Abstract
Children with optic pathway gliomas (OPGs), a low-grade brain tumor associated with neurofibromatosis type 1 (NF1-OPG), are at risk for permanent vision loss. While OPG size has been associated with vision loss, it is unclear how changes in size, shape, and imaging features of OPGs are associated with the likelihood of vision loss. This paper presents a fully automatic framework for accurate prediction of visual acuity loss using multi-sequence magnetic resonance images (MRIs). Our proposed framework includes a transformer-based segmentation network using transfer learning, statistical analysis of radiomic features, and a machine learning method for predicting vision loss. Our segmentation network was evaluated on multi-sequence MRIs acquired from 75 pediatric subjects with NF1-OPG and obtained an average Dice similarity coefficient of 0.791. The ability to predict vision loss was evaluated on a subset of 25 subjects with ground truth using cross-validation and achieved an average accuracy of 0.8. Analyzing multiple MRI features appear to be good indicators of vision loss, potentially permitting early treatment decisions.
I. INTRODUCTION
About one in five children with neurofibromatosis type 1 (NF1), a genetic condition causing tumor growth along the nerves, will develop optic pathway gliomas (OPGs). NF1-OPGs are low-grade brain tumors affecting the anterior visual pathway (AVP) that includes the optic nerves, optic chiasm and optic tracts [1]. Localized along AVP, the tumor has a large impact on the quality of life in children with NF1-OPG because it can cause permanent vision loss. Children experiencing or at risk for vision loss are treated with chemotherapy. However, less than 50% of NF1-OPGs require treatment with chemotherapy [2] and in some cases determining clinical deterioration can be very challenging. Thus, determining which children with NF1-OPGs are experiencing disease progression and when treatment with chemotherapy is needed remains a significant challenge.
A decline in visual acuity (VA) is a well-established clinical indication to initiate treatment, however this can be challenging in young children who may not reliably cooperate with testing. Instead, optical coherence tomography (OCT) images of the circumpapillary retinal nerve fiber layer (cpRNFL) have been used as an objective measure of damage to the AVP and can be complementary to standard VA testing. In this paper, we develop a fully automatic end-to-end framework using magnetic resonance imaging (MRI) to predict VA loss.
MRI is critical for the diagnosis and monitoring of NF1-OPGs. Generally, an MRI scan is performed every 3 to 6 months after diagnosis to monitor for changes in tumor size and enhancement. MRI features not measured during the clinical interpretation may contain important information relevant to decision-making. However, radiomic features in general have not been formally studied in NF1-OPGs.
Generally, radiomic analysis for clinical diagnosis and prognosis is a multi-step process [3], [4]. One of the most important steps is an accurate segmentation of a dedicated region of interest (ROI) to permit feature extraction. To avoid the inter-observer variability due to manual processing and to enable a reliable framework for integration in clinical workflows and large-scale studies, automatic segmentation of the ROI is needed. However, automatic quantification and segmentation of the AVP is difficult due to its small, elongated, and thin morphology. The segmentation is even more challenging in the presence of amorphous OPGs along the AVP.
Deep learning approaches have been used to effectively address such challenges in AVP segmentation [5]. However, deep learning-based models strongly depend on the quantity and quality of their training data. Thus, for rare diseases, such as pediatric brain tumors, it is critical to obtain a sufficiently large and diverse supply of data from multiple sources, which is both challenging and adds protocol variability to the training data. On the other hand, transformer-based models have shown great potential for downstream vision tasks such as classification, when pre-trained on larger amounts of data and then transferred to smaller datasets [6]. When processing 3D medical imaging data, transformer-based models usually need to deal with high-resolution image complexity and limited supply of data. To address these challenges and obtain a high-quality segmentation for VA loss prediction, we pretrained a variation of Swin transformer for 3D images (SwinUNETR) [7], [8], [9] on a large publicly available dataset (BraTS [10], [11]) with adult brain MRIs. Further, only a part of the pretrained weights were transferred to be finetuned on our in-house OPG dataset, to remedy for large domain gap between these two datasets.
Our work is also inspired by previous reports that demonstrated that overall tumor size assessed using volumetric measures of the AVP is associated with vision outcomes in children with NF1-OPGs [12]. Thus, the automated volumetric analysis of NF1-OPG could advance the clinical care of impacted children. Other indicators related to imaging intensity features (diffusion tensor imaging) have also been studied [13]. However, features from the most common clinical MRI sequences are not established. To address this gap in clinical knowledge, in this work, we perform automated NF1-OPG segmentation with comprehensive analysis of features including shape, size, dimension, and intensity from multi-sequence MRI (T1-weighted, T2-weighted and T2-FLAIR) to define predictive scores on VA loss.
The main contributions of this work are: 1) fully automatic deep learning-based framework for vision outcome prediction using MRI; 2) 3D Swin transformer model with adapted transfer learning to tackle the segmentation of small and amorphous structures like AVP; 3) analysis of correlation of MRI radiomic features and their predictive performance on vision outcome.
II. METHODS
A. Image acquisition and preprocessing
This study includes brain MRI of 75 children with NF1-OPG at the Children’s Hospital of Philadelphia. Data were acquired using Siemens scanner (Erlangen, Germany). Each subject in the dataset contains multi-sequence MRI: high-resolution (HR) T1-weighted volumetric sequence, low-resolution (LR) anisotropic T2-weighted sequence and LR anisotropic T2-FLAIR sequence (Table I). The AVP ground truth was manually segmented on unprocessed T1 sequences by medical experts. This study was conducted retrospectively using human subject data and received ethical approval from Institutional Review Boards.
TABLE I.
Summary of acquisition protocols: mean ± standard deviation of the resolution across 75 subjects. SR, CR, AR refer to sagittal, coronal, axial resolution, respectively.
| Sequence | T1 | T2 | T2-FLAIR |
|---|---|---|---|
| SR (mm) | 0.91 ± 0.04 | 0.54 ± 0.05 | 0.60 ± 0.14 |
| CR (mm) | 0.83 ± 0.05 | 0.69 ± 0.46 | 0.61 ± 0.14 |
| AR (mm) | 0.83 ± 0.06 | 1.93 ± 0.54 | 3.69 ± 0.96 |
Standard image preprocessing was applied, including N4 bias field correction and intra-subject co-registration of the three sequences to T1. Preprocessing was carried out using the Advanced Normalization Tools [14]. All registered images were resampled to an isotropic resolution of 0.84 × 0.84 × 0.84 mm, close to the finest average resolution of T1.
A subset of 25 children among the above subjects underwent neuro-ophthalmic evaluation to determine the presence or absence of VA loss as ground truth. OCT images were acquired using a spectral-domain OCT device (Spectralis; Heidelberg Engineering, Heidelberg, Germany). The VA outcome per-eye (normal or abnormal vision) was determined by both VA (≥ 0.2 logarithm of the minimum angle of resolution (logMAR)) and cpRNFL thickness (< 80 microns). If abnormal vision occurred in any of the two eyes, the subject was labeled as positive for VA loss, which resulted in a balanced dataset of 13 positive and 12 negative cases.
The dataset of the BraTS Challenge 2021 [15], [16] was used for supervised pretraining of the deep learning model, which includes multi-institutional multi-sequence brain MRI. 1251 adult cases with 3 glioblastoma labels (necrotic area, peritumoral edematous, enhancing tumor) are available. Each case has 4 co-registered sequences (T1, contrast enhanced T1, T2, T2-FLAIR), all resampled to an isotropic resolution of 1 mm3 and 240 × 240 × 155 voxels.
B. Transformer-based segmentation network
The principal aim of this paper is to develop a fully automatic end-to-end framework for VA loss prediction and seamlessly provide VA loss-related risks from input MRI sequences (T1, T2, T2-FLAIR) in real time. The framework is composed of the following parts: i) transformer-based segmentation network; ii) radiomic feature extraction and selection using statistical analysis; iii) estimator for VA loss prediction (Fig. 1).
Fig. 1.
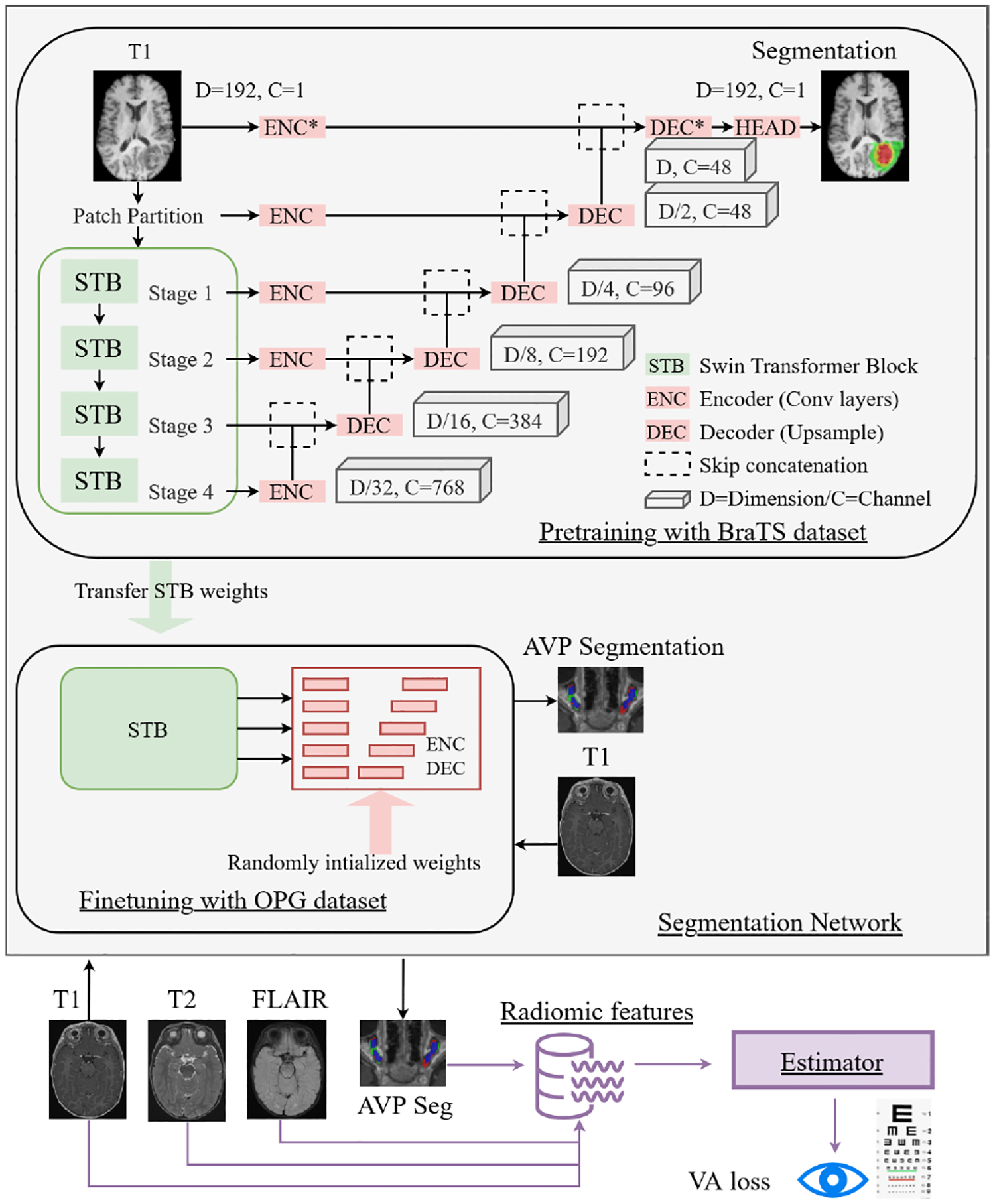
Our proposed end-to-end framework for AVP ROI segmentation and VA loss prediction using statisitically signficant radiomic features from the ROI in pediatric brain MRIs.
The transformer-based model was pretrained on the large BraTS dataset available through the challenge and then finetuned on our specific in-house OPG dataset. This was done to facilitate transfer learning of knowledge and achieve efficient AVP segmentation with the limited number of available subjects with rare NF1-OPG condition (n=75). To make the pretraining samples consistent with our NF1-OPG training data, only the 1251 T1 sequences in the BraTS dataset containing whole tumor labels were used, so that our network can benefit from pretraining on the same MRI sequence and output, but on a larger dataset.
However, using pretrained weights for all the network layers may not be helpful due to the distinct domain gap between images and labels (adult vs. pediatric, glioblastoma vs. AVP/OPG). In the pretrained Swin transformer, features learned by later-stage convolutional neural network (CNN) layers are highly conditioned by the BraTS segmentation task, which could prevent a better fine-tuning on OPG. Thus, partially pretraining the transformer (only green blocks in Figure 1) should be more optimal. The effect of training from scratch, full pretraining, and partial pretraining is further investigated in the results section (Section III-A).
Another well-known challenge with transformer models is related to HR images and the gradient corruption problem in a later stage of training. There are several reasons for transformer models to be unstable compared to CNN, such as mixed precision, learning rate, and batch size [17]. In our segmentation task, the input patch size of a training image plays a critical role. A small patch size in each dimension can only cover a very small portion of the 3D T1 image, which leads to insufficient contextual information for the network to learn. On the other hand, the whole MR image cannot be fed into the network due to limitation of computational resources available in a typical research and clinical environment. However, it has been demonstrated that increasing patch size can improve the stability and performance [18]. Thus, we selected a patch size of 192 × 192 × 192 voxels to cover the largest feasible portion of the input images for training on a state-of-the-art GPU and to ensure stable training. In addition, the BraTS images were upsampled to match the resolution of our training images and the same patch size was applied. The SwinUNETR model was implemented in PyTorch-based framework MONAI (https://monai.io).
C. Radiomics and feature selection
Using the segmented AVP volume of interest, we extracted 293 radiomic features using the PyRadiomics package [19]. These features are divided into four groups: shape-based features (n=14) and three groups of intensity-based features, associated with the three image sequences (n=93 features per image sequence). Three additional numerical features were also included: brain volume, AVP volume normalized by brain volume (NormVol), and age of child. Thus, we ended up with a total of 296 imaging and demographic features. The brain volume was computed after skull-stripping.
The following univariate analyses were applied to identify 10 representative features that predict VA loss most accurately. Empirically, 10 features are sufficient for a sample size of 25 for a prediction task. We used leave-one-out cross-validation to investigate the stability of the feature selection process. Within each fold, univariate statistical tests (ANOVA F-test) were performed to rank the 296 features based on VA labels. After the selection of 10 highest-ranked features, strongly correlated features were removed and a smaller set of features was used to predict VA loss, as presented in Section III-B.
D. Visual acuity loss prediction
Following feature selection, a linear support vector machine (SVM) was used to predict VA loss (normal or abnormal vision). To prevent overfitting and estimate unbiased performance, leave-one-out cross-validation was also applied (25 folds). We used the Platt scaling for probability calibration to compute the receiver operating characteristic (ROC) curve. Univariate analysis, SVM and probability calibration were implemented in scikit-learn [20].
III. RESULTS
A. Anterior visual pathway segmentation
The dataset (n=75) with T1 images and labels was randomly split for 5-fold cross-validation into a training (80%) and a validation set (20%). The segmentation network was separately trained and validated within each fold, and metrics were aggregated. Randomly cropped 3D volumes were used to feed the network. The training was performed on an NVIDIA RTX A6000 48G GPU using batch size of 1 and 350 epochs.
The models were evaluated on three metrics. The first is Dice similarity coefficient (DSC). The second is normalized surface distance (NSD) [21], and its implementation in [22] was used. In our study, the AVP volume is an important clinical feature for patient prognosis, thus the metric evaluated was the relative volumetric error (RVE): RV E(S,T) = ǁVS − VTǁ/VT, where VS and VT are the volumes of the segmentation and ground truth, respectively.
Fig. 2 illustrates qualitative results of the segmented AVP. Table II summarizes quantitative results that demonstrate an overall high-quality segmentation. To put these results in perspective, in another independent study [5], the human inter-observer variability defined by average of DSC was reported to be 0.75 ± 0.06. Previous work on automatic AVP segmentation [24] showed a DSC of 0.602±0.201 and a RVE of 0.373 ± 0.293, but on a different and smaller dataset.
Fig. 2.
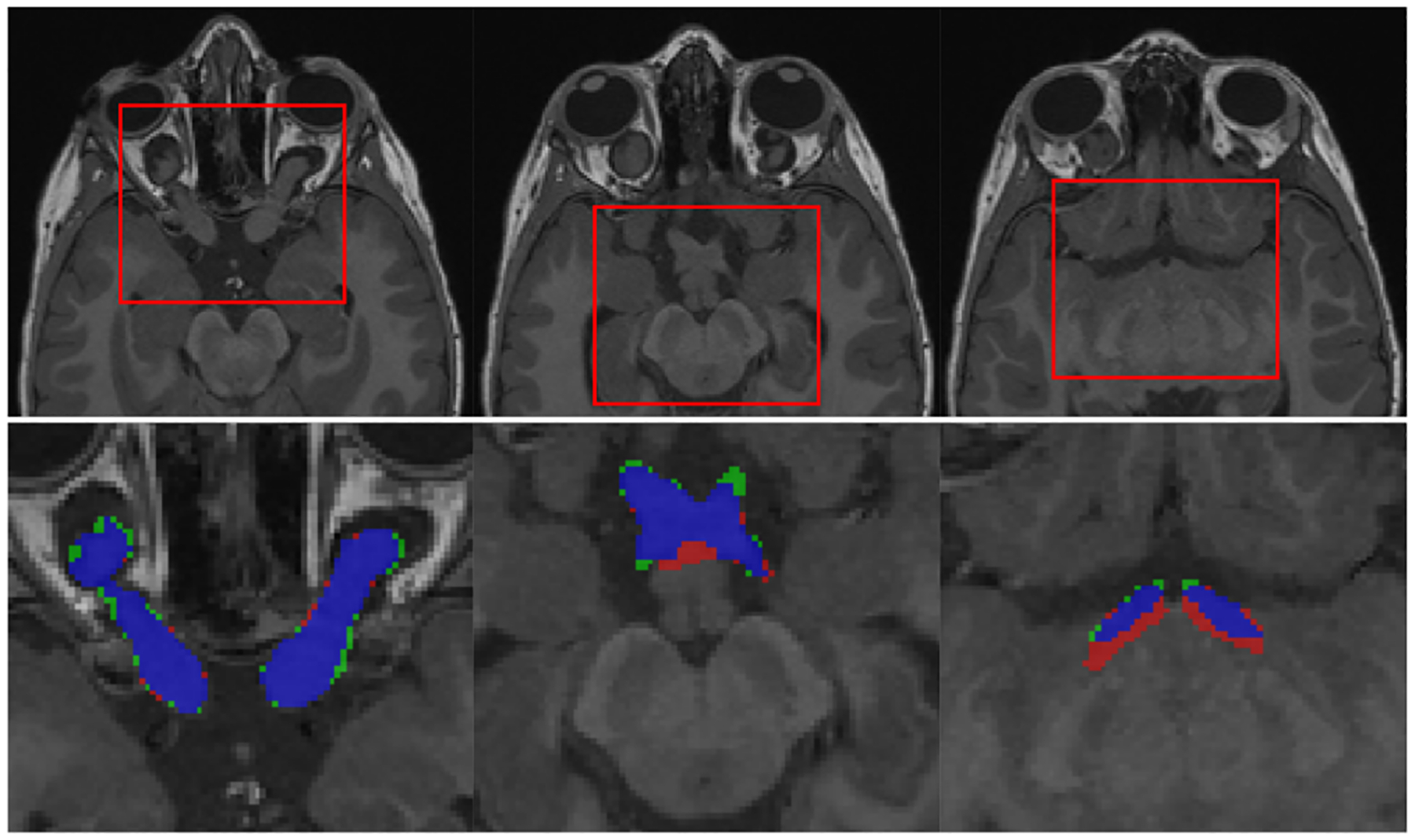
Qualitative results of AVP segmentation: selected case has large OPGs (AVP volume=5.095 ml, DSC=0.877). From left to right: 3 parts of AVP (optic nerves, chiasm and optic tracts) shown on different slices of T1 sequence. Labels: segmentation-red, ground truth-green, overlap-blue.
TABLE II.
Quantitative results of AVP segmentation. Baseline refers to a CNN model: SegResNet [23]. Wilcoxon paired test was used to compute p-value when comparing DSC of Baseline against three other models. Moreover, partially pretrained model outperformed fully pretrained one with a p-value of 0.005.
| Model (validation n=75) | DSC | NSD (mm) | RVE | DSC p-value | |
|---|---|---|---|---|---|
| Baseline | 0.776 ± 0.075 | 0.372 ± 0.186 | 0.182 ± 0.178 | N/A | |
| SwinUNETR | without pretraining | 0.786 ± 0.091 | 0.348 ± 0.208 | 0.174 ± 0.186 | < 0.001 |
| fully pretrained | 0.780 ± 0.074 | 0.368 ± 0.255 | 0.178 ± 0.154 | 0.083 | |
| partially pretrained | 0.791 ± 0.075 | 0.335 ± 0.179 | 0.176 ± 0.183 | < 0.001 | |
B. Statistical analysis of vision loss prediction
Based on the averaged cross-validated ranks of features, we identified the following 10 features to be the most predictive of vision loss. Table III summarizes these features (shape- and intensity-based) with associated p-values (rounded to 0.001) using ANOVA F-tests, the p-values show greater statistical significance of the observed difference in each feature between normal and abnormal vision outcome.
TABLE III.
Summary of top ranking features: rank, type, and p-values computed using ANOVA F-tests are presented.
| Rank | Type | p-value | Description |
|---|---|---|---|
| 1 | Shape | < 0.001 | sphericity |
| 2 | Shape | 0.001 | surface/volume ratio |
| 3 | Intensity | 0.003 | gray level run length non uniformity in T2 |
| 4 | Intensity | 0.003 | local homogeneity in T2 measured by inverse difference moment normalized |
| 5 | Shape | 0.003 | maximum 2D diameter |
| 6 | Shape | 0.004 | NormVol |
| 7 | Intensity | 0.004 | local homogeneity in T2 measured by invesrse difference normalized |
| 8 | Shape | 0.01 | volume measured from the mesh |
| 9 | Shape | 0.01 | volume measured from voxels |
| 10 | Intensity | 0.01 | gray level run length non uniformity in T1 |
Multiple features were highly correlated with each other and needed to be eliminated. After eliminating lower ranking features with Pearson correlation coefficient larger than 0.8, two final features were included in the classification, namely the sphericity and local homogeneity in T2 measured by inverse difference moment normalized. The quantitative prediction of our cross-validation resulted in an accuracy of 0.8 with 0.69 sensitivity, 0.92 specificity and ROC of 0.77.
IV. DISCUSSION
The segmentation results confirmed that applying directly pretrained model weights may not perform well due to a large domain gap between general adult brain tumors and pediatric cases with NF1-OPG. In some cases, the knowledge acquired on one dataset for one task cannot be transferred directly to another dataset for a different downstream task. Using partially randomly initialized weights in the pretrained model can be a remedy for a successful transfer learning.
To put our results in perspective, previous work [12] using manually segmented images has validated the association of normalized total AVP volume with VA loss. Our study confirms this conclusion. Sphericity, which was strongly correlated with volume, was the top-ranking feature in the classifier. In addition, we identified new intensity-based features from T2-weighted images that may improve the prediction of vision loss. Importantly, our approach is automatic and reproducible compared to the variable manual segmentations of the AVP, which are unreliable and impractical in clinical practice.
A limitation of this work is the size of the dataset, which was acquired at a single institution. This is a general limitation for the advancement of studies of rare diseases. Nevertheless, the quality of data curation was a great advantage in this study that demonstrated good potential to have an impact on the clinical outcome of children with NF1-OPG. In future, we will evaluate the framework on datasets from more institutions that enroll patients with NF1-OPG in clinical trials.
V. CONCLUSIONS
The decision on whether to start chemotherapy treatment for children with NF1-OPGs is critical and challenging due to the competing risks of treatment versus permanent vision loss caused by tumor progression. In this paper, we proposed a deep learning-based automatic framework to assist the clinical decision-making. The framework predicts vision loss from multi-sequence MRI by performing automatic segmentation of AVP using a transformer-based model, followed by radiomic features extraction and analysis. Through this work, we identified new shape- and intensity-based features of NF1-OPG that have the potential to accelerate and guide the treatment of affected children.
Clinical relevance—
Accurately determining which children with NF1-OPGs are at risk and hence require preventive treatment before vision loss remains challenging, towards this we present a fully automatic deep learning-based framework for vision outcome prediction, potentially permitting early treatment decisions.
ACKNOWLEDGMENT
This work was supported by The National Cancer Institute (UG3CA236536) and US Department of Defense (W81XWH1910376).
References
- [1].Avery RA, Fisher MJ, and Liu GT, “Optic pathway gliomas.,” J. Neuroophthalmol, vol. 31, no. 3, pp. 269–278, 2011. [DOI] [PubMed] [Google Scholar]
- [2].de Blank PMK et al. , “Optic pathway gliomas in neurofibromatosis type 1: An update: Surveillance, treatment indications, and biomarkers of vision,” J. Neuroophthalmol, vol. 37, no. Suppl 1, pp. S23–S32, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].van Timmeren JE et al. , “Radiomics in medical imaging—“how-to” guide and critical reflection,” Insights into Imaging, vol. 11, no. 91, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Madhogarhia R et al. , “Radiomics and radiogenomics in pediatric neuro-oncology: A review,” Neuro-Oncology Advances, vol. 4, no. 1, 05 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mansoor A et al. , “Deep learning guided partitioned shape model for anterior visual pathway segmentation,” IEEE Transactions on Medical Imaging, vol. 35, no. 8, pp. 1856–1865, 2016. [DOI] [PubMed] [Google Scholar]
- [6].Dosovitskiy A et al. , “An image is worth 16×16 words: Transformers for image recognition at scale,” in International Conference on Learning Representations, 2021. [Google Scholar]
- [7].Hatamizadeh A et al. , “Swin UNETR: Swin transformers for semantic segmentation of brain tumors in mri images,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Crimi A and Bakas S, Eds. 2022, pp. 272–284, Springer, Cham. [Google Scholar]
- [8].Tang Y et al. , “Self-supervised pre-training of swin transformers for 3d medical image analysis,” 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 20698–20708, 2022. [Google Scholar]
- [9].Hatamizadeh A et al. , “UNETR: Transformers for 3d medical image segmentation,” in IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 2022, pp. 574–584. [Google Scholar]
- [10].Menze BH et al. , “The multimodal brain tumor image segmentation benchmark (brats),” IEEE Transactions on Medical Imaging, vol. 34, no. 10, pp. 1993–2024, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bakas S et al. , “Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features,” Scientific Data, vol. 4, no. 1, pp. 170117, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Avery RA et al. , “Optic pathway glioma volume predicts retinal axon degeneration in neurofibromatosis type 1,” Neurology, vol. 87, no. 23, pp. 2403–2407, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pisapia JM et al. , “Predicting pediatric optic pathway glioma progression using advanced magnetic resonance image analysis and machine learning,” Neuro-Oncology Advances, vol. 2, no. 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Avants BB et al. , “The insight toolkit image registration framework,” Frontiers in Neuroinformatics, vol. 8, pp. 44, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baid U et al. , “The RSNA-ASNR-MICCAI brats 2021 benchmark on brain tumor segmentation and radiogenomic classification,” CoRR, vol. abs/2107.02314, 2021. [Google Scholar]
- [16].Jiang Z, Zhao C, Liu X, and Linguraru MG, “Brain tumor segmentation in multi-parametric magnetic resonance imaging using model ensembling and super-resolution,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Crimi A and Bakas S, Eds., Cham, 2022, pp. 125–137, Springer International Publishing. [Google Scholar]
- [17].Chen X, Xie S, and He K, “An empirical study of training self-supervised vision transformers,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021, pp. 9640–9649. [Google Scholar]
- [18].Hamwood J, Alonso-Caneiro D, Read SA, Vincent SJ, and Collins MJ, “Effect of patch size and network architecture on a convolutional neural network approach for automatic segmentation of OCT retinal layers,” Biomedical Optics Express, vol. 9, no. 7, pp. 3049–3066, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van Griethuysen JJM et al. , “Computational Radiomics System to Decode the Radiographic Phenotype,” Cancer Research, vol. 77, no. 21, pp. e104–e107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pedregosa F et al. , “Scikit-learn: Machine learning in Python,” Journal of Machine Learning Research, vol. 12, pp. 2825–2830, 2011. [Google Scholar]
- [21].Nikolov S et al. , “Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy,” arXiv:1809.04430, 2021. [DOI] [PMC free article] [PubMed]
- [22].Antonelli M et al. , “The medical segmentation decathlon,” arXiv:2106.05735, 2021. [DOI] [PMC free article] [PubMed]
- [23].Myronenko A, “3d mri brain tumor segmentation using autoencoder regularization,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. 2019, pp. 311–320, Springer, Cham. [Google Scholar]
- [24].Tor-Diez C et al. , “Unsupervised mri homogenization: Application to pediatric anterior visual pathway segmentation,” in Machine Learning in Medical Imaging, Liu M et al. , Ed. 2020, pp. 180–188, Springer, Cham. [DOI] [PMC free article] [PubMed] [Google Scholar]


