Abstract
We examined the ability of a live, attenuated deletion mutant of simian immunodeficiency virus (SIV), SIVmac239Δ3, which is missing nef and vpr genes, to protect against challenge by heterologous strains SHIV89.6p and SIVsmE660. SHIV89.6p is a pathogenic, recombinant SIV in which the envelope gene has been replaced by a human immunodeficiency virus type 1 envelope gene; other structural genes of SHIV89.6p are derived from SIVmac239. SIVsmE660 is an uncloned, pathogenic, independent isolate from the same primate lentivirus subgrouping as SIVmac but with natural sequence variation in all structural genes. The challenge with SHIV89.6p was performed by the intravenous route 37 months after the time of vaccination. By the criteria of CD4+ cell counts and disease, strong protection against the SHIV89.6p challenge was observed in four of four vaccinated monkeys despite the complete mismatch of env sequences. However, SHIV89.6p infection was established in all four previously vaccinated monkeys and three of the four developed fluctuating viral loads between 300 and 10,000 RNA copy equivalents per ml of plasma 30 to 72 weeks postchallenge. When other vaccinated monkeys were challenged with SIVsmE660 at 28 months after the time of vaccination, SIV loads were lower than those observed in unvaccinated controls but the level of protection was less than what was observed against SHIV89.6p in these experiments and considerably less than the level of protection against SIVmac251 observed in previous experiments. These results demonstrate a variable level of vaccine protection by live, attenuated SIVmac239Δ3 against heterologous virus challenge and suggest that even live, attenuated vaccine approaches for AIDS will face significant hurdles in providing protection against the natural variation present in field strains of virus. The results further suggest that factors other than anti-Env immune responses can be principally responsible for the vaccine protection by live, attenuated SIV.
Infection by live, attenuated deletion mutants of simian immunodeficiency virus (SIV) has afforded rhesus monkeys strong protection against subsequent challenge by wild-type, disease-causing strains of SIV (4, 7, 18, 36). While it has been argued that this protection could result from some sort of blocking or interference phenomenon (13, 32, 34), several lines of circumstantial evidence suggest that the protection may be immune mediated (16). Protection has been shown to be time dependent: the longer the time interval, the better the protection, despite the fact that vaccine virus loads are highest during the initial weeks following immunization (4, 36). While the most solid protection has been observed in monkeys with apparent sterilizing immunity, some animals have exhibited long-term protective effects despite a transient take of the challenge virus (18, 36), again apparently more consistent with immune-mediated protection than with viral interference. Monkeys that control replication of SIVmac239Δnef least effectively are the least protected upon subsequent challenge (23). Finally, CD4+ cells in peripheral blood mononuclear cells (PBMC) of vaccinated animals are perfectly capable of supporting the replication of SIV in culture (11).
Despite these arguments in support of immune-mediated mechanisms for the protection, no clear evidence has been forwarded regarding what these immune responses might be. There is considerable interest in the scientific community in identifying the responses that are protective in this system because even if the live, attenuated approach is never forwarded for trials in humans, such research in monkeys may be able to tell us what types of immune responses are needed for protection. In addition, it is not known how broadly protective the live, attenuated vaccine approach can be against pathogenic, heterologous virus challenge in this rhesus monkey model. We have addressed these issues by vaccinating rhesus monkeys with the attenuated strain SIVmac239Δ3 (9, 36) and subsequently challenging them with pathogenic, uncloned, heterologous strains SHIV89.6p (28) and SIVsmE660 (12, 15).
MATERIALS AND METHODS
Animals.
Rhesus monkeys were received from the Oregon Regional Primate Research Center, Beaverton, Oreg., or Laboratory Animal Breeders and Services, Yemassee, S.C. Upon receipt, the monkeys underwent a 6-week quarantine. During this period, the animals received three intradermal tuberculin tests, sampling for hematology and serum chemistry profiles, and rectal swabs for bacterial culture. Feces were also analyzed for occult blood, ovum, and parasite determinations. In addition, each animal was screened for antibody status with respect to SIV, simian type D virus, simian T-cell lymphotropic virus type 1, herpesvirus B, and measles virus. All animals were antibody negative for SIV, simian type D virus, and simian T-cell lymphotropic virus type 1. All animal care and use procedures conformed to the revised Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Vaccination and challenge.
The preparation of SIVmac239Δ3 vaccine stock has been described previously (9, 36). By intravenous inoculation, four male rhesus monkeys were vaccinated with SIVmac239Δ3 containing 0.01 ng of p27 and four were vaccinated with SIVmac239Δ3 containing 1.1 ng of p27. These amounts of this stock represent 23 tissue culture infectious doses and 5 rhesus monkey infectious doses in the first group and 2.3 × 103 tissue culture infectious doses and 5 × 102 rhesus monkey infectious doses in the second group. The preparations of the SIVsmE660 challenge stock (provided by Vanessa Hirsch) and of the SHIV89.6p challenge stock (provided by Norman Letvin) have been described (12, 15, 28). Monkeys in the first group were challenged intravenously with 20 tissue culture infectious doses of SIVsmE660 28 months after the time of vaccination. Monkeys in the second group were challenged intravenously with SHIV89.6p containing 0.019 ng of p27 37 months after the time of vaccination.
Flow cytometry.
Whole blood collected in EDTA was analyzed for expression of CD4 (OKT4a [Ortho] and Anti-Leu 3a [Becton Dickinson]), CD8 (Anti-Leu 2a [Becton Dickinson]), and CDw29 (4B4 [Coulter Immunology]) by a whole-blood lysis technique previously described (36). Briefly, antibody (volume dependent upon the specific antibody) was added to 100 μl of whole blood and incubated for 10 min in the dark. Lysing solution (Becton Dickinson) was added and the samples were incubated for 10 min at room temperature. Stained cells were fixed with 0.5% paraformaldehyde. Samples were analyzed on a Becton Dickinson FACScan cytometer.
Cell-associated virus loads.
Cell-associated virus loads were determined by quantitating the numbers of infectious cells in PBMC as previously described (9). Twelve serial 1:3 dilutions of PBMC, beginning with 106 cells, were cocultured in duplicate with 105 CEM×174 cells per well in 24-well plates. Supernatant samples were collected after 21 days of culture and stored frozen at ≤−70°C until analysis for p27 antigen with the Coulter p27 antigen assay kit.
Antiviral antibodies by ELISA.
Enzyme-linked immunosorbent assay (ELISA) plates were coated with purified lysed SIVmac251, SIVmac239, or SIVsmE660 as previously described (6, 9). Human immunodeficiency virus type 1 (HIV-1) and SIV envelope proteins were purchased from Immunodiagnostics (Bedford, Mass.) and also used to coat ELISA plates. Dilutions of sera from the monkeys were assayed for antibody binding by using an alkaline phosphatase-conjugated goat anti-human immunoglobulin G (heavy and light chain) in accordance with our previously described procedures (6, 9).
Plasma RNA loads.
Citrated plasma samples were assayed for SIV RNA levels as described previously (9, 35).
DNA PCR.
Genetic analysis of mutant versus wild-type SIV sequences was performed by using a PCR method previously described (21, 36), with slight modifications. One-half microgram of chromosomal DNA was used per 100-μl reaction. DNA was added to the reaction mixture minus Taq polymerase and denatured at 95°C for 10 min and immediately placed on ice. The DNA-reaction mix was briefly centrifuged and returned to ice. The DNA-reaction mix was placed in a thermocycler preheated to 72°C for 1 to 2 min; Taq polymerase (2.5 U) was added and the PCR was carried out for 35 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min 45 s. The analyses for SIVmac239Δ3 and E660 sequences were conducted separately. The primer pairs for SIVmac239Δ3 were as previously described (36). The sequences of the forward primers for E660 were PCR1 (5′-ACA ACA AAA CAT GGA TGA TGT GG) and PCR2 (5′-AGT CCC CTT AAG GGC CAT GAC ATA).
HIV-1 env sequences were amplified similarly using HIV-1 89.6 env-specific primer pairs. The expected size for the first round of PCR was 962 bp (primers were F1 [GCAACCACCACTCTATTTTGTGC] and R1 [CCTCCTGAGGATTGATTAAAGGC]). The expected size for the nested PCR was 453 bp (primers were F2 [GGATGAAAGCCTAAAGCCATGTG] and R2 [AGCAGTTGAGTTGACACCACTGG]). Ten microliters was electrophoresed in a 1.5% agarose gel containing ethidium bromide.
Virus neutralization assays.
Neutralizing antibodies were measured in either CEM×174 cells (SIVmac251LP and SIVsmE660) or MT-2 cells (HIV-1 MN and SHIV-89.6p) by a reduction in virus-induced cell-killing effects as described previously (4, 15). Assay stocks of SIVmac251LP, SIVsmE660, and HIV-1 MN were prepared in H9 cells and SHIV-89.6p was prepared in human PBMC. Titers are reported as the reciprocal serum dilutions at which 50% of cells were protected from virus-induced killing. This cutoff usually corresponds to a >90% reduction in viral Gag antigen synthesis.
Assay of CTL activity.
SIV-specific cytotoxic T lymphocytes (CTL) were analyzed following antigen-specific stimulation of PBMC as described previously (17). Briefly, PBMC were stimulated by using autologous herpesvirus papio-transformed B lymphoblastoid cell lines (B-LCL) infected with a recombinant vaccinia virus (vAbt388; provided by D. Panicali, Therion Biologics, Cambridge, Mass.) containing the SIVmac251 gag and pol genes and the SIVmac239 env gene, which were inactivated with UV psoralen after overnight incubation. After 10 to 12 days of culture, stimulated PBMC were used as effector cells in a standard 51Cr-release assay. Target cells consisted of autologous B-LCL infected with recombinant vaccinia viruses expressing SIV proteins. Recombinant vaccinia viruses used to infect target cells include vAbt252 (encoding the SIVmac251 p55 Gag and protease proteins; Therion), rVV-239 (encoding the SIVmac239 envelope [Env]) (30), and the control vaccinia virus NYCBH. Cold targets consisting of unlabeled autologous B-LCL infected with the control vaccinia virus NYCBH were used at a cold/hot target ratio of 15:1 to decrease background lysis. Chromium released was assayed after a 5-h incubation at 37°C in a 5% CO2 incubator, and the percent cytotoxicity was calculated as follows: (test release − spontaneous release)/(maximum release − spontaneous release) × 100. SIV-specific release was then calculated by subtracting lysis of control NYCBH-infected target cells from that of target cells expressing SIV antigens. Based on examination of 10 naive control animals not infected with SIV, SIV-specific lysis of greater than 5% seen at more than one effector/target ratio was interpreted as significant.
RESULTS
Vaccine phase.
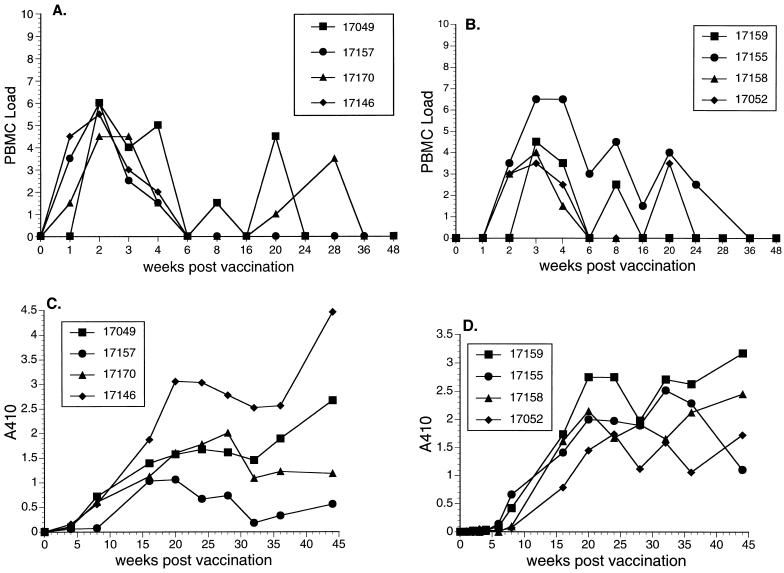
Eight monkeys were vaccinated by intravenous inoculation with SIVmac239Δ3, which contains two deletions in nef and a deletion in vpr (9). The response of rhesus monkeys to the SIVmac239Δ3 was similar to what has been described previously (9). There was an early spike in virus load around 2 to 3 weeks after inoculation, which subsequently resolved (Fig. 1A and B). Anti-SIV antibody responses were readily detected (Fig. 1C and D) and these persisted for the duration of the vaccine phase. There were no statistically significant differences in either peak viral PBMC loads or levels of SIV antibodies at 44 weeks between the groups of monkeys vaccinated with SIVmac239Δ3 containing 0.01 versus 1.1 ng of p27 (P > 0.3; Mann-Whitney test).
FIG. 1.
Vaccine phase. (A) Cell-associated vaccine virus loads following vaccination in monkeys that were subsequently challenged with SHIV89.6p. (B) Cell-associated vaccine virus loads following vaccination in monkeys that were subsequently challenged with SIVsmE660. Code for PBMC load: 0, virus was not recovered with 106 or fewer PBMC; 1, virus was recovered with 106 but not fewer PBMC; 2, 333,333 PBMC; 3, 111,111 PBMC; 4, 37,037 PBMC; 5, 12,345 PBMC; 6, 4,115 PBMC; 7, 1,371 PBMC; 8, 457 PBMC. (C) Anti-SIV antibody responses as detected by ELISA following vaccination in monkeys that were subsequently challenged with SHIV89.6p. (D) Anti-SIV antibody responses as detected by ELISA following vaccination in monkeys that were subsequently challenged with SIVsmE660. A410, absorbance at 40 nm.
SHIV89.6p challenge.
SHIV89.6 is a recombinant virus which contains gag and pol sequences from SIVmac239 and env sequences from HIV-1 isolate 89.6 (29). SHIV89.6p is a highly aggressive derivative that causes acute declines in CD4+ cell numbers and death with AIDS over a time course that is usually less than 12 weeks (28). Thus, challenge in this case was with an uncloned strain of virus very closely matched in gag and pol sequences (19) but totally mismatched in env sequences (Table 1). Challenge was performed by the intravenous route with SHIV89.6p containing 0.019 ng of p27gag antigen 37 months after the time of vaccination. Two naive rhesus monkeys served as controls for challenge.
TABLE 1.
Sequence divergence and protection
| Challenge virus | % Amino acid identity in gag-pola | % Amino acid identity in enva | Protection |
|---|---|---|---|
| SHIV89.6p | 99.5 | <30 | +++b |
| SIVsmE660 | 89–92 | ∼82 | +c |
Compared to vaccine strain SIVmac239Δ3.
37 months after the time of vaccination. +++, strong but incomplete.
28 months after the time of vaccination. +, weak.
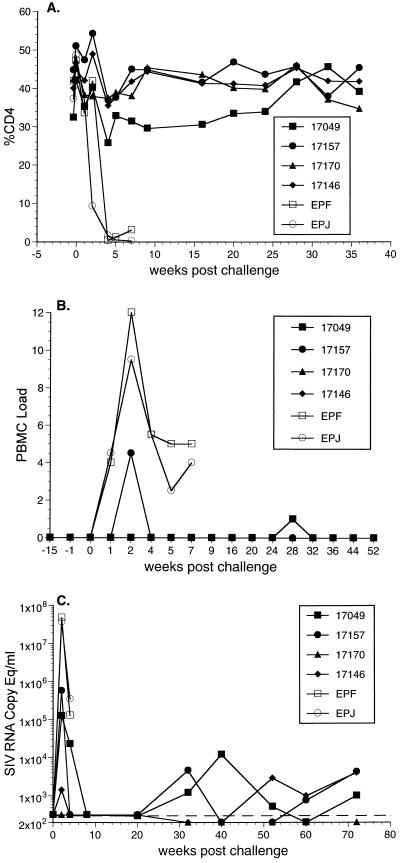
Both unvaccinated monkeys inoculated with SHIV89.6p developed rapid, severe declines in CD4 cell numbers (Fig. 2A). CD4+ cell counts were reduced to near zero by 5 weeks (Fig. 2A). These control animals were euthanized at 8 weeks postinoculation because of deteriorating clinical condition. At necropsy, one animal had Epstein-Barr virus esophagitis and lymphoid hyperplasia and the other animal had lymphoid follicular involution and inflammatory infiltrates in the gastrointestinal tract. This acute clinical course is typical of pathogenic SHIV89.6p infections that we have observed in four other monkeys inoculated with the same stock of virus. In contrast to these control animals, all four of the vaccinated monkeys were completely protected against any CD4 cell depletion (Fig. 2A).
FIG. 2.
Outcome of challenge with SHIV89.6p. (A) CD4+ T lymphocytes. (B) Cell-associated virus loads measured as the number of infectious cells in PBMC. Code is as used previously (9, 36): 2, 333,333 PBMC; 3, 111,111 PBMC; 4, 37,037 PBMC; 5, 12,345 PBMC; 6, 4,115 PBMC; 7, 1,371 PBMC; 8, 457 PBMC. (C) Viral RNA levels in plasma. Open symbols used for control animals EPF and EPJ. Closed symbols are for vaccinated animals. The detection limit, approximately 300 copy equivalents (Copy Eq) per ml, is indicated by the dashed line.
Vaccine virus was not recovered from PBMC of any of the four vaccinated monkeys at the time of challenge (Fig. 2B) or during the weeks preceding challenge, even when 106 PBMC were used for cocultivation. These findings are consistent with previous descriptions of the attenuated nature of SIVmac239Δ3 (9). Cell-associated virus loads, measured as the numbers of infectious cells in PBMC, peaked in the control animals around 2 weeks after challenge and, following a slight decline, remained at moderate or high levels until the time of death (Fig. 2B). Virus was recovered from only one of the four vaccinated animals at week 2 postinoculation. In fact, cell-associated viral loads remained low in all four vaccinated animals throughout the remaining course of measurements (Fig. 2B).
Viral loads were also evaluated after challenge by quantitating the amounts of virion-associated viral RNA in plasma. Viral RNA was detected in plasma for three of the four vaccinated monkeys at week 2 following challenge (Fig. 2C). However, the levels of viral RNA at week 2 postchallenge were 2 to 4 logs lower in these three vaccinated animals than in the two controls (Fig. 2C). The levels of viral RNA declined to undetectable levels by week 8, but they persisted at fluctuating levels of 1,000 to 10,000 RNA copy equivalents per ml of plasma in three of the four vaccinated animals between 30 and 72 weeks postchallenge (Fig. 2C). The one animal (17170) that had no detectable viral RNA in plasma in the initial weeks following challenge is the same animal that maintained undetectable levels of plasma RNA 30 to 72 weeks after challenge (Fig. 2C).
DNA was prepared from PBMC obtained at day 15 and weeks 20, 32, and 70 from the SHIV-challenged monkeys. DNA was used for genetic analysis of the presence of HIV-1 89.6 env sequences by PCR. As expected, day 15 PBMC from both unvaccinated control animals yielded a fragment consistent with the presence of SHIV89.6 challenge virus (Table 2). The two vaccinated monkeys with significant viral loads at 2 weeks after the SHIV challenge (17157 and 17049) also showed HIV-1 env sequences with the day 15 sample (Table 2). The other two vaccinated monkeys did not have detectable HIV-1 env sequences in the day 15 PBMC sample (Table 2). None of the four vaccinated monkeys had detectable HIV-1 env sequences in the week 20 PBMC sample. However, all four of the previously vaccinated monkeys had HIV-1 env sequences detectable in their PBMC at both 32 and 70 weeks after the SHIV89.6p challenge (Table 2).
TABLE 2.
HIV-1 env sequences in PBMC after SHIV89.6p challengea
| Monkey | Result at:
|
|||
|---|---|---|---|---|
| Day 15 | Week 20 | Week 32 | Week 70 | |
| EPF | + | NA | NA | NA |
| EPJ | + | NA | NA | NA |
| 17049 | + | − | + | + |
| 17157 | + | − | + | + |
| 17146 | − | − | + | + |
| 17170 | − | − | + | + |
NA, sample not available because of death of the animal. EPF and EPJ are the control unvaccinated monkeys. Each sample was analyzed in triplicate and triplicate samples were either all positive (+) or all negative (−) as indicated. Primers specific for a conserved region of HIV 89.6 env were used as described in Materials and Methods.
SIVsmE660 challenge.
SIVsmE660 (12) is an independent isolate of SIVsm, unlinked by any recent history to SIVmac239 or SIVmac251. Infection of rhesus monkeys with SIVsmE660 results in consistently high viral loads and eventual progression to AIDS and death (15). Thus, challenge in this case was with an uncloned strain of SIV within the same grouping of primate lentiviruses as SIVmac but with natural sequence variation throughout its genome. Although SIVsmE660 has not been directly sequenced, it is closely related to SIVsmH4 and SIVsmE543-3 since all were derived from the same original infected animal (14, 15). SIVsmH4 and SIVsmE543-3 have been completely sequenced; they are very similar, and the relatedness of their sequences to SIVmac239 has been previously calculated (14). We can thus estimate 89 to 92% amino acid identity in gag-pol and about 82% identity in env between SIVsmE660 and SIVmac239 (Table 1). Challenge was performed by the intravenous route with 20 tissue culture infectious doses of virus 28 months from the time of SIVmac239Δ3 vaccination. Two naive rhesus monkeys again served as controls for the challenge.
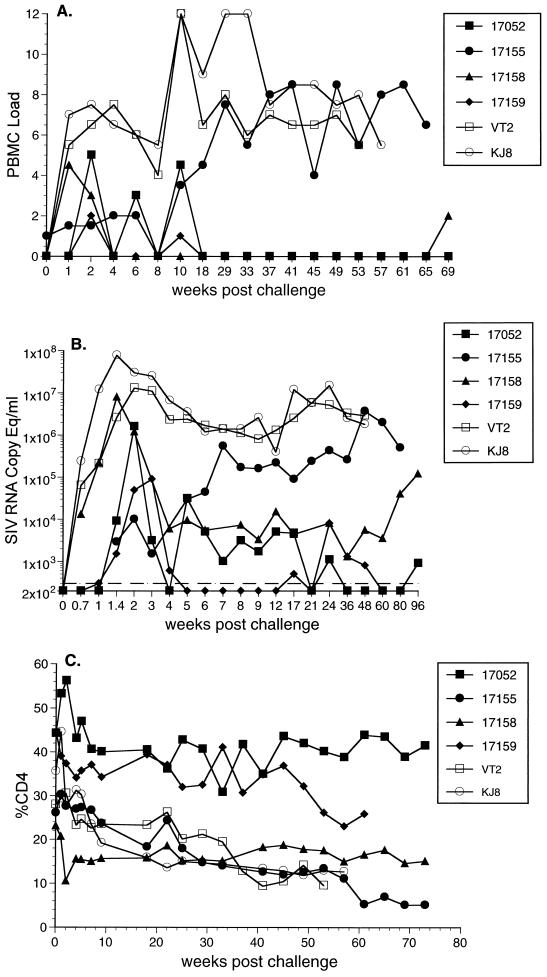
Vaccine virus was not recovered from three of the four vaccinated monkeys around the time of challenge, and it was recovered from the fourth only when 106 PBMC were used (Fig. 3A). Cell-associated virus loads in the control animals reached high levels in the weeks immediately following challenge and remained at high levels until the time of their deaths with AIDS 50 to 60 weeks following inoculation (Fig. 3A). SIV was recovered from three of the four vaccinated animals on multiple occasions over the first 10 weeks with 333,333 or fewer PBMC (Fig. 3A). Cell-associated loads subsequently declined to low levels, with no virus recovered even with 106 PBMC in three of the four vaccinated monkeys between 18 and 65 weeks postchallenge. The fourth vaccinated monkey (17155), however, maintained high cell-associated viral loads for the duration of the measurements.
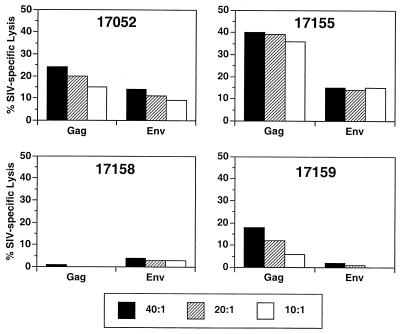
FIG. 3.
Outcome of challenge with SIVsmE660. (A) Cell-associated virus loads measured by the number of infectious cells in PBMC. Code for PMBC load: 2, 333,333 PBMC; 3, 111,111 PBMC; 4, 37,037 PBMC; 5, 12,345 PBMC; 6, 4,115 PBMC; 7, 1,371 PBMC; 8, 457 PBMC. (B) Viral RNA loads in plasma. (C) CD4+ T lymphocytes. Open symbols are for control unvaccinated animals VT2 and KJ8. Closed symbols are for vaccinated animals.
The results of plasma RNA measurements in this study also agreed well with the cell-associated viral load measurements. Plasma RNA in 17155 increased to levels approximating those in the control animals (Fig. 3B). Two of the three remaining vaccinated animals (17052 and 17158) maintained persistently detectable plasma RNA levels for at least 24 weeks, but the levels were approximately 2 to 3 logs lower than those seen in the unvaccinated controls (Fig. 3B).
Both of the unvaccinated controls exhibited declines in CD4 cell numbers up until the time of their deaths (Fig. 3C). At necropsy, neither of the two control animals had opportunistic infections. However, animal KJ8 had diffuse follicular hyperplasia with multiple lymphoid nodules characteristic of lymphoproliferative disease, and animal VT2 had lymphoproliferative disease and SIV arteriopathy. Only one of the four vaccinated animals (17052) has remained alive with normal CD4 cell numbers (Fig. 3C). Three of the four vaccinated animals were euthanized due to clinical deterioration 50 to 60 weeks following inoculation. All three of these animals had decreased CD4 cell percentages (Fig. 3C). At necropsy, animal 17155 had marked diffuse follicular hyperplasia and disseminated lymphoid nodules characteristic of lymphoproliferative disease. Animal 17158 had an opportunistic infection with cryptosporidiosis in the small and large intestine as well as lymphocytic meningoencephalitis. The third vaccinated animal that died, 17159, had glomerulonephritis, thymic dysinvolution, SIV arteriopathy, and rare multinucleate giant cells in the pleura of the lung. The decreases in CD4 numbers, clinical condition, and necropsy findings are compatible with SIV-induced disease.
Nested PCR was used to examine viral DNA sequences present in PBMC at 15 days, 22 weeks, and 49 weeks after challenge. Primers spanning the nef gene specific for SIVmac239 and SIVsmE660 sequences were used. For the control unvaccinated animals, fragments corresponding to full-length challenge virus were all that were detected (Table 3). Using PBMC from the four vaccinated monkeys, SIV239-specific primers detected either no viral sequences or viral sequences corresponding to the length of the vaccine strain in 11 of the 12 samples examined (Table 2). The SIVsmE660-specific primers detected challenge virus in all four vaccinated and challenged animals at one or more of the three time points examined (Table 3). Vaccinated monkey 17159, which maintained the lowest viral loads after challenge (Fig. 3B), was negative for the detection of viral sequences with both sets of primers at weeks 22 and 49 (Table 3).
TABLE 3.
Viral sequences in PBMC after SIVsmE660 challenge
| Primer and monkey | Resultb at:
|
||
|---|---|---|---|
| Day 15 | Week 22 | Week 49 | |
| SIV239 primersa | |||
| 17052 | Neg | Neg | Neg |
| 17155 | Δ3 | WT | Δ3 |
| 17158 | Δ3 | Δ3 | Neg |
| 17159 | Δ3 | Neg | Neg |
| VT2 | ND | WT | WT |
| KJ8 | ND | WT | WT |
| SIVE660 primersa | |||
| 17052 | WT | WT | WT |
| 17155 | Neg | WT | WT |
| 17158 | WT | Neg | Neg |
| 17159 | WT | Neg | Neg |
| VT2 | ND | WT | WT |
| KJ8 | ND | WT | WT |
PCR primers corresponding to sequences specific for SIVmac239 and SIVsmE660 that span nef were used (see Materials and Methods).
Neg, viral sequences were not detected. ND, not done. WT, PCR product corresponding to full-length, nondeleted, wild-type challenge virus was detected.
Immune responses.
Serum taken just prior to the time of challenge was used for measurement of SIV-specific antibody responses. Ability to neutralize a laboratory-passaged stock of SIVmac251, an uncloned virus closely related in sequence to SIVmac239 (3), around the time of challenge was similar in the two groups: a median of 1:459 in the SHIV-challenged group and 1:685 in the SIVsmE660-challenged group (Table 4). Three of the four animals challenged initially with SIVsmE660 showed weak neutralizing activity against SIVsmE660 at the time of challenge (Table 4). The four animals challenged with SHIV89.6p showed no detectable neutralizing activity against SHIV89.6p or against HIV-1MN (Table 4). The lack of neutralizing activity against HIV-1 and SHIV is not surprising given the very low level of amino acid similarity in env compared to SIVmac239Δ3 (Table 1).
TABLE 4.
Virus-neutralizing activity in sera from SIVmac239Δ3-vaccinated monkeys at the time of challengea
| Challenge and monkey | Result following vaccination with:
|
|||
|---|---|---|---|---|
| SIVmac251LPb | SIVsmE660 | HIV-1MN | SHIV89.6p | |
| SHIV89.6p | ||||
| 17049 | 466 | NTc | <20 | <20 |
| 17146 | 345 | NT | <20 | <20 |
| 17157 | 451 | NT | <20 | <20 |
| 17170 | 1,410 | NT | <20 | <20 |
| SIVsmE660 | ||||
| 17052 | 1,054 | 28 | NT | NT |
| 17155 | 8,344 | 31 | NT | NT |
| 17158 | 291 | <10 | NT | NT |
| 17159 | 316 | 32 | NT | NT |
Sera taken within 24 h prior to challenge were used in virus neutralization assays. Activities are presented as the reciprocal of serum dilutions at which 50% of the cells were protected.
SIVmac251LP, a laboratory-passaged stock of SIVmac251.
NT, not tested.
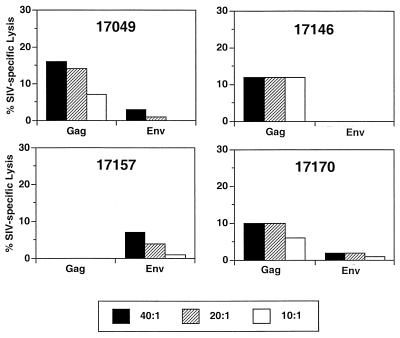
CTL activity against SIVgag and SIVenv antigens was also assessed by using blood samples taken on the day of challenge. Three of the four SHIV-challenged animals showed significant CTL activity to SIVmac251 antigens on the day of challenge as assessed by percent specific lysis (Fig. 4). 17157, the animal without measurable anti-Gag CTL activity, is the one animal with a spike in recoverable virus at week 2 after challenge (Fig. 2B) and this animal also had HIV-1 env sequences detectable in PBMC at week 2 after challenge (Table 2). CTL activity against SIVmac251 antigens in the SIVsmE660-challenged animals assessed on the day of challenge was as high as or higher than in the SHIV-challenged animals (Fig. 5). The animal with the lowest level of SIV-specific CTL activity prechallenge (17158) had the highest peak level of viremia, while the animal with the highest CTL activity (17155) had the lowest peak viremia. CTL activity against SIVsmE660 antigens was not assessed prior to challenge.
FIG. 4.
CTL activity to SIVmac antigens (Gag and Env) on the day of challenge with SHIV89.6p. Assays were performed following antigen-specific stimulation of PBMC (17) at the indicated effector/target ratios.
FIG. 5.
CTL activity against SIVmac antigens (Gag and Env) on the day of challenge with SIVsmE660.
DISCUSSION
Our results demonstrate significant but incomplete protection afforded by SIVmac239Δ3 against challenge with SHIV89.6p. All four vaccinated animals were strongly protected against the rapid CD4 declines and the development of disease. One of the four vaccinated and challenged monkeys exhibited viral load set points below the limits of detection and the other three exhibited fluctuating levels between cutoff (∼300 copy equivalents of viral RNA per ml of plasma) and 10,000 copy equivalents of viral RNA per ml of plasma. None of the vaccinated animals exhibited apparent sterilizing immunity against the SHIV89.6p challenge. While there have been previous reports of protection against SHIV by live, attenuated SIV, most have used nonpathogenic SHIV (1, 5, 33). Our results, and those in a very recent publication (23), extend the descriptions of protective effects of live, attenuated SIV to more highly stringent challenge with an aggressive, pathogenic SHIV. Our findings are consistent not only with the recent report of Lewis et al. (23) but also with reports on the ability of nonpathogenic SHIVs to protect against challenge by pathogenic SIVmac or SIVsm (24, 27).
Vaccination with SIVmac239Δ3 was also only partially protective against SIVsmE660 challenge when performed in a similar format. Protection against SIVsmE660 was actually less impressive than protection against SHIV89.6p in the limited number of animals studied. No significant protection was observed in one of the vaccinated, E660-challenged animals and the other three exhibited 2- to 4-log reductions in viral load at set point compared to control unvaccinated animals. Nonetheless, only one of these four vaccinated, E660-challenged monkeys remains alive with normal CD4 counts. The level of protection afforded by SIVmac239Δ3 against E660 challenge appears to be much lower than what has been observed previously for similar intravenous challenge with SIVmac251 (36). SIVmac251 exhibits a pattern of disease and time course in naive rhesus monkeys similar to that of SIVsmE660, but SIVmac251 is much more closely related in sequence to the vaccine strain, SIVmac239Δ3, than is SIVsmE660. SIVmac251 is related in passage history to SIVmac239 (8, 20) and is only slightly heterologous (3). Thus, as difficult as it will be to match the live, attenuated approach for protective efficacy, even the live, attenuated approach will face significant hurdles in providing protection against the natural variation present in field strains of virus.
Because gp120/gp41 Env proteins are the only virus-encoded proteins on the surface of viral particles and since neutralizing antibodies are directed to them, it has generally been assumed that anti-Env immune responses would be important for providing protection. Our results indicate that factors other than anti-Env immune responses can be principally responsible for the vaccine protection by SIVmac239Δ3, at least against SHIV challenge. The sequences of the env genes of HIV-1 and those of SIVmac/SIVsm are highly divergent. Inspection of their predicted amino acid sequences reveals identical stretches no greater than six amino acids in length. We found that antibodies raised to the SIV vaccine strain did not react appreciably to HIV-1 env (data not shown) and did not neutralize HIV-1 infectivity detectably (Table 4), similar to previous reports (25, 31, 33). In contrast, antibodies raised to the SIV vaccine strain did react well with SIVsmE660 (data not shown) and they were able to neutralize SIVsmE660 infectivity at least to some extent (Table 4). Nonetheless, protection against SIVsmE660 was no better than, and perhaps even worse than, the protection against SHIV89.6p. In contrast to the minimal degree of matching of env sequences, the gag-pol sequences of SHIV89.6 are >99.5% identical with gag-pol sequences of SIVmac239Δ3 (Table 1) (19).
The results described in this report and in another recent manuscript from our group (18) focus attention on the potential of cellular responses, particularly CTL, to Gag and Pol for controlling SIV and SHIV infection. In the recent study (18), monkeys were vaccinated with SIVmac239Δ3X and SIVmac239Δ4 and were challenged vaginally. The development of early or stronger SIV-specific CTL responses appeared to be associated with protection and, in the Δ4-immunized monkeys, protection was observed in the absence of detectable neutralizing antibodies and in one case with only extremely low levels of binding antibodies detectable only with sensitive tests (18). The importance of CTL responses is consistent with the extreme difficulty in neutralizing primary isolates of SIV and HIV (2), with evidence suggesting a role for CTL in controlling viremia during natural infection (22, 26), and with the ability of vaccine-induced anti-Nef CTL to suppress viral replication in some cases following challenge (10). This of course does not exclude the possibility that antibodies could provide complete protection on their own in some circumstances or that antibodies could contribute to the protective capacity of live, attenuated vaccine approaches.
ACKNOWLEDGMENTS
We thank María García-Moll of Bio-Molecular Technology, Inc., for the genetic analyses; Michael Piatak, Jr., Li Li, and Tom Parks for plasma RNA analyses; and Rhona Glickman for the CTL assays. We thank Vanessa Hirsch and Norman Letvin for providing the challenge virus stocks. We also thank Susan Czajak for technical assistance and help with figures and Joanne Newton and Jane FitzPatrick for manuscript preparation.
This work has been supported by PHS grants AI35365 and RR 00168, NIAID contract N01-AI-65303, and federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-C0-56000.
REFERENCES
- 1.Bogers W M, Niphuis H, ten Haaft P, Laman J D, Koornstra W, Heeney J L. Protection from HIV-1 envelope-bearing chimeric simian immunodeficiency virus (SHIV) in rhesus macaques infected with attenuated SIV: consequences of challenge. AIDS. 1995;9:F13–F18. [PubMed] [Google Scholar]
- 2.Burton D R. A vaccine for HIV type 1: the antibody perspective. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi W S, Colignon C, Thiriart C, Burns D P W, Stott E J, Kent K A, Desrosiers R C. Effects of natural sequence variation on recognition by monoclonal antibodies that neutralize simian immunodeficiency virus infectivity. J Virol. 1994;68:5395–5402. doi: 10.1128/jvi.68.9.5395-5402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connor R I, Montefiori D C, Binley J M, Moore J P, Bonhoeffer S, Gettie A, Fenamore E A, Sheridan K E, Ho D D, Dailey P J, Marx P A. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cranage M P, Whatmore A M, Sharpe S A, Cook N, Polyanskaya N, Leech S, Smith J D, Rud E W, Dennis M J, Hall G A. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology. 1997;229:143–154. doi: 10.1006/viro.1996.8419. [DOI] [PubMed] [Google Scholar]
- 6.Daniel M, Letvin N, Sehgal P, Schmidt D, Silva D, Solomon K R, Hodi F S, Jr, Ringler D, Hunt R D, King N W, Desrosiers R C. Prevalence of antibodies to 3 retroviruses in a captive colony of macaque monkeys. Int J Cancer. 1988;41:601–608. doi: 10.1002/ijc.2910410421. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live-attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 8.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 9.Desrosiers R C, Lifson J D, Gibbs J S, Czajak S C, Howe A Y M, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallimore A, Cranage M, Cook N, Almond N, Bootman J, Rud E, Silvera P, Dennis M, Corcoran T, Stott J, McMichael A, Gotch F. Early suppression of SIV replication by CD8+ nef-specific cytotoxic T cells in vaccinate macaques. Nat Med. 1995;1:1167–1173. doi: 10.1038/nm1195-1167. [DOI] [PubMed] [Google Scholar]
- 11.Gauduin M-C, Glickman R L, Means R, Johnson R P. Inhibition of simian immunodeficiency virus (SIV) replication by CD8+ T lymphocytes from macaques immunized with live attenuated SIV. J Virol. 1998;72:6315–6324. doi: 10.1128/jvi.72.8.6315-6324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein S, Elkins W R, London W T, Hahn A, Goeken R, Martin J E, Hirsch V M. Immunization with whole inactivated vaccine protects from infection by SIV grown in human but not macaque cells. J Med Primatol. 1994;23:75–82. doi: 10.1111/j.1600-0684.1994.tb00105.x. [DOI] [PubMed] [Google Scholar]
- 13.Heilman C A, Baltimore D. HIV vaccines—where are we going? Nat Med. 1998;4(Suppl.):532–534. doi: 10.1038/nm0598supp-532. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch V, Adger-Johnson D, Campbell B, Goldstein S, Brown C, Elkins W R, Montefiori D C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Goldstein S, Piatak M J, Montefiori D C, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson R P, Desrosiers R C. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr Biol. 1998;10:436–443. doi: 10.1016/s0952-7915(98)80118-0. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R P, Glickman R L, Yang J Q, Kaur A, Dion J T, Mulligan M J, Desrosiers R C. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J Virol. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, R. P., J. D. Lifson, S. C. Czajak, K. S. Cole, K. H. Manson, R. Glickman, J. Yang, D. C. Montefiori, R. Montelaro, M. Wyand, and R. C. Desrosiers. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relation of degree of protection with level of attenuation. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 19.Karlsson G B, Halloran M, Li J, Park I-W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin N L, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 21.Kirchhoff F, Kestler III H W, Desrosiers R C. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J Virol. 1994;68:2031–2037. doi: 10.1128/jvi.68.3.2031-2037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis M G, Yalley-Ogunro J, Greenhouse J J, Brennan T P, Jiang J B, VanCott T C, Lu Y, Eddy G A, Birx D L. Limited protection from a pathogenic chimeric simian-human immunodeficiency virus challenge following immunization with attenuated simian immunodeficiency virus. J Virol. 1999;73:1262–1270. doi: 10.1128/jvi.73.2.1262-1270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller C J, McChesney M B, Lu X, Dailey P J, Chutkowski C, Lu D, Brosio P, Roberts B, Lu Y. Rhesus macaques previously infected with simian/human immunodeficiency virus are protected from vaginal challenge with pathogenic SIVmac239. J Virol. 1997;71:1911–1921. doi: 10.1128/jvi.71.3.1911-1921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montefiori D C, Cornell R J, Zhou J Y, Zhou J T, Hirsch V M, Johnson P R. Complement control proteins, CD46, CD55, and CD59, as common surface constituents of human and simian immunodeficiency viruses and possible targets for vaccine protection. Virology. 1994;205:82–92. doi: 10.1006/viro.1994.1622. [DOI] [PubMed] [Google Scholar]
- 26.Ogg G S, Jin X, Bonhoeffer B, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 27.Quesada-Rolander M, Makitalo B, Thorstensson R, Zhang Y-J, Castanos-Velez E, Biberfeld G, Putkonen P. Protection against mucosal SIVsm challenge in macaques infected with a chimeric SIV that expresses HIV type 1 envelope. AIDS Res Hum Retroviruses. 1996;12:993. doi: 10.1089/aid.1996.12.993. [DOI] [PubMed] [Google Scholar]
- 28.Reimann K A, Li J T, Veazey R, Halloran M, Park I-W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimann K A, Waite B C D, Lee-Parritz D E, Lin W, Uchanska-Ziegler B, O’Connell M J, Letvin N L. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J Virol. 1996;70:3198–3206. doi: 10.1128/jvi.70.5.3198-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritter G D J, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 31.Robert-Guroff M, Aldrich K, Muldoon R, Stern T L, Bansal G P, Matthews T J, Markham P D, Gallo R C, Franchini G. Cross-neutralization of human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus isolates. J Virol. 1992;66:3602–3608. doi: 10.1128/jvi.66.6.3602-3608.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rud E W, Oglivie L, Clarke B E, Almond N, Kent K, Chan L, Page M, Kitchin P, Stott J, Cook N, Sharpe S, Ashworth T, Dennis M, Hall G, Cranage M P. A naturally attenuated SIVmac32H vaccine or viral interference? In: Norrby E, Brown F, Chanock R M, Ginsberg H S, editors. Vaccines ’94. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 217–223. [Google Scholar]
- 33.Shibata R, Siemon C, Czajak S C, Desrosiers R C, Martin M A. Live, attenuated simian immunodeficiency virus vaccines elicit potent resistance against a challenge with a human immunodeficiency virus type 1 chimeric virus. J Virol. 1997;71:8141–8148. doi: 10.1128/jvi.71.11.8141-8148.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stebbings R, Stott J, Almond N, Hull R, Lines J, Silvera P, Sangster R, Corcoran T, Rose J, Cobbold S, Gotch F, McMichael A, Walker B. Mechanisms of protection induced by attenuated simian immunodeficiency virus II. Lymphocyte depletion does not abrogate protection. AIDS Res Hum Retroviruses. 1998;14:1187–1198. doi: 10.1089/aid.1998.14.1187. [DOI] [PubMed] [Google Scholar]
- 35.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load by determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 36.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]