Abstract
Objective
Hepatocellular carcinoma (HCC) poses a significant clinical challenge because the long-term benefits of immune checkpoint blockade therapy are limited. A comprehensive understanding of the mechanisms underlying immunotherapy resistance in HCC is imperative for improving patient prognosis.
Design
In this study, to systematically investigate the characteristics of cancer-associated fibroblast (CAF) subsets and the dynamic communication among the tumor microenvironment (TME) components regulated by CAF subsets, we generated an HCC atlas by compiling single-cell RNA sequencing (scRNA-seq) datasets on 220 samples from six datasets. We combined spatial transcriptomics with scRNA-seq and multiplexed immunofluorescence to identify the specific CAF subsets in the TME that determine the efficacy of immunotherapy in HCC patients.
Results
Our findings highlight the pivotal role of POSTN+ CAFs as potent immune response barriers at specific tumor locations, as they hinder effective T-cell infiltration and decrease the efficacy of immunotherapy. Additionally, we elucidated the interplay between POSTN+ CAFs and SPP1+ macrophages, whereby the former recruits the latter and triggers increased SPP1 expression via the IL-6/STAT3 signaling pathway. Moreover, we demonstrated a spatial correlation between POSTN+ CAFs and SPP1+ macrophages, revealing an immunosuppressive microenvironment that limits the immunotherapy response. Notably, we found that patients with elevated expression levels of both POSTN+ CAFs and SPP1+ macrophages achieved less therapeutic benefit in an immunotherapy cohort.
Conclusion
Our research elucidates light on the role of a particular subset of CAFs in immunotherapy resistance, emphasizing the potential benefits of targeting specific CAF subpopulations to improve clinical responses to immunotherapy.
Keywords: Tumor microenvironment - TME, Hepatocellular Carcinoma, Immunotherapy, Immunosuppression
WHAT IS ALREADY KNOWN ON THIS TOPIC.
WHAT THIS STUDY ADDS
Here we discovered that POSTN+ CAFs play a crucial role as potent immune response barriers at specific tumor sites, facilitating remodeling of the stromal microenvironment. This ultimately suppresses effector T-cell infiltration and decreases the efficacy of immunotherapy. Furthermore, we revealed that the interaction between POSTN+ CAFs and SPP1+ macrophages within the TME promotes the development of an immunosuppressive milieu that limits the immunotherapy response by activating the IL-6/STAT3 signaling pathway.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study revealed that POSTN+ CAFs were associated with adverse clinical outcomes, including poor overall survival and decreased response to immunotherapy in hepatocellular carcinoma patients. This finding underscores the significance of devising approaches aimed at targeting POSTN+ CAFs to improve the tumor response to immunotherapy.
Introduction
Hepatocellular carcinoma (HCC) is a prevalent and deadly malignancy that is diagnosed at an advanced stage and has a poor 5-year survival rate.1 2 Recurrence or metastasis still occurs within 5 years in the majority of patients even after surgical resection.3 The application of immune checkpoint blockade (ICB), such as PD-1/L1 inhibitors and CTLA4 inhibitors, has led to breakthroughs in the treatment of liver cancer patients.4,6 However, only a small proportion of patients can benefit from these treatments, indicating significant challenges in the field of immunotherapy for HCC.7 8 Cancer-associated fibroblasts (CAFs), which are critical components of the tumor microenvironment (TME), promote immune evasion, tumor metastasis, and therapy resistance by remodeling the extracellular matrix (ECM), secreting growth factor and cytokines.9 10 Genetic depletion or pharmacological targeting of distinct CAF populations leads to different outcomes,11 12 highlighting the CAFs are highly heterogeneous in types and functions affecting cancer progression and immunomodulation.
CAF populations are diverse and dynamic and originate from various cell sources, which contributes to their functional diversity.13 Understanding the heterogeneity and subpopulation characteristics of CAFs is essential for targeting protumorigenic CAFs or interference with their activity as potential strategies in anticancer therapy. Recent advancements in techniques such as single-cell sequencing and flow cytometry have enabled precise analysis of CAF subtypes and their biological functions in different cancer types.13,16 These studies provide evidence supporting the presence of two distinct functional populations of CAFs: procancer CAFs and anticancer CAFs. Procancer CAFs orchestrate ECM remodeling and tumor-promoting inflammation, and modulate the immune microenvironment toward immunosuppression,17 such as myofibroblastic CAFs and inflammatory CAFs (iCAFs). Conversely, anticancer CAFs are able to stimulate T-cell activation associated with MHC class II expressing to improve immunotherapeutic strategies, such as antigen-presenting CAFs (apCAFs).18 CAFs are highly interrelated with sensitivity to anticancer therapies. Neoadjuvant chemotherapy can remodel CAFs to enhance immune activation and inhibit tumor progression.11 However, CAFs can also remodel the ECM, making it denser and potentially forming a physical barrier that impedes the delivery of chemotherapy and immunotherapy drugs.19 Overall, various CAF subtypes may activate distinct molecular pathways to influence the TME and resistance to immunotherapy and chemotherapy.
However, the intricate crosstalk between CAFs, HCC cells, and immune cells, particularly within different CAF subtypes, remains unclear. A comprehensive understanding of the multidimensional interactions between CAFs and infiltrating immune cells in the TME would help us elucidate the mechanisms through which CAFs induce immunosuppression. Further exploration of these interactions may reveal potential molecular targets for CAF-targeted therapy, providing new strategies for subsequent immunotherapy.
We integrated single-cell RNA sequencing (scRNA-seq) datasets for HCC tumors. Our analysis led to the identification of six common subtypes of CAFs in HCC tumors: STMN1+ CAFs, CXCL12+ CAFs, MYH11+ CAFs, SEPT7+ CAFs, POSTN+ CAFs, and CD36+ CAFs. Among these subtypes, we specifically focused on the newly discovered POSTN+ CAF subpopulation. POSTN+ CAFs play a crucial role in promoting the progression of HCC through the activation of the ECM, hypoxia and transforming growth factor beta (TGF-β) signaling pathways. Furthermore, they contribute to the formation of an immune barrier that suppresses the infiltration of CD8+ T cells into the TME. Additionally, POSTN+ CAFs interact with SPP1+ macrophages via the IL-6/STAT3 axis. Our study revealed that the abundance of POSTN+ CAFs can serve as a reliable predictor of the response to immunotherapy in HCC patients. This indicates that targeting POSTN+ CAFs could enhance the effectiveness of immunotherapeutic approaches for treating HCC. In summary, our study elucidated the novel intercellular communication network between POSTN+ CAFs and SPP1+ macrophages and between POSTN+ CAFs and CD8+ T cells. These findings suggest that POSTN+ CAFs play a significant role in promoting the development of an immunosuppressive TME.
Material and methods
Human liver samples
Liver samples were obtained from patients who underwent hepatectomy or percutaneous liver biopsy. Clinical information is summarized in online supplemental table 1.
Cell culture
The human myeloid leukemia mononuclear cell line (THP-1) was obtained from the Cell Resource Center at the Chinese Academy of Medical Sciences and authenticated by the same. THP-1 cells were cultured in RPMI1640 supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Invitrogen, USA). To conduct differentiation assays, THP-1 cells were differentiated into macrophages through 24-hour culturing with 100 ng/mL phorbol myristate acetate (Sigma). CAFs transfected with si-CREB3L1 or si-NC for 48 hours were seeded in six-well plates and cultured under hypoxia or normoxia for 24 hours. siRNAs targeting CREB3L1 are listed in online supplemental table 2.
Plasmid transfections
The pcDNA3-based plasmid encoding human POSTN was acquired from Gentlegen (Suzhou, China). CAFs were seeded and allowed to grow in 24-well culture plates for 48 hours before transfection with the plasmids using Lipofectamine 3000 (Invitrogen).
Chemotaxis assay
Macrophages at a concentration of 4×104 cells per well were plated in the upper chamber with 0.1% gelatin-coated membranes, while CAFs were plated in the lower chamber at a concentration of 1×105 cells per well. Tocilizumab (200 µg/mL) was added to the upper chamber. After incubating for 24 hours at 37°C, the cells were washed with 1x PBS, fixed in 10% paraformaldehyde for 30 min, and stained with 0.05% crystal violet for another 30 min. Finally, the number of cells that had migrated to the bottom of the membrane was assessed by counting them in four different regions.
Protein extraction and western blot
The cells were lysed using RIPA solution (Beyotime Biotechnology, China) supplemented with protease and phosphatase inhibitors (Beyotime Biotechnology). Western blot analysis was conducted following previously described protocols.20 The protein concentration in cell lysates was determined using a BCA assay (Beyotime Biotechnology). Subsequently, the cell lysates were subjected to 10% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA) for antibody incubation.
Antibodies
Primary antibodies used in this study were as follows: rabbit anti-POSTN mAb (ab215199, Abcam); rabbit anti-CD8 mAb (245118, Abcam); rabbit anti-MYH11 mAb (ab133567, Abcam); rabbit anti-STMN1 mAb (ab52630, Abcam); rabbit anti-CD36 mAb (ab252923, Abcam); rabbit anti-α-SMA mAb (19 245S, Cell Signaling Technology); rabbit anti-STAT3 mAb (A1192, abclonal Technology); rabbit anti-IL-6 mAb (A1570, abclonal Technology); rabbit anti-SPP1 mAb (8242S, Cell Signaling Technology); rabbit anti-p-STAT3 mAb (9145S, Cell Signaling Technology); rabbit anti-Osteopontin mAb (A1499, abclonal Technology); rat anti-CD68 mAb (ab53444, Abcam); rabbit anti-Osteopontin mAb (ab214050, Abcam); rabbit anti-α-SMA mAb (19 245S, Cell Signaling Technology); rabbit anti-CREB3L1 mAb (11 235–2-AP, Proteintech); mouse anti-β-actin mAb (3700S, Cell Signaling Technology).
Secondary antibodies used in this study were as follows: HRP-conjugated goat anti-rabbit IgG (ab205718, Abcam); HRP-conjugated goat anti-rat IgG (ab205720, Abcam); Alexa Fluor 488-conjugated goat anti-rabbit IgG (A-11008, Invitrogen); Alexa Fluor 594-conjugated goat anti-mouse IgG (A-11005, Invitrogen).
Quantitative real-time PCR
Total RNA was extracted from the samples using TRIzol reagent (Invitrogen) and then precipitated with isopropyl alcohol. The concentration and quality of the RNA were assessed using a NanoDrop system. Complementary DNA was synthesized using a commercially available kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. Real-time quantitative PCR was performed using SYBR-green-based kits (Vazyme) to quantify mRNA levels. The relative expression of the target gene was normalized to β-actin in each sample. The following primer sequences were used: β-actin (forward, 5′-CTCGCCTTTGCCGATCC-3′; reverse, 5′-ATCCTTCTGACCCATGCCC-3′), POSTN (forward, 5′-CTCATAGTCGTATCAGGGGTCG-3′; reverse, 5′-ACACAGTCGTTTTCTGTCCAC-3′), IL-6 (forward, 5′-TGAGGAGACTTGCCTGGTGAA-3′; reverse, 5′-CAGCTCTGGCTTGTTCCTCAC-3′), CREB3L1 (forward, 5′-GGAGAATGCCAACAGGACC-3′; reverse, 5′-GCACCAGAACAAAGCACAAG-3′), COL1A1 (forward, 5′-CCCCTGGAAAGAATGGAGATG-3′; reverse, 5′-TCCAAACCACTGAAACCTCTG-3′), COL3A1 (forward, 5′-AGGAAATGATGGTGCTCCTG-3′; reverse, 5′-GTTCCCCAGGTTTTCCATTT-3′), COL5A1 (forward, 5′-TCGCTTACAGAGTCACCAAAG-3′; reverse, 5′-GTTGTAGATGGAGACCAGGAAG-3′).
Animal studies
All mice were housed in an SPF level animal facility with controlled temperature and humidity on a 12–12 hours light-dark cycle with food and water supplied ad libitum.
Adeno-associated virus (AAV)-mediated gene transfer for knockdown POSTN in vivo, the following AAV was used in this study: AAV8-control and AAV-shPOSTN (Shanghai Genechem). AAV8-shPOSTN was injected into 6 weeks old male C57BL/6 mice via the tail vein at a dose of 1×1099 infectious units (IFU) per 200 µL per mouse 2 weeks before orthotopic tumor transplantation. For orthotopic implantation, 3×106 Hepa1-6 cells were injected into the left lobes of the livers of C57BL/6 mice. Mice were treated with either 10 mg/kg anti‐mouse PD‐1 (BE0146, BioXCell) or isotype IgG control (BE0089, BioXCell) through intraperitoneal injection twice weekly starting on day 7. Mice were sacrificed on day 21. shRNAs targeting POSTN are listed in online supplemental table 2.
Tissue processing and fibroblast isolation
Primary HCC tissues were minced using a scalpel in a tissue culture dish. Subsequently, the minced tissues were enzymatically dissociated in 10 mL of PBS containing 0.1% collagenase I at 37°C for 1 hour with gentle agitation. The resulting suspension was neutralized with a complete medium and then centrifuged at 300×g for 5 min. The collected cells were plated in culture dishes and allowed to grow. After 48 hours, the non-adherent cells and tissue debris were removed by washing the dishes twice with PBS. The adherent fibroblasts were further incubated for 6–10 days until they reached 80%–90% confluence.
Flow cytometry
Briefly, the cells were resuspended in the staining buffer and incubated with the following antibodies on ice for 30 min: anti-CD45 (368521, Biolegend, USA), anti-EMCAM (324221, Biolegend), Live/Dead blue fluorescent dye (L34963, Thermo Scientific) and anti-CD29 (303023, Biolegend). Among live cells, EpCAM and CD45 were used among live cells to eliminate epithelial and immune cells, respectively, while CD29 was indicative of stromal cells. For intracellular staining, the cells were fixed with fixation buffer (Biolegend) on ice for 15 min and then washed twice with an Intracellular Staining Permeabilization Wash Buffer. Antibodies against POSTN (sc-398631, Santa Cruz) were added and incubated for 1 hour on ice.
Multiplex immunohistochemistry staining
Multiplex immunohistochemistry (mIHC) staining was employed to visualize the expression of α-SMA, STMN1, MYH11, CD36, POSTN, CD8, and CD68 in tumor tissues. Paraffin blocks were used to obtain three consecutive sections, each 3 µm thick, with one section designated for H&E staining. The remaining two FFPE tumor slides, also 3 µm thick, underwent a 3-hour dehydration process at 70°C. Subsequently, the paraffin sections were then de-paraffinized using xylene and rehydrated with alcohol. Heat-induced antigen retrieval was performed in EDTA buffer at pH 9.0 using a microwave oven. The sections were then blocked with a commercially available blocking buffer for 10 min. Following this, the slides were sequentially incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies, followed by tyramide signal amplification (TSA). After each TSA round, the slides underwent antigen retrieval and antibody stripping. Finally, nuclei were stained with DAPI. This comprehensive procedure facilitated the visualization of multiple antigens in the tumor tissues, thereby enabling the assessment of expression patterns for the targeted proteins.
Data collection
The public scRNA-seq datasets used in this research were obtained from various sources. Specifically, the datasets GSE149614,21 GSE151530,22 GSE156625,23 GSE189903,24 and GSE20264225 were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Additionally, dataset HRA00174826 was obtained from the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001748). For the analysis of normalized gene expression data and microarray datasets of HCC samples, publicly available data from The Cancer Genome Atlas (TCGA) data portal (http://gdac.broadinstitute.org/) and GEO were employed. The specific datasets accessed from GEO include GSE10143,27 GSE192912,28 GSE109211,29 GSE10186,30 and GSE54236.31 To acquire the expression matrix and clinical information of the TCGA-LIHC dataset and GSE14520,32 the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse14520) was accessed. Furthermore, expression profile and clinical information from the Fudan HBV-HCC cohort33 were collected and collated from the biosino NODE database (OEP000321,33 https://www.biosino.org/node/project/detail/OEP000321).
Spatial transcriptome sequencing data of 13 samples including HCC tumors, normal tissues, and leading tissues were acquired from the study conducted by Wu et al34 (http://lifeome.net/supp/livercancer-st/data.htm). Furthermore, spatial transcriptome data from tumor sections of eight HCC patients who underwent anti-PD-1 treatment (five non-responders and three responders) were downloaded from the study conducted by Liu et al35 (https://data.mendeley.com/datasets/skrx2fz79n/1).
Single-cell transcriptome analysis and differential expression analysis
CellRanger (V.6.0.2) was used for read mapping and gene expression quantification. Seurat package (V.4.1.1) was employed for downstream analysis. Cells with fewer than 1000 UMIs or greater than 15% mitochondria genes were excluded from the analysis. Doublets were assessed using the DoubletFinder (V.2.0.3) algorithm each sample. To address batch effects and prevent the analysis from being dominated by individual patient characteristics, the Harmony algorithm was applied to remove batch effects between samples in the single-cell data. To perform dimensionality reduction and identify cell subtypes, the top 4000 most variable genes were selected using the FindVariableFeatures function and the data were scaled accordingly. Principal component analysis (PCA) was conducted using these variable genes. Nearest neighbors for graph clustering based on the top 50 principal components were determined using the FindNeighbors function. The resulting clusters were obtained using the FindCluster function, and cell visualization was achieved using the uniform manifold approximation and projection (UMAP) algorithm. Furthermore, gene signatures specific to various cell types were scored in the identified clusters. These gene signatures included markers for B cells (CD79A and CD79B), endothelial cells (PECAM1 and VWF), hepatocytes (ALB and APOA2), fibroblasts (COL1A1 and COL1A2), myeloid cells (CSF3R, C1QB, CD1C, and CLEC9A), plasma cells (MZB1 and IGHG1), and T/NK cells (CD3D, CD3E, KLRD1, and KLRC1). Differential gene expression analysis between clusters was performed using the “FindAllMarkers” function. The parameters used for this analysis were min.pct=0.15, logfc.threshold=0.15, and only.pos=TRUE. The Wilcoxon’s rank-sum test with the Benjamini-Hochberg method was employed to obtain p values and adjusted p values for the comparisons.
Gene sets level analysis for scRNA-seq data
Single-cell signature scoring was conducted employing the RunAUCell function in pochi R package (V.0.1.0). Differential signature score enrichment between groups was determined using a two-sided Wilcoxon rank-sum test with Benjamini-Hochberg FDR correction. The gene sets used for this analysis were provided in online supplemental table 3.
Pathway and cytokine signaling signatures analysis
We performed pathway and cytokine signaling analyses on CAF subgroups using PROGENy and CytoSig, respectively. Scores were computed using the run_aucell function from decoupleR package (V.2.2.2) based on PROGENy network model and Cytosig python packages with default parameters. The top 1000 target genes of the progeny model were used, as recommended for single-cell data.
Function enrichment and gene set enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment and gene set enrichment analysis (GSEA) of differentially expressed gene sets were implemented in the clusterProfiler R package (V.4.7.1.2). Enriched pathways with adjusted p value below 0.05 were considered as significantly enriched by DEGs.
Trajectory analysis
To better understand the cellular differentiation among various subtypes of CAFs, we used the Monocle 2 algorithm V.2.24.1. Initially, we honed in on the top 100 differentially expressed genes distinguishing between the CAF subtypes. Employing the reduceDimension function with the DDRtree method and limiting it to a maximum of two components facilitated dimensionality reduction. Subsequently, we used the “differentialGeneTest” function to identify genes exhibiting significant differential expression correlated with pseudotime values. Additionally, we used branch expression analysis modeling (BEAM) to unravel gene expression patterns influenced by branch fate. For visualizing the expression dynamics of branch-specific genes along the differentiation trajectories, we employed the plot_genes_branched_pseudotime function, generating curves that depicted the Loess-smoothed expression profiles of genes along each trajectory.
Transcription factor regulon analysis
We used pySCENIC for the analysis of the regulatory network and regulon activity. Through pySCENIC analysis, we identified subgroup specific transcription factors (TFs) using the Wilcoxon rank-sum test. Additionally, we generated regulons-associated specific score (RSS) within each specific cell type by computing Jensen-Shannon divergence, facilitated by philentropy (V.0.6.0) package.
Cell–cell communications
To explore potential interactions among different cell types within the HCC TME, we conducted cell–cell communication analysis using the CellPhoneDB Python package (V.2.1.1). This approach relies on a publicly available repository of curated receptors and ligands along with their interactions. Enriched receptor-ligand pairs between two cell types were predicted based on the expression of a receptor by one cell type and the corresponding ligand by another. By default, we considered only ligands and receptors expressed in at least 10% of cells within a given cell type. Additionally, we selected significant pairs with a p value <0.05 and a mean value ≥1 for subsequent analyses.
To further identify the key mediators of two cell subgroups, we applied Nichenet package to infer the interaction between POSTN+ CAFs and SPP1+ macrophages (Mphs). For ligands and receptor interactions, genes which are expressed in larger than 10% cells of clusters were considered. Top 20 ligands and top 100 targets of differential expressed genes of “sender cells” and “receive cells,” were extracted for paired ligand-receptor activity analysis. We used the active_ligand_target_links function to compute the potential intensity of regulation between the ligand and target. Activity scores ranged from 0 to 1. The expression of differential expressed ligands and receptors were also shown in heatmap by calculating the average genes expression in indicated cell types and scaled across indicated subtypes.
Cell types deconvolution for RNA-seq datasets
To assess the function of cell types in larger compendiums of samples, we applied the online tool CIBERSORTx to generate a reference signature matrix from our single-cell RNA-seq dataset and estimate cell-type proportions from the public bulk RNA-seq and microarray datasets based constructed cell-type reference. For creating signature matrices, CIBERSORTx was run with quartile normalization was disabled for RNA-seq datasets and was enabled for microarray datasets, and all other parameters with their default settings. For the imputation of cell fractions, the quartile normalization was disabled for RNA-seq datasets and was enabled for microarray datasets, the permutation parameter was set to 500 times, and all other parameters were kept at their default settings. Spearman’s correlation analysis was performed to assess the relationship among the proportions of cell-type infiltration and was plotted by corrplot package (V.0.92).
Survival analysis
Survival analysis was performed by R package survival. HR was calculated by Cox proportional hazards model and 95% CI was reported, and Kaplan-Meier survival curve was modeled by the survfit function. The two-sided long-rank test was used to compare Kaplan-Meier survival curves. The comparison of the percentage of patients who respond to ICB treatment between different groups was determined by the χ2 test.
Spatial transcriptomics analysis
The gene-spot matrices generated after ST data processing from ST and Visium samples were analyzed with the Seurat package (V.4.1.1) in R. Spots were filtered for minimum detected gene count of 200 genes while genes with fewer than 10 read counts or expressed in fewer than three spots were removed. Normalization across spots was performed with the LogVMR function. Dimensionality reduction and clustering were performed with independent component analysis (PCA) with the first 30 PCs. To better exhibit spatial expression of features, the spots were enhanced using the “spatialEnhance” function of the BayesSpace package (V.1.6.0), the expression features were enhanced with the “enhanceFeatures” function. The signature score derived from the scRNA-seq dataset was added to the “metadata” of the ST dataset with the “AddModulScore” function with default parameters in Seurat. Spatial feature expression plots were generated with the “SpatialFeaturePlot” function in the Seurat package. We applied the SpaGene method to identify ten spatial gene expression patterns for each sample and assessed the similarity between patterns based on the Jaccard index. The result was shown using the ComplexHeatmap package (V.2.12.1).
Statistical analysis
Statistical analyses were performed as described in Seurat (V.4.1.1) in R (V.4.2.0) software. Spearman correlation was used to estimate correlations. Survival analysis was measured using the Kaplan-Meier method. Statistical significance was determined by the Kruskal-Wallis test and Wilcoxon’s rank-sum test with the Benjamini-Hochberg method for multiple comparison correction.
Results
Generation of an HCC single-cell transcriptomic atlas
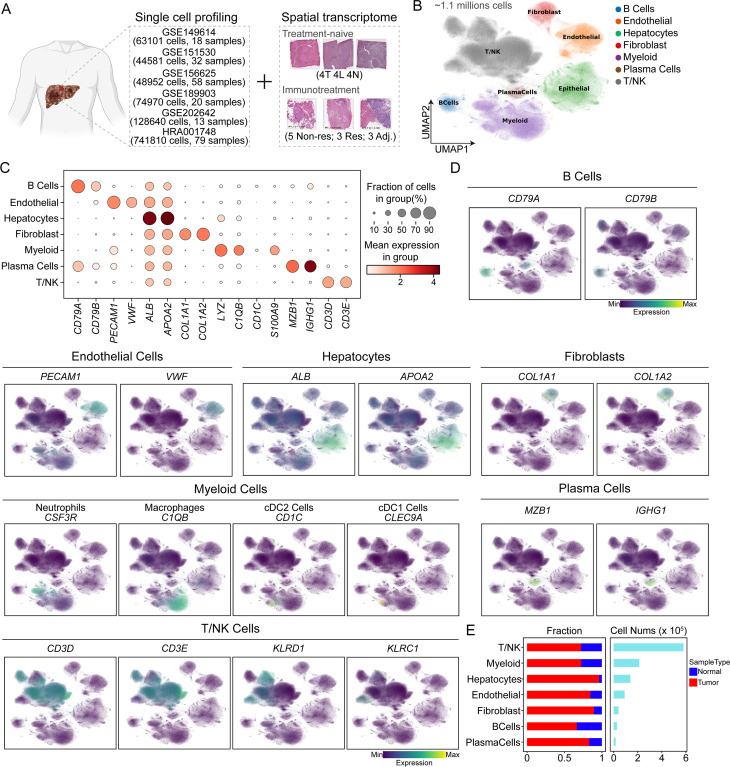
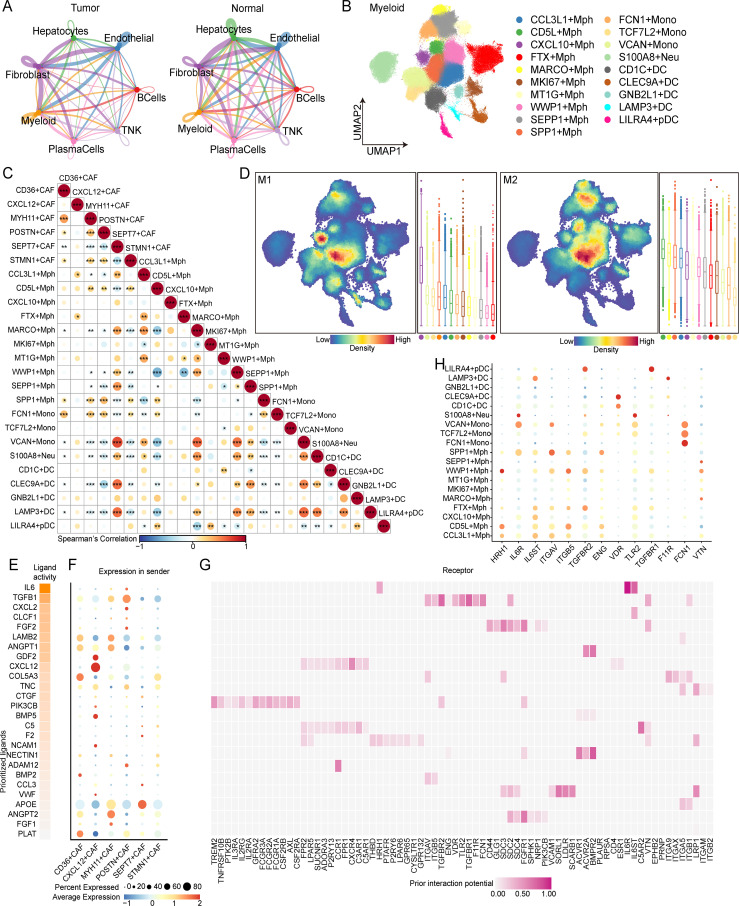
To systematically investigate the cellular composition of HCC, we compiled an HCC atlas by compiling scRNA-seq datasets from six datasets for a total of 220 samples (figure 1A and online supplemental figure S1B–C). This comprehensive scRNA-seq atlas of HCC incorporates quality-controlled and preanalyzed transcriptomic data from publicly available datasets on tumor samples and adjacent normal tissues. We employed the Harmony algorithm to correct for batch effects in scRNA-seq datasets from tumor and adjacent normal tissues. Overall, we retained nearly 1.1 million cell transcriptomes for subsequent analysis, and the cells were clustered into 32 distinct clusters representing seven major cell subtypes. These subtypes were identified based on classical single-cell marker expression. Specifically, we identified 25,946 B cells marked by CD79A and CD79B, 90,809 endothelial cells marked by PECAM1 and VWF, 137,809 hepatocytes marked by ALB and APOA2, 39,230 fibroblasts marked by COL1A1 and COL1A2, 212,308 myeloid cells marked by LYZ and C1QB, 15,986 plasma cells marked by MZB1 and IGHG1, and 579,944 T/NK cells marked by CD3D and CD3E (figure 1B–E and online supplemental figure S1A). Additionally, a subset of myeloid cells expressed CSF3R (neutrophil marker), CD1C (cDC2 marker), and CLEC9A (cDC1 marker), while a subset of T/NK cells expressed KLRD1 and KLRC1 (NK cell markers) (figure 1D). Although all seven major cell types were present in both tumor and adjacent normal tissues, their infiltration levels varied, possibly reflecting different stages of HCC progression. Remarkably, in addition to hepatocytes, a greater proportion of fibroblasts originated from tumor tissue to other liver cells (figure 1E), suggesting a potential role for CAFs in the progression of HCC.
Figure 1. Panoramic scRNA-seq analysis of liver samples of hepatocellular carcinoma patients. (A) Schematic representation of the data collection process. (B) Uniform manifold approximation and projection (UMAP) plot showing the seven major cell types. Dots represent individual cells, and colors represent different cell populations. (C) Dot plot showing the top two differentially expressed genes of major cell types. (D) UMAP plots showing the expression of classical molecular markers of corresponding cell types; the color represents the marker expression value. (E) Proportion of the sample types in seven major cell types shown in bar plots (left panel) and the total cell number of each cell type (right panel) are shown.
Identification and distinct features of six CAF subtypes in human HCC
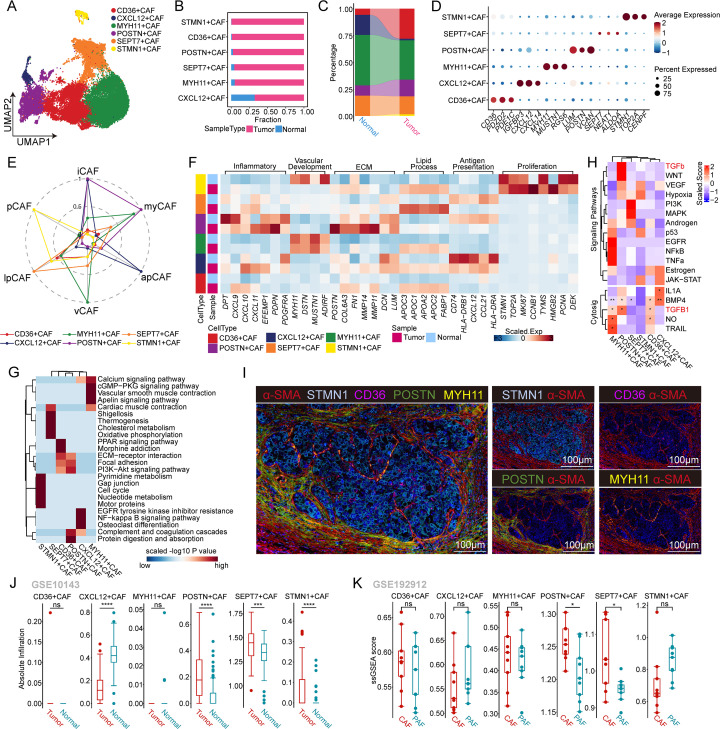
The heterogeneity and plasticity of CAFs play pivotal roles in tumor progression. In our study, we identified six subpopulations of CAFs based on their gene expression profiles: CD36+ CAFs, CXC12+ CAFs, MYH11+ CAFs, POSTN+ CAFs, SEPT7+ CAFs, and STMN1+ CAFs (figure 2A,D and online supplemental figure S2A). CD36+ CAFs, POSTN+ CAFs, and STMN1+ CAFs were significantly more abundant in tumors than in non-tumor tissues, while the opposite trend was observed for CXC12+ CAFs, MYH11+ CAFs, and SEPT7+ CAFs (figure 2B,C and online supplemental figure S2B). In addition to changes in cell abundance, scRNA-seq revealed distinct cellular characteristics of different CAF subtypes. According to the literature, we reclassified CAFs from HCC patients into iCAFs, myofibroblast-like CAFs (myCAFs), apCAFs, vascular CAFs (vCAFs), lipid process-associated CAFs (lpCAFs), and proliferative CAFs (pCAFs) based on their molecular features. POSTN+ CAFs exhibited characteristics of iCAFs and myCAFs, as they expressed inflammatory signature genes (CXCL9, CXC10, and CXCL11) and ECM-associated signature genes (POSTN, MMP14, and MMP11). Notably, POSTN+ CAFs in tumor tissues expressed higher levels of inflammatory and ECM signatures than those in normal tissues. MYH11+ CAFs were identified as vCAFs due to their high expression of vascular-related signatures (MYH11, MUSTN1, and DSTN). CXCL12+ CAFs were designated apCAFs and exhibited high levels of antigen presentation signature genes (CD74, CXCL12, and HLA-DRB1). Interestingly, CXCL12+ CAFs in tumor tissues showed lower levels of antigen presentation signature genes than those in normal tissues, suggesting a potential tumor-suppressive phenotype. CD36+ CAFs and SEPT7+ CAFs were categorized as lpCAFs due to their high expression of lipid metabolism-related genes (APOC3, APOC1, and FABP1). In tumor tissues, these CAF subtypes showed even higher levels of lipid process signature genes than did normal tissues. Finally, the STMN1+ CAFs were classified as pCAFs because they exhibited high expression of genes associated with cell proliferation (STMN1, TOP2A, and MKI67). This finding was consistent with the results of the cell cycle analysis (figure 2E,F and online supplemental figure S2B–D).
Figure 2. Heterogeneity and plasticity analysis of cancer-associated fibroblasts in hepatocellular carcinoma (HCC). (A) Uniform manifold approximation and projection plot showing the classification of cancer-associated fibroblasts (CAFs), with different colors representing different subtypes. (B) Proportion of the sample types in six CAF subtypes shown in bar plots, with different colors representing different sample types. (C) Sankey plot showing variation in the proportions of different CAF subtypes between the sample types, with different colors representing different CAF subtypes. (D) Dot plot showing the expression of the top three different expressed genes in the six CAF subtypes. The color depth represents the average expression value and the size of the point represents the percentage of the gene expression. The Wilcoxon rank-sum test was used to assess differences. (E) Radar plot showing the mean expression of six CAF subgroup signatures calculated by the AUCell algorithm in the reclassified CAF subtypes. (F) Heatmap showing the mean expression of associated gene signatures in each CAF subtype. (G) Kyoto Encyclopedia of Genes and Genomes terms of differentially expressed genes significantly enriched in each CAF subtype. The colors represent the scaled value of the −log10 p value. (H) Differential activation of (up) PROGENy pathways and (bottom) CytoSig cytokine signaling signatures in each CAF subtype. The heatmap colors indicate the deviation from the overall mean. The black dots indicate significance at different p value thresholds. Only cytokine signatures with a p value ≤0.05 in at least one group are shown. (I) Representative IF staining of human HCC tissue. The levels of α-SMA (red), STMN1 (silver), CD36 (purple), POSTN (green), and MYH11 (yellow) in individual and merged channels are shown. Bar, 100 µm. (J) Box plots showing the absolute infiltration of each CAF subtype in tumor and normal samples in GSE10143. (K) Box plots showing the ssGSEA score of each CAF subtype in paired CAFs (n=9) and para-cancer fibroblasts (PAFs, n=9) in GSE192912. apCAFs, antigen-presenting CAFs; ECM, extracellular matrix; iCAFs, inflammatory CAFs; lpCAFs, lipid process-associated CAFs; myCAFs, myofibroblast-like CAFs; pCAFs, proliferative CAFs; vCAFs, vascular CAFs.
To investigate the functionality of different CAF subpopulations, we conducted KEGG functional enrichment analysis on each subpopulation. Notably, KEGG analyses revealed significant enrichment of the PPAR signaling pathway and cholesterol metabolism pathway in CD36+ CAFs and SEPT7+ CAFs, consistent with the findings of the lpCAF signature. Moreover, pathways related to ECM-receptor interactions, focal adhesion, and PI3K-Akt signaling were found to be significantly enriched in POSTN+ CAFs. In line with the vCAF signature, MYH11+ CAFs were significantly enriched in pathways associated with vascular smooth muscle contraction, calcium signaling, and cardiac muscle contraction. CXCL12+ CAFs exhibited significant enrichment in pathways related to cytokine–cytokine receptor interactions, complement and coagulation cascades, and viral protein interaction with cytokines and cytokine receptors. Furthermore, STMN1+ CAFs, referred to as pCAFs, were found to be significantly enriched in the cell cycle and DNA replication pathways based on KEGG pathway enrichment analysis (figure 2G and online supplemental figure S2E).
Interestingly, the TGF-β pathway, which has been implicated in CAF-mediated cancer progression involving cell proliferation, invasion, and metastasis, was significantly upregulated in POSTN+ CAFs (figure 2H). Consistently, the CytoSig tool revealed significant upregulation of the TGF-β1 cytokine signature in POSTN+ CAFs, and genes in this signature are known to stimulate growth and ECM production in fibroblasts (figure 2H). This finding suggested that POSTN+ CAFs may play a crucial role in tumor progression and fibrosis through TGF-β signaling. To validate the presence of major CAF subsets in human HCC samples, we performed mIHC staining (figure 2I). Remarkably, POSTN+ CAFs were more highly enriched in the peripheral region of HCC tumors than in the core region (figure 2I and online supplemental figure S2H). This finding was further supported by the analysis of a liver tissue array dataset and fibroblast transcriptome sequencing data, which both confirmed the specific enrichment of POSTN+ CAFs in tumor tissues (figure 2J,K).
Cell-state transition trajectory of different CAF subpopulations
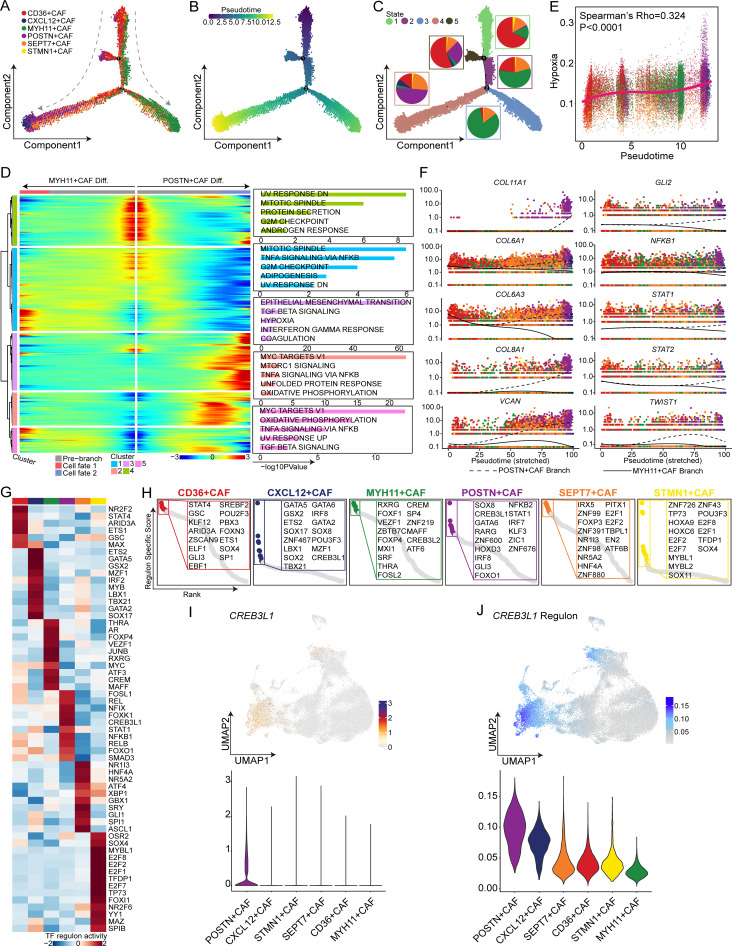
To gain insights into the dynamic processes underlying CAF subtypes at the single-cell level, we derived the pseudotime cell trajectory of the various CAF subtypes based on the Monocle 2 algorithm (figure 3A). Remarkably, statistical analyses of the combination of pseudotime and inferred state data revealed two distinct trajectories, with CD36+ CAFs (lpCAFs) representing the progenitor state and subsequently evolving into POSTN+ CAFs or MYH11+ CAFs separately (figure 3B,C and online supplemental figure S3A). Additionally, the majority of proliferative STMN1+ CAFs were present in the initiation state, while CXCL12+ CAFs were present in the terminal state within the differentiation branch, leading to differentiation of POSTN+ CAFs. Moreover, we explored gene expression patterns associated with the two differentiation branches involved in CAF state transitions. As depicted in the smoothed heatmap, the functional enrichment of highly expressed genes in the early stage of the trajectory (cluster 4) in both differentiation branches was enriched in protein secretion and G2M checkpoint pathways. In contrast to the MYH11+ CAF differentiation branch, the terminal state of the POSTN+ CAF differentiation branch exhibited higher expression levels of cluster 3 genes, which are implicated in epithelial-mesenchymal transition (EMT), TGF-β signaling, and hypoxia pathways known to be strongly associated with tumor progression. Nevertheless, the terminal state of the MYH11+ CAF differentiation branch displayed increased expression of cluster 1 genes, which were enriched in TNF-α signaling via the NF-KB, G2M checkpoint, and adipogenesis pathways (figure 3D).
Figure 3. Trajectory analysis of cancer-associated fibroblast (CAF) subtypes in hepatocellular carcinoma. (A) The developmental trajectory of CAFs was inferred by monocle 2 and colored according to the subtype. (B) The pseudotime of the developmental trajectory. The colors from purple to yellow indicate values from low to high, respectively. (C) The inferred states of the trajectory. The proportions of CAF subtypes are shown in the circles with different colors representing different CAF subtypes, and each circle color represents a different state. (D) Expression heatmap of significant (q<1e-3) genes based on branch expression analysis comparing the two CAF cell fates (left). Box plots showing the top five significantly enriched hallmark pathways in each gene cluster (right). (E) Dot plot showing Spearman’s correlation of the pseudotime and hypoxia scores, with different colors representing different CAF subtypes. (F) Pseudotime projections of transcriptional changes in extracellular matrix-associated genes and hypoxia-associated genes between the two trajectory branches. (G) Heatmap showing the mean activity of the top differentially activated regulons in each CAF subtype inferred by pyscenic. (H) Dot plots showing the top 15 specific activated transcription factors ranked by the regulon-specific score in each CAF subtype. (I) Uniform manifold approximation and projection (UMAP) plot (up) and violin plot (bottom) showing the expression of CREB3L1. (J) UMAP plot (up) and violin plot (bottom) showing the activity of the CREB3L1 regulon.
We further evaluated the correlation between the pseudotime axis and a set of progression-related pathways, including the EMT, TGF-β signaling, and hypoxia pathways. Interestingly, the hypoxia pathway showed a significant positive correlation with pseudotime (Spearman’s correlation test, rho=0.324, p<0.001) and was specifically activated in POSTN+ CAFs (figure 3E and online supplemental figure S3B,C). Hypoxia is a critical characteristic of the TME and plays a role in regulating CAF heterogeneity and promoting tumor progression. To identify potential differentially expressed genes between the two branches, we performed BEAM. We observed that antigen presentation-related genes (CD74 and CXCL12) and cytokines (CCL19, CCL21, and IL32) were significantly upregulated in the POSTN+ CAF differentiation branch (online supplemental figure S3D) and increased expression of ECM-related genes (COL11A1, COL6A1, COL6A3, COL8A1, and VCAN) and hypoxia-associated features (GLI2, NFKB1, STAT1, STAT2, TWIST1, and CREB3L1) in the POSTN+ CAF differentiation branch (figure 3F). Moreover, to assess the spatial organization of POSTN+ CAFs and hypoxic regions, spatial transcriptomics analysis of tumor-derived sections from two HCC patients was performed. The results revealed colocalization of POSTN+ CAFs and malignant cells in hypoxic zones within the same spot (online supplemental figure S3E). This finding suggested that hypoxia might stimulate CAF-mediated collagen secretion to modulate ECM properties.
To further determine the master regulator of POSTN+ CAFs, we conducted pySCENIC analysis to evaluate the most highly expressed TFs and their activities in the TF regulatory network. Specifically, we observed that CREB3L1 exhibited high expression and activity levels in the regulatory network of POSTN+ CAFs, and we constructed an RSS model for the POSTN+ CAF cell type using Jensen-Shannon divergence and identified CREB3L1 as one of the 15 most prevalent TFs based on the RSS, suggesting that CREB3L1 may serve as the key TF driving this differentiation pathway (figure 3G–J). Similarly, we found that hypoxia stimulated CAFs expressing POSTN and CREB3L1 (online supplemental figure S3F). Additionally, we next observed that the protein level of CREB3L1 was significantly greater in OE-POSTN-CAFs than in OE-NC-CAFs and that POSTN expression was strongly correlated with CREB3L1 expression in the TCGA-LIHC cohort (online supplemental figure S3G,H).
A previous study demonstrated that CREB3L1 is highly correlated with ECM production,36 which facilitates remodeling of the tumor stromal microenvironment. We next detected the mRNA levels of ECM-related genes in CAFs treated with si-NC or si-CREB3L1 under hypoxia, and the results showed that CREB3L1 promoted collagen secretion to modulate ECM properties (online supplemental figure S3I). These findings suggested that CREB3L1 may be a key molecule driving this differentiation pathway through its regulation of downstream signaling pathways.
Tumor-specific POSTN+ CAFs are associated with HCC progression
The differences in the proportions of infiltrated cell types between tumor-adjacent tissues and tumor tissues indicate that the TME undergoes dynamic remodeling, which plays a crucial role in HCC progression. CAFs have long been suggested to be a critical stromal cell type involved in the regulation of tumorigenesis and cancer progression. However, the heterogeneous nature of CAF populations has not been fully elucidated. To further investigate this possibility, we used publicly available data on HCC to validate the presence of tumor-specific POSTN+ CAFs. We compared the differential infiltration of POSTN-expressing fibroblasts between tumor and adjacent normal tissues from each donor. Tissue immunofluorescence staining revealed significantly greater POSTN expression in tumor tissues than in non-tumor tissues (online supplemental figure 4A). This phenomenon was also verified at the cellular level through immunofluorescence analysis and flow cytometry (online supplemental figure 4B,C). Notably, HCC patients with more POSTN+ CAFs had shorter overall survival and progression-free survival (online supplemental figure 4D), indicating that POSTN+ CAFs may promote tumor progression and adversely impact patient prognosis.
High infiltration of POSTN+ CAFs is associated with T-cell exclusion
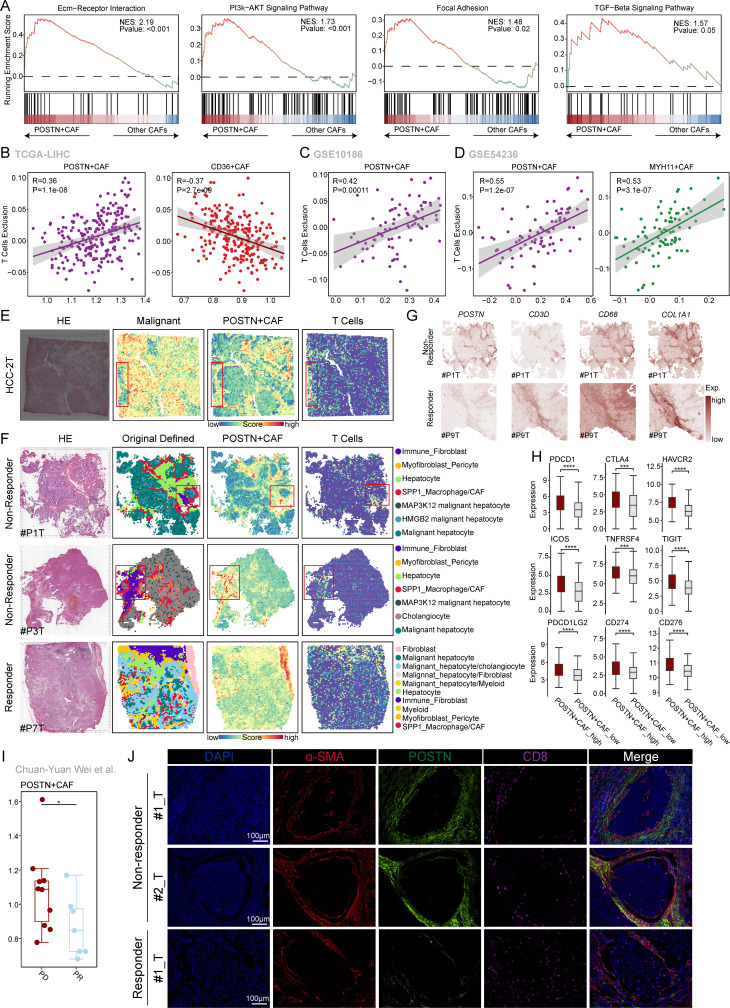
To gain further insight into the potential functional signals that induce POSTN+ CAFs, we conducted an analysis to identify genes that differentially expressed between POSTN+ CAFs and other CAF subgroups. Subsequently, we performed GSEA to examine the enriched pathways. online supplemental figure S5A illustrates the results of this analysis. The upregulated genes in POSTN+ CAFs were significantly enriched in several pathways associated with ECM remodeling in the TME, including the ECM-receptor interaction, the PI3K-AKT signaling pathway, focal adhesion, and the TGF-β signaling pathway (figure 4A). Numerous studies have highlighted the crucial role of ECM-related CAFs in promoting resistance to immunotherapy by facilitating T-cell exclusion.37,39 To validate this finding, we assessed the correlation between the infiltration of POSTN+ CAFs and T-cell exclusion in three independent cohorts downloaded from the Tumor Immune Dysfunction and Exclusion Database (http://tide.dfci.harvard.edu/) (figure 4B–D). Our findings revealed a significant positive correlation between the infiltration of POSTN+ CAFs and T cell exclusion. Furthermore, we observed a significant negative correlation between the infiltration of POSTN+ CAFs and CD8+ T cell infiltration (online supplemental figure S5B–C). To investigate the spatial organization of POSTN+ CAFs and CD8+ T cells, we performed spatial transcriptomic analysis. Interestingly, our observations revealed that POSTN+ CAFs were responsible for excluding T cells from the malignant area within HCC tumor tissue sections(figure 4E). Similar to the findings reported by Liu et al, POSTN+ CAFs formed a tumor immune barrier structure that excluded T cells in non-responders to ICB therapy but not in responders (figure 4F). Bayesian-enhanced spatial data highlighted that regions with high POSTN expression exhibited elevated levels of CD68 and COL1A1 in non-responders, while regions with high CD3D expression demonstrated lower levels of POSTN in responders (figure 4G). In addition, we explored the differences in the expression of immune checkpoint-associated genes between POSTN+ CAFs with high infiltration and POSTN+ CAFs with low infiltration in the TCGA-LIHC cohort. The results suggested that the presence of POSTN+ CAFs is associated with immunotherapy efficacy (figure 4H). Importantly, analysis of patients with progressive disease (PD) compared with those with a partial response (PR), based on the dataset from Chuan-Yuan Wei et al40, revealed significantly greater infiltration of POSTN+ CAFs in PD patients. This suggests that patients with a high proportion of POSTN+ CAFs exhibit a significantly poorer response to anti-PD-1 treatment than other patients (figure 4I). Additionally, we analyzed the response to sorafenib, a first-line targeted therapy for advanced HCC. Compared with responders and patients treated with placebo, non-responders to sorafenib showed greater infiltration of POSTN+ CAFs (based on the GSE109211 dataset) (online supplemental figure S5D). Furthermore, we performed mIHC staining to validate our findings. We observed significantly greater levels of POSTN+ CAFs with colocalization of POSTN and α-SMA protein expression in non-responders than in responders. Importantly, the colocalization of POSTN+ α-SMA+ CAFs correlated with the exclusion of CD8+ cells in non-responders (figure 4J), suggesting potential crosstalk between POSTN+ CAFs and CD8+ T cells associated with immunotherapy efficacy. To further validate whether POSTN knockdown could promote PD-1 mAb therapy in vivo, we established an orthotopic HCC implantation model for anti-PD-1 therapy. We observed that AAV8-shPOSTN and IgG cotreatment significantly inhibited tumor proliferation compared with the AAV8-control and IgG co-treatment group. More importantly, cotreatment with AAV8-shPOSTN and PD-1 mAb further impaired tumor proliferation compared with treatment with AAV8-shPOSTN or PD-1 mAb alone (online supplemental figure S6A–C). These findings provide valuable insights into the role of POSTN+ CAFs in HCC and offer potential immunotherapeutic targets and predictive biomarkers.
Figure 4. POSTN+ CAFs were associated with T-cells exclusion. (A) Gene set enrichment analysis (GSEA) of the ECM-receptor interaction, PI3K-AKT signaling, focal adhesion, and TGF-beta signaling pathways between POSTN+ CAFs and other CAFs. The genes were ranked by fold change in expression between these two conditions. (B–D) The correlation between the ssGSEA score of each CAF subtype and T-cell exclusion score in the TCGA-LIHC cohort (B), GSE10186 (C), and GSE54236 cohort (D). (E) Spatial HE staining and spatial feature plots of the signatures of malignant cells, POSTN+ CAFs and T cells in HCC-2T tissue sections (from left to right). (F) HE staining of tumor tissues from immune checkpoint blockade (ICB) non-responders and responders at ST spots (left). Original defined cell types in ICB-treated non-responder and responder tumor tissues (middle). Spatial feature plots of the signatures of POSTN+ CAFs and T cells in the tumor tissue of ICB-treated non-responders and responders (right). (G) Enhanced spatial feature plots showing the expression of POSTN, CD3D, CD68, and COL1A1 in ICB non-responder and ICB responder tumor tissues. (H) Box plots showing the expression of immune checkpoint-associated genes between the POSTN+ CAFs with high infiltration group and the POSTN+ CAFs with low infiltration groups in the TCGA-LIHC cohort. The Wilcoxon rank-sum test was used to assess the differences. (I) Box plots showing the difference in the POSTN+ CAF ssGSEA score between the PD and PR anti-PD1 treatment groups. The Wilcoxon rank-sum test was used to assess the differences, and the p value was ≤0.05.* (J) Representative IF staining of anti-PD1 responder and non-responder tissues. DAPI (blue), α-SMA (red), POSTN (green), and CD8 (purple) are shown in individual and merged channels. Bar, 100 µm. CAFs, cancer-associated fibroblasts; ECM, extracellular matrix; PD, progressive disease; PR, partial response; TGF-beta, transforming growth factor beta.
Cell–cell interactions between POSTN+ CAFs and SPP1+ macrophages may contribute to the immunosuppressive TME
To explore the mechanisms underlying the regulation of the immune microenvironment in HCC by POSTN+ CAFs, we performed cell–cell interaction analysis using the CellPhoneDB method to examine the interplay among seven major cell types. Compared with those in normal samples, stronger interactions were observed between fibroblasts and myeloid cells in tumor samples (figure 5A), suggesting that POSTN+ CAFs may promote the development of an immunosuppressive microenvironment by modulating myeloid cells. Based on the expression of specific marker genes, we identified 19 subtypes within the myeloid lineage: nine macrophage subtypes (CCL3L1+ Mph, CXCL10+ Mph, FTX+ Mph, MKI67+ Mph, MT1G+ Mph, WWP1+ Mph, SEPP1+ Mph, and SPP1+ Mph), three monocyte subtypes (FCN1+ Mono, TCF7L2+ Mono and VCAN+ Mono), one neutrophil subtype (S100A8+ Neu), two Kupffer cell subtypes (CD5L+ Mph and MARCO+ Mph), and five dendritic cell subtypes (CD1C+ DC, CLEC9A+ DC, GNB2L1+ DC, LAMP3+ DC and LILRA4+ pDC) (figure 5B and online supplemental figure S7A,B). We found that SPP1+ macrophages were predominantly enriched in tumor tissues (online supplemental figure S7B). We employed CIBERSORTx to assess the infiltration of 25 cell clusters identified by scRNA-seq, including the above-mentioned subtypes, in the TCGA-LIHC cohort and calculated pairwise Spearman correlations for pairs of cell clusters (figure 5C). We found that POSTN+ CAFs and SPP1+ macrophages exhibited the strongest correlation among the cell populations in the TCGA-LIHC cohort (figure 5C).
Figure 5. Characterization of myeloid cells in normal and hepatocellular carcinoma tissues. (A) Chord diagrams showing the cell‒cell interactions between seven major cell types in tumor (left) and normal (right) tissues. (B) Uniform manifold approximation and projection (UMAP) plot of individual myeloid cells. Each dot denotes one cell; the color represents the myeloid subtype. (C) Dot plots showing the Spearman correlation for the pairwise infiltration of cancer-associated fibroblast (CAF) subtypes and myeloid subtypes in the TCGA-LIHC cohort. Dot size and color represent Spearman’s correlation, and black dots indicate significance at different p value thresholds, p≤0.05*, 0.05<p≤0.01**, p≤0.001***. (D) UMAP plots showing the density distribution of M1 and M2 macrophages and the corresponding box plots showing the M1 and M2 scores of each macrophage and monocyte subtype. (E) Top-ranked ligands inferred to regulate SPP1+ macrophages via POSTN+ CAFs according to NicheNet. (F) Dot plots showing the expression percentage (dot size) and intensity (dot intensity) of the top-ranked ligands (E) in each CAF subtype. (G) Ligand–receptor pairs showing interactions between SPP1+ macrophages and POSTN+ CAFs ordered by ligand activity (E). (H) Dot plots showing the percentage (dot size) and intensity (dot intensity) of the expression of IL-6 or TGFB1-targeted receptors in each myeloid subtype.
By calculating the M1/M2 polarization scores for CXCL9-SPP1 macrophages and assessing the expression profiles of M1/M2 CXCL9-SPP1 macrophages, we found that SPP1+ Mphs were more prevalent in anti-inflammatory responses and skewed toward the M2 phenotype (figure 5D and online supplemental figure S7C). To identify key regulators of the interaction between POSTN+ CAFs and SPP1+ macrophages in HCC patients, we employed the NicheNet package to examine the interplay mechanisms between these cell types. We observed that POSTN+ CAFs exhibited high levels of ligand activity for IL-6, TGFB1, FGF2, and CXCL2, along with relatively elevated gene expression of IL-6, TGFB1, FGF2, and CXCL2 (figure 5E,F). In addition, the protein encoded by IL-6 bound to the receptors IL-6R, IL6ST, and THBD expressed on SPP1+ macrophages (figure 5G), while SPP1+ macrophages expressed high levels of IL-6R (figure 5H), suggesting that POSTN+ CAFs and SPP1+ macrophages may regulate the formation of an immunosuppressive environment via the IL-6/IL-6R axis. We further explored the interplay mechanisms between SPP1+ macrophages and POSTN+ CAFs and observed that SPP1+ macrophages expressed cytokines (TGFB1, IL1A, and IL1B) that interacted with corresponding receptors expressed on POSTN+ CAFs (online supplemental figure S7D). Collectively, our findings suggest the formation of a mutually supportive and functional interaction network between POSTN+ CAFs and SPP1+ macrophages, which may play crucial roles in ECM remodeling and promote the formation of an immunosuppressive environment in HCC.
POSTN+ CAFs promote macrophage SPP1 expression via the IL-6/IL-6R axis
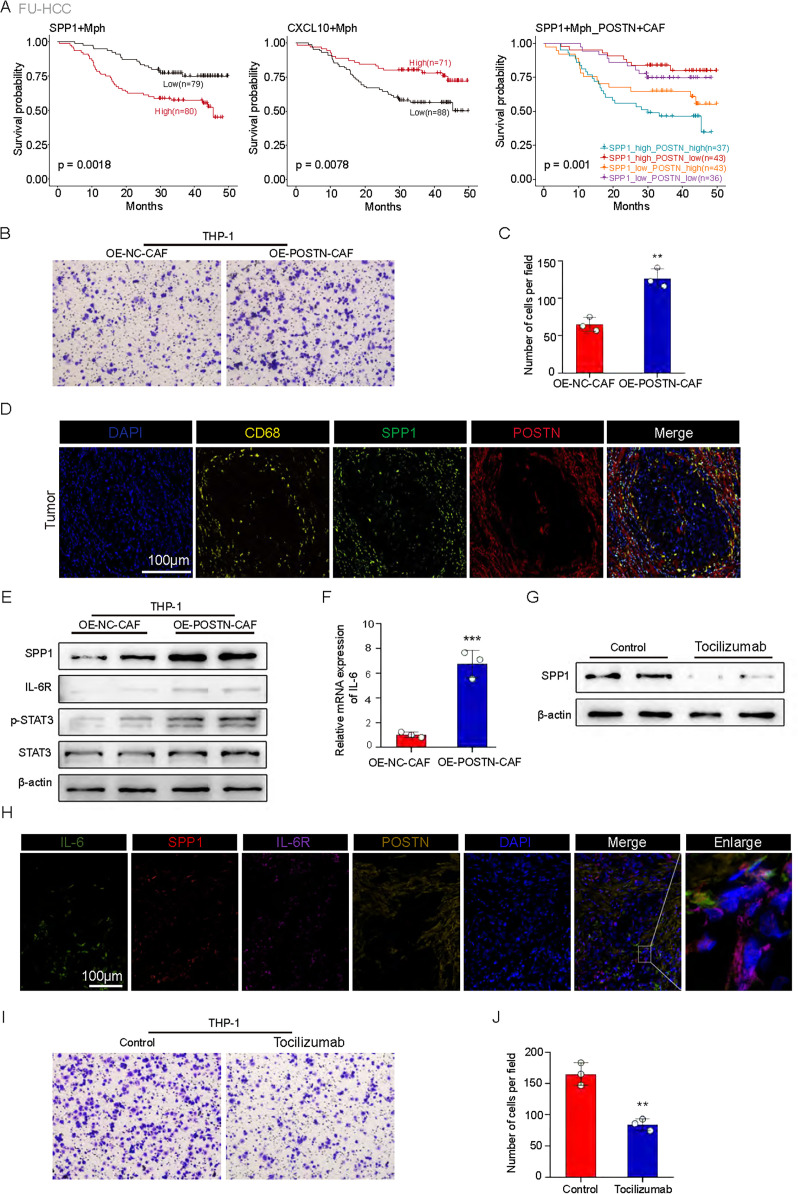
The correlation between SPP1+ macrophages and POSTN+ CAFs suggests that these cells may work together to create an immunosuppressive microenvironment in HCC and contribute to tumor progression. Our study aimed to explore the clinical significance of this correlation and investigate the mechanisms involved. We observed that HCC patients with high levels of POSTN+ CAFs and SPP1+ macrophages had the shortest OS (figure 6A), indicating synergistic promotion of tumor progression by these two cell types. To further understand the relationship between POSTN+ CAFs and SPP1+ macrophages, we performed experiments to examine their interaction. We overexpressed POSTN in CAFs and found that the ability of these cells to recruit macrophages was significantly greater than that of control CAFs (figure 6B,C). This recruitment phenomenon was also observed in HCC patient tissues (figure 6D).
Figure 6. High infiltration of POSTN+ cancer-associated fibroblasts (CAFs) and SPP1+ macrophages was associated with poor patient survival. (A) Survival analyses for patients with low and high infiltration of SPP1+Mph (left), CXCL10+ Mph (middle), and SPP1+ Mph/POSTN+ CAF (right) in the FU-HCC cohort. (B) Transwell assay of chemotactic macrophages (n=3). (C) Statistical data. ***p<0.001. (D) Representative IF staining of tumor tissues. DAPI (blue), CD68 (yellow), SPP1 (green), and POSTN (red) are shown in individual and merged channels are shown. Bar, 100 µm. (E) Western blot analysis of the protein levels of SPP1, IL-6R, p-STAT3, and STAT3. OE-NC-CAFs: macrophages treated with conditioned medium from control CAFs; OE-POSTN-CAFs: macrophages treated with conditioned medium from CAFs overexpressing POSTN. (F) IL-6 levels were examined by qRT-PCR analysis (n=3). (G) Western blot analysis of the protein levels of SPP1. Control: macrophages treated with conditioned medium from CAFs overexpressing POSTN and PBS; tocilizumab: macrophages treated with conditioned medium from CAFs overexpressing POSTN and tocilizumab. (H) Representative IF staining of tumor tissues. DAPI (blue), IL-6 (green), SPP1 (red), IL-6R (purple), and POSTN (yellow) are shown in individual and merged channels. Bar, 100 µm. (I) Transwell assay of chemotactic macrophages (n=3). (J) Statistical data. **p<0.01; ***p<0.001.
Previous studies have indeed demonstrated that secreted POSTN can bind to the surface ligand integrin-αvβ5 on macrophages, thereby facilitating their recruitment.41 Our results align with these findings, as we observed a high expression of ITGB5 and ITGAV in SPP1+ macrophages (figure 5H and online supplemental figure S6E). Our findings support the role of POSTN secreted by POSTN+ CAFs in macrophage chemotaxis. Furthermore, we investigated whether POSTN+ CAFs promote high SPP1 expression in recruited macrophages. Compared with macrophages cultured in control medium, macrophages cultured in medium derived from OE-POSTN-CAFs exhibited significant upregulation of SPP1, IL-6R, and p-STAT3 (figure 6E). Previous studies have indicated that IL-6, which is secreted by CAFs, mediates the activation of STAT3 to promote chemotherapeutic resistance in gastric carcinoma.42 Moreover, it has been demonstrated that p-STAT3 can bind to the promoter region of SPP1, enhancing its transcription and facilitating melanoma progression.43 Consistent with these findings, we observed significant upregulation of IL-6 mRNA expression in the OE-POSTN-CAF group compared with the control group (figure 6F). Additionally, tocilizumab treatment led to a decrease in the protein expression level of SPP1 (figure 6G). Consistent with our previous findings, our mIHC results of HCC patient tissues revealed that IL-6 was mainly localized in POSTN+ CAFs, IL-6R was mainly localized in SPP1+ macrophages and that SPP1+ macrophages were also concentrated in the same region as POSTN+ CAFs (figure 6H). Additionally, when we blocked the effect of IL-6 with tocilizumab, macrophage recruitment was inhibited (figure 6I,J). This led us to hypothesize that POSTN+ CAFs recruit macrophages and promote their high SPP1 expression through IL-6-IL-6R signaling. In conclusion, our study suggested that the interaction between POSTN+ CAFs and SPP1+ macrophages play a crucial role in the immunosuppressive microenvironment of HCC. Further investigation into the mechanisms involved in this interaction could reveal the potential to identify therapeutic targets for the treatment of HCC.
High infiltration of POSTN+ CAFs and SPP1+ macrophages is correlated with immunotherapy resistance
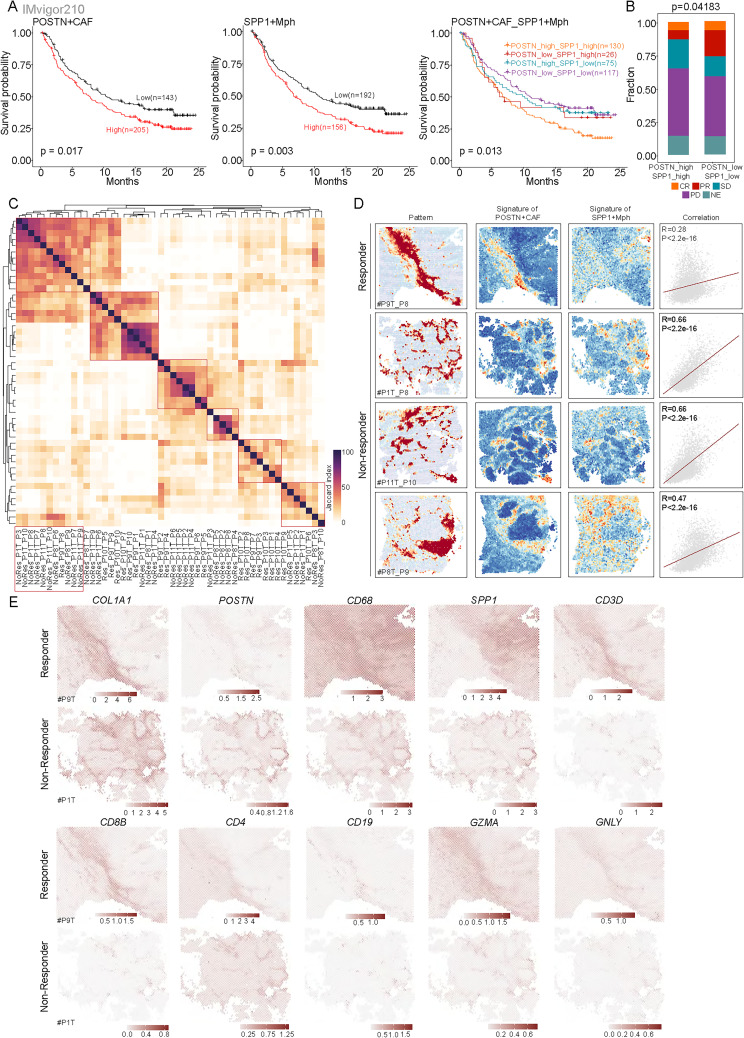
The immunosuppressive “cold” TME is the main cause of immunotherapy resistance in HCC patients. Currently, the conventional type of immunotherapy treatment is checkpoint inhibitors, which mobilize T cells to induce T-cell mediated responses. Checkpoint inhibitors have little effect on cold-induced tumors. The analysis of survival data from the IMvigor210 dataset revealed that patients treated with anti-PD-L1 therapy who had high infiltration of POSTN+ CAFs or SPP1 macrophages exhibited worse OS than those without these features (figure 7A). Moreover, anti-PD-L1 antibody-treated patients with high infiltration of POSTN+ CAFs and SPP1+ macrophages had shorter survival times than those with other infiltration profiles (figure 7A). Importantly, patients with high infiltration of these cells had lower complete response and PR rates and a greater PD rate than patients with low infiltration of these cells (figure 7B). This suggests that patients with TMEs enriched in POSTN+ CAFs and SPP1+ macrophages have a significantly poorer response to anti-PD-L1 therapy.
Figure 7. High infiltration of POSTN+ cancer-associated fibroblasts (CAFs) and SPP1+ macrophages is correlated with immunotherapy resistance. (A) Survival analyses for patients with low and high infiltration of POSTN+ CAFs alone (left), SPP1+ macrophages alone (middle), and both (right) in the anti-PD-L1 immunotherapy cohort using Kaplan-Meier curves (IMvigor210 cohort). (B) Bar plots showing that patients with both low infiltration of both POSTN+ CAFs and SPP1+ macrophages were more likely to respond to anti-PD-L1 treatment. Fisher’s exact test was used to assess differences. (C) Heatmap showing the Jaccard index of gene programs in each sample identified by SpaGene calculated based on the overlap of top-ranked genes. (D) Spatial feature plots of the signature score of the specific gene pattern, POSTN+ CAFs and SPP1+ macrophages in ICB-treated non-responders and responder tumor tissues (from left to right), and dot plots showing the Spearman’s correlation of the signature score of POSTN+ CAFs and SPP1+ macrophages. (E) Enhanced spatial feature plots showing the expression of COL1A1, POSTN, CD68, SPP1, CD3D, CD8B, CD4, CD19, GZMA, and GNLY in immune checkpoint blockade (ICB) non-responder and ICB responder tumor tissues.
To further assess the spatial gene expression patterns associated with immunotherapy responsiveness, we used the SpaGene algorithm to identify ten spatial gene expression patterns for each sample and assessed the similarity between patterns based on the Jaccard index (figure 7C). The patterns highlighted with red circles in figure 7C and online supplemental figure S8 A–D were differentially expressed between immunotherapy-sensitive and immunotherapy-resistant HCC patients and were associated with ECM-receptor interactions and the focal adhesion pathways, suggesting that these patterns might be closely linked to POSTN+ CAFs. To test this hypothesis, we quantified the specific gene expression pattern, the signature score for POSTN+ CAFs, and the signature score for SPP1+ macrophages in each spot in the corresponding samples. Notably, we observed that an ECM-associated pattern was present in both immunotherapy-responsive and immunotherapy-resistant patients and colocalized with POSTN+ CAFs (figure 7D). Interestingly, we observed stronger spatial colocalization between POSTN+ CAFs and SPP1+ macrophages in non-responders than in responders (figure 7D). This further supports the finding that patients with enriched POSTN+ CAFs and SPP1+ macrophages have a significantly poorer response to anti-PD-1 therapy. Similarly, mIHC staining revealed that non-responders exhibited greater infiltration of POSTN+ CAFs or SPP1+ macrophages than responders (online supplemental figure S9). Bayespace-enhanced spatial data showed that most POSTN+ CAFs and SPP1+ macrophages were colocalized in the same region, whereas T cells or B cells were excluded from the tumor core (figure 7E). Collectively, these results indicate that the interaction between POSTN+ CAFs and SPP1+ macrophages limit the infiltration of immune cells into the tumor core. These two cell types may determine the efficacy of immunotherapy in HCC patients.
Discussion
Despite significant advancements in immunotherapy for tumor treatment, not all patients exhibit a positive response, and the underlying mechanisms of non-response remain incompletely understood.44,46 Limited information is available regarding the prediction of immunotherapy response in clinical trials involving HCC patients treated with ICB. In our study, we conducted a comprehensive analysis using publicly available scRNA-seq and bulk RNA sequencing datasets, as well as ST data from HCC patients undergoing ICB treatment. These findings were further validated using mIHC staining and other techniques. Our study provides valuable insights into the distinct subpopulations of CAFs in HCC and their properties at the single-cell level. Moreover, we demonstrated that the interactions between POSTN+ CAFs and CD8+ T cells, as well as between POSTN+ CAFs and SPP1+ macrophages, could play a crucial role in the development of an immunosuppressive microenvironment.
The heterogeneity of CAFs plays a crucial regulatory role in the immune microenvironment.47 Recent advancements in scRNA-seq have allowed for a deeper understanding of CAF heterogeneity across various tumor types.48 Consequently, biomarker genes that define different potential CAF subgroups have been identified.49 In our study, we systematically deciphered the heterogeneity and characteristics of CAFs in HCC and identified six unique CAF subpopulations: POSTN+ CAFs, STMN1+ CAFs, MYH11+ CAFs, CD36+ CAFs, CXCL12+ CAFs, and SEPT7+ CAFs.
Previous studies have shown that interactions between CAFs and HCC cells occur through multiple molecular mechanisms.50 51 However, the different activation states, stress responses, and CAF sources lead to functional heterogeneity.52 Thus, the role of specific functional CAF subpopulations in regulating HCC progression is unclear. Our analysis revealed that among human CAFs, the marker gene POSTN is specifically enriched in HCC tumors. Further analysis demonstrated that ECM/TGF-β/hypoxia signaling was highly enriched in POSTN+ CAFs, and that these signaling pathways played key roles in tumor progression and the immune microenvironment.53,55 POSTN is a secreted protein that has been well studied in tumor cells. However, its role in CAFs is poorly understood. Our study revealed that POSTN+ CAFs are located mainly in the peripheral regions of tumors and dominate ECM remodeling compared with other CAF subpopulations. ECM remodeling is an important factor in tumor progression and creates a physical barrier that inhibits the recruitment of immune cells, especially T cells, to cancer sites.56 57 Collagen density in the ECM also determines T-cell distribution.58 Our data suggest that POSTN+ CAFs suppress CD8+ T-cell infiltration, suppress antitumor immunity, promote the development of an immunosuppressive microenvironment, and affect the response to immunotherapy. Our findings indicated a significant increase in the number of POSTN+ CAFs in tumor tissues, which was strongly correlated with poor patient prognosis. By identifying key CAF subpopulations and their functions, our study provides new insights for developing therapeutic strategies targeting the immune microenvironment in HCC.
Identification of factors contributing to TME immunosuppression is crucial for the development of effective immunotherapies. Interactions between CAFs and immune cells, including tumor-associated macrophages, have been shown to modulate the immune microenvironment and suppress antitumor immune responses.59 60 In our study, we investigated the role of CAFs and macrophages in HCC and their impact on the immune microenvironment.
We observed a close interaction between CAFs and macrophages, with macrophages being prominent in the dense zone of CAFs. CAFs were found to promote the recruitment and differentiation of monocytes into M2-type macrophages via various regulatory molecules.61 These macrophages, in turn, impair effector T-cell responses and induce an immunosuppressive state in the TME.62 SPP1+ macrophages were identified as the most relevant cell subtype interacting with POSTN+ CAFs and were associated with poorer OS in HCC patients. We further characterized the immune microenvironment in HCC and focused on the key cell subtypes regulating immune evasion. Through single-cell and spatial transcriptomics, immunofluorescent labeling, and analysis of other datasets, we investigated the interactions between POSTN+ CAFs and SPP1+ macrophages. We predicted that POSTN+ CAFs promote the overexpression of SPP1 in macrophages through IL-6/STAT3 signaling, leading to the differentiation of macrophages toward the SPP1+ phenotype. We also mapped the spatial distribution of SPP1+ macrophages, which interact with POSTN+ CAFs. In regions enriched with POSTN+ CAFs and SPP1+ macrophages, we observed reduced infiltration of lymphocytes, suggesting the formation of an immune-excluded microenvironment. Furthermore, high expression of POSTN and/or SPP1 was predictive of poorer OS and a reduced response to anti-PD-1 therapy, supporting the presence of an immunosuppressive microenvironment.
Our findings provide insights into the mechanisms involved in the formation of the immunosuppressive microenvironment in HCC. Specifically, we highlighted the crucial interaction between POSTN+ CAFs and SPP1+ macrophages. Targeting this interaction holds promise as a potential strategy for HCC immunotherapy. Our findings indicate that the interaction between POSTN+ CAFs and SPP1+ macrophages plays a crucial role in remodeling the ECM and creating a microenvironment that promotes fibroblast proliferation. This microenvironment suppresses the infiltration of lymphocytes into the core of the tumor, thereby reducing the effectiveness of PD-1 therapy. We propose a possible mechanism for this interaction: The induction of POSTN+ CAFs by hypoxia leads to the expression of CREB3L1, subsequently facilitating CREB3L1 nuclear transport, thereby influencing ECM remodeling. Additionally, POSTN+ CAFs secrete IL-6, which activates the STAT3 signaling pathway in macrophages, leading to increased expression of SPP1. Furthermore, our results suggest that SPP1+ macrophages may influence POSTN+ CAFs through the action of TGFβ1. This interaction promotes the secretion of matrix metalloproteinases and collagen, which contributes to ECM remodeling. However, further investigation is necessary to fully elucidate the underlying biological mechanisms involved. Our study revealed that the proportion of POSTN+ CAFs determines the efficacy of immunotherapy in HCC patients. This highlights the importance of developing strategies that target POSTN+ CAFs to overcome immunosuppression and enhance the tumor response to immunotherapy.
supplementary material
Footnotes
Funding: This study was supported by grants from the National Natural Science Foundation of China (81971495), the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (No. 2019-I2M-5-035), the Foundation Research Project of Jiangsu Province (BRA2017533, BK20191490), the Fundamental Research Funds for the Central Universities (3332023118).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Consent obtained directly from patient(s)
Ethics approval: This study involves human participants and the study design and sample collection protocols were approved by the Ethical Board of Peking Union Medical College Hospital (ethical approval number: JS-3502D). All patients provided informed consent at the time of recruitment. All animal study protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (serial number: IACUC-2209059).
Data availability free text: The public scRNA-seq datasets used in this research were obtained from the datasets GSE149614, GSE151530, GSE156625, GSE189903, and GSE202642 were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The specific datasets GSE14520, GSE10143, GSE192912, GSE109211, GSE10186, and GSE54236 were accessed from GEO. Dataset HRA001748 was obtained from the Genome Sequence Archive (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001748). The publicly available data of HCC samples from the Cancer Genome Atlas (TCGA) data portal (http://gdac.broadinstitute.org/). The Fudan HBV-HCC cohort (FU-HCC) were collated from the biosino NODE database (OEP000321, https://www.biosino.org/node/project/detail/OEP000321). Spatial transcriptome sequencing data were acquired from the study conducted by Wu et al. (http://lifeome.net/supp/livercancer-st/data.htm) and Yao Liu et al. (https://data.mendeley.com/datasets/skrx2fz79n/1). All data used in this study are publicly available for download. All data are available on request from the authors.
Contributor Information
Hao Wang, Email: wyknyd@163.com.
Yuan Liang, Email: 230229240@seu.edu.cn.
Zheng Liu, Email: drlzheng@163.com.
Rui Zhang, Email: zhangrui1@njmu.edu.cn.
Jiashuo Chao, Email: chaojiashuo2@163.com.
Mingming Wang, Email: dr_wangmm@163.com.
Mu Liu, Email: liumu@njmu.edu.cn.
Lei Qiao, Email: qiaolei@stu.njmu.edu.cn.
Zhengfeng Xuan, Email: xuanzhengfeng2022@163.com.
Haitao Zhao, Email: zhaoht@pumch.cn.
Ling Lu, Email: lvling@njmu.edu.cn.
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.Sugawara Y, Hibi T. Surgical treatment of hepatocellular carcinoma. Biosci Trends. 2021;15:138–41. doi: 10.5582/bst.2021.01094. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y-Y, Yang X, Wang Y-C, et al. Clinical outcomes of Lenvatinib plus Transarterial Chemoembolization with or without programmed death Receptor-1 inhibitors in Unresectable hepatocellular carcinoma. World J Gastroenterol. 2023;29:1614–26. doi: 10.3748/wjg.v29.i10.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–72. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 6.Xu F, Jin T, Zhu Y, et al. Immune Checkpoint therapy in liver cancer. J Exp Clin Cancer Res. 2018;37:110. doi: 10.1186/s13046-018-0777-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sangro B, Sarobe P, Hervás-Stubbs S, et al. Advances in Immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525–43. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinter M, Jain RK, Duda DG. The current landscape of immune Checkpoint blockade in hepatocellular carcinoma: A review. JAMA Oncol. 2021;7:113–23. doi: 10.1001/jamaoncol.2020.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18:99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 10.Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor Microenvironment: new findings and future perspectives. Mol Cancer. 2021;20:131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin P, Chen H, Wang Y, et al. Cancer-associated fibroblasts undergoing Neoadjuvant chemotherapy suppress Rectal cancer revealed by single-cell and spatial Transcriptomics. Cell Rep Med . 2023;4:101231. doi: 10.1016/j.xcrm.2023.101231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mucciolo G, Araos Henríquez J, Jihad M, et al. EGFR-activated Myofibroblasts promote metastasis of Pancreatic cancer. Cancer Cell. 2024;42:101–18. doi: 10.1016/j.ccell.2023.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Lavie D, Ben-Shmuel A, Erez N, et al. Cancer-associated fibroblasts in the single-cell era. Nat Cancer . 2022;3:793–807. doi: 10.1038/s43018-022-00411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer Y, Hocine HR, Gentric G, et al. Single-cell analysis reveals fibroblast clusters linked to Immunotherapy resistance in cancer. Cancer Discov. 2020;10:1330–51. doi: 10.1158/2159-8290.CD-19-1384. [DOI] [PubMed] [Google Scholar]
- 15.Luo H, Xia X, Huang L-B, et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor Microenvironment. Nat Commun. 2022;13:6619. doi: 10.1038/s41467-022-34395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyai Y, Esaki N, Takahashi M, et al. Cancer-associated fibroblasts that restrain cancer progression: hypotheses and perspectives. Cancer Sci. 2020;111:1047–57. doi: 10.1111/cas.14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteran L, Erez N. The dark side of fibroblasts: cancer-associated fibroblasts as mediators of immunosuppression in the tumor Microenvironment. Front Immunol. 2019;10:1835. doi: 10.3389/fimmu.2019.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman G, Levi-Galibov O, David E, et al. Cancer-associated fibroblast compositions change with breast cancer progression linking the ratio of S100A4(+) and PDPN(+) Cafs to clinical outcome. Nat Cancer . 2020;1:692–708. doi: 10.1038/s43018-020-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer Stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol. 2020;17:487–505. doi: 10.1038/s41575-020-0300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao J, Wang H, Ni M, et al. Fstl1 promotes liver fibrosis by Reprogramming macrophage function through Modulating the intracellular function of Pkm2. Gut. 2022;71:2539–50. doi: 10.1136/gutjnl-2021-325150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Yang A, Quan C, et al. A single-cell Atlas of the Multicellular Ecosystem of primary and metastatic hepatocellular carcinoma. Nat Commun. 2022;13:4594. doi: 10.1038/s41467-022-32283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su X, Zhao L, Shi Y, et al. Clonal evolution in liver cancer at single-cell and single-variant resolution. J Hematol Oncol. 2021;14:22. doi: 10.1186/s13045-021-01036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A, Seow JJW, Dutertre C-A, et al. Onco-fetal Reprogramming of endothelial cells drives immunosuppressive Macrophages in hepatocellular carcinoma. Cell. 2020;183:377–94. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Heinrich S, Wang L, et al. Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer. Nat Commun. 2022;13:7533. doi: 10.1038/s41467-022-35291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu G-Q, Tang Z, Huang R, et al. Cd36(+) cancer-associated fibroblasts provide immunosuppressive Microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor. Cell Discov. 2023;9:25. doi: 10.1038/s41421-023-00529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue R, Zhang Q, Cao Q, et al. Liver tumour immune Microenvironment subtypes and neutrophil heterogeneity. Nature. 2022;612:141–7. doi: 10.1038/s41586-022-05400-x. [DOI] [PubMed] [Google Scholar]
- 27.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eun JW, Yoon JH, Ahn HR, et al. Cancer-associated fibroblast-derived secreted Phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and Lenvatinib. Cancer Communications . 2023;43:455–79. doi: 10.1002/cac2.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinyol R, Montal R, Bassaganyas L, et al. Molecular predictors of prevention of recurrence in HCC with sorafenib as adjuvant treatment and Prognostic factors in the phase 3 STORM trial. Gut. 2019;68:1065–75. doi: 10.1136/gutjnl-2018-316408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oishi N, Kumar MR, Roessler S, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200C and epithelial-Mesenchymal transition in Intrahepatic Cholangiocarcinoma. Hepatology. 2012;56:1792–803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villa E, Critelli R, Lei B, et al. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular Carcinomas and worst survival. results from a prospective study. Gut. 2016;65:861–9. doi: 10.1136/gutjnl-2014-308483. [DOI] [PubMed] [Google Scholar]
- 32.Roessler S, Jia H-L, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–12. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Q, Zhu H, Dong L, et al. Integrated Proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–77. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 34.Wu R, Guo W, Qiu X, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021;7:eabg3750. doi: 10.1126/sciadv.abg3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Xun Z, Ma K, et al. Identification of a tumour immune barrier in the HCC Microenvironment that determines the efficacy of Immunotherapy. J Hepatol. 2023;78:770–82. doi: 10.1016/j.jhep.2023.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Pan Z, Xu T, Bao L, et al. Creb3L1 promotes tumor growth and metastasis of Anaplastic thyroid carcinoma by remodeling the tumor Microenvironment. Mol Cancer. 2022;21:190. doi: 10.1186/s12943-022-01658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnamurty AT, Shyer JA, Thai M, et al. Lrrc15(+) Myofibroblasts dictate the Stromal Setpoint to suppress tumour immunity. Nature. 2022;611:148–54. doi: 10.1038/s41586-022-05272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins L, Jungwirth U, Avgustinova A, et al. Cancer-associated fibroblasts suppress Cd8+ T-cell infiltration and confer resistance to immune-Checkpoint blockade. Cancer Res. 2022;82:2904–17. doi: 10.1158/0008-5472.CAN-21-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feig C, Jones JO, Kraman M, et al. Targeting Cxcl12 from FAP-expressing carcinoma-associated fibroblasts Synergizes with anti-PD-L1 Immunotherapy in Pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20212–7. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei CY, Zhu MX, Zhang PF, et al. Pkcalpha/Zfp64/Csf1 axis Resets the tumor Microenvironment and fuels anti-Pd1 resistance in hepatocellular carcinoma. J Hepatol. 2022;77:163–76. doi: 10.1016/j.jhep.2022.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Allard DE, Wang Y, Li JJ, et al. Schwann cell-derived Periostin promotes autoimmune peripheral polyneuropathy via macrophage recruitment. J Clin Invest. 2018;128:4727–41. doi: 10.1172/JCI99308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ham I-H, Oh HJ, Jin H, et al. Targeting Interleukin-6 as a strategy to overcome Stroma-induced resistance to chemotherapy in gastric cancer. Mol Cancer. 2019;18:68. doi: 10.1186/s12943-019-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng G, Zeng F, He Y, et al. Eef2K silencing inhibits tumour progression through repressing Spp1 and Synergises with BET inhibitors in Melanoma. Clin Transl Med. 2022;12:e722. doi: 10.1002/ctm2.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ladd AD, Duarte S, Sahin I, et al. Mechanisms of drug resistance in HCC. Hepatology. 2024;79:926–40. doi: 10.1097/HEP.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z, Wang Y, Gao P, et al. Immune Checkpoint inhibitor resistance in hepatocellular carcinoma. Cancer Lett. 2023;555:216038. doi: 10.1016/j.canlet.2022.216038. [DOI] [PubMed] [Google Scholar]
- 46.Harkus U, Wankell M, Palamuthusingam P, et al. Immune Checkpoint inhibitors in HCC: cellular, molecular and systemic data. Semin Cancer Biol. 2022;86:799–815. doi: 10.1016/j.semcancer.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Wu F, Yang J, Liu J, et al. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct Target Ther. 2021;6:218. doi: 10.1038/s41392-021-00641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutton C, Heider F, Blanco-Gomez A, et al. Single-cell analysis defines a Pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell. 2021;39:1227–44. doi: 10.1016/j.ccell.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster DS, Januszyk M, Delitto D, et al. Multiomic analysis reveals conservation of cancer-associated fibroblast phenotypes across species and tissue of origin. Cancer Cell. 2022;40:1392–406. doi: 10.1016/j.ccell.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhee H, Kim H-Y, Choi J-H, et al. Keratin 19 expression in hepatocellular carcinoma is regulated by fibroblast-derived HGF via a MET-Erk1/2-Ap1 and Sp1 axis. Cancer Res. 2018;78:1619–31. doi: 10.1158/0008-5472.CAN-17-0988. [DOI] [PubMed] [Google Scholar]
- 51.Liu C, Liu L, Chen X, et al. Lsd1 stimulates cancer-associated fibroblasts to drive Notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 2018;78:938–49. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 52.Chhabra Y, Weeraratna AT. Fibroblasts in cancer: unity in heterogeneity. Cell. 2023;186:1580–609. doi: 10.1016/j.cell.2023.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng D, Fu M, Wang M, et al. Targeting TGF-beta signal Transduction for fibrosis and cancer therapy. Mol Cancer. 2022;21:104. doi: 10.1186/s12943-022-01569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Najafi M, Farhood B, Mortezaee K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J of Cellular Biochemistry. 2019;120:2782–90. doi: 10.1002/jcb.27681. [DOI] [PubMed] [Google Scholar]
- 55.Schwörer S, Cimino FV, Ros M, et al. Hypoxia potentiates the inflammatory fibroblast phenotype promoted by Pancreatic cancer cell-derived Cytokines. Cancer Res. 2023;83:1596–610. doi: 10.1158/0008-5472.CAN-22-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun X, Wu B, Chiang H-C, et al. Tumour Ddr1 promotes collagen fibre alignment to instigate immune exclusion. Nature. 2021;599:673–8. doi: 10.1038/s41586-021-04057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Z, Tan Y, Cai Y, et al. Nanodrug removes physical barrier to promote T-cell infiltration for enhanced cancer Immunotherapy. Journal of Controlled Release. 2023;356:360–72. doi: 10.1016/j.jconrel.2023.02.029. [DOI] [PubMed] [Google Scholar]
- 58.Kuczek DE, Larsen AMH, Thorseth M-L, et al. Collagen density regulates the activity of tumor-infiltrating T cells. J Immunother Cancer. 2019;7:68. doi: 10.1186/s40425-019-0556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang PC-T, Chung JY-F, Xue VW-W, et al. Smad3 promotes cancer-associated fibroblasts generation via macrophage-Myofibroblast transition. Adv Sci (Weinh) 2022;9:2101235. doi: 10.1002/advs.202101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeh C-R, Slavin S, Da J, et al. Estrogen receptor alpha in cancer associated fibroblasts suppresses prostate cancer invasion via reducing Ccl5, Il6 and macrophage infiltration in the tumor Microenvironment. Mol Cancer. 2016;15:7. doi: 10.1186/s12943-015-0488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho H, Seo Y, Loke KM, et al. Cancer-stimulated Cafs enhance monocyte differentiation and Protumoral TAM activation via Il6 and GM-CSF secretion. Clin Cancer Res. 2018;24:5407–21. doi: 10.1158/1078-0432.CCR-18-0125. [DOI] [PubMed] [Google Scholar]
- 62.Timperi E, Gueguen P, Molgora M, et al. Lipid-associated Macrophages are induced by cancer-associated fibroblasts and mediate immune suppression in breast cancer. Cancer Res. 2022;82:3291–306. doi: 10.1158/0008-5472.CAN-22-1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request.