Abstract
Secondary and tertiary structures in the 3′ untranslated region (UTR) of plus-strand RNA viruses have been postulated to function as control elements in RNA replication, transcription, and translation. Here we describe a 54-nucleotide (nt) hairpin-type pseudoknot within the 288-nt 3′ UTR of the bovine coronavirus genome and show by mutational analysis of both stems that the pseudoknotted structure is required for the replication of a defective interfering RNA genome. The pseudoknot is phylogenetically conserved among coronaviruses both in location and in shape but only partially in nucleotide sequence, and evolutionary covariation of bases to maintain G · U pairings indicates that it functions in the plus strand. RNase probing of synthetic transcripts provided additional evidence of its tertiary structure and also identified the possible existence of two conformational states. These results indicate that the 3′ UTR pseudoknot is involved in coronavirus RNA replication and lead us to postulate that it functions as a regulatory control element.
Coronaviruses are enveloped, nonsegmented, intracytoplasmically replicating, plus-strand RNA viruses with the largest known viral RNA genome (approximately 30 kb) (reviewed in references 31 and 38). During replication, a 3′-coterminal nested set of subgenomic mRNAs are synthesized by an unresolved mechanism of discontinuous transcription (3, 51, 61) that places a common 5′-terminal leader sequence (only part of the 5′ untranslated region [UTR]) on each mRNA. It has been suggested that the common 5′ and 3′ termini on genomic and subgenomic mRNAs enable these molecules to amplify by a replication mechanism (57). Such a pathway would explain the existence of minus-strand copies of subgenomic mRNAs (20, 21, 56, 57) and of subgenomic mRNA-length replicative intermediates (50, 52). Replication of coronavirus RNA molecules from input plus strands, however, has been demonstrated for only the viral genome (when extracted from virions and transfected into uninfected cells [7, 53]) and defective interfering (DI) RNAs (when synthesized in vitro from cDNA clones and transfected into helper virus-infected cells [11, 29]), leaving unresolved by direct proof the degree of replicability of coronavirus subgenomic mRNAs. If coronavirus subgenomic mRNAs are lacking in signals for replication, it is unlikely that they would map within the 3′ UTR since this region is identical among the genome and subgenomic mRNAs (8).
Several reports have provided evidence for higher-order structural elements in the 3′ UTRs of plus-strand RNA viruses that are thought to function in RNA replication or translation by interacting with viral or cellular proteins. Stem-loop structures in the 3′ UTR of rubella virus (42), West Nile virus (4), and hepatitis C virus (5, 25) represent recognition sites for cellular proteins. The recruitment of a trans-acting factor(s) to postulated higher-order structures in other animal viruses including picornaviruses (26, 40, 41, 45, 46, 64), flaviviruses (59), and coronaviruses (24) has not been elucidated. Among plant viruses, alfalfa mosaic virus coat protein binds a 3′ UTR with extensive secondary structure (2) and cellular proteins recognize higher-order motifs within the 3′ termini (14, 15, 19) and in upstream sequences (33). The functional relevance of a tertiary interaction involving the poly(A) tail and 3′ UTR of bamboo mosaic virus has not yet been characterized (65).
The use of coronavirus DI RNAs to define replication signaling elements within genomic termini would seem reasonable since the 5′ and 3′ UTRs on known DI RNAs, the only available cloned coronavirus replicons, are identical to those on cognate virus genomes (8). Thus, in the bovine coronavirus (BCV) genome and DI RNA, the 5′ UTR is 210 nucleotides (nt) in length, inclusive of the common 65-nt leader sequence, and the 3′ UTR is 288 nt, exclusive of the poly(A) tail (11, 32). To investigate the potential for signaling structural elements within the BCV 3′ UTR, a computer-based RNA-folding algorithm was used to predict thermodynamically stable secondary structures. We then analyzed a potential pseudoknot beginning 63 nt downstream from the termination codon of the N gene by genetic and biochemical approaches, including phylogenetic comparisons, site-directed mutagenesis in a cloned DI RNA followed by Northern blotting, and enzymatic structure probing of in vitro-transcribed RNA. These findings strongly suggest that the pseudoknot plays a role in coronavirus RNA replication.
MATERIALS AND METHODS
Computer prediction of secondary structure.
The Microgenie sequence analysis program (Beckman Instruments) was used to identify inverted repeats and to calculate the free energy of secondary structures by following the rules of Tinoco (63).
Virus and cells.
A DI RNA-free stock of the Mebus strain of BCV at 4.5 × 108 PFU/ml was prepared and used as a helper virus (11). The human rectal tumor cell line HRT-18 was used in all experiments (11).
Construction of mutant DI RNAs.
Mutant DI RNAs were all modifications of pDrep1, a cloned naturally occurring DI RNA modified to contain a 30-nt reporter insert (Fig. 1A) (11). Mutations in the two stems of the pseudoknot were introduced by a method based on the technique of gene splicing by overlap extension (23, 55). The restriction endonuclease sites used in making the mutant DI RNAs are shown in Fig. 1A. To construct the single-mutant pS1L, the gel-purified 532-nt PCR product from a pDrep1-templated reaction with primers BCV#3(−) and BCV3′end(+) and the gel-purified 397-nt PCR product from a pDrep1-templated reaction with primers 5′S1(−) and Reverse(+) were combined in an overlap extension reaction with primers BCV#3(−) and Reverse(+) to form a product of 786 nt. From this, the 566-nt NsiI-MluI fragment was used to replace the corresponding region in pDrep1. All other mutant DI RNAs were similarly constructed, except for the use of primers 3′S1(−), 5′S2(−), and 3′S2(−) in the first PCR mutagenesis reaction (resulting in 365-, 377-, and 352-nt products, respectively) to generate the single mutants pS1R, pS2L, and pS2R, respectively, and the use of pS1L and pS2L template DNA in the 532-nt PCR product reaction to generate the double mutants pS1L/R and pS2L/R, respectively. All transferred PCR fragments were completely sequenced to exclude off-site mutations.
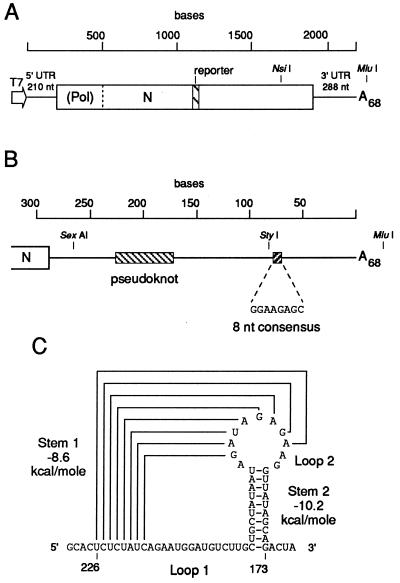
FIG. 1.
Proposed pseudoknot and its location within the BCV 3′ UTR. (A) Cloned reporter-containing DI RNA (pDrep1) of BCV used to test the RNA replication function of the pseudoknot. The cloned DI RNA is under control of a T7 RNA polymerase promoter in plasmid vector pGEM3Zf(−) (Promega). The reporter is an in-frame 30-nt sequence from the N gene of TGEV (11). The numbering starts at the 5′ end of the transcript. (B) Schematic location of the pseudoknot and of an 8-nt consensus sequence (32) in the genomic 3′ UTR. The numbering starts at the base of the poly(A) tail. (C) Proposed pseudoknot structure.
Assay for DI RNA replication.
The assay for DI RNA replication was performed as previously described (11), except that cells at approximately 3.4 × 106 per 35-mm-diameter dish at 80% confluency were infected with BCV at a multiplicity of 25 PFU per cell and transfected with 600 ng of transcript. For passage of progeny virus, supernatant fluids were harvested at 48 h posttransfection and 500 μl was used to directly infect freshly confluent cells in a 35-mm dish. Following Nonidet P-40 lysis, proteinase K digestion, and ethanol precipitation, a set volume from each of the preparations was centrifuged, and the cytoplasmic RNA (typically 2.5 to 5 μg) was applied to individual lanes in a formaldehyde-agarose gel. Approximately 1 ng of transcript was loaded per lane when used as marker. Northern blots were probed with the oligonucleotide TGEV#8(+) 5′-end labeled with 32P to specific activities ranging from 5.5 × 105 to 8.8 × 105 cpm/pmol (Cerenkov counts) and exposed to Kodak XAR-5 film in the presence of an intensifying screen for 6 to 24 h at −80°C. The replication of pDrep1 transcripts was assayed in parallel in all experiments (not shown).
Total RNA extraction and RT-PCR analysis.
Fresh cells were infected with supernatant fluids collected at 48 h posttransfection, and total RNA was isolated with TRIzol reagent (GIBCO BRL) at 6 h postinfection, a time of peak minus-strand RNA synthesis (21). The conditions for cDNA synthesis and PCR were those described by the manufacturer of SuperScript II reverse transcriptase (GIBCO BRL). A 20 μl reverse transcription (RT) reaction was performed with approximately 5 μg of total RNA and the primer BCVDI420(−), complementary to sequences in gene 1a (polymerase) and specific to minus-strand DI and genomic RNAs (11). The cDNA then served as template in a nested-PCR strategy with primers located within the 3′ UTR and the DI reporter sequence. The first PCR, with the outer primers BCVDI420(−) and BCV3′end(+), used 2 μl of cDNA solution and consisted of 40 cycles of DNA amplification (30 s at 94°C, 1 min at 55°C, and 2 min at 72°C), and the second PCR, with the inner primers TGEV#7(−) and BCV3′UTR(+), used 5 μl of the first PCR mixture and consisted of 30 cycles of DNA amplification (30 s at 94°C, 1 min at 55°C, and 1 min at 72°C). Both PCR mixtures contained Taq DNA polymerase (Promega) in a reaction volume of 50 μl, and the reactions included an initial denaturation for 3 min at 94°C and a final extension for 10 min at 72°C. A PCR product with the expected size of 938 bp was gel purified and used in a ligation reaction as specified in the instructions supplied with the TOPO XL PCR cloning kit (Invitrogen).
Enzymatic probing of in vitro-transcribed RNA.
To synthesize a short RNA encompassing the pseudoknot region, a stretch of the 3′ UTR was placed under the T7 promoter. pDrep3 (11), a DI RNA with a 5′ terminus identical to the first 22 nt from the multiple-cloning site of pGEM3Z (Promega), was cut with EcoRI and SexAI (Fig. 1B), filled in with T4 DNA polymerase (New England Biolabs), and religated. The resulting construct, p3NS, was confirmed by sequencing through the junction region. RNA transcribed in vitro from StyI-linearized p3NS with T7 RNA polymerase (Promega) contained 9 nt derived from the vector plus 188 nt of the BCV 3′ UTR.
The method of enzymatic probing was performed as previously described (12), except that 2 μg of template DNA was included in the transcription reaction and RNA was renatured by slow cooling at room temperature. Digestion reactions with RNase T1 (GIBCO BRL) (results not shown), RNase T2 (GIBCO BRL) and RNase V1 (Pharmacia) were done at 4°C for 30 min. The primer BCV3′end(+), complementary to sequences at the 3′ end of the RNA, was 5′-end labeled with 32P, annealed to one-half of the treated RNA, and extended with avian myeloblastosis virus reverse transcriptase (Promega). The products were separated on a 6% polyacrylamide–8.33 M urea gel.
Synthetic oligonucleotides.
The oligonucleotides used in this study are described in Table 1.
TABLE 1.
Synthetic oligonucleotides in this study
| Oligonucleotidea | Sequenceb | Binding regionc |
|---|---|---|
| BCVDI420(−) | 5′GTTGTGTGCAGTCTAGCCTAATAC3′ | 419–442 |
| TGEV#8(+) | 5′CATGGCACCATCCTTGGCAACCCAGA3′ | 1099–1128 |
| TGEV#7(−) | 5′TCTGGGTTGCCAAGGATGGTGCCATG3′ | 1099–1128 |
| BCV#3(−) | 5′TGGGAATCTTGACGAGCCCCAGAAGG3′ | 1542–1567 |
| 5′S1(−) | 5′AAGGCACTCTTGTACAGAATGGATG3′ | 1931–1955 |
| 5′S2(−) | 5′GGATGTCTTGCTCGATTAATAGATAGAG3′ | 1951–1978 |
| 3′S1(−) | 5′GCTATAATAGTACAAGCAGGTTATAGC3′ | 1963–1989 |
| 3′S2(−) | 5′GAGAAGGTTAATCGAGACTATAGATTAATTAG3′ | 1976–2007 |
| BCV3′UTR(+) | 5′CCAACACTATACATTACCAC3′ | 2021–2040 |
| BCV3′end(+) | 5′TCGGCAATTACTTCCGCAAG3′ | 2054–2073 |
| Reverse(+) | 5′CACAGGAAACAGCTATGACC3′ |
Oligonucleotides bind to either plus-strand RNA, as indicated by (+), or to minus-strand RNA, as indicated by (−).
Underlined bases indicate differences from the genomic sequence.
Numbers correspond to the pDrep1 plus-strand sequence, except for oligonucleotides TGEV#8(+) and TGEV#7(−), for which numbers refer to the TGEV N gene plus-strand sequence (11). Reverse(+) binds to pGEM3Zf(−) DNA just downstream of the multiple cloning region.
RESULTS
A predicted pseudoknot in the 3′ UTR of BCV is highly conserved among coronaviruses.
To explore the RNA-folding pattern of the 288-nt 3′ UTR of BCV, computer-assisted secondary-structure analysis was performed with the Tinoco algorithm (63). This thermodynamically based approach predicted the existence of two stem regions between 226 and 173 nt from the base of the poly(A) tail (Fig. 1B) with high negative free-energy values (ΔG = −8.6 and −10.2 kcal[1 cal = 4.184 J]/mol for stems 1 and 2, respectively) and the potential to form a hairpin-type (or classical) pseudoknot (Fig. 1C) (68). A classical pseudoknot is a tertiary interaction involving base pairing between a single-stranded region in a hairpin loop and unpaired bases outside of the loop (reviewed in references 47 and 62). When folded, the base-pairing loop region becomes adjacent to the other stem, leading to coaxial stacking of the two stem regions and formation of a quasi-continuous double helix. The proposed pseudoknot is defined by stems 1 and 2 (8 and 10 bp, respectively), connecting loops 1 and 2 (15 and 2 nt, respectively), and a single intervening nucleotide between the two stems.
The pseudoknot identified in this analysis differs from a classical hairpin pseudoknot in two respects. (i) In a classical pseudoknot, loop 2 is generally larger than loop 1. Loop 1 crosses the deep and narrow major groove of stem 2, and loop 2 crosses the shallow and wide minor groove of stem 1. The BCV pseudoknot has a relatively lengthy loop 1, implying that these sequences have a biological relevance. On the other hand, loop 2, 3 nt long with the fraying of the 182G-194U base pair at the top of stem 2, is not sufficient to bridge the minor groove of an 8-bp (stem 1) A-form helix (48). (ii) The stem regions in a classical pseudoknot are contiguous. The BCV pseudoknot has an insertion of 1 nt, A193, between the two stem regions, which may prevent a linear arrangement of the stems. The presence of the intervening nucleotide, along with the steric constraints caused by the short length of loop 2, probably results in a bent conformation of the pseudoknot with stems 1 and 2 tilting toward each other (58).
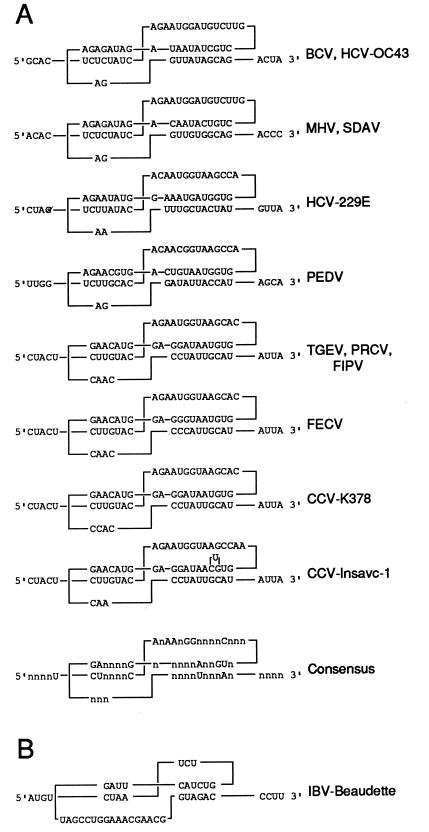
To determine whether this pseudoknotted structure is supported by phylogenetic analysis, the 3′ UTRs of all sequenced coronaviruses were examined. These comparisons revealed a similar pseudoknot in the same relative location for each coronavirus. Among the mammalian coronaviruses (Fig. 2A), structure at the secondary and tertiary levels is maintained despite differences in RNA sequences. Only minor variations are found in stem and loop lengths, and a number of residues are absolutely conserved (note the consensus structure in Fig. 2A). An analogous structure in the avian coronavirus infectious bronchitis virus (IBV) (Fig. 2B) diverges more considerably in stem and loop dimensions but retains some of the conserved features of the mammalian coronaviruses (e.g., a consensus motif nAnnnnGGnnnnnnnnn in IBV loop 2 found in mammalian coronavirus loop 1). Taken together, these observations led to the working hypothesis that the pseudoknot functions in the coronavirus life cycle.
FIG. 2.
Comparison of the phylogenetically conserved pseudoknots among coronaviruses. Stems 1 and 2 are shown on the left and right, respectively. (A) Members of coronavirus serogroups 1 (HCV-229E [54], PEDV [9], TGEV [28], PRCV [49], FIPV [13], FECV [67] and CCV [22, 67]), and 2 (BCV [32], HCV-OC43 [27], MHV [44], and SDAV [30]) are shown. (B) IBV (6), the only member of coronavirus serogroup 3, is shown. Abbreviations: HCV, human coronavirus; SDAV, rat sialodacryoadenitis virus; PEDV, porcine epidemic diarrhea virus; PRCV, porcine respiratory coronavirus; FIPV, feline infectious peritonitis virus; FECV, feline enteric coronavirus; CCV, canine coronavirus.
Base pairing in both stems of the pseudoknot is required for BCV DI RNA replication.
Previous studies have described the cloning of a 2.2-kb reporter-containing BCV DI RNA replicon (pDrep1) (11). The DI RNA is a fusion between the 5′ and 3′ termini of the virus genome, resulting in a single fused open reading frame (Fig. 1A). When synthetic uncapped transcripts from MluI-linearized pDrep1 were transfected into BCV (helper virus)-infected cells, replication was observed as monitored by Northern analysis with a strand-specific probe for the reporter sequence (11). Plus-strand DI RNA accumulated in transfected cells with kinetics paralleling that of helper virus genome, and it appeared during passage 1 of progeny virus on fresh cells.
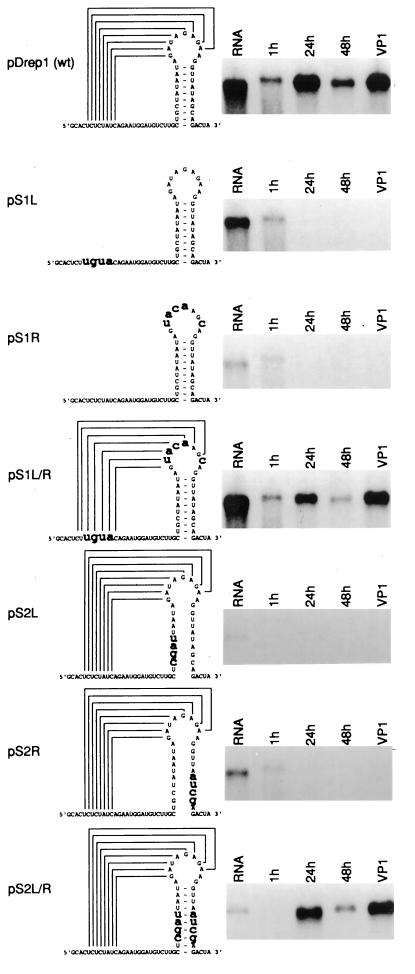
To test whether the pseudoknot plays a role in RNA replication, site-directed mutagenesis was undertaken to disrupt and then restore base pairing in both stems in pDrep1. Multiple base substitutions were designed to create mismatches predicted to destroy the thermodynamic stability of each stem (data not shown). The mutations were derived from the analogous stem in porcine transmissible gastroenteritis virus (TGEV) (for stem 1 single mutants pS1L and pS1R) and from the other side of the stem (for stem 2 single mutants pS2L and pS2R). Double mutants for stems 1 and 2 (pS1L/R and pS2L/R, respectively) incorporating both sets of mutations were also generated, and all mutant constructs were assayed for replication.
The effects of these mutations were demonstrated by Northern analysis and are shown in Fig. 3. The single mutants with disrupted stems showed no evidence of replication. On the other hand, the double mutants with restored stems replicated at or near wild-type levels and became packaged into virions as evidenced by successful passaging. A lower negative free-energy value (ΔG = −5.8 kcal/mol) for the 7-bp TGEV-mimicked stem in pS1L/R and alterations in the highly conserved bases A198, G195, and U178 in the restored stem in pS2L/R were tolerated by the DI RNA. The latter finding indicates that the primary sequence in these regions is not a requirement for replication. In summary, these experiments establish a correlation between the maintenance of the two stem regions in the pseudoknot and BCV DI RNA replication.
FIG. 3.
Importance of stems 1 and 2 in DI RNA replication. Synthetic transcripts of pDrep1 and six mutants were transfected in BCV-infected cells, and cytoplasmic RNA was extracted at 1, 24, and 48 h posttransfection and at 48 h after the first virus passage (VP1). Extracted RNA was separated by electrophoresis on a formaldehyde-agarose gel and probed in a Northern blot with radiolabeled oligonucleotide for detection of the plus strand of the reporter sequence. An increase in the amount of DI RNA after 1 h posttransfection is evidence of replication. RNA shown in the first lane is an aliquot of the in vitro transcript used as marker.
The mutated sequences in pS1L/R and pS2L/R are retained after a viral passage.
To evaluate the integrity of the mutations in the replicating transcripts from pS1L/R and pS2L/R, RT-PCR analysis was performed on minus-strand templates in total RNA extracted after the first viral passage. This approach should ensure that the product was derived from only molecules that had undergone replication and packaging. A DI RNA-specific product was amplified and cloned, and five insert-containing colonies for each construct were sequenced across the region of the pseudoknot (data not shown). The number of colonies containing matched pairs of mutations for pS1L/R and pS2L/R were two and four, respectively. The remaining sequences were of wild-type origin, indicating that the VP1 bands on the Northern blots (Fig. 3) represent a mixture of DI RNA molecules, presumably the consequence of recombination events between the helper virus and the transfected DI RNA. The results also show that the double mutants undergo replication and become packaged in supernatant virus.
Enzymatic probing yields evidence for both open and closed forms of the pseudoknot.
To obtain direct biochemical evidence for the presence of the pseudoknot, the structure of an in vitro-transcribed RNA was probed by enzymatic treatment. The 197-nt RNA substrate, including 40 nt upstream and 94 nt downstream of the pseudoknot, was digested with RNases specific for single-stranded regions (RNase T1 [G specific] and RNase T2 [nonspecific with a preference for A]) and helical regions (cobra venom RNase V1). The positions of cleavage sites in the RNA were determined by analyzing the primer extension products in parallel with a dideoxy sequencing ladder generated from untreated RNA and the same primer. Figure 4A, an autoradiograph of a representative gel, shows that increasing the amount of enzyme in the digestion mixture yields specific banding patterns. A summary of all enzymatic probing data superimposed on the pseudoknot is given in Fig. 4B.
FIG. 4.
Enzymatic probing of the pseudoknot in synthetic RNA transcripts. (A) Electrophoretic analysis of digestion products. Lanes: 1 to 4, RNase T2 digestion with 0.65, 0.065, 0.0065, and 0 U, respectively; 5 to 8, sequencing ladder generated from the same primer; 9 to 12, RNase V1 digestion with 0, 0.01, 0.1, and 1.0 U, respectively. The region encompassed by the dashed bracket may show both single-stranded and double-stranded properties (see the text). Results from digestion with RNase T1 are not shown. ss, single-stranded; ds, double-stranded. (B) Schematic summary of enzymatic probing.
Three major areas were susceptible to RNase V1: those between C225 and G217, G209 and C207, and C200 and A196. RNase V1 recognizes RNA sequences in a double-stranded region (a canonical helix) and, on occasion, in a single-stranded region adopting an approximately helical conformation (37). An interpretation that the first and third of these sets of cleavages occur in double-helical RNA is consistent with the base pairing predicted for both stems 1 and 2. The second set might arise during an altered conformation of the pseudoknot (postulated below). The reactive sites between C219 and G217, located outside stem 1, may result from the ability of RNase V1 to cleave a stacked single-stranded region immediately adjacent to a stem (1). Note that only one strong banding pattern is observed for each stem. An explanation for this result is that RNase V1 cleavage on the 5′ side of a stem is generally greater than that on the 3′ side of a stem (1).
RNase T2-sensitive areas, on the other hand, were found between A218 and U210 and between A193 and G183. Digestion with RNase T1 yielded similar results, with strong cleavages between G213 and A211, between G188 and A187, and between G186 and A185 (data not shown). These single-stranded regions include not only loops 1 and 2 but also the side of stem 1 between G192 and A185, in apparent contradiction to the predicted pseudoknot. These data are most compatible with a model in which the base pairings in stem 1 are present in a “breathing” (open and closed) interaction. Therefore, the pseudoknot may exist in equilibrium with the hairpin formed by stem 2. An “open” pseudoknot also may account for the area of RNase V1 reactivity between G209 and C207 by allowing these bases to interact with another single-stranded region.
An RNA sequence able to form a pseudoknot also has the potential to form two hairpin structures (stem 1 alone or stem 2 alone). Experiments with oligonucleotides have provided evidence that pseudoknots with coaxially stacked stems are only marginally more stable than either of the constituent hairpins and that equilibria among secondary and tertiary structures are sensitive to changes in ionic conditions, temperature, loop sequence, and loop size (69). A decrease in the size of pseudoknot loop 2, for example, shifts the equilibria among conformations by stabilizing the stem 2 hairpin relative to the pseudoknot. In the proposed pseudoknot, the small size of loop 2 (2 nt) and the higher negative free energy of stem 2 (compared to stem 1) potentially contribute to the observed labile conformation. In addition, the single nucleotide at the stem-stem junction, A193, may not allow coaxial stacking of the stems, resulting in a greater degree of breathability (58).
DISCUSSION
From the experiments presented here, we conclude that a phylogenetically conserved hairpin-type pseudoknot in the 3′ UTR of BCV is a cis-acting signal for DI RNA replication and, by extension, viral genome replication. We postulate, furthermore, that the structure functions in the plus strand, since a large number of G · U pairs are found in stem 2 of the mammalian coronaviruses (ranging from one in BCV to three in human coronavirus 229E [Fig. 2]) and bases on the minus strand fail to covary to maintain base pairings. Consistent with our conclusions are studies on DI RNA of mouse hepatitis virus (MHV), another serogroup 2 coronavirus, for which it has been shown that the minimal sequences at the 3′ end of the genome required for replication of DIssE (29), DIssF (34), and MIDI (66) include the corresponding region of the pseudoknot. In addition, deletions that remove regions within the pseudoknot have been found to abolish DI RNA replication (29, 34). These mutant constructs, however, also remove all or part of a proposed signaling bulged stem-loop mapping immediately upstream of the pseudoknot (24) and eliminate a 26-nt sequence downstream of the pseudoknot shown to be important in both RNA-protein binding and DI RNA replication (36, 70). Except for a preliminary report of this work (68), a pseudoknotted structure has not been identified as a coronavirus 3′-UTR RNA replication signal.
Although 3′-UTR pseudoknotted tertiary structures in plus-strand animal RNA viruses have been postulated to function as regulatory elements in RNA replication, to our knowledge the only other example of such a structure supported by mutational analyses is the phylogenetically conserved pseudoknot of enteroviruses (40, 41, 46). In this case, however, the pseudoknot was noted not to be a conventional one, since relatively long-distance loop-loop interactions are needed to form the stems. How this element functions mechanistically in enterovirus RNA replication was not determined.
How might the BCV 3′ UTR pseudoknot exert an effect on RNA synthesis? Despite its superficial similarity to the well-characterized hairpin-type pseudoknot active in the ribosomal frameshifting at the coronavirus gene 1a/1b junction (60), there is no indication from inspection of the sequence that it is involved in a similar frameshifting event or stop codon readthrough that might yield a novel cis-acting protein. It also seems unlikely, based on a recent demonstration that the minimal 3′-terminal sequence requirement for minus-strand synthesis in MHV DI RNA is approximately 50 nt (exclusive of the poly (A) tail) (35) and on the finding that the 3′ UTRs of BCV and MHV DI RNAs can be exchanged without loss of replicability (unpublished data), that the pseudoknot is part of the signal directing minus-strand synthesis.
An answer to the question of 3′-UTR pseudoknot function may lie in precedents established by studies of positive-strand plant RNA viruses. Two separate but poorly understood regulatory pathways have been described. In the first, pseudoknots mapping just downstream of the open reading frame in the nonpolyadenylated tobacco mosaic virus genome (the so-called upstream pseudoknots in tobacco mosaic virus) have been shown to functionally mimic the poly(A) tail as an enhancer of translation (17). Inasmuch as translation is a cis-acting requirement for RNA replication in many plus-strand RNA viruses (43), regulation of translation could be a means of controlling replication. The replication of BCV DI RNA is also strictly dependent on the translatability of its open reading frame (10), making pseudoknot-mediated translational regulation an attractive model to test.
In the second example, the 3′-terminal pseudoknots in plant virus genomes comprise the acceptor arm in cis-acting tRNA-like structures (TLSs) (reviewed in reference 16). The mechanistic contribution of the TLSs to genome replication is not understood, but one hypothesis envisions an interaction with components of the translation apparatus, such as tRNA synthetases and elongation factors, to form the enzymatically active RNA replication complex (18). The coronavirus 3′ UTR pseudoknot, therefore, may function as an independent upstream element to recruit protein factors as part of a replication complex. It should be noted that a tRNA-like element need not be aminoacylatable or even 3′-terminal for recognition by its cognate synthetase, as shown for the TLS within the upstream region of the threonyl-tRNA synthetase mRNA from Escherichia coli (reviewed in reference 39). Also, the identity features of a canonical tRNA molecule or TLS that signal synthetase recognition can be contained entirely within the acceptor arm (16). It will therefore be important to determine whether the coronavirus 3′ UTR pseudoknot can be recognized by cellular synthetases and elongation factors.
Three-dimensional modeling and further mutagenesis studies will be required to delineate more precisely the structure and function of the pseudoknot. In addition, it will be important to determine whether similar tertiary structural elements function in the genomes of toroviruses and arteriviruses, the other genera within the order Nidovirales.
ACKNOWLEDGMENTS
We thank Jennifer Black and Savithra Senanayake for valuable discussions and David Draper (Johns Hopkins University) and Cornelis Pleij (University of Leiden) for critical comments on the manuscript.
This work was supported by Public Health Service grant AI14367 from the National Institutes of Health and by funds from the University of Tennessee College of Veterinary Medicine Center of Excellence Program for Livestock Diseases and Human Health.
REFERENCES
- 1.Auron P E, Weber L D, Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982;21:4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- 2.Baer M L, Houser F, Loesch-Fries L S, Gehrke L. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 1994;13:727–735. doi: 10.1002/j.1460-2075.1994.tb06312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric R S, Stohlman S A, Lai M M C. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight K J, Rice C M. Secondary-structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursnell M E G, Binns M M, Foulds I J, Brown T D K. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J Gen Virol. 1985;66:573–580. doi: 10.1099/0022-1317-66-3-573. [DOI] [PubMed] [Google Scholar]
- 7.Brian D A, Dennis D E, Guy J S. Genome of porcine transmissible gastroenteritis virus. J Virol. 1980;34:410–415. doi: 10.1128/jvi.34.2.410-415.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brian D A, Spaan W J M. Recombination and coronavirus defective interfering RNAs. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridgen A, Duarte M, Tobler K, Laude H, Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhoea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J Gen Virol. 1993;74:1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- 10.Chang R-Y, Brian D A. cis-requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang R-Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang R-Y, Krishnan R, Brian D A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeGroot R J, Andeweg A C, Horzinek M C, Spaan W J. Sequence analysis of the 3′-end of the feline coronavirus FIPV 79-1146 genome: comparison with the genome of porcine coronavirus TGEV reveals large insertions. Virology. 1988;167:370–376. doi: 10.1016/0042-6822(88)90097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felden B, Florentz C, Giege R, Westhof E. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA. 1996;2:201–212. [PMC free article] [PubMed] [Google Scholar]
- 15.Felden B, Florentz C, Westhof E, Geige R. Non-canonical substrates of aminoacyl-tRNA synthetases: the tRNA-like structure of brome mosaic virus genomic RNA. Biochimie. 1993;75:1143–1157. doi: 10.1016/0300-9084(93)90014-j. [DOI] [PubMed] [Google Scholar]
- 16.Florentz C, Giege R. tRNA-like structures in plant viral RNAs. In: Soll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: American Society for Microbiology; 1995. pp. 141–164. [Google Scholar]
- 17.Gallie D R, Walbot V. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990;4:1149–1157. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- 18.Giege R. Interplay of tRNA-like structures from plant viral RNAs with partners of the translation and replication machineries. Proc Natl Acad Sci USA. 1996;93:12078–12081. doi: 10.1073/pnas.93.22.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giege R, Florentz C, Dreher T W. The TYMV tRNA-like structure. Biochimie. 1993;75:569–582. doi: 10.1016/0300-9084(93)90063-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann M A, Brian D A. The 5-prime end of coronavirus minus-strand RNAs contains a short poly(U) tract. J Virol. 1991;65:6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsburgh B C, Brierley I, Brown T D. Analysis of a 9.6 kb sequence from the 3′ end of canine coronavirus genomic RNA. J Gen Virol. 1992;73:2849–2862. doi: 10.1099/0022-1317-73-11-2849. [DOI] [PubMed] [Google Scholar]
- 23.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 24.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamahora T, Soe L H, Lai M M C. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989;12:1–9. doi: 10.1016/0168-1702(89)90048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapke P A, Brian D A. Sequence analysis of the porcine transmissible gastroenteritis coronavirus nucleocapsid protein gene. Virology. 1986;151:41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunita S, Mori M, Terada E. Sequence analysis of the nucleocapsid protein gene of rat coronavirus SDAV-681. Virology. 1993;193:520–523. doi: 10.1006/viro.1993.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lapps W, Hogue B G, Brian D A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leathers V, Tanguay R, Kobayashi M, Gallie D R. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y-J, Liao C-L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Yu W, Leibowitz J L. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- 37.Lowman H B, Draper D E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- 38.Luytjes W. Coronavirus gene expression: Genome organization and protein synthesis. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Publishing Corp.; 1995. pp. 33–54. [Google Scholar]
- 39.Mans R M W, Pleij C W A, Bosch L. tRNA-like structures: structure, function and evolutionary significance. Eur J Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- 40.Melchers W J G, Hoenderop J G J, Bruins Slot H J, Pleij C W A, Pilipenko E V, Agol V I, Galama J M D. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakhasi H L, Rouault T A, Haile D J, Liu T-Y, Klausner R D. Specific high-affinity binding of host cell proteins to the 3′ region of rubella virus RNA. New Biol. 1990;2:255–264. [PubMed] [Google Scholar]
- 43.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 44.Parker M M, Masters P S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilipenko E V, Maslova S V, Sinyakov A N, Agol V I. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 1992;20:1739–1745. doi: 10.1093/nar/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilipenko E V, Poperechny K, Maslova S V, Melchers W J G, Bruins Slot H J, Agol V I. cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (‘kissing’) interactions. EMBO J. 1996;15:5428–5436. [PMC free article] [PubMed] [Google Scholar]
- 47.Pleij C W A. RNA pseudoknots. Curr Opin Struct Biol. 1994;4:337–344. [Google Scholar]
- 48.Pleij C W A, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasschaert D, Duarte M, Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J Gen Virol. 1990;71:2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- 50.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawicki S G, Sawicki D L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Med Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 52.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schochetman G, Stevens R H, Simpson R W. Presence of infectious polyadenylated RNA in the coronavirus avian bronchitis virus. Virology. 1977;77:772–782. doi: 10.1016/0042-6822(77)90498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schreiber S S, Kamahora T, Lai M M C. Sequence analysis of the nucleocapsid protein gene of human coronavirus 229E. Virology. 1989;169:142–151. doi: 10.1016/0042-6822(89)90050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senanayake S D, Brian D A. Precise large deletions by the PCR-based overlap extension method. Mol Biotechnol. 1995;4:13–15. doi: 10.1007/BF02907467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sethna P B, Hung S-L, Brian D A. Coronavirus subgenomic minus-strand RNA and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen L X, Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 59.Shi P-Y, Brinton M A, Veal J M, Zhong Y Y, Wilson W D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 60.Somogyi P, Jenner A J, Brierley I, Inglis S C. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Ziejst B A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ten Dam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 63.Tinoco I, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 64.Todd S, Semler B L. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′ non-coding region. Nucleic Acids Res. 1996;24:2133–2142. doi: 10.1093/nar/24.11.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai C-H, Cheng C-P, Peng C-W, Lin B-Y, Lin N-S, Hsu Y-H. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J Virol. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Most R G, Luytjes W, Rutjes S, Spaan W J M. Translation but not the encoded sequence is essential for the efficient propagation of defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vennema H, Rossen J W, Wesseling J, Horzinek M C, Rottier P J. Genomic organization and expression of the 3′ end of the canine and feline enteric coronaviruses. Virology. 1992;191:134–140. doi: 10.1016/0042-6822(92)90174-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams G D, Chang R-Y, Brian D A. Evidence for a pseudoknot in the 3′ untranslated region of the bovine coronavirus genome. Adv Exp Med Biol. 1995;380:511–514. doi: 10.1007/978-1-4615-1899-0_81. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt J R, Puglisi J D, Tinoco I. RNA pseudoknots: stability and loop size requirements. J Mol Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 70.Yu W, Leibowitz J L. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J Virol. 1995;69:2016–2023. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]