Abstract
Human herpesvirus 8 (HHV-8) has been associated with classical, endemic (African), and AIDS-related Kaposi’s sarcoma (KS), body cavity-based primary effusion lymphomas, and multicentric Castleman’s disease (MCD). HHV-8 encodes a functional homologue of interleukin-6 (IL-6), a cytokine that promotes the growth of KS and myeloma cells and is found at elevated levels in MCD lesions and patient sera. We have previously reported that the viral IL-6 (vIL-6) gene product can support the growth of the IL-6-dependent murine hybridoma cell line, B9, and that the gp80 (IL-6 receptor [IL-6R]) component of the IL-6 receptor-signal transducer (gp180) complex plays a role in mediating this activity. However, it has been shown by others that vIL-6 can function in human cells independently of IL-6R. Here we have extended our functional studies of vIL-6 by identifying transcription factors and pathways used in human Hep3B cells, investigating the utilization of gp130 and IL-6R by vIL-6, and undertaking mutational analyses of vIL-6 and gp130. The data presented here establish that vIL-6, in common with its endogenous counterparts, can mediate signal transduction through gp130 and activate multiple transcription factors, map residues within the vIL-6 protein that are and are not important for vIL-6 signalling, and identify a gp130 mutant that is nonfunctional with respect to vIL-6 signalling in the absence of IL-6R but that retains the ability to mediate vIL-6 and human IL-6 (hIL-6) signal transduction when IL-6R is coexpressed. The data presented demonstrate functional and mechanistic similarities between vIL-6 and endogenous IL-6 proteins but also highlight differences in the structural and receptor-binding properties of vIL-6 relative to its human counterpart.
Human herpesvirus 8 (HHV-8) is the most recently discovered human herpesvirus; sequences of the virus were originally identified through the application of representational difference analysis applied to Kaposi’s sarcoma (KS) and non-KS tissue (10). HHV-8 DNA was subsequently detected in KS lesions of all types (16, 24, 54), in body cavity-based primary effusion lymphoma (BCBL; PEL) (9), and in multicentric Castleman’s disease (MCD) (58). More recently there have been reports that HHV-8 may also be associated with multiple myeloma (7, 44, 45).
The genomes of two strains of HHV-8 have been sequenced, and the resulting data have demonstrated that HHV-8 is a gamma-2 herpesvirus closely related to herpesvirus saimiri (37, 47). Despite their general colinearity, however, there are major regions of divergence between HHV-8 and herpesvirus saimiri, as there are between gammaherpesvirus genomes in general, and one of these loci, divergent locus B (DL-B), in HHV-8 contains a unique cluster of genes that includes a functional homologue of interleukin-6 (IL-6), vIL-6, that is able to support the growth of the IL-6-dependent murine B9 hybridoma cell line and human myeloma cells and to induce acute-phase gene expression and STAT activation in hepatic cell lines (8, 36, 38, 39, 47). The presence of vIL-6 in HHV-8 is significant because of the potential role of IL-6 in the development and progression of human diseases with which HHV-8 has been associated. IL-6 promotes the growth of KS and myeloma cells and is found at elevated levels in MCD lesions and patient sera (27, 28, 33, 62). Furthermore, it has recently been reported that IL-6 is produced by and is an important autocrine growth factor for PEL cells (4).
Endogenous IL-6-mediated activation of acute-phase genes is effected through STAT and C/EBP transcription factors (STAT1, STAT3, and C/EBPβ) that are activated, in response to signal transduction via the gp130 protein (which forms part of the IL-6 receptor), through Janus and mitogen-activated protein kinase pathways, respectively (1, 2, 26, 43). The promoters of the acute-phase genes contain binding sites for C/EBP and STAT transcription factors, for each of which there are several known types with similar DNA binding specificities (C/EBPα through C/EBPδ and STAT1 through STAT6 [25, 40]), and these binding sites are important in the activation of acute-phase genes by IL-6 and other inducers of the acute-phase response (12, 29, 42, 55). The STAT binding site is especially important for induction (29, 55). The specificity of STAT induction by tyrosine phosphorylation is determined by the particular signal transducer used during activation and thus is a reflection of the ability of individual Jak-activated receptor complexes to recruit specific STATs rather than of Jak specificity of STAT activation (25, 26). Signal transduction through gp130 predominantly activates STAT3, although STAT1 also is a target of activated gp130 (2, 25, 26). Studies of vIL-6-mediated activation in human HepG2 hepatoma cells have demonstrated the activation of STAT1 and STAT3 by an apparently IL-6 receptor (IL-6R)-independent mechanism and determined that BAF130 cells expressing gp130 in the absence of IL-6R are responsive to vIL-6 (35).
The motivation for the present study was to further characterize the function of vIL-6 by investigating its ability to induce acute-phase gene expression generally and to activate transcription factors in Hep3B cells and determining similarities and differences of vIL-6 and human IL-6 (hIL-6) with respect both to the receptors used for signal transduction and ligand and receptor structural requirements for signalling. These studies have identified STAT1, STAT3, and C/EBP transcription factors as targets of vIL-6 signalling in Hep3B cells and indicated that gp130 is sufficient for vIL-6 but not hIL-6 signalling. However, the IL-6R component does augment vIL-6 activity and enables signal transduction by vIL-6 through a gp130 variant that is otherwise nonfunctional with respect to vIL-6 signalling. Generation and utilization of altered forms of vIL-6 specifically mutated in highly conserved residues, some of which are known to be important for hIL-6 function or are present in regions corresponding to receptor binding domains of hIL-6, have provided data suggesting that there are significant differences in the structural requirements of vIL-6 and hIL-6 for receptor binding and signal transduction.
MATERIALS AND METHODS
Cell culture and transfections.
Hep3B cells were grown at 37°C as monolayers in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 0.1 mM nonessential amino acids, and 1 mM pyruvate. Hep3B cells were passaged 12 to 24 h before transfection to produce monolayers of 40 to 60% confluency in 80-cm2 tissue culture flasks. Transfections were carried out by the calcium phosphate-DNA coprecipitation method with HEPES-buffered saline, and cells and medium were harvested 48 h posttransfection. For acute-phase gene induction experiments and the preparation of vIL-6 stocks and negative controls, 10 μg of pvIL-6, pSVvIL-6, pvIL-6neg, or pSG5 was used. Cells and medium were harvested for mRNA and for use in C/EBP and STAT induction experiments, respectively. Conditioned media were applied to serum-starved (15 to 18 h) Hep3B cell monolayers for assays of transcription factor induction. The functional activities of specifically altered vIL-6 proteins were determined by assays involving cotransfection of 15 μg of each pSG5-based vIL-6 expression construction with 5 μg of the reporter plasmid pα2MCAT (53) into Hep3B cells. Cotransfection assays involving IL-6R and/or gp130 expression constructions were performed with 2.5 μg of each, together with 15 μg of pSVvIL-6, pSG5-hIL-6, or pSG5 and 1 μg of pα2MCAT.
Plasmids and oligonucleotides.
The vIL-6 expression plasmids used contain coding sequences cloned downstream of either the human cytomegalovirus MIE promoter-enhancer (pvIL-6) or the simian virus 40 early promoter-enhancer (pSVvIL-6) and have been described elsewhere (38). Plasmid pvIL-6neg (38) contains the vIL-6 sequences cloned in the inverse orientation relative to pvIL-6, while pSG5 (Stratagene, La Jolla, Calif.) represents the empty-vector counterpart of pSVvIL-6. The human IL-6R and human gp130 expression vectors pEFBOS–hIL-6R and pEFBOS-hgp130 were kindly provided by M. Narazaki and T. Kishimoto and comprise the receptor coding sequences cloned between the XbaI (IL-6R) or SacI and BamHI sites of the pEF-BOS eukaryotic expression vector (34). Specifically altered vIL-6 coding sequences were derived from M13 clones by PCR amplification of mutated sequences with primers directed to the 5′ and 3′ ends of the vIL-6 coding sequence (38) and containing added 5′ restriction sites (BamHI or SmaI [5′, vIL-6] and BamHI or BglII [3′, vIL-6] sites), to enable subsequent cloning of these sequences into the corresponding sites in pSG5 (or a polylinker-altered derivative). Double-stranded radiolabelled oligonucleotides used in the electrophoretic mobility shift assays (EMSAs) contained either C/EBP (C/EBP-wt: GATCCATTGCGCAATAATTCG-3′) or STAT (APRF-wt: 5′-AGCTTCCTTCTGGGAATTCCT-3′) binding sites (underlined) (1, 2). An altered version of APRF-wt, APRF-mut., containing changes within the core STAT binding sequences was used in competition assays with the APRF-wt probe. The APRF-mut. sequence is 5′-AGCTTCCTTagtttcATTCCT-3′, where the lowercase letters correspond to altered bases with respect to the APRF-wt sequence. Oligonucleotides (HpxA-wt and HpxA-mut.) correspond to the −129 to −106 region of the hemopexin promoter, previously shown to bind C/EBP (42), and were used in competition assays with the C/EBP probe. HpxA-wt and HpxA-mut. (containing mutations in the core C/EBP binding site) sequences are 5′-GATCCTATTTGCAGTGATGTAATCAGCG-3′ and 5′-GATCCTATTTGCAaaGcTtTAgTCAGCG-3′, respectively.
Northern analysis.
Extraction of cellular RNA, size fractionation on denaturing agarose gels, transfer to nitrocellulose, and nucleic acid hybridizations were carried out by standard methods (49). 32P-labelled DNA probes corresponding to selected acute-phase genes (hemopexin, haptoglobin, and complement factor B) were generated from cDNA-containing plasmids (Genome Systems) by nick translation in the presence of [α-32P]dATP.
EMSAs.
Nuclear extracts of cells treated with vIL-6-conditioned medium (or negative controls) or treated with recombinant hIL-6 (Gibco-BRL, Gaithersburg, Md.) were made essentially by the method of Dignam et al. (15). 5′-Dephosphorylated double-stranded oligonucleotide probes for use in EMSAs were labelled with [γ-32P]ATP by using T4 nucleotide kinase. In binding assays, 1 ng of probe, 2 μg of nonspecific competitor [poly(dI-dC)], and 2 to 5 μg of nuclear extract was incubated at room temperature for 30 min in binding buffer (10 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, 0.5 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 5% glycerol). For competition assays, 100 ng (100-fold excess over probe) of unlabelled double-stranded oligonucleotides was included in the incubation mixture. Complexes were run on 6% 0.25× Tris-borate-EDTA (TBE) nondenaturing polyacrylamide gels.
Western blotting.
For the preparation of cell extracts for Western blot analysis, cells were lysed in RIPA buffer (1% Nonidet P-40, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 60 mM sodium deoxycholate, 2 mM EDTA) containing proteinase inhibitors (4 mM NaVO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 4 μM leupeptin, 3 μM antipain, 3 μM pepstatin A) and cell debris was removed by centrifugation. Proteins in supernatants were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose membranes. These were blocked in TBS-T (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 6% bovine serum albumin (for α-PY-STAT1 and α-PY-STAT3 antibodies) or 5% nonfat milk, prior to addition of primary antibody (0.5 to 1.0 μg/ml). Antibodies used for the detection of STAT1, phospho-STAT1, STAT3, and phospho-STAT3 were obtained from Santa Cruz Biotechnology, Santa Cruz, Calif. (sc-346, sc-482, and sc-7199); Upstate Biotechnology (06-657); and New England BioLabs, Beverly, Mass. (9131 and 9132). After the samples were washed in TBS-T containing 5% nonfat milk, horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G secondary antibody (Bio-Rad no. 170-6515; diluted 1:2,000) was used to detect filter-bound primary antibody. Filter-bound HRP on TBS-washed filters was visualized by the enhanced chemiluminescence assay (Amersham International). Analogous procedures were used to identify vIL-6 in conditioned media and bacterial extracts on dot blots; here, primary antibody comprised rabbit antisera directed to vIL-6 C-terminal peptide sequence (residues 192 to 204 [38]) and HRP-conjugated goat anti-rabbit immunoglobulin G secondary antibody was used for detection.
Bacterially produced vIL-6.
Glutathione S-transferase (GST)–vIL-6 fusion protein, used on dot blots, was purified from pGEX–vIL-6-transformed bacteria by passage of sonicated cell extracts over Sepharose 4B-glutathione columns (Pharmacia Biotech, Piscataway, N.J.). His6–vIL-6 fusion protein was derived from pTrcHisB–vIL-6-transformed bacteria and enriched by passage over HiTrap affinity columns (Pharmacia Biotech). The vectors pGEX and pTrcHisB were obtained from Pharmacia Biotech and Invitrogen (San Diego, Calif.), respectively.
Site-directed mutagenesis.
Mutations within the vIL-6 coding sequence (see Table 2) were introduced by using mutagenic primers directed to 15 positions within the vIL-6 open reading frame and comprising 15-nucleotide complementary flanking sequences with either redundant or specifically altered (mutant 28) nucleotides corresponding to the targeted codon(s). Five regions of gp130 were also targeted for specific mutagenesis (see Table 1). Mutagenesis was performed essentially by the method of Kunkel (30). Single-stranded, uracil-containing M13 template corresponding to vIL-6 sequences cloned into M13mp18 was prepared from pelleted bacteriophage after growth in Escherichia coli CJ236 cultures in 2× TY broth supplemented with uridine (0.25 μg/ml) and chloramphenicol (25 μg/ml). A 10-ng portion of each 5′-phosphorylated primer was annealed with approximately 1 μg of template by slow cooling from 65 to 35°C in annealing buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 50 mM NaCl), and the primers were extended at 37°C in ligation buffer (50 mM Tris-HCl [pH 7.8], 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP) with Klenow DNA polymerase (5 U) in the presence of 2 mM each deoxynucleoside triphosphate and 1 U of DNA ligase. One-tenth (2 μl) of each extension/ligation reaction mixture was then transfected into competent TG.1 cells and plated onto Luria agar for overnight incubation at 37°C. Plaques were picked into diluted TG.1 overnight cultures for growth of bacteriophage, which were then harvested for single-stranded DNA. These templates were sequenced to identify mutated vIL-6 sequences.
TABLE 2.
Mutated residues in vIL-6
| Position(s) | Residue(s) | Change(s) | Mutant designation | Activity | Comments | Reference(s) |
|---|---|---|---|---|---|---|
| 13 | G | W | 1 | +++ | Conserved | |
| 41, 43 | W, L | F, G | 19 | +++ | Site II, gp130 interactions (hIL-6) | 50, 52 |
| 54 | C | D | 2 | +++ | Conserved | |
| G | 3 | +++ | ||||
| 60 | C | S | 4 | +++ | Conserved | |
| L | 5 | +++ | ||||
| 75 | P | S | 7 | +++ | Conserved | |
| 83 | C | G | 8 | +++ | Conserved, functional (hIL-6) | 46, 57 |
| 88 | F | L | 20 | +++ | Site I, important for IL-6R binding (hIL-6) | 59 |
| 93 | C | G | 9 | +++ | Site I, conserved, functional (hIL-6) | 46, 57 |
| W | 10 | +++ | ||||
| 103–105 | EFE | EGG | 22 | +++ | Site I, E-103 conserved, important for IL-6R binding (hIL-6) | 59 |
| GVA | 23 | +++ | ||||
| 129 | T | L | 11 | +++ | Site II, conserved | |
| A | 12 | +++ | ||||
| 151 | P | F | 13 | +++ | Conserved | |
| S | 14 | +++ | ||||
| 167 | W | G | 15 | + | Site III, conserved | |
| R | 16 | +++ | ||||
| 167, 172 | W, A | R, P | 24 | + | Site III, required for hexameric (functional) complex formation | 13 |
| 183 | F | W | 17 | +++ | Conserved, F to A in hIL-6 shows reduced cell surface binding | 31 |
| G | 18 | +++ | ||||
| E | 29 | +++ | ||||
| L | 31 | +++ | ||||
| 189 | R | L S C | 25 26 27 | +++ +++ +++ | Site I, conserved, positively charged residue at this position in hIL-6 important for binding and activity | 20, 31 |
| 195 | P | Stop | 28 | +++ | C-tail deletion, 195–204 |
TABLE 1.
Mutations in the gp130 coding sequence
| Designation | Position | Residues | Changes |
|---|---|---|---|
| PM1 | 236–238 | LSS | DDD |
| PM2 | 287 and 288 | QD | SV |
| PM5 | 190–192 | YFV | AAA |
| PM7 | 251 and 252 | SV | AA |
RESULTS
Induction of acute-phase genes by vIL-6.
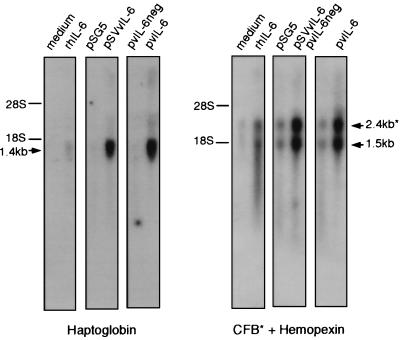
To investigate whether vIL-6 could activate acute-phase genes other than α1-acid glycoprotein (shown previously by us to be induced by vIL-6 [38]), we undertook Northern analysis of RNA from vIL-6-transfected or control cells. Hep3B (human hepatocarcinoma) cells were transfected with either of two vIL-6-encoding plasmids (pvIL-6 and pSVvIL-6) for transient expression of vIL-6 or with negative controls (pvIL-6neg and pSG5). The cells were harvested after 48 h, and total RNA was prepared for Northern analysis. The filter-immobilized, size-fractionated RNA was probed with radiolabelled DNA corresponding to cDNA sequences of the acute-phase genes hemopexin, haptoglobin, and complement factor B. The results are shown in Fig. 1 and demonstrate that all of the assayed acute-phase genes are activated in vIL-6-transfected Hep3B cultures. Thus, vIL-6, in common with endogenous IL-6 proteins, can coordinately activate acute-phase gene expression.
FIG. 1.
Acute-phase gene induction by vIL-6. Hep3B cell monolayers were transfected with pSG5 (empty vector), pSVvIL-6 promoter (simian virus 40 promoter-driven vIL-6 expression vector), pvIL-6 (cytomegalovirus MIE-driven vIL-6 expression vector), or pvIL-6neg (vIL-6 in negative orientation relative to MIE). Other Hep3B cells were either untreated (“medium”) or treated with rhIL-6 (500 U/ml). After 48 h, the cells were harvested for RNA, and 5 μg of RNA per sample was analyzed by size fractionation and Northern blot techniques (see Materials and Methods). 32P-radiolabelled haptoglobin, hemopexin, and complement factor B (CFB) probes were generated from the respective cloned cDNA sequences. The positions of 28S and 18S rRNA markers and the estimated sizes of the detected acute-phase transcripts are indicated.
vIL-6 induction of C/EBP and STAT DNA binding activities.
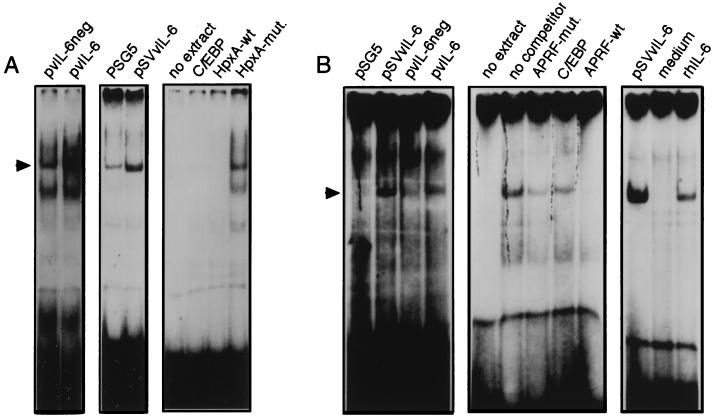
IL-6 induction of acute-phase gene expression is mediated via activation of factors belonging to the C/EBP and STAT families of transcriptional activators. Specifically, the C/EBPβ (CRP2, NF-IL6) and STAT3 (APRF) proteins have been implicated in the IL-6 signal transduction pathway that is initiated through the gp80/gp130 membrane receptor (1, 2). vIL-6 also induces phosphorylation and activation of STATs, a function that is mediated through gp130 but is not dependent on IL-6R (35). To determine whether vIL-6 could induce C/EBP and STAT activities in Hep3B cells, we performed experiments to identify DNA binding by nuclear proteins to double-stranded C/EBP- or STAT-specific probes (C/EBP-wt and APRF-wt [see Materials and Methods]) containing a consensus binding site for these factors. Hep3B cells were transfected with the vIL-6 expression plasmid pvIL-6 or pSVvIL-6 (38) or with negative controls (pvIL-6neg or pSG5), and the medium from these cultures was harvested to provide sources of vIL-6 or appropriate negative controls. These media were then applied to fresh Hep3B cell cultures for 15 min, and cells were harvested for the preparation of nuclear extracts. These nuclear extracts were used in EMSAs with the 32P-labelled C/EBP-wt or APRF-wt probes. The results of these assays are shown in the left two panels of Fig. 2A and the left panel of Fig. 2B and indicate that C/EBP- and STAT-like DNA binding activities were induced as a function of vIL-6 expression. For C/EBP, the different bands observed in the EMSAs are likely to reflect different homodimeric and heterodimeric complexes of C/EBP isoforms (e.g., see references 3 and 61).
FIG. 2.
C/EBP- and STAT-related DNA binding activities induced by vIL-6. (A) Hep3B cells were transiently transfected with either pSVvIL-6, pvIL-6, pSG5, or pvIL-6neg, and growth media were harvested 48 h posttransfection. These were applied to fresh, serum-starved Hep3B cell cultures for 15 min before harvesting of cells for the preparation of nuclear protein extracts. Equal amounts of nuclear protein were used in EMSAs with the C/EBP-wt probe (see Materials and Methods). Induction of C/EBP binding activity was detected in the pvIL-6- and pSVvIL-6-transfected cells (left and middle panels, respectively [arrow]). Competition assays (right panel) were carried out with a 100-fold molar excess of unlabelled C/EBP-wt oligonucleotide, HpxA-wt (corresponding to sequences containing the C/EBP binding site in the −129 to −106 region of the hemopexin promoter [42]), and HpxA-mut. (containing base changes in the C/EBP core binding sequences). (B) Similar EMSAs were carried out for the detection of STAT binding activities with a probe (APRF-wt) derived from the α2-macroglobulin promoter (see Materials and Methods). vIL-6 induced complexes were evident (left panel), comigrated with complexes induce by rhIL-6 (500 U/ml) (right panel), and could be competed with a 100-fold molar excess of unlabelled APRF-wt but not with APRF-mut. (containing changes in the STAT binding site) or unrelated C/EBP-wt (C/EBP) oligonucleotides (middle panel).
To confirm the presence of C/EBP in the shifted complexes, competition assays were carried out with the pSVvIL-6-activated extract (Fig. 2A, middle panel) and unlabelled double-stranded C/EBP oligonucleotide or oligonucleotides corresponding to native (HpxA-wt) or C/EBP-site-altered (HpxA-mut.) versions of −129 to −106 hemopexin A promoter sequences, previously shown to bind C/EBP (42). The results of these assays (Fig. 2A, right) demonstrated that competition for formation of the vIL-6-activated and other DNA-protein complexes on the C/EBP-wt probe was dependent on the C/EBP-binding core sequences of the HpxA oligonucleotide, indicating that C/EBP factors are present in the observed complexes.
Analogous competition assays were carried out to confirm the presence of STATs in the APRF-wt–protein complexes. These complexes could be competed with a 100-fold excess of unlabelled APRF competitor (APRF-wt), but not with an altered version (APRF-mut.) containing alterations in the STAT recognition sequences or with the C/EBP-wt oligonucleotide that contains unrelated sequences (Fig. 2B, middle). Furthermore, the vIL-6-induced complex comigrated with a complex induced by treatment of cells with rhIL-6 (Fig. 2B, right), indicating that the nuclear factors activated by vIL-6 do indeed represent the same STAT-related factors known to be induced by hIL-6 (2).
Identification of vIL-6-induced STATs.
Since several types of STAT transcription factors are known to be present and expressed together in many cell types, we next sought to identify the types of STAT factors induced by vIL-6 in Hep3B cells. Signalling through gp130 can be mediated by a number of cytokines in addition to IL-6, and STAT1 and STAT3 are the STAT factors that are activated by this signal transducer in association with gp130-binding tyrosine kinases.
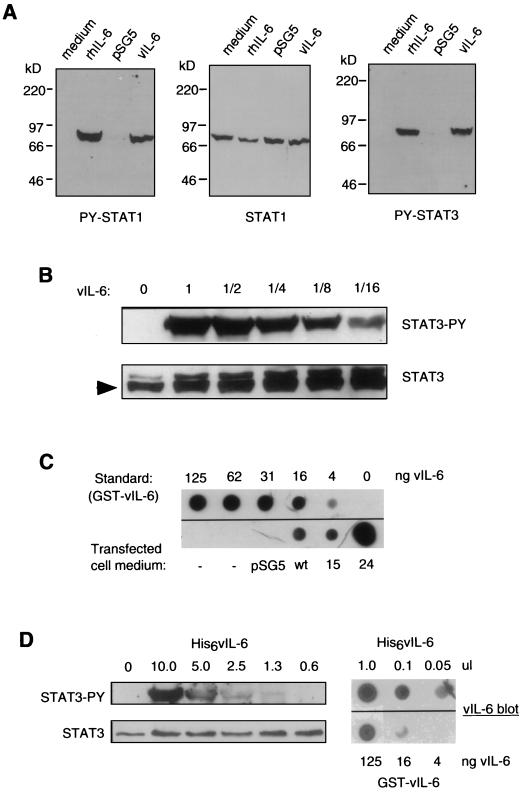
To determine whether STAT1 and STAT3 were activated in Hep3B cells, we used peptide antisera directed against phosphorylated (activated) STAT1 or STAT3 in Western analyses of extracts of vIL-6- or rhIL-6-treated Hep3B cells. Figure 3A shows the results of one such experiment. Extracts of cells treated with rhIL-6, vIL-6 (derived from pSVvIL-6-transfected Hep3B cells), or negative control medium, comprising either fresh medium without added rhIL-6 or conditioned medium from Hep3B cells transfected with pSG5, were probed for the presence of either phospho-STAT1 (left panel) or phospho-STAT3 (right panel). The blot probed for phospho-STAT1 was stripped and reprobed with STAT1 peptide antiserum to determine the total levels of STAT1 present in each of the cell extracts to confirm equivalent protein loading of the lanes (middle panel). The data shown in Figure 3A are representative of those obtained from several experiments of this type and demonstrate that STAT1 and STAT3 are targets of vIL-6 signal transduction in Hep3B cells. Similar experiments demonstrated that STAT5 was not induced by vIL-6 or hIL-6 (data not shown); STAT5 is not activated through gp130.
FIG. 3.
Characterization of vIL-6-mediated STAT induction in Hep3B cells. (A) Medium containing vIL-6 (from pSVvIL-6-transfected Hep3B cells) or hrIL-6 (500 U/ml) or control medium (fresh or derived from pSG5-transfected Hep3B cells) was applied to serum-starved Hep3B cultures (15 min), and the cells were harvested for the preparation of whole-cell extracts. These extracts were analyzed by Western blot analysis to detect induced (phosphorylated) STAT1 and STAT3 (left and right panels) or total STAT1 (middle panel; stripped, reprobed PY-STAT1 blot). (B) Various dilutions of pSVvIL-6-transfected cell medium were tested for STAT3-inducing activity. Levels of induced STAT3 (top panel) and total STAT3 (bottom panel) in Hep3B cell cultures treated with pSVvIL-6 (1 to 1/16) or pSG5 (0) transfected cell medium were determined by Western analysis. (C) The amount of vIL-6 present in the vIL-6 transfected cell medium was determined by dot blot analysis with a vIL-6 peptide antiserum (see Materials and Methods). The signal from 3 ml of undiluted vIL-6 stock (corresponding to the maximum amount of vIL-6 used for STAT3 induction in panel B) was compared to signals obtained from a dilution series of purified GST–vIL-6 fusion protein to determine the amount of vIL-6 present. The calculated amounts of the vIL-6 component of the GST-vIL-6 fusion protein at each dilution is indicated. Control medium (from pSG5-transfected Hep3B cells) gave no background signal. Also shown are the results of analyses of the expression of vIL-6 variants 15 and 24 (Table 2) secreted from transfected Hep3B cells. (D) The activity and concentration of bacterially synthesized vIL-6 (see Materials and Methods) were determined by STAT3 induction and dot blot assays analogous to those used for Hep3B-expressed vIL-6. Different amounts (in microliters) of bacterial extract (His6vIL-6) were used for each; amounts (in nanograms) of the vIL-6 component of blotted GST–vIL-6 are indicated.
Medium containing vIL-6, derived from Hep3B cell cultures transfected with pSVvIL-6, was used at various dilutions to determine the amount required to induce STAT3 phosphorylation in fresh Hep3B cells (Fig. 3B) and was also analyzed by dot blot procedures to quantify the amount of vIL-6 present in the conditioned medium (Fig. 3C). vIL-6 protein was detected by a vIL-6 peptide rabbit antiserum (see Materials and Methods), and quantitation of the amount of vIL-6 present in the conditioned medium was made possible by inclusion on the blot of a dilution series of purified GST–vIL-6 fusion protein as a standard. The results of this analysis showed that vIL-6 present at less than 0.2 ng/ml (the lowest dilution of vIL-6 stock used) was sufficient to effect detectable STAT3 induction in Hep3B cells, as measured by Western analysis of cell extracts (Fig. 3B). Higher levels of phosphorylated STAT3 were detected when vIL-6 was applied at higher doses, with maximum induction being achieved with the highest two concentrations of vIL-6 used (approximately 1.5 and 3.0 ng/ml). By using bacterial cell extracts containing the His6–vIL-6 fusion protein (see Materials and Methods) in analogous STAT3 induction and dot blot assays (Fig. 3D), it was found that the levels of the bacterially produced vIL-6 required to effect STAT3 induction were at least 103-fold higher than the active levels of vIL-6 derived from transfected Hep3B cells.
Utilization of gp130 and IL-6R receptor subunits by vIL-6.
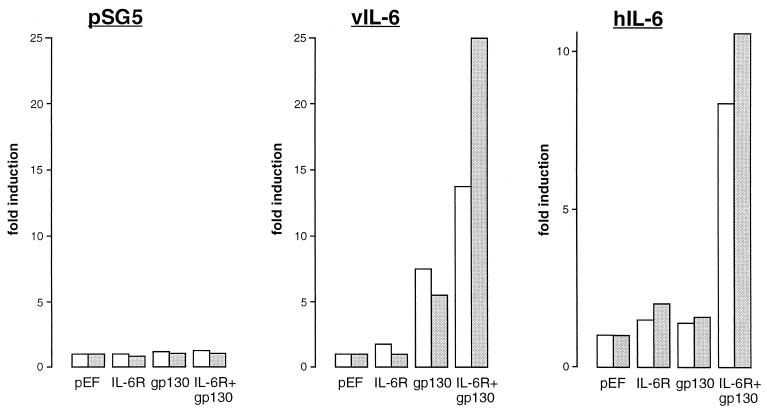
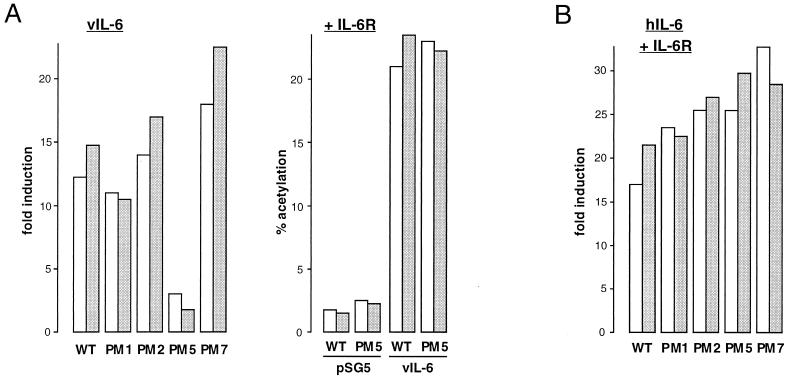
To investigate the functional utilization of IL-6R and gp130 by vIL-6, we used IL-6R and gp130 expression vectors and a promoter-reporter construction, pα2MCAT, comprising sequences of the α2-macroglobulin promoter linked to the chloramphenicol acetyltransferase (CAT) gene (53), in transient-transfection assays. IL-6 and gp130 expression plasmids comprising the respective coding sequences cloned in the powerful eukaryotic expression plasmid pEF-BOS (34) were used in these transfections to ensure very high levels of receptor expression in transfected cells, thereby minimizing potential effects of the naturally expressed receptors in Hep3B cells. Hep3B cells were cotransfected with pα2MCAT together with pEF-BOS, pEFBOS–IL-6R, pEFBOS-gp130, or pEFBOS–IL-6R plus pEFBOS-gp130 in the absence or presence of vIL-6 or hIL-6 (produced from cotransfected pSVvIL-6 and pSVhIL-6; pSG5 was used as the negative control). Cells were subsequently harvested for determinations of relative CAT activities in cell extracts to determine the contribution of each of the receptor components to signalling by vIL-6 and hIL-6. The results of these experiments, performed in duplicate, are shown in Fig. 4. The results obtained in the pSVvIL-6 cotransfection experiments demonstrated that vIL-6 could effect signalling through gp130 in the absence of cotransfected IL-6R, in contrast to hIL-6, which required the coexpression of transfected gp130 and IL-6R. These results indicate that vIL-6, but not hIL-6, can signal independently of IL-6R. While we cannot exclude, on the basis of these data, the formal possibility that the low endogenous levels of IL-6R expressed in Hep3B cells were required for vIL-6 signalling through overexpressed (transfected) gp130, the inability of hIL-6 to signal under the same conditions argues strongly against this possibility (also see below). The presence of overexpressed IL-6R together with gp130 in these assays led to an increase (between two- and fivefold) in the activation of pα2MCAT by vIL-6, suggesting that functional vIL-6–gp130–IL-6R complexes may form and enhance vIL-6 signalling through gp130.
FIG. 4.
Receptor utilization by vIL-6. (A) Hep3B cells were transfected with pSG5, pSVvIL-6, or pSVhIL-6 in the presence of IL-6R and/or gp130 expression vectors (pEFBOS-IL-6R and pEFBOS-gp130) or empty vector (pEF-BOS) and pα2MCAT. CAT activities were determined in cell extracts after 48 h. The results, expressed as fold induction above CAT activities obtained with pEF-BOS vector controls, of duplicate experiments are shown.
Identification of gp130 E-F loop mutation important for vIL-6 signalling.
We introduced several mutations onto the cytokine binding domain of gp130 based on the residues in the homologous region of IL-6R known to be important for hIL-6 binding, for interactions of hIL-6/IL-6R with gp130, and for function (48, 60). Four of the alterations introduced into the gp130 coding sequences are detailed in Table 1. The PM1 and PM2 mutations correspond to previously made alterations within the analogous positions of the homologous domain of IL-6R; the IL-6R mutations (at positions 230 and 260 plus 261) lead to decreased IL-6/IL-6R association with gp130 (48, 60). The PM5 and PM7 mutations are in positions within the cytokine binding loops (E-F and B2-C2) of the cytokine binding domain of gp130, whose crystal structure has recently been published (6).
To assay for the functions of the altered gp130 proteins, we again used the pα2MCAT reporter in Hep3B transfection assays. pSVvIL-6 (alone) or pSVhIL-6 plus pEFBOS–IL-6R were cotransfected along with each of the gp130 expression plasmids (pEFBOS-gp130.PM1, pEFBOS-gp130.PM2, pEFBOS-gp130.PM5, and pEFBOS-gp130.PM7), or pEF-BOS (negative control), and pα2MCAT. CAT activities in cell extracts were determined, and fold inductions relative to the pEF-BOS controls were calculated. The data from duplicate experiments are shown in Fig. 5. While the PM1 and PM2 mutations and the B2-C2 loop mutation, PM7, had no significant effect on vIL-6 or hIL-6 signalling, the PM5 mutation led to almost complete abrogation of gp130 signalling in response to vIL-6 (Fig. 5A, left). However, inclusion of IL-6R in the cotransfection assays of vIL-6 and gp130.PM5 enabled signal transduction by vIL-6 at levels comparable to wild-type gp130 (Fig. 5A, right). These data demonstrate that gp130 residues 190 to 192 (altered in gp130.PM5) are important for IL-6R-independent vIL-6 signalling, possibly representing a ligand-gp130 contact site, and indicate that IL-6R can form part of the functional vIL-6–receptor complex. It is noteworthy (and of relevance to the preceding section) that the requirement for overexpressed (transfected) IL-6R for signalling through gp130.PM5 also demonstrates that endogenous levels of IL-6R in Hep3B cells are functionally insignificant in these cotransfection assays.
FIG. 5.
Functional properties of gp130 signal transducer variants. Each of the altered gp130 coding sequences (Table 1), cloned in expression vector pEF-BOS, was cotransfected into Hep3B cells with pα2MCAT and either pSG5–vIL-6 (alone) (A, left panel) or pSG5-hIL-6 plus pEFBOS-IL-6R (B). Cells were harvested after 48 h for determinations of CAT activities in cell extracts. Levels of induction were calculated by comparisons with CAT activities obtained from pEF-BOS-transfected cells. The data from duplicate experiments are shown. Similar cotransfection experiments were conducted with gp130.PM5 to investigate the effects of coexpressed IL-6R on signal transduction by vIL-6 (A, right panel).
Mutational analysis of vIL-6 to identify residues important for vIL-6 signal transduction.
Detailed mutagenesis and structural studies of hIL-6 have identified the regions and residues of the molecule that are important for receptor interactions and signalling through the IL-6R/gp130 complex (see, e.g., references 5, 17, 23, 41, and 51). Three main functional domains of hIL-6, sites 1, 2, and 3, have been recognized and are involved either in hIL-6 interaction with IL-6R (site 1) or in associations of IL-6/IL-6R with gp130 to effect the assembly of the hexameric, functional ligand-receptor complex (32, 56).
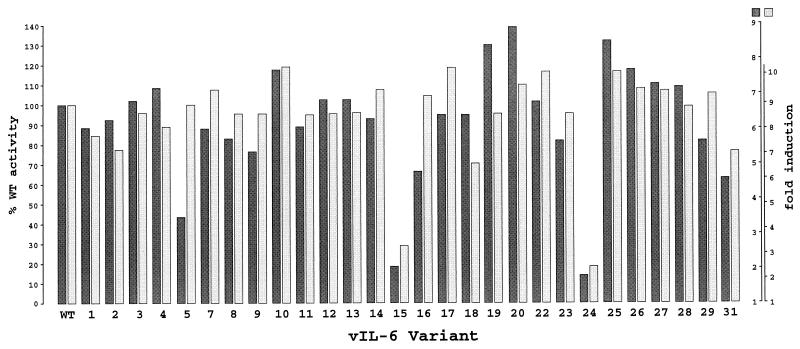
To identify residues within vIL-6 that are functionally important with respect to signalling through induction of STAT transcription factors, we generated a panel of vIL-6 mutants for use in functional assays involving cotransfection of each of the vIL-6 variants with pα2MCAT into Hep3B cells. Residues targeted for mutagenesis were chosen because they are highly conserved between species or because they correspond to residues in hIL-6 known to be important for interactions with IL-6R or gp130. Thus, many of the mutated residues fall into the regions of vIL-6 corresponding to binding site 1, 2, or 3 of hIL-6 (56) (Table 2).
Mutations were introduced into vIL-6 coding sequences by using the M13-based method of Kunkel (30) (see Materials and Methods). Except for the introduction of a stop codon at position 95 (mutant 28), mutagenesis at each position was performed with degenerate mutagenic primers to enable the introduction of multiple changes at each of the targeted amino acid positions. A total of 16 positions were targeted for mutagenesis, and 28 vIL-6 variants were selected for functional analysis. Each altered vIL-6 open reading frame cloned into pSG5 was sequenced to confirm the presence of the mutation(s). The various mutations that were generated are summarized in Table 2.
Each of the vIL-6 mutant expression vectors was cotransfected with pα2MCAT into Hep3B cells to compare their activities to native vIL-6 and to each other. The entire panel of vIL-6 mutants was used in two separate experiments (Fig. 6), and the activities of the altered vIL-6 proteins are indicated in Table 2. It is noteworthy that only two of the tested vIL-6 mutants showed significant changes in activity in this assay, although hIL-6 equivalents of many of the targeted residues are known to be important for hIL-6 function. For example, the first and second pair of cysteine residues are involved in disulfide bridging in hIL-6 (11), and such bridging between the third and fourth cysteine residues is important for function (46, 57). However, in vIL-6, disulfide bridging between the conserved cysteine residues is clearly not necessary for function, since each of the four cysteines (C54, C60, C83, and C93) could be altered without any significant effect on vIL-6 activity. Similarly, while changes in F88, E103 to E105, and R189 had no effect on vIL-6 function, alterations of equivalent residues in hIL-6, all occurring within site 1, lead to loss of IL-6R binding or function (20, 31, 59). Also, position 183 in vIL-6 contains a highly conserved phenylalanine residue which in hIL-6 is important for high-affinity cell surface binding (31); mutation of this residue in vIL-6 (to W, G, E, or L) had no significant impact on function. Two positions in vIL-6 were identified as being important for functional integrity; these are positions 167 and 172, which lie within a region equivalent to receptor interaction site 3 in hIL-6. When W167 was changed to glycine (mutant 15), the activity of vIL-6 was severely impaired (Fig. 6). An arginine substitution at this position had no significant effect. However, replacement of the arginine at position 172 by proline, together with the R167 substitution (mutant 24), greatly diminished vIL-6 activity. This altered vIL-6 protein corresponds to an hIL-6 mutant (W157T162 to R157P162, numbering from the first residue of the mature, cleaved protein) that is able to bind IL-6R with increased affinity but shows reduced bioactivity, most probably due to a decreased ability to form stable, functional complexes with gp130 (13). Such reduced stability of hIL-6–IL-6R–gp130 hexameric complex formation has been demonstrated for a comparable Q159T162-to-E159P162 hIL-6 variant (14, 22). It is likely that the effects on function of these analogous mutations in vIL-6 and hIL-6 result from similar effects on cytokine-gp130 associations. However, the combined data from our vIL-6 mutagenesis studies indicate that there are fundamental differences in the structures and receptor binding requirements of vIL-6 and hIL-6.
FIG. 6.
Functional analyses of vIL-6 mutants. Each of the altered vIL-6 coding sequences (Table 2), present in the expression vector pSG5, was cotransfected with pα2MCAT into Hep3B cells, and CAT activities in cell extracts were determined. Shown are the results of duplicate experiments, with activities of each vIL-6 variant expressed as a percentage of the wild-type (WT) vIL-6 activity (100%). Fold inductions above pSG5 negative controls are indicated for each experiment (right scales).
To rule out the possibility that vIL-6 variants 15 and 24 contain alterations other than those specifically introduced, which could account for their decrease activities, each vIL-6 coding sequence was sequenced completely; no unexpected changes were identified. To investigate the possibility that the expressed levels of the secreted proteins were reduced due to decreased stabilities of the altered proteins, the presence and levels of the vIL-6 variants relative to wild-type vIL-6 in the medium of transfected Hep3B cells were determined. This was done by dot blot analysis with our vIL-6 peptide antiserum, with a dilution series of purified GST–vIL-6 fusion protein used as a standard for quantitation. The results of this analysis (Fig. 3C) demonstrated that vIL-6 mutant 15 was present in the culture medium at a concentration (approximately 3 ng/ml) equivalent to that of wild-type vIL-6 whereas mutant 24 was present at a level well in excess of 100 ng/ml. Comparable amounts of vIL-6 variant 24 (measured as nanograms per milliliter and relative to wild-type) were seen consistently in the medium in several independent Hep3B transfection experiments (data not shown); a possible explanation is that the W167 to R and/or the A172 to P change(s) has a stabilizing effect, allowing greater accumulation of the protein in the culture medium.
DISCUSSION
The presence of vIL-6 in HHV-8, a virus that is associated with KS, PEL, and MCD, is significant in view of the findings that cytokines, particularly IL-6, are likely to play a role in the development and progression of these diseases. AIDS-KS cells have been reported to secrete several cytokines, including IL-6, basic fibroblast growth factor, and platelet-derived growth factor; IL-6 and IL-6R are present at higher levels in KS lesions in vivo than in surrounding normal dermis; and IL-6 can promote the growth of KS cells in vitro (18, 33). Furthermore, IL-6 has also been implicated in the development of MCD (with which KS is associated), with high levels of IL-6 being detected in involved tissues (27). For PEL, a recent report has noted that hIL-6 is produced by and appears to act as an intracrine mitogenic factor in at least two cell lines (4). Paracrine mechanisms effecting cell growth have been noted for HHV-8-infected endothelial cell cultures, in which a minority of HHV-8+ cells are able to support the rapid proliferation of uninfected endothelial cells (19). These kinds of findings raise the possibility that the HHV-8-specified vIL-6 protein is relevant in influencing the development or manifestation of HHV-8-associated diseases. Characterization of the functional and mechanistic properties of vIL-6 is important for the assessment of its role in vivo.
It has been reported previously that HHV-8 vIL-6 encodes a functional protein displaying mitogenic properties in murine B9 cell cultures (36, 38) and that this function is at least partly dependent on the gp80 (IL-6R) component of the IL-6 receptor complex (38). More recent studies with human HepG2 hepatoma cells identified gp130 as a vIL-6 signal transducer, but the results of experiments in which anti-hIL-6R antibody was used to block potential binding of vIL-6 to IL-6R had no effect on vIL-6 signalling through gp130 (35). The data presented here show that several acute-phase genes and C/EBP and STAT transcription factors can be induced by vIL-6 in Hep3B cells, that vIL-6 signalling in Hep3B cells utilizes gp130, that vIL-6 can signal through gp130 independently of IL-6R, and that IL-6R can enhance or enable vIL-6 signal transduction through gp130 or the variant gp130.PM5. Furthermore, the vIL-6 structure-function studies presented here identify amino acid residues important (and unimportant) for vIL-6 function, highlighting differences between vIL-6 and hIL-6.
The present data showing that IL-6R plays a role in vIL-6 signal transduction in transfected Hep3B cells is consistent with our previous investigations of the effects of vIL-6 on murine B9 cell growth, in which neutralizing antibody against murine IL-6R was able to partially inhibit vIL-6 mitogenic activity (38). Inhibitory effects of anti-hIL-6R antibody, in combination with anti-gp130 antibody, on vIL-6-stimulated human myeloma cell growth have also been reported (8). However, these investigators and Molden et al. (35) found no significant effects of anti-hIL-6R alone on vIL-6 function. The apparent inconsistencies between these findings and our previously reported results of murine B9 cell proliferation assays (38) may be the consequence of technical aspects of the experimental procedures used or of differences in the cell lines used for the various studies. For example, there may be differences in IL-6R-ligand contact sites between vIL-6 and hIL-6 resulting in ineffective blocking of vIL-6 binding by the particular “blocking” antibody used in the studies of Molden et al. (35) and Burger et al. (8), or there may be qualitative differences in hIL-6R and mIL-6R recognition by vIL-6. The data presented here show a small (two- to fivefold) positive effect of cotransfected hIL-6R on vIL-6 activity, as measured with an α2-macroglobulin promoter-CAT reporter. Furthermore, using the gp130.PM5 variant, we were able to demonstrate clearly that IL-6R can play a role in vIL-6 signal transduction, since gp130.PM5 was fully functional with respect to vIL-6 signalling in the presence of cotransfected IL-6R, but severely inhibited in its absence. The simplest interpretation of these combined results is that while vIL-6 can recognize and signal through gp130 alone, IL-6R can also interact with vIL-6 and/or gp130 (wild-type and gp130.PM5) to stabilize functional-complex formation. Resolution of the question whether vIL-6 and gp130 can indeed form functional complexes with IL-6R will require appropriate biochemical analyses to detect such ligand-receptor interactions directly.
Comparison of the amino acid sequence of vIL-6 with its sequenced endogenous counterparts has shown that it is considerably diverged from these homologues (36, 38, 39). The amino acid identity between vIL-6 and hIL-6 is approximately 25%. This degree of divergence of the primary structure is likely to affect the secondary and tertiary structures of the proteins, resulting in significant differences between vIL-6 and hIL-6. Indeed, evidence for such structural divergence is apparent from the results of the vIL-6 mutagenesis studies presented in Fig. 6 and Table 2. Only two of the mutations introduced into vIL-6 led to a significant decrease in vIL-6 activity, those at positions 167 (W to G) and 172 (A to P, within the context of a functionally neutral W-to-R mutation at position 167). Even mutations of the highly conserved third and fourth cysteine residues, previously reported to be important in hIL-6 for structural integrity and function, and of other residues with functionally important roles in hIL-6 (e.g., vIL-6 residues 88, 103 to 105, 183, and 189) had little or no effect on vIL-6 signalling. These differences between the vIL-6 and hIL-6 structure-function profiles could indicate different gp130 contact sites and reflect, at least in part, the IL-6R independence of vIL-6 signalling. Deletion of the C-terminal 10 amino acids of vIL-6, representing an “extension” relative to the endogenous IL-6 proteins, had no effect on vIL-6 function, demonstrating that this region of the protein is not involved in receptor recognition or in maintenance of overall protein structure.
Finally, a notable finding of the present study is that the concentration of vIL-6 required to mediate signal transduction, as measured by induction of STAT3, is comparable to the level of hIL-6 required for signalling, as established clearly by many published reports. The minimum active concentration of vIL-6 used for STAT3 induction experiments presented in Fig. 3A was approximately 0.2 ng/ml, with maximum STAT3 induction being achieved with a vIL-6 concentration of around 2 ng/ml. This is in marked contrast to the approximately 103-fold-higher concentration of bacterially expressed recombinant vIL-6 found to be active in our STAT3 induction assays (Fig. 3D) and reported to be required for support of B9 cell growth (8). The underlying basis for this difference in eukaryotically and bacterially expressed vIL-6 specific activity is unclear but could relate to the lack of appropriate posttranslational modification and/or the presence of the N-terminal polyhistidine tract in the recombinant vIL-6 proteins. Such a situation would distinguish vIL-6 from various cellular IL-6 proteins whose specific activities appear not to be dependent on posttranslational modifications and specific N-terminal processing.
The data presented in this report extend the results of previous studies (8, 35, 38) into the types and mechanistic basis of vIL-6 activity by identifying transcription factors activated by vIL-6 in Hep3B human hepatocarcinoma cells, providing a detailed structure-function analysis of vIL-6, identifying a region (possibly representing a vIL-6–gp130 contact site) within the E-F cytokine binding loop of gp130 that is required for vIL-6 function in the absence of IL-6R, and demonstrating the involvement of IL-6R in vIL-6 signal transduction. We hope that these data will prove useful in contributing to an overall understanding of vIL-6 function in vivo and to the development of methods, possibly employing vIL-6-receptor binding-site peptide sequences, to specifically block the function of vIL-6, an HHV-8 cytokine that may influence viral pathogenesis.
ACKNOWLEDGMENTS
We are grateful to M. Narazaki and T. Kishimoto for supplying the pEFBOS–hIL-6R and pEFBOS-hgp130 expression vectors.
This work was supported by R55 and R01 grants CA76445 from the National Institutes of Health.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of the C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 3.An M R, Hsieh C-C, Reisner P D, Rabeck J P, Scott S G, Kuninger D T, Papaconstantinou J. Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol Cell Biol. 1996;16:2295–2306. doi: 10.1128/mcb.16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asou H, Said J W, Yang R, Munker R, Park D J, Kamada N, Koeffler H P. Mechanisms of growth control of Kaposi’s sarcoma-associated herpesvirus-associated primary effusion lymphoma. Blood. 1998;91:2475–2481. [PubMed] [Google Scholar]
- 5.Brakenhoff J P J, de Hon F D, Fontaine V, Ten Boekel E, Schooltink H, Rose-John S, Heinrich P C, Content J, Aarden L A. Development of a human interleukin-6 receptor agonist. J Biol Chem. 1994;269:86–93. [PubMed] [Google Scholar]
- 6.Bravo J, Staunton D, Heath J K, Jones E Y. Crystal structure of a cytokine-binding region of gp130. EMBO J. 1998;17:1665–1674. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brousset P, Meggetto F, Attal M, Delsol G. Kaposi’s sarcoma-associated herpesvirus infection and multiple myeloma. Science. 1997;278:1972. [PubMed] [Google Scholar]
- 8.Burger R, Neipel F, Fleckenstein B, Savino R, Ciliberto G, Kalden J R, Gramatzki G. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cell lines. Blood. 1998;91:1858–1863. [PubMed] [Google Scholar]
- 9.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Clogston C L, Boone T C, Crandall B C, Mendiaz E A, Lu H S. Disulfide structures of human interleukin-6 are similar to those of human granulocyte colony stimulating factor. Arch Biochem Biophys. 1989;272:144–151. doi: 10.1016/0003-9861(89)90205-1. [DOI] [PubMed] [Google Scholar]
- 12.Dalmon J, Laurent M, Courtois G. The human beta fibrinogen promoter contains a hepatocyte nuclear factor 1-dependent interleukin-6 responsive element. Mol Cell Biol. 1993;13:1183–1193. doi: 10.1128/mcb.13.2.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Hon F D, ten Boekel E, Herrman J, Clement C, Ehlers M, Taga T, Yasukawa K, Ohsugi Y, Kishimoto T, Rose-John S, Wijdenes J, Kastelein R, Aarden L A, Brakenhoff J P J. Functional distinction of two regions of human interleukin-6 important for signal transduction via gp130. Cytokine. 1995;7:398–407. doi: 10.1006/cyto.1995.0055. [DOI] [PubMed] [Google Scholar]
- 14.de Hon F D, Ehlers M, Rose-John S, Ebling S B, Bos H K, Aarden L A, Brakenhoff J P J. Development of an interleukin (IL) 6 receptor antagonist that inhibits IL-6-dependent growth of human myeloma cells. J Exp Med. 1994;180:2395–2400. doi: 10.1084/jem.180.6.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupin N, Grandadam M, Calvez V, Gorin I, Aubin J T, Harvard S, Lamy F, Leibowitch M, Huraux J M, Escande J P, Agut H. Herpesvirus-like DNA in patients with Mediterranean Kaposi’s sarcoma. Lancet. 1995;345:761–762. doi: 10.1016/s0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 17.Ehlers M, Grötzinger J, de Hon F D, Müllberg J, Brakenhoff J P J, Liu J, Wollmer A, Rose-John S. Identification of two novel regions of human IL-6 responsible for receptor binding and signal transduction. J Immunol. 1994;153:1744–1753. [PubMed] [Google Scholar]
- 18.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, Cafaro A, Chang H-K, Brady J N, Gallo R C. Synergy between basic fibroblast growth factor and HIV-1 tat protein in induction of Kaposi’s sarcoma. Nature. 1994;371:674–680. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 19.Flore O, Rafii S, Ely S, O’Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 20.Fontaine V, Savino R, Arcone R, de Wit L, Brakenhoff J P, Content J, Ciliberto G. Involvement of the Arg179 in the active site of human IL-6. Eur J Biochem. 1993;211:749–755. doi: 10.1111/j.1432-1033.1993.tb17605.x. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, Andus T, Klapproth J, Hirano T, Kishimoto T, Heinrich P C. Induction of rat acute phase proteins by interleukin-6 in vivo. Eur J Immunol. 1988;18:717–721. doi: 10.1002/eji.1830180510. [DOI] [PubMed] [Google Scholar]
- 22.Hammacher A, Simpson R J, Nice E C. Interleukin-6 (IL-6) partial antagonist (Q159E, T162P) IL-6 interacts with the IL-6 receptor and gp130 but fails to induce a stable hexameric receptor complex. J Biol Chem. 1996;271:5464–5473. doi: 10.1074/jbc.271.10.5464. [DOI] [PubMed] [Google Scholar]
- 23.Hammacher A, Ward L D, Weinstock J, Treutlein H, Yasukawa K, Simpson R J. Structure-function analysis of human IL-6: identification of two distinct regions that are important for receptor binding. Protein Sci. 1994;3:2280–2293. doi: 10.1002/pro.5560031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi’s sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 25.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 26.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 27.Ishiyama T, Nakamura S, Akimoto Y, Koike M, Tomoyasu S, Murata Y, Sato T, Wakabayashi Y, Chiba S. Immunodeficiency and IL-6 production by peripheral blood monocytes in multicentric Castleman’s disease. Br J Haematol. 1994;86:483–489. doi: 10.1111/j.1365-2141.1994.tb04777.x. [DOI] [PubMed] [Google Scholar]
- 28.Kawano M, Hirano T, Matsuda T, Taga T, Horii Y, Iwato K, Asaoku H, Tang B, Tanabe O, Tanaka H, Kuramoto A, Kishimoto T. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature. 1988;332:83–85. doi: 10.1038/332083a0. [DOI] [PubMed] [Google Scholar]
- 29.Kordula T, Travis J. The role of Stat and C/EBP transcription factors in the synergistic activation of rat serine protease inhibitor-3 gene by interleukin-6 and dexamethosone. Biochem J. 1996;313:1019–1027. doi: 10.1042/bj3131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel T A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Rock F, Chong P, Cockle S, Keating A, Ziltner H, Klein M. Structure-function analysis of the C-terminal segment of human interleukin-6. J Biol Chem. 1993;268:22377–22384. [PubMed] [Google Scholar]
- 32.Menziani M C, Fanelli F, Benedetti P G. Theoretical investigation of IL-6 multiprotein receptor assembly. Proteins. 1997;29:528–544. [PubMed] [Google Scholar]
- 33.Miles S A, Rezai A R, Salazar-Gonzales J F, Vander Mayden M, Stevens R H, Logan D M, Mitsuyasu R T, Taga T, Hirano T, Kishimoto T, Martinez-Maza O. AIDS Kaposi’s sarcoma-derived cells produce and respond to interleukin 6. Proc Natl Acad Sci USA. 1990;87:4068–4072. doi: 10.1073/pnas.87.11.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molden J, Chang Y, You Y, Moore P S, Goldsmith M A. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 subunit. J Biol Chem. 1997;272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 36.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 37.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 39.Nicholas J, Ruvolo V R, Zong J, Ciufo D, Guo H-G, Reitz M S, Hayward G S. A single 13 kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (HHV-8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osada S, Yamamoto H, Nishihara T, Imagawa M. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem. 1996;271:3891–3896. doi: 10.1074/jbc.271.7.3891. [DOI] [PubMed] [Google Scholar]
- 41.Paonessa G, Graziani R, De Serio A, Savino R, Ciapponi L, Lahm A, Salvati A L, Toniatti C, Ciliberto G. Two distinct and independent sites on IL-6 trigger gp130 dimer formation and signalling. EMBO J. 1995;14:1942–1951. doi: 10.1002/j.1460-2075.1995.tb07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poli V, Cortese R. Interleukin 6 induces a liver-specific nuclear protein that binds to the promoter of acute-phase genes. Proc Natl Acad Sci USA. 1989;86:8202–8206. doi: 10.1073/pnas.86.21.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poli V, Mancini F P, Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- 44.Rettig M B, Ma H J, Vescio R A, Pold M, Schiller G, Belson D, Savage A, Nishikubo C, Wu C, Fraser J, Said J W, Berenson J R. Kaposi’s sarcoma-associated herpesvirus infection of bone marrow dendritic cells from multiple myeloma patients. Science. 1997;276:1851–1854. doi: 10.1126/science.276.5320.1851. [DOI] [PubMed] [Google Scholar]
- 45.Rettig M B, Said J W, Sun R, Vescio R A, Berenson J R. Kaposi’s sarcoma-associated herpesvirus infection and multiple myeloma. Science. 1997;278:1972–1973. [Google Scholar]
- 46.Rock F L, Li X, Chong P, Ida N, Klein M. Roles of disulfide bonds in recombinant interleukin-6 conformation. Biochemistry. 1994;33:5146–5154. doi: 10.1021/bi00183a018. [DOI] [PubMed] [Google Scholar]
- 47.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvati A L, Lahm A, Paonessa G, Ciliberto G, Toniatti C. Interleukin-6 (IL-6) antagonism by soluble IL-6 receptor α mutated in the predicted gp130-binding interface. J Biol Chem. 1995;270:12242–12249. doi: 10.1074/jbc.270.20.12242. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Savino R, Ciapponi L, Lahm A, Dermatis A, Cabibbo A, Toniatti C, Delmastro P, Altamura S, Ciliberto G. Regional design of a receptor superantagonist of human interleukin-6. EMBO J. 1994;13:5863–5870. doi: 10.1002/j.1460-2075.1994.tb06931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savino R, Lahm A, Giorgio M, Tramontano A, Ciliberto G. Saturation mutagenesis of the human interleukin-6 receptor-binding site: implications for its three-dimensional structure. Proc Natl Acad Sci USA. 1993;90:4067–4071. doi: 10.1073/pnas.90.9.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savino R, Lahm A, Salvati A L, Ciapponi L, Sporeno E, Altamura S, Paonessa G, Toniatti C, Ciliberto G. Generation of interleukin-6 receptor antagonists by molecular-modeling guided mutagenesis of residues important for gp130 activation. EMBO J. 1994;13:1357–1367. doi: 10.1002/j.1460-2075.1994.tb06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schalling M, Ekman M, Kaaya E E, Linde A, Bieberfeld P. A role for a new herpesvirus (KSHV) in different forms of Kaposi’s sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 55.Schumann R R, Kirschning C J, Unbehaun A, Aberle H, Knopf H-P, Lamping N, Ulevitch R J, Herrmann F. The lipopolysaccharide-binding protein is a secretory class 1 acute-phase protein whose gene is transcriptionally activated by APRF/STAT-3 and other cytokine-inducible nuclear proteins. Mol Cell Biol. 1996;16:3490–3503. doi: 10.1128/mcb.16.7.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson R J, Hammacher A, Smith D K, Matthews J M, Ward L D. Interleukin-6: structure-function relationships. Protein Sci. 1997;6:929–955. doi: 10.1002/pro.5560060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snouwaert J N, Leebeek F W G, Fowlkes D M. Role of disulfide bonds in biological activity of human interleukin-6. J Biol Chem. 1991;266:23097–23102. [PubMed] [Google Scholar]
- 58.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 59.Weiergräber O, Schneider-Mergener J, Grötzinger J, Wollner A, Küster A, Exner M, Heinrich P C. Use of immobilized synthetic peptides for the identification of contact sites between human intereleukin-6 and its receptor. FEBS Lett. 1996;379:122–126. doi: 10.1016/0014-5793(95)01482-9. [DOI] [PubMed] [Google Scholar]
- 60.Yawata H, Yasukawa K, Natsuka S, Murakami M, Yamasaki K, Hibi M, Taga T, Kishimoto T. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J. 1993;12:1705–1712. doi: 10.1002/j.1460-2075.1993.tb05815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin, M., S. Q. Yang, H. Z. Lin, M. D. Lane, S. Chatterjee, and A. M. Diehl. Tumor necrosis factor α promotes nuclear localization of cytokine-inducible CCAAT/enhancer binding protein isoforms in hepatocytes. J. Biol. Chem. 271:17974–17978. [DOI] [PubMed]
- 62.Zhang X G, Klein B, Bataille R. Interleukin-6 is a potent myeloma-cell growth factor in patients with aggressive multiple myeloma. Blood. 1989;74:11–13. [PubMed] [Google Scholar]