Abstract
In multiple sclerosis clinical trials, MRI outcome measures are typically extracted at a whole-brain level, but pathology is not homogeneous across the brain and so whole-brain measures may overlook regional treatment effects. Data-driven methods, such as independent component analysis, have shown promise in identifying regional disease effects but can only be computed at a group level and cannot be applied prospectively. The aim of this work was to develop a technique to extract longitudinal independent component analysis network-based measures of co-varying grey matter volumes, derived from T1-weighted volumetric MRI, in individual study participants, and assess their association with disability progression and treatment effects in clinical trials. We used longitudinal MRI and clinical data from 5089 participants (22 045 visits) with multiple sclerosis from eight clinical trials. We included people with relapsing–remitting, primary and secondary progressive multiple sclerosis. We used data from five negative clinical trials (2764 participants, 13 222 visits) to extract the independent component analysis-based measures. We then trained and cross-validated a least absolute shrinkage and selection operator regression model (which can be applied prospectively to previously unseen data) to predict the independent component analysis measures from the same regional MRI volume measures and applied it to data from three positive clinical trials (2325 participants, 8823 visits). We used nested mixed-effect models to determine how networks differ across multiple sclerosis phenotypes are associated with disability progression and to test sensitivity to treatment effects. We found 17 consistent patterns of co-varying regional volumes. In the training cohort, volume loss was faster in four networks in people with secondary progressive compared with relapsing–remitting multiple sclerosis and three networks with primary progressive multiple sclerosis. Volume changes were faster in secondary compared with primary progressive multiple sclerosis in four networks. In the combined positive trials cohort, eight independent component analysis networks and whole-brain grey matter volume measures showed treatment effects, and the magnitude of treatment–placebo differences in the network-based measures was consistently greater than with whole-brain grey matter volume measures. Longitudinal network-based analysis of grey matter volume changes is feasible using clinical trial data, showing differences cross-sectionally and longitudinally between multiple sclerosis phenotypes, associated with disability progression, and treatment effects. Future work is required to understand the pathological mechanisms underlying these regional changes.
Keywords: multiple sclerosis, independent component analysis, grey matter, disability, treatment effects
Colato et al. present a new network-based measure of brain atrophy using longitudinal MRI that could be used prospectively in multiple sclerosis clinical trials and found networks of co-varying grey matter volumes that differed across clinical phenotypes, were associated with disability progression and showed treatment effects.
Graphical Abstract
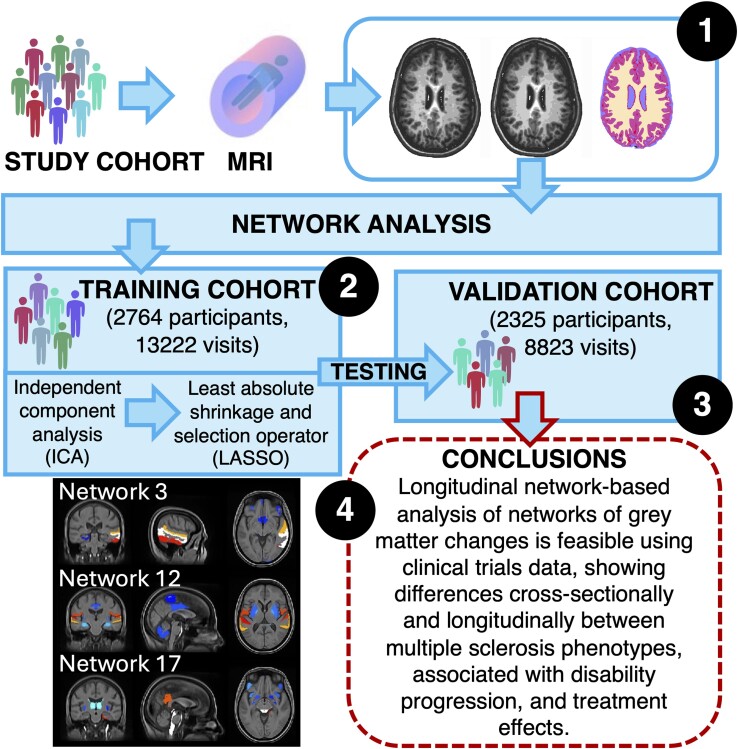
Graphical abstract.
Introduction
In multiple sclerosis, the main cause of irreversible progressive disability is thought to be neurodegeneration.1,2 While we have many treatments approved for relapsing–remitting (RR) multiple sclerosis, only a few have proven effective in progressive multiple sclerosis when there is still evidence of focal inflammatory activity. A key question for clinical trials is whether we can slow neurodegeneration that is not immediately linked with focal inflammation. Sensitive, and clinically relevant, measures of neurodegeneration are needed to address this question. Brain atrophy, measured with MRI, is the main in vivo marker of neurodegeneration used in clinical trials.3-6 However, it is usually measured at a whole-brain or whole grey matter (GM) level, but it has been shown that atrophy varies substantially between brain regions and that this differs between multiple sclerosis clinical phenotypes.7
Recently, independent component analysis (ICA) has been applied to brain MRI scans to unpick overlapping patterns, or networks, of GM atrophy.8,9 Bergsland et al.8 performed a longitudinal study using voxel-wise, network-based, ICA-based approach and found patterns including structurally or functionally related GM regions. It has been postulated that some brain networks (structurally or functionally connected brain regions) are more vulnerable to, or specifically targeted by, neurodegenerative pathologies, which leads to the complex patterns of regional brain atrophy that are seen.10 Furthermore, it has been suggested that network-based measures may better explain clinical outcomes than whole-brain measures and be more sensitive to clinically relevant changes in early-phase clinical trials.11 For example, we and others have shown that MRI measures derived from the motor network are more closely associated with Expanded Disability Status Scale (EDSS) than the whole-brain MRI measures.12 A crucial and unresolved question is whether the dynamics of structural network changes differ between multiple sclerosis phenotypes, and if so, how this relates to clinical progression. To our knowledge, no study has investigated the longitudinal evolution of GM volumes using a data-driven network-based approach.
While ICA is well established as a data-driven tool for network discovery, producing measures for individuals within a cohort, it must be computed simultaneously for the whole group. If there are new participants or time points, we need to re-estimate the whole ICA model using the new sample, which is computationally cumbersome and precludes its use prospectively in clinical trials, for example, to calculate interim outcome measures. Machine learning models are adept at mapping complex inputs and outputs at an individual level, and such models can be used to predict outcomes for new input data (supervised learning).13,14 The least absolute shrinkage and selection operator (Lasso) method is a machine learning regression method that can robustly model a large number of variables, which would not be possible with linear regression models.13 This approach is potentially well suited to map features extracted at a group level using ICA to measures that can prospectively assess individual patient network measures, without the need to refit ICA to the whole population of people when there is a new participant or time point. This in turn may be used to assess the effects of treatments on longitudinal network-based measures at an individual level.
In this study, we aimed to (i) develop a novel longitudinal measure of brain structural network changes that could be applied to previously unseen data, (ii) assess its potential utility by investigating relationships of the network-based measures with disability progression and multiple sclerosis phenotypes and (iii) determine if network measures can detect a treatment effect in clinical trials.
Materials and methods
Participants
We used longitudinal data from eight randomized, double-blind trials in RR multiple sclerosis, secondary progressive (SP) multiple sclerosis and primary progressive (PP) multiple sclerosis, collected under the auspices of the International Progressive Multiple Sclerosis Alliance and the Multiple Sclerosis-Secondary Progressive Multi-Arm Randomization Trial (MS-SMART) (Supplementary Table 1).5,15-20 We divided participants into training and application cohorts. The training sample included five clinical trials in RR multiple sclerosis, SP multiple sclerosis and PP multiple sclerosis that did not report significant treatment effects at a group level, and we used this cohort to train our models. In the application sample, we included two clinical trials in RR multiple sclerosis and one in PP multiple sclerosis that showed significant treatment effects. We divided the cohorts in this way to reduce the risks that treatment effects could distort model training and assess known treatment effects in the application cohort in greater detail.
MRI processing
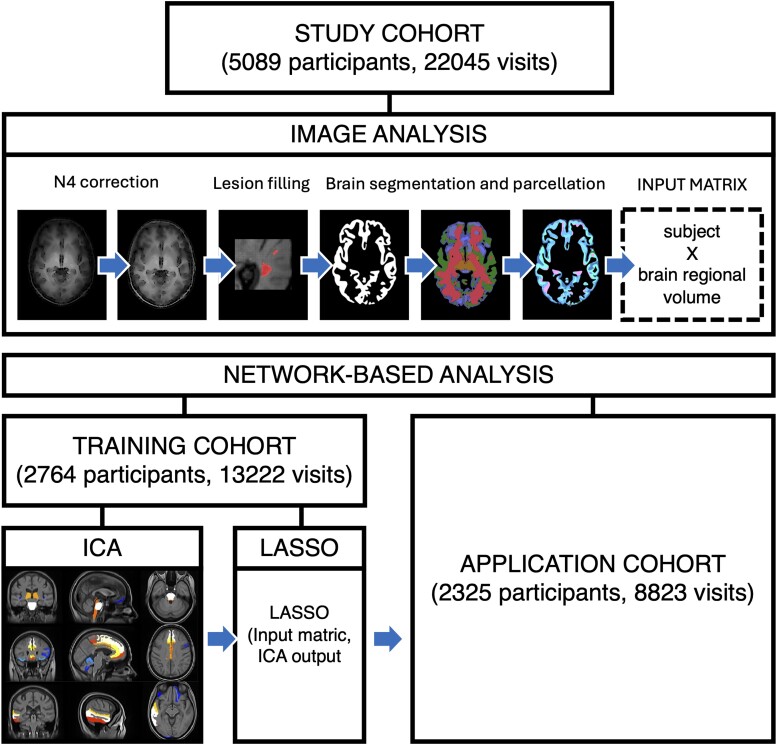
We processed MRI data from the training and application cohorts as shown in Fig. 1 and detailed below. We used a pipeline similar to our previous work21 to process T1-weighted MRI scans and obtain GM volumes. We corrected T1-weighted scans for scanner inhomogeneities using N4 bias field correction toolbox.22 We used a convolutional–neural network-based model to automatically segment hyperintense lesions on T2-FLAIR scans23; the lesion masks were manually checked and used to fill T1-weighted hypointense lesions as per Prados et al.24 after registration with the Advanced Normalization Tools25; and the resulting images were segmented using Geodesic Information Flows version 3.026 into GM and white matter, and CSF, across 125 brain GM regions based on the Neuromorphometric Atlas.27 We then obtained regional volumes by multiplying the sum of the probability of the segmented tissue voxels in each parcellated region with the voxel volume.
Figure 1.
Study design. We processed all the available images as follows. We corrected T1 images for scanner inhomogeneities, performed lesion filling on T1 images, segmented, parcellated the brain and measured brain GM region volumes. We divided the study cohort into training and application cohorts based on whether data were obtained from a positive or negative clinical trial. We fitted a spatial ICA on the regional GM volume measures data from the training cohort. We fitted a machine learning model, using Lasso, to the ICA measures in the training cohort, and then used the fitted model to obtain network-based measures for each participant and time point in the application cohort.
Network-based analysis
In the training cohort, we used spatial ICA to identify networks of co-varying GM regional volumes from GM regional volume measures. We applied the FastICA algorithm28 implemented in scikit-learn 0.23.1, allowing for up to 20 networks of co-varying brain regions volumes to be identified. We decided to include a larger number of components compared with previous studies using ICA (e.g. Bergsland et al.8 set the number of components to 8) to capture potentially interesting but less strong data patterns.29 Indeed, empirical studies have suggested that including a larger number of components, so potentially over-fitting rather than under-fitting, is better in terms of errors.29 From this, we obtained a network-based measure for each participant and each time point in the training cohort (Fig. 1).
In the training cohort, we then trained a Lasso regression model30 to obtain individual-level network-based measures for participants. We used the same regional GM volumes analysed with ICA as the input measures and the ICA network loadings as the target output measure. We split the training cohort into a training set (70% of the training cohort) and cross-validation set (the remaining 30% of the training cohort), to build and validate the Lasso model. We applied the fitted model to the application cohort to obtain individual-level network-based measures for each participant and each time point without the need to re-estimate the whole model parameters (Fig. 1).
While the training and application cohorts were deliberately separated based on treatment responses, as discussed earlier, we also repeated the ICA in the application cohort and performed pairwise spatial cross-correlations between components from the training and application cohorts to assess their stability. Specifically, we concatenated results from the training and application cohorts into two 4D images (each volume representing a distinct network) and used fslcc31 to determine spatial correlations between each volume of the two images. Additionally, a visual comparison was conducted to further validate the results.
We applied ComBat to harmonize GM network-based measures and whole-brain GM volume for the effect of different MRI vendors and sites using the neuroCombat package (version 1.0.13) in R.
Statistical analysis
We used R (version 4.2) for statistical analysis. We corrected all analyses described below for multiple comparisons using false discovery rate using Benjamini–Hochberg procedure.
ICA and Lasso model characteristics
ICA components are not necessarily parallel to the measures underlying them (for an individual ICA, both positive and negative loading can represent lower GM volumes), and so we performed Pearson correlations between the ICA and Lasso loading for each network and the whole-brain GM volume measures. To aid readability, we inverted the sign of the loading for a network so that for the final results a negative loading equated to lower GM volumes. We used R2 to evaluate model fit. We assessed intra-class correlations between the ICA network measures and measures predicted through Lasso both for the training and the application cohort.
Clinical associations with GM network-based measures
To investigate differences in GM network measures at baseline and over time across multiple sclerosis phenotypes, in the training cohort (which included RR multiple sclerosis, SP multiple sclerosis and PP multiple sclerosis), we used mixed-effect models with Lasso loading factors of each network as the dependent variable. The independent variables included multiple sclerosis phenotype with SP multiple sclerosis as reference group, time (years from baseline) and the interaction between multiple sclerosis phenotype and time as fixed effects, baseline age, sex, disease duration, total intracranial volume and treatment as covariates. We used nested random effects models, in which visit was nested within each subject. We also repeated the analysis with RR multiple sclerosis as reference group. For context, we also repeated the analysis using whole-brain GM volume measures. To confirm that the Lasso model outputs effectively mirrored the ICA loading factors, we repeated these analyses using the loading factors of networks obtained using ICA.
To determine whether network loading factors could explain disability progression, in the training cohort, we built mixed-effect models with EDSS as the dependent variable. We selected eight GM networks involving brain regions (cerebellum, basal ganglia, premotor, associative, temporal and parietal cortices, pons and brainstem) known to be impaired and associated with disability in multiple sclerosis (i.e. Networks 2, 3, 11, 12, 13, 14, 18 and 19).7,21,32-35 These models included GM network loading factor (as z-scores), time and the interaction between them: baseline age, sex, disease duration, total intracranial volume and treatment as independent, fixed-effect variables. Again, we repeated these analyses using both the Lasso model outputs and the source ICA-based loading factors to determine if they were consistent.
To determine the independent predictive ability of network-based measures, in the training cohort, we undertook a stepwise linear regression starting with all the network-based measures that were significant in the mixed-effect models, along with whole-brain GM volumes, and correcting for baseline age, sex, disease duration, treatment line, total intracranial volume and number of visits. The stepwise models were run separately for baseline EDSS scores and longitudinal EDSS changes.
Detecting treatment effects
In the application cohort, to determine whether network-based measures could detect treatment effects, we built a mixed-effect model with the interaction between time and treatment arm, baseline age, sex and total intracranial volume as fixed-effect variables and network measures as outcomes.
We repeated these analyses using Lasso- and ICA-network measures to determine their consistency. For all the significant measures, we estimated the difference in each measure between the first and last visits, computed the effect size by taking the difference between the placebo and treated and ran a sample size calculation (P = 0.05, power = 0.80, pwr package).
Results
Demographics
We included clinical and MRI data for 5089 participants with multiple sclerosis (22 045 visits). The training cohort included data from 2764 participants (1060 RR multiple sclerosis, 1282 SP multiple sclerosis and 422 PP multiple sclerosis), 13 222 visits, with a mean follow-up of 3.4 (SD = 2.5) years. The application cohort comprised data from 2325 participants (1624 RR multiple sclerosis and 701 PP multiple sclerosis), 8823 visits, with a mean follow-up of 2.1 (0.7) years. Table 1 shows the demographic and clinical characteristics of cohorts.
Table 1.
Demographics
| Entire cohort (N = 5089) | Training cohort (N = 2764; 54%) | Application cohort (N = 2325; 46%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Whole | RR multiple sclerosis (N = 1060, 39%) | SP multiple sclerosis (N = 1282, 46%) | PP multiple sclerosis (N = 422, 15%) | Whole | RR multiple sclerosis (N = 1624, 70%) | PP multiple sclerosis (N = 701, 30%) | ||
| Age (years) | 42.4 (10.4) | 45.0 (10.5) | 37.5 (9.1) | 49.5 (8.5) | 49.9 (8.9) | 39.4 (9.5) | 37.1 (9.2) | 44.6 (8.0) |
| Sex (M/F) | 1873/3216 | 966/1798 | 293/767 | 464/818 | 209/213 | 907/1418 | 554/1070 | 353/348 |
| Disease duration (years) | 6.4 (7.1) | 8.71(8.1) | 4.9 (5.2) | 13.5 (8.4) | 4.0 (4.3) | 3.6 (4.5) | 4.0 (4.9) | 2.8 (3.1) |
| Total N visits | 22 045 | 13 222 | 5197 | 6183 | 1842 | 8823 | 6072 | 2751 |
| N visits | 4.3 (1.9) | 4.8 (2.3) | 4.9 (2.7) | 4.8 (2.3) | 4.4 (1.7) | 3.8 (0.8) | 3.7 (0.8) | 3.9 (0.8) |
| Follow-up (years) | 2.8 (2.0) | 3.4 (2.5) | 5.3 (3.1) | 2.2 (0.9) | 2.2 (0.6) | 2.1 (0.7) | 2.2 (0.8) | 2.1 (0.6) |
| Baseline EDSSa | 4.0 [2.5–5.5] | 4.5 [3.0–6.0] | 2.5 [1.5–3.5] | 6.0 [5.5–6.5] | 5.0 [3.5–6.0] | 3.5 [2.0–4.5] | 2.5 [2.0–3.5] | 4.5 [3.5–6.0] |
EDSS, Expanded Disability Status Scale; PP, primary progressive; RR, relapsing–remitting; SP, secondary progressive.
aMedian and first and third interquartile.
The training and application cohorts significantly differed in baseline age, disease duration and EDSS (P < 0.0001). This result was expected because we split the cohort assigning the positive trials to the application cohort as we were interested in testing the treatment effect. Compared with the training cohort, at baseline, participants in the application cohort were significantly younger [mean 39.4 (9.5) versus mean 45.0 (10.5)], with shorter disease duration [mean 3.6 (4.5) years versus mean 8.7 (8.1) years], and more clinically impaired [median EDSS 4.5 (interquartile rage: 3.0–6.0) versus median EDSS 3.5 (2.0–4.5)], all P < 0.0001, and included more males (P < 0.005) (Table 1).
The ICA-identified components were consistent between cohorts
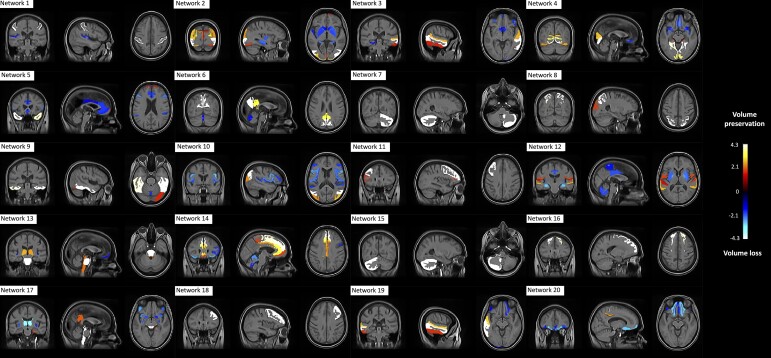
We identified 20 networks of co-varying GM regions (Fig. 2). Spatial cross-correlations between the ICA networks from the training and application cohorts ranged from 0.53 to 1.00 (see Supplementary Table 2). For 17 of these networks, the coefficient of correlation was ≥0.8, and for the remaining three, the coefficient of correlation ranged from 0.53 to 0.75. For a complete description of brain regions involved in each network, see Supplementary Table 3.
Figure 2.
Network of co-varying GM regional volumes. We identified 20 networks of co-varying GM regions. Higher loadings represent higher GM content (brain volume preservation), while lower loadings indicate lower GM content (brain volume loss).
The Lasso model replicated ICA loading factors at an individual level
Intra-class correlation coefficients for Lasso model were greater than 0.99 for all networks. We, therefore, present findings from the Lasso model outputs below, unless otherwise specified.
Group comparison of GM network measures at baseline, and changes over time, across multiple sclerosis phenotypes
See Supplementary Table 4 for the coefficients and P-values for each network. In the training cohort at baseline, the Lasso model-derived loading factors in two networks (13 and 17) significantly differed between people with SP multiple sclerosis and PP multiple sclerosis, and between people with PP multiple sclerosis and RR multiple sclerosis, but not between people with RR multiple sclerosis and SP multiple sclerosis. The loading factor of Network 5 differed between people with RR multiple sclerosis and PP multiple sclerosis and between people with RR multiple sclerosis and SP multiple sclerosis, but not between people with SP multiple sclerosis and PP multiple sclerosis. In addition, the loading factor of four networks (7, 11, 12 and 15) differed between people with RR multiple sclerosis and people with SP multiple sclerosis. Whole-brain GM volumes also differed between RR multiple sclerosis and SP multiple sclerosis and between PP multiple sclerosis and RR multiple sclerosis.
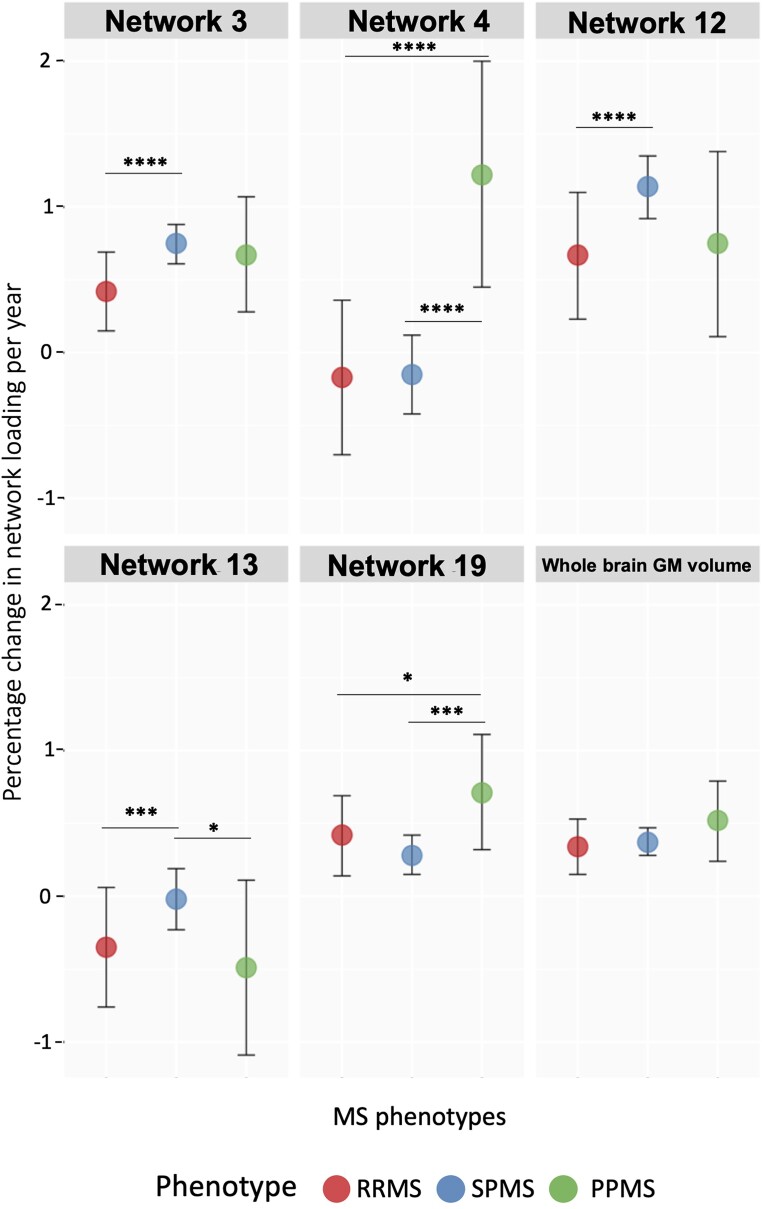
Figure 3 shows an exemplar of how the loading factor rate changes across multiple sclerosis phenotypes. Over time, three of the identified networks showed greater volume loss in people with PP multiple sclerosis than those with SP multiple sclerosis and RR multiple sclerosis (Networks 4, 9 and 19), and two showed a greater volume loss in SP multiple sclerosis than in people with RR multiple sclerosis and PP multiple sclerosis (Networks 13 and 17). In two of these networks (3 and 12), volume loss was faster in SP multiple sclerosis than in RR multiple sclerosis, and the annual change in the loading factor of Network 8 was faster in SP multiple sclerosis and RR multiple sclerosis than in PP multiple sclerosis. Network 14 should have a greater volume loss in people with RR multiple sclerosis than SP multiple sclerosis, Network 7 a faster volume loss in people with SP multiple sclerosis than in PP multiple sclerosis and Network 17 in people with RR multiple sclerosis than in PP multiple sclerosis (Supplementary Table 4, Fig. 3, and Supplementary Fig. 1).
Figure 3.
Examples of the rate of GM volume loss across multiple sclerosis phenotypes over time (see Supplementary Fig. 1 for all networks). We report here an example of the different rates of change in GM networks among multiple sclerosis phenotypes over time. To investigate differences in GM networks measures over time across multiple sclerosis phenotypes, in the training cohort, we used mixed-effect models with Lasso loading factors of each network as the dependent variable. The independent variables included multiple sclerosis phenotype, time (years from baseline) and the interaction between multiple sclerosis phenotype and time as fixed effects, baseline age, sex, disease duration, total intracranial volume and treatment as covariates. We used nested random effects models, in which visit was nested within each subject. Network 3 differed between participants with SP multiple sclerosis and RR multiple sclerosis, showing a greater annual percentage change in network loading (more GM atrophy) in people with SP multiple sclerosis than in RR multiple sclerosis (annual percentage change: 0.75% versus 0.42%, β: 0.003, CI [0.002:0.005], P < 0.0001). Networks 4 and 19 over time differed between people with PP multiple sclerosis and those with RR multiple sclerosis and SP multiple sclerosis, with a faster volume loss rate in people with PP multiple sclerosis [Network 4: annual percentage change of 1.22% versus −0.17% versus −0.15%, β: −0.014, CI (−0.018:0.009), P < 0.0001; β: −0.014, CI (−0.018:0.009), P < 0.0001; Network 19: annual percentage change of 0.72% versus 0.42% versus 28%, β: −0.004, CI (−0.007:−0.002), P < 0.005; β: −0.003, CI (−0.005:−0.001), P < 0.05]. Network 12 differed between participants with SP multiple sclerosis and RR multiple sclerosis, showing a greater annual percentage change in network loading in people with SP multiple sclerosis than in RR multiple sclerosis [Network 12: annual percentage change of 1.14% versus 0.67%, β: 0.005, CI (0.003:0.007), P < 0.005]. Network 13 differed between participants with SP multiple sclerosis and those with RR multiple sclerosis and PP multiple sclerosis, with a faster volume loss rate in people with SP multiple sclerosis [Network 13: annual percentage change of −0.02% versus −0.35% versus −0.49%, β: 0.003, CI (0.001:0.005), P < 0.005; β: 0.005, CI (0.001:0.009), P < 0.05). Whole-brain GM volume did not differ among multiple sclerosis phenotypes after correcting for multiple comparisons. ****P < 0.0001; ***P < 0.005; **P < 0.01; and *P < 0.05. β, beta coefficient; CI, confidence interval; GM, grey matter; MS, multiple sclerosis; RRMS, relapsing–remitting multiple sclerosis; SPMS, secondary progressive multiple sclerosis; PPMS, primary progressive multiple sclerosis.
Substituting the Lasso-generated network measures for the original ICA loading factors yielded very similar results (Supplementary Table 5).
GM network measures are associated with disability and clinical progression in multiple sclerosis
Using the Lasso loading factors, in the training cohort at baseline, network measures were not associated with clinical disability (Table 2), while baseline EDSS scores were associated with whole-brain GM volume [β = −0.03, 95% confidence interval (CI) (−0.05:−0.01), P < 0.05] (Table 2 and Fig. 4).
Table 2.
Cross-sectional and longitudinal association of clinical disability (EDSS) with GM network-based measures (obtained using Lasso), and whole-brain GM measures, in the training cohort
| Entire cohort (β, 95% CI, P) | ||
|---|---|---|
| Network 2 | Baseline | −0.004, −0.029:0.022, P = 0.78 |
| Rate of change | −0.009, −0.016:−0.002, P < 0.01* | |
| Network 3 | Baseline | −0.030, −0.070:0.009, P = 0.14 |
| Rate of change | −0.011, −0.018:−0.004, P < 0.005* | |
| Network 11 | Baseline | −0.017, −0.058:0.023, P = 0.40 |
| Rate of change | −0.004, −0.011:0.004, P = 0.32 | |
| Network 12 | Baseline | −0.036, −0.070:−0.002, P < 0.05 |
| Rate of change | −0.006, −0.014:0.001, P = 0.08 | |
| Network 13 | Baseline | −0.024, −0.059:0.011, P = 0.19 |
| Rate of change | −0.015, −0.022:−0.007, P < 0.0001* | |
| Network 14 | Baseline | −0.026, −0.056:0.005, P = 0.10 |
| Rate of change | −0.009, −0.016:−0.001, P < 0.05* | |
| Network 18 | Baseline | −0.017, −0.059:0.025, P = 0.43 |
| Rate of change | −0.002, −0.010:0.005, P = 0.53 | |
| Network 19 | Baseline | −0.045, −0.081:−0.009, P < 0.05 |
| Rate of change | −0.015, −0.022:−0.007, P < 0.0001* | |
| GM volume | Baseline | −0.18, −0.25:−0.11, P < 0.0001* |
| Rate of change | −0.009, −0.016:−0.001, P < 0.05* |
*P-values that remained significant after correcting for multiple comparisons with the false discovery rate using Benjamini-Hochberg procedure.
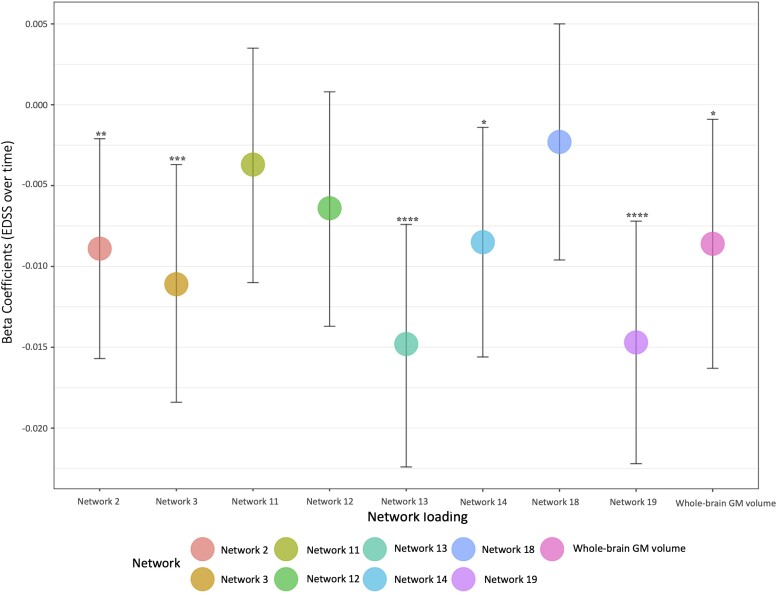
Figure 4.
Networks of co-varying GM regional volumes and whole-brain GM volume measures explain disability over time. Figure shows associations between clinical disability (EDSS) and MRI measures. To determine whether network loading factors could explain disability progression, in the training cohort, we built mixed-effect models with EDSS as the dependent variable and GM network loading factor (as z-scores), time and the interaction between them; baseline age, sex, disease duration, total intracranial volume and treatment as independent, fixed-effect variables. Over time, five GM networks (2, 3, 13, 14 and 19) and the volume of the whole-brain GM were associated with the progression of disability (higher EDSS score) [Network 2: β = −0.01, 95% CI (−0.02:−0.002), P < 0.01; Network 3: β = −0.011, 95% CI (−0.02:−0.004), P < 0.005; Network 13: β = −0.02, 95% CI (−0.02:−0.01), P < 0.0001; Network 14: β = −0.009, 95% CI (−0.02:−0.001) P < 0.05; Network 19: β = −0.015, 95% CI (−0.02:−0.01), P < 0.0001; whole-brain GM: β = −0.01, 95% CI (−0.02:−0.01), P < 0.05]. ****P < 0.0001; ***P < 0.005; **P < 0.01; and *P < 0.05. β, beta coefficient; CI, confidence interval; EDSS, Expanded Disability Status Scale; GM, grey matter; MRI, magnetic resonance imaging.
When we looked at associations between GM network measures and EDSS over time, five networks (2, 3, 13, 14 and 19), and whole-brain GM volumes, were associated with EDSS changes over time [Network 2: β = −0.01, 95% CI (−0.02:−0.002), P < 0.01; Network 3: β = −0.011, 95% CI (−0.02:−0.004), P < 0.005; Network 13: β = −0.02, 95% CI (−0.02:−0.01), P < 0.0001; Network 14: β = −0.009, 95% CI (−0.02:−0.001), P < 0.05; Network 19: β = −0.015, 95% CI (−0.02:−0.01), P < 0.0001; whole-brain GM: β = −0.01, 95% CI (−0.02:−0.01), P < 0.05] (Table 2 and Fig. 4).
Results were similar when using the ICA network measures rather than Lasso loading factors (Supplementary Table 6).
In stepwise regression models, the final model explaining the disability progression, the final model explaining the disability progression included two networks (13 and 19), whole-brain GM volume, baseline age and disease duration (adjusted R2: 0.37). A regression model including only whole-brain GM volume, baseline age and disease duration had an adjusted R2 of 0.35.
When we repeated the analyses in the application cohort, both with Lasso network measures (trained using the ICA loading factors from the training cohort) and ICA loading factors (from the ICA in the application cohort), consistently the loading factor of Network 19, and whole-brain GM measures were associated with baseline disability. Disability progression was not associated with factor loadings or whole-brain GM volume measures (Supplementary Tables 7 and 8).
GM network-based measures are sensitive to treatment effects
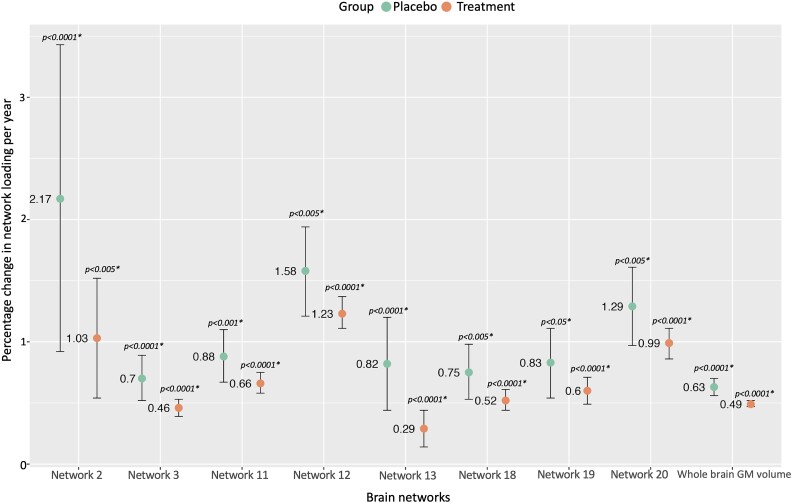
Using the Lasso network measures, in the application cohort, changes in eight GM networks, and whole-brain GM volumes, differed between the treated and the comparator groups (Fig. 5; Supplementary Table 9). Participants in the treated group, compared with comparator arm, had significantly lower annual percentage reductions in GM volumes in Network 2 (1.03% versus 2.17%), 3 (0.46% versus 0.70%), 11 (0.66% versus 0.88%), 12 (1.23% versus 1.58%), 13 (0.29% versus 0.82%), 18 (0.52% versus 0.75%), 19 (0.60% versus 0.83%) and 20 (0.99% versus 1.29%). The magnitude of treatment–placebo differences in the network-based measures was consistently greater for the GM networks compared with whole-brain GM volume measures (GM network treated versus placebo differences ranging from 1.14% to 0.22%, compared with 0.14% at a whole-brain level). Results were similar when using the loading factors derived from the ICA in this cohort (Supplementary Table 10). Sample size estimates, based on the observed differences between treated and comparator arm groups, over an average of 2.1 (0.9) years, with an alpha of 0.05 and power 80%, were lower in five of the eight Lasso-derived network-based measures (from 227 to 565 participants per arm) compared with whole-brain GM volume measures (753 participants per arm) (Supplementary Table 11).
Figure 5.
Networks of co-varying GM and whole-brain GM volume measures show a treatment effect. To determine whether network-based measures could detect treatment effects, we built a mixed-effect model with the interaction between time and treatment arm, baseline age, sex and total intracranial volume as fixed-effect variables and network measures as outcomes. Over time, there is a general decrease in GM volumes. However, eight of the identified GM networks, and whole-brain GM, showed a treatment effect: treated participants had relatively preserved GM (smaller annual percentage change) compared with those in the placebo arm. For instance, Network 13 showed a 0.29% annual percentage change in the treated group [β: −0.003, CI (−0.004:−0.001), P < 0.0001] compared with a 0.82% annual percentage change in the comparator arm [β: −0.005, CI (−0.008:−0.003), P < 0.0001]. ****P < 0.0001; ***P < 0.005; **P < 0.01; and *P < 0.05. β, beta coefficient; CI, confidence interval; GM, grey matter.
Discussion
Using ICA, we identified multiple networks of co-varying GM volumes that differ between multiple sclerosis phenotypes at baseline and evolve more rapidly over time in PP multiple sclerosis and SP multiple sclerosis than in RR multiple sclerosis. Multiple network measures were associated with baseline clinical disability and disability progression. A Lasso model, based on the ICA network measures, when prospectively applied to unseen data from positive clinical trials was able to detect treatment effects that were of greater magnitude than those seen with whole-brain measures.
We found 20 networks of co-varying GM regional volumes, 17 of which were highly (r ≥ 0.8) consistent between the training and application cohorts. In the training cohort, at baseline, the loading factors in two networks differed between people with SP multiple sclerosis and PP multiple sclerosis and between people with PP multiple sclerosis and RR multiple sclerosis. Additionally, the loading factors of five networks differed between people with RR multiple sclerosis and those with progressive multiple sclerosis, while four networks differed between participants with RR multiple sclerosis and SP multiple sclerosis. Over time, people with SP multiple sclerosis showed greater volume loss than those with RR multiple sclerosis in four networks and those with PP multiple sclerosis in four networks. Participants with PP multiple sclerosis had greater volume loss than those with SP multiple sclerosis in three networks and those with RR multiple sclerosis in other three networks. People with RR multiple sclerosis had a faster volume rate in Network 14 than people with SP multiple sclerosis and in the loading factors of two networks than those with PP multiple sclerosis. If GM atrophy can be attributed to a single process affecting the whole brain, albeit progressing at different rates regionally, then we would have expected the same networks to differentiate multiple sclerosis phenotypes cross-sectionally and to change longitudinally, but this is not what we have observed. Instead, our results suggest that, while there are some common regional effects, there are some that are at least partly independent and phenotypically specific. It is possible that tract-mediated effects of white matter lesions could explain this, and it is already known that the distribution of white matter lesion differs between multiple sclerosis phenotypes.36 It would be of interest to see if a disconnectome analysis37 reveals spatially concordant cortical GM regions that also differ between multiple sclerosis phenotypes. However, it is also possible that multiple mechanisms are at work, including some regionally targeted neurodegenerative processes.9,38,39 Further work is now required to understand the pathological processes underlying these dynamic network-based differences and whether they represent multiple sclerosis phenotype-specific treatment targets.
We found that some, but not all, networks of GM volume loss could explain disability progression, as assessed by EDSS. When we consider that the EDSS is essentially a lower limb motor score in the range covered by the cohort included in this study (median EDSS 4.0, range 2.5–5.5), we would expect pathology affecting the motor network to be most relevant. Of the networks that correlated with disability, three out of five overlapped with motor network regions, spanning the cerebellum, thalamus, putamen, pallidum, supplementary motor cortex and supramarginal and postcentral gyri. This result suggests that pathology needs to occur in a clinically eloquent network to have detectable effects, and so not all atrophy will necessarily correlate with a given clinical outcome measure.
Eight GM network-based measures showed treatment effect with a lower percentage annual change in participants from the treated arm than those in the comparator group, and for five of these, the estimated cohort sizes were lower than for whole-brain GM volumes. This was most noticeable for Networks 11 and 13 (requiring cohorts of 477 and 227, respectively, compared with 753 for whole-brain GM volumes), which span brain regions involved in sensory and motor functions, suggesting that such targeted measures are potentially substantially more sensitive treatment effects in multiple sclerosis. The whole-brain measure used in this study was obtained using Geodesic Information Flows, making it directly comparable with the network-based measures, but in many clinical studies whereas SIENA is used, which assesses volume changes directly between images, rather than computing volumes and then subtracting, and this yields potentially more sensitive measures and, therefore, reduces the required sample size. However, using SIENA for this study would not be feasible (5089 participants over 22 045 visits). This is because pairwise comparisons between all visits for each patient are required, and for example, a patient with five visits would introduce more than 10 pairwise comparisons. Using volumetric information with mixed-effect models is more statistically efficient, which is why we chose to use it.
It would be of interest to combine direct measures of change in regional cortical volumes (this is not currently a function of SIENA) and a network-based approach, to see if this further increases potential sensitivity to treatment effects.
Recalling that ICA can only be run at a whole cohort level, i.e. it cannot be applied to unseen data, we developed a Lasso model to replicate the ICA network measures from the source GM regional volumes. The Lasso model achieved intra-class correlation scores of 0.99 with the ICA network measures, and the results (multiple sclerosis phenotype differences, associations with disability and treatment effects) using both Lasso and ICA network measures were materially the same. Further, the Lasso model (trained on a separate cohort) could be applied to another to look for treatment effects, and again results were materially the same as those derived from an ICA in the application cohort. While ideally an ICA would be undertaken when the whole cohort completes a study, the results suggest that the Lasso model measures are a reasonable substitute, and they have the advantage that they can be used in interim analyses or decision-making (for example in multi-arm clinical trials40).
This study was based on clinical trial data, using multiple treatment agents, studied over many years, MRI scanners and sites. This will have added variability to the imaging and clinical data that could have reduced sensitivity to regional atrophy and clinical associations, but equally it gives us confidence that the regional networks of GM atrophy we have identified are not artefacts of specific cohorts or MRI scanners and that it is realistic to expect that the method we have developed can be used in clinical trials. The training and application cohorts were deliberately different, as considered earlier. This meant that the Lasso model built using the ICA loading factors from the training cohort would not have been optimized for the application cohort, and this could have reduced sensitivity to clinical associations. In practice, this effect appears to have been small, given that we were able to replicate all the findings from the Lasso analyses with loading factors from an ICA run in the application cohort despite the demographic differences in cohorts. Moreover, we decided to use data from clinical trials that did not report a positive effect in the training cohort and those that showed a treatment effect in the validation cohort because we did not want results to be biased by the treatment effect. While mixing the trials would provide improved performance, there is a risk that this would come from over-fitting rather than from a true improvement.41 Moreover, mixing patients from the same trial across the train and test sets would introduce several issues: (i) it affects the underlying randomization process, and this would provide biased treatment effects or even misleading ones; (ii) data leakage: by separating the train and test, our approach ensures that our models not only generalize to new groups or cohorts (generalizability) but also have ‘transportability’ meaning that they can be applied in completely new settings. We applied ICA to regional GM volume measures, rather than running voxel- and surface-based analysis. A voxel or surface-based analysis may increase sensitivity to regional effects that do not match one of the Neuromorphometric Atlas regions; however, this approach also has drawbacks. For example, they require images to be registered to a common space (which can also reduce sensitivity to regional effects due to image smoothing) and they would have required a considerably higher computational power (which would currently be impractical for clinical studies). Because we included relatively more data from participants with progressive than RR multiple sclerosis, our cohort was older than the general multiple sclerosis population and network measures might also have been influenced by the effect of ageing. While we corrected all statistical analysis for the effect of age, future studies should investigate the age-specific dynamics in network measures and address network measures after stratifying the cohort according to age.
Conclusion
In conclusion, the significant differences in GM network-based atrophy measures observed between multiple sclerosis phenotypes and their associations with clinical progression suggest that multiple pathological processes may underlie neurodegeneration in multiple sclerosis. Differences between the RR multiple sclerosis and SP multiple sclerosis groups suggest that SP multiple sclerosis is not simply a continuation of patterns of pathology seen in RR multiple sclerosis. Further work will be needed to determine the processes underlying these patterns and how they can best be targeted with treatments. Practically, this work shows that longitudinal network-based analyses of GM atrophy are feasible using clinical trials data and that they show treatment effects.
Supplementary Material
Acknowledgements
The authors are grateful to all the International Progressive MS Alliance investigators (Supplementary Appendix S1) who have contributed trial data to this study as part of EPITOME (Enhancing Power of Intervention Trials Through Optimized MRI Endpoints network). We thank the MS-SMART (Multiple Sclerosis-Secondary Progressive Multi-Arm Randomization Trial) investigators: David Miller, Lorraine Smith, Marios Yiannakas, Marcello Moccia, Almudena Garcia, Alberto Calvi, Tim Friede, Michael Hutchinson, Stanley Hawkins, Robert Swingler, John Thorpe, John O’Brien, and Allan Walker and Alan Thompson, and we thank those people with multiple sclerosis who took part in this trial.
Contributor Information
Elisa Colato, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK.
Jonathan Stutters, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK.
Sridar Narayanan, McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, Quebec, H3A 2B4, Canada.
Douglas L Arnold, McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, Quebec, H3A 2B4, Canada.
Jeremy Chataway, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC), London, W1T 7DN, UK.
Claudia A M Gandini Wheeler-Kingshott, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; Brain Connectivity Centre, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Mondino Foundation, Pavia, 27100, Italy; Department of Brain and Behavioural Sciences, University of Pavia, Pavia, 27100, Italy.
Frederik Barkhof, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC), London, W1T 7DN, UK; Department of Radiology and Nuclear Medicine, Vrije Universiteit (VU) Medical Centre, Amsterdam, 1081 HZ, The Netherlands; Centre for Medical Image Computing (CMIC), Department of Medical Physics and Biomedical Engineering, University College London, London, WC1V 6LJ, UK.
Olga Ciccarelli, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC), London, W1T 7DN, UK.
Arman Eshaghi, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; Centre for Medical Image Computing (CMIC), Department of Computer Science, University College London, London, WC1V 6LJ, UK.
Declan T Chard, Queen Square Multiple Sclerosis Centre, Department of Neuroinflammation, University College London (UCL) Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, WC1N 3BG, UK; National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC), London, W1T 7DN, UK.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
This investigation was supported in part by an award from the International Progressive Multiple Sclerosis Alliance (reference number PA-1412-02420). D.T.C., F.B. and O.C. are supported by the National Institute for Health and Care Research (NIHR) biomedical research centre at the University College London Hospitals (UCLH). The Multiple Sclerosis-Secondary Progressive Multi-Arm Randomization Trial (MS-SMART) trial was funded by the Efficacy and Mechanism Evaluation program as project number 11/30/11. MS-SMART is an investigator-led project sponsored by University College London. This independent research is awarded by and funded by the Medical Research Council, the United Kingdom Multiple Sclerosis Society and the National Multiple Sclerosis Society and is managed by the National Institute for Health Research on behalf of the Medical Research Council–National Institute for Health partnership. Additional support came from the University of Edinburgh; the National Institute for Health Research University College London Hospitals Biomedical Research Centre and University College London; and the National Institute for Health Research Leeds Clinical Research Facility (Dental Translational and Clinical Research Unit).
Competing interests
The authors report no competing interests with respect to this research. The full disclosure statement is as follows: D.L.A. has received personal compensation for serving as a Consultant for Alexion, Biogen, Celgene, Eli Lilly, EMD Serono, Frequency Therapeutics, Genentech, Merck, Novartis, Roche, Sanofi and Shionogi and holds an equity interest in NeuroRx. F.B. has received compensation for consulting services and/or speaking activities from Bayer Schering Pharma, Biogen Idec, Merck Serono, Novartis, Genzyme, Synthon BV, Roche, Teva, Jansen research and IXICO and is supported by the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre. O.C. has received research grants from the Multiple Sclerosis Society of Great Britain & Northern Ireland, the NIHR UCLH Biomedical Research Centre, EUH2020, Spinal Cord Research Foundation and Rosetrees Trust. She serves as a consultant for Novartis, Teva and Roche and has received an honorarium from the American Academy of Neurology as Associate Editor of Neurology and serves on the Editorial Board of Multiple Sclerosis Journal. D.T.C. is a consultant Hoffmann-La Roche. In the last three years, he has been a consultant for Biogen and has received research funding from Hoffmann-La Roche, the International Progressive Multiple Sclerosis Alliance, the Medical Research Council, the Multiple Sclerosis Society and the NIHR UCLH Biomedical Research Centre and a speaker's honorarium from Novartis. He co-supervises a clinical fellowship at the National Hospital for Neurology and Neurosurgery, London, which is supported by Merck. A.E. has received speaker's honoraria from Biogen and At The Limits educational programme. A.E. has received research grants from Medical Research Council, UK Research and Innovation's Innovate UK, Biogen, UCL Innovation and Enterprise and Roche. A.E. serves on the Editorial Board of Neurology. He has received travel support from the National Multiple Sclerosis Society and honorarium from the Journal of Neurology, Neurosurgery and Psychiatry for Editorial Commentaries. A.E. and F.B. have equity stake in Queen Square Analytics. J.C. has received support from the Efficacy and Evaluation (EME) Programme, a Medical Research Council (MRC) and NIHR partnership and the Health Technology Assessment (HTA) Programme (NIHR), the UK Multiple Sclerosis Society, the US National Multiple Sclerosis Society and the Rosetrees Trust. He is supported in part by the NIHR UCLH Biomedical Research Centre. He has been a local principal investigator for a trial in Multiple Sclerosis funded by the Canadian Multiple Sclerosis Society. A local principal investigator for commercial trials was funded by Ionis, Novartis and Roche and has taken part in advisory boards/consultancy for Azadyne, Biogen, Lucid, Janssen, Merck, NervGen, Novartis and Roche. E.C. and J.S. have nothing to disclose.
Data availability
Data used in this work are controlled by several pharmaceutical companies and cannot be shared directly. Applications for access should be directed to the companies controlling each dataset. The computer codes to obtain longitudinal individual-level network-based measures can be found at: https://github.com/co-el/Longitudinal_individual_level_structural_network.
References
- 1. Fisher E, Lee JC, Nakamura K, Rudick RA. Gray matter atrophy in multiple sclerosis: A longitudinal study. Ann Neurol. 2008;64(3):255–265. [DOI] [PubMed] [Google Scholar]
- 2. Roosendaal SD, Bendfeldt K, Vrenken H, et al. Grey matter volume in a large cohort of MS patients: Relation to MRI parameters and disability. Mult Scler J. 2011;17(9):1098–1106. [DOI] [PubMed] [Google Scholar]
- 3. Chataway J, Nicholas R, Todd S, et al. A novel adaptive design strategy increases the efficiency of clinical trials in secondary progressive multiple sclerosis. Mult Scler J. 2011;17(1):81–88. [DOI] [PubMed] [Google Scholar]
- 4. Chataway J, Schuerer N, Alsanousi A, et al. Effect of high-dose simvastatin on brain atrophy and disability in secondary progressive multiple sclerosis (MS-STAT): A randomised, placebo-controlled, phase 2 trial. Lancet. 2014;383(9936):2213–2221. [DOI] [PubMed] [Google Scholar]
- 5. Montalban X, Hauser SL, Kappos L, et al. Ocrelizumab versus placebo in primary progressive multiple sclerosis. N Engl J Med. 2017;376(3):209–220. [DOI] [PubMed] [Google Scholar]
- 6. Kapoor R, Furby J, Hayton T, et al. Lamotrigine for neuroprotection in secondary progressive multiple sclerosis: A randomised, double-blind, placebo-controlled, parallel-group trial. Lancet Neurol. 2010;9(7):681–688. [DOI] [PubMed] [Google Scholar]
- 7. Pagani E, Rocca MA, Gallo A, et al. Regional brain atrophy evolves differently in patients with multiple sclerosis according to clinical phenotype. Am J Neuroradiol. 2005;26(2):341–346. [PMC free article] [PubMed] [Google Scholar]
- 8. Bergsland N, Horakova D, Dwyer MG, et al. Gray matter atrophy patterns in multiple sclerosis: A 10-year source-based morphometry study. Neuroimage Clin. 2018;17:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steenwijk MD, Geurts JJG, Daams M, et al. Cortical atrophy patterns in multiple sclerosis are non-random and clinically relevant. Brain. 2016;139(1):115–126. [DOI] [PubMed] [Google Scholar]
- 10. Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–198. [DOI] [PubMed] [Google Scholar]
- 11. Chard DT, Alahmadi AAS, Audoin B, et al. Mind the gap: From neurons to networks to outcomes in multiple sclerosis. Nat Rev Neurol. 2021;17(3):173–184. [DOI] [PubMed] [Google Scholar]
- 12. Pardini M, Yaldizli Ö, Sethi V, et al. Motor network efficiency and disability in multiple sclerosis. Neurology. 2015;85(13):1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Myszczynska MA, Ojamies PN, Lacoste AMB, et al. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat Rev Neurol. 2020;16(8):440–456. [DOI] [PubMed] [Google Scholar]
- 14. Badillo S, Banfai B, Birzele F, et al. An introduction to machine learning. Clin Pharmacol Ther. 2020;107(4):871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spain R, Powers K, Murchison C, et al. Lipoic acid in secondary progressive MS. Neurol Neuroimmunol Neuroinflamm. 2017;4(5):e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chataway J, De Angelis F, Connick P, et al. Efficacy of three neuroprotective drugs in secondary progressive multiple sclerosis (MS-SMART): A phase 2b, multiarm, double-blind, randomised placebo-controlled trial. Lancet Neurol. 2020;19(3):214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hawker K, O’Connor P, Freedman MS, et al. Rituximab in patients with primary progressive multiple sclerosis: Results of a randomized double-blind placebo-controlled multicenter trial. Ann Neurol. 2009;66(4):460–471. [DOI] [PubMed] [Google Scholar]
- 18. Gold R, Kappos L, Arnold DL, et al. Placebo-Controlled phase 3 study of Oral BG-12 for relapsing multiple sclerosis. N Engl J Med. 2012;367(12):1098–1107. [DOI] [PubMed] [Google Scholar]
- 19. Hauser SL, Bar-Or A, Comi G, et al. Ocrelizumab versus interferon Beta-1a in relapsing multiple sclerosis. N Engl J Med. 2017;376(3):221–234. [DOI] [PubMed] [Google Scholar]
- 20. Kapoor R, Ho PR, Campbell N, et al. Effect of natalizumab on disease progression in secondary progressive multiple sclerosis (ASCEND): A phase 3, randomised, double-blind, placebo-controlled trial with an open-label extension. Lancet Neurol. 2018;17(5):405–415. [DOI] [PubMed] [Google Scholar]
- 21. Eshaghi A, Prados F, Brownlee WJ, et al. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol. 2018;83(2):210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 24. Prados F, Cardoso MJ, Kanber B, et al. A multi-time-point modality-agnostic patch-based method for lesion filling in multiple sclerosis. Neuroimage. 2016;139:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tustison NJ, Holbrook AJ, Avants BB, et al. The ANTs longitudinal cortical thickness pipeline. BioRxiv Preprint. 2017:1–43. 10.1101/170209 [DOI] [Google Scholar]
- 26. Cardoso MJ, Modat M, Wolz R, et al. Geodesic information flows: Spatially-variant graphs and their application to segmentation and fusion. IEEE Trans Med Imaging. 2015;34(9):1976–1988. [DOI] [PubMed] [Google Scholar]
- 27. Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hyvärinen A, Oja E. Independent component analysis: Algorithms and applications. Neural Netw. 2000;13(4–5):411–430. [DOI] [PubMed] [Google Scholar]
- 29. Esposito F, Scarabino T, Hyvarinen A, et al. Independent component analysis of fMRI group studies by self-organizing clustering. Neuroimage. 2005;25(1):193–205. [DOI] [PubMed] [Google Scholar]
- 30. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in python. JMLR. 2011;12:2825–2830. [Google Scholar]
- 31. Prestel M, Steinfath TP, Tremmel M, Stark R, Ott U. fMRI BOLD correlates of EEG independent components: Spatial correspondence with the default mode network. Front Hum Neurosci. 2018;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cordani C, Hidalgo de la Cruz M, Meani A, et al. MRI correlates of clinical disability and hand-motor performance in multiple sclerosis phenotypes. Mult Scler J. 2021;27(8):1205–1221. [DOI] [PubMed] [Google Scholar]
- 33. Geurts JJG, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. Lancet Neurol. 2012;11(12):1082–1092. [DOI] [PubMed] [Google Scholar]
- 34. Khaleeli Z, Cercignani M, Audoin B, Ciccarelli O, Miller DH, Thompson AJ. Localized grey matter damage in early primary progressive multiple sclerosis contributes to disability. Neuroimage. 2007;37(1):253–261. [DOI] [PubMed] [Google Scholar]
- 35. Solana E, Martinez-Heras E, Montal V, et al. Regional grey matter microstructural changes and volume loss according to disease duration in multiple sclerosis patients. Sci Rep. 2021;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dalton CM, Bodini B, Samson RS, et al. Brain lesion location and clinical status 20 years after a diagnosis of clinically isolated syndrome suggestive of multiple sclerosis. Mult Scler. 2012;18(3):322–328. [DOI] [PubMed] [Google Scholar]
- 37. Ravano V, Andelova M, Fartaria MJ, et al. Validating atlas-based lesion disconnectomics in multiple sclerosis: A retrospective multi-centric study. Neuroimage Clin. 2021;32:102817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chard DT, Miller DH. What lies beneath grey matter atrophy in multiple sclerosis? Brain. 2016;139(1):7–10. [DOI] [PubMed] [Google Scholar]
- 39. Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJG, Reynolds R, Martin R. Exploring the origins of grey matter damage in multiple sclerosis. Nat Rev Neurosci. 2015;16(3):147–158. [DOI] [PubMed] [Google Scholar]
- 40. Li V, Leurent B, Barkhof F, et al. Designing multi-arm multistage adaptive trials for neuroprotection in progressive multiple sclerosis. Neurology. 2022;98(18):754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Collins GS, Dhiman P, Ma J, et al. Evaluation of clinical prediction models (part 1): From development to external validation. BMJ. 2024;384:e074819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this work are controlled by several pharmaceutical companies and cannot be shared directly. Applications for access should be directed to the companies controlling each dataset. The computer codes to obtain longitudinal individual-level network-based measures can be found at: https://github.com/co-el/Longitudinal_individual_level_structural_network.