Abstract
Environmental pollution associated with the petroleum industry is a major problem worldwide. Microbial degradation is extremely important whether in the extractive process or in bioremediation of contaminants. Assessing the local microbiota and its potential for degradation is crucial for implementing effective bioremediation strategies. Herein, contaminated soil samples of onshore oil fields from a semiarid region in the Northeast of Brazil were investigated using metagenomics and metataxonomics. These soils exhibited hydrocarbon contamination and high salinity indices, while a control sample was collected from an uncontaminated area. The shotgun analysis revealed the predominance of Actinomycetota and Pseudomonadota, while 16S rRNA gene amplicon analysis of the samples showed Actinomycetota, Bacillota, and Pseudomonadota as the most abundant. The Archaea domain phylotypes were assigned to Thermoproteota and Methanobacteriota. Functional analysis and metabolic profile of the soil microbiomes exhibited a broader metabolic repertoire in the uncontaminated soil, while degradation pathways and surfactant biosynthesis presented higher values in the contaminated soils, where degradation pathways of xenobiotic and aromatic compounds were also present. Biosurfactant synthetic pathways were abundant, with predominance of lipopeptides. The present work uncovers several microbial drivers of oil degradation and mechanisms of adaptation to high salinity, which are pivotal traits for sustainable soil recovery strategies.
Keywords: Soil microbiome, 16S rRNA metataxonomics, Metagenomics, Metabolic profile, Biosurfactants, oil degradation
1. Introduction
Crude oil, formed over thousands of years ago in sedimentary rocks, is usually found at hundreds to thousands of kilometers below the surface [1], where high pressures and temperatures have transformed organic matter into liquid hydrocarbons through the process of diagenesis. Over recent decades, the exploration, drilling, extraction, and refining of this resource have significantly influenced the worldwide economy [2]. Once an economically viable oil well is identified, it usually operates for an average of 20–30 years until the resource is depleted or is no longer financially viable.
Environmental pollution caused by oil spills is considered a problem of great concern by scientists [3]. However, onshore oil contamination has not received the necessary attention, as it happens frequently, albeit on a smaller scale. Specifically, in onshore oil production, contamination can arise during extraction, storage, and transportation, causing substantial harm to both local populations and ecosystems [4].
Soil environmental pollution results in the contamination of intricate systems, thereby affecting both the ecosystem and the surrounding populace. Oliveira et al. [] examined the physicochemical characteristics of water samples collected in the Japaratuba River Basin, the second largest onshore oil field in Brazil, revealing contamination by hydrocarbons and ions in surface and groundwater [5]. The main challenges in Brazilian onshore oil-producing areas arise from pipeline leaks, from wells to processing and distribution facilities, often from produced water [6]. Oil-induced contamination induces adverse changes in soil, including reduced wettability, altered hydraulic conductivity, increased hydrophobicity, and enhanced CO2 efflux [[7], [8], [9]]. High salinity hampers microbial growth by increasing osmolarity and reducing water activity [6]. Consequently, the presence of hydrocarbons and high salinity reduces oxygen availability, as the solubility of this electron acceptor decreases with rising salt concentration [6,10,11].
Oil pollution induces significant shifts within the microbial communities [12] resulting in reduced diversity [13] and enhanced degradation activity, the most effective oil dissipation process [14]. Therefore, an in-depth understanding of microbial transformations in these environments is essential, by assessing changes in cooperative relationships and interactions among coexisting microorganisms, the stability and function of different taxa within the community [15]. Assessment of those complex microbial dynamics has been possible by application of “Omics”, such as next-generation sequencing (NGS) [16]. Studies driven by public databases and in silico tools have been extremely useful for understanding the role of microorganisms in the degradation of pollutants and identifying new genes and biochemical pathways involved in microbial remediation, shedding light onto the intricate relationships within contaminated environments [17].
While previous studies have documented the influence of salinity on degradation of polycyclic aromatic hydrocarbons (PAH) [[18], [19], [20]], the interplay within soil microbial communities and changes in degradation genes in response to increased salinity remain poorly understood. These findings highlight the importance of molecular monitoring of extensive onshore oil production sites to improve and sustain environmental conservation. Investigation and regular inspection of affected sites are essential to controlling ecological risks and the collective well-being of communities that coexist with these complex ecosystems.
The aim of our study was to explore the taxonomic and functional structure of the microbial communities present in the soils of two Brazilian onshore oil production fields with chronic contamination challenges. The impact of contamination on two distinct samples was assessed by two DNA sequencing techniques, shotgun metagenomics and 16S rRNA gene amplicon sequencing, to unravel the microbiome structure associated with these samples. The results show a comprehensive depiction of the taxonomic composition and functional profiles, providing important insights for designing effective soil recovery strategies and addressing the unique challenges posed by chronic saline and hydrocarbon contamination.
2. Materials and methods
2.1. Study sites and sample collection
Three distinct soil samples were kindly collected and provided by Petróleo Brasileiro S.A. (Petrobras), from two Brazilian onshore oil-producing fields. All of them are in the Brazilian Northeast, within the states of Rio Grande do Norte (RN) and Alagoas (AL). Both states have onshore and offshore oil production, with Rio Grande do Norte being the largest oil-producing state in onshore fields (41.47 %) and Alagoas representing 2.98 % of national onshore oil production. The first soil sample (RNC) originates from a field with chronic contamination, in a semiarid region, and was collected from a site located 300 m away from a waste containment reservoir (5°10′31.32"S; 37°19′12.38"W). This sample was collected in December 2018. The second sample (ALC) is from the state of Alagoas, which has a humid tropical climate, within a region known as "Zona da Mata", an area of coastal rainforest (9°27′15.66"S; 36° 8′42.49"W). In this field, the sample was collected in May 2019, from a production field with chronic contamination by produced water. The third soil sample (ALW), also from the state of Alagoas, was collected in November 2019 and originates from the same production field as the ALC sample, within an uncontaminated site.
The soil sampling and collection was performed as recommended by the US EPA LSASDPROC-300-R5 [21]. Briefly, for each sampled area (RNC, ALC, and ALW) six soil aliquots of 500 g were collected from 0 to 15 cm layers, sieved, homogenized, and stored in sterilized glass jars at 4 °C. The samples were transported to the laboratory and stored at −80 °C. Either sample (RNC, ALC, and ALW) represents a pool their six respective soil aliquots, and was used for chemical and physicochemical analyses, DNA extraction and metagenomic studies. All samples were stored at −80 °C until DNA extraction.
2.2. Soil physicochemical analysis and hydrocarbon characterization
Physicochemical tests were conducted to quantify the level of contamination. The soils pH was measured using a 1:1 (w/v) soil:water and analyzed according to US EPA 9045D (US EPA 2004; SevenMulti™ - Mettler Toledo). The electric conductivity was determined using a Condutivimeter LE Sensor LE438 (SevenMulti™ - Mettler Toledo) as reported by Corwin and Yemoto (2020) [22]. Total organic carbon and total nitrogen concentrations were determined according to US EPA 9060 and 351.2 methods. Chloride concentrations were determined according to Standard Methods for the Examination of Water and Wastewater [23].
The analysis of total petroleum hydrocarbons (TPH) and PAH was conducted according to US EPA 8015B and 8270C. Briefly, hydrocarbons extraction was performed by ultrasound, with dichloromethane (HPLC grade) as the solvent. Soil TPH was determined via gas chromatography with a flame ionization detector, using a Shimadzu Europe equipment (model QP5050A) equipped with a Valcobond VB-5 column. Signal integration was performed to consider the fraction in the range of gasoline, kerosene, diesel, and diesel oil, as well as the unresolved complex mixture (MCNR). Before quantification, samples were spiked with 2-fluorobisphenyl and terphenyl-d14 for recovery factor calculations. The internal standards used were C20-d42 and C24-d50. The ten-priority PAHs were determined by chromatography gas phase coupled to mass spectroscopy. Deuterated polycyclic aromatic hydrocarbons were used as internal standards (naphthalene-d8, acenaphthened10, phenanthrene-d10, chrysene-d12, and perylene-d12).
2.3. DNA extraction
Prior to DNA extraction, the soil samples were dialyzed with SnakeSkin Dialysis Tubing, 3.5K MWCO 35 mm (ThermoFisher), against a Tris-HCl-EDTA buffer (10 mM and pH 8.0). Genomic DNA was extracted from 1 g of each soil sample using the DNeasy PowerSoil Kit (QIAGEN, Germany) following the manufacturer's instructions. The quality and quantity of DNA were determined by measuring the absorbance at 260/280 nm (A260/A280) on the NanoDrop device (Thermo, Massachusetts, USA), and DNA integrity was verified by 0.8 % agarose gel electrophoresis.
2.4. 16S rRNA gene library and sequence data analysis
For PCR amplification, we used two hypervariable regions of 16S rRNA V3–V4 (Domain Bacteria) [24]and Arc V4 (Domain Archaea) [25], that were amplified using a specific primer (Table 1). All PCR reactions were carried out with Phusion High-Fidelity PCR Master Mix (Thermo Fischer, Waltham, MA, USA). The library was constructed using the Nextera XT Index Kit (Illumina Inc., San Diego, CA, USA) and size-validated by running on a Bioanalyzer 2100, then sequenced using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA) to generate paired-end (PE250) reads.
Table 1.
Universal primers set utilized in this study for 16S rRNA sequencing.
The 16S rRNA sequences were analyzed using QIIME2 (V2021.4) (https://qiime2.org) [26]. Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequences. Paired-end reads were demultiplexed and quality-filtered using the q2‐demux plugin followed by denoising with DADA2 [27], resulting in the unique amplicon sequence variants (ASVs). Bacterial and archaeal primer-specific trained Naive Bayes taxonomic classifiers, using the SILVA 138 release files, were used to classify the representative ASV sequences (qiime feature-classifier classify-sklearn) [28]. Taxonomy analysis was performed using SILVA_138_database classifier (classifier silva-138-99-nb-classifier) [29].
2.5. Shotgun metagenomic sequencing
The metagenomic DNA that was extracted underwent sequencing through the Illumina sequencing platform. The eligible DNA libraries were employed for microbiome shotgun sequencing, conducted on the Illumina HiSeq 2500 platform using a paired-end sequencing methodology, with a specific target read length of 150 base pairs. Taken together, a total of 84.602.367 read pairs of raw metagenomic reads were obtained from the three samples. Fastp was used to assess the read quality and trimmed/removed adaptor and low-quality base pairs. The reads obtained were merged using Pear 0.9.11 [30]. After merging, the reads were classified against NCBI NR (database downloaded Jun 2022) using Diamond 2.0.14.152 using default settings [31]. The output.daa files were meganized using the tool daa-meganizer supplied with the software MEGAN Community Edition version 6.24.23 [32] using the software supplied megan-map-Jan2021-ue.db database. The data was then imported to MEGAN6 and analyzed further. The aligned sequences were subjected to taxonomic and functional analysis in the MEGAN6 pipeline, and COG and SEED analysis were performed for function, identification, and classification of subsystems. Pathway analysis was done using KEGG (Kyoto Encyclopedia of Genes and Genomes) [33,34]. Furthermore, we employed an extension of MEGAN6 for functional analysis, employing the BiosurfDB database [35], which specifically targets genes related to biosurfactant production and biodegradation pathways. Statistical analysis was carried out using STAMP [36], p-values and confidence intervals were calculated using the two-sided Fisher's exact test and Newcombe-Wilson method, respectively. The correction for p-values was performed using Benjamini-Hochberg procedure [37].
2.6. Principal Component Analysis (PCA)
PCA analysis was conducted using the PCA function in R v.4.2.2 [38], with RStudio 2023.06.0 [39]. The data were pre-processed for normalization. PCA was applied to reduce the dimensionality of the data, explore, and compare sample structures. Bray-Curtis distance was used to compare data matrices across different groups, considering the relative composition of biotic and abiotic data.
3. Results
3.1. Physicochemical properties of soil samples
The physicochemical properties of three soil samples from states in the Brazilian Northeast (Alagoas and Rio Grande do Norte) were analyzed as summarized on Table 2. The two samples from Alagoas, uncontaminated control (ALW) and contaminated (ALC), presented pH values of 7.56 and 5.69, respectively. In addition, the contaminated sample from Rio Grande do Norte (RNC) presented a pH of 7.14. Both contaminated soils exhibited a high electrical conductivity (EC) and a high chloride content, while the uncontaminated soil showed a low conductivity (Table 2). The C/N ratio of the ALC and RNC soil samples are 556:10 and 4416:10, respectively, further above the recommended ratio for soil bioremediation strategies (100:10) [40,41].
Table 2.
Physicochemical parameters of soil samples.
| Parameters |
ALW |
ALC |
RNC |
|---|---|---|---|
| Site | Alagoas | Alagoas | Rio Grande do Norte |
| Time point | Nov/2019 | May/2019 | Dec/2018 |
| pH | 7.56 | 5.69 | 7.14 |
| Electrical Conductivity (EC1:5) (dS/m) | 0.98 | 14.60 | 12.04 |
| Total Nitrogen (mg/Kg) | 622.8 | 469.7 | 214.8 |
| C/N ratio | 44.9 | 55.6 | 441.6 |
| Chloride (mg/kg) | 259.3 | 77119.0 | 8063.4 |
| n-Alkanes (mg/kg) | 0.0 | 1.8 | 160.1 |
| Poly-Aromatic Hydrocarbon (PAH) (mg/kg) | 0.0 | 0.097 | 12.49 |
| TPH C8–C40 (mg/kg) | 0.0 | 22.4 | 24613.5 |
Regarding the hydrocarbon contamination, the RNC exhibited higher contamination rates than the ALC (Table 2). In contrast, the uncontaminated soil control, ALW, showed no signs of hydrocarbon contamination. The level of contaminants found in the RNC soil were as follows: 24,613.5 mg/kg of TPH, 160.1 mg/kg of n-alkane hydrocarbons, and a significantly high presence (19,494.1 mg/kg) of an unresolved complex mixture (UCM), indicating that a large part of the contaminating oil was degraded. The ALC sample presented low values of oil contamination, with less than 25 mg/kg of TPH and characteristics of degraded oil (UCM). For the ALW sample, no hydrocarbons were detected.
3.2. Taxonomic identification of metagenomic sequences
Around 29,380,112 sequences were assigned to broad taxonomic groups (Table S1). In all samples, the Bacteria domain contributed on average 95.60 %. At the phylum level, the most abundant bacterial sequences were identified as Actinomycetota, accounting for 42.05 %, 37.06 %, and 35.86 %, followed by Pseudomonadota with 34.03 %, 25.30 %, and 56.01 %, for the ALW, ALC, and RNC samples, respectively (Fig. 1A). The lowest percentages of phyla varied among each sample, where ALC sample presented 11.85 % for Bacillota, Bacteroidota (4.16 %), Acidobacteriota (3.26 %), and Chloroflexota (2.67 %). In the ALW sample, the least abundant phyla were Acidobacteriota (9.11 %), Planctomycetota (4.31 %), and Chloroflexota (2.34 %). For the RNC sample, the other most abundant phyla were Bacillota (1.80 %), Chloroflexota (1.53 %), and Bacteroidota (1.22 %). The Archaea domain contributed 10.67 % in the ALC sample, represented by phylum Methanobacteriota (10.29 %), while the other two samples did not exceed 0.5 %. Lastly, the Eukaryota domain contributed less than 1 % in all samples. Only the RNC sample showed the presence of viral genomes (0.10 %), specifically belonging to the Caudovirales order, which is classified under the phylum Uroviricota. Unassigned and unclassified sequences corresponded to 9.01 % of the average.
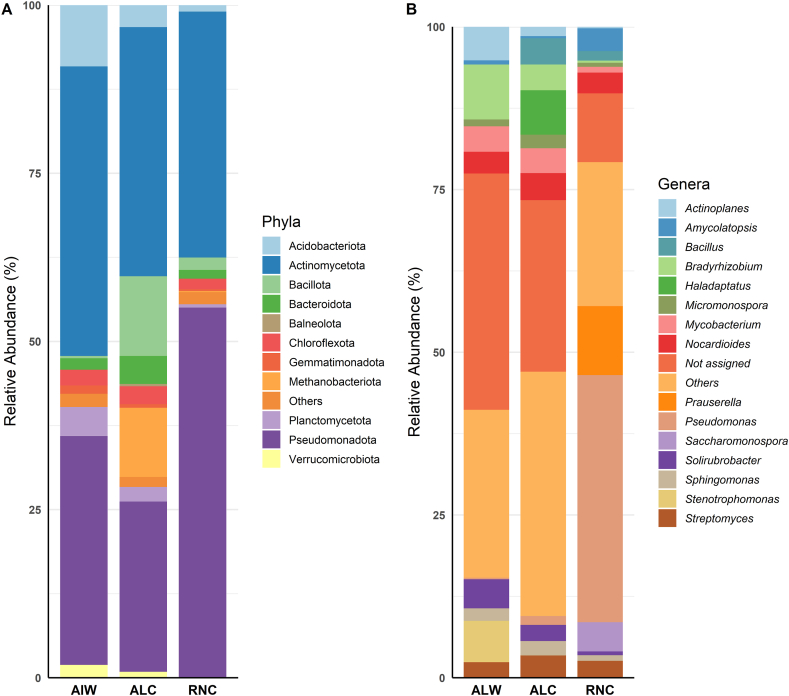
Fig. 1.
Relative abundance of bacteria in soil samples analyzed by the shotgun metagenomics. The figure shows the most abundant bacteria at phylum level (A) and genus level (B). In (A), it's possible to observe a significant relative abundance of Actinomycetota and Pseudomonadota in all soil samples. In (B), the most abundant genera are Bradyrhizobium (8.43 %) in ALW, Haladaptus (6.82 %) in ALC, and Pseudomonas (37.97 %) in RNC. Uncontaminated soil sample from Alagoas (ALW), Contaminated soil sample from Alagoas (ALC) and Contaminated soil sample from Rio Grande do Norte (RNC).
The most abundant bacterial genera found in the ALW sample were Bradyrhizobium (8.43 %), Stenotrophomonas (6.35 %), Actinoplanes (5.17 %), Solirubrobacter (4.48 %), Mycobacterim (3.92 %), and Nocardioides (3.34 %). In the ALC sample, the predominant genera were Haladaptus (6.82 %), Nocardioides (4.14 %), Bacillus (4.03 %), Bradyrhizobium (3.95 %), Mycobacterium (3.83 %), and Streptomyces (3.41 %). Finally, for the RNC sample, the genera found were Pseudomonas (37.97 %), Prauserella (10.58 %), Saccharomonospora (4.50 %), Amycolatopsis (3.48 %), and Nocardioides (3.17 %) (Fig. 1B).
Principal Component Analysis (PCA) was used to explore the relationships between environmental variables and the structure of the microbial community, as shown in Fig. S6. The first and second axes of the PCA explain 78.8 % and 21.2 % of the total data variance, respectively. TPH, PHA, n-alkanes, and pH emerged as the main determinants of the first axis, being positively related. Conductivity was positively related with the second axis, while pH was negatively related, suggesting an opposite correlation between these variables. The phyla Bacillota and Methanobacteriota were associated with environments (soils) with higher salinity, while phyla located in the third quadrant may be related to environments with lower salinity. The phylum Pseudomonadota, on the other hand, exhibited distinct characteristics compared to other phyla, showing a more positive relation with petroleum hydrocarbons and salinity.
3.3. Amplicon sequence variants (ASV) based on 16S ribosomal genes
To facilitate future comparative analyses against public databases, a more widely used analytical strategy was performed. The 16S ribosomal gene amplicons were obtained from genomic DNA of the three samples, based on regions the V3–V4 and V4, for bacteria and archaea, respectively. The V3–V4 region generated a total of 77,988 reads for ALW, 60,689 reads for ALC, and 409,220 reads for RNC, with an average sequence length of 418 bp. After quality control and removal of chimeric sequences using DADA2 in QIIME2, the sequences were reduced to 45,438, 41,081, and 320,520 reads to ALW, ALC, and RNC, respectively. The clustering analysis revealed an average count of Amplicon Sequence Variants (ASVs) within the soil samples. Specifically, in the V3–V4 region, the filtered reads were organized into 500 ASVs for ALW, 349 ASVs for ALC, and 836 ASVs for RNC.
For the V4 Archaea-specific region there were 61,753 reads to ALW, 60,529 reads to ALC, and 196,923 to RNC. After filtering, the sequences were reduced to 41,210, 44,943, and 146,111 sequences, respectively. The RNC sample showed little representation of Archaea, corresponding to less than 2 %. In the case of the V4 region, we identified 78 ASVs associated with archaea in the ALW sample, and 29 ASVs in the ALC sample. The diversity and richness of these communities were also estimated using Shannon diversity indices and rarefaction curve. The Shannon indices for the V3–V4 regions were 5.47, 4.64, and 4.74 to ALW, ALC, and RNC, respectively, while V4 was 3.26 to ALW and 2.71 to ALC.
3.3.1. Taxonomic distribution of bacterial populations
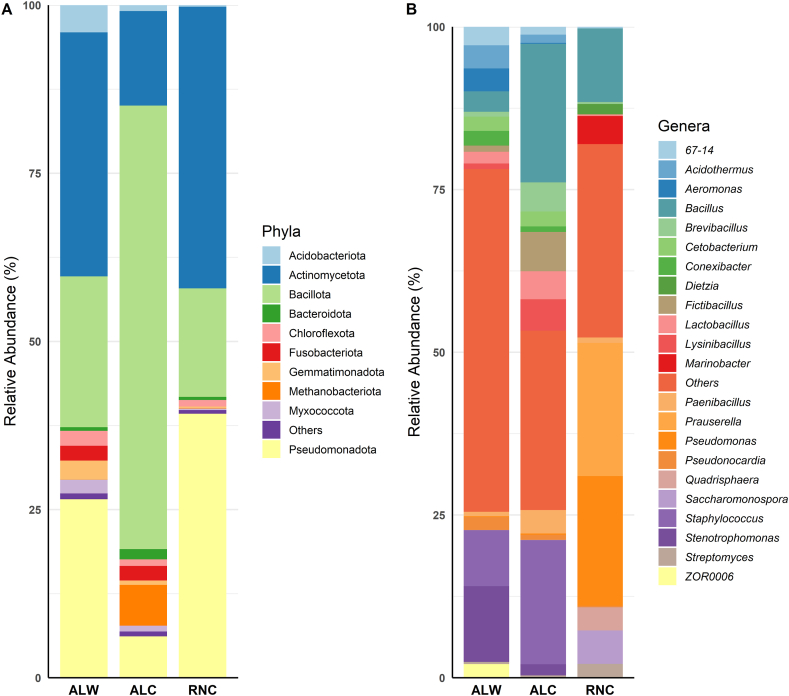
The bacterial populations using a specific primer for V3–V4 regions show the predomination of phylum Actinomycetota (36.28 %, 14.95 %, and 41.93 %), Bacillota (22.43 %, 70.21 %, and 16.15 %), and Pseudomonadota (26.55 %, 6.54 %, and 39.30 %) for ALW, ALC and RNC, respectively. Other phyla comprised less than 5 %: Acidobacteriota (4.05 %, 0.94 %, and 0.24 %), Chloroflexota (2.22 %, 1.04 %, and 0.98 %), Fusobacteriota (2.21 %, 2.30 %, and 0.00 %). At genus level, the genus Stenotrophomonas was predominant in the ALW sample (11.63 %), followed by Staphylococcus (8.60 %), Acidothermus (3.54 %), and Aeromonas (3.53 %). In the ALC sample, the most abundant genera were Bacillus (20.01 %), Staphylococcus (17.91 %), and Fictibacillus (6.04 %), while in the RNC sample, the most abundant genera were Prauserella (20.45 %), Pseudomonas (19.91 %), and Bacillus (11.31 %). Fig. 2A and B shows an overview of the distribution at phylum and genus level.
Fig. 2.
Relative abundance of bacteria in soil samples analyzed by 16S rRNA amplicon sequencing. The figure shows the most abundant bacteria at phylum level (A) and genus level (B), based on region V3–V4. The most abundant phyla, Actinomycetota, Bacillota, and Pseudomonadota, are dominant across all samples (A). At the genus level (B), each sample exhibits distinct abundance: Stenotrophomonas (11.63 %) in ALW, Bacillus (20.01 %) in ALC, and Prauserella (20.45 %) in RNC. Uncontaminated soil sample from Alagoas (ALW), Contaminated soil sample from Alagoas (ALC) and Contaminated soil sample from Rio Grande do Norte (RNC).
3.3.2. Taxonomic distribution of archaeal populations
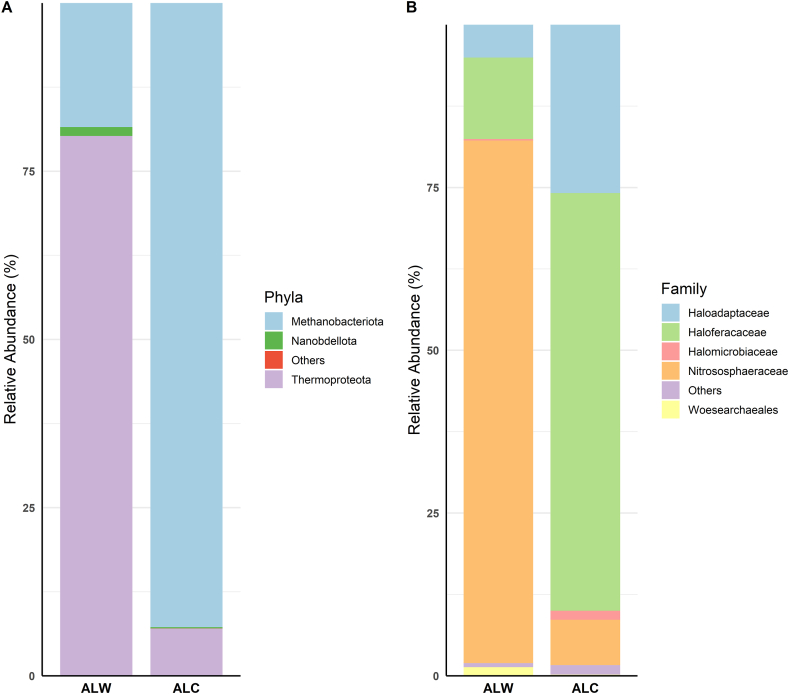
The archaeal community was compared at phylum level (Fig. 3A), revealing Thermoproteota (80.24 % and 7.02 %), Methanobacteriota (18.43 % and 92.76 %), and Nanobdellota (1.32 % and 0.18 %) in the ALW and ALC samples, respectively. Interestingly, the presence of the Methanobacteriota phylum was also assigned by the V3–V4 region in the ALC sample, although in a smaller proportion (6.05 %). The most predominant archaeal families in the ALW and ALC samples were classified (Fig. 3B), respectively, as Nitrososphaeraceae (80.24 % and 6.97 %), Haloferacaceae (12.52 % and 64.15 %), Haloadaptaceae (5.05 % and 25.86 %). The most abundant genera within Nitrososphaeraceae (56.22 % and 4.72 %), Candidatus Nitrocosmicus (7.41 % and 0.60 %) and Haladaptatus (5.05 % and 25.86 %), in ALW and ALC, respectively.
Fig. 3.
Relative abundance of Archaea in soil samples analyzed by the 16S rRNA amplicon sequencing. The figure depicts the distribution of Archaea within phyla (A) and families, based on the sequencing of V4Arc region. In this representation, we observe the relative abundance of phyla in different soil types (A), Thermoproteota and Methanobacteriota are prominent, highlighting an inversion between ALW and ALC. Analyzing the families of Archaea (B), Nitrososphaeraceae and Haloferacaceae are the most representative. Uncontaminated soil sample from Alagoas (ALW), Contaminated soil sample from Alagoas (ALC) and Contaminated soil sample from Rio Grande do Norte (RNC).
3.4. Functional and metabolic analysis based on metagenomic analyses
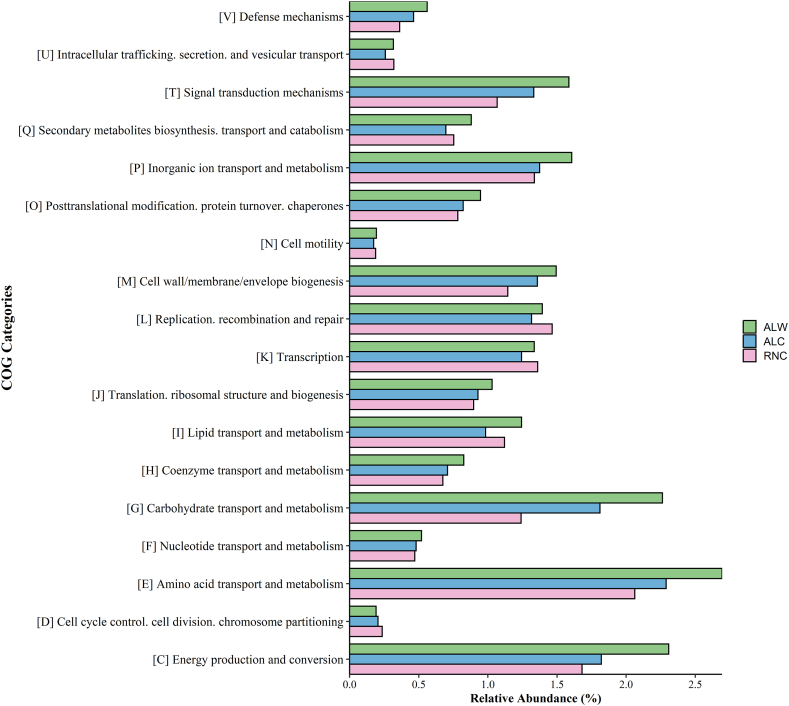
The metagenomic functional analysis of soil samples was identified using COG e SEED databases (Fig. 4 and Fig. S1). In total, 22 functional categories were identified by COG database. Most of the reads were functionally distributed within pathways of amino acid transport and metabolism [E], energy production and conversion [C], and carbohydrate transport and metabolism [G]. We observed that soil without oil contamination (ALW) presented higher relative abundance in all categories, except in DNA replication, recombination, and repair [L], transcription [K], and cell cycle control [D] (Fig. 4). These features were supported by STAMP analysis (Fig. S2).
Fig. 4.
Distribution of the most abundant functional categories among the three soil metagenomes. The colored bars indicate the relative abundance of functional orthologs in the non-contaminated soil (ALW, green), the contaminated soil from production field with chronic contamination by produced water (ALC, blue), and contaminated soil with chronic contamination by oil (RNC, pink). Uncontaminated soil sample from Alagoas (ALW), Contaminated soil sample from Alagoas (ALC) and Contaminated soil sample from Rio Grande do Norte (RNC). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Profile of genes involved in nitrogen metabolism
The coding sequences from SEED were categorizes using the KEGG database to detect the functional genes involved in nitrogen metabolism. We identified 53 groups of orthologs (KO) related to the enzymes involved in nitrogen metabolic pathways (Table S3). Genes in the KO nitrogen metabolic pathways have been identified for ALW (27), ALC (31), and RNC (51). ALC soil exhibited a reduced proportion of genes associated with nitrogen metabolism compared to ALW, particularly genes related to nitrate reduction (narH, narJ, and narI) as well genes involved in the denitrification of nitrous oxide to nitrogen (nosZ) (Fig. S3). Therefore, nitrogen metabolism was more required in contaminated soils, with 15 (KO) found exclusively in RNC. Furthermore, genes associated with nitrogen transport (nrtA, nrtB, and nrtC) were more prevalent in the RNC sample. The biological nitrogen fixation genes (nifX, nifZ, nifQ, nifB, and nifT) and those constituting the nitrogenase complex (nifD, nifK, and nifH) were pronounced in RNC. Genes involved in biological N2 fixation, such genes related to nitrification (nxrA and nxrB) and nitrate reduction (narG, narH, narJ, and narI) were present in all soil samples, with RNC showing the highest abundance.
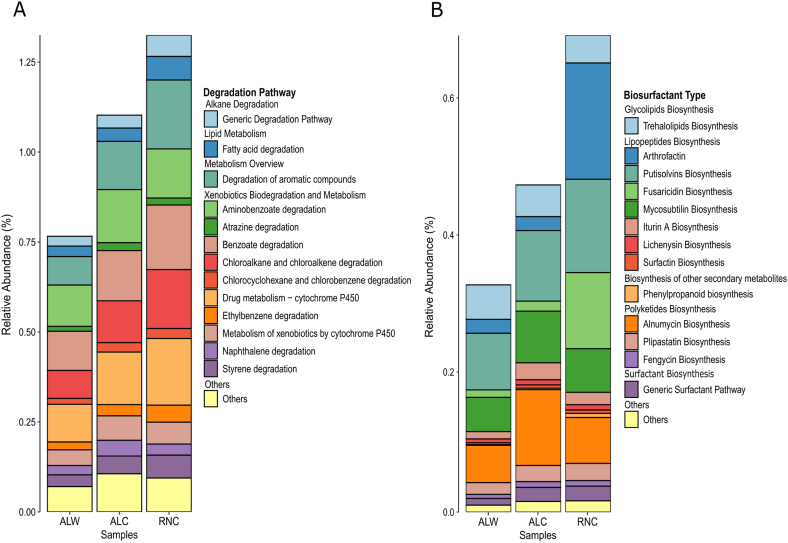
3.6. Hydrocarbon degradation and biosynthesis of biosurfactants
To obtain the functional analysis of genes related to hydrocarbon degradation and biosynthesis of biosurfactants, we employed the integrated BioSurfDB and DIAMOND + MEGAN pipeline. Data analysis within BiosurfDB is systematically classified into three pivotal categories: degradation, methane, and surfactants. It was identified that 0.77–1.33 % of the total reads were attributed to the degradation category (Fig. 5A). The degradation process further delineates into distinct subcategories, encompassing pathways intricately associated with the degradation of organic pollutants. The most abundant pathway subcategory is xenobiotics biodegradation and metabolism in all samples (on average 0.83 %). Within this subcategory, the most prevalent were aminobenzoate degradation, benzoate degradation, drug metabolism-cytochrome P450, and chloroalkane and chloroalkene degradation pathway, which showed a significant difference between contaminated and uncontaminated soil samples (Fig. S4). In chloroalkane and chloroalkene degradation there was a greater detection of aldehyde dehydrogenase, such as ALDH, aldB, mmsA/iolA (ALDH6A1; malonate-semialdehyde dehydrogenase (acetylating)/ethylmalonate-semialdehyde dehydrogenase), aldH (2,5-dioxopentanoate dehydrogenase) and aldH (4-(gamma-glutamylamino) butanal dehydrogenase).
Fig. 5.
Metagenomic profile of biodegradation of hydrocarbons and biosurfactant biosynthesis. Additional functional analysis with BiosurfDB pipeline was used to assess pathways specifically related to petroleum hydrocarbon metabolism. The figure displays the abundance fractions of totally annotated genes in each soil sample. The corresponding metabolic pathways are: (A) Degradation Pathway and (B) Biosurfactant Type. Uncontaminated soil sample from Alagoas (ALW), Contaminated soil sample from Alagoas (ALC) and Contaminated soil sample from Rio Grande do Norte (RNC).
Furthermore, the second most abundant subcategory comprised pathways involving degradation of aromatic compounds, accounting for 0.08–0.19 % of the total reads for the sampled regions, followed by lipids metabolism (0.03–0.07 %) and alkane degradation (0.03–0.06 %). The most frequent genes for the subcategory “Degradation of Aromatic Compounds” include transposases (IS5 family), arsB (arsenical pump membrane protein), chrA (chromate transporter), and xylA (alpha-ketoglutaric semialdehyde dehydrogenase). Comparison between ALW and ALC samples demonstrated an increase in the subcategories involved in degradation of linear hydrocarbons and PAH. However, RNC showed higher values in relation to degradation genes (Fig. 5A–S4).
Concerning the surfactant biosynthesis pathways, the following percentages were found: 0.30 % for ALW, 0.44 % for ALC, and 0.64 % for RNC. Within the surfactant classification, the most abundant subcategory was lipopeptides and polyketyde biosynthesis, with the highest ratings being attributed to putisolvins, arthrofactin, mycosubtilin, fusaricidin and alnumycin biosynthesis. Pairwise comparisons between ALW/ALC and ALW/RNC show differences between the surfactants profile. In the first comparison, the subcategory alnumycin biosynthesis is significantly higher in the contaminated soil from Alagoas, while the second comparison the subcategory arthrofactin is most prevalent in the sample ALC (Fig. S5).
4. Discussion
Oil contamination is a widespread issue, attracting major efforts to develop efficient remediation strategies, often involving application microbial systems to restore contaminated environments [42]. With NGS, it is possible to estimate the microbial population from an environmental sample without the need of isolation and culture [43]. In the present study, we assessed the physicochemical properties of oil-contaminated soil samples, along with a non-contaminated soil, collected at the Brazilian Northeast, a relevant oil-producing region in the country. Additionally, we describe the taxonomic and functional characteristics of microbial communities within these soil samples, as they respond to chronic contamination with specific industrial byproducts. Two main approaches were applied: metagenomics (shotgun metagenomic sequencing) and metataxonomics (16S rRNA amplicon sequencing).
4.1. Physicochemical properties of the soil samples
In this study, soil contaminations are different among the samples. ALC shows produced water contamination and low hydrocarbon content, while RNC displays crude oil contamination, with significant levels of hydrocarbons and produced water. Comparing ALW and ALC, there is a reduction in the pH of the ALC contaminated soil, whereas RNC maintains a neutral pH despite the hydrocarbon contamination. The acidification process was also observed in the São Paulo region (Brazil), another chronically contaminated soil [44]. The pH is crucial in controlling various chemical and biological soil processes [45]. The ideal pH range for effective biodegradation is near neutrality, and metabolic activities can be significantly inhibited with slight pH variations [46]. Liang et al. [45] studied oil fields in different geographical locations in China, emphasizing the importance of soil pH assessment for proper recovery technique selection. Interestingly, despite the acid pH in the ALC soil, there was a reduction in the Acidobacteriota phylum, suggesting that salinity, rather than pH, might be the main regulatory factor. As observed in our study, Yang et al. [47] revealed that high and extreme salinities significantly reduce the abundance of Actinomycetota and Acidobacteriota.
Salinity measurements in both ALC and RNC soils indicated a significant presence of salt in these environments (Table 2). High salinity can affect the composition and activity of soil microbial communities, reshaping their ecological roles in nutrient cycling and other essential processes [6], including soil fertility [48]. According to Gao et al., 2015 oil salinity had a suppressive effect on most bacterial populations without changing their dominance, while soil TPH influenced the bacterial diversity selectively [49]. Most laboratory experiments are conducted in soils artificially contaminated by fresh crude oil. Bioavailable carbon from petroleum could stimulate soil microbial activities, leading to depletion of bioavailable components of crude oil after long-term attenuation. Salinity as a stressful environmental factor for soil microorganisms was reported by Yuan et al. (2007), who found that EC (ranging from 0.32 to 23.05 mS cm−1) had a significantly negative exponential relationship [50]. During bioremediation of oil-contaminated saline soil, high salinity would reduce the metabolic activity of many microorganisms and inhibit microbial degradation [49].
Principal Component Analysis (PCA) identified Conductivity, TPH, PHA, and pH as the most correlated components with the soil microbial community. This observation has also been reported in other studies [[51], [52], [53]], [53]. In this study, soil conductivity, associated with salinity, showed a positive correlation with the Bacillota phylum, while pH exhibited a negative correlation. This correlation pattern was also observed for Bacillota in coastal wetland soils of the YRD region [54]. Additionally, Pseudomonadota showed positive correlations with the presence of hydrocarbons, like soils from other regions [53,55], demonstrating their adaptation to environments polluted by petroleum.
Regarding hydrocarbon contamination, chromatographic analysis show that RNC is a weathered soil (data not shown), showing 1 % of linear hydrocarbons, 79 % of UCM, and only 21 % are unsaturated and aromatic hydrocarbons, representing a challenge for degradation. Hydrocarbon contamination is present in high concentrations, especially those with 3–4 condensed rings. Despite the weathering, pristane and phytane were detected, demonstrating the presence of recalcitrant compounds.
In the weathering process, residual hydrocarbons are trapped in the soil matrix and can adsorb to soil aggregates, hindering bioaccessibility [56]. Some light hydrocarbon fractions (labile fraction) are easily degraded, while heavier fractions remain unavailable for degradation. Monoaromatic compounds (BTEX), when degraded by microorganisms, generally reach non-toxic levels. In contrast, the degradation of polycyclic aromatic hydrocarbons and cycloalkanes is slower, making them potential producers of toxic intermediate metabolites, resulting in low biodegradation efficiency [57]. The effectiveness of hydrocarbon removal from soils are inversely related with soil aging due to weathering [44]. However, other studies suggest that the addition of compounds like extracellular enzymes, light oils, and biosurfactants are alternatives that can enhance the bioavailability of these compounds [57,58].
4.2. Diversity of bacteria and archaea revealed with metataxonomics
Shotgun sequencing offers a valuable perspective for taxonomic assessment by encompassing functional information and genomic details. On the other hand, 16S sequencing also provides robust taxonomic resolution, crucial for a deeper understanding of microbial community composition [59]. To obtain a more comprehensive view of the microbial communities, it may be useful to employ both approaches and integrate the results. Therefore, we opted for 16S rRNA ASV sequencing for taxonomic analysis and WGS sequencing to obtain additional functional information.
While both the 16S rRNA ASV and shotgun (WGS) can provide taxonomic information, only the WGS can provide solid evidence of the functional and metabolic profile. Regarding the taxonomic profiling, there are numerous comparative studies between these approaches in various systems, but the application in soil studies is uncommon [60]. A relevant consideration is that the 16S rRNA ASV and WGS methods typically utilize different taxonomic databases, which may lead to discrepancies. Despite its comprehensiveness, WGS-based metagenomics remains less popular due to its complexity and higher costs [61,62]. Likewise, Ranjan et al. (2016) observed a 13 % increase in Bacteroidota abundance and a decrease in Actinomycetota, while using 16S rRNA ASV. On the other hand, they found that the relative abundance of Bacillota and Pseudomonadota was similarwhen using 16S rRNA ASV and WGS [63]. Since the use of the 16S ASV is more widespread, we find it is a more appropriate tool for comparative analyses with other data in the literature.
When comparing the 16S rRNA ASV results with WGS in our samples, we observed significant biases in the V3–V4 region of the 16S rRNA, for the major phyla. In all samples, there was an overestimation of relative abundance for the Bacillota and Gemmatimonadota phyla, in detriment of Pseudomonadota, Bacteroidota, Chloroflexota, Acidobacteriota. In sample ALC, Actinobacteriota were underestimated. This discrepancy at phylum-level underscores the importance of carefully selecting primers for environmental samples, aiming at more accurate quantification of specific taxa.
The literature also highlights the critical importance of primer selection for 16S rRNA ASV analysis [64,65]. Na et al. (2023) investigated the performance of different 16S rRNA primer sets in clinical samples to detect common microorganisms in oral microbiomes. They observed that primers exert a selective influence on species detection, potentially affecting the estimated bacterial abundance. Notably, they found that V3–V4 primers showed higher relative abundance of the Bacillota phylum compared to the V1–V2 primers, while Pseudomonadota and Actinomycetota were less abundant. Sunagawa et al. (2015) noted that the 515F/806RB primer set was less selective in detecting the Pseudomonadota phylum, dominant in oceans [66]. Additionally, Lee et al. (2023) observed that 23F/228R primer set selectively amplified Bacteroidota to a greater extent compared to other phyla. This trend of Bacteroidota amplification was consistent with results obtained with other primer sets, such as PRK341F/PRK806R, 515F/806R, 515F/806RB, and 515F–Y/926R [64].
In the oil contaminated (RNC) and non-contaminated (ALW) soil samples, 16S rRNA gene analysis revealed that Actinomycetota and Pseudomonadota phyla were dominant. Within Actinomycetota, the most abundant genus in sample RNC was Prauserella, commonly known for comprising halophilic or halotolerant species [67]. Several Prauserella members have been isolated from oil-contaminated soils [67,68], strongly suggesting that it may exhibit the ability to carry out oil degradation. Actinomycetota, in turn, are commonly known for their ability to degrade petroleum hydrocarbons [58]. Previous studies have explored the role of Actinomycetota in the degradation of hydrocarbons, including n-alkanes [69,70]. Another important feature that could potentially increase biodegradation of oil compounds is the ability to produce biosurfactants and express oxidoreductase systems, expected to facilitate bioremediation [58,71]. Another dominant genus identified in this study, Pseudomonas, has been commonly detected in oil contaminated environments, including produced water generated in the Brazilian Northeast onshore platforms, as well as oil contaminated soils [[72], [73], [74], [75]]. Therefore, Pseudomonas seems to be ubiquitous in oil-contaminated environments, thanks to its metabolic versatility and ability to degrade various petroleum hydrocarbons [75,76].
Various studies have reported the use of mixed cultures of Actinomycetota and Pseudomonadota to provide a synergistic activity in degradation of hydrocarbons [[77], [78], [79]]. Thus, suggesting that the two groups could be acting together on the degradation of a variety of hydrocarbons, including PAHs, which are widespread pollutants of major concern, corroborating the importance of metabolic cooperation between species.
In the soil impacted by produced water (ALC), Bacillota and Actinomycetota were dominant, showing a different taxonomic composition compared to samples RNC and ALW, possibly due to the increased salinity. Similarly, Yang et al. described that an increased proportion of Bacillota and Bacteroidota were positively correlated with increased salinity and a high C/N ratio (53.88) [47]. Within Bacillota, the most abundant genera were Bacillus and Staphylococcus, whereas within Actinomycetota, the majority of the ASVs were unclassified at genus level, indicating that these microbial communities are largely unknown.
Interestingly, in samples ALC and ALW Archaea comprised three main families: Nitrososphaeraceae, Haloferacaceae, and Haloadaptaceae. In the non-contaminated control soil (ALW), a relatively high abundance of Nitrososphaeraceae was found, in contrast to sample ALC (Fig. 3). These data corroborate with a previous report showing that different saline gradients in soils from the Yellow River Delta, without any petroleum pollution, displayed a dominance of Nitrososphaeraceae [80]. Family Nitrososphaeraceae belongs to an ammonia-oxidising group (AOA), which plays an important role in nitrification [81]. Moreover, AOA can grow at extremely low levels of NH3, where the bioavailability of substrates is scarce, and oxidize ammonia under acidic conditions [82].
Archaea families Haloferacaceae and Haloadaptaceae are highly abundant in sample ALC, unlike sample ALW that shows lower salinity. These families belong to the class Halobacteria, characterized by tolerance to high salt levels. Accordingly, salinity levels seem to have a more drastic impact on the abundance and variability of archaeal communities, when compared to hydrocarbon contamination [6]. In fact, variations in salinity are reported as a pivotal factor on the archaeal population in various contaminated matrices (soils, sediments, groundwater) [6,83,84]. Our results also corroborate with the findings of Navarro-Noya et al. [85] who observed that the presence of the archaeal members with very specialized functions are directly dependent on the physicochemical characteristics of the environment (e.g. pH and EC values). Such findings lead us to conclude that it is highly likely that archaeal populations participate in biogeochemical cycles through different mechanisms, according to the environment, contributing to the great adaptability of this community.
The Shannon diversity for V3V4 region between ALW and ALC is 5.47 to 4.64, respectively. Comparing ALC to RNC revealed that the latter had slightly higher diversity (4.74). In similar studies, a decrease in diversity was observed in response to salinity, in the Yellow River delta, China (from 6.50 to 5.15) [47]. In soils near our study area, in Carmópolis/Sergipe, a significant reduction in Shannon diversity index for bacteria was identified with increased oil and NaCl concentration (0–50 g kg−1) [6].
In addition, a lower variety of ASVs was observed for Archaea in sample ALC, as shown by Shannon indices (3.26 to ALW and 2.71 to ALC). Similar findings are reported by Camacho-Montealegre et al. [6], where a slight reduction in diversity was identified in increased salinity. Generally, Shannon values for Archaea vary with the environment, especially under extreme conditions. For example, in a produced water sample from a Brazilian offshore field [86], a value of 4.65 was found for Archaea. In groundwater contaminated with TPH gradients, variations in Shannon values were also observed, ranging from 5.51 to 3.98 [87].
4.3. Functional profile
Allied to the taxonomic composition of a microbial community, an important aspect of metagenomic studies is the assessment of the contribution of each component of the sample regarding the functional genes. Previous studies have documented a correlation between the identification of functional genes in polluted environments and the adaptation and reshaping of the indigenous microbial community [88,89]. Based on the COG annotations, 22 functional categories were identified in the present study. The most abundant classes across all three samples were amino acid transport and metabolism [E], energy production and conversion [C], and carbohydrate transport and metabolism [G]. These classes were also representative in other contaminated soil analyses [[90], [91], [92]]. According to the COG analysis, there is a significant reduction in most functions from ALW to ALC, this functional difference may have occurred due to the stress promoted by the contaminants in ALC. In agreement with our findings, previous reports demonstrate that high salinity and hydrocarbon concentrations may lead to restructuring in soil microbial functions and metabolic pathways [18,88,93].
Nevertheless, the RNC soil showed a higher abundance in the category transcription [K], replication, recombination, and repair [L], intracellular trafficking, secretion, and vesicular transport [U], and cell cycle control, cell division, chromosome partitioning [D] when compared to ALW and ALC samples. The high proportion of these functions in RNC soil might be attributed to defense mechanisms developed to adapt to hydrocarbon-induced stress. This aligns with functional data derived from a case study of a historically petroleum-contaminated soil [91]. Particularly noteworthy is the observation that in RNC soil, the categories replication, recombination, and repair [L] and transcription [K] exhibited a higher proportion than in the ALW sample, suggesting a potential adaptation to contamination-induced stress by enhancing mechanisms associated with core metabolism, especially DNA repair [80,91]. Another study reported increased functions related to DNA replication and repair in the microbiota of soils contaminated with high levels of heavy metals and petroleum-derived hydrocarbons [94]. It has also been reported that high salinity damages structural molecules and their biological functions, thereby stimulating cellular regeneration [95]. Additionally, petroleum reservoirs can represent extreme environments for microbial life due to high salinity and low nutrient availability [96].
The functional profile predicted by the COG analysis highlighted the metabolic potential of microbial communities in the investigated samples. These results imply that microorganisms under different environmental stress conditions, such as hydrocarbon contamination and high salt concentration from petroleum or produced water, may positively regulate energy metabolism for homeostasis and survival, while negatively affecting other pathways, such as defense mechanisms, cell cycle control, motility, and intracellular trafficking.
4.4. Profile of genes involved in nitrogen metabolism
Nitrogen limitation significantly influences the bioremediation process in soils. Functional genes encoding key enzymes in the nitrogen cycle serve as valuable indicators during the degradation process [97]. In soils, the ideal carbon/nitrogen (C/N) ratio generally employed is 100:10 [40,41]. Our results reveal an important nitrogen deficiency in the two contaminated soils, as the C/N ratio deviates from the recommended values (Table 2). In soil, C and N can act as electron donors and substrates for denitrifying bacteria. The nitrous oxide reductase (nosZ), is directly linked to the final step of denitrification process, significantly influencing the C/N ratio [98,99].
Previous studies also reported that salinity levels and petroleum hydrocarbons alter the metabolic flow of nitrogen [18,100,101], as the activity of nitrogen-fixing bacteria is generally inhibited at higher salt concentrations [102]. In fact, of the contaminated samples investigated herein, ALC presented a higher value of salinity when compared to RNC, along with lower abundance of nitrification and denitrification pathways, as revealed by the functional analysis (Table S3). The change in the abundance of genes that control nitrogen cycle, as observed between ALW and ALC, signals the functional loss of the nitrogen cycle in the soil. In general, high soil salinity is a challenge in any ecosystem, as excess salts can induce osmotic stress in plant roots, reduce water and nutrient absorption, and even plant mortality [103,104].
Nitrogen-cycling bacteria, including nitrogen-fixers, nitrifiers, and denitrifiers, play essential roles in nitrogen cycling [105]. The KEGG analysis revealed a greater demand for nitrogen metabolism in RNC soil, with 15 orthologous groups exclusively present in this soil. The nitric oxide reductase (NOR) enzyme, crucial for reducing NO to N2O in anaerobic bacterial denitrification [106,107], was identified solely in RNC (norB and norC). This aligns with the prevalence of denitrifying representatives within the abundant genus Pseudomonas in the RNC, which commonly includes denitrifying representatives. Genes such as nirS, nirK and nosZ, commonly used in denitrifying functional analysis, were detected in 14 Pseudomonas species, indicating potential N2O emissions from biological denitrification [[108], [109], [110]]. Despite their presence, these genes exhibited lower frequencies than other pathways related to the nitrogen cycle. Genes related to nitrate reduction were more abundant in RNC soil, including narG, a well-documented component in this pathway [111,112]. The increase in genes associated with the nitrogen cycle, including nirK, nosZ, narG, and norB, has been associated to improved biodegradation process [97].
The study by Scott et al. [101] suggests that PAH contamination may lead to a shift in the metabolic flow of nitrogen to denitrification pathways, potentially including the anammox pathway. Furthermore, the availability of nutrients such as nitrogen may be limited in sediments contaminated by PAHs, since most of the nitrogen in petroleum is found in aromatic heterocyclic compounds whose carbon-nitrogen bonds are difficult to break.
4.5. Hydrocarbon degradation and biosurfactants
Microorganisms can also facilitate the biodegradation and metabolism of xenobiotics to protect soils from exogenous pollutants such as oil and heavy metals [113]. Considering that the degradation pathways and regulatory enzymes of foreign compounds by microorganisms in oil-contaminated soils are central to the study, we focused on analyzing the difference between the samples, regarding hydrocarbon degradation and biosurfactant synthetic pathways. The results obtained from the Megan + BiosurfDB pipeline showed the predominance of genes related to xenobiotics biodegradation and metabolism, and degradation of aromatic compounds were higher in RNC sample (Fig. 5). This finding correlates the RNC soil's physicochemical characteristics and taxonomic profile, which contains higher quantities of TPH and PAH, allied to abundant taxonomic groups known to degrade hydrocarbons, such as Pseudomonas and Prauserella. Several members of these genera have the ability to grow in presence of a wide variety of hydrocarbons and to degrade n-alkanes and PAHs, such as fluorene, phenanthrene, and pyrene [114,115].
Comparing contaminated and non-contaminated soils revealed that genes involved in degradation and metabolism of xenobiotic compounds (except toluene, nitrotoluene, and carbon degradation) and aromatic compounds were significantly enriched in the contaminated soils. The most abundant pathways were aminobenzoate, benzoate, chloroalkane and chloroalkene degradation, drug metabolism-cytochrome P450, and degradation of aromatic compounds, demonstrating very distinct metabolic profiles between contaminated and non-contaminated samples. Successful microbial biodegradation requires several specific conditions, such as temperature, oxygen, pH, salinity, water, and nutrients [116]. Salinity and water activity could be responsible for decreasing the hydrocarbon metabolism rates due to a general decline in microbial metabolic rates, including key enzymes [117,118]. Nevertheless, even in highly saline conditions, our data showed a rich microbiome profile, indicating the potential of the microbial community for biodegradation. In addition, previous metagenomic and metatranscriptomic studies of other saline soil samples have demonstrated high transcriptional activity and abundance of genes related to the degradation and metabolism of xenobiotics, suggesting that it could be positively influenced by salinity [18,119].
Bacterial biosurfactants are highly effective at reducing surface and interfacial tension, also acting as antibiotics, biofilm modulators, signaling molecules, conferring adaptive advantages, and stimulating horizontal gene transfer within and between species [120,121]. Furthermore, biosurfactants aid in nutrient uptake by emulsifying hydrophobic compounds, thereby increasing their solubility in water, and making them more accessible [122].
In the lipopeptide class, a significant difference in abundance was found for arthrofactin, fusaricidin, mycosubtilin, and putisolvins, which were more abundant within the contaminated microbiomes (Fig. 5B). In RNC, the proportions of arthrofactin and fusaricidin were approximately eight times higher than in ALC and ALW soils. The proportion of putisolvins in RNC was also higher when compared to the other two soils. In RNC, the predominance of the Pseudomonadota was observed, while in ALW and ALC Actinomycetota were more abundant, indicating an adaptation of these microorganisms, possibly induced by the contaminants and natural soil composition. Lipopeptides have been described in several genera, from diverse phyla, such as Streptomyces, Serratia, Burkholderia, Pseudomonas, and Bacillus [[123], [124], [125]]. Interestingly, arthrofactin and putisolvins are typically produced by Pseudomonas [126], an abundant genus found in RNC. Biosynthesis of lipopeptides occurs mainly in oxygen-rich environments [127], mediating active or passive cell motility. These compounds allow the expansion and displacement of bacterial colonies [128], which also facilitates degradation of hydrocarbons, including PAHs [129].
The biosynthesis of Alnumycin, a polyketide compound, exhibited higher abundance in contaminated soils, particularly in the ALC sample. Within this functional category, there was a prevalence of genes linked to Actinomycetota. Polyketides, known for their diverse functions, are implicated in inhibiting biofilms and engaging in microbial competition, as indicated in previous studies [130].
Taken together, the present study shows a clear association between predominance of aromatic compounds degradation genes and less abundant biosurfactant categories, a similar profile to those of previously described terrestrial microbiomes. According to Oliveira et al. [131], terrestrial biomes have more degradation genes, especially for cyclic compounds, and fewer surfactant genes, when compared to aquatic biomes.
Data availability statement
The sequences obtained in this study have been submitted to the NCBI Sequence Read Archive (SRA) database under BioProject accession (PRJNA1041842).
CRediT authorship contribution statement
Danielly C.O. Mariano: Writing – original draft, Methodology, Investigation. Graciela Maria Dias: Writing – original draft, Data curation. Michele Rocha Castro: Writing – review & editing. Diogo Antonio Tschoeke: Writing – review & editing, Data curation. Fernando J.S. de Oliveira: Resources, Conceptualization. Eliana Flavia C. Sérvulo: Supervision, Conceptualization. Bianca Cruz Neves: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Petróleo Brasileiro S.A. (Petrobras), Grant No. 0050.0079375.12.9, awarded to Bianca C. Neves. We thank Hadassa Loth de Oliveira for invaluable contribution and design of the graphical abstract.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34336.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Copard Y., Amiotte-Suchet P., Di-Giovanni C. Storage and release of fossil organic carbon related to weathering of sedimentary rocks. Earth Planet Sci. Lett. 2007;258:345–357. doi: 10.1016/J.EPSL.2007.03.048. [DOI] [Google Scholar]
- 2.Mohaddes K., Pesaran M.H. Oil prices and the global economy: is it different this time around? Energy Econ. 2017;65:315–325. doi: 10.1016/J.ENECO.2017.05.011. [DOI] [Google Scholar]
- 3.Hazen T.C., Dubinsky E.A., DeSantis T.Z., Andersen G.L., Piceno Y.M., Singh N., Jansson J.K., Probst A., Borglin S.E., Fortney J.L., Stringfellow W.T., Bill M., Conrad M.E., Tom L.M., Chavarria K.L., Alusi T.R., Lamendella R., Joyner D.C., Spier C., Baelum J., Auer M., Zemla M.L., Chakraborty R., Sonnenthal E.L., D'Haeseleer P., Holman H.Y.N., Osman S., Lu Z., Van Nostrand J.D., Deng Y., Zhou J., Mason O.U. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science (1979) 2010;330:204–208. doi: 10.1126/SCIENCE.1195979/SUPPL_FILE/PAPV2.PDF. [DOI] [PubMed] [Google Scholar]
- 4.Johnston J.E., Lim E., Roh H., Johnston J.E. Impact of upstream oil extraction and environmental public health: a review of the evidence. Sci. Total Environ. 2019;657:187–199. doi: 10.1016/j.scitotenv.2018.11.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliveira L.M.C., Stefano P.H.P., Vedana L.A., Carregosa J.C., Santos M.M.N., Wisniewski A., Pereira F.M.C.C. A hydrogeological impact survey on the largest onshore oil field in Brazil: physicochemical and total petroleum hydrocarbon (TPH) analyses in the south of Japaratuba River Basin, Sergipe. Environ. Earth Sci. 2020;79:1–14. doi: 10.1007/S12665-020-09121-0/TABLES/2. [DOI] [Google Scholar]
- 6.Camacho-Montealegre C.M., Rodrigues E.M., Morais D.K., Tótola M.R. Prokaryotic community diversity during bioremediation of crude oil contaminated oilfield soil: effects of hydrocarbon concentration and salinity. Braz. J. Microbiol. 2021;52:787–800. doi: 10.1007/S42770-021-00476-5/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng P., Ma A., Wei X., Chen X., Yin J., Hu F., Zhuang X., Song M., Zhuang G. Interaction and spatio-taxonomic patterns of the soil microbiome around oil production wells impacted by petroleum hydrocarbons. Environmental Pollution. 2022;307 doi: 10.1016/J.ENVPOL.2022.119531. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Rodríguez N., Rivera-Cruz M.C., Trujillo-Narcía A., Almaráz-Suárez J.J., Salgado-García S. Spatial distribution of oil and Biostimulation through the Rhizosphere of Leersia hexandra in degraded soil. Water Air Soil Pollut. 2016;227:1–14. doi: 10.1007/S11270-016-3030-9/FIGURES/5. [DOI] [Google Scholar]
- 9.Hewelke E., Szatyłowicz J., Hewelke P., Gnatowski T., Aghalarov R. The impact of diesel oil pollution on the hydrophobicity and CO2 efflux of forest soils. Water Air Soil Pollut. 2018;229 doi: 10.1007/S11270-018-3720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das R., Kazy S.K. Microbial diversity, community composition and metabolic potential in hydrocarbon contaminated oily sludge: prospects for in situ bioremediation. Environ. Sci. Pollut. Res. Int. 2014;21:7369–7389. doi: 10.1007/S11356-014-2640-2. [DOI] [PubMed] [Google Scholar]
- 11.Sattar S., Hussain R., Shah S.M., Bibi S., Ahmad S.R., Shahzad A., Zamir A., Rauf Z., Noshad A., Ahmad L. Composition, impacts, and removal of liquid petroleum waste through bioremediation as an alternative clean-up technology: a review. Heliyon. 2022;8 doi: 10.1016/J.HELIYON.2022.E11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao S., Chen W., Wang J., Du N., Li Q., Wei G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome. 2018;6:1–13. doi: 10.1186/S40168-018-0526-0/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang J., Zhang R., Zeng Y., Dai T., Ye Z., Gao Q., Yang Y., Guo X., Li G., Zhou J. Petroleum pollution changes microbial diversity and network complexity of soil profile in an oil refinery. Front. Microbiol. 2023;14 doi: 10.3389/FMICB.2023.1193189/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon G., Stavi I., Shavit U., Rosenzweig R. Oil spill effects on soil hydrophobicity and related properties in a hyper-arid region. Geoderma. 2018;312:114–120. doi: 10.1016/J.GEODERMA.2017.10.008. [DOI] [Google Scholar]
- 15.Liu J., Meng Z., Liu X., Zhang X.H. Microbial assembly, interaction, functioning, activity and diversification: a review derived from community compositional data. Mar Life Sci Technol. 2019;1:112–128. doi: 10.1007/S42995-019-00004-3/FIGURES/3. [DOI] [Google Scholar]
- 16.Micheel C.M., Nass S.J., Omenn G.S. C. On the R. Of O.-B.T. For P.P.O. In C. Trials, B. on H.C. Services, B. on H.S. Policy, I. of Medicine. Omics-Based Clinical Discovery: Science, Technology, and Applications. 2012 https://www.ncbi.nlm.nih.gov/books/NBK202165/ [Google Scholar]
- 17.Sharma P., Kumar S., Pandey A. Bioremediated techniques for remediation of metal pollutants using metagenomics approaches: a review. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/J.JECE.2021.105684. [DOI] [Google Scholar]
- 18.Muñoz-García A., Mestanza O., Isaza J.P., Figueroa-Galvis I., Vanegas J. Influence of salinity on the degradation of xenobiotic compounds in rhizospheric mangrove soil. Environmental Pollution. 2019;249:750–757. doi: 10.1016/J.ENVPOL.2019.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Nie Y., Zhao J.Y., Tang Y.Q., Guo P., Yang Y., Wu X.L., Zhao F. Species divergence vs. functional convergence characterizes crude oil microbial community assembly. Front. Microbiol. 2016;7 doi: 10.3389/FMICB.2016.01254/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie W., Zhang C., Zhou X., Wang P. Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl. Microbiol. Biotechnol. 2014;98:7971–7982. doi: 10.1007/S00253-014-5838-9/TABLES/4. [DOI] [PubMed] [Google Scholar]
- 21.Soil Sampling | US EPA, (n.d.). https://www.epa.gov/quality/soil-sampling (accessed June 7, 2024).
- 22.Corwin D.L., Yemoto K. Salinity: electrical conductivity and total dissolved solids. Soil Sci. Soc. Am. J. 2020;84:1442–1461. doi: 10.1002/saj2.20154. [DOI] [Google Scholar]
- 23.E.W. Rice, R.B. Baird, A.D. Eaton, Standard Methods for the Examination of Water and Wastewater, 23rd Edition, (n.d.).
- 24.Yu Y., Lee C., Kim J., Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005;89:670–679. doi: 10.1002/BIT.20347. [DOI] [PubMed] [Google Scholar]
- 25.Turner S., Pryer K.M., Miao V.P.W., Palmer J.D. Investigating Deep Phylogenetic relationships among Cyanobacteria and Plastids by Small Subunit rRNA sequence Analysis1. J. Eukaryot. Microbiol. 1999;46:327–338. doi: 10.1111/J.1550-7408.1999.TB04612.X. [DOI] [PubMed] [Google Scholar]
- 26.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Bin Kang K., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J.J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8 37):852–857. doi: 10.1038/s41587-019-0209-9. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018;6:1–17. doi: 10.1186/S40168-018-0470-Z/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and “All-species Living tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/NAR/GKT1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614. doi: 10.1093/BIOINFORMATICS/BTT593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2014;12(1 12):59–60. doi: 10.1038/nmeth.3176. 2014. [DOI] [PubMed] [Google Scholar]
- 32.Huson D.H., Beier S., Flade I., Górska A., El-Hadidi M., Mitra S., Ruscheweyh H.J., Tappu R. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016;12 doi: 10.1371/JOURNAL.PCBI.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–D592. doi: 10.1093/NAR/GKAC963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/NAR/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva G.F., Gautam A., Duarte I.C.S., Delforno T.P., de Oliveira V.M., Huson D.H. Interactive analysis of biosurfactants in fruit-waste fermentation samples using BioSurfDB and MEGAN. Sci. Rep. 2022;12:1–11. doi: 10.1038/s41598-022-11753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123. doi: 10.1093/BIOINFORMATICS/BTU494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y., Hochberg Y. Controlling the False Discovery rate: a Practical and Powerful Approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. doi: 10.1111/J.2517-6161.1995.TB02031.X. [DOI] [Google Scholar]
- 38.R: The R Project for Statistical Computing (n.d.). https://www.r-project.org/(accessed June 30, 2024).
- 39.Posit | The Open-Source Data Science Company (n.d.). https://posit.co/(accessed June 27, 2024).
- 40.Liang Y., Li G., Van Nostrand J.D., He Z., Wu L., Deng Y., Zhang X., Zhou J. Microarray-based analysis of microbial functional diversity along an oil contamination gradient in oil field. FEMS Microbiol. Ecol. 2009;70:324–333. doi: 10.1111/J.1574-6941.2009.00774.X. [DOI] [PubMed] [Google Scholar]
- 41.Oh Y.S., Sim D.S., Kim S.J. Effects of nutrients on crude oil biodegradation in the Upper Intertidal Zone. Mar. Pollut. Bull. 2001;42:1367–1372. doi: 10.1016/S0025-326X(01)00166-7. [DOI] [PubMed] [Google Scholar]
- 42.Thacharodi A., Hassan S., Singh T., Mandal R., Chinnadurai J., Khan H.A., Hussain M.A., Brindhadevi K., Pugazhendhi A. Bioremediation of polycyclic aromatic hydrocarbons: an updated microbiological review. Chemosphere. 2023;328 doi: 10.1016/J.CHEMOSPHERE.2023.138498. [DOI] [PubMed] [Google Scholar]
- 43.Costa O.Y.A., Costa O.Y.A., De Hollander M., Pijl A., Liu B., Kuramae E.E. Cultivation-independent and cultivation-dependent metagenomes reveal genetic and enzymatic potential of microbial community involved in the degradation of a complex microbial polymer. Microbiome. 2020;8:1–19. doi: 10.1186/S40168-020-00836-7/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trindade P.V.O., Sobral L.G., Rizzo A.C.L., Leite S.G.F., Soriano A.U. Bioremediation of a weathered and a recently oil-contaminated soils from Brazil: a comparison study. Chemosphere. 2005;58:515–522. doi: 10.1016/J.CHEMOSPHERE.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 45.Liang Y., Zhang X., Wang J., Li G. Spatial variations of hydrocarbon contamination and soil properties in oil exploring fields across China. J. Hazard Mater. 2012;241–242:371–378. doi: 10.1016/J.JHAZMAT.2012.09.055. [DOI] [PubMed] [Google Scholar]
- 46.Kebede G., Tafese T., Abda E.M., Kamaraj M., Assefa F. Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: mechanisms and impacts. J. Chem. 2021;2021 doi: 10.1155/2021/9823362. [DOI] [Google Scholar]
- 47.Yang C., Li K., Lv D., Jiang S., Sun J., Lin H., Sun J. Inconsistent response of bacterial phyla diversity and abundance to soil salinity in a Chinese delta. Sci. Rep. 2021;11 doi: 10.1038/S41598-021-92502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy N., Crohn D.M. Effects of soil salinity and carbon availability from organic amendments on nitrous oxide emissions. Geoderma. 2014;235–236:363–371. doi: 10.1016/J.GEODERMA.2014.07.022. [DOI] [Google Scholar]
- 49.Gao Y.C., Guo S.H., Wang J.N., Li D., Wang H., Zeng D.H. Effects of different remediation treatments on crude oil contaminated saline soil. Chemosphere. 2014;117:486–493. doi: 10.1016/J.CHEMOSPHERE.2014.08.070. [DOI] [PubMed] [Google Scholar]
- 50.Yuan B.C., Li Z.Z., Liu H., Gao M., Zhang Y.Y. Microbial biomass and activity in salt affected soils under arid conditions. Appl. Soil Ecol. 2007;35:319–328. doi: 10.1016/J.APSOIL.2006.07.004. [DOI] [Google Scholar]
- 51.O'Brien F.J.M., Almaraz M., Foster M.A., Hill A.F., Huber D.P., King E.K., Langford H., Lowe M.A., Mickan B.S., Miller V.S., Moore O.W., Mathes F., Gleeson D., Leopold M. Soil salinity and pH drive soil bacterial community composition and diversity along a lateritic slope in the Avon River Critical Zone Observatory, Western Australia. Front. Microbiol. 2019;10 doi: 10.3389/FMICB.2019.01486/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaoping K., Zhiwei D., Bingchen W., Huihui W., Jialiang L., Hongbo S. Changes of sensitive microbial community in oil polluted soil in the coastal area in Shandong, China for ecorestoration. Ecotoxicol. Environ. Saf. 2021;207 doi: 10.1016/J.ECOENV.2020.111551. [DOI] [PubMed] [Google Scholar]
- 53.Wang S., Sun L., Ling N., Zhu C., Chi F., Li W., Hao X., Zhang W., Bian J., Chen L., Wei D. Exploring soil factors determining composition and structure of the bacterial communities in saline-alkali soils of Songnen plain. Front. Microbiol. 2020;10:2902. doi: 10.3389/FMICB.2019.02902/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Q., Chang H., Yang X., Wang D., Wang W. Salinity and nutrient modulate soil bacterial communities in the coastal wetland of the Yellow River Delta, China. Environ. Sci. Pollut. Control Ser. 2021;28:14621–14631. doi: 10.1007/s11356-020-11626-x. [DOI] [PubMed] [Google Scholar]
- 55.Gao H., Wu M., Liu H., Xu Y., Liu Z. Effect of petroleum hydrocarbon pollution levels on the soil microecosystem and ecological function. Environmental Pollution. 2022;293 doi: 10.1016/j.envpol.2021.118511. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Y., Brassington K.J., Prpich G., Paton G.I., Semple K.T., Pollard S.J.T., Coulon F. Insights into the biodegradation of weathered hydrocarbons in contaminated soils by bioaugmentation and nutrient stimulation. Chemosphere. 2016;161:300. doi: 10.1016/J.CHEMOSPHERE.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan M.A.I., Biswas B., Smith E., Naidu R., Megharaj M. Toxicity assessment of fresh and weathered petroleum hydrocarbons in contaminated soil- a review. Chemosphere. 2018;212:755–767. doi: 10.1016/J.CHEMOSPHERE.2018.08.094. [DOI] [PubMed] [Google Scholar]
- 58.Behera S., Das S. Potential and prospects of Actinobacteria in the bioremediation of environmental pollutants: cellular mechanisms and genetic regulations. Microbiol. Res. 2023;273 doi: 10.1016/J.MICRES.2023.127399. [DOI] [PubMed] [Google Scholar]
- 59.Jaarsma A.H., Sipes K., Zervas A., Jiménez F.C., Ellegaard-Jensen L., Thøgersen M.S., Stougaard P., Benning L.G., Tranter M., Anesio A.M. Exploring microbial diversity in Greenland Ice Sheet supraglacial habitats through culturing-dependent and -independent approaches. FEMS Microbiol. Ecol. 2023;99:1–16. doi: 10.1093/FEMSEC/FIAD119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toole D.R., Zhao J., Martens-Habbena W., Strauss S.L. Bacterial functional prediction tools detect but underestimate metabolic diversity compared to shotgun metagenomics in southwest Florida soils. Appl. Soil Ecol. 2021;168 doi: 10.1016/J.APSOIL.2021.104129. [DOI] [Google Scholar]
- 61.Khachatryan L., de Leeuw R.H., Kraakman M.E.M., Pappas N., te Raa M., Mei H., de Knijff P., Laros J.F.J. Taxonomic classification and abundance estimation using 16S and WGS—a comparison using controlled reference samples. Forensic Sci Int Genet. 2020;46 doi: 10.1016/J.FSIGEN.2020.102257. [DOI] [PubMed] [Google Scholar]
- 62.Kuczynski J., Lauber C.L., Walters W.A., Parfrey L.W., Clemente J.C., Gevers D., Knight R. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2011;13:47–58. doi: 10.1038/NRG3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranjan R., Rani A., Metwally A., McGee H.S., Perkins D.L. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016;469:967–977. doi: 10.1016/J.BBRC.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee H.B., Jeong D.H., Cho B.C., Park J.S. Comparative analyses of eight primer sets commonly used to target the bacterial 16S rRNA gene for marine metabarcoding-based studies. Front. Mar. Sci. 2023;10 doi: 10.3389/FMARS.2023.1199116/BIBTEX. [DOI] [Google Scholar]
- 65.Na H.S., Song Y., Yu Y., Chung J. Comparative analysis of primers used for 16S rRNA gene sequencing in oral microbiome studies. Methods Protoc. 2023;6:71. doi: 10.3390/MPS6040071/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A., Cornejo-Castillo F.M., Costea P.I., Cruaud C., D'Ovidio F., Engelen S., Ferrera I., Gasol J.M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B.T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Boss E., Follows M., Karp-Boss L., Krzic U., Reynaud E.G., Sardet C., Sieracki M., Velayoudon D., Bowler C., De Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M.B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S.G., Bork P. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348 doi: 10.1126/SCIENCE.1261359. [DOI] [PubMed] [Google Scholar]
- 67.Dastgheib S.M.M., Tirandaz H., Nikou M.M., Ramezani M., Shavandi M., Amoozegar M.A., Ventosa A. Prauserella oleivorans sp. nov., a halophilic and thermotolerant crude-oil-degrading actinobacterium isolated from an oil-contaminated mud pit. Int. J. Syst. Evol. Microbiol. 2017;67:3381–3386. doi: 10.1099/IJSEM.0.002124. [DOI] [PubMed] [Google Scholar]
- 68.Almutairi A. Prauserella soli sp. nov., isolated from crude oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2015;65:3060–3065. doi: 10.1099/IJS.0.000378/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 69.Quatrini P., Scaglione G., De Pasquale C., Riela S., Puglia A.M. Isolation of Gram‐positive n‐alkane degraders from a hydrocarbon‐contaminated Mediterranean shoreline. J. Appl. Microbiol. 2008;104:251–259. doi: 10.1111/J.1365-2672.2007.03544.X. [DOI] [PubMed] [Google Scholar]
- 70.Olajuyigbe F.M., Ehiosun K.I. Assessment of crude oil degradation efficiency of newly isolated actinobacteria reveals untapped bioremediation potentials. 2016. 133–143. [DOI]
- 71.Alvarez V.M., Marques J.M., Korenblum E., Seldin L. Comparative bioremediation of crude oil-Amended tropical soil Microcosms by natural Attenuation, bioaugmentation, or Bioenrichment. Appl Environ Soil Sci. 2011:1–10. doi: 10.1155/2011/156320. 2011. [DOI] [Google Scholar]
- 72.de Oliveira H.L., Dias G.M., Neves B.C. Genome sequence of Pseudomonas aeruginosa PA1-Petro—a role model of environmental adaptation and a potential biotechnological tool. Helyon. 2022;8 doi: 10.1016/J.HELIYON.2022.E11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Araujo A.S.F., Oliveira L.M. de S., Melo V.M.M., Antunes J.E.L., Araujo F.F., Mendes L.W. Distinct taxonomic composition of soil bacterial community across a native gradient of Cerrado-Ecotone-Caatinga. Appl. Soil Ecol. 2021;161 doi: 10.1016/j.apsoil.2020.103874. [DOI] [Google Scholar]
- 74.Dong W., He C., Li Y., Huang C., Chen F., Ma Y. Complete genome sequence of a versatile hydrocarbon degrader, Pseudomonas aeruginosa DN1 isolated from petroleum-contaminated soil. Gene Rep. 2017;7:123–126. doi: 10.1016/J.GENREP.2017.04.001. [DOI] [Google Scholar]
- 75.Swati P. Ghosh, Thakur I.S. Biodegradation of pyrene by Pseudomonas sp. ISTPY2 isolated from landfill soil: process optimisation using Box-Behnken design model. Bioresour. Technol. Rep. 2019;8 doi: 10.1016/J.BITEB.2019.100329. [DOI] [Google Scholar]
- 76.Medić A.B., Karadžić I.M. Pseudomonas in environmental bioremediation of hydrocarbons and phenolic compounds- key catabolic degradation enzymes and new analytical platforms for comprehensive investigation. World J. Microbiol. Biotechnol. 2022;38(10 38):1–28. doi: 10.1007/S11274-022-03349-7. 2022. [DOI] [PubMed] [Google Scholar]
- 77.Sun R., Jin J., Sun G., Liu Y., Liu Z. Screening and degrading characteristics and community structure of a high molecular weight polycyclic aromatic hydrocarbon-degrading bacterial consortium from contaminated soil. Journal of Environmental Sciences. 2010;22:1576–1585. doi: 10.1016/S1001-0742(09)60292-8. [DOI] [PubMed] [Google Scholar]
- 78.Isaac P., Martínez F.L., Bourguignon N., Sánchez L.A., Ferrero M.A. Improved PAHs removal performance by a defined bacterial consortium of indigenous Pseudomonas and actinobacteria from Patagonia, Argentina. Int Biodeterior Biodegradation. 2015;101:23–31. doi: 10.1016/j.ibiod.2015.03.014. [DOI] [Google Scholar]
- 79.Feng X., Liu Z., Jia X., Lu W. vol. 26. Transactions of Tianjin University; China: 2020. pp. 22–32. (Distribution of Bacterial Communities in Petroleum-Contaminated Soils from the Dagang Oilfield). [DOI] [Google Scholar]
- 80.Wang B., Kuang S., Shao H., Cheng F., Wang H. Improving soil fertility by driving microbial community changes in saline soils of Yellow River Delta under petroleum pollution. J Environ Manage. 2022;304 doi: 10.1016/J.JENVMAN.2021.114265. [DOI] [PubMed] [Google Scholar]
- 81.Stahl D.A., De La Torre J.R. Physiology and diversity of ammonia-oxidizing archaea. 2012. 83–101. [DOI] [PubMed]
- 82.Hatzenpichler R. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 2012;78:7501–7510. doi: 10.1128/AEM.01960-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li R., Xiao X., Zhao Y., Tu B., Zhu X. Characteristics of the archaeal communities in petroleum hydrocarbon-contaminated groundwater. Water Air Soil Pollut. 2022;233:1–11. doi: 10.1007/S11270-022-05544-6/FIGURES/5. [DOI] [Google Scholar]
- 84.Liu B.B., Govindan R., Muthuchamy M., Cheng S., Li X., Ye L., you Wang L., xian Guo S., Li W.J., Alharbi N.S., M Khaled J., Kadaikunnan S. Halophilic archaea and their extracellular polymeric compounds in the treatment of high salt wastewater containing phenol. Chemosphere. 2022;294 doi: 10.1016/J.CHEMOSPHERE.2022.133732. [DOI] [PubMed] [Google Scholar]
- 85.Navarro-Noya Y.E., Valenzuela-Encinas C., Sandoval-Yuriar A., Jiménez-Bueno N.G., Marsch R., Dendooven L. Archaeal communities in a heterogeneous Hypersaline-Alkaline soil. Archaea. 2015;2015 doi: 10.1155/2015/646820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Sousa Pires A., Dias G.M., Chagas de Oliveira Mariano D., Akamine R.N., Cruz de Albuquerque A.C., Groposo C., Soares Ribeiro C.M., de Figueiredo Vilela L., Neves B.C. Molecular diversity and abundance of the microbial community associated to an offshore oil field on the southeast of Brazil. Int Biodeterior Biodegradation. 2021;160 doi: 10.1016/J.IBIOD.2021.105215. [DOI] [Google Scholar]
- 87.Li R., Xiao X., Zhao Y., Tu B., Zhu X. Characteristics of the archaeal communities in petroleum hydrocarbon-contaminated groundwater. Water Air Soil Pollut. 2022;233:1–11. doi: 10.1007/S11270-022-05544-6/FIGURES/5. [DOI] [Google Scholar]
- 88.Das N., Bhuyan B., Pandey P. Correlation of soil microbiome with crude oil contamination drives detection of hydrocarbon degrading genes which are independent to quantity and type of contaminants. Environ. Res. 2022;215 doi: 10.1016/J.ENVRES.2022.114185. [DOI] [PubMed] [Google Scholar]
- 89.Abbasian F., Lockington R., Megharaj M., Naidu R. The Biodiversity changes in the microbial population of soils contaminated with crude oil. Curr. Microbiol. 2016;72:663–670. doi: 10.1007/S00284-016-1001-4/FIGURES/4. [DOI] [PubMed] [Google Scholar]
- 90.Salam L.B., Obayori S.O., Nwaokorie F.O., Suleiman A., Mustapha R. Metagenomic insights into effects of spent engine oil perturbation on the microbial community composition and function in a tropical agricultural soil. Environ. Sci. Pollut. Control Ser. 2017;24(8 24):7139–7159. doi: 10.1007/S11356-017-8364-3. 2017. [DOI] [PubMed] [Google Scholar]
- 91.Li J., Xu Y., Song Q., Yang J., Xie L., Yu S., Zheng L. Polycyclic aromatic hydrocarbon and n-alkane pollution characteristics and structural and functional perturbations to the microbial community: a case-study of historically petroleum-contaminated soil. Environ. Sci. Pollut. Control Ser. 2021;28:10589–10602. doi: 10.1007/S11356-020-11301-1/FIGURES/4. [DOI] [PubMed] [Google Scholar]
- 92.Patel V., Sharma A., Lal R., Al-Dhabi N.A., Madamwar D. Response and resilience of soil microbial communities inhabiting in edible oil stress/contamination from industrial estates. BMC Microbiol. 2016;16:1–14. doi: 10.1186/S12866-016-0669-8/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang L., Ye J., Jiang K., Wang Y., Li Y. Oil contamination drives the transformation of soil microbial communities: Co-occurrence pattern, metabolic enzymes and culturable hydrocarbon-degrading bacteria. Ecotoxicol. Environ. Saf. 2021;225 doi: 10.1016/J.ECOENV.2021.112740. [DOI] [PubMed] [Google Scholar]
- 94.Li Q., You P., Hu Q., Leng B., Wang J., Chen J., Wan S., Wang B., Yuan C., Zhou R., Ouyang K. Effects of co-contamination of heavy metals and total petroleum hydrocarbons on soil bacterial community and function network reconstitution. Ecotoxicol. Environ. Saf. 2020;204 doi: 10.1016/J.ECOENV.2020.111083. [DOI] [PubMed] [Google Scholar]
- 95.Yang C., Zhao Y., Cao W., Xing M., Xu X., Wang Z., Sun J. Metagenomic analysis reveals antibiotic resistance genes and virulence factors in the saline-alkali soils from the Yellow River Delta, China. Environ. Res. 2022 doi: 10.1016/j.envres.2022.113823. [DOI] [PubMed] [Google Scholar]
- 96.Scheffer G., Hubert C.R.J., Enning D.R., Lahme S., Mand J., de Rezende J.R. Metagenomic investigation of a low diversity, high salinity offshore oil reservoir. Microorganisms. 2021;9:2266. doi: 10.3390/MICROORGANISMS9112266/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu H., Fu Y., Yu J., Jing W., Zhou M. Metagenomic insight on consortium degradation of soil weathered petroleum and its supplement based on gene abundance change. Enzyme Microb Technol. 2023;169 doi: 10.1016/J.ENZMICTEC.2023.110285. [DOI] [PubMed] [Google Scholar]
- 98.Tang S., Rao Y., Huang S., Xu Y., Zeng K., Liang X., Ling Q., Liu K., Ma J., Yu F., Li Y. Impact of environmental factors on the ammonia-oxidizing and denitrifying microbial community and functional genes along soil profiles from different ecologically degraded areas in the Siding mine. J Environ Manage. 2023;326 doi: 10.1016/J.JENVMAN.2022.116641. [DOI] [PubMed] [Google Scholar]
- 99.Shan J., Zhao X., Sheng R., Xia Y., Ti C., Quan X., Wang S., Wei W., Yan X. Dissimilatory nitrate reduction processes in typical Chinese Paddy soils: rates, relative contributions, and influencing factors. Environ. Sci. Technol. 2016;50:9972–9980. doi: 10.1021/ACS.EST.6B01765/ASSET/IMAGES/LARGE/ES-2016-01765S_0005.JPEG. [DOI] [PubMed] [Google Scholar]
- 100.John R.C., Itah A.Y., Essien J.P., Ikpe D.I. Fate of nitrogen-fixing bacteria in crude oil contaminated wetland Ultisol. Bull. Environ. Contam. Toxicol. 2011;87:343. doi: 10.1007/S00128-011-0320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott N.M., Hess M., Bouskill N.J., Mason O.U., Jansson J.K., Gilbert J.A. The microbial nitrogen cycling potential is impacted by polyaromatic hydrocarbon pollution of marine sediments. Front. Microbiol. 2014;5 doi: 10.3389/FMICB.2014.00108/ABSTRACT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sorokin I.D., Kravchenko I.K., Doroshenko E.V., Boulygina E.S., Zadorina E.V., Tourova T.P., Sorokin D.Y. Haloalkaliphilic diazotrophs in soda solonchak soils. FEMS Microbiol. Ecol. 2008;65:425–433. doi: 10.1111/J.1574-6941.2008.00542.X. [DOI] [PubMed] [Google Scholar]
- 103.Paul D. Osmotic stress adaptations in rhizobacteria. J. Basic Microbiol. 2013;53:101–110. doi: 10.1002/JOBM.201100288. [DOI] [PubMed] [Google Scholar]
- 104.Geilfus C.M. Chloride in soil: from nutrient to soil pollutant. Environ. Exp. Bot. 2019;157:299–309. doi: 10.1016/J.ENVEXPBOT.2018.10.035. [DOI] [Google Scholar]
- 105.Stein L.Y., Klotz M.G. The nitrogen cycle. Curr. Biol. 2016;26:R94–R98. doi: 10.1016/j.cub.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 106.Hino T., Matsumoto Y., Nagano S., Sugimoto H., Fukumori Y., Murata T., Iwata S., Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science (1979) 2010;330:1666–1670. doi: 10.1126/SCIENCE.1195591/SUPPL_FILE/PAPV2.PDF. [DOI] [PubMed] [Google Scholar]
- 107.Shiro Y. Structure and function of bacterial nitric oxide reductases: nitric oxide reductase, anaerobic enzymes. Biochim. Biophys. Acta Bioenerg. 2012;1817:1907–1913. doi: 10.1016/J.BBABIO.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Bellini M.I., Gutiérrez L., Tarlera S., Scavino A.F. Isolation and functional analysis of denitrifiers in an aquifer with high potential for denitrification. Syst. Appl. Microbiol. 2013;36:505–516. doi: 10.1016/J.SYAPM.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 109.Heylen K., Gevers D., Vanparys B., Wittebolle L., Geets J., Boon N., De Vos P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 2006;8:2012–2021. doi: 10.1111/J.1462-2920.2006.01081.X. [DOI] [PubMed] [Google Scholar]
- 110.Stres B., Mahne I., Avguštin G., Tiedje J.M. Nitrous oxide reductase (nosZ) gene Fragments differ between native and cultivated Michigan soils. Appl. Environ. Microbiol. 2004;70:301–309. doi: 10.1128/AEM.70.1.301-309.2004/ASSET/74EB1D86-D6F7-4FDA-8BAD-81D68B905168/ASSETS/GRAPHIC/ZAM0010410040004.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Levy-Booth D.J., Prescott C.E., Grayston S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014;75:11–25. doi: 10.1016/J.SOILBIO.2014.03.021. [DOI] [Google Scholar]
- 112.Luvizotto D.M., Araujo J.E., Silva M.C.P., Dias A.C.F., Kraft B., Tegetmeye H., Strous M., Andreote F.D. The rates and players of denitrification, dissimilatory nitrate reduction to ammonia (DNRA) and anaerobic ammonia oxidation (anammox) in mangrove soils. An. Acad. Bras. Cienc. 2018;91 doi: 10.1590/0001-3765201820180373. [DOI] [PubMed] [Google Scholar]
- 113.Varjani S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017;223:277–286. doi: 10.1016/J.BIORTECH.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 114.Medić A., Lješević M., Inui H., Beškoski V., Kojić I., Stojanović K., Karadžić I. Efficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. RSC Adv. 2020;10:14060–14070. doi: 10.1039/C9RA10371F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chebbi A., Hentati D., Zaghden H., Baccar N., Rezgui F., Chalbi M., Sayadi S., Chamkha M. Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolated Pseudomonas sp. strain from used motor oil-contaminated soil. Int Biodeterior Biodegradation. 2017;122:128–140. doi: 10.1016/J.IBIOD.2017.05.006. [DOI] [Google Scholar]
- 116.Al-Hawash A.B., Dragh M.A., Li S., Alhujaily A., Abbood H.A., Zhang X., Ma F. Principles of microbial degradation of petroleum hydrocarbons in the environment. The Egyptian Journal of Aquatic Research. 2018;44:71–76. doi: 10.1016/J.EJAR.2018.06.001. [DOI] [Google Scholar]
- 117.Ebadi A., Khoshkholgh Sima N.A., Olamaee M., Hashemi M., Ghorbani Nasrabadi R. Effective bioremediation of a petroleum-polluted saline soil by a surfactant-producing Pseudomonas aeruginosa consortium. J. Adv. Res. 2017;8:627–633. doi: 10.1016/J.JARE.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qin X., Tang J.C., Li D.S., Zhang Q.M. Effect of salinity on the bioremediation of petroleum hydrocarbons in a saline-alkaline soil. Lett. Appl. Microbiol. 2012;55:210–217. doi: 10.1111/J.1472-765X.2012.03280.X. [DOI] [PubMed] [Google Scholar]
- 119.Isaza J.P., Sandoval-Figueredo V., Rodelo M.C., Muñoz-García A., Figueroa-Galvis I., Vanegas J. Metatranscriptomic characterization of the bacterial community of a contaminated mangrove from the Caribbean. Reg Stud Mar Sci. 2021;44 doi: 10.1016/J.RSMA.2021.101724. [DOI] [Google Scholar]
- 120.Dobler L., Vilela L.F., Almeida R.V., Neves B.C. Rhamnolipids in perspective: gene regulatory pathways, metabolic engineering, production and technological forecasting. N Biotechnol. 2016;33:123–135. doi: 10.1016/J.NBT.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 121.Gutiérrez-Chávez C., Benaud N., Ferrari B.C. The ecological roles of microbial lipopeptides: where are we going? 2021. [DOI] [PMC free article] [PubMed]
- 122.Costa S.G.V.A.O., Nitschke M., Haddad R., Eberlin M.N., Contiero J. Production of Pseudomonas aeruginosa LBI rhamnolipids following growth on Brazilian native oils. Process Biochemistry. 2006;41:483–488. doi: 10.1016/J.PROCBIO.2005.07.002. [DOI] [Google Scholar]
- 123.Clements-Decker T., Kode M., Khan S., Khan W. Underexplored bacteria as reservoirs of novel antimicrobial lipopeptides. Front. Chem. 2022;10 doi: 10.3389/FCHEM.2022.1025979/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oni F.E., Esmaeel Q., Onyeka J.T., Adeleke R., Jacquard C., Clement C., Gross H., Barka E.A., Höfte M. Pseudomonas lipopeptide-Mediated Biocontrol: Chemotaxonomy and biological activity. Molecules. 2022;27 doi: 10.3390/MOLECULES27020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao H., Shao D., Jiang C., Shi J., Li Q., Huang Q., Rajoka M.S.R., Yang H., Jin M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017;101:5951–5960. doi: 10.1007/S00253-017-8396-0. [DOI] [PubMed] [Google Scholar]
- 126.Das P., Mukherjee S., Sen R. Genetic regulations of the biosynthesis of microbial surfactants: an overview. Biotechnol. Genet. Eng. Rev. 2008;25:165–186. doi: 10.5661/BGER-25-165. [DOI] [PubMed] [Google Scholar]
- 127.Li J.Y., Wang L., Liu Y.F., Zhou L., Gang H.Z., Liu J.F., Yang S.Z., Mu B.Z. Microbial lipopeptide-producing Strains and their metabolic roles under anaerobic conditions. Microorganisms. 2021;9 doi: 10.3390/MICROORGANISMS9102030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nadell C.D., Drescher K., Foster K.R. Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 2016;14:589–600. doi: 10.1038/NRMICRO.2016.84. [DOI] [PubMed] [Google Scholar]
- 129.Bezza F.A., Chirwa E.M.N. The role of lipopeptide biosurfactant on microbial remediation of aged polycyclic aromatic hydrocarbons (PAHs)-contaminated soil. Chem. Eng. J. 2017;309:563–576. doi: 10.1016/J.CEJ.2016.10.055. [DOI] [Google Scholar]
- 130.Oja T., Galindo P.S.M., Taguchi T., Manner S., Vuorela P.M., Ichinose K., Metsä-Ketelä M., Fallarero A. Effective antibiofilm polyketides against Staphylococcus aureus from the pyranonaphthoquinone biosynthetic pathways of Streptomyces species. Antimicrob. Agents Chemother. 2015;59:6046–6052. doi: 10.1128/AAC.00991-15/SUPPL_FILE/ZAC010154400SO1.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oliveira J.S., Araújo W.J., Figueiredo R.M., Silva-Portela R.C.B., De Brito Guerra A., Da Silva Araújo S.C., Minnicelli C., Carlos A.C., De Vasconcelos A.T.R., Freitas A.T., Agnez-Lima L.F. Biogeographical distribution analysis of hydrocarbon degrading and biosurfactant producing genes suggests that near-equatorial biomes have higher abundance of genes with potential for bioremediation. BMC Microbiol. 2017;17:1–10. doi: 10.1186/S12866-017-1077-4/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained in this study have been submitted to the NCBI Sequence Read Archive (SRA) database under BioProject accession (PRJNA1041842).