Abstract
Therapy resistance is imposing a daunting challenge on effective clinical management of breast cancer. Although the development of resistance to drugs is multifaceted, reprogramming of energy metabolism pathways is emerging as a central but heterogenous regulator of this therapeutic challenge. Metabolic heterogeneity in cancer cells is intricately associated with alterations of different signaling networks and activation of DNA damage response (DDR) pathways. Here we consider how the dynamic metabolic milieu of cancer cells regulates their DNA damage repair ability to ultimately contribute to development of therapy resistance. Diverse epigenetic regulators are crucial in remodeling the metabolic landscape of cancer. This epigenetic-metabolic interplay profoundly affects genomic stability of the cancer cells as well as their resistance to genotoxic therapies. These observations identify defining mechanisms of cancer epigenetics-metabolism-DNA repair axis that can be critical for devising novel, targeted therapeutic approaches which could sensitize cancer cells to conventional treatment strategies.
Keywords: Metabolism, resistance, epigenetic, DNA repair, breast cancer
INTRODUCTION
In initial stages many breast cancer patients respond to therapy, however, therapy-resistance develops in most cases over-time due to epigenomic (1) and metabolic (2) changes within the tumor that promotes drug inhibition, degradation and heightened DNA damage repair (3). Acquisition of treatment-resistance and subsequent tumor recurrence is thus one of the most major challenges in breast cancer management.
Metabolism is an adaptive process that remains significantly altered in tumor cells, and is considered a hallmark of cancer (4) (5) (6)(7). Thus, reprogramming of energy metabolism has been associated with the development of resistance towards different therapeutic regimens (8), (9), (10), (11). Recently, the DNA damage response (DDR) pathways have been highlighted as a pivotal cause of drug resistance (12). Interestingly, metabolic flux can hyperactivate different DDR pathways through a complex mechanism that eventually heightens cell proliferation as well as survival, thereby promoting resistance to therapies (13), (14). Notably, besides genetic mutation, epigenetic changes play a key role in regulating metabolic reprogramming as well as DNA repair (15). The axis between epigenetics, cellular metabolism, and DNA repair provides a great platform for understanding the role of this synergy in the subsequent emergence of drug resistance and thus we will trace out the underlying involved epigenetic regulatory mechanisms.
CONVENTIONAL THERAPEUTIC REGIMENS AGAINST BREAST CANCER AND THERAPY-RESISTANCE
Breast cancer is a complex and heterogeneous disease, and its therapeutic strategy and outcome critically depend on its molecular subtype. Here we discuss the clinical significance of molecular subtyping in breast cancer in reference to treatment strategy and acquisition of therapy-resistance.
Luminal type breast cancer, therapy and resistance
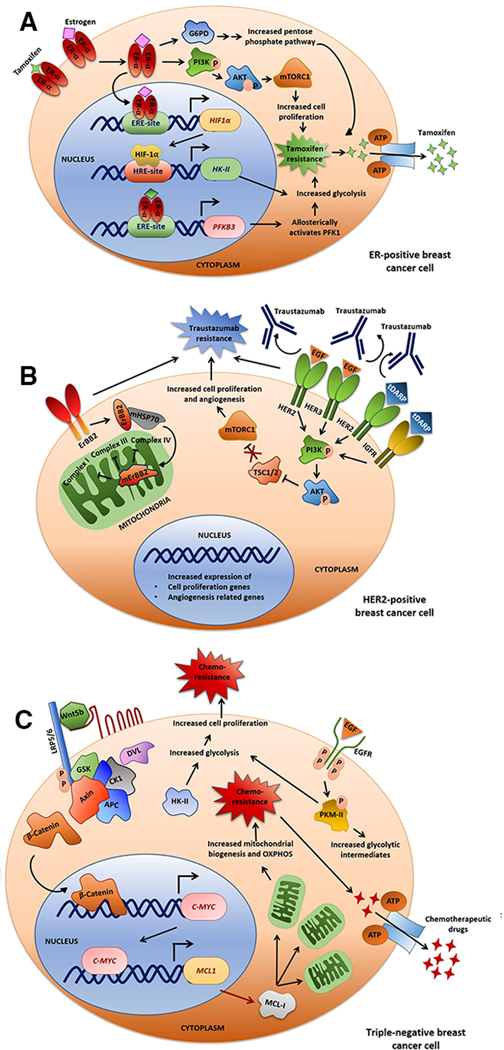
Luminal breast cancers, are generally positive for estrogen receptor (ER) and/or progesterone receptor (PR) which are subdivided based on ER, PR and HER2 receptor status and Ki67 expression (16). The mainstay treatment strategy employed against luminal breast cancer is anti-estrogen therapy using selective estrogen receptor modulator (SERM) (like Tamoxifen), selective estrogen receptor down regulators (SERD) and aromatase inhibitors (17). Despite the remarkable sensitivity of luminal tumors to anti-ER therapy, 30–50% of cases show relapse after 20 years of diagnosis (16), (18). Tamoxifen is very frequently used as adjuvant therapy along with chemotherapy in ER+ breast cancer, however about 20–30% of the patients is shown to develop resistance to tamoxifen-therapy (19). The mechanism of tamoxifen resistance has been extensively studied, and modifications in glycolysis have been identified as a major underlying mechanisms (19). PFKFB3 (6-Phosphofructokinase-2-kinase/Fructose-2,6-bisphosphatase-3), which catalyzes the formation of fructose 2,6-bisphosphate during glycolysis, is transcriptionally activated by estrogen signaling (20). HIF1α is also an ER-responsive gene that transcriptionally upregulates hexokinase II (HKII) (21), a major rate-limiting enzyme of glycolysis. Cumulatively, these events enhance glycolysis-driven tamoxifen resistance in luminal breast cancer (22) (Fig. 1A).
Fig. 1: Therapeutic regimens aginst different breast cancer subtypes and therapy-resistance.
A. ER+ breast cancer. In estrogen receptor (ER)+breast cancer, tamoxifen is used as a major therapeutic agent. Here the PI3K-AKT-mTOR-pathway plays a major role in tamoxifen-resistance. Also, Hypoxia-inducible factor 1α (HIF1α) have estrogen-response element (ERE) on its promoter where estrogen hormone-receptor complex binds and triggers its expression. HIF1α promotes hexokinaseII (HKII) expression by binding to hypoxia-response element (HRE) on its promoter. Elevated HKII leads to increased glycolysis, proliferation and subsequent acquisition of thearapy-resistance. B. HER2+ breast cancer. A major targeting therapy for HER2+breast cancer is trastuzumab, resistance to which is mediated by alterations in the PI3K-AKT-mTOR pathway. C. Triple negative breast cancer (TNBC). Chemotherapy is the mainstay therapeutic-regiman for TNBC. The Wnt-β catenin pathway plays a key role in acquisition of its chemo-resistance. β catenin promotes c-myc expression, which in turn induces mitochondrial biogenesis through MCL1, ultimately resulting in amplified oxidative phosphorylation (OXPHOS). Also HKII is elevated in TNBC, increasing glycolytic flux, further promoting cell proliferation and chemo-resistance.
HER2-positive breast cancer targeting therapy and resistance
The transmembrane tyrosine kinase receptor encoded by the human epidermal growth factor receptor 2 (HER2), intricately regulates cell proliferation and survival. About 15–20% of all breast tumors show HER2 overexpression, leading to aggressive tumor development and a poor prognosis (23). The clinical management strategies of HER2+ cancer includes usage of monoclonal antibodies (mAbs), tyrosine kinase inhibitors (TKIs) and antibody-drug conjugates (ADCs), which have shown improvement of patient prognosis at all stages of the illness (23). The mAbs trastuzumab and pertuzumab, and TKIs lapatinib in combination with capecitabine is frequently used to treat advanced HER2+ breast cancer (24). Co-treatment of trastuzumab/pertuzumab with microtubular network-disrupter docetaxel has been shown to improve Progression-free survival (PFS) (25). However, despite these targeting strategies, HER2+ breast cancer mostly remains a fatal illness with median PFS of only 1 year even after therapy (23) (26). Early HER2+ breast cancer recurs in about 26% of patients after trastuzumab therapy, whereas in advanced metastatic cases, relapse increases to about 70% within a year (27).
Trastuzumab acts by internalization and degradation of HER2 receptor, which further leads to inhibition of downstream PI3K-AKT-mTOR pathway (23). Activating mutations in the PI3K-AKT-mTOR network renders trastuzumab-resistance to therapy (28),(29) (Fig. 1B). Nearly 30% of human HER2+ tumors display mutations in PI3K-AKT-mTOR pathway genes, with about 20–23% cases showing activating mutations of PI3K-catalytic subunit-α (PI3KCA) which mediates trastuzumab-resistance (30).
Basal type or triple negative breast cancer (TNBC)
About 15% of all invasive breast cancers are basal-like, which display higher incidence of local and distant recurrence (31). TNBC, the most dominant subgroup (about 80–90%) of basal-like breast tumors are highly aggressive and show poor clinical outcome (31),(32). Successful treatment of TNBC is extremely challenging and often results in poor prognostic outcomes as endocrine therapy and HER2 targeting therapies are ineffective due to absence of ER, PR and HER2. Hence, a standard neoadjuvant chemotherapy (NACT), often basMed on anthracyclines, taxanes, and cyclophosphamides, is used against most TNBC patients (33). Adjuvant chemotherapy is also administered to patients with residual illness after the completion of NACT (34).
Being the most aggressive form, TNBC often become resistant to conventional chemotherapy and its recurrence peaks early at around 3 years after diagnosis, with distant metastasis (32). TNBC patients have lower survival rate than other subtypes of breast cancer, having a fatality rate of 40% in the first five years following diagnosis (32). About 46% of individuals with TNBC develop distant metastases, following which, the median survival period is just 13.3 months, and up to 25% of cases relapse after surgery (32).
The aberrant activation of several signaling cascades in TNBC cells promotes their aggressive nature and therapy resistance. For instance, PI3K-AKT-mTOR and EGFR signaling often gets hyper-activated in TNBC, and about 70–78% TNBC show EGFR overexpression (35), (32). TNBC also show metabolic alterations that drive their resistant nature. For instance, glycolysis is especially pronounced in TNBC, promoting heightened cell proliferation and chemoresistance (36). Also, Wnt and EGFR pathways, plays a significant role in augmenting carbohydrate metabolism in TNBC. For example, MYC and MCL1, two important downstream targets of Wnt signaling (37), is reported to cooperatively promote mitochondrial biogenesis (Fig. 1C), which in turn enhances oxidative phosphorylation, ultimately causing paclitaxel resistance in TNBC (38). TNBC often has aberrant genetic abnormalities in DNA pathways, including BRCA1/2 mutations and p53 malfunction that drives their resistant nature (39). DNA damage repair (DDR)-inhibitors alone or in combination with conventional therapies can open up potential avenues in TNBC treatment (40).
THE ROLE OF METABOLIC MILIEU OF CANCER IN DRUG RESISTANCE
Both the extracellular metabolic microenvironment and the internal molecular and metabolic milieu of the tumor cells play crucial role in acquisition of therapy-resistance.
The role of tumor microenvironment in rewiring cancer cell metabolism and mediating resistance
Tumor microenvironment (TME) is composed of tumor cells, non-malignant stromal cells like endothelial cells, cancer-associated fibroblasts (CAFs), immune cells (macrophages, lymphocytes etc.), and the non-cellular fibers such as collagen, hyaluronan, fibronectin and laminin etc. (41). A metabolic crosstalk exists among cancer cells of residing in different TME compartments. For instance, a lactate shuttle exists between hypoxic core cancer cells with the peripheral well-oxygenated cells, in which the hypoxic cancer cells produce lactate by anaerobic glycolysis and release it by MCT4 exporter, which is then taken up by the cancer cells proximal to blood vessels through MCT1 importer (42). This in turn protects the tumor cells from DNA damage and mediates resistance (43).
Breast cancer cells consume glucose and secrete lactate to sustain neighboring CAFs which in turn release exosome to help the cancer cells to cope with the nutrient deprived conditions in TME (44). Additionally, CAFs supply glutathione (GSH) and cysteine to cancer cells to balance their redox homeostasis to circumvent the chemotherapeutic challenges (45). Interestingly, the metabolites secreted by tumor cells dampens the immunosurveillance by modulating TME-immune profile. Increased lactate in hypoxic regions of tumor inhibits the differentiation of monocytes and dendrites and also block T cell activation (41). Beside this, high lactate level promotes the number of M2 macrophages via induction of VEGF and arginase which mainly produce immunosuppressive cytokines like IL10 (41), (46). Notably, M2 macrophages promote resistance against radiation therapy via IL4/IL13 mediated STAT6 phosphorylation (47). One of the key determinants of the response to radiation is the extent of DNA damage. Thus, immune-modulation of TME mediates radio-resistance (47). In a nutshell, the metabolic inter-connection and crosstalk among different cell types in the TME plays critical role in mediating therapeutic resistance.
The role of internal metabolic reprogramming in DNA repair derived resistance
The altered metabolic architecture within cancer cells is intricately associated with the acquisition of therapy-resistance linked with DNA damage response (48), (49) (50).
Role of DNA damage response in drug resistance
Sustaining genomic integrity by repair of DNA damage is a crucial cellular activity (51). A complex pathway involving sensors, transducers, and effectors, known as the DNA damage response (DDR), orchestrate this, facilitating cellular survival. Accumulating results indicate that cancer cells exhibit alterations in DNA repair pathways, which promotes their inherent and acquired resistance toward genotoxic therapies (52).
Alterations in DNA repair pathways of tumor cells lead to heightened genomic instability, augmented mutation rate and increased intra-tumor heterogeneity (53). Most conventional anti-cancer regimens target cancer cells by increasing double-strand breaks (DSB), and hence, DSB repair via non-homologous end-joining (NHEJ), and homology-directed repair (HDR) plays a significant role in the resilience of cancer cells towards therapy. Dissecting the intricate regulatory network underlying DDR and modulating them therefore can be a potential approach to sensitize tumor cells toward therapeutic strategies (54).
DNA damage and subsequent breaks promote an immediate cell-wide increase of bound NADH fraction triggered by poly (ADP-ribose) polymerase (PARP) dependent formation of poly (ADP-ribose) (PAR). PAR recruits repair proteins to DNA-damaged sites and shifts the metabolic balance to oxidative phosphorylation (OXPHOS) over glycolysis. Clinically, PARP inhibitors (PARPi) are successful for breast cancer patients with a deficiency in HDR (55), which is synthetically lethal with PARP deficiency. Following DNA repair initiation, poly (ADP-ribose) glycohydrolase (PARG) hydrolyzes the PAR ‘cloud’ into mono-nucleotide ADP-ribose (ADPr), so PARGi can re-sensitize PARPi resistant cancer cells (56).
Decade-long studies are unraveling an intricate connection between altered metabolic landscape and DDR pathways in cancer, which can in turn influence therapy-resistance (57). Heightened glucose intake or the Warburg effect is a primal characteristic of most solid tumors, which is associated with a metabolic shift from OXPHOS to anaerobic glycolysis, leading to the augmented production of lactic acid (58). Recent reports positively associate elevated lactic acid production with chemo/radio-resistance and metastasis, which in turn implicates cellular metabolic status in conferring therapy-resistance by modulating DDR (59), (60).
Oxidative stress in DNA damage and repair
A key link between metabolism and DNA damage is via redox balance. Reactive oxygen species (ROS), one of the major inducers of DNA damage, is produced by several metabolic reactions, with the electron transport chain being the chief source (61). Heightened oxidative DNA damage due to ROS burdens the DNA-repair machinery, leading to genomic instability (Fig. 2). A major ROS-DNA interaction happens by reaction of °OH with sugar, purines and pyrimidines of the DNA strands, which leads to formation of oxidative DNA lesions like 8-oxo-dG, thymine glycol, and strand-break(62), which require efficient DNA base repair (63). Incomplete DNA repair due to the increased lesion burden generates pro-mutagenic repair intermediates such as single-strand DNA breaks and abasic sites. Accumulation of these pro-mutagenic sites within a short DNA span is referred to as oxidatively generated clustered DNA lesions. These are often detected after radiotherapy, posing a major threat to genomic integrity, and promoting development of radio-resistance (64).
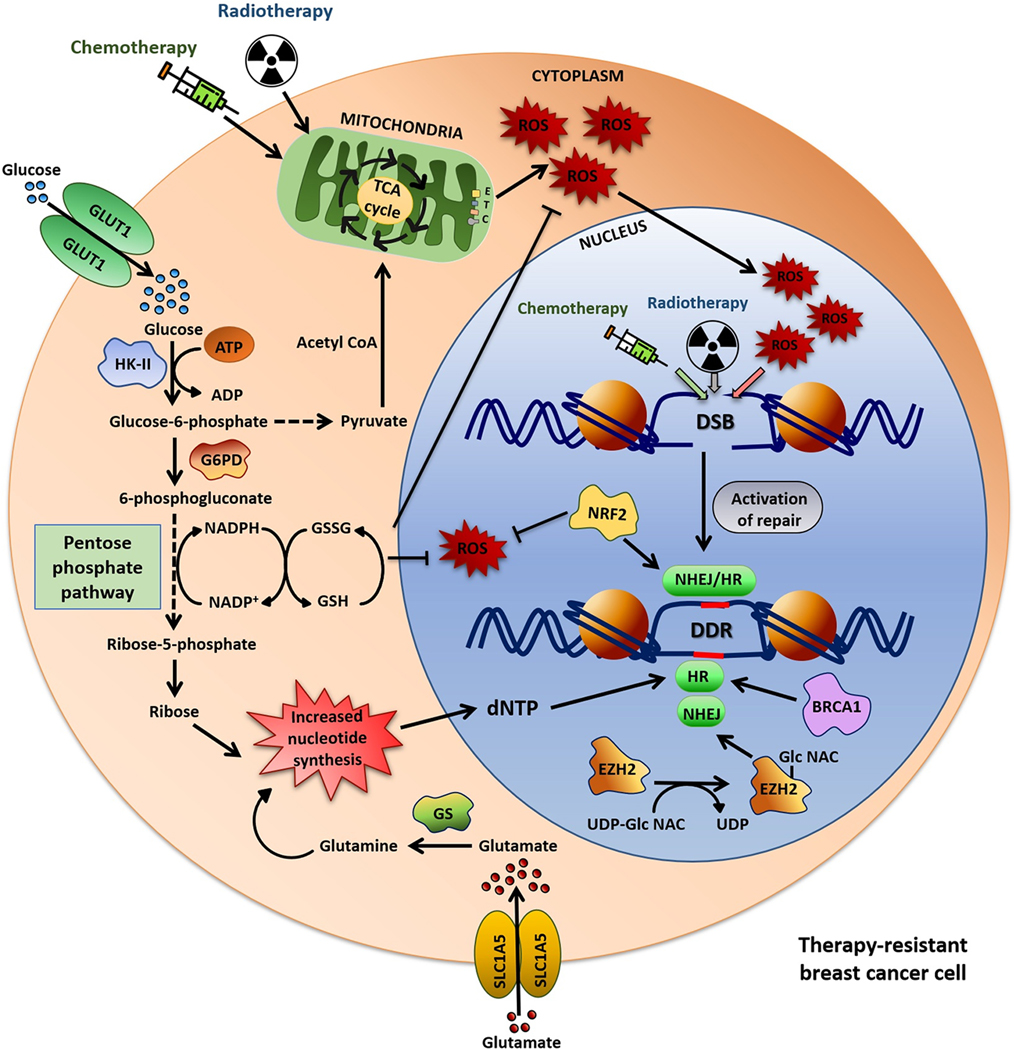
Fig. 2: Correlation between metabolic pathways and DDR-mediated therapeutic-resistance.
Chemo- and radiotherapy leads to DNA damage resulting in DSBs. Accumulation of reactive oxygen species (ROS) also leads to generation of DSB. Both free radical-scavenging pathway and pentose phosphate pathway (PPP) play crucial role in facilitating DNA repair. The ROS-scavanging pathway neutralizes the ROS generated by chemo/radiotherapy. On the other hand, increased PPP in resistant cells augments dNTP production and supply to the DNA damage repair (DDR) pathways, specially to homologous recombination (HR)-mediated DDR which requires high amount of dNTP. Glutamine synthetase (GS), which converts glutamate to glutamine, is overexpressed in resistant cancer cells, leading to usage of glutamine in PPP promoting increased nucleotide-biosynthesis. The DDR protein BRCA1 induces HR-mediated DNA repair, NRF2 promotes both NHEJ and HR-mediated DNA repair. This metabolic pathways facilitates DNA repair (indicated by red line) in chemo/radio-damaged cancer cells, thus promoting therapy-resistance.
Prolonged exposure to chemotherapy alters the overall ROS level in tumor cells, which in turn can lead to acquisition of resistance (65)(66). Elevated ROS production during chemotherapy stabilizes HIF-1α and promotes vascular endothelial growth factor (VEGF) and macrophage migration-inhibitory factor (MIF) expression, which in turn confer chemoresistance via upregulation of anti-apoptotic proteins through RAS/MAPK pathway (66). On the other hand, cancer stem cells (CSCs) display significantly lower ROS levels and augmented expression of free radical scavenging systems, which renders their resistance towards most conventional therapies (67) (68).
Multiple metabolic intermediates and pathways work in coordination to balance intracellular ROS levels. The enzyme GSH peroxidase scavenges ROS while oxidizing from GSH to GSSG. The reverse reaction, catalyzed by glutathione reductase, utilizes the reducing potential of NADPH (69). Glutathione is produced from glutamate and cysteine, with the availability of cysteine being the rate-limiting step of this reaction (70). The cystine-glutamate antiporter xCT transports cysteine into cells in exchange for glutamate (71).Culturing tumor cells in media containing cystine leads to xCT-dependent glutamine-dependency, alterations in ROS level, and sensitization of cancer cells to glutaminase-inhibition (72).
Nuclear factor-erythroid 2-related factor-2 (NRF2) is an important modulator of ROS level (73). Interestingly, the DNA-repair protein, BRCA1 regulates NRF2, thus linking ROS levels and DNA repair pathways (74). BRCA1 recruitment is facilitated by histone-acetylation specifically H4K16ac close to DNA-damage sites, interfering with the binding of NHEJ repair factors and promoting homologous recombination (HR)-mediated DNA repair (75), (76), (77), (78). Hence, targeting BRCA1 and NRF2 is a potential approach to enhance DNA repair and reverse ROS-mediated therapy-resistance in cancer cells. Activation of NRF2 through synthetic triterpenoids acts as a potential strategy to shield the normal tissues against severe ionizing radiation treatment (79).
Interestingly, the disruption of the pentose phosphate pathway (PPP) can also act as a shield against ROS. Depletion of the phosphogluconate dehydrogenase (PGD) enzyme leads to the accumulation of 6-phosphogluconate (6PG), which inhibits the glycolytic pathway and increases glucose flux through PPP that enhances NADPH production (80). NADPH synthesis is also dependent on NAD+ production and its two principal enzymes: nicotinamide phosphoribosyltransferase (NAMPT) and nicotinate phosphoribosyltransferase (NAPRT)(81). Tumor cells display enhanced NAD+ cycling rate and increased dependency on either of these two enzymes (82), inhibition of which promotes susceptibility of the cancer cells to oxidative stress (83),(84). Apart from this, S-adenosylmethionine (SAM) is one of the stimulators of GSH synthesis and hence plays an important role in GSH-mediated ROS detoxification (85).
Due to the critical involvement of ROS in rendering resistance, modulating it can be a potential strategy to disrupt the resistant phenotype of cancer cells. Targeting redox-regulating factors like NADPH oxidase, glutathione metabolism-related enzymes, xCT and NRF2 could modulate ROS levels and attenuate drug-resistance in cancer (65). For instance, ML385, an NRF2 inhibitor can sensitize resistant cancer cells to chemotherapeutic drugs like doxorubicin, carboplatin or paclitaxel (86). Another NRF2 inhibitor IM3829, which disrupts NRF2 binding to its target genes, promotes radiosensitivity of cancer cells (87). MTH1 (human MutT homologue 1, NUDT1), another protein involved in oxidative stress response, has become an attractive anti-cancer target (88). Karonudib, an MTH1 inhibitor (89) is currently undergoing clinical trials for the treatment of solid cancers. Buthionine sulfoximine, an inhibitor of γ-GCS (the first enzyme of GSH -biosynthetic pathway), is reported to suppress cisplatin-resistance in breast cancer (90).
Nucleotide pool and DNA repair potential of cancer cells
Modulation of the nucleotide pool is a central metabolic pathway that regulates DNA damage and repair (91), (92). Alterations in the dNTP pool can promote genomic instability by inducing replication stress and reducing DNA repair. Several factors are recruited to DNA damage sites that aid in dNTP generation to ensure a steady dNTP supply for successful DNA repair. For instance, rapid recruitment of RRM1 and RRM2 (ribonucleotide reductase family member 1 and 2) at DNA damage site ensures the dNTP synthesis needed for successive DNA repair (93), (94). RRM1 and RRM2 jointly constitute the Ribonucleotide reductase (RNR) enzyme complex that catalyzes the de-novo dNTP synthesis from the corresponding ribonucleotides (95). The concentration of the dNTP pool is critical in DNA repair pathways as HR requires a higher concentration of dNTPs than NHEJ (96). Hence, metabolic pathways involved in nucleotide biosynthesis play a key role in DNA repair and resistance to genotoxic drugs (Fig. 2) (91), (97), (98).
DNA damage promotes tumor cells to reprogram their metabolic pathways towards PPP (14), which plays a critical role in promoting tumor cell proliferation and survival by supplying cells with NADPH and ribose-5-phosphate (99). The multi-enzyme PPP diverges from glycolysis at glucose-6-phosphate and runs in parallel. Glucose 6-phosphate dehydrogenase (G6PDH) the rate-limiting enzyme of PPP, is associated with tumor cell survival (100). The PPP synthesizes several critical intermediates of cell metabolic pathways including ribose-5-phosphate, a key precursor for the synthesis of the ribose backbone of both purines and pyrimidines (101) (Fig. 2). The PPP intermediates can also return to glycolysis by giving rise to glyceraldehyde-3-phosphate (102). In breast cancer cells, heightened PPP is responsible for the constant production of NADPH and nucleotides (103). Also, the PPP remains significantly enhanced in several cancers, and has been connected with cancer progression, migration and drug-resistance (104), (100).
In cisplatin-resistant cancer cells, inhibition of G6PDH re-sensitizes them to cisplatin (100). Activation of ATM in response to DSBs also reportedly induces G6PDH to augment the synthesis of dNTPs to promote DNA repair (105). Another metabolic enzyme, phosphoglycerate mutase 1 (PGAM1) plays role in HR-mediated DSB repair through modulation of nucleotide pools by converting 3-phosphoglycerate to 2-phosphoglycerate and coordinating PPP and glycolysis. Inhibition of PGAM1 leads to dNTP depletion, through the accumulation of 3-phosphoglycerate and inhibition of PPP enzyme 6-phosphogluconate dehydrogenase (6PGD). This also promotes ubiquitination and degradation of CTBP-interacting protein (CtIP), an important factor of the HR pathway, by the E3 ubiquitin-ligase APC/C-Cdh1 (106). In response to ionizing radiation-mediated DNA damage, fructose-2,6-biphosphatase-3 localizes into the nucleus in an ATM-H2AX-MDC1 dependent-manner, where it induces RRM2 to promote local generation of dNTP pool. Inhibition of it thus leads to cancer cell sensitization to radiation therapy by repressing DNA repair (107).
Alterations in glutamate is also important in metabolic rewiring of cancer cells for enhancing dNTP production. Cancer cells overexpress the glutamine synthetase (GS) enzyme which converts glutamate to glutamine and utilized by cells for nucleotide biosynthesis (Fig. 2) (108), (109), (110), (111), (112). Glutamine synthetase has also been reported to be overexpressed in radio-resistant cancer cells, playing an integral role in sustaining their DNA repair and promoting their resistance(113). Inhibiting GS is thus a potential approach to combat radio-resistance in cancer cells.
Another instance of dNTP regulation in the context of DNA repair can be traced through the dNTP triphosphohydrolase enzyme SAMHD1 that breaks down dNTP molecules into deoxynucleoside and inorganic triphosphate, inhibiting usage of the dNTPs in DNA synthesis (88). The higher expression of SAMHD1 is often associated with drug-resistance because its substrates also include anticancer nucleoside analogues (88). Suppression of SAMHD1 promotes sensitivity towards the deoxyguanosine analogue chemotherapeutic drug nelarabine (114).
cGAS-STING pathway in DNA damage response
The cytosolic DNA-sensing cyclic GMP-AMP synthase (cGAS)- stimulator of interferon genes (STING) pathway (115) (116) promotes innate immune responses by production of cyclic GMP-AMP (cGAMP) (117). It is activated in response to DNA damage (116) (118) and induces expression of type 1 interferon (IFN), which suppresses cancer progression (119) (120). Several agonists of this pathway are currently in clinical trials and demonstrate therapeutic efficacy (121) (122). In a subgroup of ER+ breast tumors, high perinuclear-localized expression of STING was an independent predictor of good prognosis and increased immune response while low expression corelated with immunosuppression and oncogenic mTOR activation (123).
Metastatic tumors often hijack cGAS-STING pathway to circumvent therapeutic-stress (124) (125). Cancer cells exhibit chromosomal instability (CIN), resulting in cellular rupture that exposes genomic contents into cytosol through micronuclei, triggering chronic activation of cGAS-STING (126)(115); and induces an immune-suppressive TME (127) (123). Genomic instability and DDR also triggers inflammatory response (128). In BRCA1/2 mutant tumors, inflammatory signaling, as a consequence of CIN, actively modulates TME to escape immune-surveillance (129). Furthermore, cGAS suppresses HR-mediated repair, promotes tumor growth and has emerged as an attractive therapeutic target (130).
Inactivation of cGAS-STING signaling selectively impairs the survival of TNBC cells that display CIN and promotes tumorigenesis through an IL6-STAT3 mechanism (131). In a subset of TNBCs, epigenetic silencing of STING by binding of MYC on its promoter enhanced immune-evasion and conferred resistance against immune-checkpoint blockers (132). Inhibition of MYC, in conjunction with PDL-1 blockade, restored PDL1-inhibitor sensitivity and enhanced therapeutic response (132).
STING activation corelates with advanced cancer stage and promotes drug-resistance (133) and inhibition of this axis could be combined with traditional chemotherapy for potent anticancer effects. Doxorubicin mediated DNA damage activates the cGAS-STING that enable cell proliferation and survival through activation of NF- κB and IL6 pathways and corelates with resistance and poor patient outcome (134). Different breast tumors express varying levels of STING, that defines their response to chemotherapy (127). Absence of STING necessitates a significantly increased dose of 5-Fluorouracil for reducing tumor burden and its efficacy is dependent on activation of tumor-intrinsic STING (135), (136). Use of STING agonists increase the antitumor immunity when combined with classical therapies reversing therapeutic-resistance (137). Activation of STING suppresses NRF2 and sensitizes tumors to chemotherapy (138)(139) and reduces chemotherapy induced toxicity while potentially enhancing its antitumor effects (140) (141). Collectively, these studies highlight the complex interplay that determines the mechanistic roles of cGAS-STING in mediating pro or antitumor effects.
The epigenetic regulatory mechanisms involved in the DDR-metabolism axis
Epigenetic regulation plays an integral role in repair pathway choice (142)(143)(144)(145). Methylation and acetylation, depend on the metabolic pathways for the availability of methyl- and acetyl-group. For instance, ATP-citrate lyase (ACLY), which is responsible for the production of nuclear acetyl-CoA and oxaloacetate from citrate, localizes at the DNA damage sites and supplies acetyl-CoA to promote histone acetylation (146). ACLY remains overexpressed in several cancers and its activity reportedly increases in response to ionizing-radiation therapy (147). ACLY and acetyl-CoA mediated histone acetylation near DNA damage sites leads to the recruitment of BRCA1, promoting preferential DNA repair by HR (148). Moreover, ACYL mediates the activation of PI3K-AKT pathway which through downstream mechanisms induce double stranded break repair (DSBR) in cancer, thereby promoting drug-resistance (149) (150).
In cancer cells, elevated glucose and glutamine metabolism lead to increases in N-acetyl-glucosamine (GlcNAc) synthesis. This in turn increases protein O-GlcNAcylation in tumor cells, which can promote DNA repair-mediated therapy-resistance. The mechanism behind such an activity can be traced through the histone methyltransferase protein enhancer of zeste homolog 2 (EZH2). In cancer cells, EZH2 is activated and stabilized by O-GlcNAcylation, which then induces DNA repair by increasing H2K27 trimethylation, an important determining factor for NHEJ repair (151) (Fig. 2).
The oncogenic metabolite 2-hydroxyglutarate (2-HG), which is highly synthesized in some tumors from α-ketoglutarate by a mutated version of isocitrate dehydrogenase 1 (IDH1) (152), acts in altering DNA repair by epigenetic regulation. 2-HG inhibits lysine-specific demethylases such as KDM4A and KDM4B. Inhibition of KDM4B by 2-hydroxyglutarate promotes basal H3K9 hypermethylation, which results in masking of heightened trimethylation near the vicinity of DNA damage sites. This suppresses recruitment of DDR factors at the damage site, subsequently impairing DNA repair (153). Interestingly, IDH1 gets frequently mutated in cancer, and has controversial reports in DDR and chemo-,radio-resistance (154) (155).
The glycolytic enzyme PKM2 remains overexpressed in various cancers, playing crucial roles in tumor development and progression (156). PKM2 furthermore acts in HR-mediated DNA repair. As a response towards DNA damage induced by ionizing radiation and oxidative stress, PKM2 is phosphorylated by ATM and retained within the nucleus (157), in turn leading to phosphorylation and activation of CtIP, promoting HR-mediated DNA repair (158). PKM2 also phosphorylates H2AX serine-139 in response to DNA damage, thus generating γ-H2AX and playing an integral part in DDR. The above reports highlight the intricate association between the altered metabolic landscape of cancer cells and modulation of DNA damage repair, which is functionally interlinked with immune and inflammatory responses.
SYNOPSIS AND FUTURE IMPLICATIONS
The metabolic milieu of cancer cells is a crucial determinant of therapeutic-resistance and tumor relapse. The outcome of rewired energy metabolism branches into myriad manifestations that a resistant cancer cell exhibit. The integrated findings on epigenetic and metabolic mechanisms in conjunction with the DNA damage repair can promote identification of novel strategies to epigenetically target this vicious metabolism-DNA repair axis of cancer. In fact, several epi-drugs targeting this axis are actively being assessed in clinical trials in breast cancer. Overall, this review aims to facilitate identification of novel therapeutic strategies that can combat altered metabolism-mediated acquisition of DNA repair capacity and therefore drug-resistance of cancer cells.
Acknowledgements
C.D. acknowledges SwarnaJayanti Fellowship (DST/SJF/LSA-02/2017-18), Department of Science and Technology, S. Ramachandran National Bioscience Award for Career Development 2019 (BT/HRD-NBA-NWB/38/2019-20), Department of Biotechnology, and Basic and Applied Research in Biophysics and Material Science (RSI 4002), Department of Atomic Energy (DAE), Government of India. J.A.T. and Z.A. are supported by NIH grants P01 CA092548 and R35 CA220430, Cancer Prevention Research Institute of Texas grant (RP180813), and a Robert A. Welch Chemistry Chair.
Footnotes
Competing interests
C.D., S.A., A.B., S.C., S.S.Y., P.M., S.A., C.R.H., K.Y., S.R., J.A.T., Z.A., and T.K.P. have no competing interests.
References
- 1.Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL, Morey L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat Commun; 2021 [cited 2022 Nov 26];12. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desbats MA, Giacomini I, Prayer-Galetti T, Montopoli M. Metabolic Plasticity in Chemotherapy Resistance. Front Oncol; 2020 [cited 2022 Nov 26];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug Resistance in Cancer: An Overview. Cancers (Basel) [Internet]. Multidisciplinary Digital Publishing Institute (MDPI); 2014. [cited 2022 Apr 28];6:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Judge A, Dodd MS. Metabolism. Essays Biochem; 2020. [cited 2022 Nov 26];64:607–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv L, Yang S, Zhu Y, Zhai X, Li S, Tao X, et al. Relationship between metabolic reprogramming and drug resistance in breast cancer. Front Oncol; 2022 [cited 2022 Nov 26];12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell; 2012. [cited 2022 Nov 26];21:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov; 2012. [cited 2022 Nov 26];2:881–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M, Zhao Y, Ding Y, Liu H, Liu Z, Fodstad O, et al. Warburg effect in chemosensitivity: Targeting lactate dehydrogenase-A re-sensitizes Taxol-resistant cancer cells to Taxol. Mol Cancer [Internet]. BioMed Central; 2010. [cited 2021 Jun 11];9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K min Giltnane JM, Balko JM Schwarz LJ, Guerrero-Zotano AL Hutchinson KE, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab; 2017. [cited 2022 Mar 17];26:633–647.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radde BN, Ivanova MM, Mai HX, Alizadeh-Rad N, Piell K, Van Hoose P, et al. Nuclear respiratory factor-1 and bioenergetics in tamoxifen-resistant breast cancer cells. Exp Cell Res; 2016. [cited 2022 Jan 29];347:222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian C, Yuan Z, Xu D, Ding P, Wang T, Zhang L, et al. Inhibition of glycolysis by a novel EGFR/HER2 inhibitor KU004 suppresses the growth of HER2+ cancer. Exp Cell Res [Internet]. Academic Press Inc.; 2017. [cited 2022 Jan 29];357:211–21. [DOI] [PubMed] [Google Scholar]
- 12.Sadik H, Korangath P, Nguyen NK, Gyorffy B, Kumar R, Hedayati M, et al. HOXC10 Expression Supports the Development of Chemotherapy Resistance by Fine Tuning DNA Repair in Breast Cancer Cells. Cancer Res; 2016. [cited 2022 Nov 26];76:4443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobanski T, Rose M, Suraweera A, O’Byrne K, Richard DJ, Bolderson E. Cell Metabolism and DNA Repair Pathways: Implications for Cancer Therapy. Front Cell Dev Biol. Frontiers Media S.A; 2021;9:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moretton A, Loizou JI. Interplay between Cellular Metabolism and the DNA Damage Response in Cancer. Cancers (Basel); 2020. [cited 2022 Jan 29];12:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene; 2017. [cited 2022 Jul 2];36:3359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szostakowska M, Trębińska-Stryjewska A, Grzybowska EA, Fabisiewicz A. Resistance to endocrine therapy in breast cancer: molecular mechanisms and future goals. Breast Cancer Res Treat; 2019. [cited 2022 Nov 26];173:489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Criscitiello C, Fumagalli D, Saini KS, Loi S. Tamoxifen in early-stage estrogen receptor-positive breast cancer: overview of clinical use and molecular biomarkers for patient selection. Onco Targets Ther [Internet]. Dove Press; 2011. [cited 2022 Jul 8];4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ignatiadis M, Sotiriou C. Luminal breast cancer: From biology to treatment. Nat Rev Clin Oncol. 2013;10:494–506. [DOI] [PubMed] [Google Scholar]
- 19.Ali S, Rasool M, Chaoudhry H, Pushparaj PN, Jha P, Hafiz A, et al. Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation; 2016. [cited 2022 Nov 26];12:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imbert-Fernandez Y, Clem BF, O’Neal J, Kerr DA, Spaulding R, Lanceta L, et al. Estradiol stimulates glucose metabolism via 6-phosphofructo-2-kinase (PFKFB3). J Biol Chem; 2014. [cited 2022 Mar 16];289:9440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, AlTahan A, Jones DT, Buffa FM, Bridges E, Interiano RB, et al. Estrogen receptor-α directly regulates the hypoxia-inducible factor 1 pathway associated with antiestrogen response in breast cancer. Proc Natl Acad Sci U S A; 2015. [cited 2022 Nov 26];112:15172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng J, Cui Y, Xu S, Wu X, Huang Y, Zhou W, et al. Altered glycolysis results in drug-resistant in clinical tumor therapy. Oncol Lett; 2021 [cited 2022 Nov 26];21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS, et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: Current knowledge, new research directions and therapeutic perspectives. Crit Rev Oncol Hematol; 2019. [cited 2022 Nov 26];139:53–66. [DOI] [PubMed] [Google Scholar]
- 24.Schlam I, Swain SM. HER2-positive breast cancer and tyrosine kinase inhibitors: the time is now. NPJ Breast Cancer; 2021 [cited 2022 Nov 26];7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins AM, Rowland A, McKinnon RA, Sorich MJ. Predictors of long-term disease control and survival for HER2-positive advanced breast cancer patients treated with pertuzumab, trastuzumab, and docetaxel. Front Oncol. Frontiers Media S.A.; 2019;9:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pondé N, Brandão M, El-Hachem G, Werbrouck E, Piccart M. Treatment of advanced HER2-positive breast cancer: 2018 and beyond. Cancer Treat Rev; 2018 [cited 2022 Nov 26];67:10–20. [DOI] [PubMed] [Google Scholar]
- 27.Perez EA, Romond EH, Suman VJ, Jeong JH, Davidson NE, Geyer CE, et al. Four-Year Follow-Up of Trastuzumab Plus Adjuvant Chemotherapy for Operable Human Epidermal Growth Factor Receptor 2–Positive Breast Cancer: Joint Analysis of Data From NCCTG N9831 and NSABP B-31. J Clin Oncol [Internet]. American Society of Clinical Oncology; 2011. [cited 2022 Apr 26];29:3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka Y, Mukohara T, Shimada H, Saijo N, Hirai M, Minami H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann Oncol Off J Eur Soc Med Oncol [Internet]. Ann Oncol; 2010. [cited 2022 Nov 26];21:255–62. [DOI] [PubMed] [Google Scholar]
- 29.Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res; 2012. [cited 2022 Nov 26];18:6784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loibl S, Majewski I, Guarneri V, Nekljudova V, Holmes E, Bria E, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol; 2016. [cited 2022 Nov 26];27:1519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang P, Tse GM. Immunohistochemical Surrogates for Molecular Classification of Breast Carcinoma: A 2015 Update. Arch Pathol Lab Med; 2016 [cited 2022 Nov 26];140:806–14. [DOI] [PubMed] [Google Scholar]
- 32.Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res; 2020 [cited 2022 Nov 26];22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huober J, Von Minckwitz G, Denkert C, Tesch H, Weiss E, Zahm DM, et al. Effect of neoadjuvant anthracycline-taxane-based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat; 2010. [cited 2022 Nov 26];124:133–40. [DOI] [PubMed] [Google Scholar]
- 34.Masuda N, Lee S-J, Ohtani S, Im Y-H, Lee E-S, Yokota I, et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N Engl J Med [Internet]. N Engl J Med; 2017. [cited 2022 Nov 26];376:2147–59. [DOI] [PubMed] [Google Scholar]
- 35.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol; 2006. [cited 2022 Nov 26];19:264–71. [DOI] [PubMed] [Google Scholar]
- 36.Varghese E, Samuel SM, Líšková A, Samec M, Kubatka P, Büsselberg D. Targeting Glucose Metabolism to Overcome Resistance to Anticancer Chemotherapy in Breast Cancer . Cancers (Basel); 2020. [cited 2022 Nov 26];12:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, Perez AA, Fujie S, Warden C, Li J, Wang Y, et al. Wnt modulates MCL1 to control cell survival in triple negative breast cancer. BMC Cancer; 2014 [cited 2022 Nov 26];14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee K min Giltnane JM, Balko JM Schwarz LJ, Guerrero-Zotano AL Hutchinson KE, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab; 2017. [cited 2022 Nov 26];26:633–647.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J, Tao Z, Cao J, Li T, Hu X. DNA damage response inhibitors: An avenue for TNBC treatment. Biochim Biophys acta Rev cancer [Internet]. Biochim Biophys Acta Rev Cancer; 2021 [cited 2022 Nov 26];1875. [DOI] [PubMed] [Google Scholar]
- 41.Li F, Simon MC. Cancer Cells Don’t Live Alone: Metabolic Communication within Tumor Microenvironments. Dev Cell. Cell Press; 2020;54:183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front Oncol [Internet]. Front Oncol; 2019 [cited 2022 Nov 26];9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature; 2020. [cited 2022 Nov 20];577:115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang E, Wang X, Gong Z, Yu M, Wu H, Zhang D. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther 2020 51 [Internet]. Nature Publishing Group; 2020 [cited 2022 Nov 21];5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Kryczek I, Dostál L, Lin H, Tan L, Zhao L, et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell; 2016;165:1092–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature; 2014. [cited 2022 Nov 21];513:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGee HM, Jiang D, Soto-Pantoja DR, Nevler A, Giaccia AJ, Woodward WA. Targeting the Tumor Microenvironment in Radiation Oncology: Proceedings from the 2018 ASTRO-AACR Research Workshop. Clin Cancer Res; 2019 [cited 2022 Nov 21];25:2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science; 2020 [cited 2022 Nov 26];368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiaradonna F, Scumaci D. Cancer Metabolism as a New Real Target in Tumor Therapy. Cells [Internet]. Cells; 2021 [cited 2022 Nov 26];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li LY, Guan Y Di, Chen XS, Yang JM, Cheng Y. DNA Repair Pathways in Cancer Therapy and Resistance. FFront Pharmacol; 2021 [cited 2022 Nov 26];11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaziri H, Dessain SK, Eaton EN, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell; 2001. [cited 2022 Jul 21];107:149–59. [DOI] [PubMed] [Google Scholar]
- 52.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature Publishing Group; 2013. [cited 2022 Jul 21];502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature; 2013. [cited 2022 Jul 21];500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer; 2013. [cited 2022 Jul 21];13:714–26. [DOI] [PubMed] [Google Scholar]
- 55.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science; 2017. [cited 2022 Jul 21];355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houl JH, Ye Z, Brosey CA, Balapiti-Modarage LPF, Namjoshi S, Bacolla A, et al. Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat Commun; 2019 [cited 2022 Jul 21];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turgeon MO, Perry NJS, Poulogiannis G. DNA Damage, Repair, and Cancer Metabolism. Front Oncol; 2018 [cited 2022 Apr 23];8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol [Internet]. The Rockefeller University Press; 1927. [cited 2021 Jan 13];8:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turkcan S, Kiru L, Naczynski DJ, Sasportas LS, Pratx G. Lactic Acid Accumulation in the Tumor Microenvironment Suppresses 18 F-FDG Uptake. Cancer Res; 2019. [cited 2022 Apr 23];79:410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geck RC, Foley JR, Stewart TM, Asara JM, Casero RA, Toker A. Inhibition of the polyamine synthesis enzyme ornithine decarboxylase sensitizes triple-negative breast cancer cells to cytotoxic chemotherapy. J Biol Chem; 2020. [cited 2022 Apr 23];295:6263–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J [Internet]. Biochem J; 1980. [cited 2022 Apr 23];191:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res; 1992. [cited 2022 Apr 23];275:331–42. [DOI] [PubMed] [Google Scholar]
- 63.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst); 2007. [cited 2022 Jul 21];6:410–28. [DOI] [PubMed] [Google Scholar]
- 64.Georgakilas AG. Processing of DNA damage clusters in human cells: current status of knowledge. Mol Biosyst; 2008. [cited 2022 Apr 23];4:30–5. [DOI] [PubMed] [Google Scholar]
- 65.Cui Q, Wang JQ, Assaraf YG, Ren L, Gupta P, Wei L, et al. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist Updat; 2018. [cited 2022 Apr 23];41:1–25. [DOI] [PubMed] [Google Scholar]
- 66.Parekh A, Das S, Parida S, Das CK, Dutta D, Mallick SK, et al. Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo. Oncogene; 2018. [cited 2022 Apr 23];37:4546–61. [DOI] [PubMed] [Google Scholar]
- 67.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature; 2009. [cited 2021 Nov 22];458:780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis; 2016 [cited 2022 Apr 23];7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med; 2009. [cited 2022 Apr 23];30:42–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hultberg B, Hultberg M. High glutathione turnover in human cell lines revealed by acivicin inhibition of gamma-glutamyltranspeptidase and the effects of thiol-reactive metals during acivicin inhibition. Clin Chim Acta; 2004. [cited 2022 Apr 23];349:45–52. [DOI] [PubMed] [Google Scholar]
- 71.Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, et al. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal; 2013. [cited 2022 Jul 4];18:522–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muir A, Danai LV., Gui DY, Waingarten CY, Lewis CA, Vander Heiden MG. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife; 2017 [cited 2022 Apr 23];6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, et al. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A; 2012 [cited 2022 Sep 29];109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med [Internet]. J Exp Med; 2013. [cited 2022 Apr 23];210:1529–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol; 2013. [cited 2022 Apr 23];20:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horikoshi N, Sharma D, Leonard F, Pandita RK, Charaka VK, Hambarde S, et al. Pre-existing H4K16ac levels in euchromatin drive DNA repair by homologous recombination in S-phase. Commun Biol; 2019 [cited 2022 Sep 30];2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakraborty S, Singh M, Pandita RK, Singh V, Lo CSC, Leonard F, et al. Heat-induced SIRT1-mediated H4K16ac deacetylation impairs resection and SMARCAD1 recruitment to double strand breaks. iScience; 2022 [cited 2022 Sep 30];25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, et al. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res; 2013. [cited 2022 Sep 30];179:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim SB, Pandita RK, Eskiocak U, Ly P, Kaisani A, Kumar R, et al. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A; 2012 [cited 2022 Jul 8];109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dubreuil MM, Morgens DW, Okumoto K, Honsho M, Contrepois K, Lee-McMullen B, et al. Systematic Identification of Regulators of Oxidative Stress Reveals Non-canonical Roles for Peroxisomal Import and the Pentose Phosphate Pathway. Cell Rep; 2020. [cited 2022 Apr 23];30:1417–1433.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heske CM. Beyond Energy Metabolism: Exploiting the Additional Roles of NAMPT for Cancer Therapy. Front Oncol; 2020 [cited 2022 Apr 23];9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature; 2019. [cited 2022 Apr 23];569:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong SM, Park CW, Kim SW, Nam YJ, Yu JH, Shin JH, et al. NAMPT suppresses glucose deprivation-induced oxidative stress by increasing NADPH levels in breast cancer. Oncogene; 2016. [cited 2022 Apr 23];35:3544–54. [DOI] [PubMed] [Google Scholar]
- 84.Hong SM, Hwang SW, Wang T, Park CW, Ryu YM, Jung JH, et al. Increased nicotinamide adenine dinucleotide pool promotes colon cancer progression by suppressing reactive oxygen species level. Cancer Sci; 2019. [cited 2022 Apr 23];110:629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cavallaro RA, Fuso A, Nicolia V, Scarpa S. S-adenosylmethionine prevents oxidative stress and modulates glutathione metabolism in TgCRND8 mice fed a B-vitamin deficient diet. J Alzheimers Dis; 2010. [cited 2022 Apr 23];20:997–1002. [DOI] [PubMed] [Google Scholar]
- 86.Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, et al. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chem Biol; 2016. [cited 2022 Apr 23];11:3214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee S, Lim MJ, Kim MH, Yu CH, Yun YS, Ahn J, et al. An effective strategy for increasing the radiosensitivity of Human lung Cancer cells by blocking Nrf2-dependent antioxidant responses. Free Radic Biol Med; 2012. [cited 2022 Apr 23];53:807–16. [DOI] [PubMed] [Google Scholar]
- 88.Helleday T, Rudd SG. Targeting the DNA damage response and repair in cancer through nucleotide metabolism. Mol Oncol; 2022. [cited 2022 Nov 26];16:3792–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berglund UW, Sanjiv K, Gad H, Kalderén C, Koolmeister T, Pham T, et al. Validation and development of MTH1 inhibitors for treatment of cancer. Ann Oncol; 2016. [cited 2022 Nov 26];27:2275–83. [DOI] [PubMed] [Google Scholar]
- 90.Jia Y, Zhang C, Zhou L, Xu H, Shi Y, Tong Z. Micheliolide overcomes KLF4-mediated cisplatin resistance in breast cancer cells by downregulating glutathione. Onco Targets Ther [Internet]. Onco Targets Ther; 2015. [cited 2022 Apr 23];8:2319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sobanski T, Rose M, Suraweera A, O’Byrne K, Richard DJ, Bolderson E. Cell Metabolism and DNA Repair Pathways: Implications for Cancer Therapy. Front Cell Dev Biol; 2021 [cited 2022 Nov 26];9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moretton A, Loizou JI. Interplay between Cellular Metabolism and the DNA Damage Response in Cancer . Cancers (Basel); 2020. [cited 2022 Nov 26];12:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev; 2010. [cited 2022 Apr 23];24:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell; 2012. [cited 2022 Apr 23];149:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science; 1993. [cited 2022 Apr 23];260:1773–7. [DOI] [PubMed] [Google Scholar]
- 96.Burkhalter MD, Roberts SA, Havener JM, Ramsden DA. Activity of ribonucleotide reductase helps determine how cells repair DNA double strand breaks. DNA Repair (Amst); 2009. [cited 2022 Apr 23];8:1258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J; 2011. [cited 2022 Nov 26];30:546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turgeon MO, Perry NJS, Poulogiannis G. DNA Damage, Repair, and Cancer Metabolism. Front Oncol. Front Oncol; 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maitra S, Das S, Banerjee P. Diabetes, a Potential Threat to the Development and Progression of Tumor Cells in Individuals. Nutr Ther Interv Diabetes Metab Syndr. Academic Press; 2018;169–78. [Google Scholar]
- 100.Catanzaro D, Gaude E, Orso G, Giordano C, Guzzo G, Rasola A, et al. Inhibition of glucose-6-phosphate dehydrogenase sensitizes cisplatin-resistant cells to death. Oncotarget [Internet].Oncotarget; 2015. [cited 2022 Apr 23];6:30102–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patra KC, Hay N. The pentose phosphate pathway and cancer [Internet]. Trends Biochem. Sci. Elsevier Ltd; 2014. [cited 2021 May 20]. page 347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stine ZE, Altman BJ, Hsieh AL, Gouw AM, Dang CV. Deregulation of the Cellular Energetics of Cancer Cells. Pathobiol Hum Dis A Dyn Encycl Dis Mech. Academic Press; 2014;444–55. [Google Scholar]
- 103.Bokun R, Bakotin J, Milasinovic D. Semiquantitative cytochemical estimation of glucose-6-phosphate dehydrogenase activity in benign diseases and carcinoma of the breast. Acta Cytol; 1987. [cited 2022 Apr 23];31:249–52. [PubMed] [Google Scholar]
- 104.Chan B, VanderLaan PA, Sukhatme VP. 6-Phosphogluconate dehydrogenase regulates tumor cell migration in vitro by regulating receptor tyrosine kinase c-Met. Biochem Biophys Res Commun; 2013. [cited 2022 Apr 23];439:247–51. [DOI] [PubMed] [Google Scholar]
- 105.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J; 2011. [cited 2022 Apr 23];30:546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qu J, Sun W, Zhong J, Lv H, Zhu M, Xu J, et al. Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J Cell Biol; 2017. [cited 2022 Apr 23];216:409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gustafsson NMS, Färnegårdh K, Bonagas N, Ninou AH, Groth P, Wiita E, et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat Commun; 2018 [cited 2022 Apr 23];9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol; 2015. [cited 2022 Apr 23];17:1556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, et al. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A; 2008. [cited 2022 Jul 8];105:16165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blickwedehl J, Olejniczak S, Cummings R, Sarvaiya N, Mantilla A, Chanan-Khan A, et al. The proteasome activator PA200 regulates tumor cell responsiveness to glutamine and resistance to ionizing radiation. Mol Cancer Res; 2012. [cited 2022 Jul 8];10:937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo; 2006. [cited 2022 Apr 23];20:587–90. [PubMed] [Google Scholar]
- 112.Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, et al. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci U S A; 2008. [cited 2022 Sep 30];105:16165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu S, Li Z, Xiao L, Hu W, Zhang L, Xie B, et al. Glutamine Synthetase Promotes Radiation Resistance via Facilitating Nucleotide Metabolism and Subsequent DNA Damage Repair. Cell Rep; 2019. [cited 2022 Apr 23];28:1136–1143.e4. [DOI] [PubMed] [Google Scholar]
- 114.Rothenburger T, McLaughlin KM, Herold T, Schneider C, Oellerich T, Rothweiler F, et al. SAMHD1 is a key regulator of the lineage-specific response of acute lymphoblastic leukaemias to nelarabine. Commun Biol; 2020 [cited 2022 Nov 26];3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.MacKenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature; 2017. [cited 2022 Nov 26];548:461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med; 2018. [cited 2022 Nov 26];215:1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature; 2008. [cited 2022 Nov 26];455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chanut R, Petrilli V. [Cytosolic DNA sensing by the cGAS-STING pathway in cancer]. Med Sci (Paris); 2019. [cited 2022 Nov 26];35:527–34. [DOI] [PubMed] [Google Scholar]
- 119.Liu F rao, Jiang M jie, Mei Z, Lin C jing, Tian L. cGAS-STING signalings potentiate tumor progression via sustaining cancer stemness. Transl Oncol; 2022 [cited 2022 Nov 26];20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Glück S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol; 2017. [cited 2022 Jul 21];19:1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang M, Jiang M, Chen P, Chen P, Wang L, Li W, et al. cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol; 2020 [cited 2022 Nov 26];13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flood BA, Higgs EF, Li S, Luke JJ, Gajewski TF. STING pathway agonism as a cancer therapeutic. IImmunol Rev; 2019. [cited 2022 Jul 21];290:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parkes EE, Humphries MP, Gilmore E, Sidi FA, Bingham V, Phyu SM, et al. The clinical and molecular significance associated with STING signaling in breast cancer. NPJ Breast Cancer; 2021 [cited 2022 Nov 26];7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun; 2014 [cited 2022 Nov 26];5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lemos H, Mohamed E, Huang L, Ou R, Pacholczyk G, Arbab AS, et al. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res; 2016. [cited 2022 Nov 26];76:2076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science; 2019 [cited 2022 Nov 26];363. [DOI] [PubMed] [Google Scholar]
- 127.Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature; 2018. [cited 2022 Nov 26];553:467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med [Internet]. J Exp Med; 2018. [cited 2022 Jul 21];215:1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Vugt MATM Parkes EE. When breaks get hot: inflammatory signaling in BRCA1/2-mutant cancers. Trends in cancer [Internet]. Trends Cancer; 2022. [cited 2022 Jul 21];8:174–89. [DOI] [PubMed] [Google Scholar]
- 130.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature [Internet]. Nature; 2018. [cited 2022 Jul 21];563:131–6. [DOI] [PubMed] [Google Scholar]
- 131.Hong C, Schubert M, Tijhuis AE, Requesens M, Roorda M, van den Brink A, et al. cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature; 2022 [cited 2022 Jul 21];607. [DOI] [PubMed] [Google Scholar]
- 132.Lee KM, Lin CC, Servetto A, Bae J, Kandagatla V, Ye D, et al. Epigenetic Repression of STING by MYC Promotes Immune Evasion and Resistance to Immune Checkpoint Inhibitors in Triple-Negative Breast Cancer. Cancer Immunol Res; 2022. [cited 2022 Nov 26];10:829–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao H, Wang S, Zhou X, Sun J, Zhou G, Zhou D, et al. STING promotes proliferation and induces drug resistance in colorectal cancer by regulating the AMPK-mTOR pathway. J Gastrointest Oncol [Internet]. AME Publishing Company; 2022. [cited 2022 Nov 26];13:2458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vasiyani H, Mane M, Rana K, Shinde A, Roy M, Singh J, et al. DNA damage induces STING mediated IL-6-STAT3 survival pathway in triple-negative breast cancer cells and decreased survival of breast cancer patients. Apoptosis; 2022. [cited 2022 Nov 26];27:961–78. [DOI] [PubMed] [Google Scholar]
- 135.Tian J, Zhang D, Kurbatov V, Wang Q, Wang Y, Fang D, et al. 5-Fluorouracil efficacy requires anti-tumor immunity triggered by cancer-cell-intrinsic STING. EMBO J; 2021 [cited 2022 Nov 26];40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Grabosch S, Bulatovic M, Zeng F, Ma T, Zhang L, Ross M, et al. Cisplatin-induced immune modulation in ovarian cancer mouse models with distinct inflammation profiles. Oncogene [Internet]. Oncogene; 2019. [cited 2022 Nov 26];38:2380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity [Internet]. Immunity; 2014. [cited 2022 Nov 26];41:843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Syu JP, Chi JT, Kung HN. Nrf2 is the key to chemotherapy resistance in MCF7 breast cancer cells under hypoxia. Oncotarget [Internet]. Oncotarget; 2016. [cited 2022 Nov 26];7:14659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chipurupalli S, Ganesan R, Dhanabal SP, Kumar MS, Robinson N. Pharmacological STING Activation Is a Potential Alternative to Overcome Drug-Resistance in Melanoma. Front Oncol; 2020 [cited 2022 Nov 26];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yum S, Li M, Chen ZJ. Old dogs, new trick: classic cancer therapies activate cGAS. Cell Res; 2020. [cited 2022 Nov 26];30:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lohard S, Bourgeois N, Maillet L, Gautier F, Fétiveau A, Lasla H, et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat Commun; 2020 [cited 2022 Nov 26];11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karakaidos P, Karagiannis D, Rampias T. Resolving DNA Damage: Epigenetic Regulation of DNA Repair. Molecules; 2020 [cited 2022 Jul 8];25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mir US, Bhat A, Mushtaq A, Pandita S, Altaf M, Pandita TK. Role of histone acetyltransferases MOF and Tip60 in genome stability. DNA Repair (Amst); 2021 [cited 2022 Sep 30];107. [DOI] [PubMed] [Google Scholar]
- 144.Mushtaq A, Mir US, Hunt CR, Pandita S, Tantray WW, Bhat A, et al. Role of Histone Methylation in Maintenance of Genome Integrity. Genes (Basel); 2021 [cited 2022 Jul 8];12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Singh M, Bacolla A, Chaudhary S, Hunt CR, Pandita S, Chauhan R, et al. Histone Acetyltransferase MOF Orchestrates Outcomes at the Crossroad of Oncogenesis, DNA Damage Response, Proliferation, and Stem Cell Development. Mol Cell Biol; 2020 [cited 2022 Jul 8];40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV., Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science; 2009. [cited 2022 Apr 23];324:1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zaidi N, Swinnen JV., Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res; 2012. [cited 2022 Apr 23];72:3709–14. [DOI] [PubMed] [Google Scholar]
- 148.Sivanand S, Rhoades S, Jiang Q, Lee JV., Benci J, Zhang J, et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol Cell; 2017. [cited 2022 Apr 23];67:252–265.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei X, Shi J, Lin Q, Ma X, Pang Y, Mao H, et al. Targeting ACLY Attenuates Tumor Growth and Acquired Cisplatin Resistance in Ovarian Cancer by Inhibiting the PI3K-AKT Pathway and Activating the AMPK-ROS Pathway. Front Oncol; 2021 [cited 2022 Nov 26];11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De P, Sun Y, Carlson JH, Friedman LS, Leyland-Jones BR, Dey N. Doubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-ness. Neoplasia; 2014. [cited 2022 Nov 26];16:43–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Efimova E V, Appelbe OK, Ricco N, Lee SSY, Liu Y, Wolfgeher DJ, et al. O-GlcNAcylation Enhances Double-Strand Break Repair, Promotes Cancer Cell Proliferation, and Prevents Therapy-Induced Senescence in Irradiated Tumors. Mol Cancer Res; 2019. [cited 2022 Apr 23];17:1338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature; 2009. [cited 2022 Apr 23];462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sulkowski PL, Oeck S, Dow J, Economos NG, Mirfakhraie L, Liu Y, et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature [Internet]. Nature; 2020. [cited 2022 Apr 23];582:586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chou FJ, Liu Y, Lang F, Yang C. D-2-Hydroxyglutarate in Glioma Biology. Cells; 2021 [cited 2022 Nov 26];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Núñez FJ, Mendez FM, Kadiyala P, Alghamri MS, Savelieff MG, Garcia-Fabiani MB, et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Transl Med; 2019 [cited 2022 Nov 26];11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zahra K, Dey T, Ashish, Mishra SP, Pandey U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front Oncol; 2020. [cited 2022 Apr 23];10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sizemore ST, Zhang M, Cho JH, Sizemore GM, Hurwitz B, Kaur B, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res; 2018. [cited 2022 Jul 8];28:1090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sizemore ST, Zhang M, Cho JH, Sizemore GM, Hurwitz B, Kaur B, et al. Pyruvate kinase M2 regulates homologous recombination-mediated DNA double-strand break repair. Cell Res; 2018;28:1090–102. [DOI] [PMC free article] [PubMed] [Google Scholar]