Abstract
We have recently introduced a novel procedure for the construction of herpesvirus mutants that is based on the cloning and mutagenesis of herpesvirus genomes as infectious bacterial artificial chromosomes (BACs) in Escherichia coli (M. Messerle, I. Crnković, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski, Proc. Natl. Acad. Sci. USA 94:14759–14763, 1997). Here we describe the application of this technique to the human cytomegalovirus (HCMV) strain AD169. Since it was not clear whether the terminal and internal repeat sequences of the HCMV genome would give rise to recombination, the stability of the cloned HCMV genome was examined during propagation in E. coli, during mutagenesis, and after transfection in permissive fibroblasts. Interestingly, the HCMV BACs were frozen in defined conformations in E. coli. The transfection of the HCMV BACs into human fibroblasts resulted in the reconstitution of infectious virus and isomerization of the reconstituted genomes. The power of the BAC mutagenesis procedure was exemplarily demonstrated by the disruption of the gpUL37 open reading frame. The transfection of the mutated BAC led to plaque formation, indicating that the gpUL37 gene product is dispensable for growth of HCMV in fibroblasts. The new procedure will considerably speed up the construction of HCMV mutants and facilitate genetic analysis of HCMV functions.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus group, is a pathogen of worldwide importance that can cause severe disease in newborns and immunocompromised individuals (5). The linear, double-stranded DNA genome of HCMV (230 kb) is the largest among mammalian DNA viruses and has the potential to encode more than 200 different proteins (7, 8). Our understanding of the genetic program of HCMV is rather incomplete, owing to the difficulties in constructing HCMV mutants. Disruption of herpesvirus genes is conventionally achieved by the integration of a marker gene into the viral genome through homologous recombination in permissive cells (18, 35, 43, 48). This procedure is especially cumbersome with HCMV, since the slow replication kinetics and cell association of HCMV impede the isolation of viral mutants. The reconstitution of replication-competent genomes from overlapping fragments cloned as cosmids represents a recently achieved improvement (25, 33). This procedure leads to the reconstitution only of mutant virus and circumvents the tedious selection procedure against the parental virus. However, it still relies on several recombination events in eukaryotic cells that cannot be controlled.
We have recently developed a completely new approach for the construction of herpesvirus mutants that is based on cloning and mutagenesis of herpesvirus genomes as bacterial artificial chromosomes (BACs) in Escherichia coli (32). This approach has clear advantages. The powerful methods of bacterial genetics allow the introduction of any kind of mutation (deletion, insertion, or point mutation) into the cloned viral genome. Manipulated genomes can be characterized prior to the reconstitution of virus. The transfection of the mutated genomes into permissive cells results in the reconstitution only of viral mutants that are free of contamination with parental virus.
Since its original description for mutagenesis of mouse cytomegalovirus (MCMV) the BAC technique has been adopted for other herpesviruses (14, 46, 49). The application of the technique to the mutagenesis of HCMV was highly desirable. There are, however, differences in genome arrangement between MCMV and HCMV. The MCMV genome is represented by one unique sequence with only a few small direct and indirect repeats, whereas the HCMV genome has a type E sequence arrangement (45). That is, two unique segments, the unique long (UL) and the unique short (US) components, are flanked by large inverted repeat regions. The unique sequences can invert relatively to each other, yielding four isomeric forms of the HCMV genome. It was unclear whether the inverted repeats would have an effect upon the cloning and propagation of the HCMV genome as a BAC plasmid in E. coli.
Here we describe the cloning of the HCMV AD169 strain genome as infectious BAC plasmids in E. coli. The HCMV BACs could be stably maintained in bacteria. After transfection of the HCMV BACs into human fibroblasts, infectious virus progeny was recovered. To assess the feasibility of mutagenesis of the cloned HCMV genome, a viral gene (gpUL37) was disrupted. The transfection of mutated BAC DNA into permissive cells led to plaque formation, indicating that gpUL37 is nonessential for viral growth in fibroblasts. Thus, we have successfully introduced the BAC mutagenesis technique for the construction of HCMV mutants.
MATERIALS AND METHODS
Virus and cells.
The HCMV strain used was ATCC AD169 (kindly provided by E. S. Mocarski, Stanford University, Stanford, Calif.). The genome of this strain contains the 11.3-kbp HindIII M fragment (13, 36). Human foreskin fibroblasts (HFF) and MRC-5 cells (human embryonic lung fibroblasts; ATCC CCL-171) were cultured in Dulbecco’s modified Eagle medium (GIBCO-BRL) supplemented with 5% fetal calf serum (GIBCO-BRL), 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml. For the preparation of virus stocks, HFF were infected with 0.01 PFU/cell and cultured until a marked cytopathic effect was seen. Virus stocks were prepared from sonicated cells and sedimented virions (in a Sorvall model SLA-1500 rotor for 2 h at 11,000 rpm and 4°C) and stored at −70°C in a 1:1 mixture of medium and autoclaved 9% nonfat milk. Virus titers were determined by standard plaque assay on HFF cultured in a medium containing human gamma globulin (200 μg/ml). For growth curves, HFF cells plated in six-well dishes were infected at a multiplicity of infection (MOI) of 0.1. After adsorption for 1 h, the inoculum was replaced with fresh culture medium. Supernatants of three independent samples were harvested at each time point and stored at −70°C. Infectious virus was quantitated by plaque assays on HFF cells.
Generation of the HCMV recombinant.
Recombinant HCMV was generated by homologous recombination in HFF cells according to published protocols (18). Briefly, 30 μg of the recombination plasmid pEB1097 was linearized by XcmI digestion and electroporated into 1.5 × 107 HFF cells at 960 μF and 220 V with a Gene Pulser unit (Bio-Rad). One day posttransfection cells were infected with AD169 at an MOI of 3. At 100% cytopathic effect (CPE) supernatant was transferred to a new flask of HFF cells and selection was applied (100 μM mycophenolic acid and 25 μM xanthine). Three rounds of selection were performed for the amplification of recombinant viruses before circular virus DNA was prepared from infected cells. Circular virus DNA was isolated by the method of Hirt (20, 32) and electroporated into E. coli DH10B as described previously (47).
Plasmids.
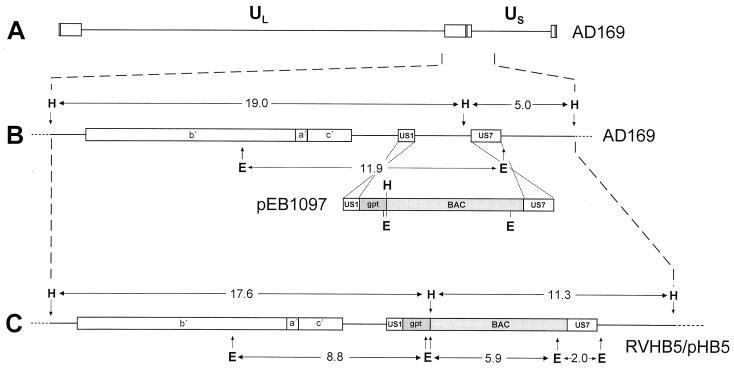
Plasmid pON2244 (19, 31) contains US1-US2 and US6-US7 sequences from HCMV AD169 (nucleotides [nt] 192648 to 193360 and 195705 to 197398 of the published HCMV sequence [8]), a tk-gpt-simian virus 40 poly(A) cassette (18) and a luciferase reporter gene. A 3.44-kbp BglII/PshAI fragment comprising the luciferase gene was excised from plasmid pON2244 and replaced by a 23-bp oligonucleotide linker containing a PacI restriction site (5′-gat cgc agc ctt aat taa cgt gg-3′). The BAC vector pKSO (32), a derivative of pBAC108L (47), was linearized with PacI and cloned into the PacI site of modified pON2244, resulting in recombination plasmid pEB1097 (Fig. 1).
FIG. 1.
Strategy for generation of the HCMV BAC plasmid. (A) The structure of the HCMV AD169 genome with the UL and US components is shown at the top. The terminal and internal repeat sequences are indicated as open boxes. (B) The BAC vector and the selection marker gpt were integrated between the US1 and US7 genes in the US region of the viral genome by homologous recombination in fibroblasts with the recombination plasmid pEB1097. (C) Genomic structure of the resulting reconstituted virus RVHB5 and the corresponding BAC plasmid pHB5. The sizes of expected HindIII (H) and EcoRI fragments (E) in the parental virus genome and in the RVHB5 genome and BAC plasmid pHB5 are indicated. b′, a′, and c′ mark components of the internal repeat sequences. The illustration is not drawn to scale.
To construct the mutagenesis plasmid pSH37b the polylinker of pBluescript II KS (Stratagene) was removed by KpnI/SacI digestion and replaced with a 24-bp BglII adapter (5′-ggc gcc tag atc tcg gtc aga gct-3′). A 9.7-kbp BglII fragment comprising the UL37 gene (nt 47366 to 57120 of ATCC AD169 [13, 36]) was excised from cosmid pCM1017 (15) and cloned into the BglII site of the modified pBluescript II KS. A 382-bp fragment of the gpUL37 open reading frame (ORF) (nt 50395 to 50777 [8]) was deleted by SnaBI digestion and replaced by a 2.6-kbp tetracycline resistance gene from pCP16 (9). An 8.9-kbp BglII/DraI fragment comprising the tetracycline cassette and flanking viral sequences (nt 47366 to 50395 and 50777 to 54041 [8]) was excised from the resulting plasmid (see Fig. 5) and cloned into the shuttle plasmid pST76K_SacB. pST76K_SacB is a derivative of the shuttle plasmid pST76K (42) that contains the Bacillus amyloliquefaciens sacB gene (16). The sacB gene was utilized to select for clones that have resolved the cointegrates and lost the shuttle plasmid by streaking the E. coli bacteria on agar plates containing 5% sucrose (4) (see below).
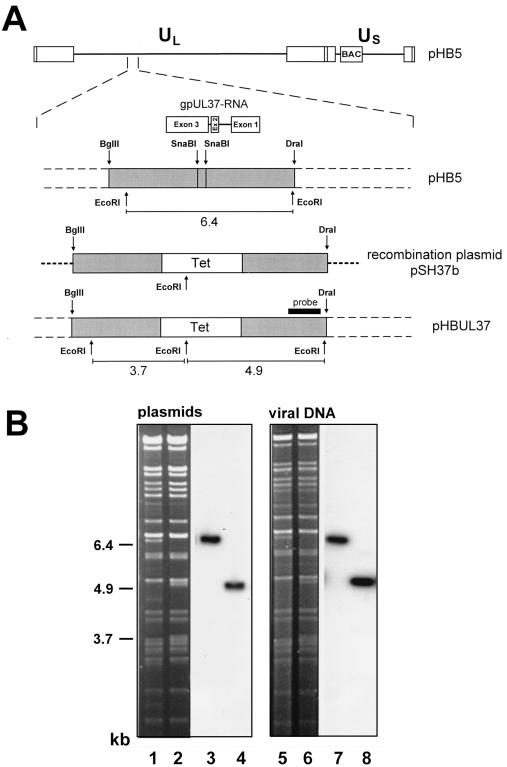
FIG. 5.
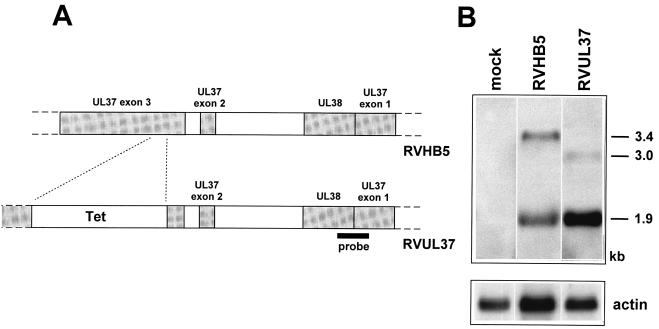
Construction scheme of the gpUL37 mutant (A) and structural analysis of the mutated BAC plasmid and mutant genome (B). (A) The top line depicts the genomic structure of the HCMV BAC plasmid pHB5, with the region encoding the gpUL37 RNA expanded below. Following recombination in E. coli between BAC plasmid pHB5 and recombination plasmid pSH37b, a 382-bp SnaBI fragment in exon 3 of the gpUL37 gene was replaced by a tetracycline resistance marker. The insertion disrupts the gpUL37 ORF after 199 codons and creates a new stop codon after an additional four codons. The sizes of the EcoRI fragments in the parental BAC plasmid pHB5 and mutant BAC plasmid pHBUL37 are indicated. The probe used for Southern blot analysis is depicted as a black bar. (B) DNA of BAC plasmids pHB5 (lane 1) and pHBUL37 (lane 2) and of the reconstituted viruses RVHB5 and RVUL37 (lanes 5 and 6) was digested with EcoRI and separated on a 0.5% agarose gel. Bands were visualized with ethidium bromide, transferred to nylon filters, and hybridized to the radiolabeled probe (lanes 3, 4, 7, and 8). The sizes of the EcoRI Q fragment (6.4 kbp) in the parental genome and of the new EcoRI fragments (3.7 and 4.9 kbp) in the mutant genome are indicated at the left margin.
BAC mutagenesis.
Mutagenesis of the HCMV BAC plasmid was performed by a two-step replacement procedure as described previously (2, 32, 38). Briefly, shuttle plasmid pSH37b was electroporated into E. coli CBTS bacteria (26) that already contained the HCMV BAC plasmid. The E. coli CBTS strain is recA positive at 30°C and recA negative at temperatures higher than 37°C (the strain was constructed by Michael O’Connor, University of California, Irvine). Transformants were selected at 30°C on Luria-Bertani (LB) agar plates containing chloramphenicol (12.5 μg/ml) and tetracycline (10 μg/ml). Clones containing cointegrates were identified by streaking the bacteria onto new LB plates with chloramphenicol and tetracycline and incubating them at 43°C. To allow resolution of the cointegrates clones were streaked onto LB plates containing chloramphenicol and incubated at 30°C. To select for clones that had resolved the cointegrate and that contained the mutant BAC plasmid, bacteria were restreaked onto LB plates containing chloramphenicol, tetracycline, and 5% sucrose. Resolution of the cointegrate was confirmed by testing for the loss of the kanamycin marker encoded by the shuttle plasmid. BAC plasmid DNA was isolated from 10-ml overnight cultures by the alkaline lysis procedure (30) and characterized by restriction enzyme analysis. Large preparations of HCMV BAC plasmids were obtained from 100-ml E. coli cultures by using Nucleobond PC 100 columns (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer.
Reconstitution of HCMV BAC virus.
MRC-5 cells (4 × 105 cells/well) were seeded into six-well dishes 1 day before transfection. About 0.5 to 1 μg of HCMV BAC plasmid DNA and 1 μg of plasmid pcDNApp71tag encoding the HCMV tegument protein pp71 were cotransfected by using the Superfect transfection reagent (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The pp71 expression plasmid pcDNApp71tag was constructed and kindly provided by B. Plachter, University of Mainz, Mainz, Germany. After transfection cells were propagated in a normal culture medium for 7 days. The cells were then split (1:3) and cultivated until plaques appeared. The supernatant of these cultures was used to infect new cells for the preparation of virus stocks.
Viral nucleic acid isolation and analysis.
Infected cells were harvested when cultures reached 100% CPE and collected by centrifugation. Cells were lysed in a solution containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.5% sodium dodecyl sulfate (SDS), and proteinase K digestion (500 μg/ml) was performed overnight at 56°C. Total DNA was extracted with phenol-chloroform and precipitated with isopropanol. After restriction enzyme digestion, DNA fragments were separated by electrophoresis on 0.5% agarose gels in 1× Tris-borate-EDTA buffer for 14 to 18 h at 2.5 V/cm. DNA fragments were visualized by ethidium bromide staining, denatured, and transferred to nylon membranes. DNA probes were radiolabeled with [α-32P]dCTP (Amersham) by using a random prime labeling kit (Pharmacia). Prehybridization was performed at 65°C for 1 h with 0.5% blocking reagent (Boehringer Mannheim) in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% N-lauroylsarcosine–0.02% SDS. Filters were hybridized with radiolabeled probes in the same solution at 65°C for about 12 h. Washing of the filters was done twice for 10 min each in 3× SSC–0.1% SDS, twice in 1× SSC–0.1% SDS, and twice in 0.3× SSC–0.1% SDS at 65°C. Results of the Southern blot experiments were obtained by autoradiography on Kodak BIOMAX MR films. DNA fragments for Southern hybridization were excised from plasmids by using appropriate restriction enzymes and purified from agarose gels: the US1-specific probe was isolated as a 750-bp SalI-NotI fragment (nt 192648 to 193360 of AD169 [8]) from pON2244, and a 1.4-kbp EcoRI-XbaI fragment from pKSO (32) was used as a BAC-specific probe. An 870-bp PvuI-EcoRI fragment (nt 53147 to 54014 of AD169 [8]) from shuttle plasmid pSH37b was used to characterize the structure of the mutant BAC plasmid.
Total RNA was purified from infected cells by using the RNeasy-Kit (Qiagen) and following the instructions of the manufacturer. When used, cycloheximide (Sigma; 50 μg/ml) was added to the culture medium commencing 1 h prior to infection. For Northern blot analysis, 3-μg RNA samples were separated by electrophoresis on denaturing formaldehyde–0.9% agarose gels, transferred to nylon membranes, and UV cross-linked. A 1.17-kbp BamHI fragment (nt 51463 to 52631 of AD169 [8]) was excised from plasmid pSH37b and labeled with the North2South biotin random prime labeling kit (Pierce, Rockford, Ill.). Hybridization of the filters to the probes was performed by using the North2South chemiluminescent nucleic acid hybridization and detection kit (Pierce) according to the manufacturer’s instructions. Filters were washed three times for 20 min at 55°C in 2× SSC–0.1% SDS. Results were visualized by chemiluminescence on Kodak BIOMAX MR films.
RESULTS
Strategy for cloning the HCMV genome as a BAC plasmid.
To clone the HCMV genome as a BAC in E. coli, we chose the same approach that we recently used for the successful cloning of the MCMV genome (32). This approach requires as a first step the insertion of the BAC vector sequences into the HCMV genome. BAC vector integration is achieved by homologous recombination between a recombination plasmid and the viral genome in human fibroblasts. After the generation and amplification of the recombinant virus, circular virus DNA is isolated from infected cells and electroporated into E. coli. Once the HCMV genome is cloned as a BAC plasmid in E. coli, it can be manipulated by the powerful methods of bacterial genetics. The transfection of mutated BAC plasmids into permissive cells leads to the reconstitution of viral mutants.
Generation of the HCMV BAC genome.
We decided to insert the BAC vector sequences between the US1 and US7 genes of the US component of the HCMV genome (Fig. 1), since ORFs IRS1 to US11 are nonessential for the replication of HCMV in cell culture (23, 24, 27). To this end, we constructed the recombination plasmid pEB1097 (Fig. 1B) that contained the BAC vector sequences and the selection marker guanine phosphoribosyl transferase (gpt) (18) flanked by viral sequences comprising the US1 and US7 genes. The flanking viral sequences provided the substrate for homologous recombination between the recombination plasmid and the HCMV genome. The BAC vector sequences comprise a chloramphenicol resistance gene and elements from the E. coli F factor that are required for the stable maintenance and partition of large plasmids in E. coli (47). The linearized recombination plasmid pEB1097 was electroporated into human fibroblasts, followed by infection with HCMV strain AD169. The correct insertion of the BAC vector should result in the formation of a recombinant HCMV BAC genome (Fig. 1C). Recombinant viruses were amplified by selection with xanthine and mycophenolic acid by utilizing the selection marker gpt (18). Finally, circular intermediates of the recombinant viral genome were isolated from infected cells by following the Hirt extraction procedure (20) and electroporated into E. coli DH10B (47).
Characterization of the HCMV BAC plasmids.
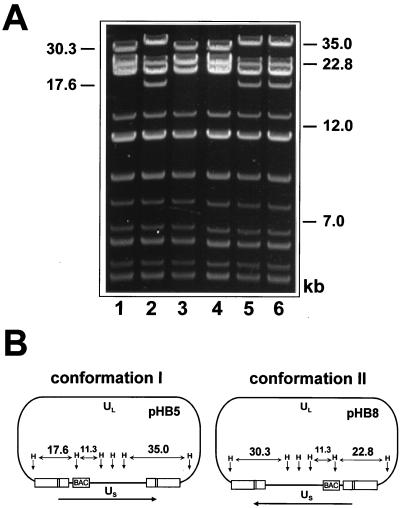
BAC plasmids were isolated from chloramphenicol-resistant bacterial clones and characterized by HindIII digestion. Figure 2 shows the HindIII pattern of the BAC plasmids of six representative clones. Two different HindIII patterns were observed. The sizes of the HindIII fragments and the expected alterations resulting from the insertion of the BAC vector (Fig. 1C) were predicted from the sequence of the HCMV AD169 genome (8). The BAC plasmids in clones 1, 3, and 4 contained a unique fragment of 30.3 kbp (Fig. 2A, lanes 1, 3, and 4), whereas unique fragments of 35 and 17.6 kbp were observed in clones 2, 5, and 6 (Fig. 2A, lanes 2, 5, and 6). In virions the linear HCMV genome is found in four different isomeric forms (34). Therefore, we supposed that the different HindIII patterns of the BAC plasmids reflect different isomers of the HCMV genome. After circulation of the HCMV genome only two isoforms are possible (Fig. 2B). Since there are no terminal fragments in a circular molecule, it is impossible to decide whether the US and UL region are inverted relative to each other. For conformation I, represented by BAC plasmid pHB5, two unique HindIII fragments of 35.0 and 17.6 kbp were predicted. For conformation II, represented by BAC plasmid pHB8, unique HindIII fragments of 30.3 and 22.8 kbp were expected (Fig. 2B).
FIG. 2.
Characterization of HCMV BAC plasmids. (A) BAC plasmids were isolated from six independent E. coli clones, digested with HindIII, and separated by agarose gel electrophoresis. The sizes of some DNA fragments and positions of DNA size markers are indicated. (B) Predicted structures of the two different conformations of the HCMV BAC plasmid. The positions of some HindIII sites (H) and the sizes of the unique HindIII fragments are shown.
Size variation between some clones was observed for the fragment with a predicted size of 17.6 kbp (data not shown). This fragment contains the α sequence which is known to occur in variable copy numbers within the terminal and internal repeats (34). The size variation of the fragment can be explained by different copy numbers of the α sequence in individual HCMV BAC plasmids. A new 22.8-kbp fragment predicted for conformation II (Fig. 2B) comigrates with the 22.8-kbp HindIII D fragment and therefore results in a double band in lanes 1, 3, and 4 of Fig. 2A. Altogether, the expected HindIII patterns were confirmed by the observed patterns of the BAC plasmids. Thus, we obtained clones with the two circular isomeric forms of the HCMV genome as BAC plasmids.
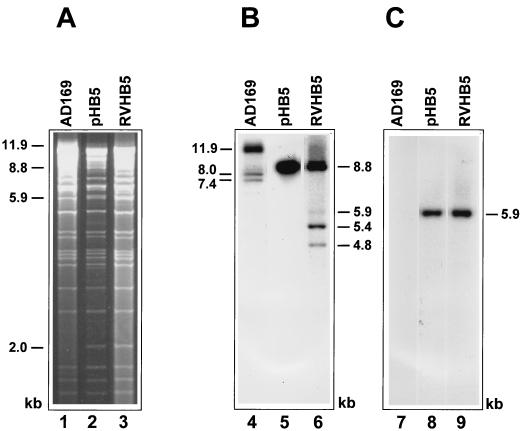
Additional characterization of the HCMV BAC plasmids was performed by EcoRI digestion. Digestion with EcoRI does not permit the discrimination between different isomeric forms. Therefore, only the data for the BAC plasmid pHB5 are shown. The EcoRI pattern of BAC plasmid pHB5 is almost identical to the pattern of HCMV AD169 DNA which was isolated from infected cells (Fig. 3A; compare lanes 1 and 2). One additional EcoRI fragment of 2.0 kbp was seen in the BAC plasmid pHB5. This new fragment resulted from the correct insertion of the BAC vector sequences in the HCMV genome (Fig. 1C). On another gel the EcoRI-digested DNAs were separated for a longer time, and Southern blot analysis was performed to visualize the additional two EcoRI bands (5.9 and 8.8 kbp) that were expected after the correct integration of the BAC vector (Fig. 1C). A US1-specific probe detected the 11.9-kbp EcoRI H fragment and fragments of 8.0 and 7.4 kbp in AD169 DNA (Fig. 3B, lane 4). The latter two bands represent expected terminal fragments of the linear AD169 genome with and without an a sequence, respectively (8, 15). An 8.8-kbp band was detected only in the BAC plasmid pHB5 as expected (Fig. 3B, lane 5; compare Fig. 1C). Hybridization with a BAC vector-specific probe showed a distinct signal at 5.9 kbp only with DNA of BAC plasmid pHB5 but not with the parental HCMV DNA (Fig. 3C, lanes 7 and 8). Characterization with US2- and US7-specific probes provided further evidence that the BAC vector was integrated at the expected site of the viral genome (data not shown). These results demonstrate the successful cloning of the HCMV genome as a BAC plasmid in E. coli.
FIG. 3.
Structural analysis of the HCMV BAC plasmid pHB5 and of the genome of reconstituted virus RVHB5. (A) DNA of HCMV AD169 (lane 1), BAC plasmid pHB5 (lane 2), and reconstituted virus RVHB5 (lane 3) was digested with EcoRI and separated on a 0.8% agarose gel. Note that the 2.0-kbp fragment resulting from the integration of the BAC vector is present only in the BAC plasmid pHB5 and in the RVHB5 genome (compare to Fig. 1). (B and C) Southern blot analysis was performed with a US1 probe (lanes 4, 5, and 6) and a BAC-specific probe (lanes 7, 8 and 9). The sizes of relevant EcoRI fragments are indicated.
Reconstitution of infectious virus from the HCMV BAC plasmids.
To reconstitute infectious virus the BAC plasmid pHB5 was transfected into MRC-5 cells. Viral plaques typically appeared 7 to 10 days after transfection. Cotransfection of a pp71 expression plasmid led to a 30-fold enhancement in plaque formation as described previously (3). Supernatants from the transfected cultures were used to infect monolayers of HFF cells, and total DNA was isolated from infected cells when 100% CPE was observed. The isolated DNA was digested with EcoRI, and the restriction enzyme pattern (Fig. 3, lane 3) was compared to the EcoRI pattern of HCMV AD169 DNA (Fig. 3, lane 1) and that of the BAC plasmid pHB5 (Fig. 3, lane 2). The EcoRI restriction patterns of the genomes of reconstituted virus RVHB5 and of BAC plasmid pHB5 showed no difference (Fig. 3, compare lanes 2 and 3). The 2.0-kbp band characteristic of the BAC plasmid pHB5 and the RVHB5 virus genome was clearly present, and the existence of the 8.8- and 5.9-kbp fragments that resulted from integration of the BAC vector was confirmed by Southern blot analysis (Fig. 3, lanes 5 and 6 and lanes 8 and 9, respectively). Additional bands of 4.8, 5.4, and 5.9 kbp were detected in the EcoRI-digested DNA of RVHB5 (Fig. 3, lane 6). These bands represent expected terminal fragments (containing zero, one, and two copies of the a sequence, respectively) of the linear DNA of RVHB5 that arise after inversion of the US component of the genome. These data demonstrate that the viral genomes were reconstituted from the BAC plasmid pHB5. Reconstitution was highly reproducible. Ten independent transfections were performed, and each transfection led to plaque formation. The EcoRI patterns of the reconstituted viral genomes were identical and indistinguishable from the EcoRI pattern of the BAC plasmid pHB5 (data not shown). The reconstituted virus RVHB5 was passaged six times on human fibroblasts to test the stability of the virus genome. After each round of infection DNA was prepared from infected cells and analyzed by EcoRI digestion. We did not observe any alterations of the DNA restriction patterns (data not shown). Thus, the genome of the recombinant virus RVHB5 is stable in cell culture, and the BAC vector sequence remains integrated in the virus genome. Comparable results were obtained after the transfection of the BAC plasmid pHB8 (data not shown).
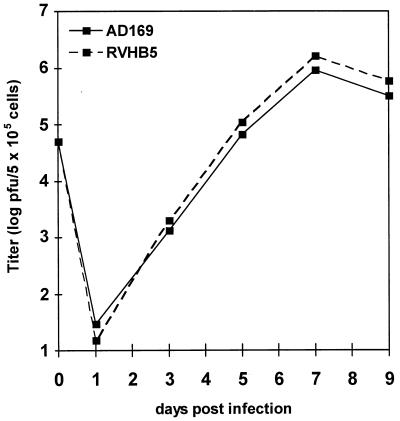
By microscopical inspection the recombinant virus RVHB5 and the parental virus strain AD169 showed the same plaque morphology. To characterize the growth properties of RVHB5, multiple-step growth curve analysis of the RVHB5 and AD169 viruses was performed (Fig. 4). After infection of fibroblasts at an MOI of 0.1, RVHB5 exhibited the same growth kinetics as the AD169 virus, and comparable titers of infectious virus were obtained in the supernatants of both the RVHB5- and AD169-infected cultures. These data confirm that the US2 to US6 genes are nonessential and that insertion of the BAC vector sequence does not exert a deleterious effect on the growth of HCMV in fibroblasts.
FIG. 4.
Multiple-step growth curve analysis of the reconstituted virus RVHB5 and of the parental HCMV strain AD169. HFF cells seeded in six-well dishes (5 × 105 cells/well) were infected with an MOI of 0.1. At the indicated time points (days postinfection) supernatants from the infected cultures were harvested, and total PFU of infectious virus in the culture supernatants were determined by plaque assay on HFF cells. Each data point represents the average of three independent wells. Day 0 titers represent input inocula.
Isomerization of the genome conformations in cells.
In cell cultures the HCMV genome occurs in four isomeric forms. Even after plaque purification, separation of the different isoforms was not possible (34). In E. coli, however, the conformation of the HCMV genome was fixed in the BAC plasmids because the inversion of the unique genome components does not occur in recombination-deficient E. coli DH10B. The transfection of both BAC plasmids pHB5 and pHB8 led to plaque formation, confirming that both genome conformations give rise to infectious virus. To analyze whether an isomerization of the genome conformations takes place in fibroblasts, circular viral DNA was prepared from cells infected with the progeny of either pHB5- or pHB8-transfected cells and electroporated into E. coli. BAC plasmids were isolated from individual bacterial clones and analyzed by HindIII digestion. Of 39 BAC plasmids obtained from the progeny of pHB5-transfected cells, 20 had conformation I and 19 had conformation II. Both isomeric forms were also found in clones obtained from the progeny of pHB8-transfected cells (25 BAC plasmids with conformation I and 8 BAC plasmids with conformation II). Thus, the isomerization of the different conformations occurs rapidly in infected cells.
Mutagenesis of the HCMV BAC plasmid.
In order to prove that the BAC-cloned HCMV genome can be manipulated by mutagenesis in E. coli, we decided to disrupt an ORF within the HCMV genome. Only a few gene loci are transcribed during the immediate-early (IE) phase of infection: UL36-38, UL122-123, UL115-119, TRS1/IRS1, and US3 (10). It is assumed that IE genes play important roles in the initiation of the HCMV infection. However, some of these genes (US3 and IRS1) have been shown to be nonessential for replication in cell cultures (23, 27), and the ie1 gene (UL123) is required only under certain conditions of infection (17, 33). Essential functions of the UL36-38, UL122, and UL115-119 genes have not been tested so far. Transients transfection studies have shown that gene products of the UL36-38 locus are required for oriLyt-dependent DNA replication (21, 39, 40). These results have not been confirmed with HCMV mutants, and it is not known which one of the UL36-38 gene products is essential for DNA replication. The UL36-38 transcription unit gives rise to several overlapping transcripts, some of them being multiply spliced. At least three transcripts are expressed at IE times of infection, a 1.8-, a 1.9-, and a 3.4-kb transcript (28, 50, 51). The 3.4-kb transcript encodes a type I transmembrane glycoprotein, gpUL37, consisting of 487 amino acids (aa). Since gpUL37 traffics to the endoplasmic reticulum and the Golgi apparatus (1), we speculated that this protein may not be essential for the initiation of viral transcription. Exon 1 of the gpUL37 ORF is shared with the UL37×1 ORF, whereas exons 2 and 3 are not part of the other transcripts encoded within the UL36-38 transcription unit (28, 50). For the selective and exclusive disruption of the gpUL37 ORF we decided to delete a 382-bp SnaBI fragment in the third exon of the UL37 gene and to replace it with a 2.6-kbp tetracycline resistance marker gene (Fig. 5A). Insertion of the tetracycline marker interrupts the gpUL37 ORF after codon 199 and creates a new stop codon after four additional codons. The truncated ORF can encode only a product of 199 aa that shares the amino-terminal 162 aa with the product of the 1.9-kb transcript. The expression of the C-terminal part of gpUL37 comprising the glycosylation, transmembrane and cytosolic domains should be abrogated.
The mutation was introduced into a 6.7-kbp BglII-DraI fragment (nt 47366 to 54041 of AD169 [8]) and subsequently transferred to the BAC plasmid pHB5 by homologous recombination in E. coli CBTS as described in Materials and Methods. The structure of the mutant BAC plasmid pHBUL37 was characterized by EcoRI digestion and compared to the parental BAC plasmid pHB5. The 6.4-kbp EcoRI Q fragment in BAC plasmid pHB5 (Fig. 5B, lane 1) was not present in the mutant BAC plasmid. Instead, two new bands of 4.9 and 3.7 kbp were observed (Fig. 5B, lane 2). These new bands result from an additional EcoRI site in the tetracycline resistance gene (Fig. 5A). Southern blot analysis was performed with a PvuI-EcoRI fragment specific for the right part of the EcoRI Q fragment in order to confirm successful mutagenesis. As expected, this probe recognized the 6.4-kbp fragment in the parental BAC plasmid pHB5 (Fig. 5B, lanes 1 and 3), whereas in the mutated BAC plasmid pHBUL37 a signal was detected at 4.9 kbp (Fig. 5B, lanes 2 and 4). The correct insertion of the mutation was also tested by sequencing relevant parts of the mutated BAC plasmid. The data confirmed that the intended alteration was introduced into the HCMV BAC plasmid.
The gpUL37 gene is nonessential for replication of HCMV in fibroblasts.
To examine whether the gpUL37 gene is essential for replication of HCMV the mutant BAC plasmid pHBUL37 was transfected into MRC-5 cells. Plaques appeared 7 to 10 days posttransfection. The supernatant of the transfected cells was used to infect new fibroblasts, and total cell DNA was isolated from infected cells. EcoRI digestion of the viral DNA showed that the mutation was maintained in the genome of the reconstituted virus mutant RVUL37 (Fig. 5B, lane 6). The 6.4-kbp EcoRI Q fragment in the RVHB5 genome was replaced by two fragments of 4.9 and 3.7 kbp in the RVUL37 genome (Fig. 5B, compare lanes 5 and 6). In order to provide evidence that the RVUL37 mutant was reconstituted from the BAC plasmid pHBUL37, Southern blot analysis of the virus genomes was performed as described for the BAC plasmids. Hybridization signals were observed at 6.4 and 4.9 kbp for RVHB5 and RVUL37 as expected (Fig. 5B, lanes 7 and 8). Thus, the gpUL37 ORF was disrupted by the tetracycline resistance gene.
Northern blot analysis was performed in order to analyze the effect of the gene disruption on transcription of the UL37 gene. A probe was used that detects the 1.9-kb UL37 exon 1 transcript and the 3.4-kb gpUL37 transcript (Fig. 6A). In accordance with previous results (28, 50), the 3.4 kb gpUL37 transcript could be detected only in RNA from RVHB5-infected cells that were treated with cycloheximide for 8 h (Fig. 6B, lane 2). The 1.9-kb UL37 exon 1 transcript was detected in RNA from cells that were infected with the parental virus RVHB5 or with the mutant virus RVUL37, irrespective of whether the cells were treated with cycloheximide (Fig. 6B, lanes 2 and 3) or not (data not shown). The abundance of the 1.9-kb transcript was higher in RVUL37-infected cells than in RVHB5-infected cells. In addition, a novel transcript with a size of about 3 kb was found in RNA from cells infected with the mutant virus RVUL37 after treatment with cycloheximide for 8 h (Fig. 6B, lane 3). This transcript was not seen when the RVUL37-infected cells remained untreated (data not shown). The abundance of this transcript is lower than that of the 3.4-kb transcript in RVHB5-infected cells. Thus, the UL37 gene disruption results in altered transcription of the UL37 gene.
FIG. 6.
Transcript analysis of the RVUL37 mutant. (A) Genomic structure of the RVHB5 and RVUL37 viruses in the UL37 region. Exons are depicted in gray. In the genome of RVUL37 exon 3 of the UL37 gene is disrupted by the insertion of the tetracycline resistance gene (Tet). The probe used (black bar) is specific for the 1.9-kb UL37 exon 1 transcript and the 3.4-kb gpUL37 transcript (28, 50). (B) Northern blot of whole cell RNA isolated from mock infected HFF cells (lane 1) and from cells 8 h after infection with 3 PFU/cell of the parental virus RVHB5 (lane 2) or the mutant virus RVUL37 (lane 3) in the presence of cycloheximide. Molecular sizes (in kilobases) of the detected transcripts are shown at the right margin. The blot was rehybridized with a probe specific for β-actin as an internal RNA control. The actin band detectable in all lanes is shown on the bottom panel.
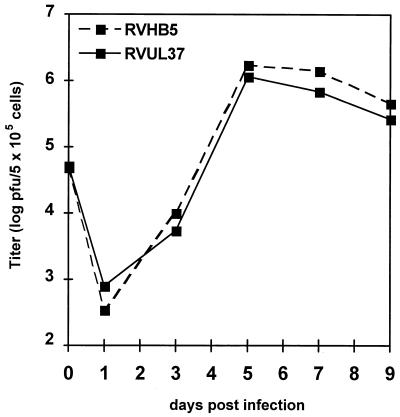
To test the consequences of the gpUL37 gene disruption on the growth properties of the UL37 mutant, a multiple-step growth curve analysis was performed. HFF cells were infected at an MOI of 0.1 with either the parental virus RVHB5 or the mutant virus RVUL37, and the virus titers in the extracellular fluid of infected cultures were determined. No significant difference was found between the growth kinetics of the two viruses (Fig. 7). We conclude from these data that the gpUL37 gene product is nonessential for the replication of HCMV in human fibroblasts.
FIG. 7.
Multiple-step growth curves of the mutant virus RVUL37 and of the parental virus RVHB5. HFF cells (5 × 105) were infected at an MOI of 0.1. Virus titers in the supernatants of infected cells were determined as described in the legend for Fig. 4.
DISCUSSION
In this study we describe the cloning and mutagenesis of the HCMV genome as an infectious BAC in E. coli. For generation of the HCMV BAC plasmid the BAC vector sequence was integrated into the HCMV genome by homologous recombination in HFF cells. Circular intermediates of the recombinant viral genome were isolated from infected cells and transformed into E. coli DH10B. The HCMV BAC plasmids could be stably maintained in E. coli and gave rise to infectious virus following transfection into human fibroblasts. A selected viral gene (gpUL37) was disrupted through homologous recombination in E. coli to demonstrate that the HCMV BAC genome can be easily and rapidly mutated in bacteria.
Following the transformation of the circular HCMV BAC genomes into E. coli we obtained BAC plasmids that contained the HCMV genome frozen in the different isomeric states. Inversion of the HCMV genome components does not occur in recombination-deficient E. coli DH10B. In eukaryotic cells fixation of the HSV and HCMV genomes in a defined conformation could be achieved only after the deletion of the internal repeat sequences (22, 34, 41). These virus genomes, which were locked in one of the four possible conformations, have been shown to be infectious. Our data confirm these previous observations by demonstrating that both isomeric forms of the HCMV BAC are infectious. When circular DNA of a virus population, which was reconstituted from a plasmid with a given conformation, was transformed back into E. coli, BAC plasmids representing the different isomeric forms were obtained. Thus, isomerization of the conformations seems to be a frequent event in human fibroblasts, and the kinetics of the isomerization process can now be analyzed in detail.
The transfection of HCMV BAC plasmids into human fibroblasts reproducibly led to a reconstitution of the desired virus genomes and recombinant viruses. The cotransfection of a pp71 expression plasmid enhanced the infectiousness of the HCMV BAC plasmids as described previously for HCMV DNA (3). This effect is probably mediated by the transactivating properties of pp71 on the major IE promoter and other viral promoters (3, 29). We never observed any rearrangements or deletions in the reconstituted viral genomes. Thus, we conclude that the integrity of the HCMV BAC is unaffected by the transfection procedure. After the reconstitution of virus from the BAC plasmids the vector sequences remain stable in the virus genome. The presence of the vector sequences will not interfere with most analyses. For MCMV, we have devised a strategy that allows the excision of the vector sequences during reconstitution of the virus genome (52). Flanking of the BAC vector sequences in the HCMV BAC plasmids with target sites for excision is under construction.
The transient expression of recombinogenic activities is required for targeted mutagenesis of the HCMV BAC plasmid. Mutagenesis was performed in E. coli CBTS, which is recA positive at 30°C and virtually recA negative at temperatures higher than 37°C. recA was expressed only as long as required. The recombinogenic activity in E. coli CBTS is low even at 30°C since large regions of homology (2 to 3 kbp) are required to achieve homologous recombination in this strain. The low-level recombinogenic activity guarantees, however, a high-level stability of the BAC plasmids. We never observed adventitious deletions or rearrangements in the HCMV BAC plasmids during propagation in E. coli CBTS or DH10B.
In principle, the large inverted repeats of the HCMV genome should provide optimal substrates for recombination. Homologous recombination would result in the inversion of the unique segments located between the repeats. Inversion of the UL and US segments of the HCMV BAC plasmid does indeed occur in recombination-proficient E. coli CBTS. However, such events seem to occur only very rarely since they could be detected only by Southern blot analysis (data not shown). The large distance between the inverted repeats and the low-level recombinogenic activity in this E. coli strain probably reduce the chance for recombination.
Recombination between direct repeats leads to the deletion of intervening sequences and to a reduction in the copy number of repeated sequences. The HCMV genome contains a number of short direct repeats, e.g., the a sequence (34). There is a natural variation in the copy number of the a sequence (34), suggesting that herpesviruses cannot keep the number of repeated sequences constant. The HCMV BAC plasmid pHB5 probably contains just one a sequence within the terminal and internal repeat sequences. One copy of the a sequence is sufficient for cleavage and packaging of the viral genome (34).
The probability for recA-mediated recombination is dependent on the length of the homologous sequences and on the presence of Chi sites (5′-GCTGGTGG-3′) within these sequences (37). The directly repeated sequences in the HCMV genome are generally short and do not contain Chi sites. Therefore, the risk of acquiring undesired deletions in the BAC plasmid by propagation in E. coli strains with low-level recombinogenic activity is considered to be low. Nevertheless, new phenotypes resulting from mutagenesis of the viral genome will require construction of revertant viruses in order to unequivocally attribute the phenotypes to the specifically introduced mutations. This requirement is independent of the mutagenesis procedure, i.e., whether the mutant viruses are generated by conventional recombination techniques in eukaryotic cells or by the BAC technique in E. coli.
Gene products encoded by ORFs UL36-38 are required for the initiation of viral DNA replication (39, 40). It is assumed that the UL36-38 proteins mediate this effect by their ability to transactivate HCMV early genes that encode members of the replication machinery (21). So far it has not been dissected which of the proteins encoded by the UL36-38 transcription unit is essential for viral replication. After the transfection of the mutant UL37 BAC plasmid, plaque formation was observed. Analysis of viral DNA confirmed that the gpUL37 gene was disrupted by the tetracycline resistance gene in the genome of the viral mutant. The UL37 mutant expresses an altered UL37 transcript with a size of about 3.0 kb. We assume that this transcript encodes a truncated gpUL37 protein. Further experiments are required to characterize this protein.
The UL37 mutant showed a pattern of growth kinetics which was comparable to that of the parental virus. We conclude from these data that gpUL37 is nonessential for the replication of HCMV in vitro. Our results are in agreement with a recent study which reported that the UL37×1 protein is much more effective than gpUL37 in the transactivation of HCMV early promoters and that the unique carboxy-terminal domain of gpUL37 downstream of aa 199 can be deleted without abrogating transactivation (11). The function of gpUL37 remains elusive. In fibroblasts the gpUL37 transcript is seen only under special infection conditions. Perhaps the gpUL37 gene function is apparent in cells other than fibroblasts. Further studies with the UL37 mutant in various cell types may help to elucidate the function of gpUL37.
Since its original description for MCMV (32) the method of cloning viral genomes as infectious BAC plasmids has been applied to several other herpesvirus genomes (14, 46, 49). This technique has major advantages over conventional recombination techniques performed with permissive eukaryotic cells. Site-directed and random mutagenesis (6) can be rapidly performed with the cloned herpesvirus genomes. Even the introduction of point mutations is feasible (32). Multiple alterations can be introduced into the cloned genome by successive rounds of mutagenesis without the need to reconstitute viral intermediates. The modified virus genome can be characterized prior to the reconstitution of virus progeny. In contrast, conventional recombination techniques allow the characterization of the mutant genome only at the very end of the mutagenesis procedure after several rounds of selection and plaque purification.
Although the BAC technique allows the construction of viral genomes with mutations in essential genes (6), complementing cell lines are still required for the reconstitution and propagation of viral mutants. The general feasibility of constructing complementing cell lines for CMV mutants has recently been demonstrated (12, 17, 33, 44), but the generation of such CMV mutants has not been achieved yet. The BAC technique will also be advantageous for this purpose, because it circumvents selection against the parental virus. Until now, the generation of viruses mutated in essential genes was dependent on the quality of the complementing cell line, which was difficult to determine. Since the construction of mutant genomes by the BAC technique in E. coli is independent of the phenotype of the viral mutant, BAC constructs mutated in essential genes may also serve as useful tools for evaluating the functionality and efficacy of complementing cell lines.
In summary, we have successfully extended the BAC cloning and mutagenesis technique to HCMV. The procedure will considerably speed up the construction of HCMV mutants. In addition, the generation of defined CMV vaccine strains now becomes feasible. In principle, the cloning and analysis of any CMV strain will be possible by the described technique. We expect that the new approach will facilitate genetic analysis of CMV functions and studies on CMV pathogenesis.
ACKNOWLEDGMENTS
Eva-Maria Borst and Gabriele Hahn contributed equally to this work.
We thank Edward Mocarski for providing HCMV ATCC AD169 and plasmid pON2244, Bodo Plachter for plasmid pcDNApp71tag, and György Pósfai for the shuttle plasmids.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF) (project 01GE9614), the Wilhelm Sander-Stiftung, and Sanderforschungsbereich SFB 455.
REFERENCES
- 1.Al-Barazi H O, Colberg-Poley A M. The human cytomegalovirus UL37 immediate-early regulatory protein is an integral membrane N-glycoprotein which traffics through the endoplasmic reticulum and Golgi apparatus. J Virol. 1996;70:7198–7208. doi: 10.1128/jvi.70.10.7198-7208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E., G. Pósfai, M. Wagner, and M. Messerle. Unpublished data.
- 5.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 6.Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski U H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 7.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchinson C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD 169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 10.Colberg-Poley A M. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL 36-38, UL 115-119, TRS1/IRS1 and US 3 loci. Intervirology. 1996;39:350–360. doi: 10.1159/000150506. [DOI] [PubMed] [Google Scholar]
- 11.Colberg-Poley A M, Huang L, Soltero V E, Iskenderian A C, Schumacher R-F, Anders D G. The acidic domain of pUL37×1 and gpUL37 plays a key role in transactivation of HCMV DNA replication gene promoter constructions. Virology. 1998;246:400–408. doi: 10.1006/viro.1998.9212. [DOI] [PubMed] [Google Scholar]
- 12.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dargan D J, Jamieson F E, Maclean J, Dolan A, Addison C, McGeoch D J. The published DNA sequence of human cytomegalovirus strain AD169 lacks 929 base pairs affecting genes UL42 and UL43. J Virol. 1997;71:9833–9836. doi: 10.1128/jvi.71.12.9833-9836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delecluse H-J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from procaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleckenstein B, Müller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 16.Gay P, LeCoq D, Steinmetz M, Ferrari E, Hoch J A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, G., M. A. McVoy, and E. S. Mocarski. Unpublished data.
- 20.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins F J, Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J Virol. 1986;59:494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones T R, Muzithras V P. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J Virol. 1992;66:2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones T R, Muzithras V P, Gluzman Y. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J Virol. 1991;65:5860–5872. doi: 10.1128/jvi.65.11.5860-5872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemble G, Duke G, Winter R, Spaete R. Defined large-scale alteration of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J Virol. 1996;70:2044–2048. doi: 10.1128/jvi.70.3.2044-2048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollert-Jöns A, Bogner E, Radsak K. A 15-kilobase-pair region of the human cytomegalovirus genome which includes US1 through US13 is dispensable for growth in cell culture. J Virol. 1991;65:5184–5189. doi: 10.1128/jvi.65.10.5184-5189.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 29.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.McVoy M A, Mocarski E S. Tetracycline-mediated regulation of gene expression within the human cytomegalovirus genome. Virology. 1999;258:295–303. doi: 10.1006/viro.1999.9724. [DOI] [PubMed] [Google Scholar]
- 32.Messerle M, Crnković I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1491aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 35.Mocarski E S, Post L E, Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980;22:243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- 36.Mocarski E S, Prichard M N, Tan C S, Brown J M. Reassessing the organization of the UL42-UL43 region of the human cytomegalovirus strain AD169 genome. Virology. 1997;239:169–175. doi: 10.1006/viro.1997.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers R S, Stahl F W. Chi and the RecBC D enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 38.O’Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 39.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pari G S, Kacica M A, Anders D G. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poffenberger K L, Tabares E, Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci USA. 1983;80:2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pósfai G, Koob M D, Kirkpatrick H A, Blattner F R. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol. 1997;179:4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 44.Prichard M N, Gao N, Jairath S, Mulamba G, Krosky P, Coen D M, Parker B O, Pari G S. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J Virol. 1999;73:5663–5670. doi: 10.1128/jvi.73.7.5663-5670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 46.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 47.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stavropoulos T A, Strathdee C A. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenney D J, Colberg-Poley A M. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 51.Tenney D J, Colberg-Poley A M. Human cytomegalovirus UL36-38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J Virol. 1991;65:6724–6734. doi: 10.1128/jvi.65.12.6724-6734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner M, Jonjić S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]