ZNF397 is a molecular switch between AR-driven, therapy-sensitive prostate cancer and TET2-driven, lineage plastic, therapy-resistant cancer, highlighting TET2 and epigenetic rewiring as targets to overcome resistance.
Abstract
Cancer cells exhibit phenotypical plasticity and epigenetic reprogramming that allows them to evade lineage-dependent targeted treatments by adopting lineage plasticity. The underlying mechanisms by which cancer cells exploit the epigenetic regulatory machinery to acquire lineage plasticity and therapy resistance remain poorly understood. We identified zinc finger protein 397 (ZNF397) as a bona fide coactivator of the androgen receptor (AR), essential for the transcriptional program governing AR-driven luminal lineage. ZNF397 deficiency facilitates the transition of cancer cell from an AR-driven luminal lineage to a ten-eleven translocation 2 (TET2)-driven lineage plastic state, ultimately promoting resistance to therapies inhibiting AR signaling. Intriguingly, our findings indicate that a TET2 inhibitor can eliminate the resistance to AR-targeted therapies in ZNF397-deficient tumors. These insights uncover a novel mechanism through which prostate cancer acquires lineage plasticity via epigenetic rewiring and offer promising implications for clinical interventions designed to overcome therapy resistance dictated by lineage plasticity.
Significance: This study reveals a bifurcated role of ZNF397, and a TET2–driven epigenetic mechanism regulating tumor lineage plasticity and therapy response in prostate cancer, enhances the understanding of drug resistance, and unveils a new therapeutic strategy for overcoming androgen receptor-targeted therapy resistance.
Introduction
Emerging evidence has identified lineage plasticity as a major hallmark of cancer (1). Cancer cells possess the ability to alter their established lineage by reverting to a stem-like and multilineage state and subsequently redifferentiate into an alternative lineage, thereby evading therapies targeting their original lineage-directed survival programs (2–5). This plasticity has been demonstrated to cause resistance to many standard-of-care cancer therapies in various types of human cancers, including prostate, breast, lung, and pancreatic cancers, as well as melanoma (3, 5–11). Although numerous genomic aberrations have been associated with the acquisition of lineage plasticity (3, 5, 6, 8, 12–18), only a limited number of tumors exhibit these known alterations (19), suggesting that a significant proportion of patients experience resistance due to lineage plasticity through unidentified mechanisms. More importantly, the underlying mechanism by which cancer cells initiate the lineage transition to a plastic state remains largely unclear. Lastly, no effective therapies currently exist to target lineage plasticity–driven resistance in tumors, emphasizing the urgent need to identify key modifiers of lineage plasticity and, consequently, druggable targets to reverse it.
In addition to genomic changes, non-mutational epigenetic reprogramming has been identified as a crucial factor in cellular lineage decisions and therapy response in various cancers (20). This includes epigenetic modifications mediated by BET family proteins, EZH2, CHD1, LSD1, and the SWI/SNF complex (6, 8, 21–26). Among the many important epigenetic modifiers associated with lineage regulation, one member of the ten-eleven translocation (TET) proteins, the methylcytosine dioxygenase TET2, has a key role in regulating cell fate and lineage decisions during embryonic development and carcinogenesis (27–32). Importantly, both TET2 expression and TET2-dependent oxidation of methylated cytosine (5mC) to 5-hydroxymethylcytosine (5hmC) are repressed by androgen receptor (AR) signaling, which is the luminal epithelial lineage-specific survival factor and drives the growth of primary prostate cancer (33–35). In contrast, TET2 expression and 5hmC levels are functionally enriched during the development of stem-like and neuronal lineages (32, 36–42) that are two alternative lineages known to confer AR-targeted therapy resistance in prostate cancer. However, the exact molecular function of TET2 in mediating lineage plasticity, as well as the mechanism by which cancer cells hijack the TET2-mediated epigenetic regulation machinery to acquire lineage plasticity and therapy resistance, remains poorly understood.
Here, we have identified zinc finger protein 397 (ZNF397) as a key coactivator of the AR, which is necessary to maintain AR signaling, luminal lineage, and AR-dependent tumor growth in prostate cancer. The frequent loss of ZNF397 in prostate cancer is a driving event that promotes the transition of cancer cells from an AR-driven luminal lineage to a TET2-driven, multilineage, and lineage plastic state expressing epithelial-to-mesenchymal transition (EMT)-like, stem-like, and neuroendocrine (NE)-like lineages, which are now AR-independent and unresponsive to AR-targeted therapies. Contrary to the conventional view that TET2 functions as a tumor suppressor in primary prostate cancer, we demonstrated that TET2 and TET2-driven epigenetic rewiring are crucial mediators of lineage plasticity–driven therapy resistance in metastatic castration-resistant prostate cancer (mCRPC). Both genetic and pharmacological inactivation of TET2 overturned the resistance to AR-targeted therapies in ZNF397-deficient tumors. Collectively, these findings reveal a cell-intrinsic molecular switch that dictates tumor lineage plasticity and suggest that targeting TET2 has the potential to overcome resistance to AR-targeted therapy, ultimately benefiting patients.
Results
ZNF397 Deficiency Confers Resistance to AR-Targeted Therapy in Prostate Cancer
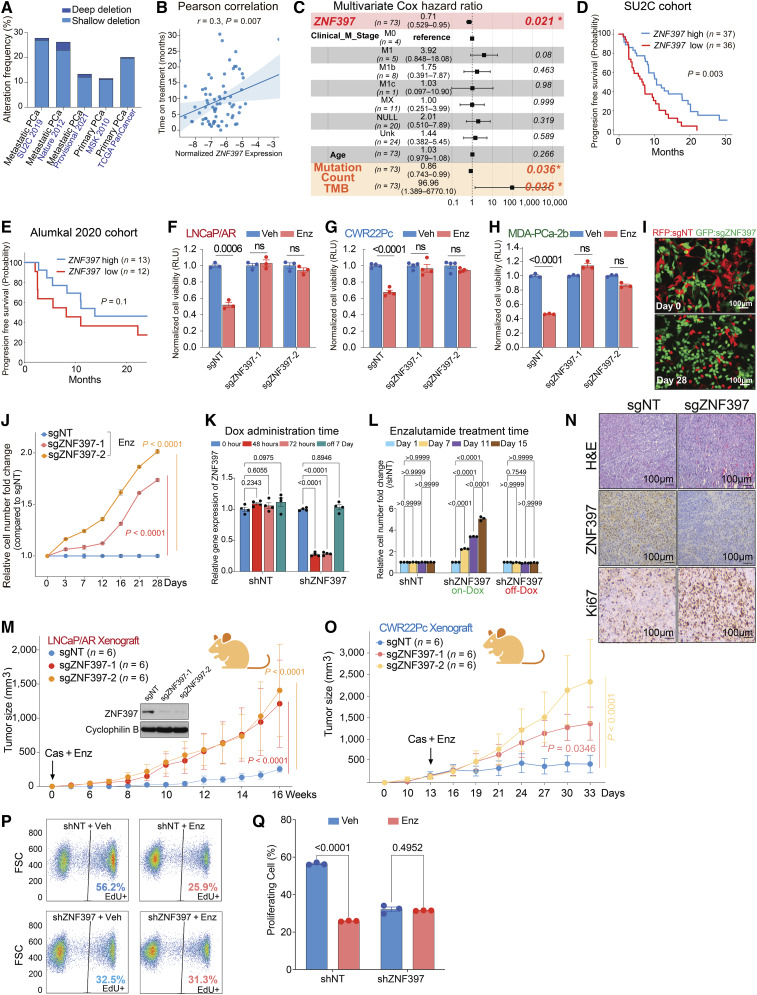
Zinc finger proteins constitute the most extensive family of transcription factors, functioning as either transcriptional repressors or activators for numerous downstream genes that play critical roles in diverse physiological and carcinogenic processes. ZNF397, a member of the classical Cys2His2 group of SCAN-zinc-finger proteins, was initially identified as a mammalian centromere protein essential for centromere localization (43). Although ZNF397 depletion (both deep and shallow) has been frequently observed in around 10% to 25% of patients with prostate cancer (Fig. 1A), its role in tumorigenesis remains elusive. Interestingly, loss of ZNF397 emerged as one of the top candidate events responsible for AR-targeted therapy resistance in our previous in vivo library screening (Supplementary Fig. S1A), according to the ranking algorithm MAGeCK (8, 44). Notably, a Pearson correlation analysis of a mCRPC cohort (a well-characterized subcohort of SU2C, n = 73), which includes associated longitudinal clinical outcomes (8, 19), revealed that the expression level of ZNF397 is significantly correlated with the progression-free survival time of patients with mCRPC undergoing AR-targeted therapies (Fig. 1B). In line with this correlation, we conducted an unbiased, multivariate-corrected Cox hazard analysis of the SU2C cohort to identify the most significant risk factors associated with resistance to AR-targeted therapy. Remarkably, only two risk factors, ZNF397 expression and tumor mutation count/mutational burden, displayed a significant correlation with resistance (Fig. 1C). To further validate the link between ZNF397 expression and AR therapy resistance, we divided the SU2C cohort into two groups based on ZNF397 expression (above or below median) and observed that the patients with low ZNF397 expression developed AR-targeted therapy resistance significantly faster than the patients with high ZNF397 expression (Fig. 1D). This correlation was further validated in another, albeit smaller, mCRPC cohort, the Alumkal 2020 cohort, which also includes longitudinal clinical outcome data (Fig. 1E; ref. 45). Collectively, these clinical correlations between ZNF397 expression and resistance to targeted therapies pose the intriguing hypothesis that ZNF397 may play a pivotal role in mediating resistance to AR-targeted therapy in mCRPC.
Figure 1.
ZNF397 deficiency confers resistance to AR-targeted therapy. A, Stacked bar plot shows the percentage of cancer samples with genomic alterations within the ZNF397 locus in different patient cohorts with prostate cancer, as created using cbioportal.org. B, Pearson correlation analysis shows the relationship between ZNF397 mRNA and time of treatment on Abi/Enz/Apa in the SU2C mCRPC patient subcohort. Abi, abiraterone; Enz, enzalutamide; Apa, apalutamide. ZNF397 expression was normalized to a collection of housekeeping genes. C, Multivariate Cox hazard ratio analysis represents the significant risk factors associated with resistance to Abi/Enz/Apa in the subcohort of patients with SU2C mCRPC. The P value was calculated with the log-rank test. D and E, Kaplan–Meier curve represents the treatment duration on AR-targeted therapies of patients with high (above median) or low (below median) expression of ZNF397 in (D) SU2C and (E) Alumkal 2020 cohorts. The P value was calculated with the log-rank test. F–H, Bar plots represent the relative cell viability of LNCaP/AR (F), CWR22Pc (G), and MDA-PCa-2b cells (H) transduced with Cas9 and annotated CRISPR guide RNAs, measured as values of relative luminescence units and normalized to vehicle-treated conditions. Enz denotes enzalutamide treatment (10 µmol/L for LNCaP/AR and MDA-PCa-2b, 1 µmol/L for CWR22Pc) for several days (7 days for LNCaP/AR, 5 days for CWR22Pc, and MDA-PCa-2b). Veh denotes vehicle, DMSO. I, Fluorescence microscope imaging shows the cell mixtures of sgZNF397 cells (green) and sgNT cells (red) on day 0 and day 28 of the FACS-based competition assay cultured with 10 µmol/L enzalutamide; representative pictures of n = 3 independently treated cell cultures are shown. J, Relative cell number fold change of LNCaP/AR cells transduced with Cas9 and annotated CRISPR guide RNAs, measured by FACS-based competition assay. Enz denotes 10 µmol/L enzalutamide treatment for 28 days. K, Relative gene expression levels of ZNF397 in the inducible shZNF397 LNCaP/AR cells treated with doxycycline (Dox) for various lengths of time. Data are normalized to 0 hour. L, Relative cell number fold change of LNCaP/AR cells transduced with inducible shZNF397. On-Dox cells were consistently exposed to Dox during the Enz treatment period, while Off-Dox cells were treated with Dox for 3 days and removed from Dox for 7 days before growth measurement. Data are normalized to day 1, measured by FACS-based competition assay. M, Tumor growth curve of xenografted LNCaP/AR cells transduced with annotated guide RNAs in castrated mice. Enz denotes enzalutamide treatment at 10 mg/kg from day 1 of grafting. Cas denotes castration. The number of tumors in each group was annotated. N, H&E and immunohistochemical staining of ZNF397 and Ki67 on sgNT and sgZNF397 xenograft tumor slides. Scale bar represents 100 μm. O, Tumor growth curve of xenografted CWR22Pc cells transduced with annotated guide RNAs in intact mice, which were castrated and treated with enzalutamide when average tumor burden reached 300 mm3. Enz denotes enzalutamide treatment at 1 mg/kg. Cas denotes castration. The number of tumors in each group was annotated. P, Representative FACS results showing the percentage of activated proliferating LNCaP/AR cells (EdU+) transduced with annotated hairpins under various treatment conditions, measured by a FACS-based EdU-incorporation assay. Q, Bar plots represent the quantification of panel P, displaying the percentage of EdU-positive and actively proliferating cells under annotated conditions. Veh denotes DMSO, and Enz denotes Enzalutamide. For all panels unless otherwise noted, n = 3 independently treated cell cultures and mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. Schematic figure was created with BioRender.com. See also Supplementary Fig. S1.
To investigate the potential role of ZNF397 in mCRPC, we first knocked out (KO) ZNF397 in a panel of prostate cancer cell line models that are sensitive to the AR antagonist enzalutamide, including LNCaP/AR, CWR22Pc, and MDA-PCa-2b cells, using multiple independent stable CRISPR/Cas9 guide RNAs (Supplementary Fig. S1B). Consistent with clinical observations, ZNF397-KO led to significant enzalutamide resistance in all 3 cell line models, as measured by cell viability assay (Fig. 1F–H) and FACS-based competition assays (Fig. 1I and J; Supplementary Fig. S1C and S1D). To further dissect the dynamics of this resistance, we utilized a doxycycline-inducible short hairpin RNA (shRNA) system to knockdown (KD) ZNF397 and observed that the ZNF397-KD conferred enzalutamide resistance was both rapid and reversible (Fig. 1K and L). Furthermore, ZNF397-KO confers significant resistance to other widely used AR antagonists, apalutamide and darolutamide (Supplementary Fig. S1E and S1F), as well as to an AR protein degrader, AC67 (Supplementary Fig. S1G; ref. 46), suggesting that the resistance in ZNF397-KO cells is not specific to enzalutamide. To assess the effect of ZNF397-KO in AR-targeted therapy resistance in vivo, we first examined the well-characterized LNCaP/AR xenograft model, which exhibits AR amplification (compared to LNCaP cells) and parallels the clinical progression of patients with mCRPC characterized by aberrant AR signaling (47). This unique feature makes this in vivo model resistant to androgen deprivation therapy (ADT/castration) but still sensitive to antiandrogens like enzalutamide (Supplementary Fig. S1H). Importantly, the clinical relevance of this model has been demonstrated through its fundamental role in the development of second-generation antiandrogens, enzalutamide and apalutamide (48). Strikingly, although wild-type (sgNT) LNCaP/AR tumors responded to enzalutamide very well, ZNF397-KO led to complete resistance to enzalutamide in castrated mice (Fig. 1M). Immunohistochemistry (IHC) staining confirmed that ZNF397-KO tumors had substantially increased Ki67 signal compared to control tumors (Fig.1N), indicating ZNF397-KO protects mCRPC tumors from AR-targeted therapy-induced inhibition of proliferation. To further corroborate these findings, we employed a second in vivo model, the CWR22Pc xenografts, where we allowed tumors to establish before castration (ADT) and enzalutamide treatment due to the high sensitivity of this in vivo model. Consistent with the results in the LNCaP/AR xenografts, ZNF397-KO CWR22Pc tumors exhibited significantly higher resistance to AR-targeted therapies (Fig. 1O). Collectively, all of these in vitro and in vivo results demonstrated that ZNF397-KO led to significant resistance to AR-targeted therapies.
To dissect the mechanism underlying the observed resistance, we initially examined the cell cycle dynamics of LNCaP/AR cells under both vehicle and enzalutamide-treated conditions. As expected, enzalutamide treatment significantly reduced the proportion of proliferating cells in the shNT control group (Fig.1P and Q). In contrast, the percentage of proliferating cells remained unchanged in shZNF397 cells treated with either vehicle or enzalutamide (Fig.1P and Q), indicating that ZNF397-KO-protected tumor cells against the proliferation inhibition caused by enzalutamide. Surprisingly, a substantial decrease in proliferating cell was also observed when comparing ZNF397-KO with wild-type cells treated with vehicle (Fig.1P and Q), in contrast to the results under enzalutamide-treated conditions. This unexpected outcome in the cell cycle analysis was consistent across cell proliferation assays in all three AR-dependent prostate cancer cell lines (LNCaP/AR, CWR22Pc, MDA-PCa-2b) under vehicle-treated conditions, showing that ZNF397-KO slowed cell growth (Supplementary Fig. S1I–S1K). These in vitro results were corroborated in vivo, whereby ZNF397-KO tumors progressed slightly slower compared to control tumors in both intact and castrated mice, in the absence of enzalutamide challenge (Supplementary Fig. S1L and S1M). These findings collectively suggest that the impact of ZNF397-KO on prostate cancer tumors not challenged by an AR antagonist differs from its effect on prostate cancer tumors under AR inhibition. This indicates that the role of ZNF397 in prostate cancer may be bifurcated and specifically associated with the regulation of AR signaling, which will be further discussed in the following sections.
ZNF397 is a Pivotal Coactivator for the AR-Driven Transcriptional and Survival Program
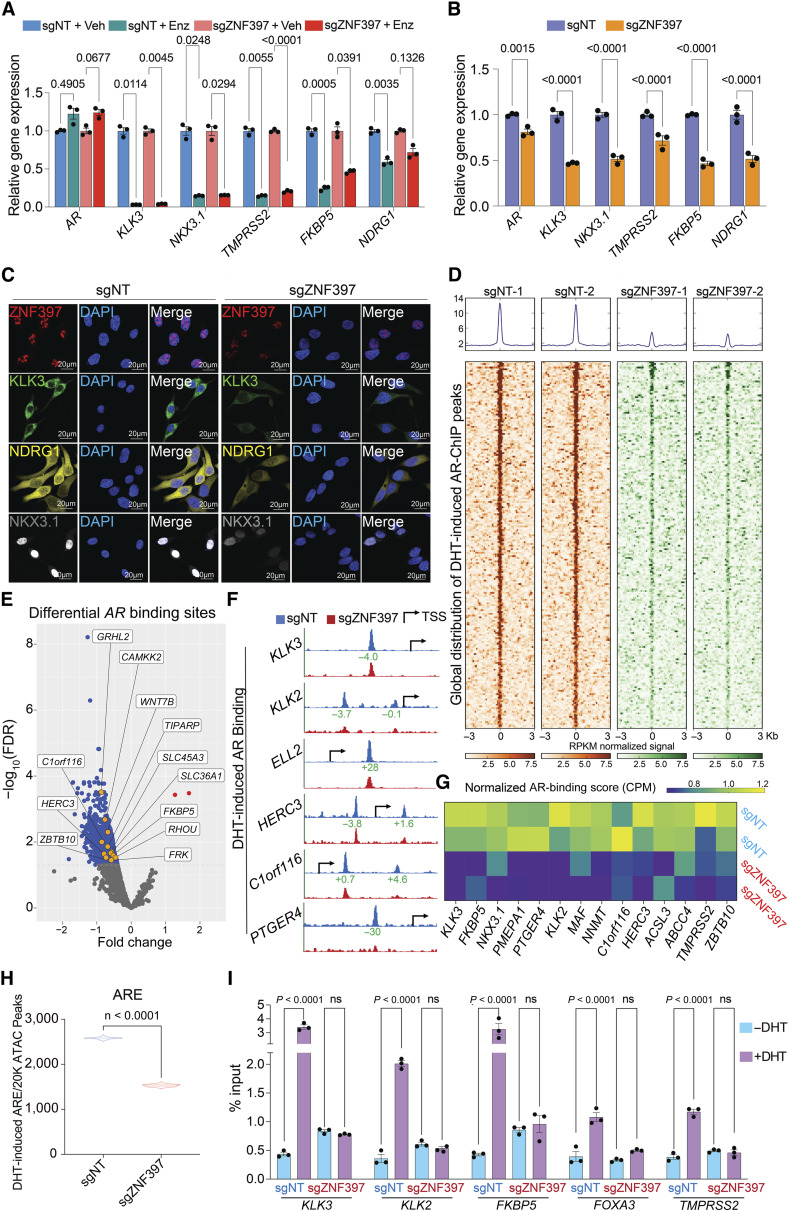
The contrasting impact of ZNF397-KO on prostate cancer tumors, depending on whether they are challenged with AR signaling inhibition, prompts us to hypothesize that the role of ZNF397 is intricately related to the regulation of AR signaling. To explore this hypothesis, we first analyzed the expression of canonical AR target genes and observed persistent inhibition of AR signaling in enzalutamide-resistant ZNF397-KO tumors (Fig. 2A), contrary to the conventional resistance mechanism of AR signaling restoration in many resistant mCRPC tumors. Remarkably, the expression of canonical AR target genes was already downregulated in ZNF397-KO cells even prior to enzalutamide treatment (Fig. 2B). This was further validated by the downregulation of protein levels of AR target genes, evidenced by both Western blots and immunofluorescent (IF) staining (Fig. 2C; Supplementary Fig. S2A). Interestingly, ZNF397-KO did not directly impact expression of AR protein itself, nor affect its cellular localization (Supplementary Fig. S2B–S2D). These results indicate that ZNF397-KO may impair the transcriptional activation of canonical AR-driven program and activate alternative transcriptional programs that relieve prostate cancer from its dependency on AR signaling.
Figure 2.
ZNF397-KO impairs AR-driven signaling and alters the AR cistrome. A, Relative gene expression levels of AR and AR target genes in LNCaP/AR cells transduced with Cas9 and annotated gRNAs and treated with vehicle (Veh, DMSO) or enzalutamide (Enz, 10 µmol/L) for 7 days, normalized to sgNT + Veh group. B, Relative gene expression levels of AR and AR target genes in LNCaP/AR cells transduced with Cas9 and annotated guide RNAs, normalized to sgNT + Veh group. P values were calculated using multiple t tests with Benjamini correction. C, Representative IF staining images of LNCaP/AR cells transduced with Cas9 and annotated guide RNAs with annotated antibodies; n = 3 independent treated cell cultures. D, Global distribution of AR binding peaks based on AR ChIP-seq results. Reads from two independently cultured cell samples were plotted. E, Volcano plot represents the genomic loci with most significantly depleted or gained AR peaks, in ZNF397-KO cells compared to the control cells. Significantly changed gene loci were annotated as blue dots and identified AR target genes were annotated as yellow dots. Reads from two independently cultured cell samples were pooled for analysis. F, Representative AR binding sites in the genomic loci of canonical AR gene loci in the LNCaP/AR cell transduced with Cas9 and annotated guide RNAs, based on AR ChIP-seq analysis. The binding peak distances (kilo base pair) to TSS (Transcriptional Start Site) are annotated in green. G, Heatmap represents the AR binding peak score (CPM, see “Methods”) in the genomic loci of AR Score genes (14 of 20) in the sgZNF397 cells compared to sgNT cells. Reads from two independent cell cultures/guides, matching input controls were used for analysis. H, Violin plot represents the number of DHT-induced ARE peaks per 20k ATAC-seq peaks in annotated cells and treatment conditions. P values were calculated using two-tailed t test. I, AR ChIP-qPCR of the genomic loci of canonical AR target genes in LNCaP/AR cells transduced with annotated constructs, treated with Veh (−DHT) or DHT (+DHT). For all panels unless otherwise noted, n = 3 independently treated cell cultures and mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. See also Supplementary Fig. S2, S3 and Supplementary Table S1.
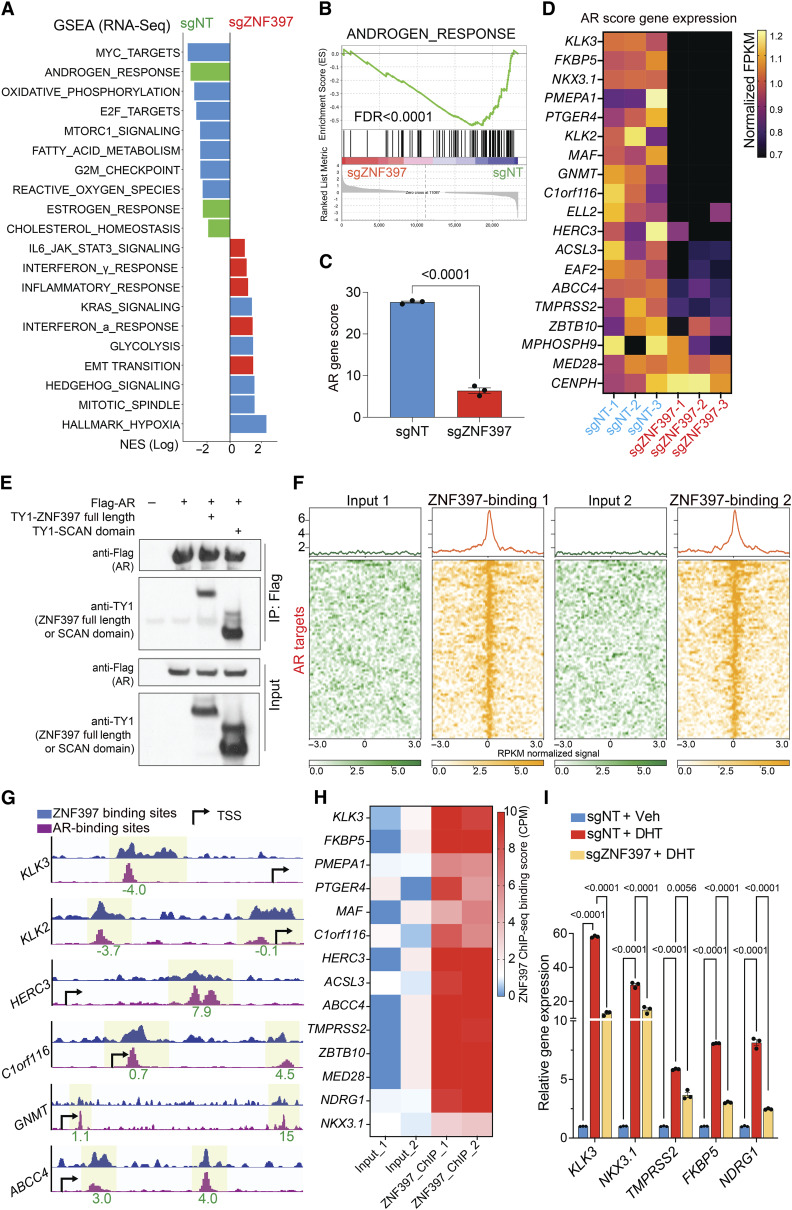
To comprehensively examine the impact of ZNF397-KO on AR-driven transcriptional programs, we conducted genome-wide analyses using AR chromatin immunoprecipitation sequencing (ChIP-seq), Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), RNA sequencing (RNA-seq), and histone marker ChIP-seq (H3K4me3, H3K27ac), to examine the changes in the cistromic and transcriptomic landscapes induced by ZNF397-KO. AR ChIP-seq globally revealed a profound loss of more than 40% of canonical AR binding peaks in ZNF397-KO cells compared to control single-guide NT (sgNT) cells (Fig. 2D and E; Supplementary Fig. S2E; Supplementary Table S1), a reduction comparable to the impact seen with the loss of known AR coactivator FOXA1 (49). This significant reduction underscores the pivotal role of ZNF397 in AR signaling, further evidenced by the profound loss of AR binding at canonical AR target gene loci (Fig. 2F and G: 14 of the 20 AR score genes; ref. 50). To further elucidate the impact of ZNF397-KO on the AR cistrome, we analyzed global chromatin accessibility changes in ZNF397-KO cells treated with AR ligand dihydrotestosterone (DHT), using ATAC-seq with a specific focus on the enrichment of androgen response element (ARE) motifs. While DHT stimulation resulted in a significant number of ARE motif enrichments in sgNT cells, this enrichment was markedly diminished (by approximately 40%) in ZNF397-KO cells (Fig. 2H; ref. 51). This observation is in line with AR ChIP-seq outcomes, indicating that ZNF397-KO compromises the optimal AR binding and AR signaling activation. This finding is further supported by the observation that the motifs most frequently diminished were directly associated with AR signaling pathways, including those for AR, GR, PR/PGR, and FOXA1 (Supplementary Fig. S2F). These genome-wide sequencing results were further validated using AR ChIP-qPCR, which showed a substantial reduction in DHT-induced AR binding to its canonical target genes in ZNF397-KO cells (Fig. 2I), supporting the crucial role of ZNF397 in maintaining AR cistrome. Interestingly, ZNF397-KO also resulted in decreased FOXA1 binding at various sites, as evidenced by both ATAC-seq and FOXA1 ChIP-qPCR results (Supplementary Fig. S2G–S2I). This observation aligns with the profound impact of ZNF397-KO on AR-related transcriptional activation. Correlating with shifts in the AR cistrome, RNA-seq analysis also unveiled global gene expression alterations following ZNF397-KO (Supplementary Fig. S3A; Supplementary Table S2). In line with diminished AR binding to its canonical targets, transcriptomic and gene set enrichment analysis (GSEA) identified a significant suppression of the canonical AR signaling gene signature (Fig. 3A and B; Supplementary Table S3), further evidenced by the marked downregulation of canonical AR score genes upon ZNF397-KO (Fig. 3C and D). This impaired AR-driven transcriptional program, as demonstrated by RNA-seq, was further supported by a significant reduction in histone markers indicative of transcriptional activation, such as H3K4me3 and H3K27ac, at canonical AR target gene sites via histone marker ChIP-seq (Supplementary Fig. S3B–S3E).
Figure 3.
ZNF397 acts as a transcriptional coactivator essential for AR-driven signaling. A, GSEA Pathways analysis shows cancer-related signaling pathways significantly altered in ZNF397-KO cells compared to wild-type cells, lineage specific pathways were highlighted with color: green-AR dependent and luminal lineage pathways, red-lineage plastic signaling pathways. B, GSEA analysis of androgen response gene expression in ZNF397-KO cells compared to wild-type cells. C, AR gene score based on the expression of canonical AR target genes (AR Score Gene) in ZNF397-KO and wild-type cells. mean ± SEM are presented. P values were calculated using two-tailed t test. D, Heatmap represents the relative gene expression of AR Score genes in ZNF397-KO cells compared to wild-type cells, measured by RNA-seq. E, Co-IP of AR (with Flag tag) and full length or the SCAN domain of ZNF397 (with TY1 tag) in HEK293T cells. F, Global distribution of ZNF397 binding peaks at known AR binding sites (as determined by AR ChIP-seq), based on ZNF397 ChIP-seq in LNCaP/AR cells. Reads from two independently cultured cell samples were plotted and matching input controls were used for analysis. G, Representative AR-binding sites (as determined by AR ChIP-seq) and ZNF397-binding sites (as determined by ZNF397 ChIP-seq) in the genomic loci of canonical AR gene loci in the LNCaP/AR cell. The binding peak distances (kilo base pair) to TSS (transcriptional start site) are annotated in green. H, ZNF397-binding peak score (CPM, see “Methods”) in the genomic loci of AR Score genes (14 of 20) in LNCaP/AR cells. Reads from two independent cell cultures/guide were plotted, matching input controls were used for analysis. I, Relative gene expression levels of AR target genes in LNCaP/AR cells transduced with Cas9 and annotated guide RNAs, and treated with Veh (EtOH) or 10 nmol/L DHT for 12 hours, normalized and compared to sgNT + Veh group. For all panels unless otherwise noted, mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. See also Supplementary Fig. S4 and Supplementary Table S2.
Given the significant impact of ZNF397-KO on AR-driven signaling, we inquired into the molecular mechanisms by which ZNF397-KO impairs AR cistrome and AR-driven transcription. Initially, through coimmunoprecipitation (co-IP) assays, we observed robust interactions of AR with both the full-length ZNF397 and its SCAN domain (Fig. 3E), suggesting ZNF397 might be an essential transcriptional coactivator for AR. To further explore this hypothesis and analyze the genome-wide binding pattern of ZNF397, we conducted ZNF397 ChIP-seq in LNCaP/AR cells and integrated this with AR ChIP-seq results. Remarkably, we noted a significant overlap between ZNF397- and AR-binding peaks (Fig. 3F), particularly at canonical AR target genes (AR score genes; Fig. 3G and H), supporting the overlapped binding patterns of ZNF397 and AR. To functionally validate the necessity of ZNF397 in AR-driven signaling, we treated LNCaP/AR cells with DHT and the DHT-induced AR signaling activation was significantly impaired in ZNF397-KO cells compared to controls (Fig. 3I). This finding is further supported by the evidence showing ZNF397-KO protected prostate cancer cells from the growth inhibition typically induced by excessive AR signaling activation with high doses of DHT (Supplementary Fig. S4A; ref. 51), which was further validated through both cell apoptosis and cell proliferation assays (Supplementary Fig. S4B–S4E). Aligning with these observations, ZNF397-KO cells also exhibited minimal responses to R1881, a synthetic androgen (Supplementary Fig. S4F), underscoring the pivotal role of ZNF397 in AR-driven signaling pathways. Finally, we overexpressed ZNF397 in parental LNCaP/AR cells, which resulted in significantly increased expression of AR downstream genes (Supplementary Fig. S4G), supporting the role of ZNF397 in promoting AR-driven transcription. Collectively, those functional results, coupled with the significant reduction of AR binding and AR signaling activation in ZNF397-KO prostate cancer cells, support the role of ZNF397 as a pivotal AR transcriptional coactivator required for optimal AR-driven signaling in prostate cancer.
ZNF397-KO Promotes Lineage Plasticity and Multilineage Transcriptional Programs
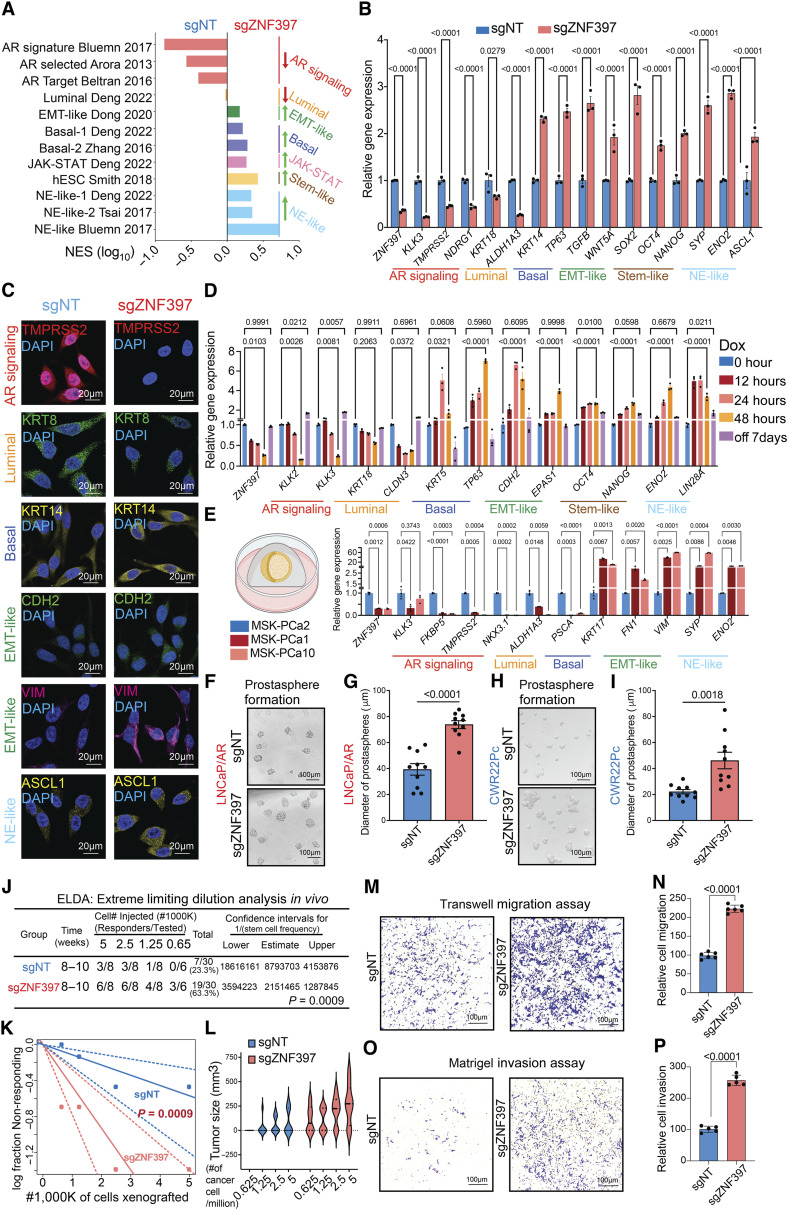
The results from both genome-wide multiomics and functional experiments underscore the critical role of ZNF397 as a key coactivator and explain the observed inhibitory effect of ZNF397-KO on AR-dependent prostate cancer tumor growth in the absence of AR antagonist enzalutamide (Supplementary Fig. S1I–S1M). Conversely, given that ZNF397-KO tumors exhibit significant resistance and AR-independent growth when faced with AR signaling inhibition, we postulated that ZNF397-KO activates alternative transcriptional programs concurrently. This shift relieves prostate cancer tumors from their dependency on AR signaling and luminal lineage, thereby conferring resistance to AR-targeted therapies. Emerging evidence suggests that lineage plasticity is a major mechanism underlying the transition to AR independence (2), characterized as a fast and reversible shift from a luminal-dominant transcriptional program to a mixed, multilineage transcriptional program, including EMT-like, stem-like, basal, and NE-like programs (2, 3, 17, 52). To reveal the global changes in the expression of various canonical lineage-specific gene signatures, we performed GSEA analysis and observed profound downregulation of the AR-dependent signaling pathways and luminal gene signatures (Fig. 4A), while there were substantial upregulations of other nonluminal, multilineage, and lineage plastic gene signatures, including basal-like, EMT-like, stem-like, NE-like, and JAK-STAT gene signatures (Fig. 4A; refs. 3, 17). The correlation between ZNF397 and the transition from an AR-driven transcriptional program to multilineage programs was further validated in a mCRPC single-cell RNA-seq (scRNA-seq) dataset (3, 53), where we identified two major clusters of prostate cancer cell subpopulations expressing either high or low ZNF397 in two of the five patients with mCRPC (Supplementary Fig. S5A and S5B). Consistent with the observations in the prostate cancer cell line, the ZNF397-low subpopulations exhibited significantly higher expression of stem-like, EMT-like, and JAK-STAT gene signatures.
Figure 4.
ZNF397-KO promotes lineage plasticity and multilineage transcriptional programs. A, GSEA pathway analysis shows lineage-specific gene signatures significantly altered in ZNF397-KO cells compared to wild-type cells, lineage-specific pathways were highlighted with various colors. B, Bar plots represent relative gene expression levels of canonical lineage-specific marker genes in LNCaP/AR cells transduced with Cas9 and annotated guide RNAs, as measured by qPCR assay. Data are normalized to sgNT cells. C, IF staining of LNCaP/AR cells transduced with Cas9 and annotated guide RNAs using antibodies against lineage-specific markers; representative images from n = 3 independent treated cell cultures are shown. D, Bar plots represent relative gene expression of canonical lineage-specific marker genes in inducible shZNF397 LNCaP/AR cells treated with Dox for varying lengths of time. Data are normalized to 0 hour. E, Bar plots represent relative gene expression of canonical lineage-specific marker genes in ZNF397-high PDOs compared to ZNF397-low PDOs, measured by qPCR. F, Representative images of LNCaP/AR cell prostasphere formation assay across independently treated cell cultures. G, Quantitative analysis presenting the number of LNCaP/AR prostaspheres formed from independently treated cell cultures for each annotated cell line. H, Representative images of a CWR22Pc cell prostasphere formation assay across independently treated cell cultures. I, Quantitative analysis presenting the number of CWR22Pc prostaspheres formed from independently treated cell cultures for each cell line. J, Number of tumors generated by xenografting various dilutions and numbers of annotated LNCaP/AR cells. Estimates for ELDA 1/(stem cell frequency), top and bottom confidence intervals, and P-value are presented. K, Log-fraction plot represents the ELDA limiting dilution model fitted to the data in J. The slope of the line indicates the log-active cell fraction. Dotted lines represent the 95% confidence interval. L, Violin plot represents the tumor sizes of tumors generated by xenografting various dilutions and numbers of annotated LNCaP/AR cells. All data points and P-values in J–L were calculated using established ELDA software, as described in the experimental procedures. M, Representative images of an LNCaP/AR cell transwell migration assay from three independently treated cell cultures. N, Quantification of migrated cell numbers from representative images, taken from three independently treated cell cultures for each of the cell lines. O, Representative images of an LNCaP/AR cell invasion assay from three independently treated cell cultures. P, Quantification of the numbers of invading cells from representative images, taken from three independently treated cell cultures for each of the cell lines. For G, I, N, and P, P-values were calculated using a two-tailed t test. For all panels unless otherwise noted, mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. Schematic figure was created with BioRender.com. See also Supplementary Figs. S5, S6 and Supplementary Table S3.
The global switch from a luminal-only transcriptional program to a mixed, multilineage program was validated using qPCR, Western blots, and IF staining of lineage marker genes (Fig. 4B and C; Supplementary Fig. S5C). Additionally, treatment of ZNF397-KO cells with enzalutamide led to further induction of lineage plastic genes (Supplementary Fig. S5D), suggesting that ZNF397-KO cells are predisposed to the induction of lineage plasticity. This transition to a multilineage and lineage plastic state was further confirmed in another two human prostate cancer models, CWR22Pc and MDA-PCa-2b cells (Supplementary Fig. S5E and S5F). As one of the key characteristics of the acquisition of lineage plasticity is the fast and reversible transition from an initial luminal-only transcriptional program to the coexpression of multilineage programs, we next deciphered the dynamics of lineage-specific gene expression in doxycycline-inducible ZNF397-KD cells and revealed that the loss of the luminal lineage program and acquisition of lineage plastic programs occurred within only 12 hours upon doxycycline administration and KD of ZNF397 (Fig. 4D). Remarkably, the expression of these multilineage marker genes was fully reversed to wild-type levels upon the restoration of ZNF397 expression following doxycycline removal (Fig. 4D), thereby providing evidence for the reversibility of this phenotype and its consistency with the characterization of lineage plasticity. This correlation between ZNF397 and lineage plastic marker gene expression was further validated in a collection of patient-derived organoids (PDO; ref. 54). Notably, the enzalutamide-sensitive MSK-PCa2 PDO, which exhibits a high level of ZNF397, showed significantly higher AR and luminal signature genes but much lower lineage plasticity genes compared to the two ZNF397-low and enzalutamide-resistant PDOs, MSK-PCa1 and MSK-PCa10 (Fig. 4E).
To functionally validate the induction of multilineage and lineage plastic programs, we specifically examined the cellular phenotype related to stem-like and EMT-like lineages. To examine the induction of the stem-like phenotype, we first conducted prostasphere formation assays in both LNCaP/AR and CWR22Pc cells and revealed a robust induction of prostasphere formation ability upon ZNF397-KO (Fig. 4F–I). To validate the induction of the stem-like phenotype in vivo, extreme limiting dilution analysis (ELDA) xenograft experiments were conducted to examine the tumor initiation capability of ZNF397-KO tumor cells (55, 56). Strikingly, ZNF397-KO cells demonstrated a significantly higher ability to initiate enzalutamide-resistant tumors, compared to wild-type cells (Fig. 4J–L). To functionally validate the induction of an EMT-like lineage program, we conducted various cellular assays to examine the migratory and invasive abilities of prostate cancer cells. Remarkably, ZNF397-KO cells exhibited significantly increased migration and invasiveness compared to control cells, as demonstrated by migration, invasion, and wound healing assays (Fig. 4M–P; Supplementary Fig. S6A and S6B). To further validate this EMT induction, we accelerated the EMT transition with TGF-β + EGF (57, 58). Remarkably, TGF-β + EGF treatment significantly induced more “spindle-like” morphological changes in ZNF397-KO cells and elevated the expression of EMT marker genes (Supplementary Fig. S6C–S6E), indicating that ZNF397-KO cells are more “primed” for EMT transition.
Collectively, the genome-wide expression data in both prostate cancer tumor and clinical models, combined with in vitro and in vivo functional assays, demonstrate the global and profound transition from a luminal-specific transcriptional program to mixed, multilineage, and lineage-plastic transcriptional programs upon ZNF397-KO. This swift and reversible transition in lineage programs corroborates the acquisition of lineage plasticity in ZNF397-KO prostate cancer.
TET2 is the Crucial Driver of Lineage Plasticity and AR-Targeted Therapy Resistance
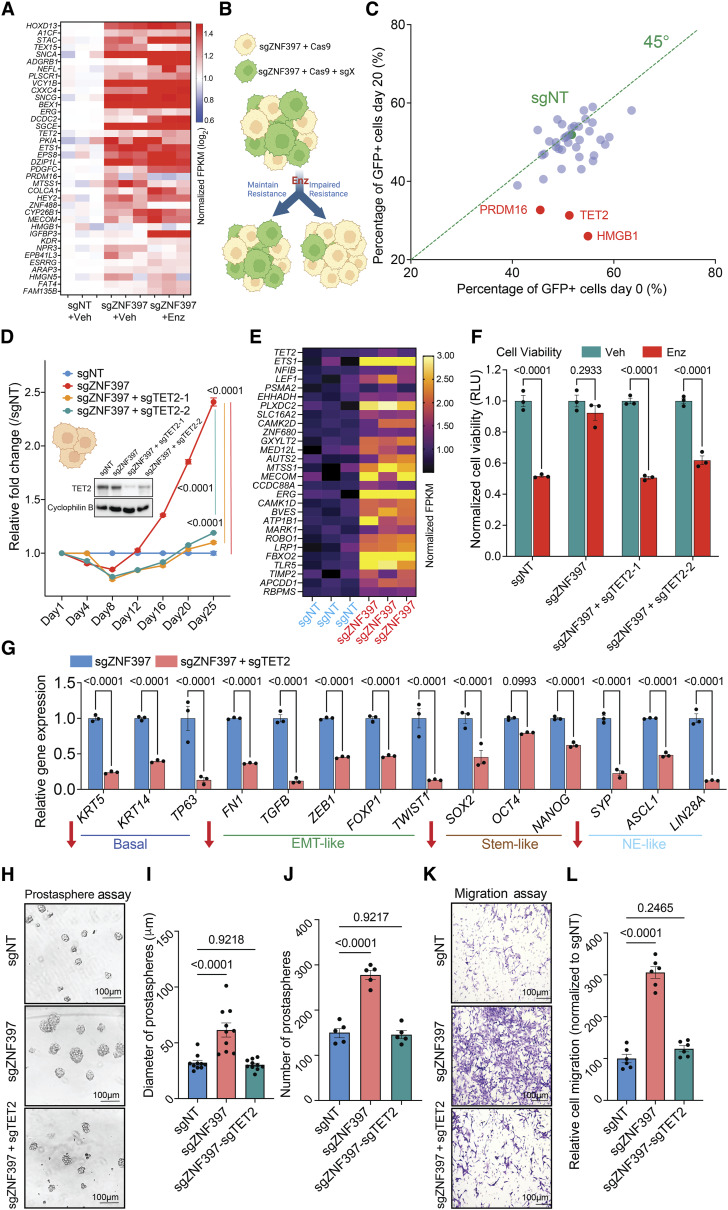
Given that ZNF397-KO impairs AR signaling and induces multilineage transcriptional programs, we hypothesized that ZNF397-KO activates an alternative transcriptional program, promoting lineage plasticity and relieving mCRPC tumors from AR dependence. To identify these alternative lineage-plastic survival programs, we integratively analyzed our genome-wide multiomics results, including transcriptomic and cistromic profiling, focusing on the transcriptional programs activated in enzalutamide-resistant ZNF397-KO tumors. We identified the 38 upregulated genes following ZNF397-KO (Fig. 5A; Supplementary Table S4) as candidate alternative resistance drivers in the context of ZNF397 loss. To explore the functional roles of these candidates in driving resistance, we utilized CRISPR deletion of each of these 38 genes to determine whether their deletion would impede the growth of enzalutamide-resistant ZNF397-KO prostate cancer cells (GFP+; Supplementary Table S4). As assessed through a FACS-based competition assay (Fig. 5B; refer to Methods for details), the KO of three candidate resistance driver genes notably eliminated enzalutamide resistance in ZNF397-KO prostate cancer cells (Fig. 5C; Supplementary Table S4). These genes included TET2, PR domain containing 16 (PRDM16), and high mobility group box 1 (HMGB1). To validate the results of the CRISPR library screening, we knocked out TET2, PRDM16, and HMGB1 in the ZNF397-KO cells and significantly inhibited the resistant growth of those cells in vitro (Fig. 5D; Supplementary Fig. S7A), corroborating their crucial roles in mediating resistance to AR-targeted therapy.
Figure 5.
TET2 is the crucial driver of lineage plasticity and AR-targeted therapy resistance. A, Unsupervised hierarchical clustering of normalized expression of differentially expressed genes whose expression was changed in ZNF397-KO cells treated with vehicle or enzalutamide, comparing to sgNT + Veh group. B, Schematic representation of the functional CRISPR library screen in ZNF397-KO LNCaP/AR cells. sgZNF397 cells (GFP negative) were transduced with Cas9 and sgRNAs targeting individual candidate resistance driver genes (GFP positive). Then these sgZNF397 + Cas9 + sgX (GFP positive) cells were mixed with sgZNF397 (GFP negative) cells to achieve a cell mixture of 40%–70% GFP positive cells. C, Scatter plot summarizing the results of the screen. Each dot represents gRNAs targeting a specific gene. The X axis is the percentage of GFP cells at day 0 and the Y axis is the percentage at day 20. The green dot identifies the sgNT control. Genes that scored positive in the screen are highlighted in red and labeled. Green dotted represents the 45° line. D, Relative cell number fold change of LNCaP/AR cells transduced with Cas9 and annotated CRISPR guide RNAs, measured by FACS-based competition assay. Enz denotes 10 µmol/L enzalutamide treatment for 25 days. E, Relative gene expression levels of canonical TET2 target genes in the LNCaP/AR cells transduced with Cas9 and annotated guide RNAs, measured by RNA-seq analysis. Data are normalized to sgNT. F, Bar plots represent the relative cell viability of LNCaP/AR cells transduced with Cas9 and annotated CRISPR guide RNAs, measured as values of RLU and normalized to vehicle-treated conditions. Enz denotes 10 µmol/L enzalutamide treatment for 7 days and Veh denotes DMSO. G, Bar plots represent the relative gene expression levels of canonical lineage specific marker genes in the LNCaP/AR cells transduced with Cas9 and annotated guide RNAs, measured by qPCR assay, normalized to sgZNF397. H, Representative images of a LNCaP/AR cell prostasphere formation assay across independently treated cell cultures. I, Quantitative analysis presenting the diameter of LNCaP/AR prostaspheres formed from each annotated cell line. J, Quantitative analysis presenting the number of LNCaP/AR prostaspheres formed from each annotated cell line. K, Representative images of an LNCaP/AR cell transwell migration assay of independently treated cell cultures. L, Quantification of the number of migrated cells based on representative images from three separate treated cell cultures for each cell line. For all panels unless otherwise noted, n = 3 independently treated cell cultures and mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. Schematic figure was created with BioRender.com. See also Supplementary Fig. S7 and Supplementary Table S4.
Among those three validated resistance drivers, the epigenetic and 5hmC modifier, TET2, plays a critical role in cell fate and lineage decision during both normal development and carcinogenesis (27–29, 40). Importantly, while TET2 is repressed by AR signaling in primary prostate cancer, the TET2-catalyzed 5hmC modifications are highly enriched in stem-like and NE-like lineages during normal prostate and prostate cancer development (36, 59). Therefore, the observed lineage transition from canonical luminal lineage to a mixed, multilineage, and lineage plastic state in ZNF397-KO tumors raised the possibility that TET2 is a master regulator conferring lineage plasticity and AR-targeted therapy resistance in prostate cancer. Interestingly, while the expression of TET2 itself is only moderately induced upon ZNF397-KO, the expression of canonical TET2 target genes was substantially upregulated in ZNF397-KO cells compared to sgNT cells, as shown in RNA-seq transcriptional results (Fig. 5E; Supplementary Fig. S7B; refs. 60, 61). These results indicate that ZNF397-KO led to a profound induction of the TET2-driven transcriptional program, without directly promoting the expression of TET2 itself.
To assess whether TET2 is required for AR-targeted therapy resistance, we depleted TET2 from ZNF397-KO cells and revealed that TET2-KO completely abolished the enzalutamide resistance conferred by ZNF397-KO, as demonstrated in both FACS-based competition assays and cell viability assays (Fig. 5D and F). To examine whether TET2 is specifically required for the resistant survival of ZNF397-KO cells, we KO TET2 in wild-type LNCaP/AR cells treated with vehicle and did not observe any significant growth difference (Supplementary Fig. S7C). Furthermore, TET2-KO in wild-type cells treated with enzalutamide did not contribute any additional growth inhibition compared to enzalutamide alone (Supplementary Fig. S7D), suggesting that TET2 emerges as a dominant resistance driver only in ZNF397-KO prostate cancer cells. This hypothesis is further supported by the observation that overexpression of TET2 cysteine-rich domain (TET2-C-OE) or catalytic domain (TET2-CD-OE) in wild-type LNCaP/AR cells did not demonstrate a significant growth benefit in the presence of ZNF397 (Supplementary Fig. S7E), supporting that ZNF397 acts as a potent blocker of TET2, and TET2 only becomes a resistance driver when ZNF397 is lost. These results were corroborated by observations in a series of well-established patient-derived explant (PDE) models (3, 44, 46). Significant downregulation of ZNF397 and AR target genes, along with upregulation of TET2 and its target ETS1, was observed in PDE samples treated with enzalutamide, as shown in both qPCR and Western blot assays (Supplementary Fig. S7F–S7H).
Having established the specific necessity of TET2 in the enzalutamide-resistance phenotype in ZNF397-KO prostate cancer, we then examined whether it is required for the induction of mixed, multilineage, and lineage-plastic transcriptional programs. Remarkably, TET2 KO largely reversed the upregulation of basal, EMT-like, stem-like, and NE-like marker genes in the ZNF397 and TET2 double KO cells (sgZNF397+sgTET2; Fig. 5G), demonstrating its crucial role in maintaining lineage plastic transcriptional programs. To functionally validate the impaired induction of stem-like and EMT-like transcriptional programs, we assessed the prostasphere formation ability of the ZNF397/TET2 double-KO cells. Intriguingly, TET2 KO largely abolished the induction of prostasphere formation seen with ZNF397-KO (Fig. 5H), as evidenced by the number and size of prostaspheres (Fig. 5I and J). Furthermore, the enhanced migratory abilities of ZNF397-KO cells were completely reversed upon TET2 KO (Fig. 5K and L), supporting an impaired induction of the EMT-like lineage program. Collectively, these results demonstrated that TET2 is required for both the induction of a mixed, multilineage transcriptional program and AR-targeted therapy resistance.
ZNF397-KO Results in TET2-Driven 5hmC and Epigenetic Rewiring
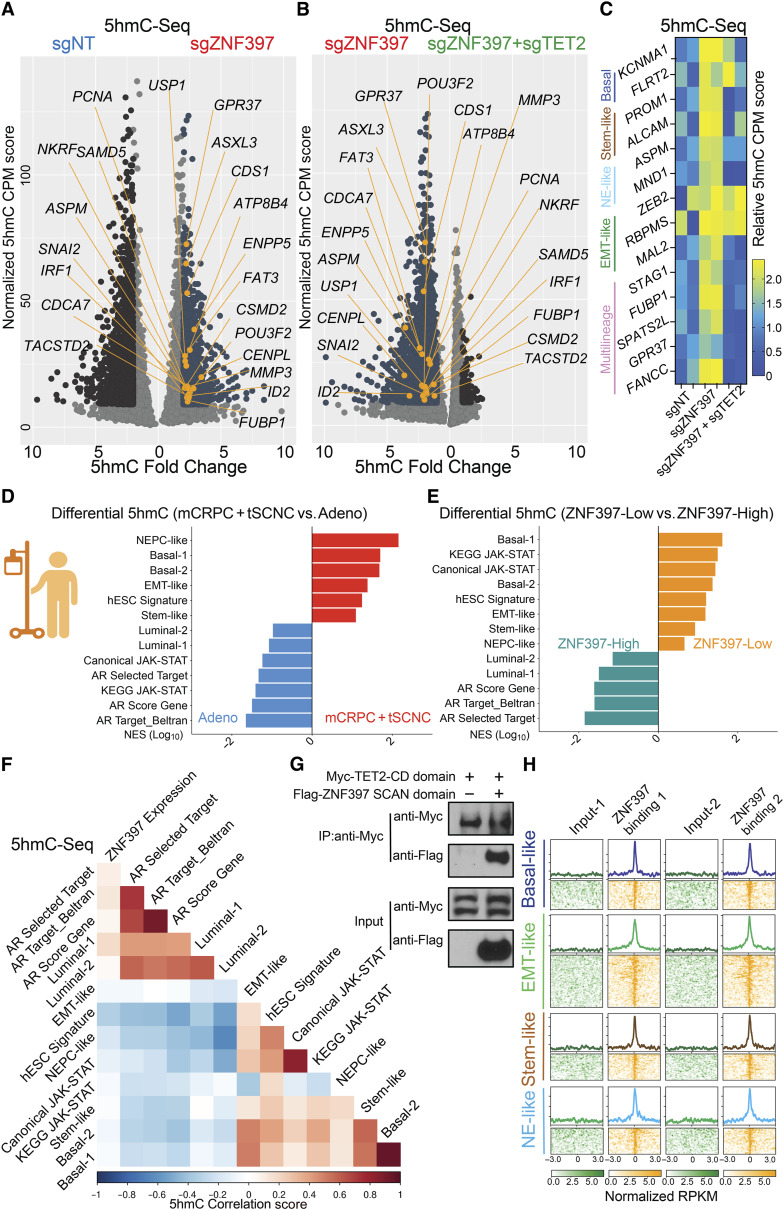
Considering the high correlation of TET2-dependent 5hmC modifications with prostate cancer lineage plasticity, we hypothesized that ZNF397-KO might induce multilineage and lineage-plastic programs by promoting TET2-driven 5hmC modifications at specific lineage plastic gene loci, thereby activating these multilineage transcriptional programs. To test this hypothesis, we performed genome-wide 5hmC-selective chemical labeling sequencing (5hmC-seal seq) to examine the changes of 5hmC landscape following ZNF397-KO and ZNF397/TET2 double-KO (46, 62). Remarkably, we observed a significant and widespread increase in 5hmC at canonical lineage plastic marker gene loci after ZNF397-KO, which was largely reversed in ZNF397/TET2 double-KO cells (Fig. 6A–C), suggesting that ZNF397-KO leads to significant changes of TET2-driven 5hmC modifications at lineage-plastic gene loci. To examine whether this induction in 5hmC was consistent with TET2 binding, we examined an existing TET2 ChIP-seq dataset from the LNCaP-derived cell line C4-2, focusing on TET2-related bindings in enzalutamide-resistant C4-2Enz-R compared to enzalutamide-sensitive control C4-2CON cells (59). Remarkably, we observed widespread and profound induction of TET2 ChIP peaks on lineage plasticity signature genes in enzalutamide-resistant C4-2Enz-R compared to the control C4-2CON cell line (Supplementary Fig. S8A and S8B), highly consistent with the induction of TET2-driven 5hmC on those lineage plastic genes.
Figure 6.
ZNF397-KO results in TET2-driven epigenetic and 5hmC rewiring. A, Volcano plot represents the genomic loci with most significantly enriched or depleted 5hmC modification, in ZNF397-KO cells compared to the control sgNT cells, according to genomic 5hmC-seal seq. Significantly changed gene loci were annotated as dark blue dots, and lineage-plastic signature genes with most significantly enriched 5hmC were annotated as yellow dots. Reads from two independently cultured cell samples were pooled for analysis. B, Volcano plot represents the genomic loci with most significantly enriched or depleted 5hmC modifications, in ZNF397/TET2-double-KO (sgZNF397 + sgTET2) cells compared to the ZNF397-KO (sgZNF397) cells, according to genomic 5hmC–seal seq. Significantly changed gene loci were annotated as dark blue, and lineage-plastic signature genes with most significantly depleted 5hmC were annotated as yellow dots. Reads from two independently cultured cell samples were pooled for analysis. C, Heatmap represents the 5hmC enrichment score (CPM, see “Methods”) in the genomic loci of lineage plastic marker genes in the sgNT, sgZNF397 and sgZNF397 + sgTET2 cells. D, Bar plot presents the lineage gene signatures with most enriched or depleted 5hmC modification in patients with AR-independent mCRPC and t-SCNC compared to patients with AR-dependent adenocarcinoma in a cohort of patients with prostate cancer, based on the results of genomic 5hmC-seq. E, Bar plot presents the lineage gene signatures with most enriched or depleted 5hmC modifications in patients with ZNF397-high (ZNF397 expression above median) compared to patients with ZNF397-low (ZNF397 expression below median) in a cohort of patients with prostate cancer, based on the results of genomic 5hmC-seq. F, Heatmap represents the correlation between ZNF397 expression and the 5hmC modification in the genomic loci of those marker genes of lineage-specific gene signatures, based on the results of genomic 5hmC-seq. G, Co-IP of ZNF397 SCAN domain and the TET2 CD domain with a Flag or Myc tag in HEK293T cells. H, Global distribution of ZNF397 binding peaks at known lineage-plastic and multilineage signature gene loci, based on ZNF397 ChIP-seq in LNCaP/AR cells. Reads from two independently cultured cell samples were plotted and matching input controls were used for analysis. Schematic figure was created with BioRender.com.See also Supplementary Figs. S8, S9 and Supplementary Table S5.
A recent landscape study of prostate cancer 5hmC provided an opportunity to validate the clinically relevant role of ZNF397 in global 5hmC modification and lineage plasticity (36). Using this study, we initially assessed the global distribution of 5hmC modifications in prostate cancer resistant to AR-targeted therapy, including mCRPC and treatment-emergent small cell neuroendocrine cancer (t-SCNC), compared to AR-dependent adenocarcinomas. As anticipated, 5hmC was significantly enriched at the signature genes driving lineage plastic transcriptional programs, including basal, EMT-like, stem-like, and NE-like lineages (Fig. 6D; Supplementary Table S5) in AR therapy–resistant tumors (mCRPC and t-SCNC). Conversely, 5hmC modifications were significantly depleted at the signature genes associated with an AR-dependent luminal lineage (Fig. 6D), indicating that a 5hmC-related lineage switch from AR dependency. We then categorized patients into two groups based on ZNF397 expression and evaluated the global distribution of 5hmC modifications. Notably, 5hmC modifications were significantly enriched at lineage plastic gene loci but depleted at AR targets and luminal lineage gene loci in ZNF397-low prostate cancer compared to ZNF397-high prostate cancer (Fig. 6E), indicating that ZNF397 deficiency correlates with 5hmC modifications at lineage plastic transcriptional programs. Correlation analysis revealed that low ZNF397 expression is correlated with 5hmC enrichment at the lineage plastic signature genes, while high ZNF397 expression is correlated with 5hmC enrichment at the AR-driven luminal lineage signature genes (Fig. 6F). These comprehensive 5hmC-seq findings from both prostate cancer cell lines and patient cohorts confirmed the profound impact of ZNF397-KO in mediating 5hmC rewiring at lineage-plastic transcriptional programs.
To further elucidate the molecular mechanism underlying how ZNF397-KO mediates TET2-driven 5hmC modifications at lineage-plastic transcriptional programs, we initially conducted co-IP assays and observed strong interactions between the SCAN domain of ZNF397 and the CD of TET2 (Fig. 6G), which was further validated through a reverse co-IP of those two proteins (Supplementary Fig. S8C). This observation aligns with a prior study indicating that the ZNF397 SCAN domain blocks the activity of TET2 CD domain, which is required for TET2-mediated 5hmC modification (63). These findings imply that the interaction of the ZNF397 SCAN domain may inhibit TET2-driven 5hmC modification at those lineage plastic gene loci. To test this hypothesis further, we revisited the ZNF397 ChIP-seq data and noted significant and widespread ZNF397 binding at multilineage and lineage-plastic marker gene sites (Fig. 6H). Additionally, histone marker ChIP-seq results revealed a marked increase in histone markers indicative of transcriptional activation, such as H3K4me3 and H3K27ac, at lineage-plastic gene sites following ZNF397-KO (Supplementary Fig. S8D and S8E), which supports the transcriptional activation at these multilineage and lineage-plastic programs. These results are consistent with ATAC-seq results, demonstrating an elevation in motifs of transcription factors known to be associated with TET2 recruitment, including KLF5, SLUG, and ERG motifs in ZNF397-KO cells (Supplementary Fig. S8F–S8H; refs. 64, 65). Finally, the inhibitory role of ZNF397 was validated by reintroducing the SCAN domain of ZNF397, which effectively reversed the enzalutamide resistance caused by ZNF397-KO (Supplementary Fig. S8I).
Given the divergent correlation of ZNF397 with 5hmC in primary prostate cancer (high ZNF397) versus mCRPC/t-SCNC (low ZNF397; Fig. 6E and F), we postulated that ZNF397-KO might affect AR-dependent primary prostate cancer and AR-independent mCRPC differently. Noting low ZNF397 expression correlates with higher chance of resistance to AR-targeted therapy in mCRPC (Fig. 1B–E), we analyzed primary prostate cancer from The Cancer Genome Atlas (TCGA) cohort (66). In line with ZNF397 acting as an essential AR coactivator, elevated ZNF397 levels, indicating greater AR signaling, were linked to more aggressive tumor growth in primary prostate cancer (Supplementary Fig. S9A). Moreover, multivariate correction analysis demonstrated that ZNF397 expression is positively correlated with a higher risk of progression in primary prostate cancer (Supplementary Fig. S9B). These results are consistent with preclinical models showing ZNF397-KO reduces AR-dependent prostate cancer tumor growth without the challenges from AR signaling inhibitors (Supplementary Fig. S1I–S1K). Further validation of the bifurcated and contrasting roles of ZNF397 in primary prostate cancer versus mCRPC came from integrating RNA-seq data from TCGA and SU2C cohorts (Supplementary Fig. S9C), where ZNF397 expression was notably higher in primary prostate cancer than benign tissue (Supplementary Fig. S9D), supporting its role as an AR coactivator. Conversely, ZNF397 levels were markedly lower in mCRPC compared to primary prostate cancer (Supplementary Fig. S9D), suggesting ZNF397 deficiency may facilitate AR-independent mCRPC growth. This contrasting pattern of ZNF397 level was confirmed by IHC staining of matched FFPE samples (Supplementary Fig. S9E). Furthermore, TET2 protein levels were higher in resistant CRPC samples than in sensitive primary prostate cancer (Supplementary Fig. S9E), consistent with its role in driving AR therapy resistance. These results not only highlight the bifurcated role of ZNF397 in AR-dependent versus AR-independent prostate cancer but also suggest its utility as a biomarker for predicting lineage plasticity-driven resistance.
Targeting TET2-Driven Epigenetic Rewiring to Overcome Lineage Plasticity and Resistance
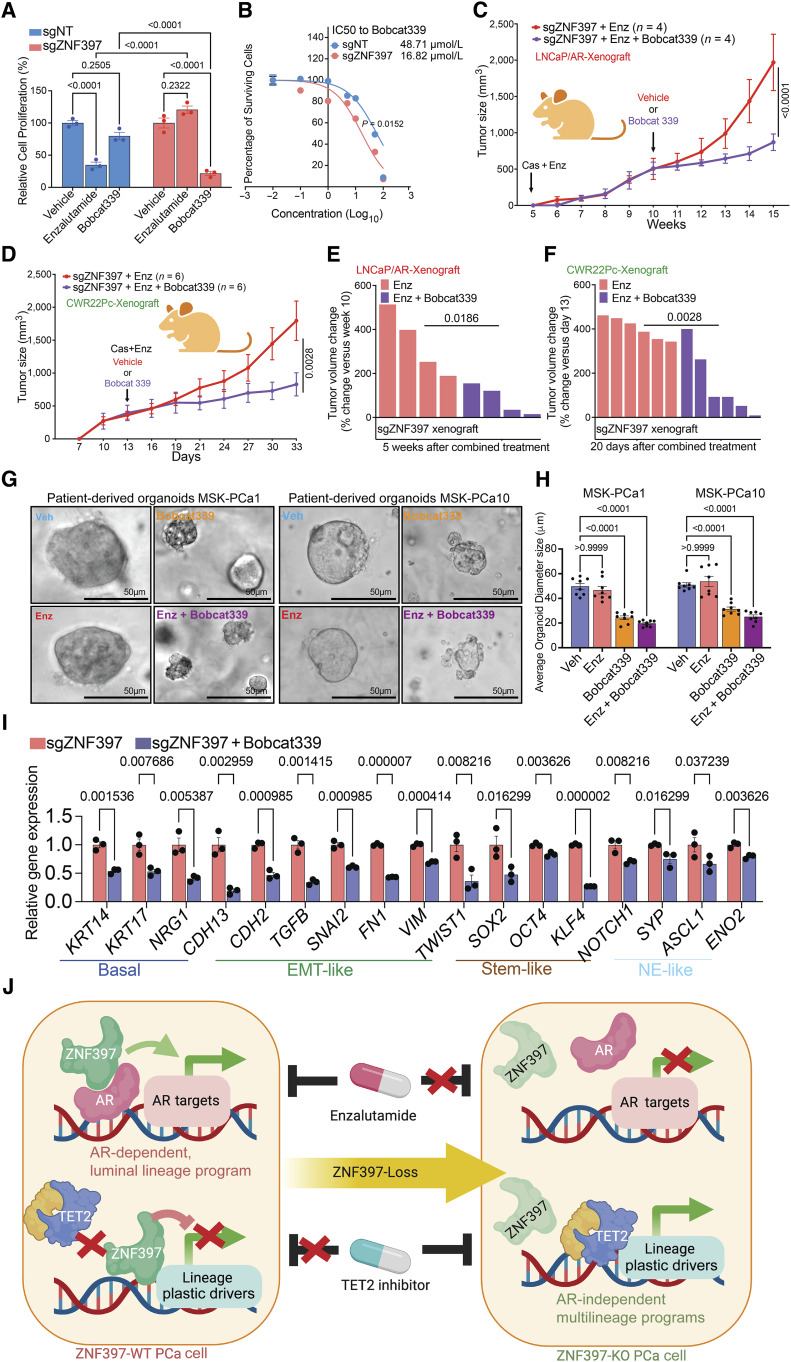
Identification of TET2 as a critical mediator in the development of lineage plasticity-driven resistance raises the hope that targeting TET2 and TET2-dependent 5hmC could overcome AR-targeted therapy resistance, thereby providing a potential benefit to patients with advanced prostate cancer. Strikingly, the combination treatment of enzalutamide and the TET2 inhibitor, Bobcat339 (67–70), significantly impaired the growth of enzalutamide-resistant ZNF397-KO cells, as demonstrated in an in vitro cell viability assay (Fig. 7A). Interestingly, Bobcat339 treatment did not lead to significant growth inhibition in wild-type cells (Fig. 7A), aligning with the hypothesis that TET2 becomes the dominant resistance driver only upon loss of ZNF397. Dose–response measurements (IC50) confirmed that ZNF397-KO cells were significantly more sensitive to Bobcat339 compared to wild-type cells (Fig. 7B). These in vitro findings were further corroborated in vivo in both LNCaP/AR and CWR22Pc xenograft experiments, where the combined treatment of Bobcat339 and enzalutamide halted the growth of enzalutamide -resistant ZNF397-KO tumors (Fig. 7C and D) and triggered profound tumor regressions compared to enzalutamide treatment (Fig. 7E and F). Interestingly, treatment with Bobcat339 in ZNF397-KO cells resulted in significant growth inhibition even without the coadministration of enzalutamide (Fig. 7A). These outcomes align with the hypothesis that loss of ZNF397 leads to a transition from an AR-dependent luminal state to an AR-independent/TET2-dependent lineage plastic state, now solely relying on TET2 for survival. This hypothesis is further validated by genetic perturbation experiments, where TET2 KO in ZNF397-KO cells resulted in slower cell growth even under vehicle-treated conditions (Supplementary Fig. S10A).
Figure 7.
Targeting TET2-driven epigenetic rewiring to overcome resistance. A, Relative cell proliferation of LNCaP/AR cells transduced with annotated guide RNAs and treated with vehicle (DMSO), enzalutamide (10 µmol/L for 7 days), or Bobcat339 (10 µmol/L for 7 days). B, Dose–response curve of ZNF397-KO and wild-type cells treated with TET2 inhibitor Bobcat339. P values were calculated by non-linear regression with extra sun-of-squares F test. C, Tumor growth curve of xenografted ZNF397-KO LNCaP/AR cells in castrated mice treated with enzalutamide. The mice were randomly separated into two groups when their tumors reached 500 mm3 and treated with vehicle or Bobcat339. D, Tumor growth curve of xenografted ZNF397-KO CWR22Pc cells in intact mice. The mice were randomly separated into two groups and castrated when their tumors reached 500 mm3 and treated with vehicle + enzalutamide or Bobcat339 + enzalutamide. E, Waterfall plot displaying changes in tumor size of xenografted ZNF397-KO LNCaP/AR cells after 5 week of combined treatments. F, Waterfall plot displaying changes in tumor size of xenografted ZNF397-KO LNCaP/AR cells after 20 days of combined treatments. For C–F, Enz denotes enzalutamide treatment at 10 mg/kg. Bobcat 339 denotes Bobcat339 treatment at 10 mg/kg. n = number of independent xenografted tumors in each group. G, Bright field pictures represent the 3D-cultured patient derived organoid models, treated with vehicle (Veh), 5 µmol/L enzalutamide (Enz), 10 µmol/L Bobcat339 or enzalutamide + Bobcat339 for 7 days. H, Bar plots represent the size of the 3D-cultured patient derived organoids, treated with vehicle (Veh), 5 µmol/L enzalutamide (Enz), 10 µmol/L Bobcat339 or enzalutamide + Bobcat339 for 7 days. I, Relative gene expression levels of canonical lineage specific marker genes in ZNF397-KO and wild-type LNCaP/AR cells treated with vehicle (DMSO) or Bobcat339 (10 µmol/L for 5 days), measured by qPCR assay. Data are normalized to vehicle-treated cells. J, A schematic figure illustrates the bifurcated role of ZNF397 as a critical coactivator of AR and a suppressor of TET2-dependent lineage plasticity. ZNF397 deficiency in prostate cancer facilitates the transition of prostate cancer cells from an AR-driven luminal lineage to a TET2-driven, multilineage, and lineage-plastic state, which no longer responded to AR-targeted therapy. A yet-to-be identified TET2 cofactor is depicted as a yellow protein. For all panels unless otherwise noted, n = 3 independently treated cell cultures and mean ± SEM are presented. P values were calculated using two-way ANOVA with Bonferroni multiple-comparison test. Schematic figure was created with BioRender.com. See also Supplementary Fig. S10.
To further validate the clinical relevance of these results, we employed two well-established PDO models, MSK-PCa1 and MSK-PCa10 (Fig. 7G; ref. 54), characterized by high TET2 expression. These 3D-cultured PDOs were treated with enzalutamide, Bobcat339, or a combination of both agents. Remarkably, the PDOs resistant to enzalutamide responded to Bobcat339 or the combined treatment of enzalutamide and Bobcat339 (Fig. 7G and H), corroborating the effectiveness of targeting TET2 to overcome enzalutamide resistance. These outcomes further align with the AR independence and TET2 dependence of these PDOs, as the combination of Bobcat339 and enzalutamide resulted in similar growth inhibition as Bobcat339 alone (Fig. 7G and H). Subsequently, we investigated the impact of Bobcat339 on the induction of multilineage transcriptional programs and found that Bobcat339 treatment significantly abolished the induction of lineage plastic marker genes triggered by ZNF397-KO, including basal, EMT-like, stem-like, and NE-like lineages (Fig. 7I). These findings in LNCaP/AR cells were also confirmed in PDOs treated with Bobcat339, as it considerably reversed the upregulation of lineage plastic genes, as evidenced by both qPCR and IF staining (Supplementary Fig. S10B and S10C). Collectively, our studies unveil the pivotal role of ZNF397 as a coactivator of AR and the bifurcated and contrasting effects of ZNF397-KO in AR-dependent versus AR-independent prostate cancer. The loss of ZNF397 in prostate cancer acts as a molecular driver event, facilitating the transition of cancer cells from an AR-driven luminal lineage state to a TET2-driven lineage plastic state, subsequently rendering them unresponsive to AR-targeted therapy (Fig. 7J). These discoveries reveal a cell-intrinsic molecular and epigenetic switch that governs tumor lineage plasticity and resistance to AR therapy.
Discussion
In the last decade, new targeted therapies have significantly advanced the treatment of various cancers, including AR-targeted therapy for advanced prostate cancer. However, resistance in metastatic prostate cancer often develops swiftly, accelerating disease progression and compromising patient outcomes. Lineage plasticity has emerged as a key mechanism of resistance across multiple cancers such as prostate, breast, lung, pancreatic cancers, and melanoma (1). mCRPC stands as one of the most prominent illustrations of lineage plasticity, whereby luminal-only prostate cancer can transition to a mixture of multilineage transcriptional programs that are no longer sensitive to AR-signaling blockage, such as NEPC, double-negative prostate cancer, or stem-like phenotypes (2). This process often involves marked changes in epigenetic regulation, with several epigenetic modifiers playing crucial roles in mCRPC. Yet, the detailed mechanism of epigenetic-driven lineage transitions remains largely unclear, and effective treatments for lineage plasticity-driven cancers are lacking, emphasizing the urgent need to discover actionable molecular targets driving lineage plasticity. Therefore, identifying ZNF397 as a pivotal molecular switch that controls epigenetic rewiring and lineage plastic transition is a major contribution of this work. Our results reveal that ZNF397 is a previously unrecognized and key coactivator of the AR-driven luminal lineage transcriptional and survival program. Interestingly, ZNF397 appears to be essential for a specific subset of AR-binding peaks (exceeding 40%), as no further increase of lost AR peaks was observed when employing less-stringent cutoffs in the examination of the AR ChIP-seq results. More importantly, ZNF397 deficiency has contrasting impacts in AR-dependent primary prostate cancer (where ZNF397 acts as an oncogene) versus AR-independent mCRPC (where ZNF397 serves as a tumor suppressor) due to its bifurcated role as an AR coactivator and lineage plasticity blocker. Unveiling the pivotal but contrasting role of ZNF397 in primary prostate cancer versus mCRPC not only enhances our understanding of the origins and development of therapy resistance but also suggests the utility of ZNF397 as a biomarker for predicting lineage plasticity-driven resistance, which could identify patients for early clinical intervention strategies to prevent or delay resistance.
In addition to the various genomic alterations already implicated in lineage plasticity, the dysregulation of the epigenetic regulation machinery and epigenetic rewiring have been recognized as the predominant determinants of lineage plasticity and therapy resistance in various cancers (6, 8, 21–24, 26). Among the many epigenetic modifications, cancer-associated DNA demethylation and 5hmC conversion, frequently mediated by TET2, play a bifurcated role in the differentiation and progression of advanced cancers, presenting a paradoxical picture. On one hand, TET2 and TET2-dependent 5hmC oxidation are known to counteract the transcriptional signaling driven by AR, which impairs the progression of primary prostate cancer and is correlated with better clinical outcomes (33). On the other hand, TET2-mediated 5hmC transition is highly enriched in the establishment of stem-like and neuronal lineages during development and carcinogenesis of various cancers and, consequently, associated with poor clinical outcomes (41, 42). This seemingly confusing role underscores a significant knowledge gap, suggesting that the dual effects of TET2-mediated modifications on cancer progression might be governed by unknown mechanisms. Contrary to the conventional view of its tumor suppressor role in primary prostate cancer, our findings reveal that TET2-driven epigenetic reprogramming is a powerful mechanism that promotes the transition of mCRPC cancer cells from an AR-driven luminal lineage to a TET2-driven lineage plastic state expressing EMT-like, stem-like, and NE-like lineages. Our results also demonstrate that TET2 only emerges as the dominant resistance driver in ZNF397-deficient and AR-independent mCRPC, which for the first time provides a clear explanation for the bifurcated and contrasting roles of TET2 in AR-dependent primary prostate cancer versus AR-independent mCRPC. These findings not only fill the knowledge gap on how TET2-driven epigenetic rewiring regulates the transition and acquisition of lineage plasticity but also suggest it as a potential therapeutic target for reversing lineage plasticity and resistance. These results parallel reports illustrating the context-dependent oncogenic role of TET2 in melanoma (71). As TET2 does not directly bind to DNA, future work will be necessary to identify the cofactors that facilitate the TET2-driven epigenetic rewiring in ZNF397-deficient prostate cancer.
Despite the clinical success of next-generation therapeutic agents targeting AR signaling in prostate cancer, resistance to these agents, often driven by lineage plasticity, inevitably arises and significantly limits the clinical outcomes for patients with advanced diseases (72, 73). However, effective therapeutic strategies specifically targeting lineage plasticity are currently lacking, highlighting the urgent need for novel approaches to reverse lineage plasticity and overcome resistance. The identification of TET2 as a key mediator of lineage plasticity and therapy resistance in mCRPC may represent a significant advancement in the management of prostate cancer. The potential to reverse lineage plasticity-driven resistance through genetic or pharmacological targeting of TET2 opens new avenues for developing effective therapeutic strategies for patients with advanced prostate cancer. These findings could pave the way for clinical trials to evaluate the efficacy of TET2 inhibitors in treating patients with mCRPC and overcoming resistance, thereby contributing to improved clinical outcomes and overall survival. Furthermore, our model suggests that ZNF397 deficiency provides prostate cancer clones with a selective advantage during AR therapy, leading to the enrichment of ZNF397-deficient tumor cells in enzalutamide-resistant tumors. This model is supported by results showing that acute AR inhibition does not directly downregulate ZNF397 expression (Supplementary Fig. S10D and S10E), indicating that these effects are unlikely to result from a prolonged selection process. These insights further support the potential of using ZNF397 genomic status as an early biomarker for predicting lineage plasticity–driven resistance and identifying patients more likely to develop resistance and benefit from TET2 inhibition.
Methods
Ethics Statement
Animals were housed under humidity- and temperature-controlled conditions with a 12 hours light/12 hours dark cycle in the pathogen-free facilities at UT Southwestern Medical Center by the Animal Resource Center and were monitored closely to minimize discomfort, distress, pain, or injury throughout the course of the in vivo experiments. Animals were removed from the study and euthanized if any signs of pain and distress were detected or if the tumor volume reached 2,000 mm3. The in vivo xenograft experiments were performed as previously described (3, 8) and described in detail below. All animals were divided into each experimental group at random, without prior designation. The xenograft tumor cells injection assays and follow-up tumor treatments were performed by one researcher, while tumor measurement and data analysis were performed by a different researcher to ensure the studies were run in a blinded manner. All procedures adhered to the guidelines provided by the Panel on Euthanasia of the American Veterinary Medical Association and the animal protocol was reviewed and approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center (protocol#2018-102461 and 2019-102493). Male C.B-lgh-1b/lcr Tac-Prkdcscid SCID mice were obtained from Taconic Biosciences.
Cell Lines and Organoids
Human LNCaP/AR and CWR22Pc cell lines were obtained from the laboratory of C.L. Sawyers at Memorial Sloan Kettering Cancer Center. LNCaP/AR and CWR22Pc were cultured in RPMI-1640 medium (Life Technologies 11875135) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific 26400044), 1% GlutaMAX Supplement (Gibco 35050061), 1% penicillin-streptomycin (Sigma-Aldrich P0781), 1% HEPES (Thermo Fisher Scientific 15630080), and 1% sodium pyruvate (Fisher Scientific 11360070). Cells were passaged every 3 to 5 days at 1:3 and 1:2 as previously described (3, 8). For hormone-starving, LNCaP/AR cells were cultured in RPMI-1640 medium containing 10% charcoal-stripped serum (CSS, Thermo Fisher Scientific 26400044), 1% GlutaMAX Supplement (Gibco 35050061), 1% penicillin-streptomycin (Sigma-Aldrich P0781), 1% HEPES (Thermo Fisher Scientific 15630080), and 1% sodium pyruvate (Fisher Scientific 11360070). Human prostate cancer cell line MDA-PCa-2b was purchased from ATCC (CRL-2422, RRID: CVCL_4748) and were cultured in Ham’s F12 K (Kaighn’s) medium supplied with 20% FBS, 1% penicillin-streptomycin (Sigma-Aldrich P0781), 25 ng/mL Cholera toxin (Sigma C8052), 10 ng/mL mouse epidermal growth factor (Fisher CB-40010), 0.005 mmol/L phosphoethanolamine (Sigma P0503), 0.1 ng/mL hydrocortisone (Sigma H0135), 45 nmol/L sodium selenite (Sigma, S9133), and 5 μg/mL human recombinant insulin (Fisher 12-585-014) and were passaged every 3 to 5 days at 1:4 as previously described (3, 8). HEK293T was purchased from ATCC (CRL-3216, RRID:CVCL_0063) and cultured in high-glucose DMEM (Thermo Fisher Scientific 11965126) containing 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% HEPES, and 1% sodium and were passaged every 3 to 5 days at 1:3 and 1:2. The identity of all cell lines were verified by human short tandem repeat profiling cell authentication at UT Southwestern genomic sequencing core every year and compared to ATCC profiles. All cells were routinely tested for mycoplasma using MycoAlert PLUS Mycoplasma Detection kit (Lonza, LT07-710) every month. Human organoids were obtained from the laboratory of Yu Chen at Memorial Sloan-Kettering Cancer Center. Organoids were cultured in 3D Matrigel according to the previously described protocol (74). The morphologies of organoids were assessed under microscope and their growth and mycoplasma contamination were tested monthly.
CRISPR-Cas9, shRNA and Overexpression Plasmid
All the plasmids used in this study were lentiviral-based constructs and modified as described before (3, 8). Specifically, CRISPR-Cas9 and guide RNAs (gRNA) were generated from all-in-one lentiCRISPRv2 (Addgene#52961, RRID: Addgene_52961), LentiCRISPRv2-GFP (Addgene#82416, RRID: Addgene_82416), LentiCRISPRv2-mCherry (Addgene#99154, RRID: Addgene_99154), pLKO5.sgRNA.EFS.RFP (Addgene#57823, RRID: Addgene_57823) and pLKO5.sgRNA.EFS.GFP (Addgene#57822, RRID: Addgene_57822) plasmids. A nontargeting gRNA was used as empty control. Benchling (RRID: SCR_013955, https://benchling.com) was used to design sgRNAs. shRNA sequences were cloned into shRNA constructs SGEP (pRRL-GFP-miRE-PGK-PuroR, Addgene#111170), SCEP (pRRL-mCherry-miRE-PGK-PuroR), LT3CEPIR (pRRL-TRE3G-mCherry-miRE-PGK-PuroR-IRES-rtTA3), and LT3GEPIR (pRRL-TRE3G-GFP-miRE-PGK-PuroR-IRES-rtTA3, Addgene#111177), which were originally obtained from the laboratory of J. Zuber at the Research Institute of Molecular Pathology. The sequence of sgRNAs/shRNA is listed in Supplementary Table S6 Key Resources Table. ZNF397-full length and ZNF397-SCAN were amplified from HEK293T cDNA; TET2-CD domain and TET2-C domain were amplified from the TET2 full-length construct (59). The ZNF397-SCAN overexpression plasmid used for rescue assays was mutated at the PAM sequence with a synonymous mutation. All the sequences were cloned into pcDNA3.1 or pLenti-CMV-P2A-Blast, a gift from Dr. Robert C. Orchard II lab at UT Southwestern Medical Center.
In Vivo Xenograft Tumor Formation and ELDA
All animal experiments were performed in compliance with the guidelines of the Animal Resource Center of UT Southwestern Medical Center. The xenograft experiments were performed as previously described (3, 8). Specifically, 2 × 106 LNCaP/AR cells were suspended in the injection solution, 50% Matrigel (BD Biosciences, 356237) and 50% growth medium. For limiting dilution tumor initiation assay, serial dilutions of cells (0.625 × 106, 1.25 × 106, 2.5 × 106, and 5 × 106 per injection) were prepared. Cell mixture was then subcutaneously injected into the flanks of 7-week-old castrated male C.B-Igh-1b/Icr Tac-Prkdcscid SCID mice on both sides. All 6- to 7-week-old SCID mice were purchased from Taconic Biosciences and separated into each experimental group randomly without prior designation. Mice were then treated daily with 10 mg/kg enzalutamide or vehicle (1% carboxymethyl cellulose, 0.1% Tween 80, 5% DMSO) by gavage feeding 1 day after the injection. Tumor size was measured weekly by using digital caliper when the tumor became measurable. For experiments depicted in Fig. 7, 10 mg/kg enzalutamide (daily) and/or 10 mg/kg Bobcat339 (daily) were given after 6 weeks (for LNCaP/AR) or 5 days (for CWR22Pc) of enzalutamide -only administration when tumors averaged around 500 mm3 in size. For ELDA assay, data were analyzed by using http://bioinf.wehi.edu.au/software/elda/ as previously described (55, 56). The tumor cell injection and follow-up tumor treatment were performed by one researcher, while tumor measurement and data analysis were performed by a different researcher to ensure the studies were run in a blinded manner. No statistical method was used to predetermine sample size, which was decided based on previously established protocol (8).
Lentivirus Production and CRISPR /shRNA/Overexpression Cell Lines Construction
Genomic modified cells were constructed by lentiviral infection as previously described with little modifications (3, 8). 24 hours before transfection, 1.5 × 106 HEK293T cells per well were seeded in 6-well plate. Lentiviral plasmids encoding Cas9/sgRNAs, shRNAs, ZNF397 or TET2, packaging plasmid psPAX2 (Addgene#12260, RRID: Addgene_12260) and the envelope plasmid pVSV-G (Addgene#138479, RRID: Addgene_138479) were mixed in Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific, 31985062). Meanwhile, lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, 11668500) was diluted in Opti-MEM. The diluted plasmid and lipofectamine 2000 were then carefully mixed and incubated at room temperature for 20 minutes. The mixture was added to HEK293T cells drop-wise (RRID:CVCL_0063). The medium was changed 8 to 16 hours after transfection. 24 and 48 hours after the medium change, the virus-containing supernatants were filtered with a 0.45 µmol/L syringe filter and were saved for infection. A total of 4 × 105 LNCaP/AR cells per well were seeded in 6-well plate 1 day before. The culture medium was then changed twice with 50% fresh virus + 50% fresh medium + 5 μg/mL polybrene both 24 and 48 hours after cell seeding. Cells were then selected with 2 μg/mL puromycin (Fisher Scientific, ANT-PR-1) or 10 μg/mL blasticidin S (Thermo Fisher Scientific, A1113903) for 5 days. Doxycycline -inducible shRNA cells were treated with 1 mg/mL doxycycline accordingly. All the shRNAs and sgRNAs sequence are listed in Supplementary Table S6 Key Resources Table.
Cell Dose–Response Curve, Cell Viability, and Cell Growth Assay
Cell dose–response and cell viability were measured using CellTiter-Glo luminescent cell viability assay kit (Promega, 7570) according to the manufacturer’s instructions. LNCaP/AR (1,000 cells per well) or CWR22Pc (20,000 cells per well) or MDA-PCa-2b (4,000 cells per well) cells were seeded in 96-well plates and treated with enzalutamide or vehicle. For 5α- DHT–treated cell viability assay, cells were starved in RPMI-1640 containing 10% CSS medium for 3 days. Cells were seeded in 96-well plate and treated with increasing concentration of DHT or vehicle (EtOH) for 7 days. Then, 100 μL of CellTiter-Glo Reagent was added to each well and mixed for 12 minutes on an orbital shaker. The luminescence was recorded by the SpectraMax iD3 Multi-Mode Microplate Reader, to ensure the results were not biased by prior knowledge of the treatment groups. For the cell growth assay, LNCaP/AR (10,000 cells per well) or CWR22Pc (50,000 cells per well) cells or MDA-PCa-2b (100,000 cells per well) cells were seeded in 24-well plates and treated with enzalutamide or vehicle (DMSO). LNCaP/AR were treated with 10 µmol/L enzalutamide for 7 days, MDA-PCa-2b were treated with 10 µmol/L enzalutamide for 5 days, and CWR22Pc were treated with 1 µmol/L enzalutamide for 5 days. Cell numbers were counted by a Countess II FL automatic cell counter (Invitrogen). Treatments were conducted in three independent cell cultures and all experiments were repeated at least twice and achieved similar conclusions. No data points were excluded. Three independent cell cultures were used and mean ± SEM were reported.
FACS-Based Competition Assay
Cell resistance to enzalutamide was tested by FACS-based competition as previously described (3, 8). CRISRP-Cas9-sgZNF397-GFP or doxycycline-inducible shZNF397-GFP LNCaP/AR cells were mixed with sgNT or shNT-mCherry cells. The mixed cells were treated with either 10 µmol/L enzalutamide or vehicle and collected on indicated days. The percentage of GFP-positive cells was measured by a flow cytometer Attune NxT (version 4.2.1627.1). Relative cell number fold change was calculated as previously described (3, 8). Doxycycline-inducible cells were maintained in 250 ng/mL doxycycline throughout the whole experiment. All experiments were repeated at least twice and achieved similar conclusions. No data points were excluded. Three independent cell cultures were used and mean ± SEM were reported.
EdU Assay
The EdU assay were performed using Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Invitrogen, C10425) according to the manufacturer’s protocol. Briefly, LNCaP/AR cells with different genomic modifications were treated with vehicle or enzalutamide for 5 days, 0.5 × 106 cells per well were then plated in 6-well plates. The next day, cells were incubated with 10 µmol/L EdU for 3 hours. After fixation and permeabilization, cells were proceeded to Click-iT Reaction using Alexa Fluor 488 azide for 30 minutes. Finally, the cell number and percentage were automatically measured by the Attune NxT Acoustic Focusing Cytometer. All experiments were repeated at least twice and achieved similar conclusions. No data points were excluded. Three independent cell cultures were used and mean ± SEM were reported.
In Vitro Migration, Invasion, Wound Healing Assays, and Induction of EMT Assay
The migration and invasion assays were performed in 24-well plates with 8.0 µm transparent PET membrane chambers (Corning, 353097). For the invasion assay, chambers were coated with 30 μg extracellular matrix gel (Corning, 354234). 20,000 LNCaP/AR cells with different genomic modification were resuspended in 300 µL Serum-Free RPMI and seeded into the chambers. 700 µL RPMI with 10% FBS serum was added into the 24-well plate. Cells were cultured for 48 hours at 37°C with 5% CO2. Chambers were washed by PBS once, fixed in 4% PFA for 5 minutes and then methanol for 20 minutes. Cells were then stained in 1% crystal violet and sealed on slides for further quantification. For wound healing assay, 1.5 × 106 LNCaP/AR cells with different genomic modification were seeded in 6-well plates and cultured for 24 hours at 37°C with 5% CO2. 200 μL tips were used to generate the wound scratch. Cell scratches were then captured at 96 hours. All the pictures were captured by Leica DMi8 inverted microscope and quantified by using ImageJ (version 2.0.0, RRID: SCR_003070). The induction of EMT assay was performed as previously described with small modifications (57, 58). LNCaP/AR cells were plated and treated with 10 ng/mL TGF-β (Prepotech, 100-21) and 100 ng/mL EGF (Prepotech, AF-100-15). Cells were collected and imaged at 24 and 48 hours. RNA was extracted for qPCR. All images were quantified by using ImageJ (version 2.0.0, RRID: SCR_003070). For blinding purpose, the pictures were allotted to blind researchers prior to data analysis to avoid bias.
Prostasphere Formation Assay
The prostasphere assay was performed as previously described with small modifications (3, 16). 200 cells per well were suspended in basic organoid medium supplemented with 20 ng/mL epidermal growth factor and 10 ng/mL basic fibroblast growth factor and were seeded into 96-well ultralow attachment plate (Corning 29443). For each condition, three wells were prepared for statistical analysis. Prostaspheres were imaged at one picture per well and quantified 12 days after seeding. All images were quantified by using ImageJ (version 2.0.0, RRID: SCR_003070).
PDE and PDO Experiments
PDE models were established in the Raj laboratory, where around 1 mm3 sample were cultured on top of a gelatin sponge soaked in RPMI-1640 media with 10% FBS, 1% penicillin-streptomycin solution, 0.01 mg/mL hydrocortisone, and 0.01 mg/mL insulin (3). PDEs were treated with 10µmol/L enzalutamide or DMSO for 24 hours and snap frozen for downstream RNA and protein extraction. RT-qPCR were performed and analyzed using a standard protocol describe in “RNA Extraction and RT-qPCR”. Western blots were performed and analyzed using a standard protocol described in “Protein extraction and Western Blot”. PDOs were cultured in 3D Matrigel with typical human organoid medium as previously described (74). PDOs were passaged every 7 days at 1:3 by using trypsin or a sterile glass pipette. For treatment assay, PDOs were cultured in typical human organoid medium supplemented with 5 µmol/L Enz and/or 10 µmol/L Bobcat339. Images were captured by using Leica DMi8 microscope. All images were quantified by using ImageJ (version 2.0.0, RRID: SCR_003070). For blinding purpose, the pictures were allotted to blind researchers prior to data analysis to avoid bias.
RNA Extraction and RT-qPCR
Cells were lysed with Trizol (Ambion 15596018) and 1 μg of extracted RNA was used for cDNA synthesis by using SuperScript IV VILO Master Mix (Thermo Fisher 11766500) per manufacturer’s instructions. RT-qPCR was performed by using 2 × PowerUP SYBR Green Master Mix (Thermo Fisher, A25778). Briefly, 1 to 10 ng single-strand cDNA was used in one reaction and each reaction was conducted in triplicate. Data were analyzed by the delta delta Ct method (2−ΔΔCq) and all the expression of target genes were normalized to reference gene. Heatmaps and bars visualizing the gene differential expression were created by Graphpad Prism 9 Software (RRID: SCR_000306, https://www.graphpad.com/scientific-software/prism/) with expression fold change normalized to control cell lines (sgNT or shNT transduced LNCaP/AR). Primers used for qPCR are listed in Supplementary Table S6 Key Resources Table. All experiments were repeated at least twice and achieved similar conclusions. No data points were excluded. Three independent cell cultures were used and mean ± SEM were reported.
Protein Extraction and Western Blot
Cell pellets were resuspended in Radioimmunoprecipitation Assay lysis buffer (RIPA buffer, 150 mmol/L sodium chloride, 1.0% NP-40 or Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mmol/L Tris, pH 8.0) supplied with proteinase and phosphatase inhibitors for 20 minutes on ice. The protein supernatants were collected by centrifuge (20,000 g for 10 minutes at 4°C) and quantified using Pierce BCA Protein Assay Kit (23225). Protein lysates were denatured in 4 × Laemmli sample buffer (Biorad 1610747) with 2-mercaptoethanol at 95°C for 5 minutes and 10 μg protein per sample was loaded on SDS-PAGE gel running in 1 × NuPAGE MES SDS buffer (Invitrogen, NP0002). Separated proteins were transferred to PVDF membrane in 1 × Blot Transfer buffer (Invitrogen BT00061). Membranes were blocked in 5% non-fat milk for 30 minutes and then incubated with diluted primary antibody overnight at 4°C. Membranes were then washed in 1X TBST 3 times and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 hour at room temperature. The signal was developed into X-ray film in a dark room by using ECL (Thermofisher 32209) or SuperSignal West Pico PLUS (Thermofisher 34580). The following antibodies were used in Western blot (also listed in Supplementary Table S6 Key Resources Table): Rabbit anti-ZNF397 antibody, Sigma-Aldrich#HPA026087, RRID: AB_2677389; Psa/klk3 (D6B1) XP Rabbit mAb, CST#5365, RRID: AB_2797609; Nkx3.1 (D6D2Z) XP Rabbit mAb, CST#92998, RRID: AB_2800197; FOXA1, Abcam#ab23738, RRID:AB_2104842; Vimentin (D21H3) XP Rabbit mAb, CST#5741, RRID: AB_10695459; Wnt5a/b (C27E8) Rabbit mAb, CST#2530S, RRID: RRID:AB_2215595; Synaptophysin, CST#36406, RRID: AB_2799098; TET2 Antibody, Diagenode#C15410255-100; Brn2/POU3F2 (D2C1L), CST#12137, RRID: AB_2797827; Cyclophilin B (D1V5J) Rabbit mAb, CST#43603S, RRID: AB_2799247; Dykddddk Tag (D6W5B) Rabbit mAb, CST#14793, RRID: AB_2572291; Ty1 monoclonal antibody, Diagenode#C15200054; Myc Antibody (9E10), Santa Cruz Biotechnology#sc-40, RRID: AB_627268; Histone H3 (1B1B2) Mouse mAb, CST#14269, RRID: AB_2756816.
H&E Staining, IHC, and IF
Mice xenograft tumors were harvested and were washed once with cold PBS, then were immediately fixed in 10% neutral-buffered formalin (StatLab Medical Products, 28600-1) at 4°C overnight. Fixed specimens were processed and embedded in paraffin by the UT Southwestern Tissue Management Shared Resource core. Tumor blocks were sectioned at 5 μm on a standard rotary microtome (Leica, Germany). For H&E staining, slices were hydrated through xylene and a series of ethanol and then were stained with Hematoxylin (Sigma-Aldrich, MHS1) and Eosin (Sigma-Aldrich, 230251). For IHC staining, after deparaffinization and hydration, slides were heated in citrate sodium buffer (Sigma-Aldrich, W302600) and then were incubated in 3% hydrogen peroxide solution (Sigma-Aldrich, H1009) in methanol (Fisher Scientific, AC423950025). Slides were blocked with 3% BSA in PBST for 30 minutes at room temperature and then incubate with primary antibody (Rabbit Anti-ZNF397 antibody, Sigma-Aldrich#HPA026087, RRID: AB_2677389; Rabbit anti-Ki67, CST#9129, RRID: AB_2687446; AR (N-20), Santa Cruz Biotechnology#sc-816, RRID: AB_1563391; Nkx3.1 (D6D2Z) XP Rabbit mAb, CST#92998, RRID: AB_2800197); Anti-TET2 Antibody, EMD Millipore#MABE462, RRID: AB_2923169 overnight at 4°C. Slides were then incubated with Biotin-conjugated anti-rabbit IgG antibody (Jackson Immunoresearch, 711-065-152, RRID: AB_2340593) and peroxidase Streptavidin (Fisher Scientific#NC9705430). The brown reaction signals were produced by using ImmPACT DAB Peroxidase (HRP) Substrate kit (Vector laboratories, SK-4105). Leica DMi8 microscope was used to capture the Images. For IF staining, LNCaP/AR cells were seeded onto round glass coverslips 1 day before. Cells were then fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Subsequently, the cells were blocked with 3% bovine serum albumin in PBS and incubated with the primary antibody overnight at 4°C. The cells were then incubated with Alexa Fluor-labeled secondary antibodies for 1 hour at room temperature, and nuclei were stained with DAPI. Images were captured using a Zeiss LSM 700 confocal laser-scanning microscope. The following antibodies were used for IF staining: Rabbit anti-ZNF397 antibody, Sigma-Aldrich#HPA026087, RRID: AB_2677389; Psa/klk3 (D6B1) XP Rabbit mAb, CST#5365, RRID: AB_2797609; Ndrg1 (D8G9) XP Rabbit mAb, CST#9485, RRID: AB_2721143; Nkx3.1 (D6D2Z) XP Rabbit mAb, CST#92998, RRID: AB_2800197; TMPRSS2 Antibody (H-4), Santa Cruz Biotechnology#sc-515727, RRID: AB_2892118; Alexa Fluor 647 anti-cytokeratin 8 (EP1628Y), Abcam#ab192468, RRID: AB_2890258; Alexa Fluor 647 anti-cytokeratin 14 (EP1612Y), Abcam#ab192056, RRID: AB_869858; Alexa Fluor 647 anti-cytokeratin 5 (EP1601Y), Abcam#ab193895, RRID: AB_2728796; Vimentin (D21H3) XP Rabbit mAb, CST#5741, RRID: AB_10695459; N-cadherin Antibody (13A9), Santa Cruz Biotechnology#sc-59987, RRID: AB_781744; Anti-MASH1/Achaete-scute homolog 1 antibody, Abcam#ab211327, RRID: AB_2924270; Synaptophysin (D8F6H) XP Rabbit mAb CST#36406, RRID:AB_2799098; Alexa Fluor 647-conjugated AffiniPure goat anti-mouse IgG (H + L), Jackson ImmunoResearch#AB_2338902, RRID: AB_2338902; Alexa Fluor 647-conjugated AffiniPure goat anti-rabbit IgG (H + L), Jackson ImmunoResearch#AB_2338078, RRID: AB_2338078. All primary antibodies were diluted into 1:100 and all the secondary antibodies were diluted into 1:1,000. For blinding purpose, the pictures were allotted to blind researchers prior to data analysis to avoid bias.
Coimmunoprecipitation
A total of 5 × 105 HEK293T cells were seeded into each well of a 6-well plate. A total of 2 μg plasmids (pcDNA3.1-Flag-AR, pcDNA3.1-TY1-ZNF397-full length, pcDNA3.1-TY1-ZNF397-SCAN domain, pcDNA3.1-Flag-ZNF397-SCAN domain, or pcDNA3.1-Myc-TET2-CD domain) were mixed with 10 μL Lipofectamine 2000 in 200 μL OPTI-MEM for 20 minutes at room temperature. The mixture was added evenly to the cells. 48 hours later, cells were lysed in 200 μL ice-cold mammalian IP lysis buffer (25 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP40) supplemented with 1 mmol/L PMSF and 1 × Pierce protease and phosphatase inhibitor (Pierce, A32965) for 30 minutes at 4°C with rotating. Protein supernatant was then collected by centrifugation at 20,000 g for 10 minutes and 30 μL sample was saved as Input. The supernatant was then incubated with 2 μL of pre-washed Anti-FLAG M2 Magnetic Beads (Millipore, M8823) or primary tag antibody overnight at 4°C with rotating. The next day, for supernatant incubated with primary tag antibody, pre-washed Protein A/G magnetic beads (Thermo Fisher) were added and incubated with lysate at 4°C for another 3 hours. Magnetic beads in combination with any target were isolated by magnet (Invitrogen 12321D) and were washed three times with cold IP lysis buffer and boiled together with Input samples in 1 X SDS loading buffer+1% β-ME at 95°C for 10 minutes. Proteins were separated by Western blot and signals were developed by ECL reagent. The following antibodies were used for Co-IP: DYKDDDDK Tag (D6W5B) Rabbit mAb, CST#14793 RRID: AB_2572291; Ty1 monoclonal antibody, Diagenode#C15200054, RRID: AB_2104842; Myc Antibody (9E10), Santa Cruz Biotechnology# sc-40, RRID: AB_627268. All experiments were repeated at least twice and achieved similar conclusions.
ChIP-qPCR
ChIP experiments were performed as previously described (3). LNCaP/AR cells with different genomic modifications were starved under regular RPMI-1640 containing 10% CSS medium for 3 days. A total of 8 × 106 cells were plated in a 15 cm plate 1 day before and treated with 10 nmol/L DHT or vehicle for 4 hours. For AR, H3K4me3, H3K27ac and FOXA1 ChIP, cells then were crosslinked with 1% PFA for 10 minutes and cross-link was quenched by 0.125 mol/L glycine for 5 minutes. For GFP-tagged ZNF397 ChIP, cells were two-step cross-linked with 2 mmol/L DSG (Disuccinimidyl glutarate) for 45 minutes and 1% PFA for 10 minutes, and cross-link was stopped by 0.125 mol/L glycine for 5 minutes. Cell pellet were washed with cold PBS and resuspended in 300 μL ChIP Lysis Buffer with protease inhibitor. The DNA were sheared into 200 to 300 bp by Bioruptor Pico, 1% sample was saved as input and the rest of the samples were incubated overnight with antibody for each reaction in 4°C. Dynabeads Protein G was added to each reaction the next day and incubated for 4 hours in 4°C. The samples were washed sequentially with low salt wash buffer, high salt wash buffer, LiCl wash buffer, and TE buffer. DNA was eluted from the beads and reverse-crosslinked in 0.2mol/L NaCl at 65°C for 4 hours. The input DNA and ChIPed DNA were purified using MinElute PCR Purification Kit (Qiagen 28006) and the concentration was measured by Qubit and ready for qPCR/ChIP sequencing. For ChIP-qPCR, DNA was amplified by 2 × PowerUP SYBR Green Master Mix (Thermo Fisher, A25778). Each reaction was conducted in triplicate and the enrichment percentage to corresponding input was calculated. The following antibodies were used in ChIP (also listed in Supplementary Table S6 Key Resources Table): Anti-Androgen Receptor Antibody, Abcam#ab108341, RRID: AB_10865716); Normal Rabbit IgG Polyclonal Antibody, Millipore Sigma#12-370, RRID: AB_145841; Anti-Histone H3 (acetyl K27) antibody, Abcam#ab4729, RRID: AB_2118291; Anti-Histone H3 (tri methyl K4) antibody, Abcam#ab8580, RRID: AB_306649; FOXA1, Abcam#ab23738, RRID:AB_2104842. Primers used for ChIP-qPCR are listed in Supplementary Table S6 Key Resources Table. Biological triplicates were used and mean ± SEM were reported, and experiments were repeated at least twice and achieved similar conclusions. No data points were excluded.
Subcellular Protein Fractionation
LNCaP/AR cells with different genomic modifications were collected and washed twice with PBS and once with buffer A (10 mmol/L HEPES pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol and protease inhibitor cocktail). Cells were then resuspended in buffer A with 0.1% (v/v) N-P40 and incubated on ice for 10 minutes. After centrifugation, the supernatant containing cytoplasmic fraction was collected. The nuclear pellet was washed once in buffer A and resuspended in buffer B (20 mmol/L HEPES pH 7.9, 1.5 mmol/L MgCl2, 300 mmol/L NaCl, 0.5 mmol/L dithiothreitol, 25% (v/v) glycerol, 0.25% Triton X-100, 0.2 mmol/L EDTA and protease inhibitor cocktail). After centrifugation, the supernatant containing the nucleosolic fraction was collected. The insoluble pellet was then sonicated and treated with RNase and DNase. The above three collected fractions were then analyzed by Western blot.
Statistical Methods
Statistical details of each experiment were shown in figure legends. Two-tailed t test with Welch’s correction for unequal variances was used to compare two groups of independent datasets that fit normality and homoscedasticity. When normality and homoscedasticity were not satisfied, Mann–Whitney U Test (nonparametric Wilcoxon rank-sum test) was used when comparing gene expressions between two patient groups. For comparisons involving more than two groups, one-way or two-way ANOVA and Kruskal–Wallis nonparametric ANOVA were used as appropriate. Mean ± SEM were reported, and P values were calculated and adjusted for multiple comparisons (Bonferroni or Benjamini correction) when applicable. For survival studies, the Kaplan–Meier method was used to estimate and plot the survival curve, and the log-rank test and Cox proportional hazard ratio analysis were used to evaluate differences in survival data among different groups. For all in vitro experiments, three biological replicates were performed except as noted in figure legends.
Bulk RNA-seq
LNCaP/AR cells with different genomic modifications were treated with 10 µmol/L enzalutamide or vehicle for 6 days before the total RNA was extracted using Trizol (Ambion, Cat 15596018) following manufacturer’s instruction. The extracted total RNAs were sent to UT Southwestern Next Generation Sequencing Core to perform bulk RNA-seq following the core’s sequencing instruction. Briefly, RNA concentration was determined by Qubit fluorometer and RNA integrity was determined by using Agilent 2100 Bioanalyzer, and samples with RNA integrity number (RIN) Score 8 or higher were used for library preparation. RNA libraries were prepared by using TruSeq Stranded Total RNA LT Sample Prep Kit from Illumina. Strand-specific cDNA was synthesized from poly-A RNA and was then a-tailed and ligated with indexed adapters. Samples are amplified by PCR and purified with Ampure XP beads, following validation on the Agilent 2100 Bioanalyzer. Samples were quantified by Qubit before they were normalized and pooled. Then all samples were run as paired end read 150 nucleotides in length on the Illumina HiSeq 2500 using SBS v3 reagents. Alignment, quantification, and differential analysis were performed using the QBRC_BulkRnaSeqDE pipeline (https://github.com/QBRC/QBRC_BulkRnaSeqDE). Briefly, alignment of reads to human reference genome (GRCh38) was done using STAR (v2.7.2b) (v2.7.2b, RRID:SCR_004463). FeatureCounts (v1.6.4, RRID:SCR_012919) was then used for gene count quantification. Differential expression analysis was performed using the R package DEseq2 (v1.26, RRID: SCR_015687). Cutoff values of absolute fold change greater than 2 and FDR < 0.05 were used to select for differentially expressed genes between sample group comparisons. GSEA was carried out with the R package fgsea (v1.14.0) using the 'KEGG' and 'WikiPathways' libraries, and a custom library on transcriptional regulatory interactions (“TRRUST_Transcription_Factors_2019”) downloaded from the Enrichr database (RRID:SCR_001575; ref. 75).
ChIP-seq
The ChIPed DNA and input DNA were prepared the same as ChIP-qPCR. The DNA concentration was tested by Qubit and the samples were assessed by Agilent 2100 Bioanalyzer. 10 ng DNA was used in library preparation, following NEBNext Ultra II DNA Library Prep kit (NEB, E7103). All samples were processed by end preparation, adaptor ligation, U excision, PCR enrichment, and clean up. The final products were qualified by Qubit and assessed by Agilent 2100 Bioanalyzer. All samples were run on Illumina NextSeq 500 by 1 × 75 bp (SR75) in CRI Sequencing Facility (Children s Medical Center Research Institute at UT Southwestern Medical Center). For AR, H3K4me3 and H3K27ac ChIP-seq, the data were analyzed as the following: Raw sequencing reads were quality controlled using FastQC Tool (v0.11.9) and aligned to human reference genome assembly (GRCh38/hg38) using Bowtie 2 (v2.4.4, RRID: SCR_016368) with default parameters. Duplicated reads were removed using Picard MarkDuplicates (v2.26.11, https://broadinstitute.github.io/picard/, RRID: SCR_006525). ChIP-seq peaks were called using MACS2 (v2.1.2; ref. 76). Input DNA was used as the control for peak calling. Genome browser tracks were generated using bamCoverage (v2.4.1; ref. 77) with parameter “—normalizeUsingRPKM” and visualized using Integrative Genomics Viewer (78). Motif analysis was performed using homer (v4.11). Differential analysis of peak enrichment and generation of volcano plot were performed using the “DiffBind” R package v3.9.0 (79). The metagene analysis was performed using the computeMatrix and plotHeatmap modules in deepTools (v3.5.0, RRID:SCR_016366; ref. 77). ChIP-seq binding score is based on count-per-million values of peak regions, which were calculated using FeatureCounts (v1.5.3) and normalized for heatmap plotting. For ZNF397 ChIP-seq, the data were analyzed as following. Raw sequencing reads were trimmed and quality-controlled using trimgalore (v0.6.4) and FastQC Tool (v0.11.8), respectively. Data were aligned to human reference genome assembly (GRCh38/hg38) using Bowtie 2 (v2.4.2, RRID: SCR_016368) with the preset parameter “–very-sensitive” and maximum fragment length set to 2,000. ChIP-seq peaks were called using MACS2 (v2.1.2; https://github.com/macs3-project/MACS). Genome browser tracks were generated using bamCoverage included within deeptools (v3.5.0, RRID:SCR_016366) with parameter “—normalizeUsing RPKM” and visualized using Integrative Genomics Viewer (78). ChIP-seq binding score is based on count-per-million values of peak regions, which were calculated using FeatureCounts (v2.0.1) and normalized for heatmap plotting. The metagene analysis was performed using the computeMatrix and plotHeatmap modules in deepTools (v3.5.0). For TET2 ChIP-seq analysis, the raw fastq files were downloaded from SRA database using “sratoolkit 3.0.2”. The ChIP-seq data were processed using “nf-core/ chipseq V 2.0.0”. The software versions can be found in “software_versions.yml” file. A customized list of multi-lineage genes was generated based on the well-established gene signatures. The AR score gene signature was adapted from Hieronymus and colleagues (50), and lineage gene signatures were combined from established signatures (Supplementary Table S5; refs. 3, 5, 12, 18, 53, 80–83). The activity of lineage-specific transcriptional programs in those cells were evaluated based on the above list.
ATAC-seq
LNCaP/AR cells with different genomic modifications were starved in CSS–RPMI 1640 Medium for 72 hours and treated with either vehicle (0.1% EtOH) or DHT (10 nmol/L) for 4 hours. A total of 80,000 fresh cells per condition were used in triplicate for ATAC-seq library preparation by following ATAC-Seq Kit Manual (Active Motif, 53150). Briefly, cells were resuspended in 100 μL of ice-cold lysis buffer and the supernatant were incubated in the Tagmentation Master Mix at 37°C for 30 min. Tagmentated DNA fragments were then purified and amplified by PCR. The size of the libraries was qualified by Agilent 2100 Bioanalyzer and the concentration of the libraries were determined by Qubit. Libraries were sequenced by Children’s Research Institute (CRI) Sequencing facility at UT Southwestern Medical Center on Illumina NextSeq 2000 P2 (PE50) per manufacturer instructions. Data were analyzed as the following: FastQC Tool (v0.11.8) was used for quality control and preprocessing (https://github.com/s-andrews/FastQC). Adapter trimming was done using trimgalore (v0.6.4, https://github.com/FelixKrueger/TrimGalore). Alignment to human genome GRCh38 was performed using Bowtie 2 (v2.4.2). Accessible chromatin peaks were called using MACS3 (v3.0.0, https://genomebiology.biomedcentral.com/articles/10.1186/gb-2008-9-9-r137) with “BAMPE” mode and q-value cutoff at 0.01. Differential accessibility analysis was done using the “Diffbind” R package (v3.16; ref. 79). Motif enrichment of differential peaks was performed using the Hypergeometric Optimization of Motif EnRichment suite (Homer, v4.9). The enrichment of ARE and other motifs were analyzed using established approach (51).
5hmC Selective Chemical Labeling Sequencing
5hmC-seal seq experiments were performed as previously described (62). Genomic DNA was extracted and sheared into fragments with an average size of 200 bp using Bioruptor Bioruptor Pico according to the manufacturer’s instructions. A total of 60 μg sonicated genomic DNA (average peak 200 bp), 80 U T4 phage β-glucosyltransferase (NEB, M0357L), 100 µmol/L UDP-6-N3-Glu (Jena Biosciences, CLK-076), and NEB buffer 4 (NEB, B7004S). The glucose transfer reaction was incubated for 2 hours at 37°C. The biotinylation reaction was then performed by the addition of 1 mmol/L DBCO-S-S-PEG3-Biotin conjugate solution at 37°C for 2 hours. The 5-hmC containing DNA were captured by Dynabeads Myone Streptavidin C1 (Invitrogen 65001) and eluted by addition of 100 mmol/L DTT. DNA was then purified by Qiagen MinElute PCR Purification Kit and libraries were prepared by following NEBNext1100 Ultra II DNA Library Prep kit (NEB, E7103). All samples were run on Illumina NextSeq 2000 P2 (100 Cycles) in CRI Sequencing Facility (Children’s Medical Center Research Institute at UT Southwestern Medical Center). Raw sequencing reads were processed and analyzed using the same pipeline described in “ChIP-seq” ZNF397 ChIP-seq analysis. 5hmC sequencing of mCRPC samples were performed and processed as previously reported (36). Gene body counts for TSS to TES in Gencode v.28 (RRID:SCR_014966) including intronic regions were extracted using featureCounts from the Rsubread R package (RRID:SCR_016945) and used for differential analysis. Differential analysis was performed for protein coding genes using the DESeq2 R package (RRID:SCR_000154) adjusted for metastatic tissue site and otherwise default parameters. GSEA was performed using the preranked method implemented in the fgsea R package with genes ranked by the DESeq2 (RRID:SCR_000154) statistic for a curated set of well-established pathway signatures (Supplementary Table S5). Pathway level enrichment scores were calculated using the singscore R package with gene body counts first normalized to TPM and then ranked (84). Correlation between sample-level pathway scores, or ZNF397 expression, were calculated using spearman correlation and visualized using the corrplot R package.
Data availability
-
•
All the described bulk RNA-seq data, ChIP-seq data, ATAC-seq, and 5hmC-seal seq have been deposited in the Gene Expression Omnibus (GEO, RRID: SCR_005012) under the accession numbers GSE230602. Accession numbers are also listed in the key resources table. Original Western blot images and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All analyses in this manuscript were performed using open-source software. Bulk RNA-seq analysis was done using the QBRC Bulk RNA-seq pipeline and the codes were deposited in GitHub (https://github.com/QBRC/QBRC_BulkRnaSeqDE). GSEA statistical analysis was carried out with the R package “fgsea” (v1.14.0). Customized codes used for ChIP-seq analysis and 5hmC-seal seq were deposited in github (https://github.com/zwang0715/Xu_et_al_ChIPseq). scRNA-seq data analysis was performed using established analysis pipeline. Detailed analysis approaches were described in “Quantification and Statistical Analysis” session.
-
•
Any additional information required to reanalyze the data reported in this paper (and requests for resources and reagents) is available from the lead contact, Dr. Ping Mu (Ping.Mu@UTSouthwestern.edu), upon request.
-
•
All cell lines, plasmids, and other reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement if there is potential for commercial application.
Supplementary Material
Table S1. Significantly Altered AR ChIP-seq Peaks in ZNF397-KO Cells
Table S2. Differentially Expressed Genes in ZNF397-KO Cells
Table S3. Significantly Enriched or Depleted Signaling Pathways in GSEA Analysis
Table S4. Candidate Resistance Driver Genes in ZNF397-KO Cells
Table S5. Lineage-Specific and AR Target Gene Signatures
Table S6. Key Resources
Figure S1. ZNF397-deficiency has bifurcated effects on PCa with or without AR inhibition. Figure S2. The impact of ZNF397-deficiency on AR protein and AR cistrome. Figure S3. ZNF397 is required for the activation of canonical AR transcriptional program. Figure S4. ZNF397-KO impairs DHT-induced AR signaling activation. Figure S5. ZNF397-KO promotes multilineage and lineage plastic gene expression. Figure S6. ZNF397-KO promotes stem-like and EMT-like transcriptional programs. Figure S7. ZNF397-KO led to induction of TET2 downstream genes. Figure S8. ZNF397-KO promotes transcriptional activation of TET2 downstream genes. Figure S9. ZNF397-deficiency has bifurcated effects on AR-dependent vs AR-independent PCas. Figure S10. Targeting TET2-driven epigenetic rewiring to overcome resistance.
Acknowledgments
We thank the SU2C, TCGA, cBioPortal.org, and Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) for providing patients’ genomic and transcriptomic data. We thank Drs. Joshi Alumkal and Michael Haffner for providing dataset of the Alumkal 2020 cohort. We thank Drs. Duojia Pan, Ralph Deberardinis, Joshua Mendell, Michael Buszczak, Jun Wu, Vincent Tagliabracci, and Kathryn O’Donnell for helpful discussion. We thank ChatGPT for double-checking the final revised manuscript for typographical and grammatical errors. This work was supported or partially supported by: National Cancer Institute/National Institutes of Health: R00CA218885, R37CA258730 and R01CA288820 P. Mu, 1R01CA258584 T. Wang, T32C124334, 1F31CA261019-01A1 C.R. Tirado ; Department of Defense: W81XWH-18-1-0411 and W81XWH21-1-0520 P. Mu, W81XWH21-1-0418 Xiaoling Li; Cancer Prevention Research Institute (CPRIT): RR170050, RP220473, P. Mu, RP230363 T. Wang; RP210041 A. Mukherji; Prostate Cancer Foundation: 17YOUN12 P. Mu 21YOUN10 M. Sjöström; Welch Foundation: I-2005-20190330 P. Mu; UTSW Deborah and W.A. Tex Moncrief, Jr Scholar in Medical Research Award: P. Mu; UTSW Harold C. Simmons Cancer Center Pilot Award: P. Mu; UTSW CCSG Data Science Shared Resources: DSSR, T. Wang; Terry Fox New Frontiers Program Project Grant (PPG19-1090 to H.H. He). Zhaoning W is a DDBrown awardee of the Life Sciences Research Foundation.
Footnotes
Note Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Sjöström reports grants from the Prostate Cancer Foundation during the conduct of the study. Y. Chen reports grants from NCI and Prostate Cancer Foundation during the conduct of the study; grants from Foghorn Pharmacetuicals and personal fees and other support from ORIC Pharmaceuticals outside the submitted work. A.B. Hanker reports grants from NIH/NCI during the conduct of the study; grants from Breast Cancer Research Foundation, non-financial support from Tempus and Daiichi Sankyo; grants from Takeda and Lilly; non-financial support from Puma Biotechnology outside the submitted work. G.V. Raj reports other support from Etirarx, Bayer, and Pfizer; other support from Astellas outside the submitted work. Zhao Wang reports grants from NIGMS R01GM143380, and NHLBI R01HL162842; grants from Welch Fundation Q-2173-20230405 outside the submitted work. C.L. Arteaga reports personal fees from Sanofi, OrigiMed, TAIHO Oncology, AstraZeneca, Daiichi Sankyo, Laekna Therapeutics; grants from Pfizer; personal fees from Komen Foundation outside the submitted work. H. Liang reports H. Liang is a shareholder and scientific advisor of Precision Scientific Ltd. F.Y. Feng reports F.Y. Feng is an advisor and holds equity in Artera, and has served as an advisor to Astellas, Bayer, Blue Earth Diagnostics, BMS, ClearNote, Janssen, Myovant, Point Biopharma, Novartis, Roivant, Sanofi, SerImmune and Amgen. T. Wang reports other support from NightStar Biotechnologies, Inc. and personal fees from Merck, Inc. outside the submitted work. P. Mu reports grants from National Cancer Institute, Department of Defense, Cancer Prevention Research Institute, Prostate Cancer Foundation, and Welch Foundation during the conduct of the study; personal fees from Accutar Biotechnology outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
Y. Xu: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Yang: Data curation, software, formal analysis, visualization. Zhaoning Wang: Data curation, software, formal analysis, visualization. M. Sjöström: Data curation, software, formal analysis, visualization. Y. Jiang: Data curation, formal analysis, validation, investigation. Y. Tang: Data curation, software, formal analysis, investigation, visualization. S. Cheng: Data curation, software, formal analysis, investigation, visualization. S. Deng: Conceptualization, validation, investigation. C. Wang: Software, formal analysis, validation, investigation. J. Gonzalez: Validation, investigation, visualization, writing–review and editing. N.A. Johnson: Validation, investigation, visualization. Xiang Li: Investigation. Xiaoling Li: Investigation. L.A. Metang: Investigation, writing–review and editing. A. Mukherji: Investigation. Q. Xu: Investigation, writing–review and editing. C.R. Tirado: Investigation. G. Wainwright: Investigation. X. Yu: Investigation. S. Barnes: Data curation, software, investigation. M. Hofstad: Resources. Y. Chen: Resources. H. Zhu: Formal analysis, methodology. A.B. Hanker: Resources. G.V. Raj: Resources. G. Zhu: Data curation, software, formal analysis, investigation. H.H. He: Resources, supervision. Zhao Wang: Supervision, investigation. C.L. Arteaga: Resources, supervision. H. Liang: Supervision, investigation. F.Y. Feng: Resources, data curation, supervision. Y. Wang: Data curation, software, formal analysis, investigation, visualization. T. Wang: Data curation, software, supervision. P. Mu: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 2. Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin Cancer Res 2019;25:6916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deng S, Wang C, Wang Y, Xu Y, Li X, Johnson NA, et al. Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat Cancer 2022;3:1071–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellis L. Understanding cancer lineage plasticity: reversing therapeutic resistance in metastatic prostate cancer. Pharmacogenomics 2017;18:597–600. [DOI] [PubMed] [Google Scholar]
- 5. Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen C-C, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell 2020;37:584–98.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 2018;362:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005;436:117–22. [DOI] [PubMed] [Google Scholar]
- 11. Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer cell 2016;30:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep 2015;12:922-–36. [DOI] [PubMed] [Google Scholar]
- 15. Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 is an androgen receptor–suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 2017;7:54–71. [DOI] [PubMed] [Google Scholar]
- 16. Lo U-G, Chen Y-A, Cen J, Deng S, Luo J, Zhau H, et al. The driver role of JAK-STAT signalling in cancer stemness capabilities leading to new therapeutic strategies for therapy- and castration-resistant prostate cancer. Clin Transl Med 2022;12:e978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan JM, Zaidi S, Love JR, Zhao JL, Setty M, Wadosky KM, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 2022;377:1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 2017;171:1611–24.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sehrawat A, Gao L, Wang Y, Bankhead A 3rd, McWeeney SK, King CJ, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A 2018;115:E4179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Welti J, Sharp A, Yuan W, Dolling D, Nava Rodrigues D, Figueiredo I, et al. Targeting bromodomain and extra-terminal (BET) family proteins in castration-resistant prostate cancer (CRPC). Clin Cancer Res 2018;24:3149–62. [DOI] [PubMed] [Google Scholar]
- 23. Quintanal-Villalonga A, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol 2020;17:360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davies A, Nouruzi S, Ganguli D, Namekawa T, Thaper D, Linder S, et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat Cell Biol 2021;23:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, et al. EZH2 inhibition activates a dsRNA–STING–interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat Cancer 2021;2:444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cyrta J, Augspach A, De Filippo MR, Prandi D, Thienger P, Benelli M, et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat Commun 2020;11:5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang S. Tet2 at the interface between cancer and immunity. Commun Biol 2020;3:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia-Outeiral V, de la Parte C, Fidalgo M, Guallar D. The complexity of TET2 functions in pluripotency and development. Front Cell Dev Biol 2020;8:630754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim MR, Wu M-J, Zhang Y, Yang J-Y, Chang CJ. TET2 directs mammary luminal cell differentiation and endocrine response. Nat Commun 2020;11:4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011;8:200–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell Stem Cell 2018;23:355–69.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takayama K-I, Misawa A, Suzuki T, Takagi K, Hayashizaki Y, Fujimura T, et al. TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat Commun 2015;6:8219. [DOI] [PubMed] [Google Scholar]
- 34. Nickerson ML, Das S, Im KM, Turan S, Berndt SI, Li H, et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene 2017;36:2172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smeets E, Lynch AG, Prekovic S, Van den Broeck T, Moris L, Helsen C, et al. The role of TET-mediated DNA hydroxymethylation in prostate cancer. Mol Cell Endocrinol 2018;462:41–55. [DOI] [PubMed] [Google Scholar]
- 36. Sjostrom M, Zhao SG, Levy S, Zhang M, Ning Y, Shrestha R, et al. The 5-hydroxymethylcytosine landscape of prostate cancer. Cancer Res 2022;82:3888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y, et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 2018;554:123–7. [DOI] [PubMed] [Google Scholar]
- 38. MacArthur IC, Dawlaty MM. TET enzymes and 5-hydroxymethylcytosine in neural progenitor cell biology and neurodevelopment. Front Cell Dev Biol 2021;9:645335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bray JK, Dawlaty MM, Verma A, Maitra A. Roles and regulations of TET enzymes in solid tumors. Trends Cancer 2021;7:635–46. [DOI] [PubMed] [Google Scholar]
- 40. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 2016;30:733–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoyanova E, Riad M, Rao A, Heintz N. 5-Hydroxymethylcytosine-mediated active demethylation is required for mammalian neuronal differentiation and function. Elife 2021;10:e66973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qi J, Shi Y, Tan Y, Zhang Q, Zhang J, Wang J, et al. Regional gain and global loss of 5-hydroxymethylcytosine coexist in genitourinary cancers and regulate different oncogenic pathways. Clin Epigenetics 2022;14:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bailey SL, Chang SC, Griffiths B, Graham AN, Saffery R, Earle E, et al. ZNF397, a new class of interphase to early prophase-specific, SCAN-zinc-finger, mammalian centromere protein. Chromosoma 2008;117:367–80. [DOI] [PubMed] [Google Scholar]
- 44. Li X, Wang Y, Deng S, Zhu G, Wang C, Johnson NA, et al. Loss of SYNCRIP unleashes APOBEC-driven mutagenesis, tumor heterogeneity, and AR-targeted therapy resistance in prostate cancer. Cancer Cell 2023;41:1427–49.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alumkal JJ, Sun D, Lu E, Beer TM, Thomas GV, Latour E, et al. Transcriptional profiling identifies an androgen receptor activity-low, stemness program associated with enzalutamide resistance. Proc Natl Acad Sci U S A 2020;117:12315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodriguez Tirado C, Wang C, Li X, Deng S, Gonzalez J, Johnson NA, et al. UBE2J1 is the E2 ubiquitin-conjugating enzyme regulating androgen receptor degradation and antiandrogen resistance. Oncogene 2024;43:265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004;10:33–9. [DOI] [PubMed] [Google Scholar]
- 48. Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324:787–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jin H-J, Zhao JC, Wu L, Kim J, Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun 2014;5:3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006;10:321–30. [DOI] [PubMed] [Google Scholar]
- 51. Augello MA, Liu D, Deonarine LD, Robinson BD, Huang D, Stelloo S, et al. CHD1 loss alters AR binding at lineage-specific enhancers and modulates distinct transcriptional programs to drive prostate tumorigenesis. Cancer Cell 2019;35:817–9. [DOI] [PubMed] [Google Scholar]
- 52. Davies A, Zoubeidi A, Beltran H, Selth LA. The transcriptional and epigenetic landscape of cancer cell lineage plasticity. Cancer Discov 2023;13:1771–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong B, Miao J, Wang Y, Luo W, Ji Z, Lai H, et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun Biol 2020;3:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tang FY, Xu D, Wang SQ, Wong CK, Martinez-Fundichely A, Lee CJ, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 2022;376:eabe1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 56. den Hollander P, Joseph R, Vasaikar S, Kuburich NA, Deshmukh AP, Mani SA. Limiting dilution tumor initiation assay: an in vivo approach for the study of cancer stem cells. Methods Mol Biol 2022;2429:547–54. [DOI] [PubMed] [Google Scholar]
- 57. Basu A, Paul MK, Alioscha-Perez M, Grosberg A, Sahli H, Dubinett SM, et al. Statistical parametrization of cell cytoskeleton reveals lung cancer cytoskeletal phenotype with partial EMT signature. Commun Biol 2022;5:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gallardo A, Molina A, Asenjo HG, Lopez-Onieva L, Martorell-Marugan J, Espinosa-Martinez M, et al. EZH2 endorses cell plasticity to non-small cell lung cancer cells facilitating mesenchymal to epithelial transition and tumour colonization. Oncogene 2022;41:3611–24. [DOI] [PubMed] [Google Scholar]
- 59. He Y, Wei T, Ye Z, Orme JJ, Lin D, Sheng H, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun 2021;12:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gontier G, Iyer M, Shea JM, Bieri G, Wheatley EG, Ramalho-Santos M, et al. Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep 2018;22:1974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi J-H, et al. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol 2014;15:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song C-X, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 2011;29:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng Z-L, Zhang M-L, Lin H-P, Gao C, Song J-B, Zheng Z, et al. The Zscan4-Tet2 transcription nexus regulates metabolic rewiring and enhances proteostasis to promote reprogramming. Cell Rep 2020;32:107877. [DOI] [PubMed] [Google Scholar]
- 64. Sardina JL, Collombet S, Tian TV, Gomez A, Di Stefano B, Berenguer C, et al. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 2018;23:727–41.e9. [DOI] [PubMed] [Google Scholar]
- 65. Rasmussen KD, Berest I, Keβler S, Nishimura K, Simon-Carrasco L, Vassiliou GS, et al. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res 2019;29:564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cancer Genome Atlas Research Network . The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chua GNL, Wassarman KL, Sun H, Alp JA, Jarczyk EI, Kuzio NJ, et al. Cytosine-based TET enzyme inhibitors. ACS Med Chem Lett 2019;10:180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Parthasarathy A, Arnold A, Eberhart C, Raabe E. Abstract 5472: inhibiting the TET pathway in diffuse midline glioma reduces levels of 5-hydroxymethylcytosine and increases levels of 5-methylcytosine. Cancer Res 2022;82(Suppl 12):5472. [Google Scholar]
- 69. Lu Z, Liu R, Wang Y, Jiao M, Li Z, Wang Z, et al. Ten-eleven translocation-2 inactivation restrains IL-10-producing regulatory B cells to enable antitumor immunity in hepatocellular carcinoma. Hepatology 2023;77:745–59. [DOI] [PubMed] [Google Scholar]
- 70. Dusadeemeelap C, Rojasawasthien T, Matsubara T, Kokabu S, Addison WN. Inhibition of TET-mediated DNA demethylation suppresses osteoblast differentiation. FASEB J 2022;36:e22153. [DOI] [PubMed] [Google Scholar]
- 71. Pan W, Zhu S, Qu K, Meeth K, Cheng J, He K, et al. The DNA methylcytosine dioxygenase Tet2 sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression. Immunity 2017;47:284–97.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371:424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blatt EB, Raj GV. Molecular mechanisms of enzalutamide resistance in prostate cancer. Cancer Drug Resist 2019;2:189–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc 2012;7:1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155:1309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun 2016;7:10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep 2020;31:107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ostano P, Mello-Grand M, Sesia D, Gregnanin I, Peraldo-Neia C, Guana F, et al. Gene expression signature predictive of neuroendocrine transformation in prostate adenocarcinoma. Int J Mol Sci 2020;21:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Foroutan M, Bhuva DD, Lyu R, Horan K, Cursons J, Davis MJ. Single sample scoring of molecular phenotypes. BMC Bioinformatics 2018;19:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Significantly Altered AR ChIP-seq Peaks in ZNF397-KO Cells
Table S2. Differentially Expressed Genes in ZNF397-KO Cells
Table S3. Significantly Enriched or Depleted Signaling Pathways in GSEA Analysis
Table S4. Candidate Resistance Driver Genes in ZNF397-KO Cells
Table S5. Lineage-Specific and AR Target Gene Signatures
Table S6. Key Resources
Figure S1. ZNF397-deficiency has bifurcated effects on PCa with or without AR inhibition. Figure S2. The impact of ZNF397-deficiency on AR protein and AR cistrome. Figure S3. ZNF397 is required for the activation of canonical AR transcriptional program. Figure S4. ZNF397-KO impairs DHT-induced AR signaling activation. Figure S5. ZNF397-KO promotes multilineage and lineage plastic gene expression. Figure S6. ZNF397-KO promotes stem-like and EMT-like transcriptional programs. Figure S7. ZNF397-KO led to induction of TET2 downstream genes. Figure S8. ZNF397-KO promotes transcriptional activation of TET2 downstream genes. Figure S9. ZNF397-deficiency has bifurcated effects on AR-dependent vs AR-independent PCas. Figure S10. Targeting TET2-driven epigenetic rewiring to overcome resistance.
Data Availability Statement
-
•
All the described bulk RNA-seq data, ChIP-seq data, ATAC-seq, and 5hmC-seal seq have been deposited in the Gene Expression Omnibus (GEO, RRID: SCR_005012) under the accession numbers GSE230602. Accession numbers are also listed in the key resources table. Original Western blot images and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All analyses in this manuscript were performed using open-source software. Bulk RNA-seq analysis was done using the QBRC Bulk RNA-seq pipeline and the codes were deposited in GitHub (https://github.com/QBRC/QBRC_BulkRnaSeqDE). GSEA statistical analysis was carried out with the R package “fgsea” (v1.14.0). Customized codes used for ChIP-seq analysis and 5hmC-seal seq were deposited in github (https://github.com/zwang0715/Xu_et_al_ChIPseq). scRNA-seq data analysis was performed using established analysis pipeline. Detailed analysis approaches were described in “Quantification and Statistical Analysis” session.
-
•
Any additional information required to reanalyze the data reported in this paper (and requests for resources and reagents) is available from the lead contact, Dr. Ping Mu (Ping.Mu@UTSouthwestern.edu), upon request.
-
•
All cell lines, plasmids, and other reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement if there is potential for commercial application.