Abstract
Circular RNAs (circRNAs) constitute a class of endogenous non-coding RNAs (ncRNAs) that lack a 5’-ended cap and 3’-ended poly (A) tail and form a closed ring structure with covalent bonds. Due to its special structure, circRNA is resistant to Exonuclease R (RNaseR), making its distribution in the cytoplasm quite rich. Advanced high-throughput sequencing and bioinformatics methods have revealed that circRNA is highly conserved, stable, and disease- and tissue-specific. Furthermore, increasing research has confirmed that circRNA, as a driver or suppressor, regulates cancer onset and progression by modulating a series of pathophysiological mechanisms. As a result, circRNA has emerged as a clinical biomarker and therapeutic intervention target. This article reviews the biological functions and regulatory mechanisms of circRNA in the context of respiratory cancer onset and progression.
Keywords: circRNA, respiratory system, non-small cell lung cancer, nasopharyngeal cancer, Laryngeal squamous cell carcinoma
1. Introduction
Circular RNA (circRNA), a novel kind of non-coding RNA (ncRNA) created through the reverse splicing of precursor mRNA (pre-mRNA), lacks a 5’cap and a 3’ poly (A) tail and is converted into a closed circular molecule via covalent bonding (1, 2). This covalent closed loop structure, unlike the majority of linear messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs), is not impacted by RNA exonucleases and not readily disintegrated, making it a highly promising biomarker (3, 4).
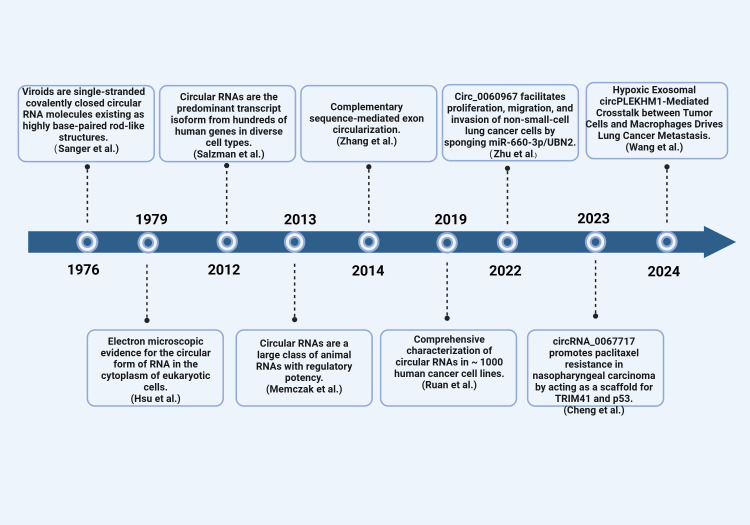
Single-stranded, thermally stable circRNA structures were first observed in 1976 by Sanger et al. (German scientists) under an electron microscope in a process that involved denaturing plant RNA viruses (5). American scientists later observed circRNA in eukaryotic (Hela) cytoplasm using an electron microscope in 1979 (5). Initially, circRNA was considered a “transcription artifact or splicing noise” and was not explored due to its low transcription abundance and technological limitations (6, 7). It was until 2012 that Salzman et al. discovered a large number of circRNAs in the human body through transcriptome sequencing of bone marrow samples from leukemia patients using high-throughput sequencing technology (8). Since then, multiple studies have confirmed the widespread existence of circRNA in various human tissues. For example, several studies have identified and characterized circRNA in various systemic cancers (9–11). Moreover, some studies have demonstrated a strong correlation between circRNA and the onset and progression of various malignant tumors of the respiratory system, including Nasopharyngeal Carcinoma (NPC) (12, 13), Laryngeal Squamous Cell Carcinoma (LSCC) (14), and Non-small Cell Lung Cancer (NSCLC) (15). Consequently, circRNA has emerged as a promising novel target for detecting and managing respiratory illnesses, as well as a significant molecular regulator in respiratory cancers. This article summarizes the biological activities and regulatory roles of circRNA in the respiratory system, aiming to inform the clinical diagnosis and treatment of respiratory cancers ( Figure 1 ).
Figure 1.
The discovery of circRNAs. Timeline of important events related to the discovery, research, and development of circRNA.
2. The biogenesis and formation mechanism of circRNA
As earlier stated, circRNA is synthesized via reverse splicing. During biogenesis, circRNA’s downstream 5’splice site is connected to the upstream 3’ splice site of the same or another exon, forming a covalently closed circular transcript and a linear transcript with selective splicing to skip the exon at the reverse splice junction (14). In other words, circRNA synthesis involves the typical spliceosome mechanism, where circRNA transcription competes with typical pre-mRNA splicing and impacts the expression of typical genes (15).
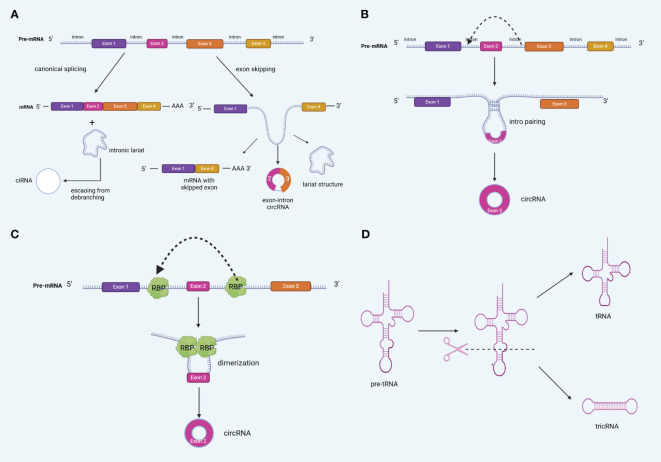
Based on their origin, circRNA formation patterns can be classified into four categories: exon-derived circRNAs (ecRNAs), intron-derived circRNAs (ciRNAs), exon-intron circRNAs (EIciRNAs; comprising both exons and introns), and intergenic circRNAs (16, 17). Notably, ecRNAs contain only reverse spliced exons and are mainly found in the cytoplasm. They are formed by turning off, jumping, and removing introns, and are the most common types of circRNAs (18). On the other hand, intron-only and exon- and intron-derived circRNAs are mainly located in the nucleus (19). Notably, different types of circRNAs are produced via different mechanisms. Presently, circRNA biogenesis is generally believed to involve four different mechanisms: LARAT-driven cyclization (exon jumping), direct reverse splicing (intron pairing-driven cyclization), RNA-Binding Protein (RBP)-mediated cyclization, and tricRNA biogenesis (intron splicing-driven cyclization) (20–22) ( Figure 2 ).
Figure 2.
Biogenesis of circRNA (A) lariat-driven circularization (exon skipping) (B) Back-splicing can be induced by intron pairing (intron pairing-driven cyclization). (C) RNA-Binding Protein (RBP)-mediated cyclization,. (D) tricRNA biogenesis.
3. Biological functions of circRNA
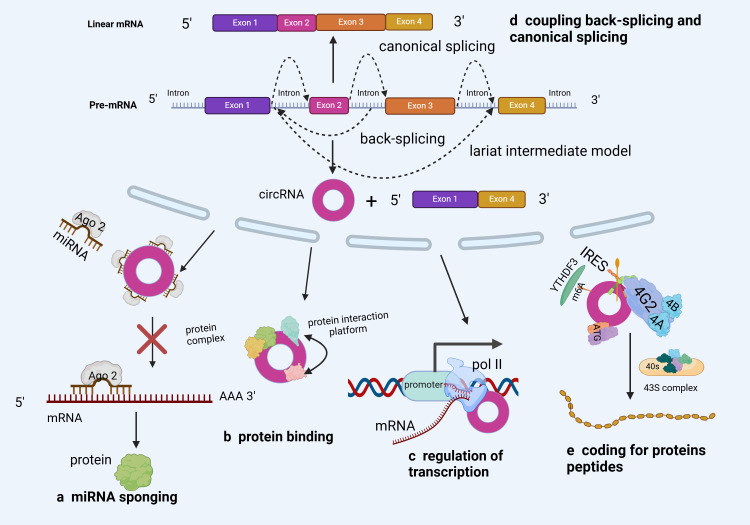
Although its biological function remains debatable (23), circRNA is widely present in eukaryotes and could function as a miRNA “sponge” (24), protein “sponge” or “scaffold” (25), and translation template (26, 27). However, compared to linear mRNAs from the same gene, most circRNAs are expressed at lower levels (28). This suggests that the majority of circRNAs might be functionally irrelevant and merely inconsequential byproducts of alternative splicing (29, 30). Therefore, although circRNAs have existed for long, research on their functions is still at infancy (31, 32). Nonetheless, most research results have shown that circRNAs have biological functions ( Figure 3 ).
Figure 3.
Biological functions of circRNA (A) The most common function of circRNAs is miRNA sponging. By sponging miRNAs, circRNAs inhibit miRNA capacity to perform their post-transcriptional inhibition. (B) circRNAs can act as RBP decoys, protein sponges or scaffolds to regulate target gene expression. (C) circRNAs can regulate parental genes in response to RNA polymerase II. (D) circRNA backsplicing can selectively compete with precursor mRNA typical splicing and affect typical gene expression rates. (E) Some circRNAs have coding potential and are translated into proteins.
3.1. Binding miRNAs as ceRNAs
The primary function of circRNAs is to bind to miRNAs as a ceRNA and block their biological activities, thus regulating the expression of target genes (33). Wu et al. (34) reported a substantial circWHSC1 expression in NSCLC cells, implying that it could be a useful biomarker for NSCLC diagnosis and prognosis ( Figure 3A ). Based on its functional mechanism, circWHSC1 could interact directly with miR-590–5p, upregulating SOX5 (a carcinogenic molecule) and accelerating malignant NSCLC development. Furthermore, some studies reported that circIFI6 may facilitate Esophageal Squamous Cell Carcinoma (ESCC) progression. Additionally, CircIFI6 regulates the expression of hub genes (HK2, MYLK, CDKN1A, CDH3, and TGFBI) by adsorbing miR-497–5p and miR-195–5p via the ceRNA mechanism, regulating the course and prognosis of esophageal cancer (35). These findings demonstrate the regulatory role of circRNAs (as ceRNAs) in tumor development, offering novel insights into circRNA-related cancer research.
3.2. Interaction with RBP
Notably, RBPs constitute a large class of proteins involved in the regulation of transcript splicing, transport, modification, stability, and translation (34, 36) ( Figure 3B ). A growing body of research has shown the significant therapeutic potential of circRNA-RBP interaction, as well as its critical involvement in the onset and progression of many cancers (37–39). For example, through luciferase and RNA pull-down assays, Tang et al. (40) confirmed that circ-0000009 suppresses Lung Adenocarcinoma (LUAD) development by upregulating PDZD2, a tumor suppressor, in a ceRNA- and RBP-dependent manner. This finding opens up new avenues for LUAD diagnosis and treatment. Moreover, the interaction of the Human antigen R (HuR) protein and circAGO2 enhanced the latter’s competitive enrichment in the 3’Untranslated Region (UTR) of the target gene. Consequently, AGO2 binding decreased, AGO2 miRNA-mediated gene silencing was suppressed, and tumor formation and invasion increased (41). In summary, circRNA’s protein sponge activity opens up new avenues for cancer prognosis monitoring, treatment, and early diagnosis.
3.3. Regulating parent gene transcription
It has been reported that circRNAs may regulate parental genes through the action of RNA polymerase II ( Figure 3C ). On the one hand, circRNA can form R-loops with its production sites, impacting DNA replication, repair, and transcription. On the other hand, circRNA could co-activate with Transcription Factors (TF) to regulate transcription (42). Ribonuclease H1 (RNase H1) is an endonuclease that degrades RNA strands in the R loop and may also degrade some ciRNAs. In a previous study, RNase H1-mediated degradation of the transcriptional R loop by cinkrd52 enhanced transcriptional elongation. Specifically, ciRNA was eliminated through RNase H1, thus facilitating transcriptional elongation (43). Furthermore (44), and STAT-3 are representative TFs that have long been associated with Cancer Stem Cells (CSCs) (45), and the absorption of miR-29a/b/c-3p by circ-0043800 in hepatoblastoma cells could induce STAT-3, thus promoting tumor growth (46).
3.4. Adjustable pre-mRNA splicing
Most anti-splicing events are generated by exons with typical splicing sites that co-express with homologous linear mRNA at their host gene sites ( Figure 3D ). In this regard, circRNA anti-splicing competes with pre-mRNA selective splicing, potentially downregulating linear mRNA (45, 47). According to research, circRNA regulates selective splicing events, which are quite common in cancer (48). Additionally, Ma et al. (49) discovered that circZFR enhances Oxidative Phosphorylation (OXPHOS) in LUAD via selective splicing regulation. Specifically, CircZFR activated the AKT mTOR pathway by stabilizing Heterogeneous Nuclear Ribonucleoprotein l (HNRNPL1), thereby regulating the selective splicing of MYO1B full-length transcript (MYO1B fl) and promoting OXPHOS and cell proliferation. These findings indicate that circZFR modulates OXPHOS to exert carcinogenic effects via selective splicing regulation, offering novel insights into cancer research.
3.5. Translation
Open Reading Frames (ORFs) in certain circRNAs have been established to encode functional proteins or peptides ( Figure 3E ). The m6A-mediated cap-independent translation start and the Internal Ribosome Entry Site (IRES) are the two putative pathways for circRNA translation (50–52). Wang et al. (53) confirmed that circSEMA4B inhibits breast cancer progression by encoding a new protein, SEMA4B-211aa, thus regulating AKT phosphorylation. Specifically, SEMA4B-211aa inhibits AKT phosphorylation by competing with p110 to bind to p85. Additionally, Zheng et al. (50) discovered that circYthdc2 has an m6A- and IRES sites-driven ORF and encodes a functional peptide comprising 170 amino acids (Ythdc2–170aa). By specifically targeting the Stimulator for Interferon Genes (STING), Ythdc2–170aa can suppress the STING-mediated antiviral immune response while promoting the replication of RNA viruses.
4. Roles and mechanisms of circRNA in respiratory cancers
Besides its involvement in various physiological and autoimmune diseases, circRNA also plays an important role in the onset and progression of various malignant tumors (54), cell proliferation (55), cell apoptosis, and metastasis (56). With advancements in bioinformatics technology, an increasing number of circRNAs have been shown to play different biological functions in various cancer systems via multiple pathways (57, 58). Some of these biological functions include the proliferation and differentiation of various digestive system malignancies, including Hepatocellular Carcinoma (HCC) (59–61), Colorectal Cancer (CRC) (62–64), and Gastric Cancer (GC) (65–67), among others. Additionally, circRNA is involved in the onset and progression of various respiratory cancers, including Laryngeal Squamous Cell Carcinoma (LSCC) (68), Nasopharyngeal Carcinoma (NPC) (69), and NSCLC (70–72),, among others. Finally, circRNA has also been reported to participate in the proliferation and migration processes of various reproductive system malignancies, including ovarian cancer (73–75), cervical cancer (76–78), and prostate cancer (79, 80). Therefore, it is plausible that circRNA has emerged as a novel and unique epigenetic regulatory factor (81). This review focused on molecular mechanisms of circRNA regulation in respiratory cancers, which is summarized in the subsequent sections.
4.1. Non-small cell lung cancer
Non-Small Cell Lung Cancer (NSCLC) is a common malignant tumor that originates from the bronchial mucosa or gland (82). NSCLC is the most common histological subtype of lung cancer, constituting about 85% of cases (82). The two primary subtypes of NSCLC are Lung Squamous Cell Carcinoma (LUSC) and Lung Adenocarcinoma (LUAD), which together account for approximately 90% of all cases (83–85). Lung Large Cell Carcinoma (LLCC) makes up about 10% of lung cancers (86). CircRNA, acting as either a driving or inhibitory factor, regulates the occurrence and progression of NSCLC through various pathological and physiological mechanisms. These include Epithelial-Mesenchymal Transition (EMT), cell proliferation, metabolism, apoptosis, drug resistance, and migration (70, 87, 88) ( Figure 4 ).
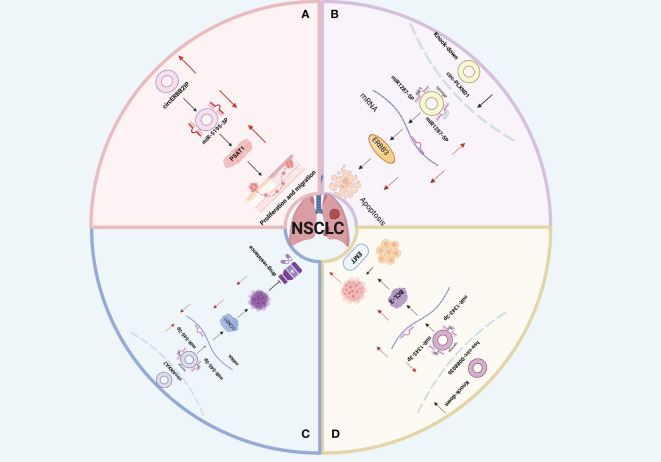
Figure 4.
(A) Schematic illustration of the circERBB2IP/miR-5195–3p/PSAT1 axis mediating non-small cell lung cancer (NSCLC) progression. (B) Downregulation of circ_PLXND1 inhibits tumorigenesis of non-small cell lung carcinoma via miR-1287–5p/ERBB3 axis4c. (C) Hsa_circ_0088036 promotes non-small cell lung cancer progression by regulating miR‐1343‐3p/Bcl‐3 axis through EMT signaling. (D) circANKRD28 inhibits cisplatin resistance in non-small-cell lung cancer through the miR-221–3p/SOCS3 axis.
4.1.1. Proliferation and migration
Forkhead box M1 (FOXM1) is a member of the conserved forkhead box (FOX) transcription factor family, recognized for its role as a multifunctional oncoprotein regulating the expression of numerous cancer-promoting genes (89). In a study by Cheng et al. (90), analysis of LUSC patient specimens revealed that circTP63 was positively correlated with tumor size and TNM stage. High circTP63 expression influenced the cell cycle progression and proliferation of LUSC cells both in vitro and in vivo. Mechanistically, circTP63 functioned as a competing endogenous RNA (ceRNA), binding to miR-873–3p and neutralizing its suppressive effect on the target gene FOXM1. This elevation of FOXM1 subsequently enhanced the expression of downstream cell cycle-related genes CENPA and CENPB, driving cell cycle progression and cell proliferation. Moreover, Facin Actin-bundling Protein 1 (FSCN1) is an important protein that regulates cancer cell migration and invasion (91). Zhang et al. (92) explored the function and mechanism of circSATB2 in NSCLC and confirmed through dual luciferase and quantitative Polymerase Chain Reaction (qPCR) assays that circSATB2 can directly bind to miR-326 to regulate FSCN1 expression and promote NSCLC progression. Additionally, Peng et al. (93) conducted animal experiments and discovered that knocking down circERBB2IP inhibited NSCLC proliferation and migration ( Figure 4A ). Furthermore, previous in vivo experiments implied some correlation between circERBB2IP and TNM grading, Lymph Node Metastasis (LNM), and tumor size in NSCLC patients. Additionally, functional analyses revealed that circERBB2IP could drive NSCLC growth by binding to miR-5195–3p to positively regulate Phosphoserine Aminotransferase 1 (PSAT1) expression. These findings highlight circRNA as a potential biomarker and therapeutic target for clinical NSCLC detection and treatment.
4.1.2. Apoptosis
Accumulating evidence suggests that (94) tumor cell apoptosis mechanisms (94). The bromodomain PHD finger transcription factor (BPTF), a core subunit of nucleosome-remodeling factor (NURF) complex, serves as a transcriptional regulator which facilitates tumor progression (95). BIRC6, as a member of the inhibitors of apoptosis protein (IAP) family, can suppress the expression of the BPTF target gene c-MYC and enhance LUAD progression by promoting apoptosis and cell cycle blockage. A study by Chen et al. (96) demonstrated that circ-100549 serves as a ceRNA by sponging miR-95–5p to upregulate BPTF expression, thus upregulating BIRC6 expression at a transcriptional level in LUAD. In addition, other studies have shown that the GSE101684 dataset from the Gene Expression Omnibus (GEO) database and discovered that circ-PLXND1, derived from the PLXND1 gene, was upregulated in NSCLC tissues. Functional deficiency tests have demonstrated that knocking down circ-PLXND1 could significantly inhibit the growth and metastasis of NSCLC cells and enhance cell apoptosis. Specifically, circ-PLXND1 silencing inhibits NSCLC progression via miR1287–5p upregulation and human Epidermal Growth Factor Receptor 3 (ERBB3) suppression (97) ( Figure 4B ). Moreover, Xia et al. (98) found that circ-0004140 was upregulated in LUAD ( Figure 4C ). Silencing circ-0004140 inhibited cell proliferation, migration, invasion, and tube formation, while inducing apoptosis in LUAD cells. These findings enhance our understanding of the regulatory functions of circRNAs in NSCLC and provide new insights into potential treatments for NSCLC.
4.1.3. Epithelial-mesenchymal Transition
Under certain physiological or pathological situations, epithelial cells can transform into mesenchymal cells, resulting in cytoskeleton alterations, decreased adhesion, and loss of original cell polarity, which could improve cell motility and migration. This process could be summarized as EMT (99). Tumor metastases account for about 90% of cancer-related deaths (100, 101). According to research, EMT is a crucial process for epithelial cells to acquire mesenchymal properties and plays a crucial role in lung cancer cell metastasis and drug resistance (99, 102). Wang et al. (103) and Ge et al. systematically analyzed exosomes derived from NSCLC under hypoxic conditions and identified exosomal circPLEKHM1 as a hypoxia-induced circRNA that significantly promotes metastasis by inducing M2 macrophage polarization. In vivo experiments using the circPLEKHM1-specific reverse splicing sequence (circPLKHM1-ASO) showed a marked reduction in the expression of M2 macrophage markers (CD206 and CD115) and OSMR in tumors from mice treated with circPLEKHM1-ASO. Overall, circPLEKHM1 promotes NSCLC metastasis by regulating OSMR-mediated macrophage polarization. Consistently, Ge et al. (104) discovered that hsa-circ-0088036 silencing upregulated miR-1343–3p, inhibiting anti apoptotic protein 3 (Bcl-3) and NSCLC proliferation, invasion, and EMT. In this regard, hsa-circ-0088036 promotes NSCLC progression and metastasis and can function as an effective target for NSCLC treatment. ( Figure 4C ). In summary, these studies reveal a new circRNA-mediated mechanism and emphasize the significance of circRNA as a prognostic biomarker and therapeutic target for lung cancer metastasis.
4.1.4. Metabolism
Besides the glycolytic metabolic pathway, circRNA also regulates NSCLC development through the glutamine metabolic pathway (105). Wang et al. (106) confirmed that circEPB41L2 induces polyubiquitination and degradation of PTBP1 by binding to its RRM1 structural domain and facilitating its interaction with the E3 ligase TRIP12. This interaction leads to the ubiquitin-mediated degradation of PTBP1, thereby blocking PTBP1-induced activation of the Pyruvate Kinase M2 subtype (PKM2). Consequently, this process inhibits glucose uptake, lactate production, and the progression and metastasis of NSCLC. Additionally, emerging evidence indicates that circRNAs play vital roles in tumor progression, including LUAD. Li et al. (107) identified a novel circRNA, circP4HB, derived from the P4HB gene, which promotes LUAD progression, accelerates aerobic glycolysis, and induces M2 macrophage polarization by interacting with PKM2. Furthermore, Glutamine, as a nitrogen source, is implicated in numerous synthetic metabolic processes in cancer. Zhu et al. (108) confirmed that circ-PDZD8 upregulates La Ribonucleoprotein 1 (LARP1) by targeting miR330–5p, thus promoting glutamine metabolism, which in turn enhances NSCLC proliferation and invasion. Taken together, these findings demonstrate that circRNAs can block NSCLC onset and progression by inhibiting metabolic pathways.
4.1.5. Drug resistance
Chemoresistance is the leading cause of poor outcomes of NSCLC. Cisplatin (DDP) is the first-line anti-cancer chemotherapy drug for NSCLC treatment (109–111). In this regard, it is plausible that NSCLC progression correlates with DDP resistance. Research has indicated that circRNA increases cisplatin resistance in NSCLC. Specifically, circ-ANXA7 upregulates CCND1 (a cell cycle-related protein) by absorbing miR-545–3p, causing abnormal proliferation of NSCLC cells and accelerating their resistance to DDP ( Figure 4D ). However, other studies have confirmed (112) that circANKRD28 overexpression could significantly suppress tumor growth in mice while increasing DDP chemotherapy sensitivity. Furthermore, miR-221–3p has been reported to regulate NSCLC cell proliferation and migration, as well as cisplatin resistance, by targeting the Suppressor of Cytokine Signaling 3 (SOCS3). However, circANKRD28 bound to the miR-221–3p/SOCS3 axis, significantly inhibiting malignant biological progression and cisplatin resistance. These findings offer novel insights into addressing DDP resistance in NSCLC by targeting circRNA.
4.2. Nasopharyngeal cancer
Nasopharyngeal Carcinoma (NPC) is a malignant tumor originating from nasopharyngeal epithelial cells. Its occurrence is mostly linked to genetic factors, EB virus infection, and environmental factors (113). Furthermore, its incidence rate is extremely uneven, with obvious regional clustering and ethnic preferences, mostly in East and Southeast Asia, especially in southern China (114). As a cancer regulatory factor, circRNA modulates NPC onset and progression via mechanisms such as proliferation and migration, drug resistance, metabolism, and signaling pathways.
4.2.1. Proliferation and migration
According to research, CircRNA is crucially involved in NPC metastasis and proliferation (115). As a major TF, C-Myc regulates over 15% of genes in the human transcriptome and can promote a range of cancer-related transcriptional processes associated with metastasis and proliferation (116, 117).Wang et al. (118)performed tests using cell and animal models and discovered that circCAMSAP1 stimulated NPC cell proliferation, invasion, and migration. Further investigations revealed that the upregulated SERPINH1 protein bound to c-Myc, blocking the latter’s ubiquitination degradation. Additionally, circCAMSAP1 bound to and stabilized the 3’ untranslated region of the molecular chaperone SERPINH1 mRNA and promoted SERPINH1 (a collagen synthesizing protein) expression. Furthermore, c-Myc stimulated SRSF10 expression and CAMSAP1 transcription, yielding pre-mRNA, whereas high SRSF10 expression stimulated pre-mRNA splicing, resulting in circCAMSAP1.Moreover C-Myc and SRSF10 jointly promoted circCAMSAP1 formation, resulting in the establishment of a positive feedback loop, leading to NPC proliferation and metastasis. These findings contribute to our understanding of the role circRNA plays in the pathogenesis of NPC ( Figure 5 ).
Figure 5.
Schematic illustration of circCAMSAP1 promoting proliferation and metastasis of NPC through a positive SERPINH1/c-Myc feedback. circCAMSAP1 bound to and stabilized the 3’ untranslated region of the molecular chaperone SERPINH1 mRNA and promoted SERPINH1 expression.Additionally,the upregulated SERPINH1 protein bound to c-Myc, blocking the latter’s ubiquitination degradation. Furthermore, c-Myc stimulated SRSF10 expression and CAMSAP1 transcription, yielding pre-mRNA, whereas high SRSF10 expression stimulated pre-mRNA splicing, resulting in circCAMSAP1.Moreover C-Myc and SRSF10 jointly promoted circCAMSAP1 formation, resulting in the establishment of a positive feedback loop, leading to NPC proliferation and metastasis.
4.2.2. Drug resistance
Chemotherapy and radiotherapy are the primary forms of treatment for patients with advanced NPC (13). Paclitaxel is often used to treat NPC, and resistance to this medication contributes significantly to the unfavorable prognosis of NPC. Cheng et al. (69) confirmed the correlation between circRNA-0067717 and NPC paclitaxel resistance, reporting significant circRNA-0067717 upregulation in paclitaxel-resistant NPC cells. Specifically, the highly expressed circRNA-0067717 lowered the p53 protein levels by binding more p53 proteins and TRIM41 (an E3 ubiquitin ligase). This process promoted the TRIM41-mediated p53 ubiquitination and destruction. Given that it blocks cell cycle progression and induces apoptosis in potentially cancerous cells, P53 is essential for cancer growth prevention (119). Therefore, circRNA-0067717 increased the resistance of NPC cells to paclitaxel treatment by acting as a scaffold for TRIM41 and p53 and promoting TRIM41-mediated p53 degradation through ubiquitination.
4.2.3. Metabolism
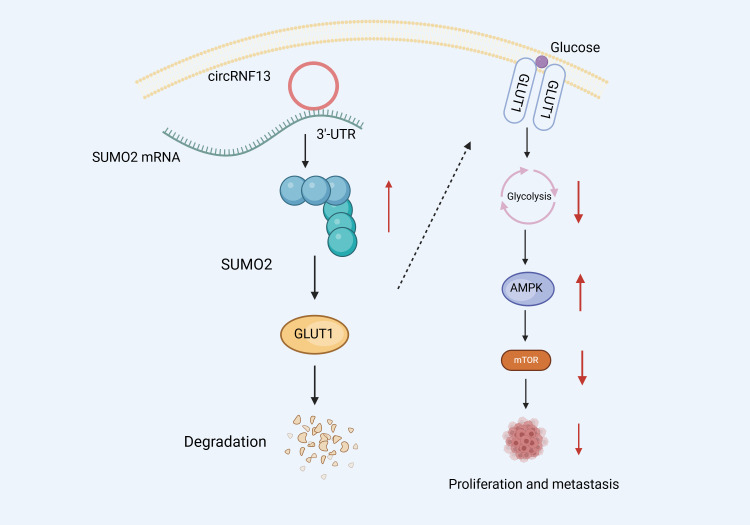
It has been established that circRNA inhibits NPC migration and invasion via glycolysis suppression. Mo et al. (114) performed RNA pull-down and dual luciferase experiments and confirmed that circRNF13 can directly bind to the 3 ‘- UTR region of Small Ubiquitin-like Modifier 2 (SUMO2), improving SUMO2 mRNA stability. Furthermore, Western Blot (WB) experiments revealed that SUMO2 overexpression could downregulate and promote the ubiquitination degradation of Glucose Transporter 1 (GLUT1), thus inhibiting the glycolysis of NPC cells. Glycolysis inhibition induces the activation and phosphorylation of the intracellular AMPK pathway. Subsequently, the activated AMPK pathway blocks protein synthesis and translation by suppressing the mTOR signaling pathway, thus inhibiting tumor growth and metastasis. These insights confirm that circRNF13 could exert tumor suppressive effects. By binding directly to SUMO2’s 3 ‘- UTR, circRNF13 could upregulate its expression, promoting GLUT1 ubiquitination degradation, which, in turn, may inhibit the glycolytic pathway and ultimately suppress NPC proliferation and migration ( Figure 6 ).
Figure 6.
Schematic diagram of the molecular mechanism of circRNF13 in NPC.circRNF13 can directly bind to the 3 ‘- UTR region of Small Ubiquitin-like Modifier 2 (SUMO2), improving SUMO2 mRNA stability. SUMO2 overexpression could downregulate and promote the ubiquitination degradation of Glucose Transporter 1 (GLUT1), thus inhibiting the glycolysis of NPC cells. Glycolysis inhibition induces the activation and phosphorylation of the intracellular AMPK pathway. Subsequently, the activated AMPK pathway blocks protein synthesis and translation by suppressing the mTOR signaling pathway, thus inhibiting tumor growth and metastasis.
4.2.4. Signaling pathways
Signaling pathway activation regulates important cellular mechanisms such as cell proliferation, invasion, survival, inflammation, and immunity (120). For example, abnormal JAK/STAT signaling has been established to promote cancer progression and metastasis (121). Furthermore, through Reverse Transcription quantitative PCR (RT qPCR), Tian et al. (122) confirmed hsa_circle_0013561 upregulation in NPC, which is closely associated with the malignant progression of NPC. By activating the JAK2/STAT3 signaling pathway, hsa_cic_0013561 promotes NPC progression and accelerates cell cycle processes. Furthermore, IL-6, as an upstream activator of the JAK2/STAT3 pathway, could over-activate the JAK2/STAT3 signaling pathway, promote the malignant progression of NPC, and shorten patients’ survival time when upregulated, thus providing a potential target for NPC treatment.
4.3. Laryngeal squamous cell carcinoma
According to pathological classifications, squamous cell carcinoma accounts for 85%-96% of laryngeal cancer cases, making it one of the most common malignant tumors of the head and neck (123). Given its involvement in LSCC proliferation and invasion, circRNA has emerged as a viable target for LSCC treatment and detection (124). Despite a low incidence rate, LSCC has demonstrated a substantial mortality rate and < 50% overall 5-year survival rate (125). Consequently, exploring the mechanism of action of LSCC is imperative.
4.3.1. Proliferation and migration
According to research, circRNA is closely associated with LSCC invasion and proliferation (126). Cyclin D1 (CCND1), a fundamental regulator of the cell division cycle, is one of the most often disrupted therapeutic targets in human cancer (127). Zang et al. (128) discovered that circ-CCND1 interacts with other molecules to prevent CCND1 mRNA decay, raising CCND1 expression after transcription, thus contributing to the development of LSCC tumors. Specifically, circ-CCND1 binds physically to the HuR protein, increasing CCND1 mRNA stability. On the other hand, circ-CCND1 could act as a sponge for miR-646, reducing its inhibitory effect on CCND1 mRNA. In summary, circ-CCND1 regulates interactions with HuR and miR-646, promoting cell proliferation, which, in turn, enhances CCND1 mRNA stability in LSCC, thus providing a theoretical basis for LSCC treatment using gene therapy.
4.3.2. Metabolism
It has been established that Malic Enzyme 1 (ME1) can oxidize and decarboxylate malic acid to pyruvate, presenting a key cellular energy metabolism mechanism (129). Xu (130) (118) confirmed circSERPINA3 upregulation in LSCC using the qRT PCR dual luciferase assay and elucidated the circSERPINA3-miR-885–5 targeting relationship. Specifically, they discovered that circSERPINA3 could promote LSCC invasion and migration by regulating the miR-885–5p/ME1 axis. According to research, Orthodenticle Homeobox 1 (OTX1), a key regulatory factor for cancer development and progression, is a potential therapeutic target for tumors (131). Through microarray analysis, Li et al. (132) discovered high circMYOF expression in the plasma of LSCC patients. Furthermore, functional analysis revealed that circMYOF silencing inhibited glucose consumption and lactate production in LSCC cells. Moreover, bioinformatics analysis showed that circMYOF knockdown inhibited cell growth, metastasis, and glycolysis through the miR-145–5p/OTX1 pathway, slowing down the malignant processes of LSCC. Therefore, circMYOF holds great promise as an oncogenic gene in LSCC treatment.
5. Conclusion and outlook
In conclusion, circRNA, as a novel ncRNA, possesses highly conserved, stable, and specific biological characteristics and participates in various molecular mechanisms in cancer tissues and illnesses. With gradual advancements in bioinformatics technology and high-throughput sequencing methods, an increasing number of circRNAs have recently been identified in respiratory system cancers. Herein, we summarized the biogenesis, biological role, and molecular mechanisms of circRNA regulation in respiratory cancers. Although circRNA has the potential to be an effective biomarker for early cancer detection, diagnosis, and prognosis, research on its interaction with other molecular mechanisms, as well as the screening and analysis of functional circRNAs in respiratory system cancers, is still in its early stages, necessitating more clinical and biological investigations. Furthermore, the interaction between circRNAs and other molecules should be validated through omics research. Future explorations in the aforementioned research areas will contribute to the clinical diagnosis and treatment of cancer.
Author contributions
NY: Writing – original draft. MJ: Writing – original draft. YZ: Writing – review & editing. SM: Writing – review & editing. LW: Visualization, Writing – review & editing. JL: Funding acquisition, Writing – review & editing.
Acknowledgments
We thank the funders of this study. Our appreciation also goes to the Gansu University of Chinese Medicine and anonymous reviewers for their assistance.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Double First-Class initiative projects of Gansu Province of China (GSSYLXM-05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Babin L, Andraos E, Fuchs S, Pyronnet S, Brunet E, Meggetto F. From circrnas to fusion circrnas in hematological Malignancies. JCI Insight. (2021) 6:e151513. doi: 10.1172/jci.insight.151513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang L, Wilusz JE, Chen L. Biogenesis and regulatory roles of circular rnas. Annu Rev Cell Dev Biol. (2022) 38:263–89. doi: 10.1146/annurev-cellbio-120420-125117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rahmani-Kukia N, Abbasi A. New insights on circular rnas and their potential applications as biomarkers, therapeutic agents, and preventive vaccines in viral infections: with a glance at SARS-cov-2. Mol Ther - Nucleic Acids. (2022) 29:705–17. doi: 10.1016/j.omtn.2022.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular rnas come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou W, Cai Z, Liu J, Wang D, Ju H, Xu R. Circular rna: metabolism, functions and interactions with proteins. Mol Cancer. (2020) 19:172. doi: 10.1186/s12943-020-01286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh D, Kesharwani P, Alhakamy NA, Siddique HR. Accentuating circrna-mirna-transcription factors axis: a conundrum in cancer research. Front Pharmacol. (2022) 12:784801. doi: 10.3389/fphar.2021.784801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng X, Zhu S, Pu K, Huang H, Chen Y, Wang W. New insight into circrnas: characterization, strategies, and biomedical applications. Exp Hematol Oncol. (2023) 12:1–91. doi: 10.1186/s40164-023-00451-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liang Y, Liu N, Yang L, Tang J, Wang Y, Mei M. A brief review of circrna biogenesis, detection, and function. Curr Genomics. (2021) 22:485–95. doi: 10.2174/1389202922666210331130722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai T, Qiu S, Gao X, Zhao C, Ge Z, Yang Y, et al. Circularrna circwnk1 inhibits the progression of gastric cancer via regulating themir -21-3p/smad7 axis. Cancer Sci. (2024) 115:974–88. doi: 10.1111/cas.16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Du R, Xiong S. Hsa_circ_0084912 drives the progression of cervical cancer via regulating mir-429/sox2 pathway. Mol Biotechnol. (2023) 65:2018–29. doi: 10.1007/s12033-023-00701-6 [DOI] [PubMed] [Google Scholar]
- 11. Tretti PC, Molin AD, Longo G, Gaffo E, Buratin A, Cani A, et al. Functional relevance of circrna aberrant expression in pediatric acute leukemia with kmt2a::aff1 fusion. Blood Adv. (2024) 8:1305–19. doi: 10.1182/bloodadvances.2023011291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, Cao H, Yang J, Wang B. Igf2bp2-m6a-circmmp9 axis recruits ets1 to promote trim59 transcription in laryngeal squamous cell carcinoma. Sci Rep. (2024) 14:3014. doi: 10.1038/s41598-024-53422-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu S, Wang X, Xie H, Liu L. Comprehensive analysis of circular rnas in nasopharyngeal cancer. Genes Genomics. (2023) 45:1339–46. doi: 10.1007/s13258-023-01438-x [DOI] [PubMed] [Google Scholar]
- 14. Ward Z, Pearson J, Schmeier S, Cameron V, Pilbrow A. Insights into circular rnas: their biogenesis, detection, and emerging role in cardiovascular disease. RNA Biol. (2021) 18:2055–72. doi: 10.1080/15476286.2021.1891393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. Circrnas: role in human diseases and potential use as biomarkers. Cell Death Dis. (2021) 12:468. doi: 10.1038/s41419-021-03743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Xu Q, Huang ZJ, Mao N, Lin ZT, Cheng L, et al. Circrnas: a new target for the diagnosis and treatment of digestive system neoplasms. Cell Death Dis. (2021) 12:205. doi: 10.1038/s41419-021-03495-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu X, Hu Z, Zhou J, Tian C, Tian G, He M, et al. Interior circular rna. RNA Biol. (2020) 17:87–97. doi: 10.1080/15476286.2019.1669391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang B, Wang Y, Zhang K. Interactions between circrna and protein in breast cancer. Gene. (2024) 895:148019. doi: 10.1016/j.gene.2023.148019 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Zhang X, Li C, Yue L, Ding N, Riordan T, et al. Circular rna profiling provides insights into their subcellular distribution and molecular characteristics in hepg2 cells. RNA Biol. (2019) 16:220–32. doi: 10.1080/15476286.2019.1565284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J, Wang C, Jia C, Zhang Y, Qing X, Zhang Y, et al. The role of circular rnas in the physiology and pathology of the mammalian ovary. Int J Mol Sci. (2022) 23:15204. doi: 10.3390/ijms232315204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tran AM, Chalbatani GM, Berland L, Cruz De Los Santos M, Raj P, Jalali SA, et al. A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular rnas. Front Cell Dev Biol. (2020) 8:50. doi: 10.3389/fcell.2020.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y, Wang Y, Zhang C, Sun X. Biological functions of circrnas and their progress in livestock and poultry. Reprod Domest Anim. (2020) 55:1667–77. doi: 10.1111/rda.13816 [DOI] [PubMed] [Google Scholar]
- 23. Zheng S, Zhang X, Odame E, Xu X, Chen Y, Ye J, et al. Circrna-protein interactions in muscle development and diseases. Int J Mol Sci. (2021) 22:3262. doi: 10.3390/ijms22063262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi X, Yang J, Liu M, Zhang Y, Zhou Z, Luo W, et al. Circular rna anapc7 inhibits tumor growth and muscle wasting via phlpp2-akt-tgf-beta signaling axis in pancreatic cancer. Gastroenterology. (2022) 162:2004–17. doi: 10.1053/j.gastro.2022.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Z, Gu S, Wu K, Li L, Dong C, Wang W, et al. Circrna-dopey2 enhances the chemosensitivity of esophageal cancer cells by inhibiting cpeb4-mediated mcl-1 translation. J Exp Clin Cancer Res. (2021) 40:361. doi: 10.1186/s13046-021-02149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun M, Yang Y. Biological functions and applications of circrnas—next generation of rna-based therapy. J Mol Cell Biol. (2023) 15:mjad031. doi: 10.1093/jmcb/mjad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kameda S, Ohno H, Saito H. Synthetic circular rna switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. (2023) 51:e24. doi: 10.1093/nar/gkac1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circrnas in human cancer. Mol Cancer. (2019) 18:6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu C, Zhang J. Mammalian circular rnas result largely from splicing errors. Cell Rep. (2021) 36:109439. doi: 10.1016/j.celrep.2021.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang F, et al. Emerging landscape of circular rnas in lung cancer. Cancer Lett. (2018) 427:18–27. doi: 10.1016/j.canlet.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 31. Xin R, Gao Y, Gao Y, Wang R, Kadash-Edmondson KE, Liu B, et al. Isocirc catalogs full-length circular rna isoforms in human transcriptomes. Nat Commun. (2021) 12:266. doi: 10.1038/s41467-020-20459-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naarmann-de Vries IS, Eschenbach J, Schudy S, Meder B, Dieterich C. Targeted analysis of circrna expression in patient samples by lexo-circseq. Front Mol Biosci. (2022) 9:875805. doi: 10.3389/fmolb.2022.875805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Belter A, Popenda M, Sajek M, Wozniak T, Naskret-Barciszewska MZ, Szachniuk M, et al. A new molecular mechanism of rna circularization and the microrna sponge formation. J Biomol Struct Dyn. (2022) 40:3038–45. doi: 10.1080/07391102.2020.1844802 [DOI] [PubMed] [Google Scholar]
- 34. Zang J, Lu D, Xu A. The interaction of circrnas and rna binding proteins: an important part of circrna maintenance and function. J Neurosci Res. (2020) 98:87–97. doi: 10.1002/jnr.24356 [DOI] [PubMed] [Google Scholar]
- 35. Dai S, Zhang C, Wei X, Wang X, Wen Y, Gao F, et al. Rna sequencing reveals the implication of the circrna-associated cerna network in oesophageal squamous cell carcinoma. Carcinogenesis. (2023) 44:596–609. doi: 10.1093/carcin/bgad050 [DOI] [PubMed] [Google Scholar]
- 36. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular rna-protein interactions: functions, mechanisms, and identification. Theranostics. (2020) 10:3503–17. doi: 10.7150/thno.42174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yan J, Wang X, Fan Z, Xu Y, Zhang Y, Liu Y, et al. Circ_0098823 binding with igf2bp3 regulates dnm1l stability to promote metastasis of hepatocellular carcinoma via mitochondrial fission. Apoptosis. (2024) 29:709–25. doi: 10.1007/s10495-023-01903-8 [DOI] [PubMed] [Google Scholar]
- 38. Yang H, He P, Luo W, Liu S, Yang Y. Circrna tata-box binding protein associated factor 15 acts as an oncogene to facilitate bladder cancer progression through targeting mir-502-5p/high mobility group box 3. Mol Carcinog. (2024) 63:629–46. doi: 10.1002/mc.23677 [DOI] [PubMed] [Google Scholar]
- 39. Gao X, Yu S, Liu S, Zhang S, Sha X, Sun D, et al. Circularrna nuclear receptor interacting protein 1 promoted biliary tract cancerepithelial-mesenchymal transition and stemness by regulating themir -515-5p/akt2 axis andpi3k/akt/mtor signaling pathway. Environ Toxicol. (2023) 38:2632–44. doi: 10.1002/tox.23898 [DOI] [PubMed] [Google Scholar]
- 40. Tan T, Ma M, Xing S. Effect of circ_0000009 on lung adenocarcinoma progression by regulating pdzd2 in a cerna- and rbp- dependent manner. Gene. (2023) 877:147555. doi: 10.1016/j.gene.2023.147555 [DOI] [PubMed] [Google Scholar]
- 41. Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular rna circago2 drives cancer progression through facilitating hur-repressed functions of ago2-mirna complexes. Cell Death Differentiation. (2019) 26:1346–64. doi: 10.1038/s41418-018-0220-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu CX, Chen LL. Circular rnas: characterization, cellular roles, and applications. Cell. (2022) 185:2016–34. doi: 10.1016/j.cell.2022.04.021 [DOI] [PubMed] [Google Scholar]
- 43. Li X, Zhang J, Lei Y, Liu X, Xue W, Zhang Y, et al. Linking circular intronic rna degradation and function in transcription by rnase h1. Sci China Life Sci. (2021) 64:1795–809. doi: 10.1007/s11427-021-1993-6 [DOI] [PubMed] [Google Scholar]
- 44. Ma J, Du WW, Zeng K, Wu N, Fang L, Lyu J, et al. An antisense circular rna circscrib enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther. (2021) 29:2754–68. doi: 10.1016/j.ymthe.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen L. The expanding regulatory mechanisms and cellular functions of circular rnas. Nat Rev Mol Cell Biol. (2020) 21:475–90. doi: 10.1038/s41580-020-0243-y [DOI] [PubMed] [Google Scholar]
- 46. Alsayed RKME, Sheikhan KSAM, Alam MA, Buddenkotte J, Steinhoff M, Uddin S, et al. Epigenetic programing of cancer stemness by transcription factors-non-coding rnas interactions. Semin Cancer Biol. (2023) 92:74–83. doi: 10.1016/j.semcancer.2023.04.005 [DOI] [PubMed] [Google Scholar]
- 47. Zhang P, Zhang XO, Jiang T, Cai L, Huang X, Liu Q, et al. Comprehensive identification of alternative back-splicing in human tissue transcriptomes. Nucleic Acids Res. (2020) 48:1779–89. doi: 10.1093/nar/gkaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng W, Wang L, Geng S, Xu T. Circythdc2 generates polypeptides through two translation strategies to facilitate virus escape. Cell Mol Life Sci. (2024) 81:91. doi: 10.1007/s00018-024-05148-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma Z, Chen H, Xia Z, You J, Han C, Wang S, et al. Energy stress-induced circzfr enhances oxidative phosphorylation in lung adenocarcinoma via regulating alternative splicing. J Exp Clin Cancer Res. (2023) 42:169. doi: 10.1186/s13046-023-02723-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong S, Feng J. Circprimer 2.0: a software for annotating circrnas and predicting translation potential of circrnas. BMC Bioinf. (2022) 23:215. doi: 10.1186/s12859-022-04705-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhong J, Wu X, Gao Y, Chen J, Zhang M, Zhou H, et al. Circular rna encoded met variant promotes glioblastoma tumorigenesis. Nat Commun. (2023) 14:4467. doi: 10.1038/s41467-023-40212-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Li H, Lu Y, Cheng L. Circular rnas in human cancer. Front Oncol. (2021) 10:577118. doi: 10.3389/fonc.2020.577118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X, Jian W, Luo Q, Fang L. Circsema4b inhibits the progression of breast cancer by encoding a novel protein sema4b-211aa and regulating akt phosphorylation. Cell Death Dis. (2022) 13:794. doi: 10.1038/s41419-022-05246-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang G, Deng L, Gong K, Zhou P, Peng L, Li C. Hsa_circ_0003528 promotes cell Malignant transformation and immune escape via increasing oncogenepdl1 through spongingmir -511-3p in non-small cell lung cancer. Environ Toxicol. (2023) 38:1347–60. doi: 10.1002/tox.23768 [DOI] [PubMed] [Google Scholar]
- 55. Tang Q, Wang X, Zhou Q, Li Q, Yang X, Xu M, et al. Fuzheng kang-ai inhibits nsclc cell proliferation via regulating hsa_circ_0048091/hsa-mir-378g/arrdc3 pathway. Phytomedicine. (2023) 114:154819. doi: 10.1016/j.phymed.2023.154819 [DOI] [PubMed] [Google Scholar]
- 56. Cheng C, Wang P, Yang Y, Du X, Xia H, Liu J, et al. Smoking-induced m2-tams, via circeml4 in evs, promote the progression of nsclc through alkbh5-regulated m6a modification of socs2 in nsclc cells. Adv Sci (Weinh). (2023) 10:e2300953. doi: 10.1002/advs.202300953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saleem A, Khan MU, Zahid T, Khurram I, Ghani MU, Ullah I, et al. Biological role and regulation of circular rna as an emerging biomarker and potential therapeutic target for cancer. Mol Biol Rep. (2024) 51:296. doi: 10.1007/s11033-024-09211-3 [DOI] [PubMed] [Google Scholar]
- 58. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circrnas in cancer and oncology. Nat Rev Clin Oncol. (2022) 19:188–206. doi: 10.1038/s41571-021-00585-y [DOI] [PubMed] [Google Scholar]
- 59. Ai J, Zhang W, Deng W, Yan L, Zhang L, Huang Z, et al. A hsa_circ_001726 axis regulated by e2f6 contributes to metastasis of hepatocellular carcinoma. BMC Cancer. (2024) 24:14. doi: 10.1186/s12885-023-11703-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu X, Chen G, Huang Y, Chen Q, Zhuo J, Su R, et al. Integrated multiomics reveals silencing of has_circ_0006646 promotes trim21-mediated ncl ubiquitination to inhibit hepatocellular carcinoma metastasis. Adv Sci (Weinh). (2024) 11(16):e2306915. doi: 10.1002/advs.202306915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Han T, Chen L, Li K, Hu Q, Zhang Y, You X, et al. Significant circrnas in liver cancer stem cell exosomes: mediator of Malignant propagation in liver cancer? Mol Cancer. (2023) 22:197. doi: 10.1186/s12943-023-01891-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zeng K, Peng J, Xing Y, Zhang L, Zeng P, Li W, et al. A positive feedback circuit driven by m6a-modified circular rna facilitates colorectal cancer liver metastasis. Mol Cancer. (2023) 22:202. doi: 10.1186/s12943-023-01848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Deng LQ, Shi CJ, Zhou ST, Zeng WQ, Xian YF, Wang YY, et al. Eif4a3-negatively driven circular rna beta-catenin (circbeta-catenin) promotes colorectal cancer progression via mir-197–3p/ctnnd1 regulatory axis. Br J Cancer. (2024) 130(9):1517–28. doi: 10.1038/s41416-024-02612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Feng P, Zhu L, Sheng N, Li X, Liu P, Chen X. Circxrn2 accelerates colorectal cancer progression through regulating mir-149–5p/macc1 axis and emt. Sci Rep. (2024) 14:2448. doi: 10.1038/s41598-024-52257-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dong X, Chen X, Zhao Y, Wu Q, Ren Y. Circtmem87a promotes the tumorigenesis of gastric cancer by regulating the mir-1276/slc7a11 axis. J Gastroenterol Hepatol. (2024) 39:121–32. doi: 10.1111/jgh.16402 [DOI] [PubMed] [Google Scholar]
- 66. Li R, Wang J, Xie Z, Tian X, Hou J, Wang D, et al. Circusp1 as a novel marker promotes gastric cancer progression via stabilizing hur to upregulate usp1 and vimentin. Oncogene. (2024) 43(14):1033–49. doi: 10.1038/s41388-024-02968-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lu C, Wu J, Li X, Huang W, Fang Y, Huang Y. Hsa_circ_0003356 suppresses gastric cancer progression via mir-556–5p/fkbp5 axis. Toxicol Vitro. (2024) 97:105787. doi: 10.1016/j.tiv.2024.105787 [DOI] [PubMed] [Google Scholar]
- 68. Fan X, Zhang Q, Qin S, Ju S. Circbrip1: a plasma diagnostic marker for non-small-cell lung cancer. J Cancer Res Clin Oncol. (2024) 150:83. doi: 10.1007/s00432-023-05558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cheng Y, Zhu Y, Xiao M, Zhang Y, Wang Z, Chen H, et al. Circrna_0067717 promotes paclitaxel resistance in nasopharyngeal carcinoma by acting as a scaffold for trim41 and p53. Cell Oncol (Dordr). (2023) 46:677–95. doi: 10.1007/s13402-023-00776-y [DOI] [PubMed] [Google Scholar]
- 70. Weidle UH, Birzele F. Circular rna in non-small cell lung carcinoma: identification of targets and new treatment modalities. Cancer Genomics - Proteomics. (2023) 20:646–68. doi: 10.21873/cgp.20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pietrzak J, Świechowski R, Wosiak A, Wcisło S, Balcerczak E. Adamts gene-derived circrna molecules in non-small-cell lung cancer: expression profiling, clinical correlations and survival analysis. Int J Mol Sci. (2024) 25:1897. doi: 10.3390/ijms25031897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lu GJ, Cui J, Qian Q, Hou ZB, Xie HY, Hu W, et al. Overexpression of hsa_circ_0001715 is a potential diagnostic and prognostic biomarker in lung adenocarcinoma. Onco Targets Ther. (2020) 13:10775–83. doi: 10.2147/OTT.S274932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, et al. Comprehensive circular rna expression profiles and the tumor-suppressive function of circhipk3 in ovarian cancer. Int J Biochem Cell Biol. (2019) 112:8–17. doi: 10.1016/j.biocel.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 74. Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. Circplekhm3 acts as a tumor suppressor through regulation of the mir-9/brca1/dnajb6/klf4/akt1 axis in ovarian cancer. Mol Cancer. (2019) 18:144. doi: 10.1186/s12943-019-1080-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Samsami M. Emerging role of circular rnas in the pathogenesis of ovarian cancer. Cancer Cell Int. (2022) 22:172. doi: 10.1186/s12935-022-02602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heydarnia E, Dorostgou Z, Hedayati N, Mousavi V, Yahyazadeh S, Alimohammadi M, et al. Circular rnas and cervical cancer: friends or foes? A landscape on circrna-mediated regulation of key signaling pathways involved in the onset and progression of hpv-related cervical neoplasms. Cell Commun Signal. (2024) 22:107. doi: 10.1186/s12964-024-01494-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang Y, Liu P, Wen D, Xiong H, Zhou Z, Yan L. Regulation of cervical cancer development by a novel circ_0000212/mir-1236–3p/grem1 cerna crosstalk. Mol Biotechnol. (2023) 65:2086–98. doi: 10.1007/s12033-023-00721-2 [DOI] [PubMed] [Google Scholar]
- 78. Zeng Q, Feng K, Yu Y, Lv Y. Hsa_circ_0000021 sponges mir-3940–3p/kpna2 expression to promotecervical cancer progression. Curr Mol Pharmacol. (2023) 17:e170223213775. doi: 10.2174/1874467216666230217151946 [DOI] [PubMed] [Google Scholar]
- 79. Wang Q, Cheng B, Singh S, Tao Y, Xie Z, Qin F, et al. A protein-encoding ccdc7 circular rna inhibits the progression of prostate cancer by up-regulating flrt3. NPJ Precis Oncol. (2024) 8:11. doi: 10.1038/s41698-024-00503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yuan H, Chen C, Li H, Qu G, Chen L, Liu Y, et al. Role of a novel circrna-cgnl1 in regulating pancreatic cancer progression via nudt4-hdac4-runx2-gamt-mediated apoptosis. Mol Cancer. (2024) 23:27. doi: 10.1186/s12943-023-01923-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circrnas in cancer. Oncogene. (2023) 42:2783–800. doi: 10.1038/s41388-023-02780-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yangyang Wang YZ, Nana R, Fangting L, Lin L, Xin Z, Zhigang ZM, et al. Repeat biopsy versus initial biopsy in term of complication risk factors and clinical outcomes for patients with non-small cell lung cancer: a comparative study of 113 ct-guided needle biopsy of lung lesions. Front Oncol. (2024) 14:1367603. doi: 10.3389/fonc.2024.1367603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ma X, Lou C, Pan J, Zhou C, Zhao X, Li N, et al. The diagnostic potential of a circrna–mirna network in non-small cell lung cancer. J Mol Med. (2023) 101:671–84. doi: 10.1007/s00109-023-02306-7 [DOI] [PubMed] [Google Scholar]
- 84. Fang K, Deng Y, Yang P, Zhang Y, Luo D, Wang F, et al. Circ_0079530 stimulates thbs2 to promote the Malignant progression of non-small cell lung cancer by sponging mir-584–5p. Histol Histopathol. (2023) 38:681–93. doi: 10.14670/HH-18-545 [DOI] [PubMed] [Google Scholar]
- 85. Shi J, Lv X, Zeng L, Li W, Zhong Y, Yuan J, et al. Circpvt1 promotes proliferation of lung squamous cell carcinoma by binding to mir-30d/e. J Exp Clin Cancer Res. (2021) 40:193. doi: 10.1186/s13046-021-01976-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ma T, Miao H, Xiong Y, Ma Y, Dong Z. Circrna circ-pdcd11 is highly expressed in lung large-cell carcinoma and predicts poor survival. Immunopharmacol Immunotoxicol. (2023) 45:89–93. doi: 10.1080/08923973.2022.2117628 [DOI] [PubMed] [Google Scholar]
- 87. Liu B, Ma H, Liu X, Xing W. Circscn8a suppresses Malignant progression and induces ferroptosis in non-small cell lung cancer by regulating mir-1290/acsl4 axis. Cell Cycle. (2023) 22:758–76. doi: 10.1080/15384101.2022.2154543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhu Z, Zhang K, Lou M, Tong J, Wu Q, Lu J, et al. Circ_0060967 facilitates proliferation, migration, and invasion of non-small-cell lung cancer cells by sponging mir-660–3p/ubn2. Mol Cell Biochem. (2023) 478:1129–40. doi: 10.1007/s11010-022-04569-z [DOI] [PubMed] [Google Scholar]
- 89. Liu C, Barger CJ, Karpf AR. Foxm1: a multifunctional oncoprotein and emerging therapeutic target in ovarian cancer. Cancers (Basel). (2021) 13:3065. doi: 10.3390/cancers13123065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, et al. Circtp63 functions as a cerna to promote lung squamous cell carcinoma progression by upregulating foxm1. Nat Commun. (2019) 10:3200. doi: 10.1038/s41467-019-11162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ruggiero C, Tamburello M, Rossini E, Zini S, Durand N, Cantini G, et al. Fscn1 as a new druggable target in adrenocortical carcinoma. Int J Cancer. (2023) 153:210–23. doi: 10.1002/ijc.34526 [DOI] [PubMed] [Google Scholar]
- 92. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular rna circsatb2 promotes progression of non-small cell lung cancer cells. Mol Cancer. (2020) 19:101. doi: 10.1186/s12943-020-01221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Peng X, Zhao L, Yao L, Dong J, Wu W, Luo T. Exosomalerbb2ip contributes to tumor growth via elevatingpsat1 expression in non-small cell lung carcinoma. Thorac Cancer. (2023) 14:1812–23. doi: 10.1111/1759-7714.14926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang Y, Mi Y, He C. 2-methoxyestradiol restrains non-small cell lung cancer tumorigenesis through regulating circ_0010235/mir-34a-5p/nfat5 axis. Thorac Cancer. (2023) 14:2105–15. doi: 10.1111/1759-7714.14993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Dai M, Hu S, Liu C, Jiang L, Yu W, Li Z, et al. Bptf cooperates with p50 nf-κb to promote cox-2 expression and tumor cell growth in lung cancer. Am J Transl Res. (2019) 11:7398–409. [PMC free article] [PubMed] [Google Scholar]
- 96. Chen F, Chen J, Yuan Y, Fang S, Xie J, Xu X, et al. Circ_100549 promotes tumor progression in lung adenocarcinoma through upregulation of birc6. Histochem Cell Biol. (2024) 161:493–506. doi: 10.1007/s00418-024-02275-z [DOI] [PubMed] [Google Scholar]
- 97. Wu J, Liu C, Yu G. Downregulation of circ_plxnd1 inhibits tumorigenesis of non-small cell lung carcinoma viamir -1287-5p/erbb3 axis. Thorac Cancer. (2023) 14:1543–55. doi: 10.1111/1759-7714.14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xia F, Xie M, He J, Cheng D. Circ_0004140 promotes lung adenocarcinoma progression by upregulatingnova2 via spongingmir -330-5p. Thorac Cancer. (2023) 14:3483–94. doi: 10.1111/1759-7714.15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhu Y, Meng X, Zhu X, Zhang J, Lv H, Wang F, et al. Circular rna mkln1 promotes epithelial-mesenchymal transition in pulmonary fibrosis by regulating the mir-26a/b-5p/cdk8 axis in human alveolar epithelial cells and mice models. Arch Toxicol. (2024) 98:1399–413. doi: 10.1007/s00204-024-03700-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li K, Guo J, Ming Y, Chen S, Zhang T, Ma H, et al. A circular rna activated by tgfβ promotes tumor metastasis through enhancing igf2bp3-mediated pdpn mrna stability. Nat Commun. (2023) 14:6876. doi: 10.1038/s41467-023-42571-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hakim SG, Alsharif U, Falougy M, Tharun L, Rades D, Jensen J. The impact of tumor budding and single-cell invasion on survival in patients with stage iii/iv locally advanced oral squamous cell carcinoma- results from a prospective cohort study. Front Oncol. (2024) 14:1404361. doi: 10.3389/fonc.2024.1404361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ning J, Luo Y, Chen L, Xiao G, Tanzhu G, Zhou R. Circrnas and lung cancer: insight into their roles in metastasis. BioMed Pharmacother. (2023) 166:115260. doi: 10.1016/j.biopha.2023.115260 [DOI] [PubMed] [Google Scholar]
- 103. Wang D, Wang S, Jin M, Zuo Y, Wang J, Niu Y, et al. Hypoxic exosomal circplekhm1-mediated crosstalk between tumor cells and macrophages drives lung cancer metastasis. Adv Sci (Weinh). (2024) 11:e2309857. doi: 10.1002/advs.202309857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ge P, Chen X, Liu J, Jing R, Zhang X, Li H. Hsa_circ_0088036 promotes nonsmall cell lung cancer progression by regulating mir-1343-3p/bcl-3 axis through tgfβ/smad3/emt signaling. Mol Carcinog. (2023) 62:1073–85. doi: 10.1002/mc.23547 [DOI] [PubMed] [Google Scholar]
- 105. Gao S, Gao T, Feng L, Li H, Dong G, Yang S. Circpkm2 aggravates the progression of non-small cell lung cancer by regulatingmtdh viamir -1298-5p. Thorac Cancer. (2023) 14:3020–31. doi: 10.1111/1759-7714.15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Y, Wang Y, Wu C, Ji Y, Hou P, Wu X, et al. Circepb41l2 blocks the progression and metastasis in non-small cell lung cancer by promoting trip12-triggered ptbp1 ubiquitylation. Cell Death Discovery. (2024) 10:72. doi: 10.1038/s41420-024-01836-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li H, Guo H, Huang Q, Wang S, Li X, Qiu M. Circular rna p4hb promotes glycolysis and tumor progression by binding with pkm2 in lung adenocarcinoma. Respir Res. (2023) 24:252. doi: 10.1186/s12931-023-02563-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhu X, Du T, Chen X, Hu P. Circ-pdzd8 promotes cell growth and glutamine metabolism in non-small cell lung cancer by enrichinglarp1 via sequesteringmir -330-5p. Thorac Cancer. (2023) 14:2187–97. doi: 10.1111/1759-7714.15006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shao N, Song L, Sun X. Exosomal circ_pip5k1a regulates the progression of non-small cell lung cancer and cisplatin sensitivity by mir-101/abcc1 axis. Mol Cell Biochem. (2021) 476:2253–67. doi: 10.1007/s11010-021-04083-8 [DOI] [PubMed] [Google Scholar]
- 110. Bonomo P, Merlotti A, Morbelli S, Berti V, Saieva C, Bergesio F, et al. Does a 6-point scale approach to post-treatment 18f-fdg pet-ct allow to improve response assessment in head and neck squamous cell carcinoma? A multicenter study. Eur J Hybrid Imaging. (2020) 4:8. doi: 10.1186/s41824-020-00077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Halmos B, Rai P, Min J, Hu X, Chirovsky D, Shamoun M, et al. Real-world outcomes on platinum-containing chemotherapy for egfr-mutated advanced nonsquamous nsclc with prior exposure to egfr tyrosine kinase inhibitors. Front Oncol. (2024) 14:1285280. doi: 10.3389/fonc.2024.1285280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Song S, Shi Y, Zeng D, Xu J, Yang Y, Guo W, et al. Circankrd28 inhibits cisplatin resistance in non-small-cell lung cancer through the mir-221-3p/socs3 axis. J Gene Med. (2023) 25:e3478. doi: 10.1002/jgm.3478 [DOI] [PubMed] [Google Scholar]
- 113. Zhou Q, Song W, Li X, Lin J, Zhu C, Cao L, et al. N6-methyladenosine reader hnrnpc-mediated downregulation of circitch prevents mir-224–3p sequestering and contributes to tumorigenesis in nasopharyngeal carcinoma. Environ Toxicol. (2024) 39(5):2893–907. doi: 10.1002/tox.24139 [DOI] [PubMed] [Google Scholar]
- 114. Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, et al. Circular rna circrnf13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via sumo2. Mol Cancer. (2021) 20:112. doi: 10.1186/s12943-021-01409-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wu P, Hou X, Peng M, Deng X, Yan Q, Fan C, et al. Circular rna circrilpl1 promotes nasopharyngeal carcinoma Malignant progression by activating the hippo-yap signaling pathway. Cell Death Differ. (2023) 30:1679–94. doi: 10.1038/s41418-023-01171-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The myc oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. (2022) 19:23–36. doi: 10.1038/s41571-021-00549-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao R, Liu Y, Wu C, Li M, Wei Y, Niu W, et al. Brd7 promotes cell proliferation and tumor growth through stabilization of c-myc in colorectal cancer. Front Cell Dev Biol. (2021) 9:659392. doi: 10.3389/fcell.2021.659392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang Y, Yan Q, Mo Y, Liu Y, Wang Y, Zhang S, et al. Splicing factor derived circular rna circcamsap1 accelerates nasopharyngeal carcinoma tumorigenesis via a serpinh1/c-myc positive feedback loop. Mol Cancer. (2022) 21:62. doi: 10.1186/s12943-022-01502-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Boutelle AM, Attardi LD. P53 and tumor suppression: it takes a network. Trends Cell Biol. (2021) 31:298–310. doi: 10.1016/j.tcb.2020.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zheng J, Deng Y, Huang B, Chen X. Prognostic implications of stk11 with different mutation status and its relationship with tumor-infiltrating immune cells in non-small cell lung cancer. Front Immunol. (2024) 15:1387896. doi: 10.3389/fimmu.2024.1387896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gutiérrez-Hoya A, Soto-Cruz I. Role of the jak/stat pathway in cervical cancer: its relationship with hpv e6/e7 oncoproteins. Cells. (2020) 9:2297. doi: 10.3390/cells9102297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kaisai T, Mantang Z, Tailei Y, Liying Z, Xiaoping C, Mingming J, et al. Hsa_circ_0013561 promotes progression of nasopharyngeal carcinoma by activating jak2/stat3 signaling pathway. Braz J Otorhinolaryngol. (2024) 90:101362. doi: 10.1016/j.bjorl.2023.101362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lv Y, Wang Y, Zhang Z. Potentials of lncrna–mirna–mrna networks as biomarkers for laryngeal squamous cell carcinoma. Hum Cell. (2023) 36:76–97. doi: 10.1007/s13577-022-00799-x [DOI] [PubMed] [Google Scholar]
- 124. Hu C, Lao Z, Yan L, Song X, Wang J. A preliminary analysis of prognostic genes in advanced laryngeal squamous cell carcinoma patients with postoperative radiotherapy. Pathol - Res Pract. (2023) 241:154229. doi: 10.1016/j.prp.2022.154229 [DOI] [PubMed] [Google Scholar]
- 125. Wang X, Wu T, Wang P, Yang L, Li Q, Wang J, et al. Circular rna 103862 promotes proliferation and invasion of laryngeal squamous cell carcinoma cells through the mir-493–5p/golm1 axis. Front Oncol. (2020) 10:1064. doi: 10.3389/fonc.2020.01064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang J, He S, Wang D, Jing Y, Shen W, Huang A, et al. The novel circular rna circtrio silence inhibits the progression of laryngeal squamous cell carcinoma. DNA Cell Biol. (2023) 42:421–32. doi: 10.1089/dna.2023.0043 [DOI] [PubMed] [Google Scholar]
- 127. Simoneschi D, Rona G, Zhou N, Jeong Y, Jiang S, Milletti G, et al. Crl4ambra1 is a master regulator of d-type cyclins. Nature. (2021) 592:789–93. doi: 10.1038/s41586-021-03445-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zang Y, Li J, Wan B, Tai Y. Circrna circ-ccnd1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating ccnd1 expression via interacting with hur and mir-646. J Cell Mol Med. (2020) 24:2423–33. doi: 10.1111/jcmm.14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Simmen FA, Alhallak I, Simmen RCM. Malic enzyme 1 (me1) in the biology of cancer: it is not just intermediary metabolism. J Mol Endocrinol. (2020) 65:R77–90. doi: 10.1530/JME-20-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Xu Q, Yu B, Chen W, Li W, Sun Y, Fang Y. Circserpina3 promoted cell proliferation, migration, and invasion of laryngeal squamous cell carcinoma by targeting mir-885-5p. Cell Biol Int. (2022) 46:1852–63. doi: 10.1002/cbin.11872 [DOI] [PubMed] [Google Scholar]
- 131. Chai J, Xu T, Yang Y, Yuan Y, Xu J, Liu J, et al. Overexpression of otx1 promotes tumorigenesis in patients with esophageal squamous cell carcinoma. Pathol - Res Pract. (2022) 232:153841. doi: 10.1016/j.prp.2022.153841 [DOI] [PubMed] [Google Scholar]
- 132. Li S, Zhang Y, He Z, Xu Q, Li C, Xu B. Knockdown of circmyof inhibits cell growth, metastasis, and glycolysis through mir-145–5p/otx1 regulatory axis in laryngeal squamous cell carcinoma. Funct Integr Genomics. (2022) 22:1–13. doi: 10.1007/s10142-022-00862-8 [DOI] [PubMed] [Google Scholar]