Abstract
Herpes simplex virus type 1 is capable of inhibiting host cell DNA synthesis following lytic infection. However, the mechanism and nature of potential effects on cell cycle progression have not been described. In this report, we characterize the dysregulation of the cell cycle following infection with the replication-incompetent virus d106, where immediate-early gene expression is restricted to infected-cell polypeptide 0 (ICP0) and the expression of all other viral genes is dramatically reduced or is not observed. Infection with d106 resulted in the accumulation of cells in both the G1/S and G2/M compartments, consistent with cell cycle arrest at both checkpoints. The isogenic variant d109, which does not express any viral proteins, failed to induce this phenotype, suggesting that the expression of ICP0 is crucial for cell cycle arrest. Analysis of global cellular gene expression patterns following infection with d106 and d109 revealed that a relatively small subset of cellular genes were induced as a consequence of ICP0 expression. A number of these genes induced in the presence of ICP0 are classically considered p53-responsive genes, including p21, gadd45, and mdm-2. However, infection with d106 of cells with both alleles of p53 deleted resulted in the same cell cycle arrest phenotype and similar cellular gene expression patterns, suggesting that the expression of ICP0 results in cell cycle arrest potentially via p53-dependent and p53-independent mechanisms. In addition, it was found that the effects of infection with d106 on viral and cellular gene expression were similar to the effects observed following treatment of cells with the histone deacetylase inhibitor trichostatin A.
The expression of herpes simplex virus type 1 (HSV-1) genes during lytic infection proceeds in a regulated cascade in which three classes of viral genes are temporally expressed: immediate-early (IE), early (E), and late (L) (40, 41). The five IE gene products, designated infected-cell polypeptide 0 (ICP0), ICP4, ICP22, ICP27, and ICP47, are the first viral proteins synthesized upon infection and encode the primary regulatory functions of the virus necessary for the efficient and timely expression of early and late gene expression (66, 70, 82). The functions of the IE proteins in the efficient expression of viral genes and the successful completion of the viral replication cycle involve the manipulation of a variety of host cell factors and metabolic pathways (2, 6, 24, 26, 31, 37, 39, 50, 54, 57, 69, 76, 78). These interactions result in virus-induced perturbations of normal cellular processes, presumably resulting in a selective advantage for viral gene expression and replication.
ICP0 is an IE protein capable of transactivating all three classes of viral promoters (4, 8, 56) and is sufficient for the reactivation of virus from latency in both in vitro and in vivo models (19, 35, 51, 71). ICP0 is considered a promiscuous activator of gene expression in transient assays (20, 32, 62, 67) and is associated with the turn on of previously silent gene expression in several heterologous systems (59, 60, 73). Thus, the role of ICP0 is generally considered to be gene activation or reactivation. Viruses lacking ICP0 function are replication competent but grow very poorly and reactivate at lower levels than wild-type viruses (9, 51, 72). These observations suggest that the host cell represses input viral genomes and that ICP0 activity leads to derepression of gene expression, increasing the probability of the lytic-cycle program. However, the exact mechanism(s) by which ICP0 may subserve these effects is not clear.
Activation of gene expression by ICP0 is considered to occur at the level of mRNA synthesis (44, 75). ICP0 does not bind DNA (27), suggesting that ICP0 may activate gene expression indirectly. Mutational analysis has demonstrated the importance of an N-terminal C3HC4 (RING finger) motif in ICP0 activity (17, 21, 25), and such domains are thought to mediate protein-protein interactions (29, 30). ICP0 interacts with a number of host cell proteins involved in a variety of cellular pathways that are potentially capable of contributing to its role as a gene activator. ICP0 has been shown to colocalize with and disrupt proto-oncogene promyelocytic leukemia protein-containing nuclear domains (ND10 or PODS) (24, 54, 55). The ability to alter ND10 structures may be a necessary early event for efficient viral gene expression of a number of DNA viruses which also elaborate proteins mediating similar interactions with ND10, including the EBNA-5 protein of Epstein-Barr virus (80), E4-orf3 and E1A of adenovirus (7, 42), and IE1 and IE2 of human cytomegalovirus (HCMV) (1). ICP0 has also been reported to interact with the protein degradation machinery of the cell (23, 26), potentially affecting the stability of cellular and viral proteins. An example of this is provided by the observation that ICP0 interacts with and is associated with the rapid degradation of the catalytic subunit of DNA-dependent protein kinase (50, 65).
It has also been reported that putative cellular functions exist during the transition of growth-arrested cells from G0 into G1 phase of the cell cycle that can functionally substitute for ICP0 transactivating ability (3, 68). Thus, ICP0 function may lead to the activation of viral gene expression by mimicking or promoting cellular conditions normally present during specific stages of the cell cycle. ICP0 may interact directly with cell cycle regulatory machinery, as it has been shown to interact with cyclin D3 (48). Infection with HSV-1 has also been linked to the inhibition of cellular DNA synthesis, presumably associated with cell cycle arrest in G1/S (12). Wu et al. have shown that a virus which minimally expresses ICP0 and ICP47 (d95) inhibits infected-cell proliferation and cellular DNA synthesis (85), suggesting that ICP0 is involved in cell cycle dysregulation. The ability to manipulate cell cycle regulatory proteins may also play a role in viral DNA replication, as HSV-1 infection has been reported to redistribute p53, pRb, proliferating cell nuclear antigen, and other cellular proteins into viral DNA replication centers (83). Cumulatively, these observations suggest a dynamic interaction between cell cycle events and viral gene expression and replication.
The mammalian cell cycle is tightly controlled at two main points: during G1, regulating the onset of S phase of DNA replication, and during G2/M, regulating the onset and completion of mitosis. Activation of the tumor suppressor protein p53 regulates cell cycle progression through both the G1/S and G2/M checkpoints in response to numerous stresses and stimuli (49, 52). Active p53 functions primarily as a transcriptional activator by binding to a specific DNA recognition element of p53-responsive genes such as p21 (15), gadd45 (47), and mdm-2 (45). p53 activation can result in cell cycle arrest or apoptosis, depending on the specific cellular genes induced and the context in which they are induced (49, 52). Importantly, many p53-inducible genes can be activated via p53-independent mechanisms such that cells maintain multiple mechanisms to regulate cell cycle progression in different situations. For example, p21 can be induced independently of p53 during differentiation processes (53, 64), in response to cytokines or growth factors (11, 58), or by inhibition of histone deacetylase (HDAC) (79). Administration of trichostatin A (TSA), a specific inhibitor of HDAC (88), results in the hyperactylation of histones. The physiologic effect of this is cell cycle arrest in G1/S and G2/M, as well as p53-independent induction of p21 (79, 87).
We recently described an HSV-1 mutant virus, d106, which is defective in the expression of all of the IE genes except that which encodes ICP0 and is therefore blocked very early in the viral replication cycle (73). Besides ICP0, the only viral protein product readily detected in d106-infected cells is ICP6, which is the product of an E gene encoding the large subunit of viral ribonucleotide reductase, which has been previously shown to have no cytotoxic effect on the host cell (43). In addition, we have constructed an isogenic variant of d106 that does not express any IE functions (73). This virus, d109, does not express any viral proteins and has no observable cytotoxic effect, even at a high multiplicity of infection (MOI) (73). We investigated the physiologic consequence of ICP0 expression and its potential interaction with cell cycle regulatory processes by comparing the effects of infection with these two viruses. A major consequence of ICP0 expression in infected cells was cell cycle arrest at both the G1/S and G2/M checkpoints. ICP0 expression resulted in the altered expression of a subset of cellular genes, including the induction of p53-responsive genes p21, gadd45, and mdm-2. ICP0 also induced these effects in the absence of cellular p53. Because the effect of ICP0 expression on cellular metabolism is very similar to the reported effect of inhibition of HDAC by TSA, we investigated whether TSA could operationally substitute for ICP0 to derepress gene expression from d109 viral genomes. Cumulatively, the effects of ICP0 on cell metabolism and on viral and cellular gene expression were very similar to those which occurred in the presence of TSA, suggesting that the two ultimately affect similar targets.
MATERIALS AND METHODS
Viruses and cells.
The viruses d92, d95, d97, d99, d100, d103, d106, and d109 (73–75, 85) have been previously described. These viruses were grown and their titers were determined on Vero-derived cells stably transfected with the appropriate trans-complementing HSV-1 IE genes as previously described (74, 75). Cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS) with appropriate antibiotic selection as previously described (13, 73). Noncomplementing HEL (CCL-137) and H1299 (CRL-5803) cells were obtained from the American Type Culture Collection.
Cell proliferation and DNA synthesis assays.
We seeded 106 HEL cells or 7 × 105 H1299 cells on 100-mm-diameter dishes. Cells were then infected or mock infected at an MOI of 10. For proliferation assays, cells were harvested by trypsinization at the indicated times after infection or mock infection and counted. DNA synthesis was monitored by determination of [3H]thymidine uptake. At the indicated time points, growth medium was removed, the cells were washed twice in 1× Tris-buffered saline, and 100 μCi (7,000 μCi/mmol) of [3H]thymidine (ICN) was added in 5 ml of HAM’s F-12 medium supplemented with 2% FBS and 1 mg of penicillin-streptomycin per ml and the mixture was incubated for 3 h. Cells were then harvested and lysed in 500 μl of digestion buffer (5 mM CaCl2, 100 mM NaCl, 10 mM Tris [pH 8], 25 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), 0.1 mg of proteinase K per ml) for 3 h at 37°C. Following phenol-chloroform-isoamyl alcohol (25:24:1) extraction, the DNA was precipitated in ethanol, resuspended in Tris-EDTA, and treated with RNase. The DNA was then re-extracted, reprecipitated, and resuspended overnight in Tris-EDTA. The purified DNA was quantified by measuring the optical density at 260 nm, and 3H content was determined by liquid scintillation spectroscopy.
Flow cytometry.
Cells were seeded and infected as described above. For synchronization, cells were incubated for 48 h prior to infection in Dulbecco modified Eagle medium supplemented with 0.25% FBS and released into medium containing 10% FBS. Cells were then harvested at the indicated time points, fixed in 50% ethanol, and stored at 4°C. The cells (106/ml) were then stained with propidium iodide (50 μg/ml) in 1× phosphate-buffered saline containing 50 μg of RNase per ml and 100 μM EDTA. Cell cycle distribution, as measured by DNA content, was determined by flow cytometry by the University of Pittsburgh Cancer Institute Flow Cytometry Facility. An equal number of cells was counted for each sample.
Expression array analysis.
Expression array analysis was conducted by using ATLAS Human cDNA Expression Arrays (Clontech) in accordance with the manufacturer’s protocol but with the following modifications. Cells (1.7 × 107) were seeded on tissue culture dishes (245 by 245 by 20 mm) for 36 h prior to infection or mock infection (MOI, 5). Total RNA was harvested by using Ultraspec reagent (Biotexc) in accordance with the manufacturer’s protocol. Poly(A)+ RNA was isolated via an oligo(dT) slurry method in accordance with the manufacturer’s (Becton-Dickinson) protocol and labeled with [α-32P]dATP (Amersham) using the reverse transcription reagents supplied with the expression arrays. Equal counts (3 × 106 cpm) of 32P-labeled cDNA were then hybridized to each filter, and the resulting autoradiograms were analyzed for differential expression.
Western blot analysis.
Cells were seeded and infected (MOI, 10) as described above. Where appropriate, cells were then treated with TSA (Sigma) at the indicated concentrations. Cells were harvested at the times indicated, pelleted, and washed in 1× phosphate-buffered saline prior to lysis in 50 μl of lysis buffer (5 mM EDTA, 10 mM Tris [pH 7.4], 10 mM sodium pyrophosphate, 20 mM NaF, 130 mM NaCl, 2 mM sodium orthovanadate, 1% Triton X-100, 1 mM dithiothreitol, 1 mM tolylsulfonyl phenylalanyl chloromethyl ketone [TLCK], 1 mM phenylmethylsulfonyl fluoride). The cells were freeze-thawed, sonicated, and cleared by centrifugation. Protein content was determined by using a Bradford-based assay (Bio-Rad) and equal amounts of total protein were separated by SDS-polyacrylamide gel electrophoresis (PAGE; 12% polyacrylamide; 29:1 polyacrylamide-bis-acrylamide). The resolved proteins were transferred to polyvinylidene difluoride membranes (Amersham) and probed with appropriate antibodies. The antibodies used included α-p53 (Calbiochem OP43), α-p21 (Pharmingen 15091), α-gadd45 (Santa Cruz sc-797), α-mdm-2 (Serotec MCA 1677), and α-ICP0 (Goodwin Institute for Cancer Research 1112). The proteins were detected with an ECL-Plus chemiluminescence assay kit (Amersham).
RESULTS
Infection with viruses that express ICP0 results in cell cycle arrest.
We have previously shown that infection with a virus that minimally expresses ICP0 and ICP47 (d95) results in decreased cell proliferation, as well as inhibition of host cellular DNA synthesis (85). This suggests that this IE protein(s) or these virion components contain activities which result in cell cycle arrest, leading to cell death. In particular, we hypothesized that ICP0 encodes functions responsible for the observed toxicity. To investigate this possibility, we studied the consequences of infection with d106. Similar to our previous observations with d95 (85), infection of HEL cells with d106 (MOI, 10) resulted in inhibition of cell proliferation (Fig. 1A). In addition, cellular DNA synthesis was markedly inhibited following d106 infection, as measured by determination of [3H]thymidine uptake (Fig. 1B). Significantly, infection with d109, which does not express any IE proteins, did not affect these parameters, indicating that components of the virus particle are not sufficient to induce this phenotype. The effect of d106 infection does not appear to be cell type specific, as we have observed similar effects on a number of different cell types (see Fig. 6 and 7; data not shown).
FIG. 1.
d106 infection results in inhibition of cellular proliferation and DNA synthesis. Monolayer cultures of HEL cells were mock infected or infected with d106 or d109 (MOI of 10) and assayed at 0, 24, and 48 h postinfection for cell number (A) and [3H]thymidine incorporation into cellular DNA (B) as described in Materials and Methods.
FIG. 6.
d106 induces cell cycle arrest in G1/S and G2/M independently of cellular p53. Flow cytometry histogram of mock-infected and d106- and d109-infected H1299 cells at 24 h postinfection. The same number of cells was counted for each sample.
FIG. 7.
p53-independent induction of mdm-2, p21, and gadd45 following d106 infection of H1299 cells. The p53 −/− cell line H1299 was mock infected (lanes M) or infected with d106 or d109 as indicated. At the indicated times (hours postinfection [hpi]), cell lysates were prepared and equal amounts of total protein were separated by SDS-PAGE. Western blot analysis was then performed as described in Materials and Methods for mdm-2, p21, gadd45, and ICP0.
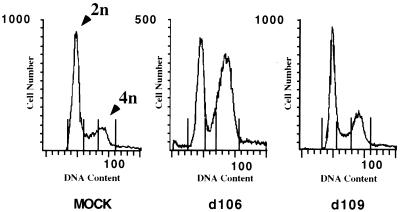
The logical explanation for the decreased proliferation of d106-infected cells and the concomitant inhibition of host DNA synthesis is that cell cycle progression is perturbed such that infected cells ultimately do not pass through S phase. To explore this possibility, we investigated the cell cycle distribution of cells infected with either d106 or d109 by flow cytometry of propidium iodide-stained cells. Infection of asynchronous cultures of HEL cells with d106 but not d109 resulted in accumulation of a lower steady-state level of cells in G1 and an increase in the accumulation of cells in G2/M (Fig. 2). Further incubation did not significantly alter this profile (data not shown). This suggests that the expression of ICP0 during d106 infection dysregulates the cell cycle and possibly causes arrest at both the G1/S and G2/M checkpoints. Infection with d109 had no discernible effect on the ability of cells to progress through the cell cycle, again emphasizing the importance of the expressed viral genes versus factors contained in the virus particle in mediating these effects.
FIG. 2.
d106 infection induces accumulation of cells in G1 and G2/M compartments. HEL cells (106) were mock infected or infected with d106 or d109 (MOI of 10). Flow cytometry of propidium iodide-stained cells was performed at 24 h postinfection as described in Materials and Methods. The same number of cells was counted for each sample. The distribution of G1, S, and G2/M phase cells is 62, 13, and 23 for mock-infected cells; 51, 7, and 37 for d106-infected cells; and 60, 12, and 21 for d109-infected cells, respectively. The peaks corresponding to 2n and 4n DNA content are indicated.
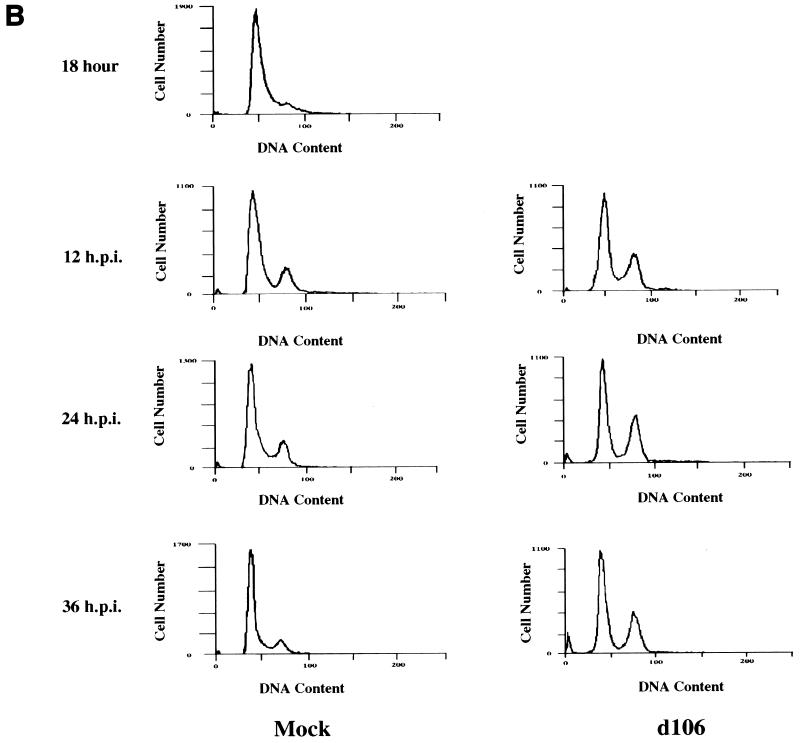
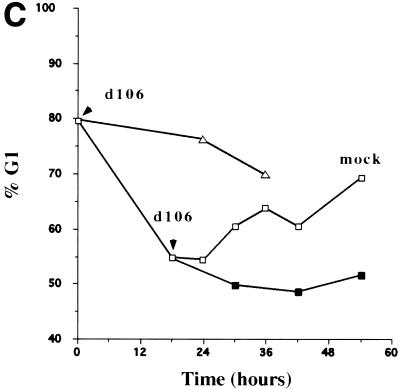
To further investigate the potential existence of two cell cycle arrest points, we infected HEL cells synchronized in G0/G1 by serum starvation at different times after release of the serum block (Fig. 3). Infection of cells shortly after release from low-serum conditions verified the presence of a G1 arrest point, as d106-infected cells remained in G1 phase and failed to enter S phase or progress to G2 for up to 36 h postinfection (Fig. 3A). Figure 3B demonstrates that infection of HEL cells with d106 18 h after release from low-serum conditions, when the majority of cells had progressed beyond the d106-induced G1/S arrest point, resulted in accumulation of cells in G2/M phase. Analysis of the percentage of cells in G1 following infection at different times after release (Fig. 3C) demonstrated that d106-infected cells failed to transit out of G1 into the subsequent S phase and failed to progress through G2/M phase into the following G1 phase. It should be noted that results identical to those shown in Fig. 2 and 3 were obtained following infection of HEL cells with d95, a different ICP4− ICP27− ICP22− virus (unpublished observations). This virus has also been shown to arrest cell division (85). Lastly, it appears that the expression of ICP0 is required for the G2/M arrest phenotype, as infection with viruses with different subsets of IE functions deleted resulted in the accumulation of cells in G2/M only when ICP0 was expressed, regardless of the status of the other IE gene products (Table 1).
FIG. 3.
Characterization of the cell cycle arrest of d106-infected cells in G1/S and G2/M. HEL cells (106) were synchronized in G0/G1 by maintenance under low-serum conditions (0.25%) as described in Materials and Methods. Cells were then infected either upon release into medium containing 10% FBS (at 0 hour) (A) or 18 h after release into 10% FBS-containing medium (B). Mock-infected and virus-infected cells were harvested at the indicated times (hours postinfection [h.p.i.]), stained with propidium iodide, and analyzed by flow cytometry. The 18-h histogram in panel B represents uninfected cells at 18 h following release. The times given for both the subsequent mock-infected and d106-infected cell samples are the times postinfection with d106 at 18 h postrelease. (C) Graphical representation of the accumulation of cells in the G1 compartment following mock infection (□), infection with d106 (▵), infection with d106 18 h (■) after release from G0/G1. The percentage of cells in the G1 fraction is plotted as a function of time following the initial release from the serum block.
TABLE 1.
Requirement of ICP0 for cell cycle arrest
| Virus | IE expression of:
|
G2/M cell cycle arresta | ||||
|---|---|---|---|---|---|---|
| ICP4 | ICP27 | ICP22 | ICP47 | ICP0 | ||
| d92 | − | − | + | + | + | Yes |
| d95 | − | − | − | + | + | Yes |
| d97 | − | − | + | + | − | No |
| d99 | + | + | + | + | − | No |
| d100 | − | + | + | + | − | No |
| d103 | − | + | − | − | + | Yes |
| d106 | − | − | − | − | + | Yes |
| d109 | − | − | − | − | − | No |
Cell cycle arrest was defined by flow cytometry of propidium iodide-stained infected cells at 24 h postinfection.
Effect of ICP0 expression on global cellular gene expression patterns.
The above-described experiments demonstrated that expression of ICP0 results in cell cycle arrest in both G1 and G2/M such that infected cells fail to enter S phase of DNA synthesis and exhibit failure to complete mitosis. Morphologically, d106-infected cells resemble d95-infected cells (85) in that the cells remain flat and adherent to the growth surface and are metabolically active for several days postinfection. However, they become enlarged with inclusions of ICP0 in the nucleus and often possess multiple subnuclei. It is clear that ICP0 has a profound effect on cell physiology when expressed from these viruses, and one might expect significant changes in cellular gene expression that are consistent with the observed cell physiology. In order to examine global changes in cellular gene expression patterns and to more narrowly define appropriate cell cycle regulatory proteins potentially involved in the cell cycle arrest phenomenon, the levels of transcription in infected versus mock-infected HEL cells were compared by using expression array analysis. The expression arrays used (ATLAS Human cDNA Expression Arrays; Clontech) consist of nylon filters spotted with cDNAs corresponding to 588 unique human genes which can be individually identified by grid position on the filter. 32P-labeled cDNA probes generated by reverse transcription of poly(A)+ RNA isolated from infected or mock-infected cells were hybridized to the filters. The resulting autoradiograms allow comparisons of cellular gene expression patterns for the 588 genes following infection with these viruses. Thus, these filters illustrate changes in gene expression occurring at the level of mRNA abundance. Alterations in the expression of a particular gene were defined as reproducible changes that were observed in replicate experiments on multiple filter sets.
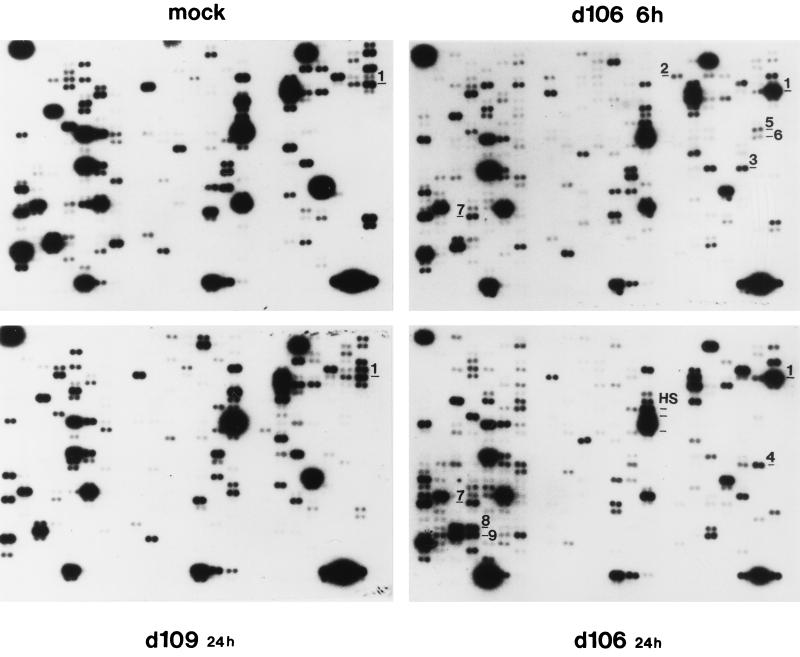
Figure 4 shows one exposure of four representative hybridizations comparing the expression of the cellular genes from mock-infected and d106 (6 and 24 h postinfection)- and d109 (24 h postinfection)-infected cells. Infection with d106 did not result in widespread qualitative changes in the expression of the cellular genes compared to mock-infected cells. Rather, only a small number of genes exhibited alterations in mRNA levels. At 6 h postinfection in d106-infected cells, gadd45 (no. 1), mdm2 (no. 2), NT-3 (no. 3), topoisomerase I (no. 5), topoisomerase IIα (no. 6), and transcription factor ETR103 (no. 7) are clearly induced relative to other genes over mock-infected cells. At 24 h postinfection in d106-infected cells, gadd45 (no. 1), interleukin-6 (no. 4), transcription factor ETR103 (no. 7), YY1 (no. 8), and TFIIB (no. 9) mRNAs were elevated in abundance. Less evident at this level of exposure is the induction of three heat shock proteins (labeled HS in Fig. 4) in d106-infected cells at 24 h postinfection.
FIG. 4.
Cellular gene expression patterns following infection with d106 and d109. Expression array analysis of mock-infected (A), d106-infected (B), and d109-infected (C) HEL cells was performed as described in Materials and Methods. Poly(A)+ mRNA from mock-infected or infected cells was used to generate [32P]dATP-labeled cDNA which was hybridized to each ATLAS filter, respectively. Changes were identified by inspection of autoradiograms resulting from multiple experiments with different filter sets. Shown are representative filters for each condition demonstrating changes in expression relative to uninfected cells in 588 genes contained on the filters. Each gene on the array is represented by two adjacent spots. The labels on the filters are described in the text.
Relative to the profile of d106-infected cells, the pattern of gene expression in d109-infected cells more closely resembled that of gene expression in mock-infected cells. We were unable to observe the reproducible induction of any gene in d109-infected cells. For both viruses, there were a number of spots of lower intensity relative to uninfected cells. This was far more pronounced in d106-infected cells. Reductions in the abundance of cellular transcripts may be a consequence of the effects of both ICP0 and components of the virion.
Induction of p53, p21, gadd45, and mdm-2 following infection with ICP0-expressing virus.
We were particularly interested in induced genes that are associated with cell cycle regulation. The induction of gadd45 and mdm2 along with p21 is often associated with the activation of p53 (15, 45, 47). Activation of p53 predominantly occurs via posttranscriptional mechanisms such as modification and/or stabilization of p53 protein, and therefore its induction was not observed on the expression arrays. ICP0 has been shown to interact with cellular translational and protein degradation pathways, possibly altering the accumulation of cellular protein products. Thus, we next examined the steady-state levels of these cell cycle regulatory proteins by Western blot analysis to confirm and extend some of the results described above at the level of protein accumulation.
Western blot analysis revealed that p53 was induced between 12 and 24 h postinfection by the ICP0-expressing virus d106 but not by d109 (Fig. 5A). In addition, d106 infection induced p21 temporally consistent with p53 induction (Fig. 5B). The induction of p21 by d106 correlated with the cell cycle arrest phenomenon observed following infection with this virus. gadd45 and mdm-2 were also induced at the protein level (Fig. 5C), consistent with alterations observed on the expression arrays. These changes are also consistent with the cell cycle arrest phenotype of d106-infected cells. The induction of gadd45 and mdm-2 occurred earlier than the induction of p21, indicating that these proteins are more sensitive to the effects of ICP0 expression. Induction of the gadd45 and mdm-2 proteins occurred prior to increases in p53 protein levels. mdm-2 has been shown to operate as a negative feedback regulator of p53 via the ability of mdm-2 to interact with p53 and target it for degradation (86). Thus, it is possible that the induction of mdm-2 serves to prevent observable increases of p53 protein at early times, but that this inhibition is overwhelmed at later times. p53 may be activated earlier after infection with d106 in a manner which does not affect the stability of the protein and/or which is not observed by Western blot analysis. Alternatively, it is possible that expression of ICP0 results in the induction of these proteins independently of p53.
FIG. 5.
Induction of p53 and p53-responsive genes gadd45, p21, and mdm-2 by d106. Western blot analysis of the indicated proteins was performed at multiple times (hours postinfection [h.p.i.]) of HEL cells with d106 or d109 to assess the steady-state levels of candidate gene products identified on the ATLAS filters. Equal amounts of total cellular protein were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and blotted with the indicated antibodies. Panels: A Western blot of cellular p53; B, Western blot of p21 and p53 of mock-infected (lanes M) versus d106-infected HEL cells; C, induction of mdm-2 and gadd45 as a consequence of ICP0 expression. Lanes 0 contained extracts from cells just prior to infection.
Induction of p21, gadd45, and mdm-2 occurs independently of p53.
The induction of p53 and p53-responsive genes gadd45, p21, and mdm-2 suggests that the cell cycle arrest which occurs as a consequence of ICP0 expression is p53 dependent. We assessed the requirement for p53 by using a human lung fibroblast cell line (H1299) with both p53 alleles deleted. H1299 cells do not express any detectable or functional p53 peptides and have a normal diploid chromosome number. H1299 cells infected with d106 failed to proliferate, and their DNA synthesis was inhibited (data not shown). Flow cytometry demonstrated that H1299 cells infected with d106 (Fig. 6) exhibited the same cell cycle arrest phenotype as d106-infected HEL cells, which contain wild-type p53 (Fig. 2). This indicates that the expression of ICP0 can potentially alter cell cycle progression via p53-independent mechanisms. Infection with d109 did not affect the progression of H1299 cells through the cell cycle.
In order to assess whether the molecular mechanisms associated with cell cycle arrest in the presence or absence of cellular p53 were similar, we analyzed the gene expression pattern of H1299 cells infected with d106 or d109 or mock infected. The expression array analysis of d106-infected H1299 cells showed a pattern of alterations similar to that described above for d106-infected HEL cells, while d109-infected H1299 cells exhibited a hybridization pattern indistinguishable from that of mock-infected cells (data not shown). Western blot analysis demonstrated that cell cycle regulatory proteins p21, gadd45, and mdm-2 were induced as a consequence of ICP0 expression independently of p53 (Fig. 7). As observed in the p53 +/+ HEL cells, induction of gadd45 as a consequence of infection with d106 occurred sooner (by 6 h postinfection) than the induction of p21 (by 24 h postinfection) in H1299 cells.
Cellular genomes and quiescent viral genomes are sensitive to TSA as well as ICP0.
The growth arrest which occurs in p53 −/− H1299 cells as a consequence of ICP0 expression is coincident with the p53-independent expression of p21. p21 has previously been reported to be induced independently of p53 following treatment with the HDAC inhibitor TSA (79). TSA is considered to derepress cellular genomes, presumably by virtue of the consequent hyperacetylation of core histones. In addition to the p53-independent expression of p21, administration of TSA also results in cell cycle arrest at both G1/S and G2/M (87). Given that derepression by ICP0 and derepression by TSA seem to have similar biologic effects, we investigated whether viral genomes were sensitive to TSA. We have previously shown that the expression of ICP0 results in the induction of the otherwise silent HCMV IE promoter-driven green fluorescent protein (GFP) transgene contained on the d109 genome, consistent with the hypothesis that ICP0 activates gene expression by derepressing viral genomes (73). We assayed the effect of TSA at increasing concentrations on the induction of the d109-encoded HCMV IE promoter-driven GFP compared to the induction of cellular p21 by Western blot analysis. Figure 8A demonstrates that viral genomes and cellular genomes were equally sensitive to TSA, as shown by the induction of GFP from the viral genome and p21 from the cellular genome at similar concentrations of TSA (100 nM). The temporal induction of p21 by 100 nM TSA was also similar to the induction of p21 by ICP0 (Fig. 8B). Thus, ICP0 appears to be operationally similar to an inhibitor of HDAC, suggesting a possible mechanism of ICP0-mediated derepression of gene expression and cell cycle arrest.
FIG. 8.
TSA and ICP0 have similar effects on gene expression from cellular and viral genomes. (A) TSA was added at increasing concentrations to H1299 cells 24 h after mock infection or infection with d109. The ability of TSA to induce expression from the cellular genome was monitored by Western blot analysis of p21 24 h after the addition of the indicated concentrations of TSA. The ability of TSA to induce expression from the resident viral d109 genome was assessed by Western blot analysis for GFP. (B) The effect of ICP0 expression was compared to the effect of TSA with respect to the induction of cellular p21. H1299 cells were mock infected (lanes M), infected with d106, or exposed to 100 nm TSA. Western blot analysis for p21 was then performed at the indicated times postinfection or after the addition of TSA. Lane 0 contained extracts from cells at the start of the experiment.
DISCUSSION
The ability to alter cellular regulatory pathways is a strategy employed by viruses to enhance viral replication. The changes induced by the virus may affect the activities or abundance of pre-existing host cell proteins or may result from virus-induced changes in cellular gene expression. ICP0 is an IE protein of HSV that has been shown to possess the ability to promiscuously induce gene expression, at least in transient assays (20, 32, 62, 67). We have previously constructed a virus in which ICP0 is the only IE protein expressed and another isogenic variant in which none of the IE proteins are expressed (73). Cells infected with the ICP0-expressing viruses do not readily round up and detach from the monolayer but rather are growth arrested and abundantly express ICP0, ICP6, and GFP from the HCMV IE promoter inserted into the virus genome for a defined period of time, until the cells eventually die (73). By contrast, a virus that does not express any IE proteins has no observable effects on host cell metabolism and its genome is relatively silent from a transcriptional standpoint in most cells. Given these two phenotypes, we used these viruses to observe how ICP0 may affect cellular gene expression and metabolism and possibly gain insight into its role in virus infection.
We report that the expression of ICP0 in the absence of other viral IE proteins results in a unique pattern of p53-independent cell cycle arrest which was coincident with the induction of certain cellular genes and consistent with the role of ICP0 as an activator of gene expression. In particular, the main findings were (i) that ICP0 expression resulted in cell cycle arrest in G1/S and G2/M independently of the p53 status of the host cell; (ii) that ICP0 expression resulted in alterations in the expression of a subset of cellular genes; (iii) that the ability to induce cell cycle arrest corresponded to the induction of cellular genes involved in cell cycle regulation, including p21, gadd45, and mdm-2 independently of cellular p53; and (iv) that ICP0 activity had effects on viral and cellular gene expression similar to those of the HDAC inhibitor TSA. The observed changes in the expression of cellular genes as a function of the expression of ICP0 is significant from several standpoints. These changes (i) may reflect the mechanism by which ICP0 induces gene expression, (ii) indicate the potential physiological effects of ICP0 on host cell metabolism, and (iii) may be indicative of the role of ICP0 in the induction of cellular genes whose protein products play a potential role in the viral life cycle.
Examination of the expression of 588 cellular genes at the mRNA level by expression array analysis suggests that only a small fraction of cellular mRNAs were affected by ICP0 expression. This is consistent with previous observations that HSV-1 infection results in the activation of a limited number of cellular promoters contained on the cellular genome (18). The changes in the expression of cellular genes may be due to the direct action of ICP0 on the gene or be a consequence of some upstream event. ICP0 has been hypothesized to act via the proteasome pathway (23, 25, 26). It is possible that ICP0 affects the stability of specific cellular gene products and that this, in turn, affects the repertoire of transcription factors in the cell or the activity of complexes that affect the metabolism of histones on the DNA. It is intriguing that TSA had an effect similar to that of ICP0 on gene expression from the HSV genome and from the cell with respect to the expression of p21. Administration of TSA also results in the induction of only a small subset of cellular genes despite its apparent global effects on the action of HDACs (81). Also similar to our observations with ICP0, TSA results in the p53-independent induction of p21 (79) and cell cycle arrest in G1 and G2/M (86). Given these similar outcomes, it is possible that ICP0 affects the higher-order packaging of DNA. Whether it acts directly in the metabolism of histones or indirectly, possibly through the proteasome pathway, by affecting the stability and hence the abundance of proteins involved in the metabolism of histones, remains to be determined. Of note is the recent report that gadd45 can alter nucleosome structure via interaction with DNA-bound histones (5). gadd45 is induced very early following ICP0 expression, suggestive of a potential indirect mechanism by which ICP0 may alter chromatin structure and thereby gene expression.
It should be noted that ICP0 was necessary for the observed effects. The molecular pathways leading to cell cycle arrest were activated very early following ICP0 expression, as induction of cell cycle regulatory proteins gadd45 and mdm-2 occurred within 6 h after infection with the ICP0-expressing virus d106. While it is possible that other viral open reading frames are expressed during infection with d106, whose protein products contribute to cell cycle arrest, accumulation of such products would be expected to be very low and occur much later than when cell cycle arrest is initiated. The effect of ICP0 on cell cycle progression was, however, independent of the expression of the other four IE proteins. In addition, d109 induced neither cell cycle arrest nor significant changes in cellular gene expression. While we cannot rule out the possibility that ICP0 acts cooperatively with components of the virion particle, it is clear that the expression of ICP0 was required to induce these effects.
These studies reiterate that the expression ICP0 can have profound effects on the metabolism and survival of cells. We have previously demonstrated that while cells expressing ICP0 can remain metabolically active for prolonged periods of time, they are growth arrested and eventually die. Everett and colleagues have demonstrated that the expression of ICP0 leads to changes in the abundance of cellular proteins by affecting the proteasome pathway (22, 23, 65), including the kinetechore-associated protein CENP-C, which may affect the function of the mitotic apparatus (22). It is not clear that cells infected with d106 die by apoptosis. Genes involved in apoptosis, such as bax and bcl-2, are not induced as inferred from the present array analysis. A more extensive analysis aimed at resolving this issue is under way. The extent of the effects of ICP0 on the cell may also be a function of the level of ICP0 expression and the cell type. ICP0 is abundantly expressed from d106. It is possible that a lower level of ICP0 expression results in a more subtle phenotype. Likewise, other cell types, such as nondividing cells, may respond differently to the presence of ICP0. Regardless, viruses like d106 are not completely nontoxic to cells and care should be taken when proposing the use of such viruses as gene therapy vectors.
It is of interest that a number of cellular genes induced as a consequence of ICP0 expression may play a role in the successful completion of the viral replication cycle. A number of DNA viruses, such as adenovirus, simian virus 40, and human papillomavirus, which require cellular DNA replication proteins for viral replication, encode functions to promote cellular S phase. Thus, the cell cycle dysregulation that occurs following infection with these viruses functions to induce or maintain intracellular environments permissive for the efficient completion of the viral replication cycle. While HSV-1 encodes a large number of genes associated with DNA synthesis, HSV-1 does not encode any known topoisomerase activity and cellular topoisomerase II has been associated with HSV-1 viral DNA replication (14, 38). Induction of the neurotrophin NT-3 during the viral replication cycle may have several implications. First, a nerve growth factor-inducible cellular activity has been shown to complement gene expression and replication of HSV-1 mutants lacking ICP0 activity. NT-3 may function similarly to nerve growth factor (90), which would have particular significance in latently infected sensory neurons, presumably to enhance and/or initiate viral gene activation during reactivation. Second, the induction of NT-3 may represent a mechanism to promote neuronal survival during reactivation of latency, as NT-3 supports the survival of a variety of neuronal cell types (89, 90), including sensory neurons of the trigeminal ganglion (16, 84).
Latently infected neurons are terminally differentiated in G0 phase, but stress stimuli have been shown to induce entry into G1 (10, 36). Transition of neuronal cells from G0 to G1 can stimulate expression from IE promoters on latent viral genomes (3, 68), which may be one of many possible mechanisms leading to reinitiation of the lytic cascade. However, it has also been suggested that G0-arrested neurons restimulated to enter the cell cycle undergo apoptosis (33, 34, 61) and that activation of G1 cyclin/cdk activity results in neuronal cell death via apoptotic pathways (63, 77). Expression of cdk inhibitors such as p21 have been shown to protect against this type of neuronal cell death (63). Of note is the fact that NT-3 has also been proposed to play a role in supporting neuron survival by preventing apoptosis (16, 28, 46). Thus, in this case, the cell cycle arrest that occurs following ICP0 expression may serve a dual function, i.e., (i) to activate viral gene expression by promoting a permissive intracellular milieu and (ii) to prolong infected-neuron metabolism. These functions may differ slightly in importance, depending on whether the infected-cell population is initially dividing or nondividing. These issues remain to be resolved.
ACKNOWLEDGMENTS
We thank Jeffery Stelzer for expert technical assistance.
This work was supported by NIH grant AI44812.
REFERENCES
- 1.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruni R, Roizman B. Herpes simplex virus 1 regulatory protein ICP22 interacts with a new cell cycle-regulated factor and accumulates in a cell cycle-dependent fashion in infected cells. J Virol. 1998;72:8525–8531. doi: 10.1128/jvi.72.11.8525-8531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Schaffer P A. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J Virol. 1991;65:4078–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate- early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrier F, Georgel P T, Pourquier P, Blake M, Kontny H U, Antinore M J, Gariboldi M, Myers T G, Weinstein J N, Pommier Y, Fornace A J., Jr Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrozza M J, DeLuca N A. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Silverstein S. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J Virol. 1992;66:2916–2927. doi: 10.1128/jvi.66.5.2916-2927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clements G B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latency-competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 10.Cole A J, Saffen D W, Baraban J M, Worley P F. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 11.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X F. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert S N, Subramanian D, Shtrom S S, Chung I K, Parris D S, Muller M T. Association between the p170 form of human topoisomerase II and progeny viral DNA in cells infected with herpes simplex virus type 1. J Virol. 1994;68:1010–1020. doi: 10.1128/jvi.68.2.1010-1020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.ElShamy W M, Ernfors P. A local action of neurotrophin-3 prevents the death of proliferating sensory neuron precursor cells. Neuron. 1996;16:963–972. doi: 10.1016/s0896-6273(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 17.Everett R, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett R D. Activation of cellular promoters during herpes virus infection of biochemically transformed cells. EMBO J. 1985;4:1973–1980. doi: 10.1002/j.1460-2075.1985.tb03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 20.Everett R D. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D, Barlow P, Milner A, Luisi B, Orr A, Hope G, Lyon D. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038–1047. doi: 10.1006/jmbi.1993.1657. [DOI] [PubMed] [Google Scholar]
- 22.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett R D, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. . (amended version of article previously published, EMBO J. 16:566–577, 1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett R D, Orr A, Elliott M. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 1991;19:6155–6161. doi: 10.1093/nar/19.22.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farinas I, Yoshida C K, Backus C, Reichardt L F. Lack of neurotrophin-3 results in death of spinal sensory neurons and premature differentiation of their precursors. Neuron. 1996;17:1065–1078. doi: 10.1016/s0896-6273(00)80240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freemont P S. The RING finger: a novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 30.Freemont P S, Hanson I M, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483–484. doi: 10.1016/0092-8674(91)90229-r. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.Fruh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampe R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 32.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardina S F, Cheung N S, Reid M T, Beart P M. Kainate-induced apoptosis in cultured murine cerebellar granule cells elevates expression of the cell cycle gene cyclin D1. J Neurochem. 1998;71:1325–1328. doi: 10.1046/j.1471-4159.1998.71031325.x. [DOI] [PubMed] [Google Scholar]
- 34.Gill J S, Windebank A J. Cisplatin-induced apoptosis in rat dorsal root ganglion neurons is associated with attempted entry into the cell cycle. J Clin Investig. 1998;101:2842–2850. doi: 10.1172/JCI1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon Y J, McKnight J L, Ostrove J M, Romanowski E, Araullo-Cruz T. Host species and strain differences affect the ability of an HSV-1 ICP0 deletion mutant to establish latency and spontaneously reactivate in vivo. Virology. 1990;178:469–477. doi: 10.1016/0042-6822(90)90344-q. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg M E, Greene L A, Ziff E B. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- 37.Gu B, Rivera-Gonzalez R, Smith C A, DeLuca N A. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc Natl Acad Sci USA. 1993;90:9528–9532. doi: 10.1073/pnas.90.20.9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammarsten O, Yao X, Elias P. Inhibition of topoisomerase II by ICRF-193 prevents efficient replication of herpes simplex virus type 1. J Virol. 1996;70:4523–4529. doi: 10.1128/jvi.70.7.4523-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 40.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson P A, Wang M J, Friedmann T. Improved cell survival by the reduction of immediate-early gene expression in replication-defective mutants of herpes simplex virus type 1 but not by mutation of the virion host shutoff function. J Virol. 1994;68:6347–6362. doi: 10.1128/jvi.68.10.6347-6362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 46.Karavanov A, Sainio K, Palgi J, Saarma M, Saxen L, Sariola H. Neurotrophin 3 rescues neuronal precursors from apoptosis and promotes neuronal differentiation in the embryonic metanephric kidney. Proc Natl Acad Sci USA. 1995;92:11279–11283. doi: 10.1073/pnas.92.24.11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulatory cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 50.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 53.Macleod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 54.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 55.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 56.Mavromara-Nazos P, Silver S, Hubenthal-Voss J, McKnight J L, Roizman B. Regulation of herpes simplex virus 1 genes: alpha gene sequence requirements for transient induction of indicator genes regulated by beta or late (gamma 2) promoters. Virology. 1986;149:152–164. doi: 10.1016/0042-6822(86)90117-0. [DOI] [PubMed] [Google Scholar]
- 57.McLauchlan J, Simpson S, Clements J B. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell. 1989;59:1093–1105. doi: 10.1016/0092-8674(89)90765-4. [DOI] [PubMed] [Google Scholar]
- 58.Michieli P, Chedid M, Lin D, Pierce J H, Mercer W E, Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 59.Mosca J D, Bednarik D P, Raj N B, Rosen C A, Sodroski J G, Haseltine W A, Hayward G S, Pitha P M. Activation of human immunodeficiency virus by herpesvirus infection: identification of a region within the long terminal repeat that responds to a trans-acting factor encoded by herpes simplex virus 1. Proc Natl Acad Sci USA. 1987;84:7408–7412. doi: 10.1073/pnas.84.21.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nabel G J, Rice S A, Knipe D M, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 61.Nuydens R, de Jong M, Van Den Kieboom G, Heers C, Dispersyn G, Cornelissen F, Nuyens R, Borgers M, Geerts H. Okadaic acid-induced apoptosis in neuronal cells: evidence for an abortive mitotic attempt. J Neurochem. 1998;70:1124–1133. doi: 10.1046/j.1471-4159.1998.70031124.x. [DOI] [PubMed] [Google Scholar]
- 62.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park D S, Morris E J, Padmanabhan J, Shelanski M L, Geller H M, Greene L A. Cyclin-dependent kinases participate in death of neurons evoked by DNA-damaging agents. J Cell Biol. 1998;143:457–467. doi: 10.1083/jcb.143.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells [see comments] Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 65.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pereira L, Wolff M H, Fenwick M, Roizman B. Regulation of herpesvirus macromolecular synthesis. V. Properties of alpha polypeptides made in HSV-1 and HSV-2 infected cells. Virology. 1977;77:733–749. doi: 10.1016/0042-6822(77)90495-0. [DOI] [PubMed] [Google Scholar]
- 67.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ralph W M, Jr, Cabatingan M S, Schaffer P A. Induction of herpes simplex virus type 1 immediate-early gene expression by a cellular activity expressed in Vero and NB41A3 cells after growth arrest-release. J Virol. 1994;68:6871–6882. doi: 10.1128/jvi.68.11.6871-6882.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Field’s virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 71.Russell J, Stow N D, Stow E C, Preston C M. Herpes simplex virus genes involved in latency in vitro. J Gen Virol. 1987;68:3009–3018. doi: 10.1099/0022-1317-68-12-3009. [DOI] [PubMed] [Google Scholar]
- 72.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirvan A, Ziv I, Zilkha-Falb R, Machlyn T, Barzilai A, Melamed E. Expression of cell cycle-related genes during neuronal apoptosis: is there a distinct pattern? Neurochem Res. 1998;23:767–777. doi: 10.1023/a:1022415611545. [DOI] [PubMed] [Google Scholar]
- 78.Smith C A, Bates P, Rivera-Gonzalez R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 80.Szekely L, Pokrovskaja K, Jiang W-Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 82.Watson R J, Preston C M, Clements J B. Separation and characterization of herpes simplex virus type 1 immediate-early mRNAs. J Virol. 1979;31:42–52. doi: 10.1128/jvi.31.1.42-52.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilcock D, Lane D P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 84.Wilkinson G A, Farinas I, Backus C, Yoshida C K, Reichardt L F. Neurotrophin-3 is a survival factor in vivo for early mouse trigeminal neurons. J Neurosci. 1996;16:7661–7669. doi: 10.1523/JNEUROSCI.16-23-07661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu N, Watkins S C, Schaffer P A, DeLuca N A. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 87.Yoshida M, Beppu T. Reversible arrest of proliferation of rat 3Y1 fibroblasts in both the G1 and G2 phases by trichostatin A. Exp Cell Res. 1988;177:122–131. doi: 10.1016/0014-4827(88)90030-4. [DOI] [PubMed] [Google Scholar]
- 88.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 89.Zhou X F, Cameron D, Rush R A. Endogenous neurotrophin-3 supports the survival of a subpopulation of sensory neurons in neonatal rat. Neuroscience. 1998;86:1155–1164. doi: 10.1016/s0306-4522(98)00076-1. [DOI] [PubMed] [Google Scholar]
- 90.Zhou X F, Rush R A. Functional roles of neurotrophin 3 in the developing and mature sympathetic nervous system. Mol Neurobiol. 1996;13:185–197. doi: 10.1007/BF02740622. [DOI] [PubMed] [Google Scholar]