Abstract
Gv1 is a genetic locus that coordinately regulates the expression of multiple murine leukemia virus-related endogenous proviral sequences. A quantitative nuclease protection assay for typing Gv1 inheritance has been developed. Use of this assay demonstrates that Gv1 controls transcription of polytropic but not of modified polytropic endogenous proviruses. A combination of genetic techniques were used to map Gv1; these analyses demonstrate that Gv1 lies approximately 37 centimorgans from the centromeric end of mouse chromosome 13.
Throughout evolution, the genomes of higher eukaryotes have been colonized by retroviruses and retrovirus-like sequences (2). The best-studied elements are the murine leukemia virus (MLV)-related viruses of mice. Inbred strains of mice contain multiple copies of endogenous MLVs integrated into the genome as proviruses, where they behave as any other Mendelian gene (11). Four classes of endogenous MLV-related proviruses have been isolated, and they are classified according to the structures of their env genes (34). Infectious ecotropic viruses can infect murine cells only, xenotropic viruses can infect nonmurine but not murine cells, whereas the polytropic and modified polytropic classes can infect cells of either type (2). The host mechanisms controlling endogenous retroviral gene expression, however, remain unclear, although they are likely to be complex and involve interactions between host cell factors and long terminal repeats (LTRs).
The 129 mouse is unusual among the common inbred strains in that it does not produce infectious virions from endogenous MLV sequences. This mouse strain has therefore been used to model antigen accumulation resulting from the expression of endogenous retroviruses without the confounding effects due to transcription of novel proviruses resulting from in vivo virus replication. The GIX antigen was the first demonstration of a retroviral gene product in 129 mice. A cytotoxicity assay was established, in which antiserum prepared in inbred rats against a syngeneic leukemia induced by wild-type MLV lysed the thymocytes of some strains of mice such as 129 (designated GIX+) and not others (GIX−) (12, 31). It was later discovered that GIX was in fact a type-specific antigenic determinant on gp70, the major glycoprotein component of the MLV envelope (24, 37, 38). Genetic data obtained from crosses between the prototype GIX+ strain 129 and C57BL/6 (GIX−) mice revealed that two unlinked chromosomal genes, Gv1 and Gv2, are required for the expression of the GIX antigen on normal lymphoid cells. The Gv2 locus controls the GIX phenotype in a dominant fashion, while Gv1 is semidominant, with heterozygote mice (Gv1+/−) demonstrating a 50% reduction in GIX expression compared to homozygotes (Gv1+/+) (31). Attempts have been made to genetically map Gv1, yet these studies have yielded contradictory results (30). Gv2 has been mapped to chromosome 7, although the chromosome position is based on the results of only one cross in which Gv1 was also segregating (32). By serial backcrossing to C57BL/6, an inbred congenic (129 GIX−) strain was established that only differed from its partner (129 GIX+) at the Gv1 locus (29). Unfortunately, this strain is no longer available.
Studies of the congenic strain revealed that the GIX− phenotype was not simply due to a complete absence of viral antigens, since low levels of both gp70 and the major core protein p30 were detected (17). It was also shown that gp70 molecules found at different tissue sites of 129 GIX+ mice, absent or reduced in the congenic strain, have distinct tryptic peptide maps (8), suggesting that multiple proviral genes are being affected by Gv1. To examine the extent of this transcriptional control, Moloney MLV DNA was used to probe RNA purified from various tissues from the congenic strain and 129 GIX+ mice. It was concluded that a negative allele at the Gv1 locus correlated with reduced steady-state levels of retroviral RNA and that the two strains differ in the expression of the same complement of retroviral structural genes (18). Northern blotting experiments also showed that the expression of endogenous MLV structural genes appeared to be regulated in a tissue-specific manner (18). Nuclease protection with an LTR-specific probe confirmed that certain MLV transcripts were under-represented in GIX− tissues and that at least two LTR sequences were under control of Gv1 based on the pattern of protected products. It was therefore proposed that Gv1 encodes a trans-activating factor that coordinately regulates the expression of multiple proviral loci (19).
Despite this work, the mechanism by which Gv1 controls proviral expression and the role of Gv2 remain unclear. To investigate these questions and with the ultimate goal of positionally cloning Gv1, we have carried out a genetic analysis of Gv1. A reliable nuclease protection assay for the gene has been established, allowing a clearer definition of the targets for Gv1 action, an assessment of the relative roles of Gv1 and Gv2 in controlling proviral transcription, and the genetic mapping of Gv1.
MATERIALS AND METHODS
Mice.
F1 and backcross mice were bred from stocks of 129/SvEv, BALB/cJ, and C57BL/6J animals maintained in the Biological Services division of the National Institute for Medical Research. The 129 mice are Gv1+ Gv2+; the C57BL/6 mice are Gv1− Gv2−, and the BALB/c mice are Gv1− Gv2+ (30, 31).
Nucleic acid purification.
Genomic DNA was prepared from fresh or snap-frozen spleen tissue as previously described (1). Total RNA was prepared from fresh or snap-frozen thymus samples as previously described (3).
Nuclease protection.
Constructs containing proviral sequences from the polytropic and modified polytropic classes of MLV were subcloned from clones pMX40 (35) and pMX33 (34) for the synthesis of riboprobes. LTR sequences were amplified from these clones by using the primers PLO4 (5′-CGCAATTAACCCTCACTAAAGGG-3′) and PLO5 (5′-GCGAATTCATTGGCAGACAC-3′). The PCR products were digested with PstI and EcoRI and subcloned into the pGEM 3Zf(+) vector. A 424-bp polytropic-specific sequence was amplified from the env region of pMX40 by using the primers PLO33 (5′-GGACTAAGACTGTACCGATC-3′) and PLO34 (5′-CTTCGGACAGGGTCAGCTTG-3′). The amplified products were cloned into the pCRII-TOPO vector (Invitrogen) as described by the manufacturer. Riboprobes were synthesized by in vitro transcription in a reaction containing 250 ng of linearized template; 1× Transcription Optimized Buffer (Promega); 0.5 mM ATP, CTP, and GTP; 25 to 125 μM UTP; 10 mM dithiothreitol; 50 μCi of [α-32P]UTP (specific activity, 800 Ci/mmol); 10 U of rRNasin RNase inhibitor (Promega); and 20 U of T7 RNA polymerase (Promega). Riboprobes were gel purified on denaturing polyacrylamide gels, and nuclease protection was carried out according to the RPA II method (Ambion). The β-actin control plasmid supplied was also used for the synthesis of riboprobes. Supplementation of the in vitro transcription reaction with up to 500 μM UTP allowed riboprobes with lower specific activities to be made for reactions in combination with the env probe to avoid interference of the β-actin-protected fragment with smaller bands of interest. Typically, 25 μg total RNA was coprecipitated with 1 × 105 to 3 × 105 cpm of labelled MLV probe plus the β-actin riboprobe as an internal control. Hybridization and RNaseA-T1 digestion was carried out as described by the manufacturer. Protected products were resolved on 5% Sequagel denaturing polyacrylamide gels (National Diagnostics), dried onto filter paper, and analyzed by autoradiography or by using a phosphorimager.
Probe-specific signals were quantified by integrating pixel intensities over defined signal volumes by using ImageQuant software (Molecular Dynamics). Relative intensities were expressed as ratios [(probe signal − background)/(β-actin signal − background)] to correct for variation in RNA content, loading, or coprecipitation during the nuclease protection procedure.
Mapping by microsatellite amplification from pooled DNA samples.
Genome scanning was carried out by using the DNA pooling strategy described by Taylor et al. (36). DNA from individual backcross mice was prepared as described above, and each sample was diluted to 25 ng/μl. Pools were made by mixing DNA from 50 animals typed by nuclease protection. PCR amplification of simple sequence repeats was carried out essentially as previously described (36), although the reaction volume was reduced to 15 μl and the optimal MgCl2 concentrations were determined empirically where necessary. Eighty-five microsatellite primer pairs, which span 97% of the genome, were purchased from Research Genetics, Inc. PCR products were run on 3% NuSeive (Flowgen) 1% agarose gels stained with ethidium bromide. For those products that could not be resolved on conventional gels, 10 pmol of one primer was end labelled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs), and 0.1 pmol was added per reaction to the standard PCR mix. PCR products were resolved on 5% denaturing polyacrylamide gels.
REVEAL-PCR.
Repeat element-viral element amplified locus PCR (REVEAL-PCR) was carried out essentially as previously described (16). First, 10 pmol of an intracisternal A-type particle (IAP)-specific oligonucleotide (JS139 or JS140) was end labelled with [γ-32P]ATP by using T4 polynucleotide kinase and combined in a PCR reaction with one of three B1 or B2 short interspersed element (SINE) primers (JS134, JS135, or JS136). Labelled PCR products were resolved on 5% denaturing polyacrylamide gels which were dried onto filter paper and exposed to X-ray film overnight.
REVEAL products of interest (REVEAL-1 and REVEAL-2) were excised from dried polyacrylamide gels and eluted in RPA II probe elution buffer (Ambion) overnight at 37°C. The products were reamplified with the same IAP-SINE repeat primer pair used previously without the addition of an end-labelled oligonucleotide, and the PCR products were cloned into the pCRII-TOPO vector as described by the manufacturer (Invitrogen). Inserts were sequenced by standard methods on the 377 automated sequencer (Perkin-Elmer) for internal primer design.
Radiation hybrid mapping.
PCR analysis of the T31 mouse radiation hybrid panel (Research Genetics) was carried out by using the REVEAL-1-specific primers JS139 (5′-GTTTACCACTTAGAACACAG-3′) and PLO16 (5′-GGAAATAGTAAGTCACACC-3′). First, 20 ng of all 100 hybrid clone DNA samples were amplified in 15 μl of PCR reactions containing 1× PCR Buffer II (Perkin-Elmer), 2.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate (dNTP), 2 pmol of primers, and 0.5 U of AmpliTaq DNA polymerase (Perkin-Elmer). PCR conditions were identical to the REVEAL-PCR protocol, and all reactions were carried out in triplicate. Based on the analysis of products on 2% agarose gels, the hybrid clones were scored according to the presence or absence of a mouse-specific band. The screening results were processed by the software Map Manger QT (20) at The Jackson Laboratory.
Microsatellite mapping.
Markers were chosen based on polymorphism between the 129 and the C57BL/6 or BALB/c strains where data were available. The sequence of primers for microsatellites in the region of interest were obtained from The Jackson Laboratory Mouse Genome Informatics World Wide Web site (23). DNA samples (50 ng) were amplified in 15-μl PCR reactions containing 1× PCR buffer I (Perkin-Elmer), 0.2 mM concentrations of each dNTP, 2 pmol each primer, and 0.2 U of AmpliTaq DNA polymerase. The initial denaturation step was 2 min at 94°C, followed by 45 cycles of denaturation for 20 s at 94°C, annealing for 30 s at 55°C, and extension for 30 s at 72°C before a final 10-min extension at 72°C. PCR products were resolved on conventional or polyacrylamide gels as described above. Linkage and quantitative trait locus (QTL) data were analyzed by using the Map Manager QT program (20).
RESULTS
Characterization of Gv1 controlled transcription.
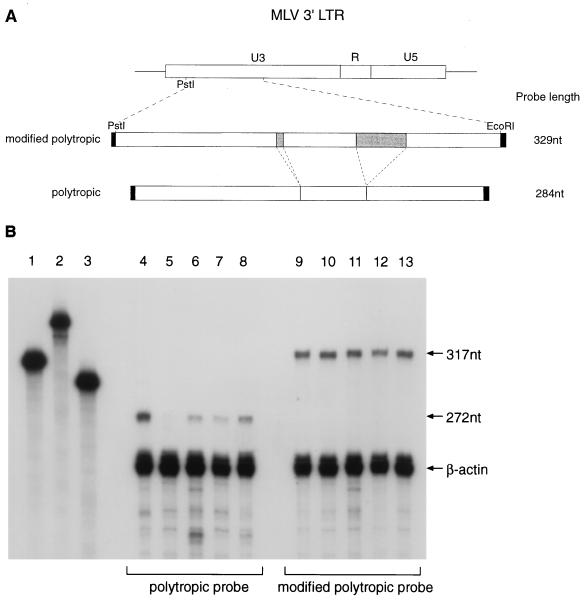
To allow studies of the inheritance of Gv1, we set out to devise a reliable nuclease protection assay for quantifying proviral expression. U3 probes were designed to allow the independent analysis of both the polytropic and modified polytropic classes of endogenous MLV that are present in multiple copies in all inbred strains of mice. Previous studies (34) have shown that the LTRs of modified polytropic proviruses contain two inserts, of 5 and 42 nucleotides, compared to polytropic proviruses in the U3 region (Fig. 1A). Probes to this region will therefore yield clearly distinguishable full-length protected fragments in nuclease protection experiments. Total thymus RNA from 129, C57BL/6, BALB/c, and F1 (129 × C57BL/6 and 129 × BALB/c) mice were hybridized to the polytropic and modified polytropic LTR probes by using β-actin as an internal control. Quantitative studies with the polytropic probe revealed that the full-length protected product of 272 nucleotides was clearly reduced in the Gv1− strains compared to 129 (Fig. 1B, compare lane 4 to lanes 5 and 6). Expression of this LTR sequence was consistently lower in the C57BL/6 strain than in the BALB/c strain, and this was reflected in the F1 mice, which showed an intermediate level of proviral expression compared to the 129 (Gv1+) animals. These levels of expression are consistent with the semidominant nature of Gv1 inheritance. The full-length protected fragment, with the modified polytropic probe, did not differ in intensity between the Gv1+ and Gv1− strains (Fig. 1B, lanes 9 to 13), implying that the Gv1 locus does not affect the expression of the modified polytropic class of endogenous MLV.
FIG. 1.
Quantitative nuclease protection demonstrates differential expression of the polytropic class of endogenous MLV LTR sequences in Gv1− strains of mice. (A) The MLV LTR region cloned for the synthesis of riboprobes. Shaded areas indicate two regions, of 5 and 42 bp, of the modified polytropic sequence absent in the polytropic clone. (B) Quantitative nuclease protection of thymus total RNA with an LTR probe and a β-actin internal control probe. Lanes 4 to 8, protected products with polytropic probe; lanes 9 to 13, modified polytropic probe. The strains of mice analyzed plus the relative intensities of the full-length protected products, corrected for β-actin are as follows: lane 4, 129, 0.118; lane 5, C57BL/6, 0.006; lane 6, BALB/c 0.032; lane 7, 129 × C57BL/6, 0.030; lane 8, 129 × BALB/c, 0.064; lane 9, 129, 0.108; lane 10, C57BL/6, 0.128; lane 11, BALB/c, 0.112; lane 12, 129 × C57BL/6, 0.109; and lane 13, 129 × BALB/c, 0.121. The undigested β-actin, modified polytropic, and polytropic probes (lanes 1, 2, and 3, respectively) are longer than the full-length protected products due to retention of plasmid sequence from the in vitro transcription reaction (regions shaded black in panel A).
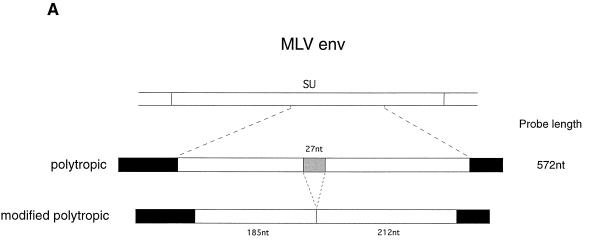
To further investigate the nature of Gv1 transcriptional control, nuclease protection experiments were carried out with an env probe designed to allow simultaneous measurement of both polytropic and modified polytropic proviral expression. These two classes of provirus can be distinguished by a 27-nucleotide deletion in modified polytropic viruses compared to polytropic viruses (34). By using a polytropic probe, polytropic transcripts will therefore protect a fragment of 424 nucleotides, whereas modified polytropic sequences will protect two fragments of 212 and 185 nucleotides (Fig. 2A). Figure 2B shows the result of such an experiment to examine env gene expression in thymus. Gv1− strains express less polytropic env mRNA than 129 as judged by the intensity of the 424-nucleotide full-length protected fragment. The patterns of expression observed are comparable to the results obtained when using the polytropic LTR probe. By contrast, the intensities of the 212- and 185-nucleotide protected fragments do not vary between the different strains. These results confirm that expression of the polytropic but not of the modified polytropic class of endogenous MLV is regulated by Gv1.
FIG. 2.
Quantitative nuclease protection demonstrates differential expression of the polytropic class of endogenous MLV env sequences in Gv1− strains of mice. (A) The region of MLV env cloned for the synthesis of riboprobes. The shaded area indicates a 27-bp region absent in the modified polytropic clone. Solid black regions indicate plasmid sequence retained in the probes from the in vitro transcription reaction. (B) Quantitative nuclease protection of thymus total RNA with a polytropic env probe and a β-actin internal control probe. The strains of mice analyzed and the relative intensities of the full-length protected products, corrected for β-actin are as follows: lane 1, 129, 0.214; lane 2, C57BL/6, 0.013; lane 3, BALB/c, 0.032; lane 4, 129 × C57BL/6, 0.096; and lane 5, 129 × BALB/c, 0.123. The relative intensities of the 212- and 185-nucleotide modified polytropic-specific fragments are as follows (respectively): lane 1, 0.086 and 0.110; lane 2, 0.080 and 0.114; lane 3, 0.075 and 0.108; lane 4, 0.091 and 0.122; and lane 5, 0.093 and 0.127. No β-actin probe was added to the 129 × C57BL/6 RNA sample in lane 6.
Genetic mapping of Gv1.
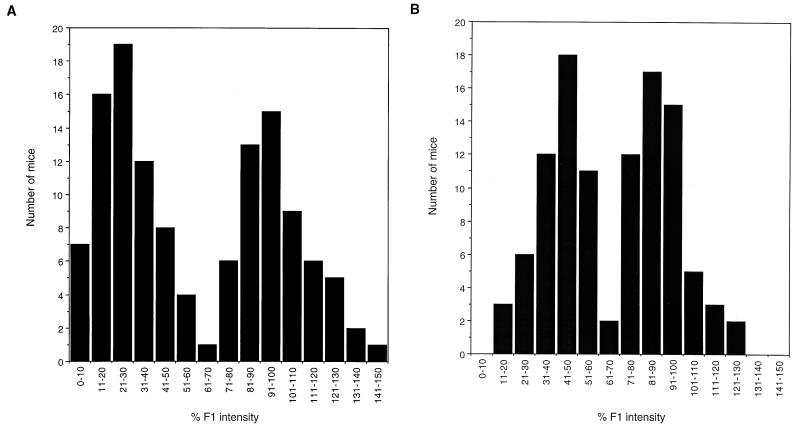
C57BL/6 and BALB/c backcrosses were set up to follow the segregation of Gv1. Nuclease protection data was collected from 230 animals from the backcross populations by using the polytropic LTR riboprobe (Table 1). In each set of reactions, the corrected intensity of the full-length protected product was compared to a reaction by using RNA from the appropriate F1 mouse. If the band intensity was over 75% of the F1 value, the mouse was classified as heterozygous at the Gv1 locus, whereas Gv1− animals were those that demonstrated intensity values of <55%. An example from the C57BL/6 backcross is shown in Fig. 3. The nuclease protection data from both backcross populations show a bimodal distribution (Fig. 4). Of the 230 animals typed for Gv1 by using the assay, 111 (48%) carry the 129 allele, which is a finding consistent with the segregation of a single gene. These results also indicate that Gv2 does not play a role in regulating proviral expression since essentially equivalent distributions were seen whether (129 × C57BL/6) or not (129 × BALB/c) the gene was segregating in the cross.
TABLE 1.
Nuclease protection data inferring Gv1 genotype from two backcross populations
| Gv1 genotype | C57BL/6 backcross | BALB/c backcross | Total |
|---|---|---|---|
| Gv1−/− | 66 | 50 | 116 |
| Gv1+/− | 57 | 54 | 111 |
| Unknowna | 1 | 2 | 3 |
| Total no. of animals | 124 | 106 | 230 |
Values from these animals lay exactly at the cutoff point between Gv1−/− and Gv1+/−.
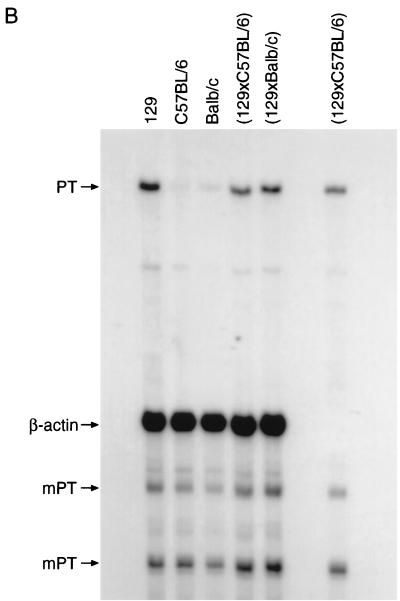
FIG. 3.
Nuclease protection of C57BL/6 backcross mice indicates Gv1 genotype. Thymus total RNA from the mouse strains indicated was analyzed by quantitative nuclease protection by using the polytropic LTR riboprobe and a β-actin internal control probe. Relative intensity of the 272-nucleotide fragments corrected for β-actin as follows: lane 1, 0.225; lane 2, 0.065; lane 3, 0.009; lane 4, 0.076; lane 6, 0.016; lane 6, 0.005; lane 7, 0.056; lane 8, 0.009; lane 9, 0.066; lane 10, 0.018; and lane 11, 0.015. Mice were classified as Gv1+/− (lanes 4, 7, and 9) by similarity (>75%) of the band intensity to RNA from the F1 animal (lane 2) or as Gv1−/− (lanes 5, 6, 7, 10, and 11) if the value obtained was under 55% of the F1 protected product intensity.
FIG. 4.
A single locus is responsible for the proviral expression levels observed in two backcross populations. A distribution of the quantitative nuclease protection data with the polytropic LTR riboprobe from the two backcross populations indicated is shown. Data are shown as a percentage of the relative intensity of the full-length protected product compared to RNA from F1 mice. The bimodal distribution of the data suggests that a single locus is controlling the proviral expression levels observed.
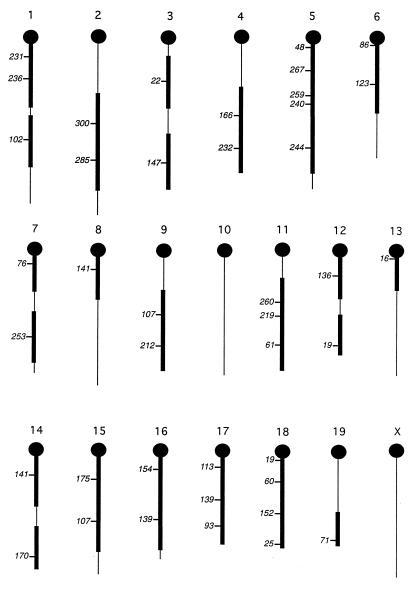
To map Gv1 in the backcross populations, a genome scan with 85 microsatellite markers was carried out based on the method of Taylor et al. (36). The phenotypic pooling method relies on the detection of markers in a segregating population, grouped together according to their phenotype at a target locus. Linked markers are identified by a change in the relative intensity of microsatellite PCR products from each DNA pool. Linkage can be identified with a 20-centimorgan (cM) distance from each marker typed. Linkage to Gv1 could be excluded from 74% of the genome from the data obtained from 41 of the primer pairs (Fig. 5). The remaining microsatellites could not be typed due to failure of one or both of the alleles to amplify or lack of polymorphism between the strains (data not shown). It was therefore decided to attempt REVEAL-PCR (16) as an alternative mapping method.
FIG. 5.
Linkage of Gv1 is excluded from 74% of the mouse genome. Chromosomal locations of the 41 microsatellite markers successfully amplified and analysed by the DNA pooling method are shown. The Mit number (6) of each marker is shown, and the centromeric ends of the chromosomes are indicated by black circles. Linkage to Gv1 has been excluded from the regions represented by thickened lines, which correspond to 74% of the mouse genome.
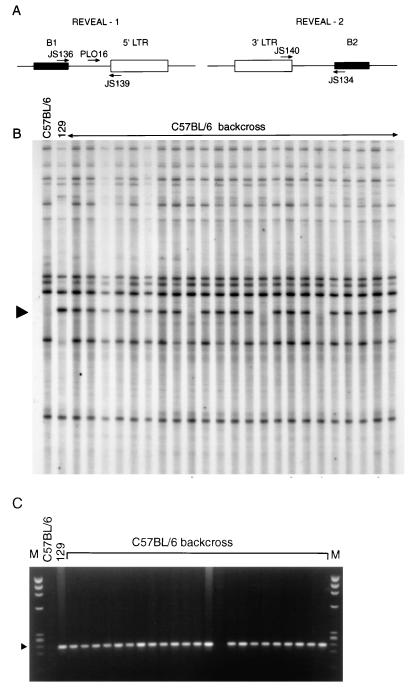
This multilocus mapping approach examines the inheritance of IAP elements and their proximity to SINEs in a PCR-based assay (Fig. 6A). To find a marker linked to Gv1, REVEAL products from 23 C57BL/6 backcross mice typed as heterozygous at the Gv1 locus were generated with all six combinations of IAP and SINE repeat primers (16). PCR products of 129 origin that were present in most of the backcross DNA samples were considered as possible mapping candidates. Of approximately 30 REVEAL products derived from the JS140/134 primer pair, one band (REVEAL-2) was present in 20 of the 23 animals, indicating linkage (Fig. 6B). Closer linkage was demonstrated by a product amplified by using the JS139/136 primer pair, REVEAL-1, with only one backcross mouse showing the absence of the band at approximately 360 bp. Both REVEAL products were excised from polyacrylamide gels, reamplified, cloned, and sequenced. Internal primers to REVEAL-1 were designed, allowing animals to be screened for the presence of this marker by PCR on conventional gels (Fig. 6C). Linkage was confirmed when REVEAL-1 was amplified from the remaining 93 C57BL/6 backcross progeny, with only seven further animals showing apparent (see below) recombination between the marker and Gv1. Linkage was also established to the REVEAL-1 marker in the BALB/c backcross (15 apparent recombinants in 100 animals tested), confirming that the same locus was responsible for the proviral expression phenotype observed.
FIG. 6.
Two IAP proviral markers are linked to Gv1. (A) Relative positions of the primer pairs used to generate two sets of REVEAL-PCR products. (B) REVEAL-PCR products amplified with JS140 and JS134, in the presence of end-labelled primer JS140, from the genomic DNA samples indicated. The arrow indicates the position of REVEAL-2, which is absent in C57BL/6 mice but was amplified from 129 mice. Of 23 backcross animals predicted to be heterozygous at the Gv1 locus, 20 show the presence of this band, indicating linkage. (C) Genomic DNA from the same 23 C57BL/6 backcross mice as in panel B was amplified by using primers JS139 and PLO-16 derived from the marker REVEAL-1. Products were run on a 2% agarose gel against the φx174/HaeIII marker (M). The arrow indicates the position of the D13Nimr1 product at 229 bp. Of 23 backcross animals, 22 show the presence of the band, indicating linkage.
To map Gv1, it was first necessary to identify the chromosomal position of the REVEAL-1 marker. For this purpose, we used the technique of whole genome radiation hybrid mapping. The T31 mouse whole-genome panel comprises 100 hybrid cell lines derived by fusing irradiated 129 embryonic stem cells with cells from a hamster fibroblast line (22). Different hybrids retain different fragments of mouse chromosomes, and these have been extensively characterized by microsatellite mapping (13). Scoring REVEAL-1 on the T31 radiation hybrid panel showed that it mapped to mouse chromosome 13. The marker, now termed D13Nimr1, was placed between the microsatellites D13Mit157 and D13Mit66 with LOD scores of 11.8 and 9.0, respectively (data not shown). This region of chromosome 13 is part of the fraction of the mouse genome which could not be excluded by the pooling method (Fig. 5).
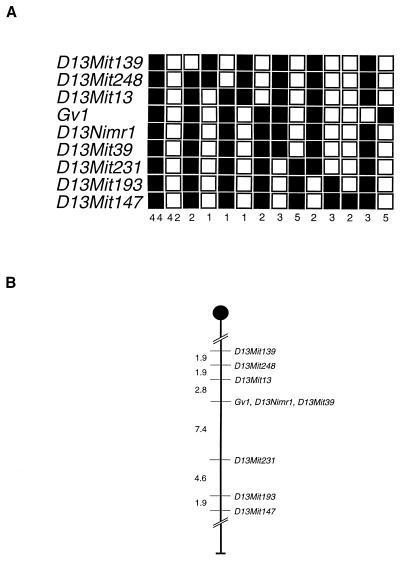
To characterize the map position of Gv1, microsatellites flanking D13Nimr1 were tested on the C57BL/6 backcross DNA pools. Of the 32 chromosome 13 markers analyzed that have been mapped to between 32 and 49 cM, only 7 appeared to be polymorphic between the 129 and C57BL/6 strains. Linkage to Gv1 was confirmed by using this method, and all 116 backcross mice were consequently screened with the same seven markers. The haplotypes of the animals are shown in Fig. 7A.
FIG. 7.
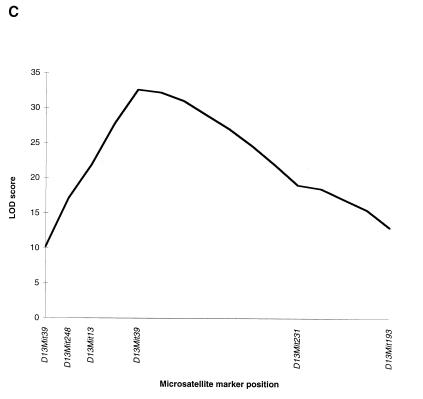
Gv1 maps to chromosome 13. (A) Haplotype analysis of the 116 C57BL/6 backcross progeny typed for markers flanking the REVEAL-PCR product, REVEAL-1. Each column represents the allele inherited from the (129 × C57BL/6)F1 parent. The number of offspring inheriting each allele is indicated at the bottom of each column; shaded boxes indicate the presence of a 129 allele, and the open boxes indicate the presence of a C57BL/6 allele. (B) Partial linkage map of chromosome 13. The relative position of Gv1 with respect to the linked markers indicated was determined by using the Map Manager QT program, without taking into account the double-recombinant animals. Map distances (in cM) are indicated on the left. (C) QTL analysis of all 116 backcross animals based on proviral expression levels observed.
Eight mice in the C57BL/6 backcross appeared to be recombinant between Gv1 and both the closest flanking markers, D13Mit13 and D13Mit39. These double recombinants in a very short genetic interval are indicative of mis-typing in the assay used to genotype the backcross animals (26). The proviral expression levels of these eight mice, based on the nuclease protection data, lie towards the center of the bimodal distribution, between 60 and 80% of the heterozygote figure. This was also the case for the 15 apparent recombinants from the BALB/c cross. This suggests that errors in the quantification method are large enough to account for the incorrect genotyping of a small percentage of animals. Assuming this to be the case, Gv1 cosegregates with both D13Nimr1 and D13Mit39, placing the gene at 37 cM from the centromere (Fig. 7B). To confirm linkage to this region of chromosome 13, the nuclease protection and backcross mapping data from all 116 mice of the C57BL/6 backcross, including the 8 animals which we believe were mis-typed, were combined and analyzed as a QTL (Fig. 7C). The highest LOD score was obtained around the chromosomal position of D13Nimr1 and D13Mit39, suggesting strong linkage between a locus at approximately 37 cM on mouse chromosome 13 and the proviral expression levels observed in the backcross population, thereby confirming the map location derived from the haplotype data.
DISCUSSION
Despite intensive studies, we know little about the factors controlling expression of endogenous retroviruses (2). Elucidation of the mode of action of Gv1 might be expected to shed light on these processes. The genetic mapping of Gv1 reported here represents the first step towards the eventual cloning and characterization of the gene.
Throughout this study we have assumed that the gene we have mapped to chromosome 13 does indeed correspond to the locus controlling GIX antigen expression that was originally described. Of particular concern was the possibility that we might have been following merely the inheritance of a single provirus rather than the trans-acting Gv1. Unfortunately, the congenic GIX− strain is no longer available, so direct tests for the presence of chromosome 13 markers of C57BL/6 origin in the congenic strain are not possible. Nevertheless, our assumption of identity does not seem unreasonable since the properties of our gene appear concordant with those of Gv1.
GIX was described as a thymus antigen expressed by 129 mice but not by BALB/c or C57BL/6 mice (31). Inheritance was semidominant. We have monitored the thymic expression of polytropic MLV sequences in crosses between 129 (positive) and BALB/c or C57BL/6 (negative), and the transcription levels are consistent with the inheritance of a single, semidominant gene. The protected products that appeared absent in the thymus RNA from congenic 129 mice in the experiments of Levy et al. (19) are in fact consistent with the use of a modified polytropic probe for detecting polytropic LTR sequences. We note that one polytropic provirus, Pmv41 (10), from DBA mice, has been shown to lie within the interval containing the gene we have mapped. However, this provirus is not present in 129 mice, and no other polytropic proviruses have been mapped to this region of chromosome 13 (9). Hybridization studies with the polytropic-specific oligonucleotide probe JS5 (35) have confirmed the absence of Pmv41 in 129/SvEv mice (33). Analysis of a small panel of backcross mice revealed no linkage between 129 proviruses and polytropic virus expression (33), a result consistent with the proposition that Gv1 encodes a trans-acting gene product.
The observation that polytropic but not modified polytropic sequences are responsive to Gv1 are consistent with the predicted GIX phenotypes of these classes of MLV sequence. The appearance of the GIX antigen is correlated with the absence of a glycosylation signal in the env gene of ecotropic MLV (7); sequence data (34) from the equivalent region of polytropic and modified polytropic MLV reveals that the former, but not the latter, class of sequence also lacks a glycosylation signal. Polytropic MLV (bases 1277 to 1285 [34]) is predicted to encode Asp-Leu-Thr, whereas modified polytropic MLV (bases 1250 to 1258) encodes Asn-Leu-Thr.
The expression of the GIX antigen is controlled by two genes, Gv1 and Gv2, and it has been hypothesized that both genes play a role in proviral transcription. Although there is a slight difference in the distribution of the nuclease protection data from the C57BL/6 and BALB/c backcrosses, this study suggests otherwise. It has been shown here that only Gv1 has any transcriptional control activity, as a bimodal distribution was observed in both backcross populations. This suggests that Gv2 must act posttranscriptionally and may play a role in MLV surface protein expression.
One of the major functions of the LTR is to regulate the transcription of retroviral genes by providing signals recognized by cellular transcriptional control machinery. Positive and negative regulatory elements have been identified in the LTR, and many transcription factors are known to bind to the U3 region directly. The factor recognition sequences are usually short and may be repeated several times in each LTR, and the signals themselves have also been shown to have different effects in different cell types (4). Since polytropic and modified polytropic MLVs differ in their responsiveness to Gv1, U3 sequences polymorphic between the two classes may therefore be a target for such a trans-activating factor. Attempts will be made to identify possible binding sites by using a gel mobility assay; a factor present in the 129 mice but not in the C57BL/6 or BALB/c strains that binds to polytropic-specific U3 probes could be a good candidate for the gene product of Gv1.
Based on the map position of Gv1, we have attempted to identify possible candidate genes within the interval between D13Mit13 and D13Mit231 (28). A number of potential DNA binding factors, for example, a number of zinc-finger-related genes, Zfp68-rs1, Zfp71-rs1, and Zfp85-rs1 (21), as well as sequences related to the gene encoding the high-mobility-group chromatin protein, Hmg-1 (25), were identified; however, none provides a compelling candidate for Gv1. Possibly more interesting are two genes with trans effects, mdac, modifier of Dac, (15) and Rsl, regulator of sex-limited protein, Slp (14). The latter case is particularly intriguing because a potential target for Rsl is the LTR of an ancient endogenous retrovirus which imposes androgen control over the expression of Slp (27). We are currently refining the map position of Gv1 with the intention of cloning the gene by using the positional or positional candidate approach (5).
ACKNOWLEDGMENTS
We thank Lucy Rowe (The Jackson Laboratory, Bar Harbor, Maine) for advice with the radiation hybrid mapping and Paul Le Tissier for valuable discussions.
This work was supported by the United Kingdom Medical Research Council.
REFERENCES
- 1.Aljanabi S M, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia: Lippincott-Raven Publishers; 1996. .:pp 1767–1847. [Google Scholar]
- 5.Collins F S. Positional cloning moves from the perditional to the traditional. Nat Genet. 1995;9:347–350. doi: 10.1038/ng0495-347. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O’Connor T J, Evans C A, DeAngelis M A, Levinson D M, Kruglyak L, Goodman N, Copeland N G, Jenkins N A, Hawkins T L, Stein L, Page D C, Lander E S. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 7.Donis-Keller H, Rommelaere J, Ellis R W, Hopkins N. Nucleotide sequences associated with differences in electrophoretic mobility of envelope glycoprotein gp70 and with GIX antigen phenotype of certain murine leukemia viruses. Proc Natl Acad Sci USA. 1980;77:1642–1645. doi: 10.1073/pnas.77.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder J H, Jensen F C, Bryant M L, Lerner R A. Polymorphism of the major envelope glycoprotein (gp70) of murine C-type viruses: virion associated and differentiation antigens encoded by a multi-gene family. Nature. 1977;267:23–28. doi: 10.1038/267023a0. [DOI] [PubMed] [Google Scholar]
- 9.Frankel W N, Coffin J M, Cote M, Seyfried T N, Rise M, Rajan T V, Nelson F K, Selsing E, Gerstein R. Backcross data for endogenous Mpmv, Pmv, and Xmv proviruses. Mouse Genome. 1991;91:266–270. [Google Scholar]
- 10.Frankel W N, Stoye J P, Taylor B A, Coffin J M. Genetic identification of endogenous polytropic proviruses by using recombinant inbred mice. J Virol. 1989;63:3810–3821. doi: 10.1128/jvi.63.9.3810-3821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel W N, Stoye J P, Taylor B A, Coffin J M. A genetic linkage map of endogenous murine leukemia viruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geering G, Old L J, Boyse E A. Antigens of leukemias induced by naturally occurring murine leukemia virus: their relation to the antigens of Gross virus and other murine leukemia viruses. J Exp Med. 1966;124:753–772. doi: 10.1084/jem.124.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson Laboratory RH Panel. 23 February 1999, revision date. [Online.] http://www.jax.org/resources/documents/cmdata/rhmap/rh.html. [1 March 1999, last date accessed.]
- 14.Jiang P P, Frederick K, Hansen T H, Miller R D. Localization of the mouse gene releasing sex-limited expression of Slp. Proc Natl Acad Sci USA. 1996;93:913–917. doi: 10.1073/pnas.93.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson K R, Lane P W, Ward-Bailey P, Davisson M T. Mapping the mouse dactylaplasia mutation, Dac, and a gene that controls its expression, mdac. Genomics. 1995;29:457–464. doi: 10.1006/geno.1995.9981. [DOI] [PubMed] [Google Scholar]
- 16.Kaushik N, Stoye J P. Intracisternal A-type particle elements as genetic markers: detection by repeat element viral element amplified locus-PCR. Mamm Genome. 1994;5:688–695. doi: 10.1007/BF00426074. [DOI] [PubMed] [Google Scholar]
- 17.Lerner R A, Wilson C B, Del Villano B C, McConahey P J, Dixon F J. Endogenous oncornaviral gene expression in adult and fetal mice: quantitative, histologic, and physiologic studies of the major viral glycoprotein, gp70. J Exp Med. 1976;143:151–166. doi: 10.1084/jem.143.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy D E, Lerner R A, Wilson M C. A genetic locus regulates the expression of tissue-specific mRNAs from multiple transcription units. Proc Natl Acad Sci USA. 1982;79:5823–5827. doi: 10.1073/pnas.79.19.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy D E, Lerner R A, Wilson M C. The Gv-1 locus coordinately regulates the expression of multiple endogenous murine retroviruses. Cell. 1985;41:289–299. doi: 10.1016/0092-8674(85)90082-0. [DOI] [PubMed] [Google Scholar]
- 20.Map Manager QT. 17 February 1999, revision date. [Online.] http://mcbio.med.buffalo.edu/mmQT.html. [1 March 1999, last date accessed.]
- 21.Marine J C, Gilbert D J, Bellefroid E J, Martial J A, Ihle J N, Copeland N G, Jenkins N A. Chromosomal location of fifteen unique mouse KRAB-containing zinc finger loci. Mamm Genome. 1996;7:413–416. doi: 10.1007/s003359900123. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy L C, Terrett J, Davis M E, Knights C J, Smith A L, Critcher R, Schmitt K, Hudson J, Spurr N K, Goodfellow P N. A first-generation whole genome-radiation hybrid map spanning the whole genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouse Genome Informatics, The Jackson Laboratory. 1996, copyright date. [Online.] http://www.informatics.jax.org/index.html. [1 March 1999, last date accessed.]
- 24.Obata Y, Ikeda H, Stockert E, Boyse E A. Relation of GIX antigen of thymocytes to envelope glycoprotein of murine leukemia virus. J Exp Med. 1975;141:188–197. doi: 10.1084/jem.141.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauken C M, Nagle D L, Bucan M, Lo C W. Molecular cloning, expression analysis, and chromosomal localization of mouse Hmg1-containing sequences. Mamm Genome. 1994;5:91–99. doi: 10.1007/BF00292334. [DOI] [PubMed] [Google Scholar]
- 26.Paulson R F, Bernstein A. A genetic linkage map of the mouse chromosome 9 region encompassing the Friend virus susceptibility gene 2. Mamm Genome. 1998;9:381–384. doi: 10.1007/s003359900774. [DOI] [PubMed] [Google Scholar]
- 27.Stavenhagen J B, Robins D M. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell. 1988;55:247–254. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson D A, Lueders K K. Mouse Chromosome 13. Mamm Genome. 1998;8:S258–S274. doi: 10.1007/s003359900658. [DOI] [PubMed] [Google Scholar]
- 29.Stockert E, Boyse E A, Obata Y, Ikeda H, Sarkar N H, Hoffman H A. New mutant and congenic mouse stocks expressing the murine leukemia virus-associated thymocyte surface antigen GIX. J Exp Med. 1975;142:512–517. doi: 10.1084/jem.142.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stockert E, Boyse E A, Sato H, Itakura K. Heredity of the GIX thymocyte antigen associated with murine leukemia virus: segregation data simulating genetic linkage. Proc Natl Acad Sci USA. 1976;73:2077–2081. doi: 10.1073/pnas.73.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stockert E, Old L J, Boyse E A. The GIX system. A cell surface allo-antigen associated with murine leukemia virus: implications regarding chromosomal integration of the viral genome. J Exp Med. 1971;133:1334–1355. doi: 10.1084/jem.133.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stockert E, Sato H, Itakura K, Boyse E A, Old L J, Hutton J J. Location of a second gene required for expression of the leukemia-associated mouse antigen GIX. Science. 1972;178:862–863. doi: 10.1126/science.178.4063.862. [DOI] [PubMed] [Google Scholar]
- 33.Stoye, J. P., and P. L. Oliver. Unpublished data.
- 34.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia viruses: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoye J P, Fenner S, Greenoak G E, Moran C, Coffin J M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 36.Taylor B A, Navin A, Phillips S J. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 37.Tung J-S, Fleissner E, Vitetta E S, Boyse E A. Expression of murine leukemia virus envelope glycoprotein gp69/71 on mouse thymocyte. Evidence for two structural variants distinguished by presence vs. absence of GIX antigen. J Exp Med. 1975;142:518–523. doi: 10.1084/jem.142.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung J-S, Vitetta E S, Fleissner E, Boyse E A. Biochemical evidence linking the GIX thymocyte surface antigen to the gp69/71 envelope glycoprotein of murine leukemia virus. J Exp Med. 1975;141:198–205. doi: 10.1084/jem.141.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]