Summary
In vitro fertilization (IVF) has revolutionized infertility treatment, benefiting millions of couples worldwide. However, current clinical practices for embryo selection rely heavily on visual inspection of morphology, which is highly variable and experience dependent. Here, we propose a comprehensive artificial intelligence (AI) system that can interpret embryo-developmental knowledge encoded in vast unlabeled multi-modal datasets and provide personalized embryo selection. This AI platform consists of a transformer-based network backbone named IVFormer and a self-supervised learning framework, VTCLR (visual-temporal contrastive learning of representations), for training multi-modal embryo representations pre-trained on large and unlabeled data. When evaluated on clinical scenarios covering the entire IVF cycle, our pre-trained AI model demonstrates accurate and reliable performance on euploidy ranking and live-birth occurrence prediction. For AI vs. physician for euploidy ranking, our model achieved superior performance across all score categories. The results demonstrate the potential of the AI system as a non-invasive, efficient, and cost-effective tool to improve embryo selection and IVF outcomes.
Keywords: in vitro fertilization, IVF, artificial intelligence, AI, embryo selection, live-birth outcomes, multi-modal contrastive learning
Graphical abstract
Highlights
-
•
The embryo-selection process in the IVF cycle is variable and experience dependent
-
•
Current AI emphasizes embryo-selection tasks with limited clinical applicability
-
•
Our system makes full use of multi-modal and unlabeled data using contrastive learning
-
•
Our system accurately and reliably predicts the embryo status and live-birth outcome
The bigger picture
In the in vitro fertilization (IVF) process, embryos are usually selected based on morphological characteristics or genetic test results, which are highly variable, experience dependent, and time consuming. To tackle data heterogeneity and labeling limitations, we propose an artificial intelligence (AI) framework system that evaluates embryo images and videos during the assessment of the IVF cycle. This research highlights the potential of AI models to serve as non-invasive, efficient, and cost-effective tools for the advancement of reproductive medicine in general, but specifically for embryo-selection tasks during IVF.
In this paper, the authors developed a unified AI system to assist embryo selection covering the in vitro fertilization cycle. With the multi-modal contrastive learning framework, this model shows superior performance in the prediction of morphology grading, euploidy status, and live-birth potential using embryos’ static images or time-lapse videos. By leveraging large amounts of unlabeled and labeled data, the model shows superior performance and marks a significant advancement in the field of embryo selection.
Introduction
The prevalence of infertility has become a global concern, with over 80 million couples suffering from infertility.1 In the field of assisted reproductive technology, in vitro fertilization (IVF) has revolutionized treatment for infertility, with over 10 million babies having been born from IVF since its invention.2,3 During IVF cycles, the newly generated embryos are fertilized in the lab and can be transferred into the uterus on either day 3 or day 5 of incubation, cryopreserved for subsequent transfers, or discarded based on the evaluation of embryo viability by an embryologist.4 The majority of embryos are selected to transfer based on a morphological score system on day 3 or day 5; others are transferred according to pre-implantation genetic testing for aneuploidy (PGT-A) diagnosis reports.
Traditional methods of embryo selection are required for improving live-birth rates, relying on visual inspection of embryo morphology, and are experience dependent and highly variable.5,6,7 For the non-invasive embryo assessment toolkit, microscopic visualization has been used for scoring embryos from the very beginning of IVF treatment. Skilled embryologists must perform and incorporate complex assessments such as zona pellucida thickness variation, number of blastomeres, degree of cell symmetry and cytoplasmic fragmentation, ploidy, and maternal conditions. Furthermore, suboptimal outcome predictions based on traditional human performance severely limit the impact of the IVF technology.8,9 Because of these factors, achieving a favorable live-birth outcome is still very challenging, with an average success rate of 20%–40%. Another non-invasive tool for implantation evaluation is based on time-lapse videos to assess the embryos’ morphological and morphokinetic information. By enabling embryo-safe recording inside the incubator, time-lapse videos can provide us with plentiful spatial and temporal information to be stored about embryo-development dynamics. To improve the success rate of embryo transfer and pregnancy outcomes, pre-implantation genetic testing (PGT) is currently used for the detection of aneuploidy in fertility clinics. However, trophectoderm (TE) biopsy for PGT has several limitations including invasiveness, cost of DNA sequencing, and inaccuracy in detecting mosaicism. In addition, only a limited number of blastocysts can be selected for PGT. Thus, the demand for a comprehensive automated system that utilizes non-invasive methods to evaluate these factors for the selection/ranking of embryos remains of paramount importance.
Artificial intelligence (AI) has shown potential for revolutionizing healthcare and improving outcomes10,11,12,13 in various domains, such as disease detection and prognosis evaluation.14 Recently, the use of deep neural networks (DNNs) has facilitated the development of efficient and intelligent tools for embryo morphological rating15 and/or implantation probability outcome evaluation using static images in IVF.16 Moreover, researchers have trained DNN-based tools to provide predictions of the embryo ploidy,17,18 blastulation, and implantation outcomes15 using embryo images extracted from time-lapse videos. Despite these recent advances, previous methodologies often rely on extensive and labeled medical data for training, posing challenges in domains such as ploidy prediction (euploids vs. non-euploids) and the subsequent determination of live-birth rates, as acquiring relevant labels can be both costly and time consuming. How to make full use of the large and unlabeled datasets to build a comprehensive automated system to predict embryo ploidy and the outcome of a live birth remains challenging.
Moreover, as mentioned above, embryo data have been accumulated using a variety of methods (e.g., static images or temporal videos) across diverse clinical practices and are therefore rather heterogeneous. Previous methods have so far been limited in representing embryo-development information thoroughly to incorporate the heterogeneous data sources and extract the spatial and temporal information embedded in static images and videos. For example, previous automated deep-learning models have limitations in using embryos screened at specific time points (e.g., 110 h after intracytoplasmic sperm injection19) during their culture, which can ignore vital information regarding embryo-development dynamics and hence hinder their clinical application.20 Therefore, the integration of heterogeneous data sources by AI models to extract embryonic developmental knowledge for viable embryo selection, as well as leading to better reproductive outcomes (such as implantation and pregnancy rates) than a selection based on the traditional assessment alone,21 is another challenge.
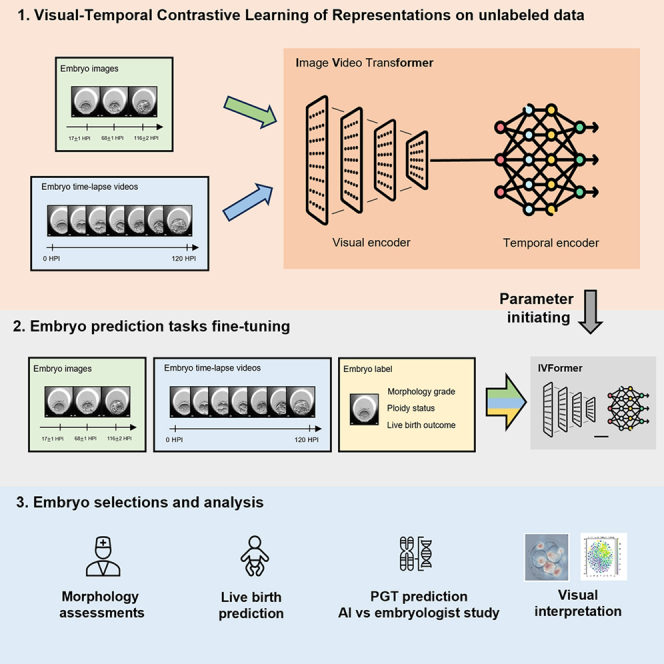
To address the above issues, we propose a novel self-supervised learning framework, named visual-temporal contrastive learning of representations (VTCLR), to learn multi-modal embryo representations from temporal videos and static images with pre-training on large unlabeled data, with a transformer-based network backbone, image video transformer (IVFormer) (Figure 1). Recent research suggests that self-supervised learning (SSL) offers a promising approach that eliminates the need for laborious manual label collections and produces deep feature representation to adapt to downstream tasks.22 Among SSL methods, contrastive learning is a simple yet effective technique without sophisticated pretext tasks, which works by extensively treating one sample as positive and the remaining ones as negative to improve feature discrimination. While contrastive learning has been successfully applied for representation learning from static images,23 it has been challenging to adapt it to video frames, which differ substantially from static images.
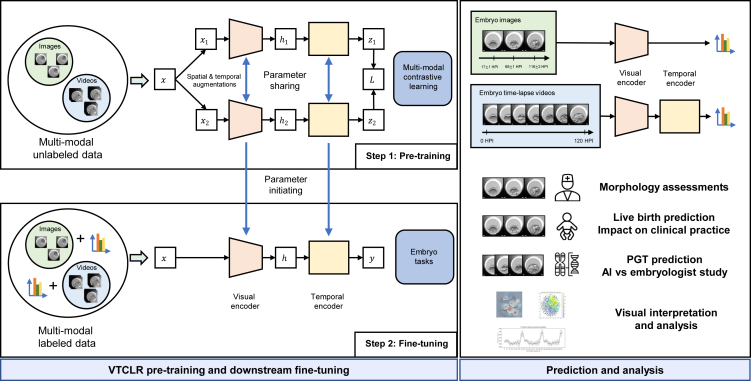
Figure 1.
Schematic illustration of the general AI system for embryo assessment and clinical outcome prediction during the whole IVF cycle
(Left) The proposed self-supervised learning VTCLR (visual-temporal contrastive learning of representations) framework and downstream fine-tuning. The whole VTCLR framework comprises a visual encoder and a temporal encoder via a novel transformer network backbone, IVFormer (image video transformer). Our pre-trained models are then fine-tuned on three downstream tasks by sharing the pre-trained parameters of the encoders and randomly initialized downstream prediction heads. (Right) An illustration of the AI system for embryo assessment to integrate multi-modal data covering the entire IVF cycle, including embryo morphological grading, ploidy prediction (euploids vs. non-euploids) using embryo time-lapse videos, and live-birth occurrence prediction using sequential images and clinical metadata. The models were further validated on independent external cohorts to ensure the AI’s generalizability. We also studied the AI vs. embryologists’ performance in euploidy ranking.
Here, we extend contrastive learning to multi-modal data with VTCLR, an SSL framework tailored for image and video synthetic augmentation. Notably, as embryo development is not a uniform process, we proposed a dynamic-aware sampling strategy that is adaptive for embryo development with contrastive learning on temporal views. To incorporate the heterogeneous data from both static images and temporal videos, we developed IVFormer, a spatial-temporal transformer-based network backbone. The transformer has currently been used to learn the long-term relations in images or videos,24 such as temporally distant segments/consecutive frames in a video.25 Our IVFormer is developed with a shared visual encoder for images and a temporal encoder for videos to capture the temporal information, thus to transfer the knowledge of embryo development between the two modalities (Figure 1).
The use of multi-modal SSL and IVFormer allows the pre-trained model to better capture the embryo-development information. We further applied the pre-trained AI model to address typical clinical scenarios that occur during the IVF process, including embryo morphology assessments, euploidy detection, and live-birth occurrence prediction (Figures 1 and S1). For embryo morphological assessment, we developed the AI models using a large dataset of 2D static embryo images via multi-task learning to extract embryo morphological information, including pronucleus type on day 1, asymmetry and severe fragmentation of blastomeres on day 3, and scoring system for blastocyst stage on day 5. Further, non-euploidies affect more than half of IVF embryos and increase with advancing maternal age and is a leading cause of implantation failure.26 Therefore, the accurate identification of euploids using non-invasive time-lapse video and clinical metadata would bring tremendous value and facilitate better outcomes in the real world. We also performed AI vs. physician validation for euploidy ranking, demonstrating a correlation between the ranking score and the observed euploidy rate. The AI demonstrated superior performance compared with the embryologists across all score categories.
Finally, the prediction of birth outcome depends on many factors including maternal age, menstrual, uterine, and cervical status, previous pregnancy, and fertility history. We utilized the embryo images and clinical metadata to fine-tune IVFormer to identify high-quality embryos with consequent live-birth outcomes using the morphological grading knowledge extracted from the embryo images and videos in pre-training. Interpretable methods were also used in order to understand what drives a prediction important for determining targeted interventions in the clinical setting. By combining embryo and maternal metrics in an ensemble AI model, we evaluated live-birth outcomes in two independent external cohorts (Figure 1). Our AI system demonstrated better performance for embryo morphological grading and blastocyst development in the euploidy ranking and live-birth occurrence prediction, being effective and interpretable for individualized embryo selection for transfer.
Results
Patient characteristics and system overview
In this study a large multi-modal embryo dataset was constructed, which consisted of embryo images, videos, maternal metadata, and clinical outcomes. The demographics and clinical information of the cohort participants are summarized in Tables S1 and S2. In the developmental dataset (EMB-Dev), a total of 41,279 embryo images and 2,136 embryo time-lapse videos were included (Table S1). These were cultured from IVF cycles between 2010 and 2021. All the two-pronuclei embryos were cultured individually and were observed to day 6 before implantation. Each embryo video covers 0 h to 140 h post insemination (HPI). We split the developmental dataset into training and tuning sets with a ratio of 90%:10% for pre-training. Pre-trained models were then fine-tuned on three downstream tasks including embryo morphological assessment, embryo ploidy prediction, and live-birth occurrence prediction.
To ensure a reliable and trustworthy AI system, the three tasks were first validated on their corresponding internal validation datasets (EMB-ME, EMB-PGT, and EMB-LBO) and further validated on three external independent datasets (Table S2). For embryo ploidy prediction, a total of 256 embryos with time-lapse videos (PGT-HE) were included in this study and were compared against human evaluation. For live-birth occurrence prediction, a total of 1,831 embryo transfers with known results were included for external validation. This included 1,343 embryo transfers using double embryo transfer in the first external validation set (LBO-DET) and 488 embryo transfers using single embryo transfer in the second external validation set (LBO-SET).
AI system overview
Our proposed AI system is a comprehensive embryo assessment platform designed to integrate multi-modal data throughout the entire IVF cycle. The system incorporates an SSL framework called VTCLR, with a novel transformer network backbone, IVFormer. Specifically, the model was trained with various lengths and frequencies of augmented temporal frames to support the model’s “understanding” of temporality. We jointly trained the video frames and static images in a unified scheme by sharing spatial augmentations such as the random resized crop, random horizontal flip, and random color jitter. After pre-training, the AI models are fine-tuned for three downstream tasks. First, it demonstrated accurate performances on embryo morphological assessment tasks, including pronucleus symmetry for pronuclear-stage embryos, number of blastomeres, asymmetry and fragmentation rate of blastomeres for cleavage-stage embryos, and grade of inner-cell mass (ICM) and grade of trophectoderm (TE) for blastocyst-stage embryos (Table S5). In addition, the pre-trained model was utilized to predict embryo ploidy (euploid vs. non-euploid) based on a combination of time-lapse image videos and clinical metadata. Finally, for the live-birth occurrence prediction task, we assessed the ability to evaluate embryo viability by using the output of the previous embryo morphology scoring results and clinical metadata.
AI system for embryo morphology assessment via multi-modal pre-training
To demonstrate the effectiveness of our SSL framework, VTCLR, for learning multi-modal representations, we benchmarked its performance on multiple challenging classification tasks for the assessment of embryo morphology grading and blastocyst development. Generally, the following parameters were used as a consensus in the selection of the good-quality embryos in IVF practice27: pronuclei morphology at the pronuclear stage, blastomere characteristics including size, symmetry, and fragmentation at cleavage stage, and grade of ICM and TE at blastocyst stage. The ground truth for embryo morphological assessment is established based on a consensus of three experienced embryologists (for more details, see experimental procedures). We compared our method with other pre-training methods (ImageNet-based pre-training, BYOL, MOCO V2) on our benchmark datasets.
At the pronuclear stage, the zygote (pronuclear) morphology is related to the growth ability for advancing to the blastocyst stage and outcomes of implantation and pregnancy. We used the Z-score system28 to grade the pronucleus symmetry of each embryo. As shown in Table 1, our AI model was able to detect abnormal pronuclear morphology with an area under the curve (AUC) of 0.833. Our AI model, pre-trained with the multi-modal VTCLR approach, outperforms other self-supervised/pre-training baseline models (such as BYOL and MOCO V2) as well as transfer learning on natural source images. For example, the superiority of VTCLR demonstrated superior performance by its 6.4% better performance compared to widely adopted ImageNet-based pre-training and 1.8% better performance compared to state-of-the-art MOCO V2. Additionally, VTCLR outperforms both VTCLR (image only) and VTCLR (video only), which were pre-trained using single-modality embryo images and time-lapse videos, respectively, by 1.6% and 1.0% AUC (Table 1). These results highlight the effectiveness of multi-modal pre-training and its ability to outperform self-supervised methods that pre-train on single-modality static images or time-lapse videos.
Table 1.
Performance comparison in the evaluation of embryos’ morphokinetic features and blastocyst development
| Models/tasks | Stages | |||||
|---|---|---|---|---|---|---|
| Pronuclear (day 1) | Cleavage (day 3) | Cleavage (day 3) | Cleavage (day 3) | Blastocyst (days 5–6) | Blastocyst (days 5–6) | |
| Nucleoli symmetry | Asymmetry | No. of abnormal cells | Severe fragmentation | Grade of ICM | Grade of TE | |
| ImageNet-based pre-training | 0.783 | 0.821 | 0.896 | 0.953 | 0.764 | 0.733 |
| BYOL | 0.808 | 0.845 | 0.919 | 0.972 | 0.799 | 0.771 |
| MOCO V2 | 0.818 | 0.851 | 0.928 | 0.979 | 0.805 | 0.780 |
| VTCLR (image only) | 0.820 | 0.855 | 0.935 | 0.981 | 0.811 | 0.789 |
| VTCLR (video only) | 0.825 | 0.859 | 0.937 | 0.983 | 0.813 | 0.797 |
| VTCLR (image and video) | 0.833 | 0.872 | 0.941 | 0.989 | 0.827 | 0.818 |
AUC showing performance of detecting abnormal pronucleus type of the day-1 embryo; morphological assessment of the day-3 embryos, including detecting blastomere asymmetry, severe fragmentation, and abnormal blastomere cell number; and grades of ICM and TE assessment of the day-5 embryos.
At the cleavage stage, we evaluated the AI model’s ability to determine the asymmetry, number of blastomeres, and fragmentation. Blastomere symmetry was defined as previously reported by Prados et al.,29 which was calculated by dividing the diameter of the smallest blastomere by that of the largest blastomere (for details see experimental procedures). We jointly trained models using different transfer and SSL methods and evaluated them on the above three evaluation metrics. The predicted scores were compared with the gold-standard scoring system.30 Our AI system demonstrated good performance with an AUC of 0.872 for the detection of asymmetry of cells, 0.989 for the binary classification tasks of severe fragmentation of blastomere detection, and 0.941 for the binary detection task of number of abnormal blastomeres (Table 1). As shown in Table 1, our VTCLR also showed substantially better evaluation abilities for embryos at the cleavage stage compared to ImageNet-based pre-training and self-supervised baselines.
At the blastocyst stage, the AI system evaluated the blastocyst morphology including ICM and TE. We evaluated the ability of our AI models and other approaches on the two assessment tasks, which are essential for the prognosis of implantation and fetal development. For ICM and TE scoring tasks, the AI system was able to recognize blastocysts with a high grade of ICM and TE and an AUC of 0.827 and 0.818, respectively (Table 1). Moreover, our VTCLR pre-training once again greatly improved the performance of the blastocyst morphology evaluation in comparison with other methods. By integrating Swin-S as the backbone in IVFormer, better performance in blastocyst grading was attained on our dataset compared to the prior state-of-the-art method employing an ImageNet pre-trained ResNet-50.31 Our model achieved an AUC of 0.827 for the ICM grade, outperforming the previous AUC of 0.751, and an AUC of 0.818 for the TE grade, surpassing the prior AUC of 0.726. This highlights the effectiveness of the representation learning approach in enabling the model to incorporate knowledge of the embryo-development dynamics for the specific task (Table S3).
Taken together, the above results demonstrated that our AI system can achieve decent performance across various embryo-selection tasks and outperforms other pre-training methods. By leveraging a multi-modal representation method that captures both morphological and temporal information about embryo-development dynamics, we extend the self-supervised model to downstream tasks, including the detection of euploids using embryo time-lapse videos and live-birth prediction using images.
Detection of non-euploids using embryo time-lapse videos and clinical metadata
Ploidy is an essential index for the assessment of embryo quality. Embryos of non-euploids, including genome aneuploidy or mosaicism, usually cannot be used for embryo transfer and will lead to extra costs in the PGT-A cycle. Thus, developing a non-invasive method to detect non-euploids is essential for achieving successful embryo transfer while reducing medical costs associated with PGT-A cycles. The decision regarding the transplantation of mosaic embryos currently lacks unified consensus, with many institutions favoring the use of fully euploid embryos over mosaic ones. Consequently, we group mosaic and aneuploid embryos as a non-euploid category, distinct from the euploid embryos, to better align with clinical requirements. We hypothesized that ploidy status can affect cell morphology and migration patterns during embryo development and is therefore amenable to detection by an AI algorithm. Here, we fine-tuned our AI model (IVFormer) to predict the ploidy status of embryos using time-lapse image videos (Table S2). Our approach leverages a transferred embryo visual and temporal encoder from a pre-trained model, with an additional multi-layer perceptron (MLP) initialized and appended to the transferred backbone. Three models for ploidy status detection were developed: a deep-learning model using time-lapse video; a random forest model using clinical metadata; and a combined AI model using both input modalities.
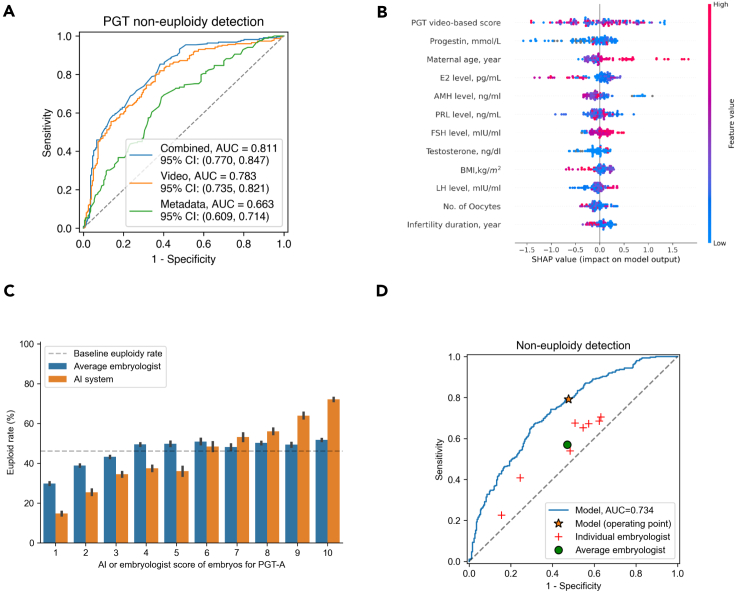
For all tasks, the combined model and the embryo video-only model performed better than the metadata-only model (Figure 2A). The AUC for detecting embryo non-euploidies was 0.663 (95% confidence interval [CI]: 0.609–0.714) for the metadata-only model, 0.783 (95% CI: 0.735–0.821) for the embryo video-only model, and 0.811 (95% CI: 0.770–0.847) for the combined model. Moreover, SSL outperformed the ImageNet-based pre-training method on predicting non-euploids vs. euploids with an AUC of 0.783 compared to 0.691, demonstrating that the representation learning approach enables the model to incorporate knowledge of the embryo-development dynamics for the relevant task (Table S3). For interpreting the effects and relative contributions of the embryo features and clinical parameters on embryo non-euploid detection, we implemented an explainer SHAP (Shapley additive explanation).32 The results showed that the embryo image features and clinical parameters such as AI video-based predicted score, maternal age, and maternal progestin contribute strongly to the detection of non-euploids (Figure 2B). These models could be used to improve the identification of non-euploid status, ultimately enhancing the IVF procedure’s success rate and clinical outcomes.
Figure 2.
Performance of our AI system in identifying ploidy (euploids/non-euploids)
(A) Receiver-operating characteristic (ROC) curves for a binary classification using the clinical metadata-only model, the embryo video-only model, and the combined model in the internal test set. The videos of embryo development are captured using time-lapse system. AUC, area under the curve.
(B) Illustration of features contributing to the progression to euploids by SHAP values. Features on the right of the risk explanation bar pushed the risk higher, and features on the left pushed the risk lower.
(C and D) Performance comparison between our AI model and eight practicing embryologists in embryos’ euploidy scoring and ranking. (C) Correlation analysis between the euploidy rate and the score groups for PGT-A ranking based on AI or embryologist score. The dashed line is the overall euploidy rate of 46.1%. AI score groups were defined by binning AI-predicted probability. (D) Performance comparison between our AI model and eight practicing embryologists in embryos’ euploidy ranking.
The AI system vs. embryologists’ ranking performance
We further conducted a trial to assess the performance of our AI algorithms compared to current standard practices for non-euploids vs. euploids ranking on the external validation set (PGT-HE). As in a euploidy screening setting, the embryologists ranked all the embryos for the probability of being euploids. The top candidate embryos would be further selected to undergo PGT-A testing. We prospectively collected 256 time-lapse videos during real-world clinical use by two IVF clinics, from which 46.1% were euploid embryos. The embryologists were asked to score the embryos from 1 to 10 by reviewing the time-lapse video, with the maternal information also provided. The AI-generated probabilities were also grouped into ten “likelihood categories” (bins) by score thresholds. In the trial, we compared the ranking performance between our AI system and eight embryologists from two fertility clinics on the euploidy rate of each ranking score. First, the consistency between the predicted euploidy probability and the observed euploidy outcomes was analyzed. As shown in Figure 2C, both the ranking score method by the AI and embryologists demonstrated a correlation between the ranking score and the observed euploidy rate. Moreover, the AI demonstrated superior performance compared with the embryologists in all score categories. For example, embryos with a score of 10 generated by our AI system demonstrated a 20.7% higher euploidy rate compared to the embryologists. Furthermore, our AI system achieved a superior AUC of 0.734 for the binary metrics evaluation compared to that of the embryologists, including both junior and senior embryologists (Figure 2D). These results demonstrate the potential of our AI system for improving the accuracy and reliability of ranking embryos for non-euploidy vs. euploidy.
Predicting live birth using embryo images and clinical metadata
To further extend the scope of our AI system for the prediction of live-birth occurrence, we fine-tuned our pre-trained model on the training and tuning sets (Tables S1 and S2). During the fine-tuning stage, we performed fully supervised learning on the target domain and developed three models: a baseline random forest model using clinical metadata; a deep-learning model using embryo images; and a combined AI model using both input modalities. Here, the embryos were transferred on day 3 or day 5/6, and the number of embryos transferred was limited to two or fewer embryos according to the guidelines published in September 2004 by the American Society for Reproductive Medicine.33
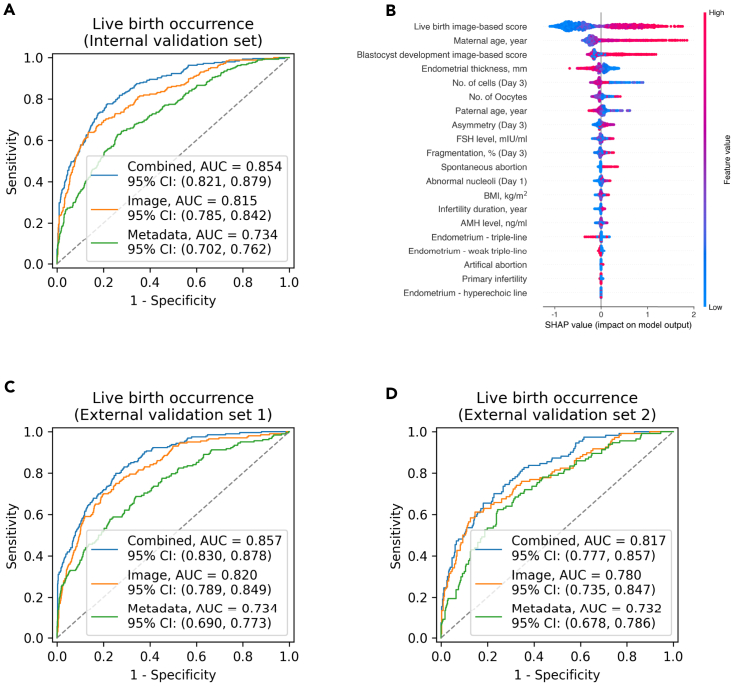
On the internal validation set, the clinical metadata alone gave an AUC of 0.734 (95% CI: 0.702–0.762), and the AI model trained using embryo images alone produced an AUC of 0.815 (95% CI: 0.785–0.842). When trained using combined clinical metadata and embryo images, the AI model achieved superior performance, with an AUC of 0.854 (95% CI: 0.821–0.879) (Figure 3A). Our SSL method demonstrates superior performance over the ImageNet-based pre-training method in predicting live birth only using embryo images, achieving an AUC of 0.815 compared to 0.744 (Table S3). Since the AI system measures many key embryological and clinical features used in IVF, we further demonstrated that it has the potential to reduce the time to grade embryos without sacrificing interpretability. Here, we used the SHAP method to demonstrate the value of the explained predictions made by the AI system and gain insight into factors that affect live-birth occurrence. Our findings indicate that the image-based score was identified as the most significant contributor to the clinical prognosis estimation. The maternal age, endometrial thickness, follicle-stimulating hormone, body mass index, and anti-Mullerian hormone were also highly associated with the live-birth rate per transfer (Figure 3B).
Figure 3.
AI models’ performance in predicting live-birth occurrence
(A) ROC curves showing performance of live-birth occurrence prediction on internal test set. The green, orange, and blue ROC curves represent using the metadata-only model, the embryo image-only model, and the combined model, respectively.
(B) Illustration of features contributing to progression to live-birth occurrence by SHAP values. Features on the right of the risk explanation bar pushed the risk higher, and features on the left pushed the risk lower.
(C and D) ROC curves showing performance of live-birth occurrence prediction on (C) external validation set 1 (double embryo transfer pregnancy) and (D) external validation set 2 (single embryo transfer pregnancy).
To evaluate the model’s performance and generalizability, we further validated these AI models using two independent external cohorts including a double embryo transfer pregnancy (LBO-DET) and a single embryo transfer pregnancy (LBO-SET). For the LBO-DET dataset, the AUC was 0.734 (95% CI: 0.690–0.773) for the clinical metadata-only model, 0.820 (95% CI: 0.789–0.849) for the embryo image model, and 0.857 (95% CI: 0.830–0.878) for the combined model (Figure 3C). The AI demonstrated similar performance for the LBO-SET dataset (Figure 3D). Taken together, these findings demonstrate not only the validity the AI model but also the potential real-life feasibility and utility of an AI-based platform.
Visualization of evidence for AI prediction
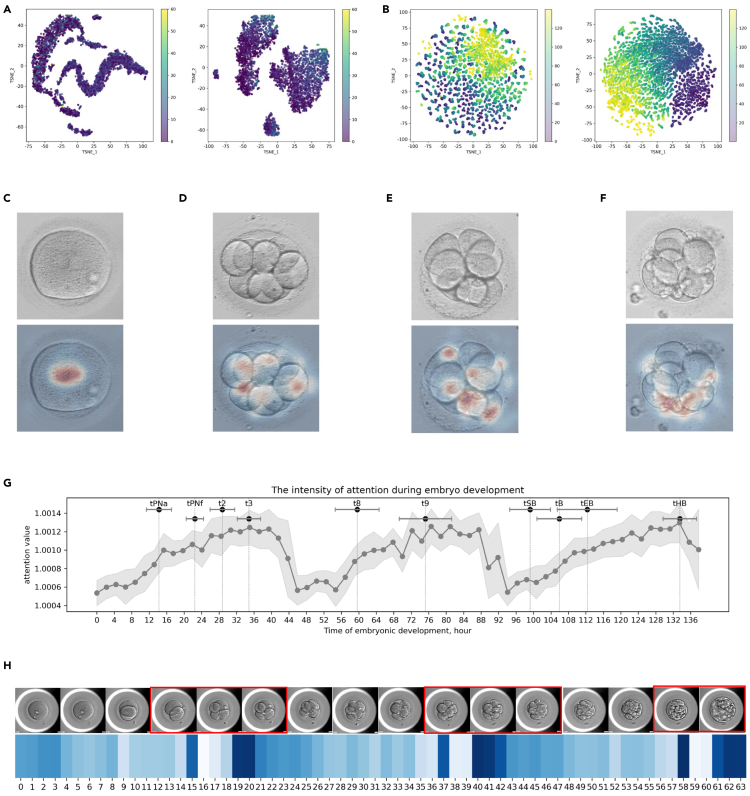
Visualization and interpretation of self-supervised learned representations are of great interest. We investigated whether it could help researchers better understand embryo development and benefit embryo selection and implantation by providing clinical correlation. Here, we visualized the embryo image representations, embryo image saliency maps, and video attention values to demonstrate the performance of our pre-trained visual and temporal encoders. First, we used t-distributed stochastic neighbor embedding (t-SNE) to analyze the representations learned by the visual encoder pre-trained by VTCLR. The t-SNE algorithm maps similar embryo-encoded feature vectors into adjacent 2D points. For representations extracted from embryo images, Figure 4A shows that our visual encoder learns to generate similar representations for embryos with similar fragmentation rates, which is essential for the morphological assessments of embryos at different stages. For representations extracted from frames of videos, Figure 4B shows that our model is also able to learn the intrinsic development dynamics of embryos, as close representations for embryos have similar HPI times. Overall, these results demonstrate that our VTCLR method effectively enables our IVFormer encoders to extract both morphological and developmental information from embryos.
Figure 4.
Visualization of evidence for embryo morphology and ploidy assessment
(A and B) Visualization of embryo representations learned by VTCLR via t-SNE. Representations were extracted from (A) embryo images and (B) frames of time-lapse videos in the pre-training dataset. Each point is colored according to its corresponding label of (A) embryo fragmentation (%) and (B) hours post insemination (HPI) on time-lapse video (hours).
(C–F) Visualization of evidence for embryo morphological assessment using the integrated gradients method. The original image (upper) and an attention map generated from our model are presented. Upper panels: the original embryo images. Lower panels: explanation method-generated saliency heatmaps. (C) Normal pronuclear type of day 1 (good); (D) blastomere cell number of day-3 embryo (normal); (E) blastomere symmetry of day-3 embryo (good); (F) fragmentation rate of day-3 embryo (normal).
(G) Average intensity of attention generated by AI during embryo development. The intensity values of attention were extracted from the pre-trained temporal encoder with all frames averaged over cases in the internal validation set. Each bar on the top represents the mean and standard deviation values of HPI of a defined time-lapse morphokinetic parameter.
(H) Case study of the temporal attention for embryo ploidy at specific frames during embryo development. Blue-colored maps indicate the attention generated by the temporal encoder of the given time-lapse video. The peaks in these maps corresponded to the temporal location of morphokinetic characteristics, which are highlighted in red boxes.
To further investigate the interpretability of the AI model for morphology assessment tasks, we applied integrated gradients (IGs) to generate saliency maps that highlight the areas of the images that were important in determining the AI model’s predictions. The saliency maps from the explanation techniques suggest that the model tends to focus on different spatial features depending on the specific embryo morphology task. For example, the model concentrates on the pronuclei for evaluating the day-1 embryo morphology (Figure 4C). For the prediction of number of blastomeres (Figure 4D) and degree of cell symmetry (Figure 4E), the model tends to focus on the spatial features around the center of day-3 embryos. Additionally, the saliency maps suggest that the AI model focuses on fragments around the cells of day-3 embryos for cytoplasmic fragmentation (Figure 4F).
Finally, to investigate the importance of different frames in time-lapse videos for ploidy prediction, we visualized the attention values generated by the temporal encoder. We marked the HPI of time-lapse morphokinetic parameters with mean and standard deviation values across time-lapse videos to indicate the development events of each frame. Our results indicate that the AI model focuses on frames at specific developmental stages, such as the tPNa, tPNf, t2, t3, t8, t9, tSB, tB, and tEB, for which the attention values produced by the AI model are higher than the average. The attention maps produced by the AI model are highly consistent with the timings/frames in time-lapse video for differentiation of non-euploids vs. euploids reported in the literature34,35 (Figures 4G and 4H). This suggests that the frames at transitions between stages and the whole blastocyst stage are the most relevant for determining ploidy. These results show that our AI model could extract developmental features to generate clinically meaningful insights for ploidy predictions and improve the assisted reproduction process.
Discussion
Progress in embryo selection is aimed at maximizing IVF success rates and reducing the time to conceive while minimizing the risk of multiple pregnancies. Current morphological grading methods rely on descriptive parameters to rank cleavage-stage embryos for transfer. In addition, the non-invasive strategy of time-lapse microscopy has been applied to human embryos, and the possible prognostic effect of morphokinetic data has been reported.36 Although interest in the use of AI to support embryo quality assessment has grown and with numerous AI algorithms having already been developed for the analysis of images or static images from time-lapse videos to aid in the selection of embryos for transfer, they focused on specific tasks for embryo selection, which limited their application in actual clinical practice. In this study, we developed a generalized AI platform on embryo evaluation and live-birth occurrence prediction for the entire IVF cycle, including an embryo morphology grading module, a non-euploidy detection module, and a live-birth prediction module. To make full use of large unlabeled multi-modal data including static images and temporal videos, we utilized an SSL VTCLR framework via a novel transformer network, IVFormer, tailored for embryo-development learning. Experiments show that our pre-trained model achieves great improvement on various benchmarks and shows generalizations in auxiliary tasks related to embryo development found in videos/images. Our results raise the possibility of AI-based selection of embryos based on subtle visual features beyond clinicians’ observational power.
Although previous studies have studied AI-assisted morphological grading37 and blastocyst prediction,38 this study has several key differences to consider. First, in clinical settings, labeled temporal video data of the embryo ploidy (euploids vs. non-euploids) and the subsequent live-birth occurrence from time-lapse technology is relatively limited. Accordingly, we bring a wealth of static images and temporal video to the learning process, aiming at learning strong representations with large-scale unannotated data. Video frames (or even uncurated image data) typically differ from static images, as they have both spatial and temporal variations (the contents of a single frame belong to the same space, and frames are collected across time). Another challenge is constructing SSL models to allow for the interpretation of multi-modal data across time. Therefore, we propose VTCLR, a novel contrast learning framework tailored for image and video synthetic augmentation. For example, as embryo development is not a uniform process, our sampling method adopted a developmental-based sampling strategy adaptive for embryo-development dynamics as temporal augmentation. This approach allowed our SSL model to take advantage of the large-scale unlabeled embryo data with both morphological and morphokinetic information. This multi-modal pre-trained framework can facilitate the model in learning the embryo-development dynamics by better utilizing the large-scale static images and valuable time-lapse videos of embryos without downstream task labels. Together with the introduction of our IVFormer model, which is designed to capture temporal and spatial embryo features, our AI system demonstrated better performance for embryo morphological grading and blastocyst development,31 the euploidy ranking,18,31 and live-birth occurrence prediction,39 being effective and interpretable for individualized embryo selection for transfer.
Oocyte40 and embryo aneuploidies, affecting more than half of embryos produced and increasing in frequency with advancing maternal age, are the main reasons for implantation failure and miscarriages in an IVF cycle, which are currently detected by successful application of an IVF PGT-A test. However, this procedure is invasive and can cause embryonal damage due to biopsy and vitrification. Furthermore, misdiagnosis or mosaicism in PGT-A may result in embryo wastage, and genomic assessment by PGT-A also means a higher cost for an IVF procedure. Significant differences in morphokinetic patterns between euploid and non-euploid embryos may exist, but since they are undetectable to human observers the clinical significance has been absent to modest at best. An alternative non-invasive method for selecting euploids based on a deep-learning method using spatial and temporal information stored in time-lapse images would be much more cost effective and could result in fewer complications. Time-lapse microscopy evaluates the embryo quality by capturing the precise timing and duration of cell divisions, which provides information on all the kinetic parameters of embryo development. These images, with corresponding clinical parameters, may reveal the genetic information encoding proper embryo development and are therefore amenable to AI-based prediction of embryo ploidy (euploids vs. non-euploids) without the use of biopsy. If we can build a non-invasive method for selecting euploids based on a deep-learning method using spatial and temporal information stored in time-lapse videos, it would be much more cost effective and could result in fewer complications. Through contrasting positive pairs against negative pairs from augmentations, VTCLR learns informative representation with IVFormer backbones. Moreover, our AI-based approach shows the potential to interpret morphokinetic features and be used as a surrogate for PGT-A to determine the chromosomal status of pre-implantation embryos.
In addition, this study has assessed the role of automated AI algorithms in improving the live-birth rate using embryo images and clinical metadata, and the selection accuracy was assessed for both single embryo transfers and double embryo transfers (Figures 3C and 3D). We further investigated our AI model’s performance compared to current clinical practitioners including the baseline rate reported from literature (Kamath et al.41), baseline rate of the external validation set (LBO-SET), and live-birth rate by PGT-A screening (Theobald et al.42), in predicting successful live-birth rate (Figure S2). On the LBO-SET dataset, compared with the baseline live-birth rate using embryos selected by embryologists and that reported in previous studies,41 the live-birth rate by AI-assisted ranking and screening of the potential embryos was significantly improved. We further evaluated our AI model’s performance with a transfer rate of selected embryos similar to that of the PGT-A test,42 demonstrating that AI-assisted evaluation could help optimize embryo selection and maximize the likelihood of viable pregnancy with an accuracy comparable to that of a PGT-A test. Moreover, the PGT-A test is invasive and limited by only allowing transplantation of blastocysts on day 5. Additionally, our AI model can yield a continuous score that represents the quality of the embryo. We showed that the AI system’s operating point can be set differently for different clinical applications, balancing the embryo-selection rate and the live-birth rate outcomes.
There are some limitations we hope to address in the future. Since our AI was trained in the Chinese population and tested in an external Chinese cohort from several different geographic areas, its generalizability in other racial populations needs to be further validated. Additionally, various non-embryo-related factors, such as the mother’s health and environmental exposures, could affect the final live-birth outcome and were not explicitly considered in this research. The integration of these data could further enhance the performance of the models in future studies. In summary, the findings presented herein could potentially provide a non-invasive, high-throughput, and low-cost screening tool to greatly facilitate embryo selection and maximize outcome performance. Such AI algorithms could also potentially assist in the standardization of embryo-selection methods across multiple clinical environments.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Guangyu Wang (guangyu.wang24@gmail.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data and any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request. All data and code access requests will be reviewed and (if successful) granted by the Data Access Committee.
The deep-learning models were developed and deployed using standard model libraries and the PyTorch framework. Custom codes were specific to our development environment and used primarily for data input/output and parallelization across computers and graphics processors. All original code has been deposited at Zenodo and is available under the terms of the Apache 2.0 license.43
Dataset characteristics
Retrospective data (embryo images and medical records) were collected from cohorts from the China Consortium of Assisted Reproductive Technology Investigation (CC-ARTI), which consists of hospitals/cohorts from Beijing, Hubei Province, Hunan Province, and Guangdong Province between March 2010 and December 31, 2021. All participants provided written informed consent and received the standard clinical treatment administered at each facility. The consent form includes a statement that the study involves research, an explanation of the research purposes, a description of the procedures, risks, and benefits, voluntary participation, and confidentiality. Institutional Review Board (IRB)/Ethics Committee approvals were obtained in all hospitals of the CC-ARTI committee, and all participating subjects signed a consent form. The work was conducted in compliance with the Chinese Health and Quarantine Law and compliance with patient privacy regulations in China and was adherent to the tenets of the Declaration of Helsinki.
IVF-ET cycles
Pronuclear stage
The oocytes were inseminated by conventional IVF before retrieval. For the day 1 (16–18 h later) embryo morphological evaluation, an embryologist scored the zygote according to the number, size, and location of the pronuclei. Scott et al.44 classified zygotes into four groups Z1–Z4 according to pronuclear morphology corresponding to their quality, using nuclear size, nuclear alignment, nucleoli alignment and distribution, and the position of the nuclei within the zygote.
Cleavage stage
Next, all the two-pronuclei embryos were cultured individually after a fertilization check. Cleavage-stage embryos were evaluated by cell number, the relative degree of fragmentation, and blastomere asymmetry, according to the Istanbul consensus (consensus 2011).27 Blastomere symmetry was defined as previously reported by Prados et al.29: embryos with blastomeres with a diameter difference of <25% were deemed symmetrical (−) and embryos with ≥25% diameter differences were deemed asymmetrical (+). This was calculated by dividing the diameter of the smallest blastomere by that of the largest blastomere.
Blastocyst stage
On the fifth day, the embryo forms a “blastocyst,” consisting of an outer layer of cells (the trophectoderm) enclosing a smaller mass (the ICM). If the embryo was cultured to blastocyst, day-5 or day-6 photographs were stored for analysis as well. Parameters, such as the ICM and TE morphology at the blastocyst stage, were used as data points in the selection of good-quality embryos. Only viable blastocysts (defined as stage ≥3, and at least one score of ICM or TE is ≥B) were selected for transfer or frozen for future use, according to Gardener scoring. The ground truth of morphokinetic features assessment was calculated based on manual evaluation by an expert panel including two independent embryologists, with a senior embryologist providing a further review.
PGT-A cycles
If an embryo was scheduled for PGT-A, a biopsy was performed on day 5 or day 6 according to the blastocyst grade, and next-generation sequencing (NGS) was employed for euploidy assessment. Here, non-euploid was defined as all abnormalities other than euploidy, including simple aneuploid, complex, and mosaic embryos. In PGT-A cycles, all the embryos went on blastocyst culture, and available blastocysts were biopsied and NGS carried out for euploidy assessment.
Live birth
Live birth was defined as the delivery of any viable neonate who was 28 weeks of gestation or older.45 The live-birth rate per embryo transfer was defined as the number of deliveries divided by the number of embryo transfers.46
Images and time-lapse video collection from IVF-ET cycles
Most of the embryos were transferred according to morphological scores on day 3 or the blastocyst stage, while in PGT-A cycles embryos were selected according to PGT-A diagnosis reports. The embryos were observed daily up to day 5/6 with each embryo having at least two photographs: at fertilization check (16–18 h after insemination) and day-3 embryo assessment (66 h after insemination). Time-lapse videos were also carried out for a portion of the patients and were also used for analysis. We used images from the Primo Vision time-lapse system, which takes an image of the embryos every 10 min at nine focal planes, at 10-μm increments (Tables S1 and S2).
Pre-training and downstream datasets
For the model development (EMB-Dev) and internal validation (EMB-Internal), we collected retrospective data from several hospitals, including the First Affiliated Hospital of Sun Yat-sen University, Yichang Central People’s Hospital, Capital Institute of Pediatrics Affiliated Children’s Hospital, and the Sixth Medical Center of the General Hospital of the People’s Liberation Army. These data were randomly split with a ratio of 2:1 for development and internal validation, respectively. The EMB-Dev dataset was used for unsupervised pre-training and fine-tuning of downstream tasks. EMB-Internal datasets (EMB-MA, EMB-PGT, and EMB-LBO) were used for validation only and were not included in any training or fine-tuning processes. External validation datasets (PGT-HE, LBO-DET, and LBO-SET) came from another hospital, Xiangya Reproductive and Genetic Hospital.
Pre-training dataset
We first pre-trained the AI models using the VTCLR method and a developmental dataset (EMB-dev) that contains 41,279 embryo static images and 2,136 embryo time-lapse videos. For VTCLR pre-training, both static images and time-lapse videos are split into a training set and tuning set with a ratio of 9:1 without labels.
Downstream dataset
The pre-trained AI models are fine-tuned on three downstream tasks for embryo morphological assessment, embryo ploidy prediction, and live-birth occurrence prediction. The downstream tasks were applied on the same training and tuning sets as the ones used for pre-training, and samples without corresponding labels were excluded. A total of six additional datasets were included for internal and external validations. The internal datasets for the three downstream tasks (EMB-MA, EMB-PGT, and EMB-LBO) were samples from the same hospitals as the development datasets and without patient-level overlapping. The embryo morphological assessment tasks including pronucleus type on day 1, abnormal number of blastomeres (number of blastomeres = 4 vs. others), asymmetry (asymmetry +/++ vs. asymmetry −), and severe fragmentation of blastomeres (fragmentation >25% vs. others) on day 3, and grade of ICM and TE on day 5, used the same patient-level dataset split with the pre-training dataset and were evaluated on an internal validation dataset. The embryo static images with known live-birth labels and clinical metadata were utilized to develop AI models for live-birth outcome prediction in groups of embryo transfer level. External validation datasets (PGT-HE, LBO-DET, and LBO-SET) came from Xiangya Reproductive and Genetic Hospital. LBO-DET and LBO-SET, consisting of 1,343 and 488 embryo transfers from 1,262 patients and 467 patients, respectively, were used for live-birth outcome prediction validations. The embryo transfers in the LBO-DET were all double embryo transfers, and the ones in the LBO-SET were all single embryo transfers. The embryo time-lapse videos with known PGT-A labels and clinical metadata were utilized for PGT-A prediction. The internal validation set was constructed with 520 embryos from 356 patients, EMB-PGT, and an additional external validation set, PGT-HE, was constructed with 256 embryos from 222 patients for human evaluation. There was no patient-level overlap between datasets (Figure S3).
Image annotation and pre-processing
During the image-grading process, all embryo images were first de-identified to remove any patient-related information. Study participants were excluded due to poor photographic quality or unreadable images. Photographs must follow certain criteria, such as: sufficient lighting such that the structures are visible; sharp focus of the zona pellucida and trophectoderm; one embryo per micrograph with no visible instruments and little or no debris in the visual field; the entire embryo shown within the limits of the image (including the zona pellucida); and text or symbols in the images not hindering the visibility of the embryos. Expected stages of embryo images were annotated based on the Istanbul consensus (Table S4). For embryo image scoring, nine senior embryologists from the two centers scored embryos according to scoring rules. In the image pre-processing stage, we used a segmentation network, U-Net, to automatically crop all embryo images with bounding boxes to reduce the bias introduced during data collection.
Overview of the AI framework
Our AI framework performs multi-modal self-supervised contrastive learning for images and video representation, named VTCLR, on top of a spatial-temporal transformer network backbone, named image video transformer (IVFormer). In recent years, rapid progress in non-invasive imaging and time-lapse techniques has generated an unprecedented amount of embryo data including both static images and temporal videos. To utilize the large-scale static images and valuable time-lapse videos of embryos without downstream task labels, our IVFormer is compatible with both modalities by a shared visual encoder for images and a temporal encoder to capture the temporal information from videos. VTCLR is applied to transfer embryo-development knowledge between the two modalities of unlabeled embryo data by alternating training shared encoders using images and videos. Temporal augmentation based on a dynamic-aware sampling strategy constructs more challenging positive and negative view pairs together with spatial augmentations to improve the quality of the embryo representations. The enhanced embryo representations are used for the downstream tasks for embryo selection covering the entire IVF cycle.
Architecture
For the compatibility of embryo images and time-lapse video, our IVFormer model consists of a visual encoder and a temporal encoder. The visual encoder for static embryo images and frames in embryo time-lapse videos can share common knowledge about embryo morphology, and the temporal encoder can extract temporal information about embryo development from time-lapse videos. For an image/frame input , the encoded image feature vector is , where is the visual encoder. Specifically, we adopted the Swin Transformer (Swin-S),47 which is a hierarchical transformer-based image backbone with shifted windows for feature extraction, as the visual encoder. For a time-lapse video that consists of multiple images , the images are first sampled with a sampling strategy as . They are then encoded into static representations with the visual encoder, . The static representations of frames are enriched with a learnable embedding to help clarify the time stamp of each frame. The video representation is further acquired as , where is the temporal encoder that consists of three attention-based blocks of the same architecture but independent parameters. In each attention-based block, the temporal enriched representations are sequentially fed into global and local relation layers. The global relation layer captures long-range relations with multi-head attention, and the local relation layer increases the feature dimensions and blends neighboring vectors by 1D convolution with rectified linear unit activation. We apply layer normalization after the global and local relation layers, after which residual connections are added to stabilize the training process. In the last layer, we apply an average pooling on the transformed temporal embeddings to produce a video representation. The model prediction is generated via an MLP as , based on the visual or temporal representation.
Multi-modal self-supervised contrastive pre-training
VTCLR is a contrastive pre-training framework to learn different but complementary visual and temporal knowledge from modalities both of image and video. Contrastive learning aims at learning representation through contrasting positive view pairs against negative view pairs. Given a set of augmented views , the training objective is to identify the positive sample among a set of unrelated noise samples for a given , where the current view and the positive sample are two different augmented views of the same data input, and the negative samples are augmented views of others. Our VTCLR is designed to learn representation from modalities, therefore consisting of two parts: spatial and temporal augmentations for images and videos and a two-stage training process.
Data augmentations
For static images and frames in time-lapse videos, all images are applied with spatial augmentations including random resized crop, random horizontal flip, and random color jitter. For time-lapse videos, we used a dynamic-aware sampling strategy to reduce redundancy in frames sampled from videos. Given the fact that embryos appear relatively static between stage transitions during development, the commonly used strategies such as continuous sampling with a fixed stride or uniform sampling along the temporal dimension will cause redundancy of morphology in the sampled frames. Therefore, we adopted a sample method based on image-level difference to increase the difference of motion magnitude between sampled frames.48 Specifically, the motion signal of frame t, is quantified with , where is the pixel value of frame , and normalized to motion salience distribution with L1-norm (i.e., ). To adjust the uniformity of the distribution , a hyper-parameter is introduced and the adjusted distribution is formulated as . To sample frames from a video, the video is segmented into parts with the same cumulated motion salience inside each part, then one frame is randomly sampled from each part to form a video sample. Specifically, we set the when sampling. By adopting the above sampling strategy, frames with more morphological changes will have a higher probability of being sampled, and the sampling process keeps sufficient randomness for temporal data augmentations.
Training process
Suppose we have a minibatch of samples and define the contrastive prediction task on pairs of augmented views derived from the minibatch, resulting in augmented views. Given a positive pair of views, we treated other views as negative views. Each view is encoded by IVFormer into a hidden vector of . A projection head of MLP is applied on the encoded vector to produce latent vector . NT-Xent loss is applied to the latent vectors to maximize the agreement of positive pairs while minimizing the agreement of negative ones as , where and are latent vectors of a positive pair, sim(.) is the cosine similarity between two vectors, and is the temperature parameter.
The training process alternates between two stages of SSL on images and videos, respectively. The consistency of cross-modality knowledge is facilitated by sharing the visual encoder in IVFormer. In the first stage, we improve the feature extraction ability of the shared visual encoder on static images. In the second stage, we further optimized the temporal encoder with time-lapse videos based on the shared visual encoder. A similar self-supervised process is applied to minibatches of sampled time-lapse videos. The positive pair is derived from two sampled video clips from the same time-lapse video with both spatial and temporal augmentations. The two training stages are alternatives with an interval of one epoch. After pre-training, the pre-trained model is further fine-tuned on the downstream tasks.
Prediction of embryo morphology scores using embryo images
To demonstrate the effectiveness of VTCLR on the shared visual encoder of IVFormer, we fine-tuned it with a joint loss of embryo morphology grading based on embryo images. Specifically, we introduce the Z score for pronuclear-stage embryos, number of blastomeres, number of blastomeres and cytoplasmic fragmentation for cleavage-stage embryos, and grade of ICM and grade of TE for blastocyst-stage embryos as the supervised embryo morphology grading losses. With the assumption of homoscedastic uncertainty, the loss of a task is weighted and factorized to for a regression task or for a classification task, where is a trainable parameter. Therefore, the combined loss function for the morphology grading can be formulated as .
Prediction of live-birth occurrence using embryo images
For the verification of the temporal encoder pre-trained with VTCLR, we fine-tuned both visual and temporal encoder in the IVFormer with the live-birth occurrence prediction task. The live birth occurrence prediction task maps a transfer with single or multiple embryos to a probability of live-birth occurrence, where is a sequence of images of embryos. To address the input with different numbers of embryos in each transfer, we adopted IVFormer to generate transfer-level live-birth occurrence by extracting features from images. We used two views of the zygote stage and cleavage stage for each embryo, and the temporal embedding of them is set according to the time stamp of each image.
Prediction of ploidy using embryo time-lapse videos
We also fine-tuned the pre-trained visual and temporal encoder with the ploidy prediction task. The ploidy prediction task predicts the embryo ploidy (euploids vs. non-euploids) using embryo time-lapse video and clinical metadata. For each time-lapse video, we first downsampled the frames of the video by uniform sampling, resulting in a total of frames, to capture morphological features and developmental kinetics of the embryo over the whole process of embryo development. The model is fine-tuned with an additional classification head for ploidy prediction. We used a 5-fold cross-validation scheme for ploidy prediction.
Training details
Embryo images/frames were resized to . The pre-processed and sampled frames in the video were stacked along the temporal axis to generate a 3D tensor, where we set . The pre-training of models by back-propagation of errors was performed for 200 epochs with an Adam optimizer,49 with a learning rate of , weight decay of , and batch size of 64. For each downstream task, fine-tuning was performed for 32 epochs, learning rate of , weight decay of , and batch size of 64.
Interpretation of AI predictions
For data in different modalities, we adopted different visualization methods for interpreting the AI predictions. There are in total three modalities of data adopted in this study. Therefore, we used the gradient-based method, attention-based method, and Shapley-value-based method for the interpretations of static images, time-lapse videos, and risk factors, respectively, in the patient metadata.
First, we used IGs50 to generate visual explanations that highlight areas contributing to the model’s prediction based on static images. Given a trained model , an input image , and an output score for class , the basic gradient-based visualization method51 generates a saliency map where the importance weight for each pixel is derived by . The IG method improves the basic method by path-integrated gradients, which quantifies the importance of each pixel as follows: , where is a baseline image. This overcomes the disadvantage of the basic method that lacks sensitivity to important features when the model output to the correct class is saturated. In this study, the baseline image used a black image with the same size of input images. The generated heatmap was filtered by a Gaussian kernel with for smooth.
Second, we used the mean of attention scores after passing the first global relation layer in the temporal decoder for the interpretations of the model’s predictions based on time-lapse videos. The attention scores can be expressed as follows: , where and are the queries and key vectors in the calculation of a global relation layer, and is the dimension of the key vector. The attention score represents the intensity of the model’s attention to different frames in the video clip. The attention scores are averaged across all time-lapse videos in the internal validation set. Finally, to display the impact of relevant risk factors on prediction for non-euploid detection and live-birth prediction, we adopted the TreeExplainer in the SHAP method. The TreeExplainer is a value-explainable tool for tree-based models, which can efficiently and exactly compute optimal local explanations, as defined by desirable properties from game theory.
Performance study of the AI system
To assess the impact of the AI system on ploidy predictions, the AI system was compared against chance (randomly assigned ploidy predictions) and eight embryologists. We conducted experiments to study the AI system vs. embryologist’s performance in the ploidy evaluation. Given an embryo, we provided the video and corresponding clinical metadata to the embryologists. The embryologists assigned a score of 1–10, with higher score indicating a greater likelihood of euploidy. Each embryo was scored twice (2 weeks after the initial reading), and the average was calculated as the final score. We then used the generated AI probabilities to calculate the ranking score for embryo evaluation and filtering for further PGT-A tests. The euploidy rate of embryos is calculated at different thresholds. Embryologists’ scores higher than 5 indicate euploidy. For the AI performance, we used receiver-operating characteristic (ROC) evaluation and operating point-based binary classification, based on the generated probability.
Statistical analysis
To evaluate the performance of regression models for continuous-values prediction in this study, we applied mean absolute error, R-squared, and Pearson correlation coefficient. We applied the Bland-Altman plot52 displaying the difference between the measured value and the predicted value of a sample against the average of the two. We evaluated the agreement of the predicted value and actual value by 95% limits of agreement and intraclass correlation coefficient. The models for binary classification were evaluated by ROC curves of sensitivity vs. 1 − specificity. The AUC of ROC curves was reported with 95% CIs. The 95% CIs of AUCs were estimated with the non-parametric bootstrap method (1,000 random resamplings with replacement). The operating point of an AI system could be set differently to balance the true-positive rate and the false-positive rate. The embryo-level models were generated using the average outputs of predictions of image level. The AUCs were calculated using the Python package of scikit-learn (version 0.22.1).
Acknowledgments
This study was funded by the National Natural Science Foundation of China (grant 62272055), the Tencent Foundation through the XPLORER PRIZE, the Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), and the Macao Young Scholars Program (AM2023024). We thank the members of the Lin, Xu, and Wang groups for their assistance. We thank many volunteers and physicians for grading embryo photographs.
Author contributions
Conceptualization, G.W. and X.L.; methodology, X.L. and K.W.; software, K.W., F.F., Z.J., T.G., and G.Y.; investigation, G.W., K.W., Y. Gao, Longbin Chen, and X.L.; resources, Longbin Chen, Y.M., S.Z., Y. Gu, G. Lu, Lei Chen, L.-S.M., Y.S., Y.X., and G. Lin; data curation, K.W., Y. Gao, and Longbin Chen; writing – original draft, G.W., K.W., Y. Gao, and X.L.; writing – review & editing, G.W., K.W., Y. Gao, Longbin Chen, Y.X., G. Lin, and X.L.; supervision, G.W., Y.X., G. Lin, and X.L.
Declaration of interests
The authors declare no competing interests.
Published: May 2, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2024.100985.
Contributor Information
Guangyu Wang, Email: guangyu.wang24@gmail.com.
Yanwen Xu, Email: xuyanwen@mail.sysu.edu.cn.
Ge Lin, Email: linggf@hotmail.com.
Xiaohong Liu, Email: xiaohong.liu@ucl.ac.uk.
Supplemental information
References
- 1.Ombelet W., Campo R. Affordable IVF for developing countries. Reprod. Biomed. Online. 2007;15:257–265. doi: 10.1016/s1472-6483(10)60337-9. [DOI] [PubMed] [Google Scholar]
- 2.ESHRE ART.Factsheet. 2023. https://www.eshre.eu/Europe/Factsheets-and-infographics
- 3.Pinborg A., Henningsen A.-K.A., Malchau S.S., Loft A. Congenital anomalies after assisted reproductive technology. Fertil. Steril. 2013;99:327–332. doi: 10.1016/j.fertnstert.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Wang J., Sauer M.V. In vitro fertilization (IVF): a review of 3 decades of clinical innovation and technological advancement. Therapeut. Clin. Risk Manag. 2006;2:355–364. doi: 10.2147/tcrm.2006.2.4.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter Bendus A.E., Mayer J.F., Shipley S.K., Catherino W.H. Interobserver and intraobserver variation in day 3 embryo grading. Fertil. Steril. 2006;86:1608–1615. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 6.Paternot G., Devroe J., Debrock S., D'Hooghe T.M., Spiessens C. Intra- and inter-observer analysis in the morphological assessment of early-stage embryos. Reprod. Biol. Endocrinol. 2009;7:105. doi: 10.1186/1477-7827-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storr A., Venetis C.A., Cooke S., Kilani S., Ledger W. Inter-observer and intra-observer agreement between embryologists during selection of a single Day 5 embryo for transfer: a multicenter study. Hum. Reprod. 2017;32:307–314. doi: 10.1093/humrep/dew330. [DOI] [PubMed] [Google Scholar]
- 8.Wahl B., Cossy-Gantner A., Germann S., Schwalbe N.R. Artificial intelligence (AI) and global health: how can AI contribute to health in resource-poor settings? BMJ Glob. Health. 2018;3 doi: 10.1136/bmjgh-2018-000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosny A., Aerts H.J.W.L. Artificial intelligence for global health. Science. 2019;366:955–956. doi: 10.1126/science.aay5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topol E.J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 11.Ravizza S., Huschto T., Adamov A., Böhm L., Büsser A., Flöther F.F., Hinzmann R., König H., McAhren S.M., Robertson D.H., et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat. Med. 2019;25:57–59. doi: 10.1038/s41591-018-0239-8. [DOI] [PubMed] [Google Scholar]
- 12.Norgeot B., Glicksberg B.S., Butte A.J. A call for deep-learning healthcare. Nat. Med. 2019;25:14–15. doi: 10.1038/s41591-018-0320-3. [DOI] [PubMed] [Google Scholar]
- 13.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K., Liu X., Xu J., Yuan J., Cai W., Chen T., Wang K., Gao Y., Nie S., Xu X., et al. Deep-learning models for the detection and incidence prediction of chronic kidney disease and type 2 diabetes from retinal fundus images. Nat. Biomed. Eng. 2021;5:533–545. doi: 10.1038/s41551-021-00745-6. [DOI] [PubMed] [Google Scholar]
- 15.Leahy B.D., Jang W.D., Yang H.Y., Struyven R., Wei D., Sun Z., Lee K.R., Royston C., Cam L., Kalma Y., et al. Automated Measurements of Key Morphological Features of Human Embryos for IVF. Med. Image Comput. Comput. Assist. Interv. 2020;12265:25–35. doi: 10.1007/978-3-030-59722-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silver D.H., Feder M., Gold-Zamir Y., Polsky A.L., Rosentraub S., Shachor E., Weinberger A., Mazur P., Zukin V.D., Bronstein A.M. Data-driven prediction of embryo implantation probability using IVF time-lapse imaging. arXiv. 2020 doi: 10.48550/arXiv.2006.01035. Preprint at. [DOI] [Google Scholar]
- 17.Huang B., Tan W., Li Z., Jin L. An artificial intelligence model (euploid prediction algorithm) can predict embryo ploidy status based on time-lapse data. Reprod. Biol. Endocrinol. 2021;19:185. doi: 10.1186/s12958-021-00864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang V.S., Kandula H., Thirumalaraju P., Kanakasabapathy M.K., Cherouveim P., Souter I., Dimitriadis I., Bormann C.L., Shafiee H. The use of voting ensembles to improve the accuracy of deep neural networks as a non-invasive method to predict embryo ploidy status. J. Assist. Reprod. Genet. 2023;40:301–308. doi: 10.1007/s10815-022-02707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes J., Brendel M., Gao V.R., Rajendran S., Kim J., Li Q., Malmsten J.E., Sierra J.T., Zisimopoulos P., Sigaras A., et al. A non-invasive artificial intelligence approach for the prediction of human blastocyst ploidy: a retrospective model development and validation study. Lancet. Digit. Health. 2023;5:e28–e40. doi: 10.1016/S2589-7500(22)00213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milewski R., Ajduk A. Time-lapse imaging of cleavage divisions in embryo quality assessment. Reproduction. 2017;154:R37–R53. doi: 10.1530/REP-17-0004. [DOI] [PubMed] [Google Scholar]
- 21.Siristatidis C., Komitopoulou M.A., Makris A., Sialakouma A., Botzaki M., Mastorakos G., Salamalekis G., Bettocchi S., Palmer G.A. Morphokinetic parameters of early embryo development via time lapse monitoring and their effect on embryo selection and ICSI outcomes: a prospective cohort study. J. Assist. Reprod. Genet. 2015;32:563–570. doi: 10.1007/s10815-015-0436-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan R., Rajpurkar P., Topol E.J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 2022;6:1346–1352. doi: 10.1038/s41551-022-00914-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H.-Y., Chen X., Zhang Y., Luo R., Wang L., Yu Y. Generalized radiograph representation learning via cross-supervision between images and free-text radiology reports. Nat. Mach. Intell. 2022;4:32–40. [Google Scholar]
- 24.Dai R., Das S., Kahatapitiya K., Ryoo M.S., Brémond F. MS-TCT: multi-scale temporal convtransformer for action detection. arXiv. 2022 doi: 10.48550/arXiv.2112.03902. Preprint at. [DOI] [Google Scholar]
- 25.Irmawati, Chai R., Basari, Gunawan D. Optimizing CNN Hyperparameters for Blastocyst Quality Assessment in Small Datasets. IEEE Access. 2022;10:88621–88631. [Google Scholar]
- 26.Fragouli E., Alfarawati S., Spath K., Jaroudi S., Sarasa J., Enciso M., Wells D. The origin and impact of embryonic aneuploidy. Hum. Genet. 2013;132:1001–1013. doi: 10.1007/s00439-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 27.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum. Reprod. 2011;26:1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 28.Scott L. Pronuclear scoring as a predictor of embryo development. Reprod. Biomed. Online. 2003;6:201–214. doi: 10.1016/s1472-6483(10)61711-7. [DOI] [PubMed] [Google Scholar]
- 29.Prados F.J., Debrock S., Lemmen J.G., Agerholm I. The cleavage stage embryo. Hum. Reprod. 2012;27:i50–i71. doi: 10.1093/humrep/des224. [DOI] [PubMed] [Google Scholar]
- 30.Johansson M., Hardarson T., Lundin K. There is a cutoff limit in diameter between a blastomere and a small anucleate fragment. J. Assist. Reprod. Genet. 2003;20:309–313. doi: 10.1023/A:1024805407058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T.-J., Zheng W.-L., Liu C.-H., Huang I., Lai H.-H., Liu M. Using Deep Learning with Large Dataset of Microscope Images to Develop an Automated Embryo Grading System. FandR. 2019;01:51–56. [Google Scholar]
- 32.Lundberg S.M., Erion G.G., Lee S.-I. Consistent individualized feature attribution for tree ensembles. arXiv. 2018 doi: 10.48550/arXiv.1802.03888. Preprint at. [DOI] [Google Scholar]
- 33.Practice Committee of the American Society for Reproductive Medicine. Practice Committee of the Society for Assisted Reproductive Technology Guidelines on number of embryos transferred. Fertil. Steril. 2009;92:1518–1519. doi: 10.1016/j.fertnstert.2009.08.059. [DOI] [PubMed] [Google Scholar]
- 34.Mumusoglu S., Yarali I., Bozdag G., Ozdemir P., Polat M., Sokmensuer L.K., Yarali H. Time-lapse morphokinetic assessment has low to moderate ability to predict euploidy when patient- and ovarian stimulation-related factors are taken into account with the use of clustered data analysis. Fertil. Steril. 2017;107:413–421.e4. doi: 10.1016/j.fertnstert.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Chavez S.L., Loewke K.E., Han J., Moussavi F., Colls P., Munne S., Behr B., Reijo Pera R.A. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat. Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyagi Y., Habara T., Hirata R., Hayashi N. Feasibility of deep learning for predicting live birth from a blastocyst image in patients classified by age. Reprod. Med. Biol. 2019;18:190–203. doi: 10.1002/rmb2.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leahy B.D., Jang W.-D., Yang H.Y., Struyven R., Wei D., Sun Z., Lee K.R., Royston C., Cam L., Kalma Y. Springer; 2020. Automated Measurements of Key Morphological Features of Human Embryos for IVF; pp. 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thirumalaraju P., Hsu J.Y., Bormann C.L., Kanakasabapathy M., Souter I., Dimitriadis I., Dickinson K.A., Pooniwala R., Gupta R., Yogesh V., Shafiee H. Deep learning-enabled blastocyst prediction system for cleavage stage embryo selection. Fertil. Steril. 2019;111:e29. [Google Scholar]
- 39.Liu H., Zhang Z., Gu Y., Dai C., Shan G., Song H., Li D., Chen W., Lin G., Sun Y. Development and evaluation of a live birth prediction model for evaluating human blastocysts from a retrospective study. Elife. 2023;12 doi: 10.7554/eLife.83662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minasi M.G., Colasante A., Riccio T., Ruberti A., Casciani V., Scarselli F., Spinella F., Fiorentino F., Varricchio M.T., Greco E. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum. Reprod. 2016;31:2245–2254. doi: 10.1093/humrep/dew183. [DOI] [PubMed] [Google Scholar]
- 41.Kamath M.S., Mascarenhas M., Kirubakaran R., Bhattacharya S. Number of embryos for transfer following in vitro fertilisation or intra-cytoplasmic sperm injection. Cochrane Database Syst. Rev. 2020;8:CD003416. doi: 10.1002/14651858.CD003416.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theobald R., SenGupta S., Harper J. The status of preimplantation genetic testing in the UK and USA. Hum. Reprod. 2020;35:986–998. doi: 10.1093/humrep/deaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G., Wang K., Gao Y., Chen L., Ma Y., Jiang Z., Gao T., Yang G., Feng F., Zhang S., et al. Zenodo; 2024. A generalized AI system for human embryo selection covering the entire IVF cycle via multi-modal contrastive learning Zenodo. [DOI] [Google Scholar]
- 44.Scott L., Alvero R., Leondires M., Miller B. The morphology of human pronuclear embryos is positively related to blastocyst development and implantation. Hum. Reprod. 2000;15:2394–2403. doi: 10.1093/humrep/15.11.2394. [DOI] [PubMed] [Google Scholar]
- 45.Shi Y., Sun Y., Hao C., Zhang H., Wei D., Zhang Y., Zhu Y., Deng X., Qi X., Li H., et al. Transfer of Fresh versus Frozen Embryos in Ovulatory Women. N. Engl. J. Med. 2018;378:126–136. doi: 10.1056/NEJMoa1705334. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson J., Roberts S.A., Showell M., Brison D.R., Vail A. No common denominator: a review of outcome measures in IVF RCTs. Hum. Reprod. 2016;31:2714–2722. doi: 10.1093/humrep/dew227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z., Lin Y., Cao Y., Hu H., Wei Y., Zhang Z., Lin S., Guo B. 2021 IEEE/CVF International Conference on Computer Vision (ICCV) 2021. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows; pp. 9992–10002. [Google Scholar]
- 48.Zhi Y., Tong Z., Wang L., Wu G. Mgsampler: An explainable sampling strategy for video action recognition. arXiv. 2021 doi: 10.48550/arXiv.2104.09952. Preprint at. [DOI] [Google Scholar]
- 49.Kingma D.P., Ba J. Adam: A method for stochastic optimization. arXiv. 2014 doi: 10.48550/arXiv.1412.6980. Preprint at. [DOI] [Google Scholar]
- 50.Sundararajan M., Taly A., Yan Q. Axiomatic attribution for deep networks. arXiv. 2017 doi: 10.48550/arXiv.1703.01365. Preprint at. [DOI] [Google Scholar]
- 51.Simonyan K., Vedaldi A., Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv. 2013 doi: 10.48550/arXiv.1312.6034. Preprint at. [DOI] [Google Scholar]
- 52.Giavarina D. Understanding Bland Altman analysis. Biochem. Med. 2015;25:141–151. doi: 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request. All data and code access requests will be reviewed and (if successful) granted by the Data Access Committee.
The deep-learning models were developed and deployed using standard model libraries and the PyTorch framework. Custom codes were specific to our development environment and used primarily for data input/output and parallelization across computers and graphics processors. All original code has been deposited at Zenodo and is available under the terms of the Apache 2.0 license.43