empirically determined from the data. Using the optimal values of these parameters, trees with similar likelihood were collected and the tree topology stability around the likelihood maxima was investigated by calculating the 50% majority-rule consensus. For an additional comparison, we used a fast tree search algorithm (quartet puzzling) for estimating maximum likelihood trees from PUZZLE version 3.1 (33), which automatically assigns estimation of support of each internal node. Additionally, to investigate the robustness of tree resolution under maximum parsimony, a bootstrapping analysis was done with 1,000 random resamplings with the nearest-neighbor-interchange perturbation algorithm within PAUP. For the shortest data set (i.e., the 211-bp L amplicon), a 2% jackknife was used as an alternative resampling method.
To determine topological incongruence among estimates obtained from different data sets for the same set of taxa, we checked whether the best topology for each data set is statistically worse when reconstructed with an alternative data set by using the Kishino and Hasegawa test implemented in PAUP 4.0. Additionally, to test for reassortment among viruses from different lineages, constraint trees were constructed; in these, the taxa were assigned to a given part of the tree, generating alternative topologies to be tested against the ones obtained from the unconstrained data. Finally, to allow the inspection of the topologies estimated from different genomic segments, the TreeMap program (22) was used to construct parallel phylogenies.
Nucleotide sequence accession numbers.
The sequences of the strains listed in Table 1 and corresponding to the L, G2, and N protein coding regions have been assigned GenBank accession no. AF134782 to AF134801, AF134492 to AF134508, and AF134530 to AF134551, respectively. The sequences of the NSs protein coding region were deposited in the EMBL database (accession no. Y12739 to Y12756) as already reported (29).
RESULTS
Phylogeny of different RVFV segments.
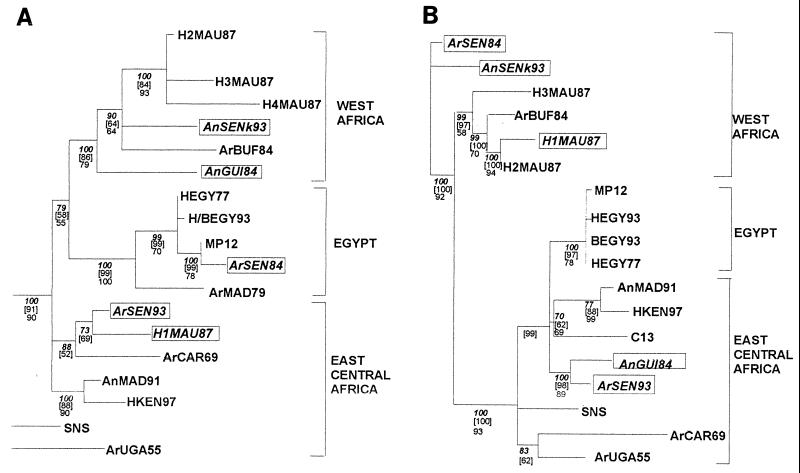
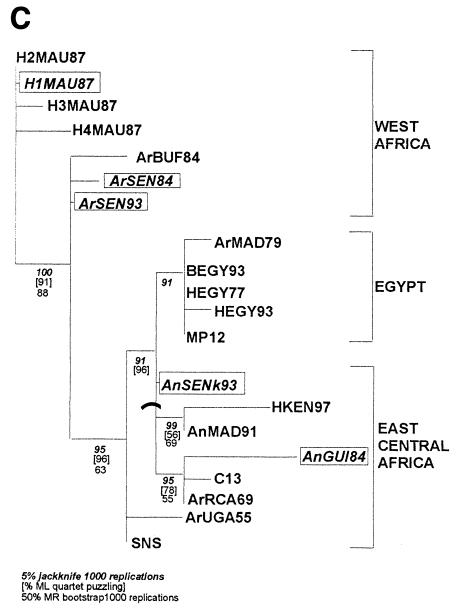
In a previous paper, phylogenetic analysis carried out with 20 isolates and derived from the 669-nucleotide-long sequences of the NSs protein coding region in the S segment showed the existence of three distinct lineages: Ia and Ib in sub-Saharan Africa and II in Egypt (29). Additional sequencing of the M and L segments was performed after RT-PCR amplification of an 809-nucleotide-long DNA fragment located within the region coding for the G2 protein and a 212-nucleotide-long DNA fragment located in the L protein. Phylogenetic analyses of these sequences using the maximum likelihood and maximum parsimony methods confirmed the distribution pattern within the three lineages, which were renamed Central-East Africa, West Africa, and Egypt for Ia, Ib, and II, respectively (see Fig. 1). The support for different nodes on the tree was evaluated by various methods (i.e., bootstrap, jackknife, or maximum likelihood topology consensus) and showed strong support for the three phylogenies.
FIG. 1.
Maximum likelihood trees for the NSs (A), G2 (B), and L (C) coding region sequences on the S, M, and L segments, respectively. Values below the different nodes indicate their robustness by 5% jacknife (indicated in boldface and italic type), maximum likelihood quartet puzzling (indicated in brackets), and bootstrapping (indicated in roman type) methods. Putative reassortant strains are boxed.
These phylogenetic analyses raised some general comments. First, some of the strains in the Central-East Africa and Egypt groups (SNS and ArUGA-55, AnMAD-91 and HKEN-97, HEGY-77 and HEGY-93) are closely related, despite their dates of isolation, suggesting the existence of an endemic-enzootic maintenance cycle of the virus in these areas. Second, no obvious grouping based on the host species was observed. This could be explained by frequent exchange of viruses between different hosts. Third, in each group, strains isolated during epidemic-epizootic and endemic-enzootic periods were found adjacent to each other, suggesting that the virus circulates alternatively in enzootic-endemic form through a maintenance cycle or epidemic-epizootic mode, both cycles providing each other with viral strains. However, despite similar distributions of each tree into three clusters, the assignment of some isolates (ArSEN-93, AnSEN-K-93, ArSEN-84, H1MAU-87, and AnGUI-84) to one particular group does not remain constant within the three phylogenies. Construction of the reconciling parallel trees using the three phylogenies emphasized that the groupings of the strains in the S, M, and L segment trees correlate quite well but clearly show some incongruences. For instance, H1MAU-87 and ArSEN-84 possess the L and M segments of the West African genotype and the S segment from the Central-East African or Egyptian lineage, respectively. AnGUI-84 has the L and M segments of the Central-East African genotype, whereas the S segment belongs to the West African lineage. Finally, two strains isolated during the same period in two different places in Senegal, Barkedji (North) and Kolda (Casamance, South), ArSEN-93 and AnSEN-K93, have a different distribution within clusters: AnSEN-K93 was located within the Western African lineage with regards to its S and M segments and within the Central-East African lineage with regards to its L segment. Reciprocally, in the case of ArSEN-93, its S and M segments belong to the Central-East African lineage and the L segment belongs to the Western African lineage (Fig. 1).
The possibility that incongruences might be due to cross contamination among isolates seems highly improbably given the following extreme precautions that were taken to avoid such artifacts. (i) RNA extraction and RT were done separately for each isolate under a laminar-flow containment hood which was decontaminated between each manipulation. (ii) PCR mix preparation, PCRs, and electrophoresis were performed in separate rooms. (iii) For each isolate, PCR products derived from independent RNA extraction and RT-PCR were sequenced several times and led to identical sequences, many of them being processed in laboratories at least 4,000 km apart (Paris, France, and Dakar, Senegal).
Given that trees for different genes are clearly incongruent in their topologies, it could be assumed that these incongruences are due to genetic exchange. Regarding the segmentation of the RVFV genome, the most probable device to explain the exchange of genetic material is RNA segment reassortment.
Tests for topological structure as indicative of reassortment.
Two approaches were used to evaluate the incongruences observed among specific isolates of different trees. First, we constructed constrained trees and tested their topologies by using likelihood against the unconstrained trees shown in Fig. 1. The likelihood value for the unconstrained tree resulting from the use of the NSs sequences was better than the one obtained for the constrained tree where the ArSEN-93 and H1MAU-87 isolates were forced into the West African group (−lnL = 1,774.76, difference of lnL = 57.21, P < 0.001). Similarly, the values obtained for the G2 and L sequence phylogenies (for G2, difference of lnL = 42.95, P = 0.0002; for L region, difference of lnL = 9.74, P = 0.06), in which ArSEN-93 and AnGUI-84 or AnSEN-K-93 and AnGUI-84, respectively, were forced into the West African group, led to the same conclusion. Second, we used the Kishino and Hasegawa test on the difference of the likelihoods for trees obtained for taxa common to both G2 and NSs data sets. The maximum likelihood tree for G2 (−lnL = 1,684.40) had a lower likelihood (difference of lnL = 30.35, P = 0.0603) under the NSs data set than the NSs tree did (−lnL = 1,654.06), and the NSs tree (−lnL = 1,842.69) had a lower likelihood (difference of lnL = 51.99, P < 0.0001) under the G2 data set than the G2 tree did (−lnL = 1,790.69). Although the difference of likelihood (lnL difference = 30.35) for the G2 tree under the NSs data set may not be statistically different under the 95% confidence level, likelihood tests using either constrained trees or the Kishino and Hasegawa test support the notion that the incongruences observed among trees are due not to poor phylogenetic topology inferences but rather to genetic exchange.
In light of the recent report of recombination in hantavirus evolution (32), it could not be excluded that the incongruences in the RVFV tree topologies could be due to intramolecular recombination. Although such events appear, so far, to be very rare among negative-strand RNA viruses (26), we considered this possibility and analyzed the NSs, G2, and L data with the split decomposition method, by using the program Splits version 1.0 (3), but we did not find any clear-cut indication of recombination, i.e., networked evolution (data not shown), within the fragments sequenced. Thus, reassortment events generating combination between L, M, and S segments of RVFV natural isolates appear to be the most likely explanation.
DISCUSSION
This study indicated that 5 of the 20 (25%) isolates, ArSEN-93, AnSEN-K-93, ArSEN-84, H1MAU-87, and AnGUI-84, appear to result from a reassortment event. This percentage may be underestimated since the method used herein to identify reassortants took into account only reassortment events between the three major lineages (Egypt, West Africa, and Central-East Africa). Interestingly, all the strains identified as reassortants, ArSEN-93, AnSEN-K-93, ArSEN-84, H1MAU-87, and AnGUI-84, were isolated from West Africa (Senegal, Guinea, and Mauritania) and involved reassortment with strains of the Central-East African or Egyptian lineage (ArSEN-84) (Table 1). In addition, these reassortants have retained combinations of homologous L plus M segments (ArSEN-84, AnGUI-84, and H1MAU-87) and M plus S segments (AnSEN-K-93 and ArSEN-93) (Table 1). In dual infection of BHK-21 cells with La Crosse and Snowshoe hare viruses, some segment associations appeared to be preferred (39). Investigations with a larger sample of RVFV field isolates would be needed to indicate whether generation and selection of reassortants in nature are random or not.
Replication errors such as base substitution and deletions or insertions are the most common mechanism of RNA virus evolution. However, major changes of viral genotype may involve exchanges of RNA segments (genetic shift), as exemplified by influenza virus (42). Comparison of RVFV isolates sampled from different geographical localities showed that evolution of this virus in nature not only appears to be due to point mutations, the percentage of base substitution varying from 0 to 9.6% in the S segment (29), but also may occur by genetic exchange. Indeed, the apparent contradictions in the groupings of the different segments within the three distinct lineages could be explained by reassortment and strongly suggest that this evolution mechanism is a common trait in RVFV natural history. Arboviruses apparently evolve approximately 10-fold more slowly than RNA viruses that have only a single vertebrate host (4, 41). Thus, the evolution of RVFV driven by reassortment and by point mutations may depend on the host number.
Reassortment under natural conditions implies the existence of at least two different strains present at the same time, in the same area, and infecting the same host. One additional requisite for finding reassortants is that, once generated, these chimeric viruses were viable and their host did not exert a negative selection against them. We do not know whether the strains identified as reassortants were host restricted, but they were isolated from different hosts, including humans, mosquitoes, bats (An GUI-84), and zebu (AnSEN-K93). With regard to cocirculation of different strains which can potentially undergo reassortment, Senegal is a very instructive example as an area where at least two different lineages circulated at the same time in 1993. Since RVFV is an arbovirus, reassortment among strains can occur in a dually infected mosquito or vertebrate host. Reassortment was demonstrated experimentally in hamsters as well as in mosquitoes that were dually infected with two RVFV natural isolates (38). It seems likely that vertebrate hosts can be naturally infected with different strains of RVFV since a human isolate from the Republic of Central Africa was shown to be composed of a heterogenous population of viral clones with different biological properties (21). Mosquitoes can be dually infected by interrupted feeding or feeding on a dually infected vertebrate host. Both mechanisms have been demonstrated experimentally for RVFV with a natural mosquito vector, Culex pipiens, suggesting that under natural conditions, the process is probably the same. There exists a third theoretical possibility for a mosquito to be dually infected: a female infected transovarially or venereally with one strain could become superinfected during a bloodmeal with another strain. However, due to homologous interference, vertically infected mosquitoes may be refractory to superinfection with the same virus species. Therefore, both mosquitoes and vertebrate hosts may act as a site for RVFV reassortment in nature but their relative contribution in the natural process of reassortment remains to be determined.
The occurrence of reassortment among natural isolates from West Africa and Central-East Africa or, less frequently, Egypt suggests two possibilities: (i) one strain was transported to a place where another strain was circulating; or (ii) the virus had to make its way through the rain forest and to adapt to the different fauna and ecotopes, establishing a sylvatic cycle. Although the existence of a sylvatic cycle would have to be demonstrated, the circulation of RVFV in the rain forest has been revealed by the high antibody prevalence in wild animals and pygmy populations in Central African Republic (6) and multiple strain isolations from mosquitoes in the primary forest of Perinet in Madagascar (15). In addition, data recently published by Pretorius et al. (25) indicated that rodents were involved as a potential reservoir of RVFV in South Africa. It is likely that different strains of RVFV circulate in the rain forest area among wild mosquitoes (16) and vertebrates like monkeys (23) and bats (5), depending on the dynamic of the interaction among hosts and virus populations. Therefore, the search and characterization of a sylvatic cycle of RVFV are of utmost importance because this would lead to better understanding and control of the epidemiology of Rift Valley fever and its potential emergence.
The existence of reassortment as a mechanism of evolution raises the question of the epidemic potential of these viruses and their pathogenicity. Isolation of a potential reassortant from a fatal case during the 1987 Mauritanian epidemic leaves this question open, even though other strains which were not identified as reassortants were isolated from fatal cases. A similar question concerning the outbreak of Sin Nombre virus was raised (11), but in spite of the evidence of reassortment among Sin Nombre virus isolates (11, 14, 27), there was no indication that the emergence of hantavirus pulmonary syndrome was due to a novel chimeric virus. On the other hand, reassortment appeared to be involved in the triggering of an influenza virus pandemic (42).
In conclusion, the high rate of reassortment among RVFV isolates raises interesting issues on vaccination with live attenuated vaccines during epidemics when virulent strains are circulating. Since reassortants containing one attenuated and two virulent segments were shown to be attenuated (30), reassortment between the wild and vaccine strains would generate attenuated viruses with protective effects against the disease, provided attenuation markers were present in each genomic segment of the live attenuated strain. Therefore, attenuated vaccine strains with several attenuation markers present in each segment would be recommended for control of RVFV. This safety strategy also stands a better chance to minimize the probability of reversion toward virulence.
ACKNOWLEDGMENTS
We are grateful to A. Billecocq, B. Le Guenno, and C. Préhaud for fruitful discussions, to M. Diallo for critically reading the manuscript, and to R. Swanepoel for providing us with the 1997 Kenyan strain. We are indebted to ORSTOM and the WHO collaborating center for arbovirus and hemorrhagic fever virus research (CRORA), whose collaborative work allowed us to obtain most of the RVFV isolates analyzed in this study. The excellent technical expertise of P. Vialat is acknowledged.
P.M.A.Z. was funded by a PQ Scholarship from CNPq and a PRONEX/PADCT grant endowed by the Brazilian Government.
REFERENCES
- 1.Anonymous. An outbreak of Rift Valley fever, Eastern Africa, 1997–98. WHO Weekly Epidemiol Rep. 1998;73:105–112. [PubMed] [Google Scholar]
- 2.Arthur R R, el-Sharkawy M S, Cope S E, Botros B A, Oun S, Morrill J C, Shope R E, Hibbs R G, Darwish M A, Imam I Z. Recurrence of Rift Valley fever in Egypt. Lancet. 1993;342:1149–1150. doi: 10.1016/0140-6736(93)92128-g. [DOI] [PubMed] [Google Scholar]
- 3.Bandelt H J, Dress A W. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 4.Beaty B J, Borucki M, Farfan J, White D. Arbovirus-vector interactions: determinants of arbovirus evolution. In: Saluzzo J F, Dodet B, editors. Factors in the emergence of arbovirus disease. Paris, France: Elsevier; 1997. pp. 23–35. [Google Scholar]
- 5.Boiro I, Konstaninov O K, Numerov A D. Isolement du virus de la fièvre de la vallée du Rift à partir de chéiroptères en République de Guinée. Bull Soc Pathol Exot Fil. 1987;80:62–67. [PubMed] [Google Scholar]
- 6.Digoutte J P, Cordellier R, Robin Y, Pajot F X, Geoffroy B. Le virus Zinga (Ar B 1976), nouveau prototype d’arbovirus isolé en République Centrafricaine. Ann Inst Pasteur Microbiol. 1974;125B:107–118. [PubMed] [Google Scholar]
- 7.Digoutte J P, Peters C J. General aspects of the 1987 Rift Valley fever epidemic in Mauritania. Res Virol. 1989;140:27–30. doi: 10.1016/s0923-2516(89)80081-0. [DOI] [PubMed] [Google Scholar]
- 8.Fields B N, Henderson B E, Coleman P H, Work T H. Pahayokee and Shark River, two new arboviruses related to Patois and Zegla from the Florida Everglades. Am J Epidemiol. 1969;89:222–226. doi: 10.1093/oxfordjournals.aje.a120932. [DOI] [PubMed] [Google Scholar]
- 9.Gargan T P, 2nd, Clark G G, Dohm D J, Turell M J, Bailey C L. Vector potential of selected North American mosquito species for Rift Valley fever virus. Am J Trop Med Hyg. 1988;38:440–446. doi: 10.4269/ajtmh.1988.38.440. [DOI] [PubMed] [Google Scholar]
- 10.Giorgi C. Molecular biology of phleboviruses. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 105–128. [Google Scholar]
- 11.Henderson W W, Monroe M C, St. Jeor S C, Thayer W P, Rowe J E, Peters C J, Nichol S T. Naturally occurring Sin Nombre virus genetic reassortants. Virology. 1995;214:602–610. doi: 10.1006/viro.1995.0071. [DOI] [PubMed] [Google Scholar]
- 12.Klimas R A, Thompson W H, Calisher C H, Clark G G, Grimstad P R, Bishop D H. Genotypic varieties of La Crosse virus isolated from different geographic regions of the continental United States and evidence for a naturally occurring intertypic recombinant La Crosse virus. Am J Epidemiol. 1981;114:112–131. doi: 10.1093/oxfordjournals.aje.a113158. [DOI] [PubMed] [Google Scholar]
- 13.Laughlin L W, Meegan J M, Strausbaugh L J, Morens D M, Watten R H. Epidemic Rift Valley fever in Egypt: observations of the spectrum of human illness. Trans R Soc Trop Med Hyg. 1979;73:630–633. doi: 10.1016/0035-9203(79)90006-3. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Schmaljohn A L, Anderson K, Schmaljohn C S. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology. 1995;206:973–983. doi: 10.1006/viro.1995.1020. [DOI] [PubMed] [Google Scholar]
- 15.Mathiot C, Ribot J J, Clerc Y, Coulanges P, Rasolofonirina N. Fièvre de la vallée du Rift et virus Zinga: un arbovirus pathogène pour l’homme et l’animal, nouveau pour Madagascar. Arch Inst Pasteur Madag. 1984;51:125–133. [PubMed] [Google Scholar]
- 16.Meegan J M, Bailey C J. Rift Valley fever. In: P. M T, editor. The arboviruses: epidemiology and ecology. IV. Boca Raton, Fla: CRC Press Inc.; 1989. pp. 51–76. [Google Scholar]
- 17.Morvan J, Rollin P E, Laventure S, Rakotoarivony I, Roux J. Rift Valley fever epizootic in the central highlands of Madagascar. Res Virol. 1992b;143:407–415. doi: 10.1016/s0923-2516(06)80134-2. [DOI] [PubMed] [Google Scholar]
- 18.Morvan J, Rollin P E, Roux J. La fièvre de la vallée du Rift à Madagascar en 1991. Enquête séro-épidémiologique chez les bovins. Rev Elev Med Vet Pays Trop. 1992a;45:121–127. [PubMed] [Google Scholar]
- 19.Morvan J, Saluzzo J F, Fontenille D, Rollin P E, Coulanges P. Rift Valley fever on the east coast of Madagascar. Res Virol. 1991;142:475–482. doi: 10.1016/0923-2516(91)90070-j. [DOI] [PubMed] [Google Scholar]
- 20.Muller R, Poch O, Delarue M, Bishop D H L, Bouloy M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol. 1994;75:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- 21.Muller R, Saluzzo J F, Lopez N, Dreier T, Turell M, Smith J, Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 22.Page R D M. Parallel phylogenies: reconstructing the history of host-parasite assemblages. Cladistics. 1995;10:155–173. [Google Scholar]
- 23.Pelissier A, Rousselot R. Enquête sérologique sur l’incidence des virus neurotropes chez quelques singes de l’Afrique équatoriale française. Bull Soc Pathol Exot. 1954;47:228–230. [PubMed] [Google Scholar]
- 24.Peters C J, Linthicum K J. Rift Valley fever. In: Steele B G W and J H., editor. Handbook of zoonoses, section B. Viral. 2nd ed. Boca Raton, Fla: CRC Press; 1994. pp. 125–138. [Google Scholar]
- 25.Pretorius A, Oelofsen M J, Smith M S, van der Ryst E. Rift Valley fever virus: a seroepidemiologic study of small terrestrial vertebrates in South Africa. Am J Trop Med Hyg. 1997;57:693–698. doi: 10.4269/ajtmh.1997.57.693. [DOI] [PubMed] [Google Scholar]
- 26.Pringle C R. Genetics and genome segment reassortment. In: Elliott R M, editor. The viruses. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 189–226. [Google Scholar]
- 27.Rodriguez L L, Owens J H, Peters C J, Nichol S T. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. 1998;242:99–106. doi: 10.1006/viro.1997.8990. [DOI] [PubMed] [Google Scholar]
- 28.Sall A A, de Zanotto A Z P M, Vialat P, Sene O K, Bouloy M. Origin of 1997–98 Rift Valley fever outbreak in East Africa. Lancet. 1998;352:1596–1597. doi: 10.1016/s0140-6736(05)61043-4. [DOI] [PubMed] [Google Scholar]
- 29.Sall A A, de Zanotto A Z P M, Zeller H G, Digoutte J P, Thiongane Y, Bouloy M. Variability of the NSs protein among Rift Valley fever virus isolates. J Gen Virol. 1997;78:2853–2858. doi: 10.1099/0022-1317-78-11-2853. [DOI] [PubMed] [Google Scholar]
- 30.Saluzzo J F, Smith J F. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine. 1990;8:369–375. doi: 10.1016/0264-410x(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 31.Schmaljohn C. Bunyaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1447–1471. [Google Scholar]
- 32.Sibold C, Meisel H, Kruger D H, Labuda M, Lysy J, Kozuch O, Pejcoch M, Vaheri A, Plyusnin A. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takehara K, Min M K, Battles J K, Sugiyama K, Emery V C, Dalrymple J M, Bishop D H L. Identification of mutations in the M RNA of a candidate vaccine strain of Rift Valley fever virus. Virology. 1989;169:452–457. doi: 10.1016/0042-6822(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 35.Turell M J, Bailey C L, Beaman J R. Vector competence of a Houston, Texas, strain of Aedes albopictus for Rift Valley fever virus. J Am Mosq Control Assoc Bull. 1988;4:94–96. [PubMed] [Google Scholar]
- 36.Turell M J, Kay B H. Susceptibility of selected strains of Australian mosquitoes (Diptera: Culicidae) to Rift Valley fever virus. J Med Entomol. 1998;35:132–135. doi: 10.1093/jmedent/35.2.132. [DOI] [PubMed] [Google Scholar]
- 37.Turell M J, Rossi C A, Bailey C L. Effect of extrinsic incubation temperature on the ability of Aedes taeniorhynchus and Culex pipiens to transmit Rift Valley fever virus. Am J Trop Med Hyg. 1985;34:1211–1218. doi: 10.4269/ajtmh.1985.34.1211. [DOI] [PubMed] [Google Scholar]
- 38.Turell M J, Saluzzo J F, Tammariello R F, Smith J F. Generation and transmission of Rift Valley fever viral reassortants by the mosquito Culex pipiens. J Gen Virol. 1990;71:2307–2312. doi: 10.1099/0022-1317-71-10-2307. [DOI] [PubMed] [Google Scholar]
- 39.Urquidi V, Bishop D H L. Non-random reassortment between the tripartite RNA genomes of La Crosse and snowshoe hare viruses. J Gen Virol. 1992;73:2255–2265. doi: 10.1099/0022-1317-73-9-2255. [DOI] [PubMed] [Google Scholar]
- 40.Ushijima H, Clerx-van Haaster M, Bishop D H. Analyses of Patois group bunyaviruses: evidence for naturally occurring recombinant bunyaviruses and existence of immune precipitable and nonprecipitable nonvirion proteins induced in bunyavirus-infected cells. Virology. 1981;110:318–332. [PubMed] [Google Scholar]
- 41.Weaver S C, Rico-Hesse R, Scott T W. Genetic diversity and slow rates of evolution in New World alphaviruses. Curr Top Microbiol Immunol. 1992;176:99–117. doi: 10.1007/978-3-642-77011-1_7. [DOI] [PubMed] [Google Scholar]
- 42.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO. Unpublished data.