Abstract
Background:
Risk estimates for women carrying germline mutations in breast cancer susceptibility genes are mainly based on studies of European ancestry women.
Methods:
We investigated associations between pathogenic variants (PV) in 34 genes with breast cancer risk in 871 cases (307 estrogen receptor (ER)-positive, 321 ER-negative, and 243 ER-unknown) and 1,563 controls in the Ghana Breast Health Study (GBHS), and estimated lifetime risk for carriers. We compared results to those for European, Asian and African-American ancestry women.
Results:
The frequency of PV in GBHS for nine breast cancer genes was 8.38% in cases and 1.22% in controls. Relative risk estimates for overall breast cancer were: OR (95% CI)=13.70 (4.03–46.51) for BRCA1, 7.02 (3.17–15.54) for BRCA2, 17.25 (2.15–138.13) for PALB2, 5/0 cases/controls for TP53, and 2.10, (0.72–6.14) for moderate-risk genes combined (ATM, BARD1, CHEK2, RAD51C, RAD52D). These estimates were similar to those previously reported in other populations and were modified by ER status. No other genes evaluated had mutations associated at P<0.05 with overall risk. The estimated lifetime risks for mutation carriers in BRCA1, BRCA2 and PALB2 and moderate risk genes were 18.4%, 9.8%, 22.4% and 3.1%, respectively, markedly lower than in Western populations with higher baseline risks.
Conclusions:
We confirmed associations between PV and breast cancer risk in Ghanaian women and provide absolute risk estimates that could inform counseling in Ghana and other West African countries.
Impact:
These findings have direct relevance for genetic counseling in West Africa since currently available data is primarily from Western populations
Keywords: Breast Cancer, pathogenic germline variants, Ghana, absolute risk, population-based case-control study
Introduction
Gene panel testing for breast cancer predisposition is widely used in high-income countries; however, susceptibility gene risk estimates are often imprecise and derived primarily from family-based studies in European ancestry populations (1–3). Recent publications from three large studies in women of European, Asian, African-American (AA), and other racial/ethnic groups in the US have enhanced our understanding of the role genetic factors in breast cancer etiology (4–6). The Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES) study performed germline DNA panel testing for 34 putative breast cancer susceptibility genes in 60,466 cases and 53,461 controls of European and Asian ancestry (4). This study identified the most clinically useful genes for genetic testing as four high-risk (relative risk (RR) >4) genes, BRCA1, BRCA2, PALB2 and TP53; five moderate-risk (RR 2–4) genes, ATM, BARD1, CHEK2, RAD51C, and RAD51D, and provided precise RR estimates for overall and estrogen receptor (ER)-defined breast cancer. Palmer et al. investigated AA women (5,054 cases and 4,993 controls) and found similar results as reported by BRIDGES based on germline DNA panel testing of 23 genes (5). The Cancer Risk Estimates Related to Susceptibility (CARRIERS) study used the same sequencing panel as Palmer et al. to investigate a multi-ethnic US population (78% non-Hispanic White, 14% non-Hispanic Black, 4% Asian, 3% Hispanic, and 2% other) of 32,247 breast cancer cases and 32,544 controls and also reported similar results as BRIDGES(6). Notably, most AA women in Palmer et al. overlapped with the CARRIERS study population.
Although breast cancer incidence historically has been low in West African compared to Western countries, incidence rates in West Africa are rising. West African women are disproportionately affected by early-onset/ER-negative breast cancer, and low survival rates mainly due to advanced stages at diagnosis (7–9). To date, there are limited data on the association of germline mutations in breast cancer susceptibility genes with disease risk in West Africa (10–13). Population-based studies are needed to clarify the role of pathogenic mutations in breast cancer genes in African women to improve our understanding of breast cancer etiology and prevention strategies. In this report, we used the 34 gene panel from BRIDGES to sequence 871 breast cancer cases and 1,563 controls in the Ghana Breast Health Study (GBHS) (14–16). We estimated odds ratios (OR) of overall and ER-defined breast cancer for protein truncating variants (PTVs) and pathogenic missense variants, and we compared them to published estimates in European, Asian, and AA populations (4–6). We also provide population-specific lifetime breast cancer risk estimates for mutation carriers in Ghana.
Methods
Study Population:
The GBHS has been described in detail elsewhere (14–16). In brief, the GBHS is a population-based case-control study of breast cancer conducted in Accra and Kumasi, Ghana. The study enrolled breast cancer cases and frequency matched population-based controls using census-based sampling of women between the ages of 18 to 74 years of age. Cases were women recommended for a biopsy based on the suspicion of malignancy or presenting at a study hospital for treatment of pathologically documented breast cancer within the previous year. Controls included women who reported never having been diagnosed with breast cancer (14–16). We performed gene panel sequencing on 1,077 cases and 1,993 controls with an available source of germline DNA. After quality control, we included 871 breast cancer cases (829 pathologically confirmed invasive, 12 pathologically confirmed in situ, and 30 considered malignant based on clinical manifestations) and 1,563 controls (Supplementary Figure 1). The study was approved by the Special Studies Institutional Review Board of the National Cancer Institute (Rockville, MD), the Ghana Health Service Ethical Review Committee and institutional review boards at the Noguchi Memorial Institute for Medical Research (Accra, Ghana), the Kwame Nkrumah University of Science and Technology (Kumasi, Ghana), the School of Medical Sciences at Komfo Anokye Teaching Hospital (Kumasi, Ghana) and Westat (Rockville, MD). All participants provided written informed consent.
Laboratory methods, variant calling and classification:
Details on the laboratory methods, variant calling, and classification have been published (4). In brief, we analyzed a gene panel of 34 known or suspected breast cancer susceptibility genes. Library preparation was conducted using the Fluidigm Juno 192.48 system at the Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge, UK. Amplified products were combined into barcoded libraries of 768 samples, each of which was run on a single lane of an Illumina Hiseq4000. Each sample was sequenced to an average depth of 349 reads, in the target region.
Variant calling was performed using VarDict;(17) comparison with other callers indicated that this had much better specificity for this type of targeted sequencing (18). We applied the following filters at the VCF level: phred scaled sequencing quality assessment of the bases contributing to the variant (QUAL) <30, allele fraction (AF) <0.2 and mean mapping quality (MQMEAN) <60, mean number of mismatches per read (NM) >2.0, AFxBase Depth < 7.5. Variants failing any of these filters were removed. PTVs were defined as frameshifting insertions/deletions, stop/gain or canonical splice variants as classified by the Emsembl Variant Effect Predictor (VEP) (19), except for variants in the last exon of each gene, which were excluded from the primary analysis. Missense variants defined as pathogenic or likely pathogenic in ClinVar by two or more clinical laboratories (Ambry Genetics, SCRP, InVitae, GeneDX, Counsyl, InSiGHT) were considered pathogenic, the same criteria as applied by Palmer et al.in the study of AA women (20).
Statistical analysis:
We used Fisher’s Exact Test to compare carrier frequencies in GBHS to carrier frequencies in BRIDES and Palmer et al. We used logistic regression to perform burden analyses to estimate odds ratios (ORs) and 95% confidence intervals (95% CI) associated with carrying a pathogenic variant (PV) in each gene adjusting for age and family history. Because risk associations for PTVs and PVs in the moderate risk genes (ATM, BARD1, CHEK2, RAD51C, and RAD51D) were found to be similar by BRIDGES (4), and the GBHS sample size was too small to evaluate each gene separately, we tested for associations with breast cancer risk for these genes combined. Disease endpoints considered were overall (invasive or in-situ), ER-defined, and triple-negative (TN, defined as being negative for ER, PR and HER2) breast cancer. Case-only analyses were used to estimate heterogeneity P-values (Phet) in associations between carrying a PV with risk of ER-defined breast cancer. We compared frequencies and risk estimates of PTVs in breast cancer risk genes with those reported among European and Asian ancestries in the BRIDGES study (4), and the frequencies and risk estimates associated with all PVs in aggregate (defined as either PTVs and pathogenic missense variants) in breast cancer risk genes with those reported among AA and a multi-ethnic (primarily non-Hispanic White) US population by CARRIERS (5,6). For comparisons to the BRIDGES studies, we were able to compare overall breast cancer risk estimates for carrying PTVs in ATM, BARD1, BRCA1, BRCA2, CHEK2, MSH6, NF1, PALB2, PTEN, RAD51C, RAD51D, and TP53 separately to European and Asian ancestry women; BRIDGES did not report risk estimates for other genes separately by ancestry group (4). We generated z-scores and corresponding P-values to compare risk estimates across studies (P-diff). Population attributable risks (PAR) for pathogenic mutations were calculated based on the mutation frequencies in the control population and OR estimates.
Lifetime absolute risk estimates of overall and ER-defined breast cancer within the 18 to 74 years age range, the oldest age in the GBHS (female life expectancy in Ghana is 64.4 years(21)), for women carrying a PV were generated using the Individualized Coherent Absolute Risk Estimator tool (iCARE)(22). Absolute risk estimation using iCARE requires the odds ratios for disease risk, age-specific disease incidence rates and competing mortality rates and an individual level reference dataset of risk factors representing the underlying population (22). We used the estimated odds ratios for risk of breast cancer for carriers of pathogenic variants in BRCA1, BRCA2, PALB2, and the combined moderate risk genes. We leveraged the census-based sampling strategy to generate sampling fractions and estimate breast cancer incidence rates in Accra and Kumasi, as previously described in greater detail (23). The population distribution of carrying a pathogenic variant was estimated using the controls from our study, which provided the reference dataset for absolute risk estimation. The overall absolute risk estimation further accounted for competing mortality due to causes other than breast cancer using the competing mortality rates derived from published estimates of overall mortality (24) and the mortality due to breast cancer published by GLOBOCAN 2018 (25). We further derived 95% confidence intervals for lifetime absolute risk of breast cancer using bootstrap resampling. This approach assumed the disease incidence rates and competing mortality rates to be known with certainty. The log-odds ratio estimating the association of each carrier type with overall breast cancer or subtype-specific breast cancer asymptotically follows a normal distribution by the theory of maximum likelihood estimation. Each bootstrap iteration sampled the log-odds ratios and sampled the reference dataset with replacement. The confidence intervals were calculated using the 2.5 and 97.5 percentiles of the lifetime absolute risk distribution derived from 1000 bootstrap iterations.
A P-value threshold of 0.05 was used to denote statistically significant associations. Analyses were performed in R (version 3.4.2) and SAS (version 9.4).
Results
The GBHS study population included 871 breast cancer cases [307 ER-positive (48.9%), 321 ER-negative (51.1%), 243 without ER tumor status data, and 172 (28%) TN] and 1,563 controls. The mean (standard deviation) age (in years) of cases was slightly older (50.8 (12.0)) than controls (45.8 (12.7)), reflecting controls being initially frequency matched to women with a suspicion of breast cancer prior to diagnosis confirmation (Supplementary Table 1).
Frequency of PTV and missense pathogenic variants
Among the 34 genes investigated, the percentage of controls carrying at least one PTV across all genes was 3.97% and for overall, ER-positive, ER-negative, and TN cases the percentages were 11.94%, 13.68%, 12.46%, and 15.52%, respectively (Supplementary Table 2, Supplementary Figure 2). Supplementary Table 3 lists the identified PVs.
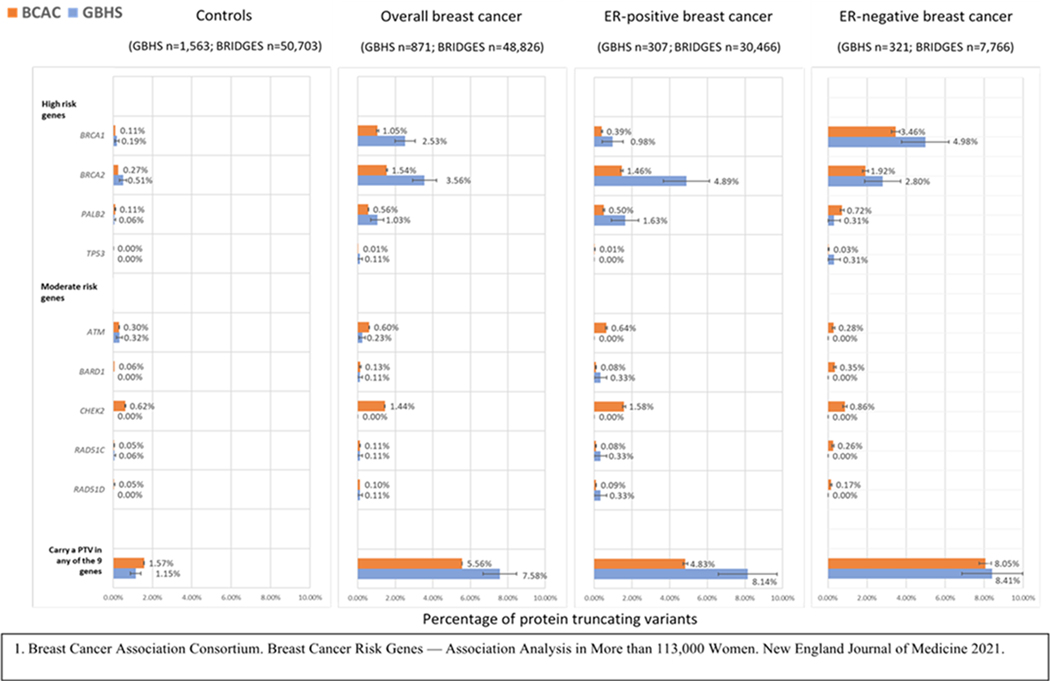
Figure 1 and Supplementary Table 4 shows PTV frequencies in GBHS and BRIDGES for the 9 genes (ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, RAD51C, RAD51D, and TP53) that were associated with breast cancer risk at a Bayesian False Discovery Probability (BFDP) <5% in BRIDGES (4). The frequency of carrying at least one PTV in these nine genes was 1.15% in controls and 7.58%, 8.14%, and 8.41% in overall, ER-positive, and ER-negative disease, respectively. Women in GBHS had approximately two-times the frequency of PTVs in BRCA1 and BRCA2 than women of European or Asian ancestry in the BRIDGES study (4) for both controls and overall breast cancer cases; however, differences were statistically significant only for overall breast cancer. For mutation frequencies in ER-defined disease the largest differences were for PTVs in ER-positive cases for BRCA2 and PALB2. There was also a higher frequency of TP53 PTVs for ER-negative cases in GBHS than BRIDGES. No controls carried a TP53 PTV, and no cases or controls carried a CHEK2 PTV. Seven cases and one control carried missense variants classified as pathogenic in ATM, BRCA1, CHEK2, and TP53 (Supplementary Tables 5-6).
Figure 1.
Comparison of the frequency of protein truncating variants (PTVs) in breast cancer genes for controls and breast cancer cases from the Ghana Breast Health Study (GBHS) and the Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES) study (European and Asian ancestries combined). Includes genes with PTVs reported associated with risk of breast cancer at a Bayesian False Discovery Probability < 5% in BRIDGES1. See Supplemental Table 4 for more details.
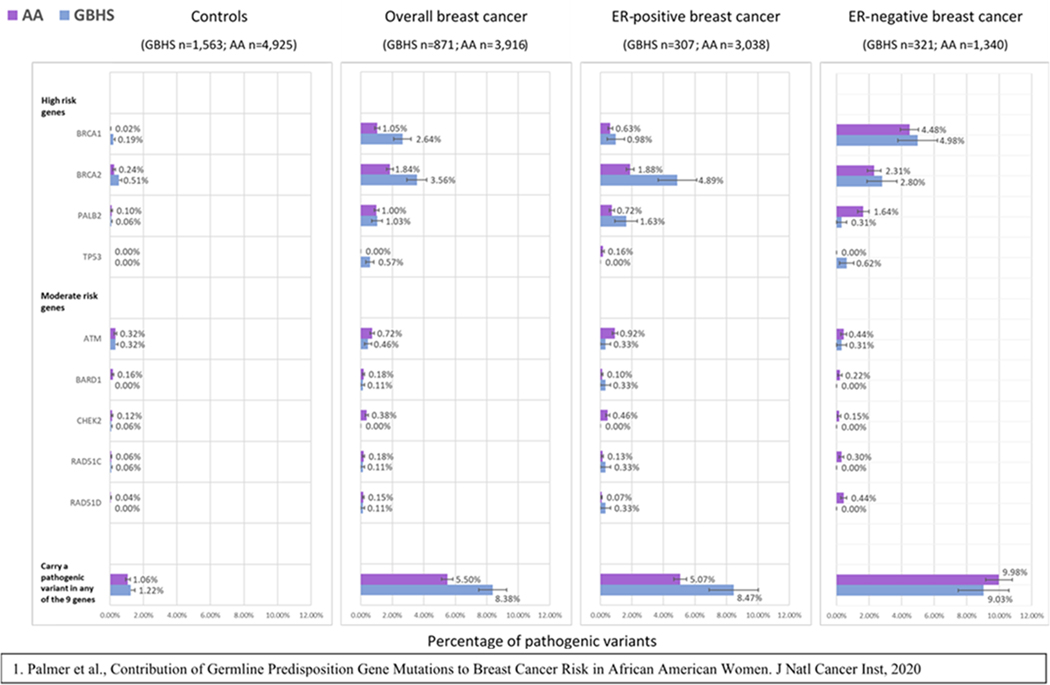
Figure 2 shows PV frequencies in the 9 breast cancer genes for GBHS and AA women in Palmer et al. (PTVs and pathogenic missense variants were combined for comparison with Palmer et al.) (5). This figure shows a higher mutation frequency in GBHS cases and controls for BRCA1 and BRCA2, in ER-positive cases for PALB2, and in ER-negative cases for TP53, like the comparisons with European or Asian ancestry women in BRIDGES. No pathogenic TP53 mutations were found in controls from either study. Notably, GBHS controls had approximately 10 times the frequency of BRCA1 pathogenic mutations compared to AA controls (Figure 2, Supplementary Table 7) (5).
Figure 2.
Comparison of the frequency of pathogenic mutations in putative susceptibility breast cancer genes in controls and breast cancer cases from the Ghana Breast Health Study (GBHS) and the CARRIERS study in African American (AA) women1. See Supplemental Table 7 for more details
Pathogenic variant carrier frequencies were higher in GBHS cases who reported a breast cancer family history in a first-degree relative at the time of interview, compared to those without such a history. Carrying a PV in BRCA1 and BRCA2 was not associated (P<0.05) with age at diagnosis for overall, ER-positive, or ER-negative disease (Supplementary Figure 3, Supplementary Tables 8-9).
Comparisons of mutation frequencies with genes not found associated with breast cancer at BFDP <5% by BRIDGES (4) are shown in Supplementary Tables 4 and 7. Comparisons of mutation frequencies between GBHS and a multi-ethnic US population in CARRIERS(6) is not shown; however, they were reported to be similar to BRIDGES (4) and Palmer et al. (5), except for BRCA1 pathogenic mutations in controls that were markedly low in AA controls (5).
Relative risk of breast cancer for mutation carriers
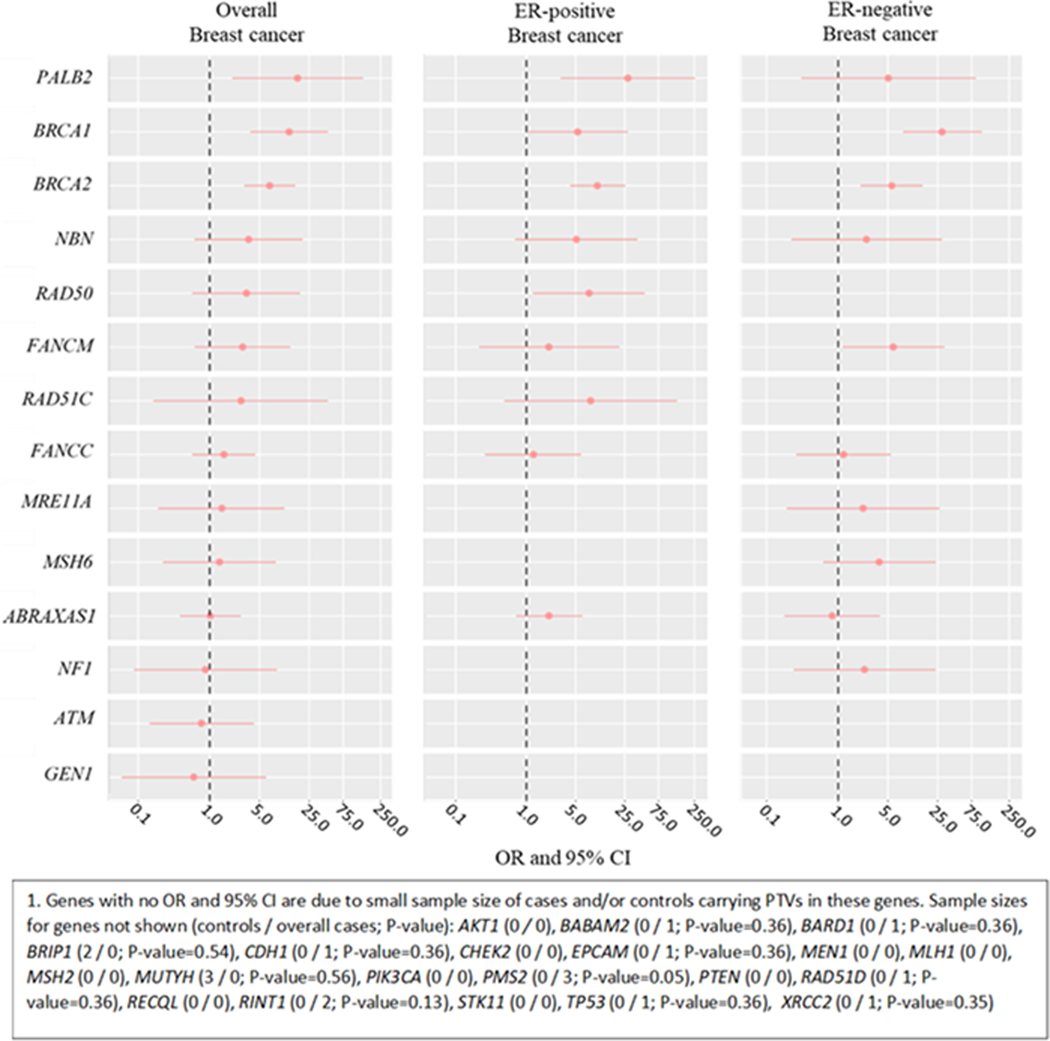
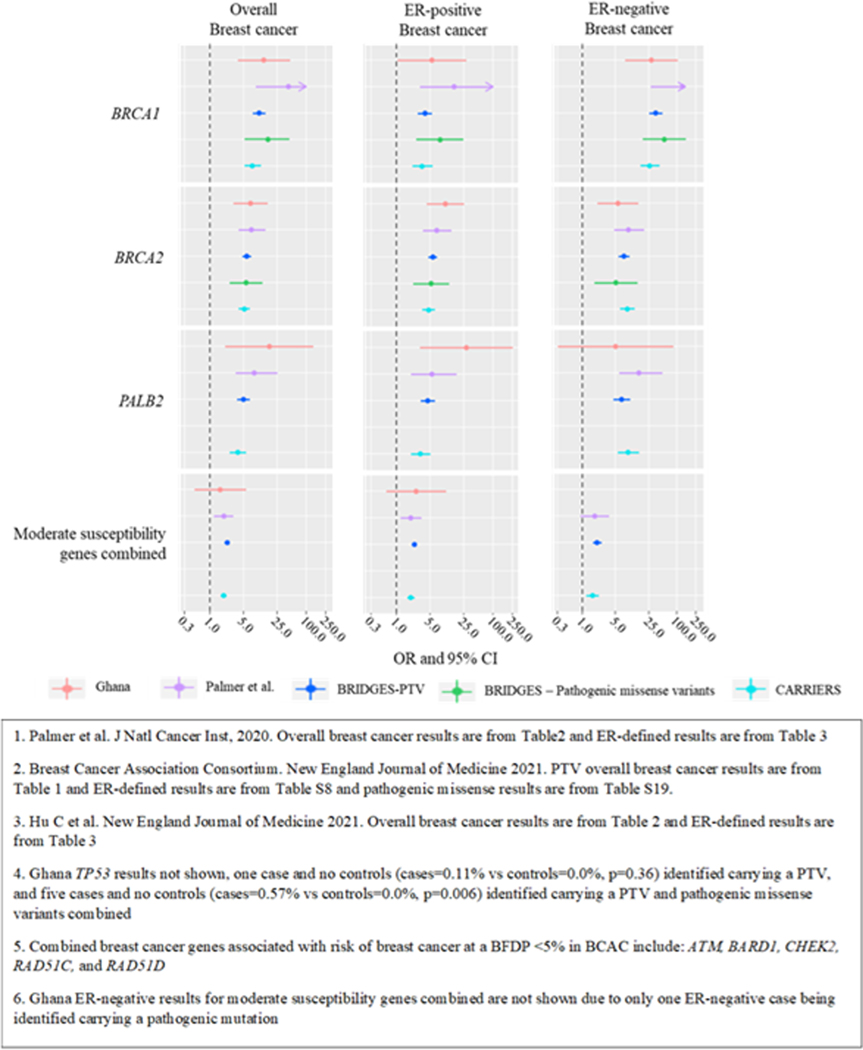
Among the 34 genes investigated we estimated associations with breast cancer risk for mutation carriers in 14 genes, as there were no carriers in cases and/or controls for the remaining 20 genes (Figure 3, Supplementary Table 2). We identified significant associations at P<0.05 between PTVs and overall breast cancer risk for three high-risk genes: BRCA1 (OR=13.17, 95% CI=3.86–44.91), BRCA2 (7.02, 3.17–15.54), and PALB2 (17.25, 2.15–138.13). The magnitude of association for these genes was like those in women of European or Asian ancestry in BRIDGES (Figure 4, Supplementary Table 10) (4). PTVs in BRCA1 were more strongly associated with ER-negative (28.62, 8.15–100.57) and TN disease (41.67, 11.53–150.54) than ER-positive (5.43, 1.08–27.43) disease (ER-positive vs ER-negative case-only Phet=0.01). PTVs in BRCA2 were more strongly associated with ER-positive (10.32, 4.27–24.96) than ER-negative (5.62, 2.12–14.88) and TN (7.96, 2.82–22.47) disease, but differences were not statistically significant (ER-positive vs ER-negative case-only Phet=0.16, Figure 4, Supplementary Tables 2). ER-specific associations for BRCA1 and BRCA2 were consistent with European and Asian populations in BRIDGES (P-diff>0.20, Figure 4, Supplementary Table 10) (4). PALB2 was more strongly associated with ER-positive than ER-negative disease (28.02, 3.17–247.74 and 5.02, 0.31–82.59, respectively), but estimates were imprecise, and differences were not statistically significant (case-only Phet=0.12; Supplementary Tables 2). PALB2 ER-specific estimates were not significantly different from those reported by BRIDGES (4). The magnitude of the associations for BRCA1, BRCA2, and PALB2 for carrying a PV in these genes was also like AA women and the multi-ethnic US population in CARRIERS. Pathogenic mutations in ATM and RAD51C were not significantly associated with breast cancer risk, although the magnitude of the estimated ORs was like those reported by BRIDGES, Palmer et al and CARRIERS (Figure 4, Supplementary Table 10). ORs could not be estimated for BARD1, CHEK2, RAD51D, and TP53 as no carriers were identified in cases or controls. Notably, five cases but no controls (cases=0.57% vs controls=0.0%, p=0.006) were identified carrying a PV in TP53. The magnitude of the estimated OR for overall breast cancer among women carrying a PV in any of the combined moderate-risk genes, ATM, BARD1, CHEK2, RAD51C, and RAD51D, was similar to other populations (2.10, 0.72–6.14) (Supplementary Table 10) (4–6). Only 1 ER-negative case carried a PV in the combined moderate-risk genes (Supplementary Table 2). Among other investigated genes, RAD50 was associated with risk of ER-positive disease (7.89, 1.29–48.20), in contrast to results from BRIDGES and CARRIERS (P-diffs=0.03 and 0.02, respectively). FANCM was associated with risk of ER-negative disease (5.91, 1.17–29.91) (4), consistent with BRIDGES and CARRIERS results (P-diff>0.08; Supplementary Figure 4, Supplementary Table 10). Evidence for associations with risk for mutations in other putative risk genes was weaker.
Figure 3.
Association between protein truncating variants (PTVs) in putative susceptibility genes and risk of breast cancer in the Ghana Breast Health Study (GBHS). The figure shows odds ratios (ORs) with 95% confidence intervals, indicated by bars. Genes are ordered by the size of estimated odds ratio for overall breast cancer. Showing 14 of 341 genes of which ORs and 95% CI could be estimated. See Supplemental Table 2 for more details.
Figure 4.
Associations between protein truncating variants (PTVs) in the Ghana Breast Health Study (GBHS), African Americans (PTVs and pathogenic missense variants combined) as reported by Palmer et al.1,Europeans and Asian ancestries in the Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES2), and in a multi-ethnic US population as reported by CARRIERS3 (PTVs and pathogenic missense variants combined) for high risk breast cancer genes (BRCA1, BRCA2, PALB2, and TP534) and five5,6 moderate susceptibility breast cancer risk genes combined. The figure shows odds ratios (ORs) with 95% confidence intervals (CI), indicated by bars. See Supplemental Table 10 for more details.
Population attributable risk and lifetime absolute risk of breast cancer for mutation carriers in 9 established breast cancer genes
Supplementary Table 11 shows the PAR estimates for BRCA1 and BRCA2 pathogenic mutations in Ghana for overall, ER-positive, and ER-negative disease, based on mutation frequencies in the control population and OR estimates. PAR estimates in Ghana might be higher than in other populations, however CIs were wide and overlapped with PAR estimates using data from previously published studies of larger sample sizes (4,5).
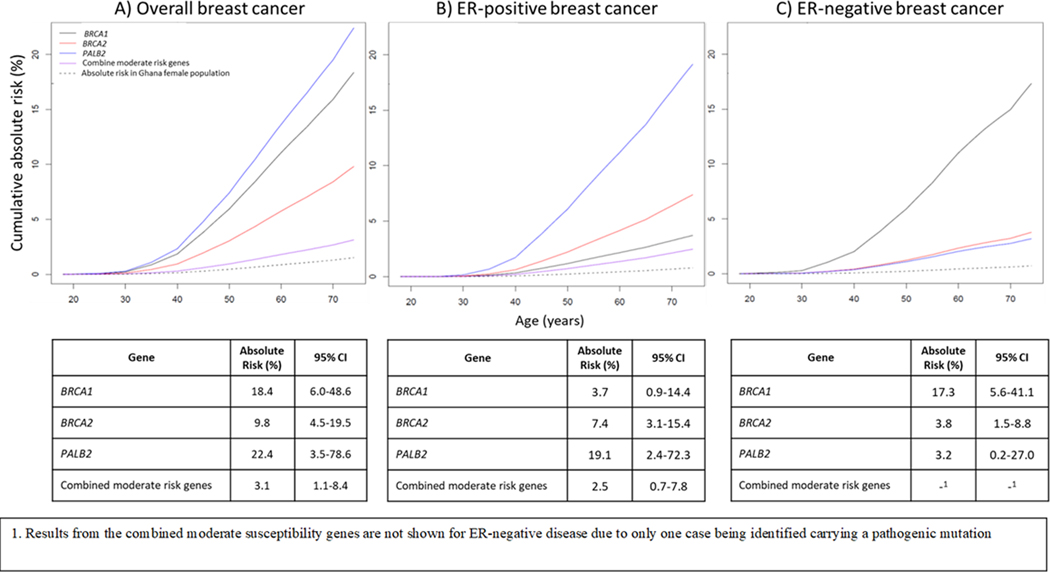
The lifetime absolute risk for overall breast cancer among women carrying a PV in BRCA1, BRCA2, PALB2, and moderate-risk genes combined (ATM, BARD1, CHEK2, RAD51C, and RAD51D) was: 18.4% (95% CI=6.0–48.6%), 9.8% (95% CI=4.5–19.5%), 22.4% (95% CI=3.5–78.6%), and 3.1% (95% CI=1.1–8.4%), respectively. BRCA1 mutation carriers had a higher lifetime risk for developing ER-negative (17.3%, 95% CI=5.6–41.1%) than ER-positive (3.7%, 0.9–14.3%) disease, while BRCA2 and PALB2 mutation carriers had higher lifetime risks for ER-positive (7.4%, 3.1–15.4% and 19.1%, 2.4–72.3%, respectively) than ER-negative (3.8%, 1.5–8.8% and 3.2%, 0.2–27.0%) disease. Mutation carriers for moderate-risk genes had a lifetime risks for ER-positive disease of 2.5% (0.7–7.8%) (Figure 5). An estimate of lifetime risk for ER-negative disease for mutation carriers in moderate-risk genes is not shown due to only 1 ER-negative case being found carrying a PV in the combined moderate-risk genes. As a sensitivity analysis, we estimated lifetime risks using OR estimates from BRIDGES while keeping Ghana age-specific disease incidence rates and competing mortality rates. Estimates were similar except for PALB2 (Supplementary Table 12), likely due to the lack of precision of the OR estimate from the GBHS.
Figure 5.
Estimates of absolute risk of A) overall, B) ER-positive, and C) ER-negative breast cancer through age 74 in women from Ghana who are carriers of pathogenic mutations in three high risk genes (BRCA1, BRCA2, PALB2), and five susceptibility genes combined (ATM, BARD1, CHEK2, RAD51C, and RAD51D). See Supplemental Table 12 for more
Discussion
In this study in Ghanaian women, we confirmed breast cancer risk associations for PVs in four established high-risk genes (BRCA1, BRCA2, PALB2, and TP53) and provided evidence consistent with moderate risk associations for ATM, BARD1, CHEK2, RAD51C, and RAD51D. Relative risks estimates were of similar magnitude to results published in large studies in women of European, Asian, AA and multi-ethnic (primarily non-Hispanic White) US population (4–6). However, Ghanaian women were about twice as likely to carry pathogenic mutations in BRCA1 or BRCA2. The estimated lifetime risks for overall breast cancer among carriers were lower for Ghanaian women than for AA(5), European ancestry women in the UK(4), and non-Hispanic Whites in the USA.(6)
By using the same gene panel, methodology and bioinformatic pipelines as in the BRIDGES study, we were able to make direct comparisons between estimates in GBHS and this large study of European and Asian ancestry women (cases=60,466 and controls=53,461 controls) (4). This gene panel was also similar to that used by Palmer et al and CARRIERS, allowing for comparisons to AA women (cases=5,054 and controls=4,993) and a multi-ethnic US population (cases=32,247 and controls=32,544), respectively (5,6). Although OR estimates for carrying a PV in established high- and moderate-risk genes were similar across populations (4–6), OR estimates in GBHS were less precise due to the smaller sample size. Notably, the OR estimates for AA BRCA1 carriers reported by Palmer et al. were also imprecise due to only 1 of 4,925 controls (0.02%) carrying a BRCA1 pathogenic mutation (5). Compared to other studies, the Carolina Breast Cancer Study (CBCS) also reported a low BRCA1 carrier frequency among controls, 1 in 1,635 (0.06%, AA=592, European-Americans=1016, other ancestry=27) (26). BRCA1 and BRCA2 carrier frequencies among GBHS controls are more like those from the Nigeria Breast Cancer Study (NBCS), which reported 3 in 997 (0.3%) and 4 in 997 (0.4%) controls to be BRCA1 and BRCA2 carrier, respectively (10). Adedokun et al. found in a study from Uganda and Cameroon 2 in 185 (1.08%) hospital-based controls to be BRCA1 carriers, while no controls were found to be BRCA2 carriers (27). Collectively, these findings suggest that women from West Africa more frequently are carriers for PVs in BRCA1 and BRCA2. However, the control populations in the GBHS, NBCS, and Adedokun et al were on average approximately 10–15 years younger than controls in BRIDGES, Palmer et al., and CARRIERS (4–6,10,27). These age differences across populations could at least partly explain differences in PV carrier frequency distributions. In addition, information on prior breast cancer screening was collected on control women in the GBHS, but they were not screened at study enrollment to identify subclinical breast cancer, which could have resulted in overestimation of pathogenic mutation frequencies.
GBHS ER-positive cases more often carried PVs in BRCA2 and PALB2 compared to ER-positive cases of European, Asian or AA ancestry (4,5). However, GBHS ER-negative carrier frequencies were similar to other populations, except for a higher TP53 carrier frequency in GBHS than AA ER-negative cases (5). Our finding suggesting PV in RAD50 being associated with risk of ER-positive disease contrasted with results from the BRIDGES and CARRIERS studies, thus this finding require replication in larger studies (4,6). Palmer et al. did not report results for RAD50 (5). Previous, reports found pathogenic mutations in BRCA1, BRCA2, and combined breast cancer genes were more common among women diagnosed at younger than older ages (4–6,10). We did not find age associated with BRCA1 and BRCA2, possibly due to our study having a smaller sample size and/or a younger, more narrower age range (median diagnosis age=51 years; range=18–74 years) than most other populations. Notably, 59.2% of NBCS cases were less than 50 years of age, which reported BRCA1 mutations were more common in women diagnosed at younger ages (10).
We were not able to obtain risk estimates for all the individual moderate risk genes due to our limited sample size, thus we combined these genes to test for associations with breast cancer risk. Among the combined moderate risk genes, we found modest evidence of an association with risk of ER-positive disease. For ER-negative cases we found only 1 case carrying a pathogenic missense variant [p.Ala2622Val] in ATM. Notably, BRIDGES, Palmer et al, and CARRIERS found RAD51C and RAD51D to be most strongly associated with risk for ER-negative disease. BRIDGES and CARRIERS also found BARD1 most strongly associated with risk of ER-negative disease, while Palmer et al. found no association with risk for BARD1 (4–6). For CHEK2, we did not identify PVs among GBHS cases; however, one control carried a pathogenic missense variant [p.Arg145Trp]. Pathogenic variants in CHEK2 were reported among 0.2% of cases from Nigeria(10) and 1.1% of cases from Cameroon, but none were the c.1100delC mutation (27). In BRIDGES, nearly 80% of CHEK2 PTVs were accounted for by c.1100delC, a founder mutation variant in north-western European populations (4,28). This variant accounted for 47% and 33% of CHEK2 PTVs found among AA cases and controls, respectively, possibly resulting from European population admixture among AAs (5).
BRIDGES estimated lifetime cumulative risks by age 80 years for BRCA1, BRCA2 and PALB2 PTV carriers in the UK of approximately 55%, 45%, and 42%, respectively (4). These results were similar to findings from CARRIERS, which reported cumulative absolute risks by age 85 for non-Hispanic Whites in the US of approximately 50% for PVs in BRCA1 or BRCA2, and 32% for variants in PALB2. Palmer et al estimated lifetime cumulative risks by age 85 for BRCA2 and PALB2 in AA women of 58% and 30%, respectively (5). Palmer et al did not report a lifetime risk for BRCA1 because only 1 control was identified carrying a PV in the gene. In contrast, we estimated much lower lifetime risks for overall breast cancer by age 74 for Ghanaian women in Kumasi and Accra carrying pathogenic mutations in BRCA1, BRCA2 and PALB2. Based on the figures reporting the lifetime absolute risk in BRIDGES (Figure 3), CARRIERS (Figure 1), and Palmer et al. (Figure 2), the absolute risk at age 74 in these populations were only slightly lower than the lifetime risks (4–6). The lower lifetime risks found in the GBHS, despite the similar OR, can be explained by the lower overall breast cancer incidence rates and higher competing mortality rates in Ghana compared to the UK and AA populations (21,29–31). Differences in rates could be partly explained by explained by younger demographics in Ghana compared to higher income countries and differences in reproductive patterns (32,33). Differences can also be explained by overdiagnosis or detection of indolent tumors in higher income countries with widespread mammographic screening programs (34–36), compared with only opportunistic screening in Ghana (14,37). This will be particularly relevant for ER-positive tumors that are more likely to be screen-detected than ER-negative tumors (38). However, we could not compare ER-specific absolute risk across populations because they were not reported for other ancestry populations (4–6).
Our estimated lifetime risks for mutation carriers could be used to inform genetic counseling of women in Ghana and other West African countries. However, our lifetime risk estimates were based on incidence rates estimates from two major cities, Kumasi and Accra, and they may not reflect estimates in more rural regions of Ghana. Other barriers for implementation of genetic testing and counseling services in these countries need to be addressed, including consideration of psycho-social and cultural factors, burden to the health care systems, and guidelines for clinical management of mutation carriers developed for local environments (39–41).
Our findings indicate that OR estimates for pathogenic mutations in established high- and moderate-risk breast cancer genes are similar across populations. However, overall lifetime risks for PV carriers were lower in Accra and Kumasi due to lower underlying rates for overall breast cancer than in Western populations. The role of other putative breast cancer genes remains unclear and larger studies in West Africa are needed to increase the precision of risk estimates.
Supplementary Material
Acknowledgments:
The authors acknowledge the research contributions of the Cancer Genomics Research Laboratory for their expertise, execution, and support of this research in the areas of project planning, wet laboratory processing of specimens, and bioinformatics analysis of generated data. This research was supported in part by funds from the intramural research program of the National Cancer Institute, National Institutes of Health (M. Garcia-Closas) and the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935; D.F. Easton) and the Wellcome Trust (grant no: v203477/Z/16/Z; D.F. Easton).
The success of this investigation would not have been possible without exceptional teamwork and the diligence of the field staff who oversaw the recruitment, interviews and collection of data from study subjects. Special thanks are due to the following individuals: Korle Bu Teaching Hospital, Accra—Dr. Adu-Aryee, Obed Ekpedzor, Angela Kenu, Victoria Okyne, Naomi Oyoe Ohene Oti, Evelyn Tay; Komfo Anoyke Teaching Hospital, Kumasi— Marion Alcpaloo, Bernard Arhin, Emmanuel Asiamah, Isaac Boakye, Samuel Ka-chungu and; Peace and Love Hospital, Kumasi—Samuel Amanama, Emma Abaidoo, Prince Agyapong, Thomas Agyei-Ansong, Debora Boateng, Margaret Frempong, Bridget Nortey Mensah, Richard Opoku, and Kofi Owusu Gyimah. The study was further enhanced by surgical expertise provided by Dr. Lisa Newman of the University of Michigan and by pathological expertise provided by Drs. Stephen Hewitt and Petra Lenz of the National Cancer Institute. Study management assistance was received from Ricardo Diaz, Shelley Niwa, Usha Singh, Ann Truelove and Michelle Brotzman at Westat, Inc. Appreciation is also expressed to the many women who agreed to participate in the study and to provide information and biospecimens in hopes of preventing and improving outcomes of breast cancer in Ghana.
Funding:
This project was funded with intramural funds from the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The BRIDGES project was funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935) and the Wellcome Trust [grant no: v203477/Z/16/Z].
Footnotes
Disclosures of potential conflicts of interest: The authors declare no potential conflicts of interest
References
- 1.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015;372(23):2243–57 doi 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee K, Seifert BA, Shimelis H, Ghosh R, Crowley SB, Carter NJ, et al. Clinical validity assessment of genes frequently tested on hereditary breast and ovarian cancer susceptibility sequencing panels. Genet Med 2019;21(7):1497–506 doi 10.1038/s41436-018-0361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsburg O, Ashton-Prolla P, Cantor A, Mariosa D, Brennan P. The role of genomics in global cancer prevention. Nat Rev Clin Oncol 2021;18(2):116–28 doi 10.1038/s41571-020-0428-5. [DOI] [PubMed] [Google Scholar]
- 4.Breast Cancer Association Consortium. Breast Cancer Risk Genes - Association Analysis in More than 113,000 Women. N Engl J Med 2021;384(5):428–39 doi 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer JR, Polley EC, Hu C, John EM, Haiman C, Hart SN, et al. Contribution of Germline Predisposition Gene Mutations to Breast Cancer Risk in African American Women. J Natl Cancer Inst 2020. doi 10.1093/jnci/djaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N Engl J Med 2021;384(5):440–51 doi 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman LA, Kaljee LM. Health Disparities and Triple-Negative Breast Cancer in African American Women: A Review. JAMA Surg 2017;152(5):485–93 doi 10.1001/jamasurg.2017.0005. [DOI] [PubMed] [Google Scholar]
- 8.Brinton LA, Figueroa JD, Awuah B, Yarney J, Wiafe S, Wood SN, et al. Breast cancer in Sub-Saharan Africa: opportunities for prevention. Breast Cancer Res Treat 2014;144(3):467–78 doi 10.1007/s10549-014-2868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;n/a(n/a) doi 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Walsh T, Gulsuner S, Casadei S, Lee MK, Ogundiran TO, et al. Inherited Breast Cancer in Nigerian Women. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(28):2820–5 doi 10.1200/jco.2018.78.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amankwaa-Frempong E, Yeboah FA, Nguah SB, Newman LA. Breast Cancer Genetic Testing Among African Patients With Breast Cancer: Deoxyribonucleic Acid Extraction From Tumor Tissue and International Multidisciplinary Partnerships. JAMA Surg 2017;152(8):800–1 doi 10.1001/jamasurg.2017.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Guo X, Long J, Ping J, Li B, Fadden MK, et al. Discovery of structural deletions in breast cancer predisposition genes using whole genome sequencing data from > 2000 women of African-ancestry. Hum Genet 2021. doi 10.1007/s00439-021-02342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambrosone CB, Zirpoli GR, Bovbjerg DH, Shankar J, Hong CC, McCann SE, et al. Associations between estrogen receptor-negative breast cancer and timing of reproductive events differ between African American and European American women. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2014;23(6):1115–20 doi 10.1158/1055-9965.epi-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinton LA, Awuah B, Nat Clegg-Lamptey J, Wiafe-Addai B, Ansong D, Nyarko KM, et al. Design considerations for identifying breast cancer risk factors in a population-based study in Africa. Int J Cancer 2017;140(12):2667–77 doi 10.1002/ijc.30688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueroa JD, Davis Lynn BC, Edusei L, Titiloye N, Adjei E, Clegg-Lamptey JN, et al. Reproductive factors and risk of breast cancer by tumor subtypes among Ghanaian women: A population-based case-control study. Int J Cancer 2020. doi 10.1002/ijc.32929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyante SJ, Biritwum R, Figueroa J, Graubard B, Awuah B, Addai BW, et al. Recruiting population controls for case-control studies in sub-Saharan Africa: The Ghana Breast Health Study. PLoS One 2019;14(4):e0215347 doi 10.1371/journal.pone.0215347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44(11):e108 doi 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandmann S, de Graaf AO, Karimi M, van der Reijden BA, Hellstrom-Lindberg E, Jansen JH, et al. Evaluating Variant Calling Tools for Non-Matched Next-Generation Sequencing Data. Sci Rep 2017;7:43169 doi 10.1038/srep43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17(1):122 doi 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer JR, Polley EC, Hu C, John EM, Haiman C, Hart SN, et al. Contribution of Germline Predisposition Gene Mutations to Breast Cancer Risk in African American Women. Journal of the National Cancer Institute 2020;112(12):1213–21 doi 10.1093/jnci/djaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghana Statistical Service, Population and Housing Census 2010 (PHC 2010), Version 1.0 of the public use dataset (December 2011). provided by the Ghana Statistical Service; 2013. [Google Scholar]
- 22.Pal Choudhury P, Maas P, Wilcox A, Wheeler W, Brook M, Check D, et al. iCARE: An R package to build, validate and apply absolute risk models. PLoS One 2020;15(2):e0228198 doi 10.1371/journal.pone.0228198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis Lynn BC, Figueroa J, Awittor FK, Ohene Oti NO, Edusei L, Titiloye N, et al. Estrogen receptor negative breast cancer incidence rates are similar in Ghanaian and Non-Hispanic Black women in the USA. medRxiv 2022:2022.02.21.22271266 doi 10.1101/2022.02.21.22271266. [DOI] [Google Scholar]
- 24.Ghana Statistical Service . Population and Housing Census 2010 (PHC 2010), Version 1.0 of the public use dataset (December 2011). provided by the Ghana Statistical Service; 2013. [Google Scholar]
- 25.Heer E, Harper A, Escandor N, Sung H, McCormack V, Fidler-Benaoudia MM. Global burden and trends in premenopausal and postmenopausal breast cancer: a population-based study. Lancet Glob Health 2020;8(8):e1027–e37 doi 10.1016/S2214-109X(20)30215-1. [DOI] [PubMed] [Google Scholar]
- 26.Walsh T, Gulsuner S, Lee MK, Troester MA, Olshan AF, Earp HS, et al. Inherited predisposition to breast cancer in the Carolina Breast Cancer Study. NPJ Breast Cancer 2021;7(1):6 doi 10.1038/s41523-020-00214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adedokun B, Zheng Y, Ndom P, Gakwaya A, Makumbi T, Zhou AY, et al. Prevalence of Inherited Mutations in Breast Cancer Predisposition Genes among Women in Uganda and Cameroon. Cancer Epidemiol Biomarkers Prev 2020;29(2):359–67 doi 10.1158/1055-9965.EPI-19-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt MK, Hogervorst F, van Hien R, Cornelissen S, Broeks A, Adank MA, et al. Age- and Tumor Subtype-Specific Breast Cancer Risk Estimates for CHEK2*1100delC Carriers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2016;34(23):2750–60 doi 10.1200/jco.2016.66.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International Agency for Research on Cancer. October 30. International Agency for Research on Cancer <https://gco.iarc.fr/today/data/factsheets/populations/288-ghana-fact-sheets.pdf>. Accessed 2020 October 30.
- 30.Cancer Research UK. October 30. Cancer Research UK <https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/breast-cancer/incidence-invasive>. Accessed 2020 October 30.
- 31.Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, et al. SEER Cancer Statistics Review, 1975–2017. Bethesda, MD: National Cancer Institute; 2020 based on November 2019 SEER data submission, posted to the SEER web site, April 2020. [Google Scholar]
- 32.Hamilton BEMJ, Osterman MJK. Births: Provisional data for 2020. Vital Statistics Rapid Release. Hyattsville, MD: National Center for Health Statistics; 2021. May 2021. [Google Scholar]
- 33.Ghana Statistical Service (GSS) GHSG, and ICF. Ghana Maternal Health Survey 2017. Accra, Ghana: GSS, GHS, and ICF; 2018. [Google Scholar]
- 34.Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. National Institute for Health and Cancer Excellence; 2013. [PubMed] [Google Scholar]
- 35.NCCN clinical practice guidelines in oncology: Genetic/familial high-risk assessment—breast, ovarian and pancreatic cancer. Version 1.2020. 2019. [DOI] [PubMed]
- 36.Mesa-Eguiagaray I, Wild SH, Rosenberg PS, Bird SM, Brewster DH, Hall PS, et al. Distinct temporal trends in breast cancer incidence from 1997 to 2016 by molecular subtypes: a population-based study of Scottish cancer registry data. British journal of cancer 2020;123(5):852–9 doi 10.1038/s41416-020-0938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biritwum R, Mensah G, Yawson A, Nadia M. Study on global AGEing and adult health (SAGE) Wave 1 The Ghana National Report. 2013. [Google Scholar]
- 38.Niraula S, Biswanger N, Hu P, Lambert P, Decker K. Incidence, Characteristics, and Outcomes of Interval Breast Cancers Compared With Screening-Detected Breast Cancers. JAMA Network Open 2020;3(9):e2018179-e doi 10.1001/jamanetworkopen.2020.18179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong A, Darren B, Dimaras H. Ethical, social, and cultural issues related to clinical genetic testing and counseling in low- and middle-income countries: protocol for a systematic review. Syst Rev 2017;6(1):140 doi 10.1186/s13643-017-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adejumo P, Aniagwu T, Oluwatosin A, Fagbenle O, Ajayi O, Ogungbade D, et al. Knowledge of Genetic Counseling Among Patients With Breast Cancer and Their Relatives at a Nigerian Teaching Hospital. J Glob Oncol 2018;4:1–8 doi 10.1200/JGO.17.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jegede AS. Culture and genetic screening in Africa. Dev World Bioeth 2009;9(3):128–37 doi 10.1111/j.1471-8847.2009.00259.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.