Abstract
We previously reported that a human immunodeficiency virus type 1 (HIV-1) envelope (Env) mutant with the whole cytoplasmic domain deleted, denoted mutant TC, is able to dominantly interfere with wild-type (wt) virus infectivity. In the present study, the feasibility of developing a dominant negative mutant-based genetic anti-HIV strategy targeting the gp41 cytoplasmic domain was investigated. Mutants TC and 427,TC, a TC derivative with a Trp-to-Ser substitution introduced into residue 427 in the CD4-binding site, and a series of mutants with deletions in the cytoplasmic domain, effectively trans-dominantly interfered with wt Env-mediated viral infectivity, as demonstrated by an env trans-complementation assay. The syncytium formation-defective 427,TC double mutant not only inhibited heterologous LAV and ELI Env-mediated viral infectivity but also interfered with syncytium formation and infectivity mediated by the Env proteins of the two primary isolates 92BR and 92US. Stable HeLa-CD4-LTR-β-gal clones that harbored Tat-controlled expression cassettes encoding the control ΔKS, which had a deletion in the env gene, wt, or mutant env gene were generated. Viral transmission mediated by laboratory-adapted T-cell-tropic HXB2 and NL4-3 viruses was greatly reduced in the TC and 427,TC transfectants compared to that observed in the control ΔKS and wt transfectants. Viral replication caused by HXB2 and NL4-3 viruses and by macrophage-tropic ConB and ADA-GG viruses was delayed or reduced in human CD4+ T cells transfected with the 427,TC env construct compared to that observed in cells transfected with the control ΔKS or TC env construct. The lack of significant interference by TC mutant was due neither to the lack of TC env gene integration into host DNA nor to the lack of TC Env expression upon Tat induction. These results indicate that this 427,TC Env double mutant has a role in the development of trans-dominant mutant-based genetic anti-HIV strategies.
The transmembrane (TM) protein gp41 cytoplasmic domain of human immunodeficiency virus type 1 (HIV-1) spans residues 706 to 856 of the envelope (Env) protein. Mutations or deletions in the cytoplasmic domain affect various steps of the virus life cycle, such as virus replication, viral infection and transmission, cytopathogenicity, Env stability, and Env incorporation into virions (9, 16, 17, 19, 24, 27, 28, 30, 34, 41, 46). In addition, this domain has been implicated in interaction with matrix (MA) protein during virus assembly and budding (15, 22, 23, 33, 45). This notion has been further supported by the demonstration of direct in vitro protein binding between the gp41 cytoplasmic domain and MA (14). It was shown that the cytoplasmic domain contains a membrane-proximal tyrosine-based, basolateral targeting signal for virus budding in polarized cells (31). The same tyrosine residue was also implicated in the high rate of internalization by endocytosis of endogenously synthesized Env protein from the plasma membrane (37). Moreover, peptide homologues mimicking lentivirus lytic peptide (LLP)-1 and LLP-2 were found to display cytolytic and calmodulin-binding and -inhibitory properties (38, 40). Because of its multifunctional role in various stages of the virus replication cycle and its involvement in cellular dysfunction and cytopathogenesis, the cytoplasmic domain of gp41 may represent an ideal target for the design of an anti-HIV strategy.
In this regard, we previously characterized an HIV-1 proviral DNA clone HXV-m and found that the Env encoded by this clone is truncated in the cytoplasmic domain (9). We also demonstrated that this mutant dominantly interferes with the infectivity of the HXB2 virus when the HXB2 and HXV-m proviruses are coexpressed. This trans-dominant defect in infectivity by the HXV-m mutant Env may be attributed to the formation of a dysfunctional complex between the wild-type (wt) and mutant Env proteins when they are coexpressed (9). Our studies suggest that a trans-dominant negative mutant-based anti-HIV strategy can be designed to target the gp41 cytoplasmic domain.
Alterations to other functional domains in the Env protein may affect the interference with wt viral infectivity by a dominant negative mutant. An NL4-3 virus-derived Env mutant 41.2 with a polar Glu or Arg substitution for the Val located at amino acid 2 of the membrane fusion domain was demonstrated to be membrane fusion defective and to exhibit a dominant interference effect with the wt viral infectivity (5, 21). Interestingly, a 12-amino-acid deletion in the CD4-binding domain greatly reduces the interference by mutant 41.2 (5). That study indicates that a functional CD4-binding region is required for the dominant interference of the 41.2 mutant. Likewise, an understanding of whether the dominant interference by the cytoplasmic domain truncation mutant requires CD4-binding ability should provide insight into what sequences in the CD4-binding domain are necessary for this dominant interference.
A consideration of a genetic anti-HIV therapeutic design is to target the genes with an intervention or therapeutic purpose to human CD4+ or other HIV-susceptible cells so that the conference of an anti-HIV state to the host cells by the target gene products can be assessed. The issue of whether previously reported Env-based dominant mutants (4, 9, 11, 21, 39) could interfere with viral replication and spread in human CD4+ T cells, which are natural host cells for HIV infection, has never been documented.
Primary or macrophage (M)-tropic isolates of HIV-1 play an important role in HIV pathogenesis throughout the lifetime of infected individuals. The prototypic subtype B strain HXB2 and primary isolates display considerable differences in their genetic, phylogenetic, and biological characteristics (25). The amino acid sequences of the env genes between members of each subtype typically vary from 3 to 23%. Utilizing trans-dominant mutants in the design of an anti-HIV approach requires that the gene products of dominant negative mutants exhibit broad cross-interference with infections caused by different viral isolates. The fusion-defective mutant 41.2 was shown to interfere in trans with syncytium formation caused by the Env of a heterologous WMJ-2 strain (21) and to inhibit viral transmission in HeLa-CD4 stable clones (4). Whether this mutant and other Env-based dominant negative mutants (4, 5, 9, 11, 21, 39) could interfere with the viral replication and spread of heterologous laboratory-adapted T-cell (T)-tropic and M-tropic viruses has not yet been addressed. In addition, whether env-targeted dominant negative mutants could be utilized in the design of genetic anti-HIV strategies to combat HIV infection is not known.
In the present study, we attempted to explore the feasibility of developing a cytoplasmic domain-based genetic anti-HIV approach. Interference with viral infectivity by an HXB2 strain-isogenic, cytoplasmic domain-truncated Env mutant (TC mutant) and its derivative, in which the CD4-binding ability of the TC Env mutant was abrogated (427,TC mutant), and by Env mutants with a series of deletions from the C terminus of the cytoplasmic tail was investigated. Both TC and 427,TC mutants confer interference with viral infection when these Tat-inducible mutant env gene constructs are stably transferred into CD4+ HeLa derivatives. However, viral replication is interfered with only in human CD4+ T cells harboring the 427,TC env gene, not in cells harboring the TC env gene. These results indicate that a functional CD4-binding domain is not required for interference by the TC mutant. Our study also provides the first demonstration of Env-based dominant interference with replication and spread caused by T-tropic and M-tropic viruses in targeted human CD4+ T cells.
MATERIALS AND METHODS
Cells and plasmids.
HeLa-CD4-LTR-β-gal, SupT1, COS-1, 293 cells, and hybridoma Chessie 8 were all previously described (11). pGEM-EB, pBaby, pBSX, pSVE7, pSVE7(ΔKS), pIIIextat, HXB2gpt, and pHXBCATΔBgl were all previously described (8–11).
Construction of plasmids.
A method that selects against a single-stranded DNA template containing uracil (29) was followed to construct env mutants (Fig. 1). Briefly, the uracil-containing single-stranded DNA template of pGEM-EB (10) was primed with the synthetic oligonucleotide 5′ GCTTGGTAGGTTTAAGAATATTTTTGCTGTACTTTCTATAG 3′ (nucleotides 8295 to 8336 with a deletion of G at position 8315 of the HXB2 sequence) to prepare the replicative form of the pGEM-EB(TC) (Fig. 1A). Oligonucleotide 5′ AAACAAATTATAAACATGTCGCAGAAAGTAGGAAAAGCA 3′ (nucleotides positioned from 7484 to 7522; the codon underlined encodes a Trp-to-Ser substitution at residue 427 in the CD4-binding site) was used to prime the uracil-containing single-stranded template of pGEM-EB(TC) to produce pGEM-EB(427,TC). The KpnI-BamHI fragments isolated from pGEM-EB(TC) and pGEM-EB(427,TC) were then substituted for the corresponding sequences in pBSX and pSVE7 to generate pBSX- and pSVE7-based mutant env-expressing vectors, respectively. pSVE7-puro (Fig. 1B), which encodes a bacterial puromycin resistance gene (puro), was constructed as follows. The 440 bp of the EcoRI fragment, which contains a simian virus 40 (SV40) ori and is located outside of the coding sequences of HIV-1 long terminal repeat (LTR) and env in pSVE7, was deleted by EcoRI digestion. The isolated vector was treated with calf intestine alkaline phosphatase, followed by filling-in with Klenow enzyme to generate a blunt-ended fragment. The puromycin resistance expression cassette, containing the SV40 early promoter, puro, and an SV40 polyadenylation signal, was excised from the pPUR vector (Clontech, Palo Alto, Calif.) by successive digestion with BamHI, mung bean nuclease, and PvuII. The isolated 1.4-kb DNA fragment was ligated to the treated 7.4-kb fragment of the pSVE7 vector, resulting in pSVE7-puro. The KpnI-BamHI sequence in the pSVE7-puro was then replaced by the corresponding DNA fragments isolated from the ΔKS and mutant pSVE7 plasmids, yielding ΔKS and mutant pSVE7-puro plasmids (Fig. 1B). pSVE7(ΔKS) is an env-defective pSVE7 with a deletion from the KpnI site to the StuI site. For construction of pSVE7-puro plasmids that encoded a series of Env mutants with deletions from the C terminus of the cytoplasmic tail (Fig. 3A), the XhoI site located at the 5′-LTR of pSVE7-puro was destroyed by limited digestion. The KpnI-XhoI sequence in this altered form of pSVE7-puro was then replaced by the homologous sequences isolated from the HXB2R3 provirus (45)-derived molecular clones TM709, TM752, TM775, TM795, TM813, and TM844 (46). For construction of a Tat-controlled LAV strain-derived env expression vector, the KpnI fragment of the env gene in pSVIII-92RW (25) was replaced by the 2.7-kb KpnI fragment (nucleotides 6343 to 9005) isolated from pNL4-3 chimera (2).
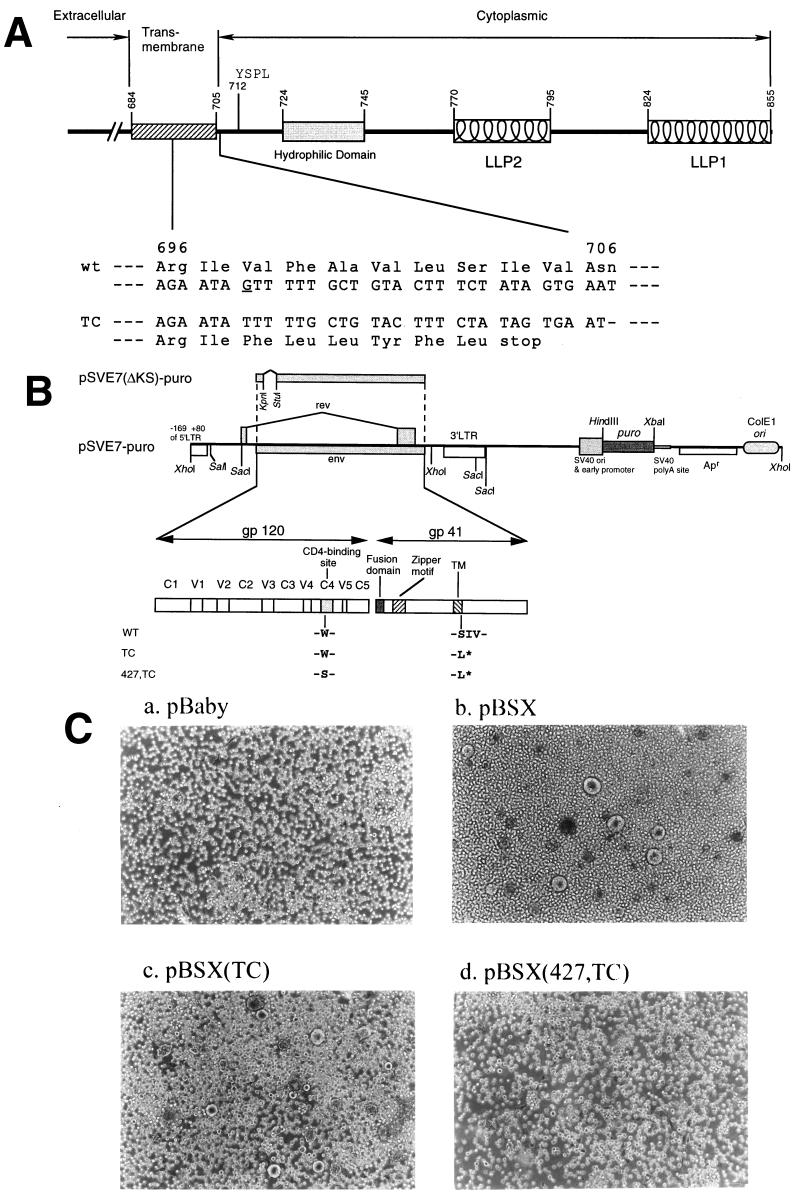
FIG. 1.
Construction and characterization of Env mutants. (A) A schematic representation of the cytoplasmic domain truncation Env mutant. Structural motifs in gp41 cytoplasmic domains are shown, such as internalization signal YSPL and the tyrosine-based basolateral-targeting signal located at residue 712, a highly hydrophilic region, and two positively charged amphipathic α-helices marked LLP-1 and LLP-2. The amino acid residues are numbered according to their positions in the Env of the HXB2 strain. The region encompassing residues from 696 to 706 and its corresponding DNA sequence are indicated. The deletion of the G base as underlined located at nucleotide 8315 of the HXB2 sequence results in a frame shift of the open reading frame and a premature termination of translation after leucine at position 703 in the TM domain. (B) Construction of wt and mutant pSVE7-puro plasmids. A puromycin resistance expression cassette was inserted into the wt pSVE7. The KpnI-BamHI fragments isolated from various mutant pSVE7 plasmids were substituted for the corresponding sequence in wt pSVE7-puro to generate various mutant pSVE7-puro plasmids. The asterisk represents the stop codon in the TM domain as described in panel A. Single-letter amino acid codes are used. (C) Analysis of the syncytium-forming ability of Env mutants. COS-1 cells were transfected with 2 μg each of pBaby, wt, or mutant pBSX as indicated. Two days after transfection, 106 SupT1 cells were added into each transfected culture, and photographs were taken 18 h after coculture under a light microscope. Magnification, ×100.
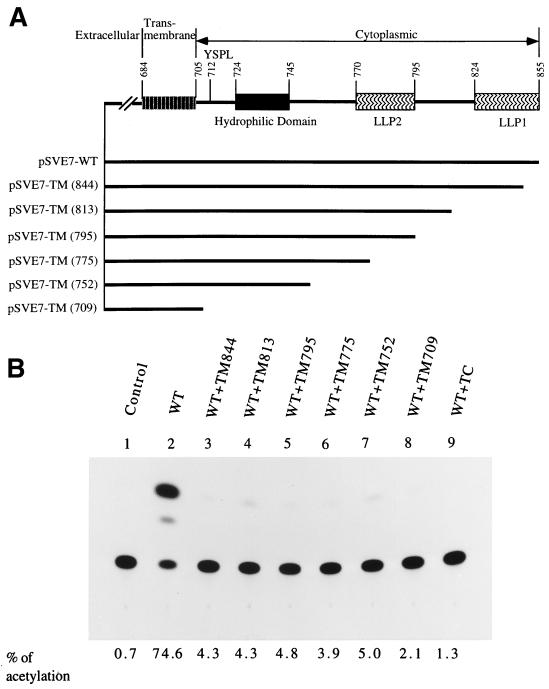
FIG. 3.
Effect of coexpression with Env deletion mutants on wt Env-mediated viral infectivity. (A) A schematic representation of a series of Env mutants with deletions from the C terminus of the cytoplasmic domain. The numbers in parentheses indicate that a stop codon was introduced after the codon of the indicated residue. (B) Interference with wt Env-mediated infectivity by deletion mutants. 293 cells were cotransfected with 7.5 μg of pHXBCATΔBgl and 7.5 μg of wt pSVE7-puro in the presence or absence of 7.5 μg of mutant pSVE7-puro plasmids that encoded deletion mutants as indicated. Cell-free viruses containing 6.5 × 104 cpm of RT activity from each viral supernatant were used to determine Env-mediated virus-to-cell transmission to HeLa-CD4-LTR-β-gal cells.
DNA sequencing.
All mutant phagemids and constructs were screened or confirmed by dideoxy-chain termination with Sequenase (Amersham, Arlington Heights, Ill.). Primers 5′ CCTCAGGAGGGGACCCAG 3′ (nucleotides 7314 to 7331) and 5′ TCCTCCAAGTCTGAAGATCTCG 3′ (nucleotides 7639 to 7618) were used for forward and reverse DNA sequencing, respectively, of the mutation in the CD4-binding site. Primer 5′ CACACGACCTGGATGGAG 3′ (nucleotides 8096 to 8113) was used to sequence the coding region encompassing the TC mutation. Primer 688f (5′ AGTAGGAGGCTTGGTAGG 3′; nucleotides 8287 to 8304), primer 734f (5′ GAAGAAGAAGGTGGAGAGAGA 3′; nucleotides 8423 to 8445), and primer 793f (5′ CTCAAATATTGGTGGAATCT 3′; nucleotides from 8600 to 8619) were used to sequence the TM series of pSVE7-puro plasmids.
Plasmid DNA transfection.
For COS-1 transfection, the DEAE-dextran method as previously described was followed (10). 293 cells were cotransfected with pHXBCATΔBgl and functional env plasmids in the presence or absence of mutant env plasmids by the calcium phosphate coprecipitation method (9, 11). The control pSVE7(ΔKS) plasmid, which does not encode a functional env gene, was added into the transfection mixtures to keep the amounts of DNA in all transfections constant. Therefore, viruses obtained from cotransfection of pHXBCATΔBgl with pSVE7(ΔKS) were used as a negative control in all env trans-complementation assays. For plasmid transfection of HeLa-CD4-LTR-β-gal env transfectants, the calcium phosphate coprecipitation method was employed. For DNA transfection of P4-R5-MAGI, the Superfect transfection method (Qiagen, Valenica, Calif.) was followed. For DNA transfection of SupT1 or env-transfected CEM-SS cells, the DEAE-dextran transfection method was used.
Establishment of stably env-transfected cells.
The env pSVE7-puro plasmids were used to transfect HeLa-CD4-LTR-β-gal cells by using the enhanced calcium phosphate coprecipitation method (3). The transfected cells were grown in medium containing puromycin, cloned, and expanded as previously described (3). The single clones were screened and confirmed by Southern hybridization and Western blotting. CEM-SS and PM1 (107 cells) were transfected with 40 μg each of pSVE7-puro constructs by electroporation by using an Electro Cell Manipulator 600 (BTX Inc., San Diego, Calif.) at 250 V and 950 μF. Transfected cells were grown in medium supplied with 0.5 μg of puromycin/ml for 3 weeks prior to viral infection studies.
Immunofluorescence microscopy.
pIIIextat-transfected HeLa-CD4-LTR-β-gal env clones grown in 4-well, gelatinized slides were fixed with phosphate-buffered saline (PBS) containing 4% paraformaldehyde on ice for 1 h and permeabilized with 1% Triton X-100 in TBS (20 mM Tris-HCl [pH 7.4] containing 137 mM NaCl) for 5 min. After blocking, the slides were successively incubated with a 1:200 dilution of mouse ascitic fluids obtained from hybridoma Chessie 13 and Chessie 8 and with a 1:100 dilution of fluorescin isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin (IgG) (Zymad Laboratories Inc., South San Francisco, Calif.). After being washed, the slides were analyzed under an Axiovert 135 fluorescence microscope (Zeiss, Jena, Germany).
Viral transmission and syncytium formation assays.
env-transfected COS-1 cells grown in 60-mm-diameter dishes were cocultured with SupT1 cells as previously described (9) to determine syncytium formation. For the env trans-complementation assay, recombinant viruses pseudotyped with T-tropic or M-tropic Env proteins and containing the amounts of reverse transcriptase (RT) activity as indicated in each experiment were used to challenge HeLa-CD4-LTR-β-gal or P4-R5 MAGI indicator cells, respectively. When env HeLa-CD4-LTR-β-gal transfectants were infected with viruses, viruses containing 5 × 105 to 5 × 106 cpm of RT activity were used. When RT activity of proviral DNA-transfected SupT1 cultures reached 105 to 106 cpm/ml, 106 of PBS-washed SupT1 cells were cocultured with HeLa-CD4-LTR-β-gal env transfectants grown in 60-mm-diameter petri dishes. Three days after infection or coculture, the cells were fixed with PBS containing 1% formaldehyde and 0.2% glutaraldehyde at room temperature for 5 min. The fixed and washed cells were stained with Accustain (Sigma, St. Louis, Mo.), which contains May-Grünwald and Giemsa stains, and cultures were then scored for syncytium formation. To assess the cytopathic effects caused by the Env proteins derived from primary isolates, P4-R5 MAGI cells were cotransfected with pIIIextat and wt pSVE7-puro and scored for syncytium formation 2 days after transfection. For infection of puromycin-selected CEM-SS or PM1 cells, virus containing 2 × 104 cpm of RT activity was used for challenge.
Enzymatic assays.
RT activity for cell-free viruses obtained from proviral DNA-transfected culture supernatants or for virus obtained from supernatants of infected cultures was determined as previously described (9, 11). Entry of the defective reporter virus into target cells was measured by chloramphenicol acetyltransferase (CAT) activity as previously described (9).
PCR amplification.
PBS-washed, env-transfected CEM-SS cells were lysed with 10 mM Tris-HCl (pH 8.0) containing 50 mM KCl, 2.5 mM MgCl2, 0.1 mg of gelatin/ml, 0.45% Nonidet P-40, and 0.45% Tween 20 at a cell density of 5 × 106 cells/ml. The cell lysates were incubated at 56°C for 2 h, followed by incubation at 95°C for 10 min. Primers 5′ AGCAGCAGGAAGCACTATGG 3′ (sense) and 5′ CCAGACTGTGAGTTGCAACAG 3′ (antisense), corresponding to the nucleotide sequences 7795 to 7814 and 7936 to 7916, respectively, of the HXB2 sequence were used in PCR to detect the env sequences in transfected CEM-SS cells. PCR with a pair of oligonucleotides complementary to the first exon of the human β-globin gene at nucleotides 14 to 33 (5′ ACACAACTGTGTTCACTAGC 3′ [sense]) and nucleotides 123 to 104 (5′ CAATTCATCCACGTTCACC 3′ [antisense]), relative to the translation initiation site was also performed to normalize the total amount of cellular DNA present in samples. The PCR buffer compositions were those recommended by Perkin-Elmer (Norwalk, Conn.). The samples were subjected to 35 cycles of amplification at 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s on a GeneAmp PCR system 2400 (Norwalk, Conn.).
RESULTS
Construction and characterization of cytoplasmic domain-truncated Env mutants.
To explore the feasibility of developing a genetic anti-HIV strategy targeting the gp41 cytoplasmic domain, an HXB2 strain (18, 36)-isogenic mutant TC clone was constructed by site-directed mutagenesis. This TC mutant encoded an Env with a premature termination of translation after leucine at position 703 in the gp41 TM region (Fig. 1A). To determine whether a functional CD4-binding domain was required for dominant interference by mutant TC, the CD4-binding ability of this mutant was abrogated. A single Ser substitution for the Trp located at residue 432 of the HIV-1 BRU Env was shown to block the receptor-binding ability of the Env and to render the virus carrying this mutation unable to replicate in SupT1 and U937 cells (13). Therefore, oligonucleotide-directed mutagenesis was performed to alter the TC env gene to encode an additional Trp-to-Ser substitution at residue 427 in the CD4-binding site (Fig. 1B). The TC and 427,TC double mutant env genes were all subcloned into pBSX- and pSVE7-based Env expression vectors. Both pBSX(TC) and pBSX(427,TC) encoded gp120 and a truncated gp160 precursor (data not shown). To determine the syncytium-forming ability of these mutants, wt or mutant pBSX-transfected COS-1 cells were cocultured with SupT1 cells, and syncytium formation was observed under a light microscope 18 h after coculture. Transfection with wt pBSX or pBSX(TC) resulted in significant syncytium formation (more than 200 giant cells in a 6-well dish) (Fig. 1C, panels b and c, respectively). Transfection with pBSX(427,TC) did not show any syncytia (Fig. 1C, panel d), consistent with the phenotype of the 427,TC mutant in that the Trp-to-Ser substitution in the CD4-binding site abolishes the ability of Env to bind to CD4 (13).
Interference with wt Env-mediated viral infectivity by cytoplasmic domain truncation mutants.
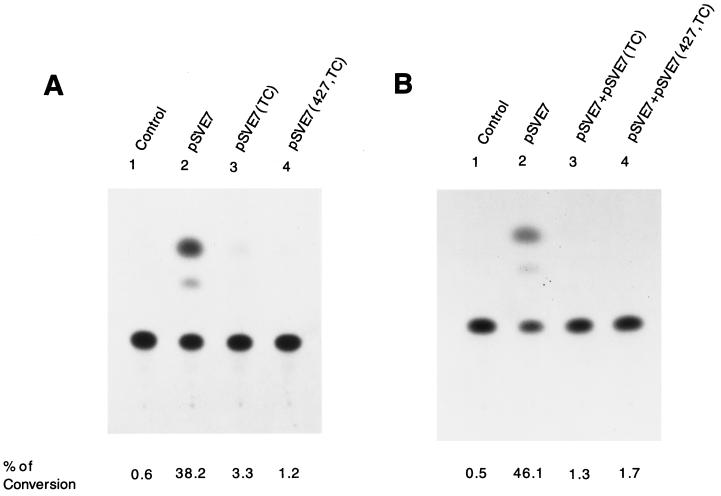
To determine whether TC and 427,TC mutants interfered with wt Env-mediated viral infectivity, a trans-complementation assay with an env-defective, HIV-1 reporter provirus pHXBCATΔBgl (26) was performed. This assay measures the ability of Env protein to mediate one round of viral replication. 293 cells were cotransfected with pHXBCATΔBgl along with the wt, TC, or 427,TC pSVE7 plasmid. Cotransfection with pHXBCATΔBgl and pSVE7(ΔKS), with the KpnI-StuI sequence deleted in the env gene (Fig. 1B), was performed in parallel. Cell-free viruses containing equal amounts of RT activity from each transfection were used to challenge HeLa-CD4-LTR-β-gal cells, and cell lysates prepared 3 days after infection were assayed for CAT activity. Env proteins encoded by the 427,TC double mutant as well as by the TC mutant did not support viral infectivity (Fig. 2A). Furthermore, the 427,TC double mutant as well as the TC mutant significantly interfered with wt Env-mediated viral infectivity when coexpressed with the wt Env protein (Fig. 2B).
FIG. 2.
Assessment of mutant Env proteins by an env trans-complementation assay. (A) Inability of mutant proteins to mediate virus-to-cell transmission. 293 cells were cotransfected with 10 μg of pHXBCATΔBgl and 10 μg each of the control ΔKS, wt, or mutant pSVE7. Cell-free viruses containing 2 × 105 cpm of RT activity from each viral stock were used to challenge HeLa-CD4-LTR-β-gal cells, and CAT activity was measured. (B) Interference with wt Env-mediated viral infectivity by Env mutants. 293 cells were cotransfected with 10 μg each of pHXBCATΔBgl and wt pSVE7 in the presence or absence of 10 μg of mutant pSVE7 as described in Materials and Methods. Viruses containing 2 × 105 cpm of RT activity from each transfection were used to determine Env-mediated virus-to-cell transmission. In panels A and B, the viruses used in the lanes marked control were produced from cotransfection of pHXBCATΔBgl and pSVE7(ΔKS). In panel B, the pSVE7(ΔKS) plasmid was added into transfection mixtures to maintain the total DNA amounts in all transfection reactions at the same levels.
Deletions in the cytoplasmic domain conferred dominant interference with wt Env-mediated viral infectivity.
To determine whether interference with viral infectivity was a general feature for deletions in the cytoplasmic domain, interference with wt Env-induced viral infectivity by a series of mutants with deletions from the C terminus of the cytoplasmic domain was tested. The env genes encoded by HXB2R3-derived proviral clones TM844, TM813, TM795, TM775, TM752, and TM709 were subcloned into the pSVE7-puro vector (Fig. 3A). These env expression plasmids encoded Env proteins with deletions after residues 844, 813, 795, 775, 752, and 709, respectively, to the C terminus of the cytoplasmic domain. Viruses encoded by these TM proviruses typically exhibit a phenotype of impaired infectivity (46). Migrations of the Env precursors and TM subunits of these deletion mutants in sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed a ladder-like pattern (data not shown). Upon challenge to HeLa-CD4-LTR-β-gal cells, viruses pseudotyped with wt and any of the deletion mutants, even with deletions as small as 12 amino acids in the C terminus (TM844), produced a much lower level of CAT than the virus pseudotyped with the wt Env alone (Fig. 3B).
Interference with functions mediated by heterologous T-tropic and M-tropic Env proteins.
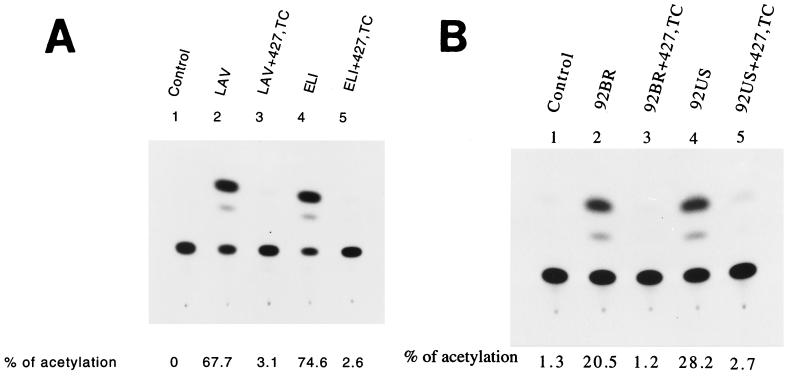
Viral infectivities mediated by the defective virus pseudotyped with the Env of the LAV or ELI strain (35), derived from two heterologous T-tropic isolates, and by the defective virus pseudotyped with the LAV or ELI Env together with the 427,TC Env mutant were compared. Coexpression with the 427,TC mutant Env strikingly interfered with viral infectivity mediated by the LAV or ELI Env protein (Fig. 4A). Interference by this double mutant on syncytium formation mediated by the 92BR and 92US Env proteins of two primary isolates (25) was then examined. The P4-R5 MAGI indicator cell line, a HeLa derivative which expresses CD4 and human CCR-5 and contains integrated copies of the β-galactosidase gene fused to the HIV-1 LTR (7), was cotransfected with pIIIextat along with pSVIII-92BR or pSVIII-92US in the presence or absence of pSVE7(427,TC). pIIIextat is an HIV-1 LTR-driven Tat expression plasmid. Cells expressing the 92BR or 92US Env in the presence of 427,TC double mutant in a 1:1 molar ratio of wt/427,TC plasmid showed a 25.3 and 30.8% reduction, respectively, in syncytium formation compared to cells expressing the 92BR or 92US Env alone (Table 1). When the ratio of wt/427,TC plasmid was set at 1:2, the reduction in syncytium formation was found to be 55.5 and 75.2%, respectively (Table 1). Moreover, defective virus pseudotyped with 92BR or 92US Env in the presence of 427,TC mutant Env significantly interfered with 92BR or 92US Env-mediated viral infectivity (Fig. 4B).
FIG. 4.
Interference by the 427,TC double mutant with viral infectivity. (A) Interference with viral infectivity mediated by heterologous T-tropic Env proteins. 293 cells were cotransfected with 7.5 μg of pHXBCATΔBgl and 5 μg each of pSVIII-LAV or pSVIII-ELI with or without 10 μg of pSVE7(427,TC). pSVIII-ELI encodes the Env of the HIV-1 ELI strain. Cell-free viruses containing 105 cpm of RT activity from each transfection were assayed for Env-mediated viral infectivity. (B) Interference with viral infectivity mediated by the Env proteins derived from primary isolates. 293 cells were cotransfected with 7.5 μg of pHXBCATΔBgl and 5 μg each of pSVIII-env expression plasmids that encoded Env proteins derived from primary isolates as indicated with or without 10 μg of pSVE7(427,TC). Viruses containing 105 cpm of RT activity from each viral supernatant were used to challenge P4-R5 MAGI indicator cells, and CAT activity was assayed.
TABLE 1.
Interference with M-tropic Env-mediated syncytia formation by the 427,TC Env double mutanta
| Transfection with | No. of syncytiab | Relative syncytium formationc |
|---|---|---|
| None | 0 | 0 |
| 92BRd | 968 | 100 |
| 92BR + 427, TC = 1:1 | 723 | 74.7 |
| 92BR + 427, TC = 1:2 | 431 | 44.5 |
| 92USd | 1,225 | 100 |
| 92US + 427, TC = 1:1 | 848 | 69.2 |
| 92US + 427, TC = 1:2 | 304 | 24.8 |
P4-R5 MAGI cells grown in 60-mm-diameter petri dishes were cotransfected with 1.5 μg of pIIIextat and 1.5 μg of pSVIII-92BR or pSVIII-92US in the presence or absence of pSVE7(427,TC). The wt/427,TC env plasmid DNA ratio was set at 1:1 or 1:2 as indicated. Two days after transfection, syncytia were scored microscopically. This experiment was performed twice, with similar results.
More than five nuclei found in a giant cell were scored as a syncytium.
Expressed as a percent of the number of syncytia found in wt and mutant cotransfection to that found in the wt transfection alone.
92BR and 92US are two pSVIII-based env expression plasmids that encode the Env proteins of two primary viruses isolated from patients in Brazil and the United States, respectively.
Establishment and characterization of env-stably transfected HeLa-CD4-LTR-β-gal clones.
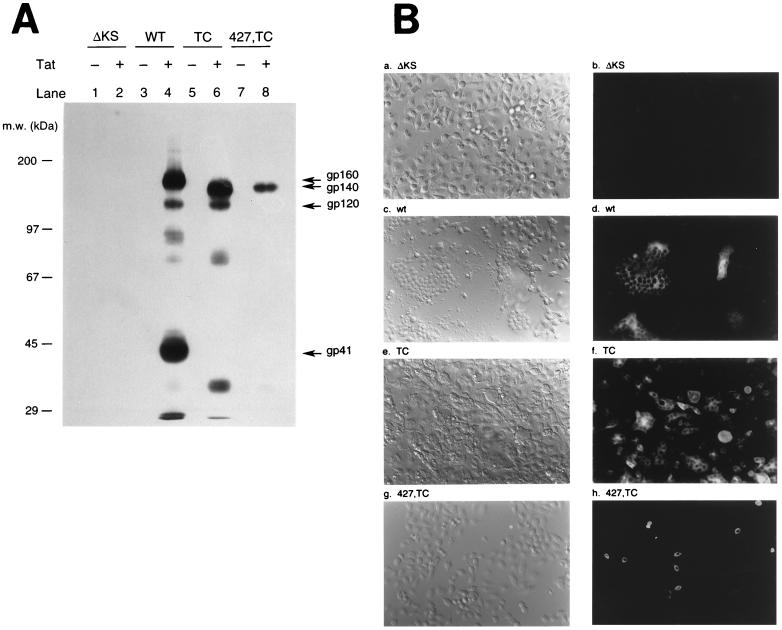
To compare interference by TC and 427,TC Env mutants in env CD4+ cells, env-stably transfected HeLa-CD4-LTR-β-gal clones that harbored silent copies of the control ΔKS, wt, or mutant pSVE7-puro env gene were generated. Approximately one copy of the control ΔKS env gene and two to four copies of the wt or mutant env gene were integrated into host chromosomal DNA, as shown by Southern hybridization by using a 32P-labeled 2.7-kb XhoI fragment that contained the entire env region linked to HIV-1 LTR derived from pSVE7(ΔKS)-puro (Fig. 1B) as a probe (data not shown). The selected env transfectants were transfected with or without pIIIextat, followed by Western blotting with mouse monoclonal antibodies (MAbs) Chessie 13 and Chessie 8 to detect Env expression. These two hybridoma map to amino acid residues 252 to 273 and 727 to 732 of the Env of HIV-1LAI, respectively (1). Tat induction in the wt env transfectant produced gp160, gp120, and gp41 (Fig. 5A, lane 4). A truncated gp160 precursor, designated gp140, and gp120 were detected in the TC transfectant upon Tat induction (Fig. 5A, lane 6). A truncated gp160 precursor was also detected in the 427,TC transfectant when Tat was expressed (Fig. 5A, lane 8), although the level of Env produced by the 427,TC transfectant was less than those produced by the wt and TC transfectants. No Env proteins could be detected in these env transfectants in the absence of Tat induction (Fig. 5A, odd-numbered lanes). Because the ΔKS transfectant harbored a defective env gene construct, this transfectant was then used as a negative control for later interference studies.
FIG. 5.
Characterization of env stable transfectants. (A) Tat-dependent HIV-1 env gene expression in stable env transfectants. The four env transfectants were transfected with or without 5 μg of pIIIextat. Equal amount of cell lysates from each transfection were subjected to an SDS–7.5% PAGE, followed by Western blotting by using MAbs Chessie 8 and Chessie 13. m.w., molecular size. (B) Analysis of Tat-induced Env expression in env transfectants by indirect immunofluorescence microscopy. The four env transfectants as indicated were transfected with pIIIextat. Three days after transfection, cells were processed for immunofluorescence analysis as described in Materials and Methods. Cells were analyzed under a differential interference contrast microscope (panels a, c, e, and g) or fluorescence microscope (panels b, d, f, and h), both at a magnification of 200×.
To determine whether mutant Env expression in these env transfectants might cause cytopathic effects and syncytium formation, the four env transfectants were transfected with pIIIextat. Three days after transfection, cells were fixed, permeabilized, and incubated with MAbs Chessie 13 and Chessie 8, followed by incubation with FITC-conjugated anti-mouse IgG. As shown in Fig. 5B, neither cytopathic effects nor immunofluorescence signals were observed in the control ΔKS transfectant (panels a and b). Tat expression in the wt env transfectant resulted in Env expression, as shown by the perinuclear localization of immunofluorescence signals (panel d). It was also noted that expression of the wt Env caused cell-to-cell membrane fusion (panel d) and an extensive cytopathic effect (panel c). An endogenous TC mutant env gene was also induced to express upon Tat expression (panel f), which was accompanied by extensive cytopathic effects (panel e). Nevertheless, syncytia smaller than those found in the wt transfectant were observed in the TC transfectant. Unlike wt and TC transfectants, induced expression of the 427,TC mutant Env did not show syncytia or cell-to-cell fusion, since immunofluorescence signals were found to be associated with individual intact cells (panel h).
Interference with exogenous wt Env-mediated cytopathic effects in mutant env HeLa-CD4-LTR-β-gal transfectants.
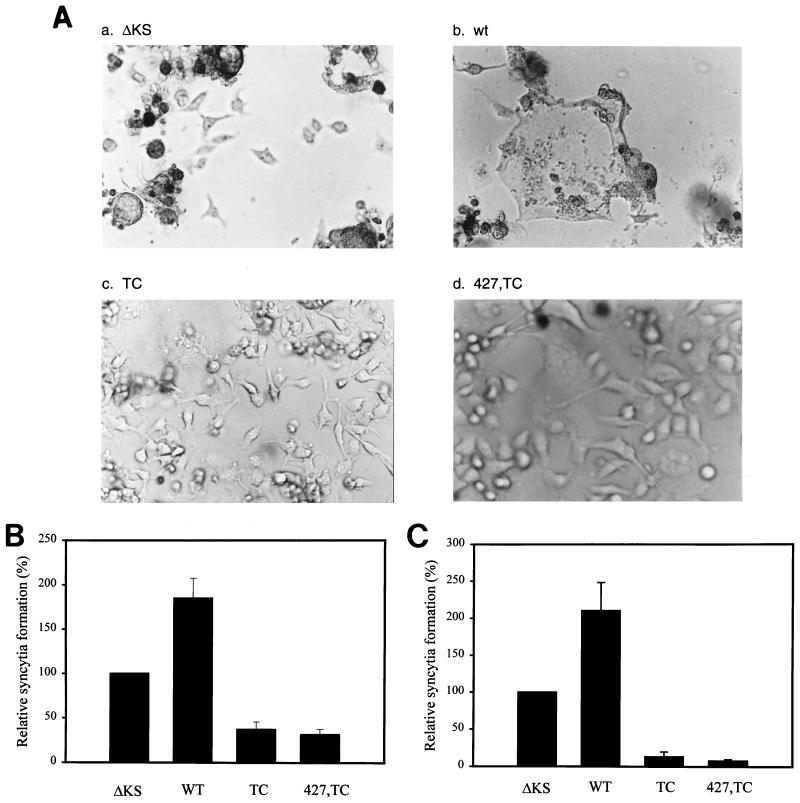
To examine whether expression of endogenous TC or 427,TC mutant Env could interfere with wt Env-mediated cytopathic effects, the four env transfectants were cotransfected with pIIIextat and wt pSVE7-puro. Cotransfection of pIIIextat with an exogenous wt env plasmid in the control ΔKS and wt env transfectants resulted in extensive cytopathic effects, including floating and dying cells, cell lysis, and syncytium formation (Fig. 6A, panels a and b, respectively). In any of the 10 fields microscopically examined, less than 10% of cells in these two cultures attached to the petri dishes. Due to extensive cytopathic effects, precise scoring for syncytia in these two transfectants was not possible. In contrast, induced expression of endogenous TC or 427,TC mutant proteins strikingly inhibited the cytopathic effect caused by the exogenous wt Env expression (Fig. 6A, panels c and d, respectively). In any of the 10 fields examined, more than 90% of cells in these two cultures remained intact, and no syncytia were visible. When the four env transfectants were transfected with a cytomegalovirus promoter-driven LAV env expression plasmid pCDNA3env(LAV), which did not encode a functional Tat protein, 950, 980, 1,050, and 940 syncytia were observed in the control ΔKS, wt, TC, and 427,TC transfectants, respectively. Taken together, these observations indicate that the lack of significant cytopathic effects and syncytia in mutant transfectants is attributable to the specific interference by the induced expression of the cytoplasmic domain truncation mutants.
FIG. 6.
Interference with wt Env-mediated viral transmission in mutant env transfectants. (A) Inhibition with exogenous Env-mediated cytopathicity. The four env transfectants as indicated were cotransfected with 10 μg of wt pSVE7-puro and 5 μg of pIIIextat. Three days posttransfection cultures were taken for photographs under a differential interference contrast microscope with a magnification of ×200. This experiment was performed three times, with similar results. Ten fields of each transfected culture were observed, all with similar results. A representative micrograph from each transfectant is shown. (B) Interference with homologous HXB2-mediated cell-to-cell-mediated transmission. The four env transfectants grown in 60-mm-diameter petri dishes with grids were cocultured with HXB2-transfected SupT1 cells. Three days after transfection, cultures were scored for syncytium formation. The degree of syncytium formation in the control ΔKS was arbitrarily set at 100%. The relative syncytium formation observed in the wt and mutant transfectants was expressed as the percentage of the numbers of syncytia observed relative to that found in the control ΔKS transfectant. The results from four individual experiments were averaged, and means and standard deviations were calculated. (C) Interference with heterologous NL4-3-mediated virus-to-cell transmission. The env transfectants as indicated were infected with the NL4-3 virus, and 3 days after infection, syncytia were scored. The diagram represents the degree of syncytium formation obtained in four independent experiments (means ± standard deviations).
Interference with viral infection mediated by T-tropic viruses in mutant env transfectants.
To quantitate interference with viral transmission by Env mutants, the four env transfectants were cocultured with HXB2-transfected SupT1 cells and scored for syncytium formation. The number of syncytia in the TC and 427,TC transfectants was reduced to 37.2 and 31.5%, respectively, of that found in the control ΔKS transfectant (Fig. 6B). Coculture of the wt transfectant showed a higher degree of syncytium formation than did the control ΔKS coculture (Fig. 6B), indicating that a higher level of Env expression in the wt transfectant results in higher levels of syncytia than those found in the control ΔKS transfectant. To assess whether expression of endogenous HXB2-derived mutant proteins interfered with viral infection caused by a heterologous T-tropic HIV-1 virus, env transfectants were infected with the NL4-3 virus, and infected cultures were scored for syncytium formation. Syncytium formation in TC and 427,TC transfectants was reduced to 13.2 and 7.8%, respectively, of that found in the control ΔKS transfectant (Fig. 6C).
Delayed replication and spread of T-tropic viruses in 427,TC mutant env-transfected CEM-SS cells.
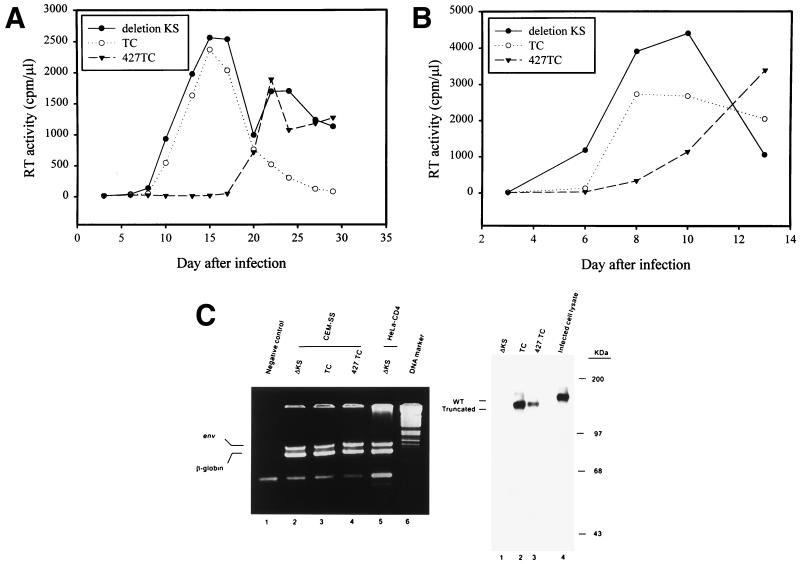
To further examine dominant interference by Env mutants, interference with viral replication in human CD4+ T cells was investigated. CEM-SS, a human T4 lymphoblastoid cell line, was transfected with the control ΔKS, TC, or 427,TC pSVE7-puro by electroporation and then grown in media containing puromycin for 3 weeks. The resultant cell populations were challenged with the HXB2 virus. Infection of CEM-SS transfected with the control ΔKS env construct showed a peak of RT production 15 days after infection (Fig. 7A). Replication of the HXB2 virus in TC env-transfected CEM-SS showed kinetics similar to those observed in the ΔKS env-transfected cells. In contrast, infection of 427,TC env-transfected cells showed a delay in viral replication with a peak of RT production occurring 22 days after infection. When the env-transfected CEM-SS cells were challenged with the NL4-3 virus, there was also a delay in viral replication in 427,TC env-transfected cells compared to viral replication in ΔKS env-transfected CEM-SS cells (Fig. 7B). Also, the delay in NL4-3 replication in TC env-transfected CEM-SS was less significant than that observed in 427,TC env-transfected cells (Fig. 7B).
FIG. 7.
Effect of Env truncation mutants on T-tropic viral replication in human CD4+ T cells. (A) Interference with viral replication and spread caused by the homologous HXB2 virus. The puromycin-selected CEM-SS cells transfected with various pSVE7-puro env constructs as indicated were infected with the HXB2 virus, and RT activity was monitored after infection. (B) Interference with NL4-3 virus-mediated viral replication and spread. CEM-SS cells transfected with env constructs as indicated in panel A were infected with the NL4-3 virus, and RT activity was monitored after infection. In panels A and B, similar results were obtained from at least three infection analyses. A representative result from each is shown. (C) Characterization of env-transfected CEM-SS cells. Left panel shows the presence of the env genes in transfected CEM-SS cells. Lysates obtained from equal volumes of env-transfected CEM-SS cells were analyzed by PCR by using env and β-globin primers as described in Materials and Methods. PCR products of each transfectant were mixed and resolved by 3% Nusieve agarose electrophoresis. The right panel shows Tat-induced Env expression in env-transfected CEM-SS cells. We transfected 107 CEM-SS cells harboring the env genes as indicated with 10 μg of pIIIextat. Three days after transfection, cell lysates were prepared and analyzed by Western blotting by using Chessie 13 and 902 MAbs. To indicate the migration position of the wt Env precursor, HIV-1-infected CEM-SS cell lysate was also analyzed in parallel (lane 4).
To determine whether these env genes were integrated into host chromosomal DNA, cell lysates obtained from the ΔKS, TC, and 427,TC env-transfected CEM-SS were analyzed by PCR by using env primers to detect the presence of env sequence from nucleotides positioned 7795 to 7936. PCR designed to amplify the human β-globin gene from nucleotides 14 to 123, relative to the translation initiation site, was also performed. The env PCR signals were detected in CEM-SS cells transfected with either the control ΔKS, TC, or 427,TC env construct (Fig. 7C, left panel). To determine whether mutant Env was expressed upon Tat induction, the control ΔKS, TC, or 427,TC env-transfected CEM-SS cells were transfected with pIIIextat by the DEAE-dextran method. Cell lysates were analyzed by Western blotting with MAbs 902 and Chessie 13. MAb 902 is specific for the gp120 V3 region of the LAV and IIIB strains of HIV-1. Truncated Env precursor was produced in TC and 427,TC env-transfected CEM-SS cells upon Tat induction (Fig. 7C, right panel, lanes 2 and 3, respectively).
Interference with replication of M-tropic isolates in 427,TC env-transfected PM1 cells.
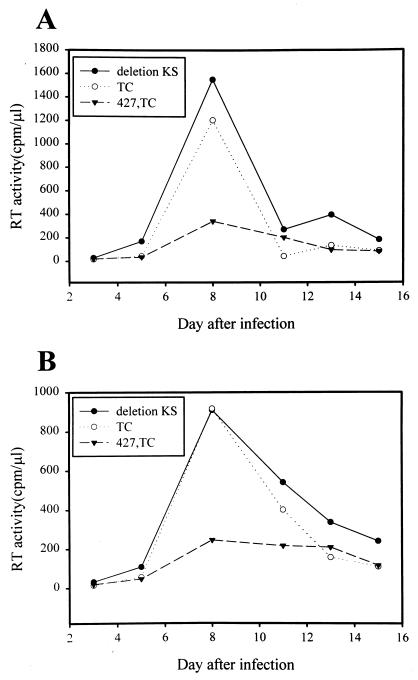
To determine whether 427,TC conferred dominant interference with infection caused by M-tropic viruses, a CD4+ T-cell line PM1 (32), a Hut 78 derivative that expresses both CXCR4 and CCR5 coreceptors, was transfected with the control ΔKS, TC, or 427,TC env construct. The puromycin-selected cells were then infected with ConB virus (43), which was produced from COS-1 cells transfected with the proviral DNA clone. The ConB virus, containing the consensus V3 sequences of HIV-1 subtype B in the backbone of the molecular clone HXB2RU3 (42), utilizes CCR5, but not CXCR4, as an entry coreceptor (43). PM1 cells harboring the 427,TC env gene showed a lower level of viral production than that observed in cells harboring the control ΔKS or TC env gene (Fig. 8A). Also, the env-transfected PM1 cells were infected with another M-tropic ADA-GG virus (43a). ADA-GG is an HXB2RU3-derived primary virus in which the sequence between the two BglI sites that encode residues 273 to 476 of gp120 was replaced by the homologous sequence from the M-tropic primary isolate ADA (44). Viral production was inhibited in 427,TC env-transfected PM1 cells as opposed to viral production observed in the control ΔKS or TC env-transfected cells (Fig. 8B).
FIG. 8.
Interference with M-tropic viral replication in mutant env-transfected PM1 cells. PM1 cells were transfected with the pSVE7-puro env constructs as indicated by electroporation. The puromycin-selected transfected cells were infected with the ConB (A) or ADA-GG (B) virus, and RT activity was monitored after infection. Each experiment was performed three times, with similar results. A representative result for each is shown.
DISCUSSION
In the present study, the feasibility of utilizing the gp41 cytoplasmic domain as a target for the design of a genetic anti-HIV approach was investigated. An HXB2 env-isogenic TC mutant lacking the whole cytoplasmic domain and the last two amino acids in the TM region as well as a series of mutants with deletions from the C terminus of the cytoplasmic domain dominantly interfere with wt Env-mediated viral infectivity (Fig. 2B and 3B). It was reported that deletions in the cytoplasmic domain affect Env stability in a cell type-dependent manner (24). Nevertheless, the incorporation of mutant Env proteins into virions appears to be normal (24). In contrast, other studies showed that truncations in the cytoplasmic domain, such as mutants TM775 and TM795, may impair Env incorporation into virions (16, 46). Viruses carrying TC, TM844, and TM813 mutations are severely impaired in infectivity, though assembly and release and Env incorporation of these viruses are not significantly altered (9, 46). The mechanism for the dominant interference by TC, TM844, and TM813 mutants may be via the ability of these mutants to trans-dominantly inhibit wt Env-mediated viral entry.
Unlike the CD4 binding-defective 41.2 double mutant (5), the syncytium-defective 427,TC mutant (Fig. 1C and 5B) dominantly interferes with viral transmission as effectively as the TC mutant (Fig. 2B and 6) and also confers interference with heterologous T-tropic Env-mediated viral infectivity (Fig. 4A). These observations indicate that interference by the TC mutant does not require a functional CD4-binding domain. In addition, the 427,TC mutant also dominantly interferes with syncytium formation and viral infectivity mediated by the Env proteins derived from two primary isolates 92BR and 92US, although less interference was observed in the syncytium assay than in the one-round viral replication assay (compare Table 1 to Fig. 4B). In the syncytium formation assay, cell-to-cell transmission is predominantly encountered, whereas in the env trans-complementation assay virus-to-cell transmission is encountered. The differential interference in these two assays may be attributable to the possibility that cell-cell and virus-cell fusion have different requirements for the Env structures and/or for the numbers of successful Env-CD4 interactions (6, 26). In env trans-complementation assays (Fig. 2B, 3B, and 4), recombinant virus produced from cotransfection of an env-defective reporter provirus with pSVE7(ΔKS) was used as a negative control. Also, pSVE7(ΔKS) was added into transfection mixtures to keep the total amount of env plasmids in all transfections the same. Therefore, the interference in viral infectivity observed in wt and mutant Env coexpression is attributed to the specific effect of the mutant Env, not to other nonspecific effects such as promoter competition.
For interference with exogenous wt Env-mediated cytopathic effects in HeLa-CD4-LTR-β-gal transfectants (Fig. 6A), two clones each of the TC and 427,TC transfectants were initially found to effectively inhibit exogenous wt Env-mediated cytopathicity. A clone selected at random from each mutant was further assessed for interference. Although the 427,TC transfectant produces a lower level of Env upon Tat induction than other transfectants (Fig. 5A), this double mutant exhibits an interfering effect as effective as the TC mutant in HeLa-CD4-LTR-β-gal transfectant analysis (Fig. 6). The reduced level of Env production in the 427,TC transfectant is not due to the reduced transfection efficiency of this particular clone as indicated by the similarity of the luciferase activity levels detected in all env transfectants when these transfectants were transfected with an env-defective, replication-incompetent pNL4-3-luc-R−E− provirus (12) (data not shown). The decreased Env production appears to be a general feature of transfectants harboring the 427,TC mutant env, since three other 427,TC transfectants also show the same phenotype (data not shown). Interference by TC and 427,TC transfectants is apparently not due to decreased cell surface CD4 expression, since all of the env transfectants show similar cell surface CD4 levels (data not shown). Because both TC and 427,TC, but not the wt transfectant, exhibit interference upon viral infection (Fig. 6), these studies indicate that the interference examined in the mutant transfectants is specific for a cytoplasmic domain truncation.
Since the dominant mutant env genes are not yet induced to express in the HeLa-CD4-LTR-β-gal stable clones or in transfected human CD4+ T cells during the first round of viral infection, no interference in viral entry by Env mutants could be anticipated in the initial infection. In fact, when the four HeLa-CD4-LTR-β-gal env transfectants were challenged with a cat-encoding, env-defective virus pseudotyped with the wt Env, similar levels of CAT activity were observed (data not shown). This observation is consistent with the finding that these env transfectants display similar levels of CD4 on the cell surface. Moreover, when the control ΔKS, TC, and 427,TC env-transfected CEM-SS were assayed for one-round viral replication, they also showed comparable levels of CAT activity (data not shown). These studies indicate that these mutant CD4+ env transfectants support single-cycle viral replication. The observations that the virus replicates efficiently in CEM-SS cells transfected with the TC mutant env gene and ultimately replicates productively in CEM-SS cells transfected with the 427,TC mutant env gene (Fig. 7A and 7B) also support this notion. The differential syncytium formation and viral replication among env transfectants (Fig. 6B and C and Fig. 7 and 8) may reflect an overall effect by Env mutants on subsequent viral infections. Tat produced by infected cells can be secreted and endocytosed by surrounding uninfected cells (20). Tat-induced expression of mutant Env proteins in initially infected or surrounding cells thus activates a defensive mechanism by forming a wt and mutant dysfunctional hetero-oligomer upon wt Env coexpression, which limits viral spread. Buchschacher et al. demonstrated that the 41.2 mutant-transfected HeLa-CD4 clone interferes with syncytium formation within 4 days following HIV infection or transfection with an HIV provirus (4). Their results show the feasibility of utilizing the HeLa-CD4 stable clone approach to address interference with viral spread by Env mutants. Moreover, the kinetic analysis of viral replication in human CD4+ T cells (Fig. 7 and 8) demonstrates the specific interference of the 427,TC mutant in multiple viral replication cycles in targeted CD4+ T cells. Since env-transfected CEM-SS and PM1 cells were selected for the puromycin resistance gene inserted in the env expression cassettes (Fig. 7 and 8), the selected cell populations represent a pool of env clones. Therefore, the interference observed in CD4+ T cells reflects the combinatory effects of individual clones, which avoids the problem of clonal heterogeneity due to differential Env expression in different clones.
The lack of effective interference by TC mutant in human CD4+ T cells (Fig. 7 and 8) cannot be attributed to the lack of TC env integration into host DNA or to the lack of TC expression upon Tat induction (Fig. 7C). An env-defective virus pseudotyped with certain cytoplasmic domain deletion mutants produced by COS-1 cells and then used to infect Jurkat cells may replicate differentially from virus whose replication is examined by using a similar transient complementation assay in transfected Jurkat cell cultures (24). Also, in Jurkat cells, some of the deletion mutants have a syncytium-forming ability equal to or greater than that of the wt Env. However, these deletion mutants may have lower syncytium-forming ability than the wt Env when they are produced in COS-1 (24). It is likely that in HeLa-CD4-LTR-β-gal transfectant, the wt-mutant TC hetero-oligomers are restricted to their mediation of viral transmission, resulting in interference with viral transmission. In human CD4+ T cells, such hetero-oligomers may still be able to mediate viral replication, resulting in less interference. The more pronounced interference effect by the 427,TC mutant compared to that of the TC mutant in human CD4+ T cells may be attributed to the absolute defective syncytium-forming nature of the 427,TC mutant.
Viral replication in CEM-SS or PM1 cells harboring the 427,TC mutant env gene is retarded but not completely blocked (Fig. 7 and 8). Early in infection, Tat-induced mutant 427,TC expression appears to be sufficient to neutralize the wt Env synthesized de novo by forming a dysfunctional hetero-oligomer. As the infection process continues de novo wt Env synthesis may accumulate to a threshold level. Beyond this level, the wt Env synthesized can no longer be neutralized by the 427,TC Env, thus resulting in a burst of virus production. Alternatively, upon infection of a cell that also expresses the 427,TC mutant, a fraction of wt homo-oligomer is present in the total Env population. The presence of a residual fraction of virus assembled with the wt homo-oligomer may ultimately result in overt productive infection.
The use of env transfectants to address Env-targeted dominant interference differs from those transient coexpression analyses. The transient coexpression analysis examines the ability of wt and mutant coexpressed proteins to mediate viral entry into CD4+ cells. In env transfectant analyses, as presented here, the mutant env genes are first directed to the CD4+ cells, and only upon viral infection is the expression of these mutant genes induced. This approach directly assesses the antiviral effect of trans-dominant env mutant genes in targeted CD4+ cells. In the present study, we demonstrate that the HXB2 env-derived 427,TC double mutant exhibits dominant interference with the Env functions of T-tropic and M-tropic viruses. Moreover, human CD4+ T cells harboring the 427,TC env gene slow T-tropic and M-tropic viral replication and spread. Thus, this design, by itself or in combination with other approaches, may have a role in the development of genetic anti-HIV strategies.
ACKNOWLEDGMENTS
The following cells, antibodies, and plasmids were obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HeLa-CD4-LTR-β-gal (from Michael Emerman), SupT1 (from James Hoxie), CEM-SS (from Peter L. Nara), PM1 (from Marvin Reitz, Jr.), hybridoma Chessie 8 and Chessie 13 (from George K. Lewis) and 902 (from Bruce Chesebro), P4-R5 MAGI (from Nathaniel Landau), goat anti-gp120 (from Michael Phelan), pNL4-3 (from Malcom Martin), and pSVIII-92RW020-5, pSVIII-92BR20.4, and pSVIII-92US715.6 (from Beatrice Hahn). pNL4-3-luc-R−E− was a gift from Nathaniel Landau (New York University Medical School, New York, N.Y.). pSVE7-ELI was obtained from Ernest F. Terwilliger (Harvard Medical School, Boston, Mass.). The ConB and ADA-GG proviral clones and HXB2R3-derived proviruses TM844, TM812, TM795, TM775, TM752, and TM709 were gifts from Ton-Hou Lee (Harvard School of Public Health, Boston, Mass.).
This work was supported by grants from the National Science Council (NSC 86-2314-B-001-017, NSC 87-2314-B-001-035, and NSC 88-2314-B-001-027) and from the Institute of Biomedical Sciences at Academia Sinica, Taipei, Taiwan, Republic of China. We are indebted to Huey-Jong Hao for technical assistance with DNA cloning and sequencing.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claasen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 4.Buchschacher G L, Jr, Freed E O, Panganiban A T. Cells induced to express a human immunodeficiency virus type 1 envelope gene mutant inhibit the spread of wild-type virus. Hum Gene Ther. 1992;3:391–397. doi: 10.1089/hum.1992.3.4-391. [DOI] [PubMed] [Google Scholar]
- 5.Buchschacher G L, Jr, Freed E O, Panganiban A T. Effects of second-site mutation on dominant interference by a human immunodeficiency virus type 1 envelope glycoprotein mutant. J Virol. 1995;69:1344–1348. doi: 10.1128/jvi.69.2.1344-1348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel P. HIV-1 reverse transcription: a termination step of the center of the genome. J Biol Chem. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 8.Chen S S-L. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1994;68:2002–2010. doi: 10.1128/jvi.68.3.2002-2010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S S-L, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 10.Chen S S-L, Lee C-N, Lee W-R, McIntosh K, Lee T-H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S S-L, Lee S-F, Hao H-J, Chuang C-K. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J Virol. 1998;72:4765–4774. doi: 10.1128/jvi.72.6.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conor R L, Chen B K, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 13.Cordonnier A, Montanier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature (London) 1989;340:571–574. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- 14.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 15.Dorfman T, Mannano F, Haseltine W A, Göttlinger H G. Role of matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubay J W, Robers S, Haln B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6016–6025. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earl P L, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher A G, Collalti E, Ratner L, Gallo R C, Wong-Staal F. A molecular clone of HTLV-III with biological activity. Nature (London) 1985;316:262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- 19.Fisher A G, Ratner L, Mitsuya H, Marselle L M, Harper M E, Broders S, Gallo R C, Wong-Staal F. Infectious mutants of HTLV-III with changes in the 3′ region and markedly reduced cytopathic effects. Science. 1986;233:655–659. doi: 10.1126/science.3014663. [DOI] [PubMed] [Google Scholar]
- 20.Frankel A D, Pabo C D. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 21.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 29.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S-J, Hu W, Fisher A G, Looney D J, Kao V F, Mitsuya H, Ratner L, Wong-Staal F. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res Hum Retroviruses. 1989;5:441–449. doi: 10.1089/aid.1989.5.441. [DOI] [PubMed] [Google Scholar]
- 31.Lodge R, Lalonde J-P, Lemay G, Cohen E A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lusso P, Cocchi F, Balotta C, Markham P D, Louie A, Farci P, Pai R, Gallo R C, Reitz M S., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannano F, Kondo E, Sodroski J, Bukovsky A, Göttlinger H G. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owens R J, Burke C, Rose J K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 36.Ratner L, Fisher M, Jagodzinski L L, Mitsuya H, Liou R-S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 37.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 38.Srinivas S K, Srinivas R V, Anantharamaiah G M, Compans R W, Segrest J P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 39.Steffy K R, Wong-Staal F. Transdominant inhibition of wild-type human immunodeficiency virus type 2 replication by an envelope deletion mutant. J Virol. 1993;67:1854–1859. doi: 10.1128/jvi.67.4.1854-1859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tencza S B, Miller M A, Islam K, Mietzner T A, Montelaro R C. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J Virol. 1995;69:5199–5202. doi: 10.1128/jvi.69.8.5199-5202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger E, Sodroski J G, Rosen C A, Haseltine W A. Effects of mutations within the 3′ orf open reading frame region of human T-cell lymphotropic virus type III (HTLV-III/LAV) on replication and cytopathogenicity. J Virol. 1986;60:754–760. doi: 10.1128/jvi.60.2.754-760.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trujillo J R, Wang W K, Lee T-H, Essex M. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology. 1996;217:613–617. doi: 10.1006/viro.1996.0158. [DOI] [PubMed] [Google Scholar]
- 43.Wang W-K, Duder T, Zhao Y-J, Brumblay H G, Essex M, Lee T-H. CCR5 coreceptor utilization involves a highly conserved arginine residue of HIV-1 type 1 gp120. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Wang, W.-K., and T.-H. Lee. Unpublished results.
- 44.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of the Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]