Abstract
Purpose
Bladder cancer (BLCA) is a prevalent malignancy. Dysregulated propionate metabolism, a key cancer factor, suggests a potential target for treating metastatic cancer. However, a complete understanding of the link between propionate metabolism-related genes (PMRGs) and bladder cancer is lacking.
Methods
From the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, we gathered BLCA patient data, which was classified into distinct subgroups using non-negative matrix factorization (NMF). Survival and pathway analyses were conducted between these clusters. The PMRGs model, created through univariate Cox and least absolute shrinkage and selection operator (LASSO) analyses, was assessed for prognostic significance using Kaplan–Meier and receiver operating characteristic (ROC) curves. A comprehensive evaluation included clinical, tumor microenvironment (TME), drug sensitivity, and immunotherapy analyses. Finally, the expression of HSD17B1 essential genes was confirmed via quantitative real-time polymerase chain reaction (qRT-PCR), with further validation through Transwell, wound healing, colony-formation, and EDU assays.
Results
We discovered two distinct subcategories (CA and CB) within BLCA using NMF analysis, with CA demonstrating significantly better overall survival compared to CB. Additionally, six PMRGs emerged as critical factors associated with propionate metabolism and prognosis. Kaplan–Meier analysis revealed that high-risk PMRGs were correlated with a poorer prognosis in BLCA patients. Moreover, significant differences were observed between the two groups in terms of infiltrated immune cells, immune checkpoint expression, TME scores, and drug sensitivity. Notably, we found that suppressing HSD17B1 gene expression inhibited the invasion of bladder cancer cells.
Conclusion
Our study proposes molecular subtypes and a PMRG-based score as promising prognostic indicators in BLCA. Additionally, cellular experiments underscore the pivotal role of HSD17B1 in bladder cancer metastasis and invasion, suggesting its potential as a novel therapeutic target.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-024-01982-6.
Keywords: Bladder cancer, Propionate, Prognosis, Risk signature, Target
Introduction
Bladder cancer (BLCA) comprises 3% of newly diagnosed cancer cases globally, standing as the eleventh most prevalent cancer type with an annual incidence of 573,000 new cases [1]. The predisposing factors for BLCA encompass genetic elements, environmental and work-related encounters, tobacco use, and conditions related to excessive body weight [2]. Non-muscle-invasive BLCA exhibits a favorable prognosis, boasting an approximate 90% 5-year overall survival (OS) rate. In contrast, metastatic BLCA is characterized by a markedly lower OS, falling below 6% [3]. Roughly half of individuals with clinically confined muscle-invasive BLCA encounter instances of disease recurrence and metastasis [4]. Hence, it is crucial to investigate biomarkers capable of accurately assessing patient prognosis.
The ability to metabolize propionate is a crucial aspect of a microorganism’s metabolic adaptability, enabling the utilization of carbon sources that generate propionyl-CoA in various environmental and host settings [5]. Recent findings indicate that propionate promotes lipid accumulation in adipocytes and demonstrates a significant correlation between serum propionate levels and human obesity [6]. Moreover, propionate exhibits antiapoptotic marker inhibition and induces proapoptotic proteins at both the gene and protein levels, suggesting its potential as a therapeutic option for cervical cancer [7]. Studies have uncovered that disrupting propionate metabolism contributes to a pro-aggressive profile in breast and lung cancer cells, enhancing their metastatic capabilities [8]. However, a research gap remains concerning propionate metabolism-related genes (PMRGs) in the context of BLCA onset and progression.
This study focuses on unraveling the role of PMRGs in BLCA pathogenesis. Leveraging comprehensive genomic datasets, such as those from TCGA and GEO, we aim to construct a robust PMRG gene model. This model promises to elucidate the intricate molecular landscape of BLCA and holds the potential to serve as a predictive tool and guide personalized therapeutic interventions for improved clinical outcomes.
Methods
Data collection
Obtain RNA-sequencing data and clinical annotations for 19 normal tissue specimens and 412 BLCA specimens from the TCGA database (https://portal.gdc.cancer.gov/). Concurrently, retrieve the gene expression dataset GSE13507 [9], consisting of 256 BLCA samples, from the GEO repository (https://www.ncbi.nlm.nih.gov/gds). Additionally, gather a set of 812 PMRGs (relevance score > 5) from the GeneCards database (https://www.genecards.org; Table S1).
Analysis of prognostic relevance and consensus clustering analysis
Initially, We used the ‘limma’ R package [10] to analyze PMRG differential expression in TCGA-BLCA and to identify significant genes. The merged TCGA and GEO datasets with survival data underwent univariate regression analysis using R packages “survival [11]” and “forest plot [12]” to identify prognostic genes. A circular plot visualized significant gene expression differences in normal and tumor tissues. Subsequently, we classified 662 tumor patients into subgroups based on 20 PMRGs using “ConsensusClusterPlus,” from Bioconductor [13] employing PCA, tSNE, and UAMP for accuracy. Kaplan–Meier curves were generated to compare survival outcomes among subtypes. Furthermore, immune-correlation analysis, GO analysis, and KEGG analysis were conducted. Immune-correlation analysis studies how various immune factors relate to each other, shedding light on their roles in diseases and treatment strategies.
Development of a prognostic signature utilizing PMRGs
Patients from TCGA and GEO databases were randomly split into control and validation groups (1:1 ratio). Lasso regression and multivariable analysis optimized the predictive risk model. The risk score for each sample was calculated using the following formula: coefficient X1 * expression X1 + coefficient X2 * expression X2 + + coefficient Xn * expression Xn. The risk score for each sample was calculated using the coefficients 1.005111879 of the regression model obtained and patients were categorized into low-risk and high-risk groups [14, 15]. Visualization depicted survival status, risk gene expressions, and scores across risk groups, with Kaplan–Meier curves illustrating OS rates.
Constructing and assessing a predictive nomogram
Through multivariate Cox regression analysis, we crafted a nomogram to depict the predictive factors succinctly. Internal validation ensued through calibration plots to verify the nomogram’s accuracy. Decision curve analysis (DCA) was employed to evaluate clinical net benefits [16]. Boxplots were utilized to visually represent the correlation between the scoring system and clinical baseline characteristics alongside pathological features.
Immune cell signature analyses
Initially, the CIBERSORT [17] algorithm within the R package CIBERSORT assessed the prevalence of 22 tumor-infiltrating immune cells in BLCA samples. Subsequently, we examined variations in immune cell infiltration between groups with high and low risk. Further analysis delved into visual representations of correlations among the 20 immune cells, the six hub genes, and risk scores. Additionally, we employed the ESTIMATE [18] method to calculate the stromal score, immune score, and overall ESTIMATE score.
Drug sensitivity analysis
We utilized the R package “pRRophetic [19]” to forecast the IC50 values for frequently prescribed drugs in BLCA, discerning variations between high- and low-score groups. This assessment aimed to gauge the distinct sensitivities to various chemotherapy drugs. Statistical significance was established at a p value below 0.05, visually depicted through box plots.
Tumor immune single cell hub (TISCH) database and validation of the HSD17B1
Access RNA-seq data for single cells from the TISCH database (http://tisch.comp-genomics.org), an online resource dedicated to the tumor microenvironment (TME) [20]. The TISCH database, employed in studying TME heterogeneity across diverse datasets and cell types, was a valuable resource. The KM plotter dataset was utilized to validate the predictive risk model. Perform a comprehensive pan-cancer analysis for HSD17B1 using the TIMER database (http://timer.cistrome.org) and evaluate its expression levels in both tumor and normal tissues (https://www.r-project.org/). Verify the protein expression patterns of prognostic genes by consulting the HPA dataset, accessible at https://www.proteinatlas.org/ [21].
Validation of HSD17B1 expression in BLCA samples
BCLA samples and adjacent tissues were procured from ten recently diagnosed BCLA patients at the First Affiliated Hospital of Nanchang University in Nanchang, China. Total RNA extraction was performed using Invitrogen TRIzol reagent, and cDNA synthesis was accomplished using the Takara PrimeScript RT reagent Kit. Real-time quantitative PCR was employed for gene expression analysis, employing SYBR Green from Roche, Switzerland, with β-actin as an endogenous reference. Each reaction underwent at least three replications. The primer sequences used were HSD17B1_F: CCTCTGTGCTGGACGTGAAT, HSD17B1_R: GCTGGCGCAATAAACGTCAT.
Cell culture and small interfering ribonucleic acid (siRNA) transfection
BLCA cell lines T24 and BIU were obtained from Procell Life Science & Technology Co., Ltd (Wuhan, China) and the cell bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), respectively. The cells were cultured in RPMI-DMEM or 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere with 5% CO2. Subsequently, cells were seeded into 6-well plates at a concentration of 40% to 60%. SiRNAs targeting HSD17B1 and corresponding controls were synthesized by GenePharma Co., Ltd. (Shanghai, China) and transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The specific sequences of the siRNAs were as follows: HSD17B1 siRNA-1 (5′-GCUUCAAAGUGUAUGCCACTT-3′) and HSD17B1 siRNA-2 (5′-GUGGGUGGCUAAUUAAGAUTT-3′).
Cell migration was assessed through wound-healing and transwell assays
T24 and BIU cells were seeded into 6-well plates and underwent siRNA transfection. Upon reaching a cell density exceeding 90%, a linear wound was created using a 200-μl pipette tip. Subsequently, the medium was substituted with a serum-free culture medium and placed in a cell culture incubator for incubation. Photographic documentation was conducted at 0 h and 24 h to compare scratch healing rates among various groups. In the Transwell assay, cells were suspended in serum-free DMEM or RPMI-1640 medium and seeded onto the upper surface of Transwell chambers, while the lower chamber was filled with medium supplemented with FBS. After a 24-h incubation period, cells attached to the lower membrane were immobilized using 4% paraformaldehyde and subsequently stained with crystal violet. Following this, optical microscope imaging was conducted to capture photographs of the cells.
Cell proliferation was evaluated via EDU and colony-formation assays
Approximately 1000 transfected cells were seeded in 6-well plates containing culture medium supplemented with 10% FBS and incubated for 7–10 days. Subsequently, the cells were treated with 4% paraformaldehyde solution and fixed for 20 min, after which they were stained with 1% crystal violet solution for 30 min. Additionally, transfected cells were plated at a density of 5000 cells per well in a 96-well plate. After 24 h of incubation, the culture medium was replaced with EdU for an additional 2 h, and staining was performed according to the instructions in the EDU assay kit (C10310-1, RiboBio). Imaging was conducted using a confocal microscope.
Statistical analysis
R software (Version 4.3.1, https://www.r-project.org/) and GraphPad Prism 8 were employed for data analysis in this study. Statistical significance was determined by two-sided p-values less than 0.05. Notably, all experiments were conducted thrice to ensure the reliability and consistency of the findings.
Results
Identification of differentially expressed PMRGs and molecular subtyping
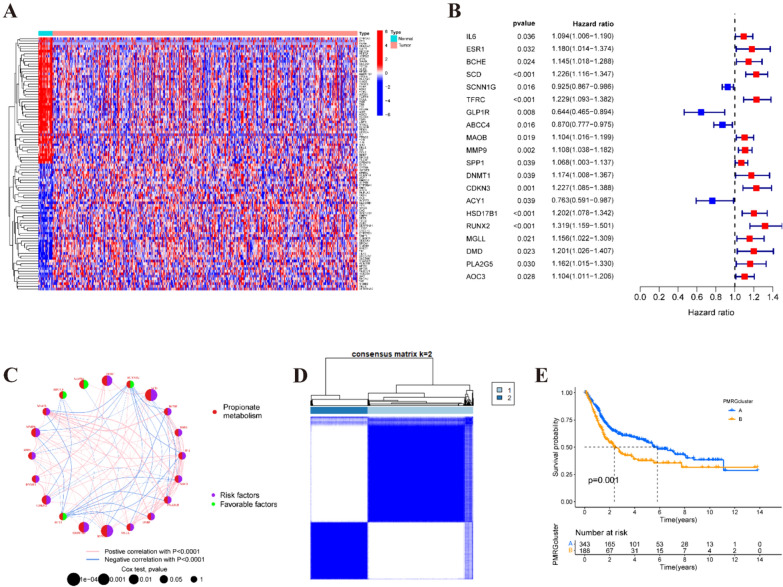
The entire protocol is delineated in Fig. S1. In the first step, a total of 812 PMRGs were retrieved from the GeneCard database (Table S1). Subsequently, RNA-seq data from the TCGA-BLCA dataset were retrieved, and a differential expression analysis was performed. Following integration with the GSE13507 dataset, 169 differentially expressed PMRGs were identified (Fig. 1A and Fig. S2A). Clinical information from 537 BLCA patients in the TCGA-BLCA database and the GSE13507 dataset was then integrated, and their survival data were utilized for univariate Cox regression analysis, resulting in the discovery of 20 PMRGs (Fig. 1B, C). To gain deeper insights into the functional role of PMRGs in BLCA, we employed the ‘Consensus Cluster Plus R program to assess the 20 PMRGs identified through univariate Cox analysis (Fig. 1D and Fig. S2B–G). Our analysis revealed the segregation of BLCA samples into two distinct subtypes when k = 2. Furthermore, survival analysis revealed a notable contrast in prognosis between these two subgroups (Fig. 1E). Notably, increased expression of the upper subset of PMRGs in cluster B was associated with a poorer prognosis in BLCA. Additionally, the efficacy of the non-negative matrix factorization method was validated through PCA, tSNE, and UMAP analyses (Fig. S2H–J).
Fig. 1.
Identification of differentially expressed PMRGs and molecular subtyping. A Heatmap of differentially expressed PMRGs. B Univariate Cox analysis demonstrated the correlation between propionate metabolism-associated genes and prognosis. C A circle plot illustrates the correlations among these PMRGs. D The consensus clustering in BLCA samples with k = 2. E Kaplan–Meier (KM) survival analysis of two subgroups
Characterizing the biological properties of PMRG subtypes
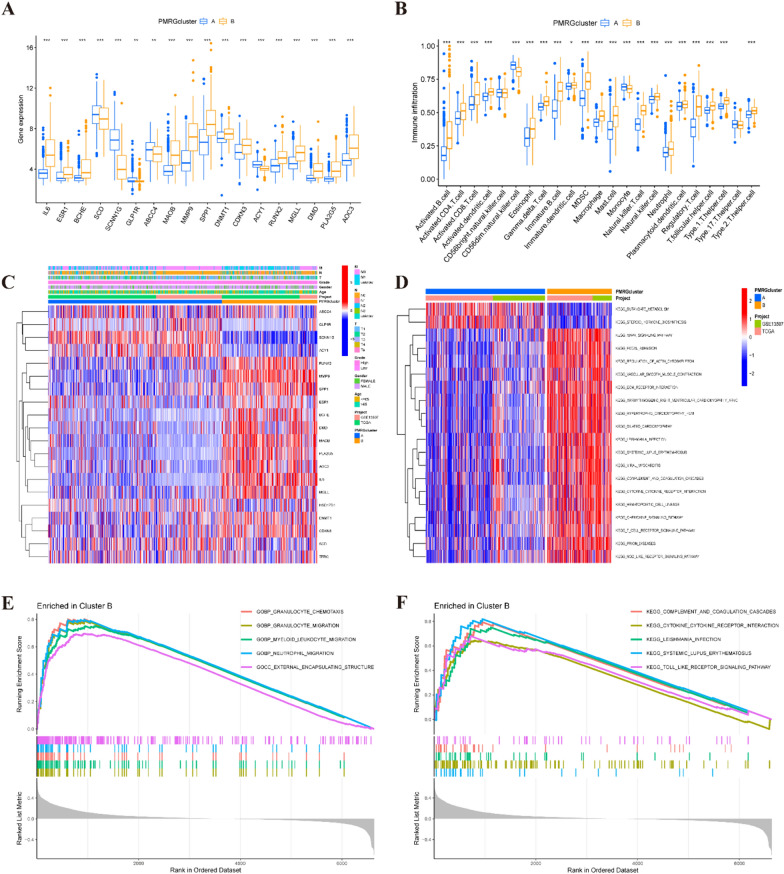
In our analysis, we identified 18 differentially expressed genes between clusters A and B of PMRGs (Fig. 2A). Additionally, we employed the CIBERSORT algorithm. We employed the CIBERSORT algorithm to estimate the levels of various cell infiltrations in BLCA samples, revealing significant distinctions in the proportions of immune cell and stromal cell infiltration ratios between clusters C1 and C2. Notably, cluster B exhibited higher levels of infiltration of activated B cells, CD4 and CD8 T cells, dendritic cells, eosinophils, macrophages, mast cells, natural killer T cells, neutrophils, regulatory T cells, and T helper cells, while cluster A showed abundance in CD56dim natural killer cells and monocytes (Fig. 2B). Furthermore, the patterns observed in PMRGs cluster B were associated with tumor stages and grades (Fig. 2C). Using gene set variation analysis (GSVA), we investigated the disparate enrichment of KEGG pathways within subgroups A and B. Notably, a robust correlation was observed between the adverse prognosis of cluster B and the heightened expression of associated pathways, including MAPK signaling, chemokine signaling, T cell receptor signaling, and NOD-like receptor signaling pathways (Fig. 2D). Additionally, gene set enrichment analysis (GSEA) revealed that enriched pathways such as granulocyte chemotaxis, external encapsulating structure, cytokine–cytokine receptor interaction, and toll-like receptor signaling pathway may also contribute to the poor prognosis of cluster B for BLCA patients (Fig. 2E, F).
Fig. 2.
The functional analysis of PMRGs. A The 18 differential expressed genes between the PMRGs cluster A and B. B Boxplots depicting the 23 immune signature ssGSEA scores of the PMRGs cluster A and B. C Difference distribution of clinicopathological features and ARGs expression among the two subtypes. D The gene set variation analysis (GSVA) of the differences in KEGG pathways within subgroups A and B. E, F GSEA between clusters A and B
Construction and validation of the PMRGs signature
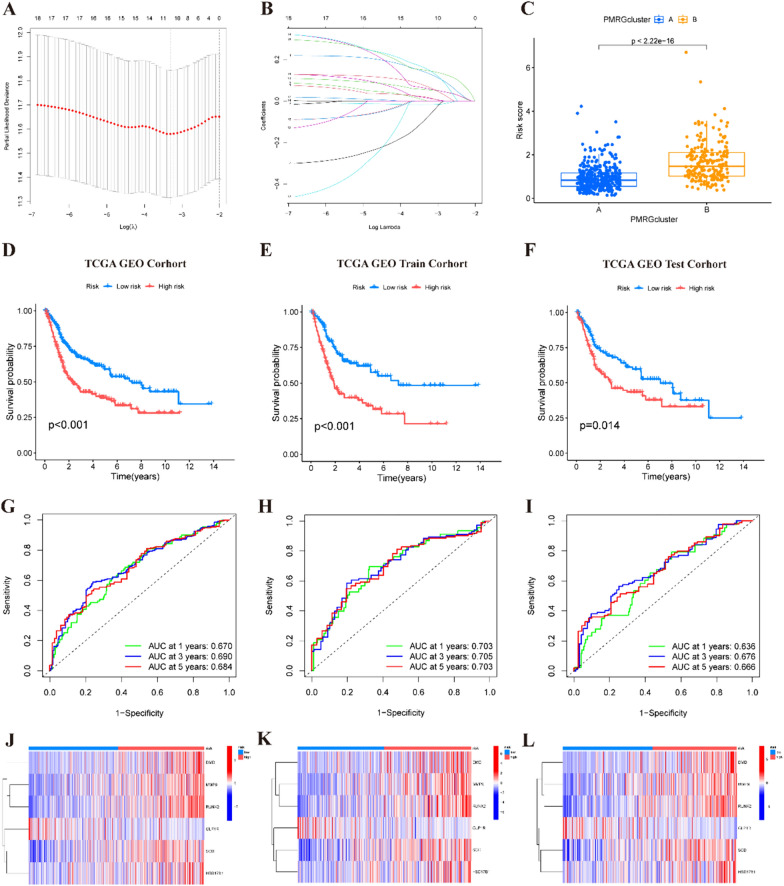
531 patients diagnosed with BLCA from the TCGA-BLCA dataset and GSE13507 were randomly divided into a training group (n = 266) and a validation group (n = 265). Through initial univariate Cox analysis followed by LASSO analysis, overfit genes were effectively eliminated, as depicted in Fig. 3A, B. Subsequently, a multivariate Cox analysis was conducted, resulting in the identification of a prognostic signature comprising six PMRGs: SCD, GLP1R, MMP9, HSD17B1, RUNX2, and DMD. Utilizing the derived formula: risk score = (0.226 × SCD expression) + (− 0.334 × GLP1R expression) + (0.091 × MMP9 expression) + (0.152 × HSD17B1 expression) + (0.219 × RUNX2 expression) + (0.322 × DMD expression), the risk scores for each patient were computed. As a result, the patients were grouped by the median risk score 1.005111879 into high- and low-risk categories. Cluster B exhibited higher risk scores than Cluster A (Fig. 3C), substantiating the poorer prognosis associated with Cluster B in BLCA patients. Kaplan–Meier survival analysis revealed a better prognosis for the low-risk group (Fig. 3D). To validate the robustness of these findings, the BLCA patients were further randomly divided into training and test cohorts. KM survival analysis on both cohorts consistently supported the earlier conclusions (Fig. 3E, F). Additionally, ROC curves for 1-, 3-, and 5-year OS yielded areas under the curve (AUC) of 0.670, 0.690, and 0.684, respectively (Fig. 3G). Notably, these results were corroborated in both the train and test cohorts (Fig. 3H, I), underscoring the predictive accuracy of the established prognostic signature across varying time frames. Moreover, the expression levels of PMRGs, including SCD, MMP9, HSD17B1, RUNX2, and DMD, were notably elevated in high-risk patients, indicative of their association with poorer prognosis. Conversely, GLP1R expression exhibited a protective effect as a prognostic predictor (Fig. 3J–L). In pursuit of precise survival risk predictions, we further investigated the efficacy of the risk signature in predicting BLCA progression. Our exploration into the relationship between the risk score and clinicopathological characteristics revealed significant distinctions among different groups concerning age, grade, and TNM stage (all p < 0.05). However, no discernible correlation was observed between the risk score and gender (p > 0.05; Fig. S3A–F). These findings collectively emphasize the effectiveness of the established prognostic signature in predicting BLCA patient survival across different time points, highlighting its clinical relevance and potential utility in guiding patient management strategies.
Fig. 3.
Establishment of a risk score signature based on PMRGs. A Plots of the coefficient profiles of six prognostic PMRGs. B The least absolute shrinkage and selection operator (LASSO) analysis identified six prognostic genes with tenfold cross-validation. C The risk score of clusters A and B. D–F The Kaplan–Meier survival curve of the OS rate in different subgroups of patients with BLCA. G–I 1-, 3-, and 5-year ROC curves of the all training and testing set. J–L The heat map of 6 PMRGs expression in each subgroup
Establishment of a nomogram
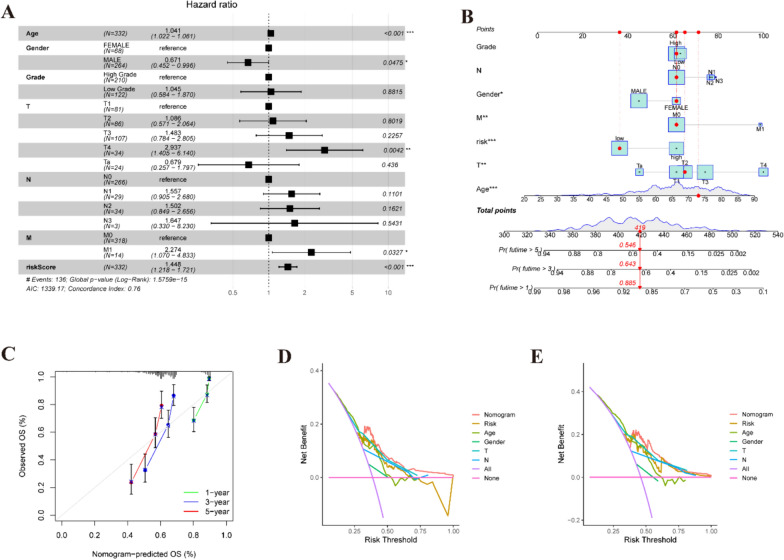
The outcomes of the multivariate Cox analysis underscored a significant correlation between the risk score derived from the propionate metabolism-related gene signature (PMRS) and prognosis, as illustrated in Fig. 4A, affirming PMRS as an independent prognostic determinant. To refine prognostic evaluation, we devised a novel nomogram amalgamating PMRS with clinical features, showcased in Fig. 4B, which demonstrated commendable predictive accuracy validated by calibration curves (Fig. 4C). Furthermore, DCA underscored the favorable prognostic performance of the nomogram across 3- and 5-year intervals (Fig. 4D, E). In summary, our findings accentuate the effectiveness of PMRS in precisely forecasting outcomes for BLCA patients, thereby furnishing crucial insights for clinical prognosis assessment.
Fig. 4.
An analysis of the predictive value of risk scores in BLCA patients from the TCGA and GEO datasets. A Assessment of clinical features and risk scores using a multivariate Cox analysis. B Predictive nomogram for survival over 1, 3, and 5 years based on risk groupings and clinical characteristics. ***P < 0.001. C Comparison of actual and predicted outcomes at 1, 3, and 5 years based on calibration curves. The DCA of risk scores and clinical features at 3 D and 5 E years
Immune landscape
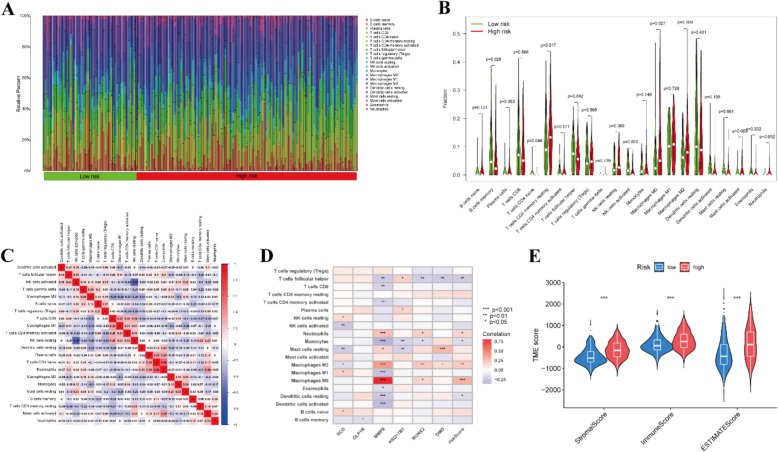
We conducted an evaluation and visualization of the proportions of 22 immune cell types in both high- and low-risk categorized patients, as depicted in Fig. 5A. Comparison of immune cell abundance revealed higher levels of T cells CD4 memory resting, T cells follicular helper, and macrophages in high-risk patients. In contrast, memory B cells and NK cells activated were more prevalent in the low-risk group (Fig. 5B). Furthermore, correlation analysis unveiled inverse relationships between most immune cell types (Fig. 5C). Spearman correlation analyses highlighted significant associations between 6 molecular entities and immune cell composition (Fig. 5D and Fig. S3G–L). Particularly noteworthy was the strong positive correlation between neutrophils, M0 macrophages, and M2 macrophages with most genes, while T cells follicular helper and monocytes showed an inverse correlation (p < 0.05). In addition, TME scores based on stromal score, immune score, and estimated score of the high-risk group surpassed those of the low-risk group (Fig. 5E). In summary, our risk model accurately predicts immune response activity in BLCA patients and provides valuable insights into the efficacy of immune checkpoint inhibition therapy.
Fig. 5.
Tumor microenvironment cell infiltration. A–C An illustration of the fractions of 22 types of infiltrating immune cells for low and high-risk patients. D A relation between infiltrating immune cells and six prognostic PMRGs, along with the risk score. E TME scores between the low and high-risk patients
Drug sensitivity
We explored differences in drug sensitivity between two BLCA patient groups based on PMRS scoring to improve treatment outcomes. As depicted in Fig. S4A–I, the IC50 values of cyclophosphamide, dasatinib, and staurosporine were lower in the high-scoring group compared to the low-scoring group, suggesting heightened sensitivity to these drugs in the former. Conversely, the IC50 values of afatinib, leflunomide, oxaliplatin, palbociclib, selumetinib, and trametinib were lower in the low-scoring group relative to the high-scoring group, indicating greater sensitivity to these drugs in the latter. This nuanced understanding of drug sensitivity profiles offers valuable insights for optimizing treatment strategies tailored to individual patient characteristics.
Single-cell analysis and validation of the HSD17B1 from external databases
Given the prognostic significance and distinctive distribution of HSD17B1 in BLCA tissues, we delved into its enrichment across specific cell types utilizing scRNA-seq data from the GSE145281_aPDL1 dataset. This analysis revealed 16 cell clusters and 5 cell types within BLCA tissues, as depicted in Fig. S5A–D, with corresponding marker genes delineated in Fig. S5E. Notably, HSD17B1 demonstrated significant enrichment in CD4Tconv, CD8T, macrophage, and NK cell populations (Fig. S5F). To validate the expression patterns of vital prognostic PMRGs, we leveraged the TISCH database to scrutinize HSD17B1 expression in both BLCA and pan-cancer contexts, as illustrated in Fig. S5G and I. Analysis using the KM plotter dataset unveiled an association between high HSD17B1 expression and poorer patient prognosis (Fig. S5H). Additionally, we corroborated these findings using the HPA database, which demonstrated elevated protein expression of HSD17B1 in BLCA compared to normal tissue, as showcased in Fig. S5J.
Utilizing HSD17B1 as the core gene of the scoring system and conducting in vitro validation
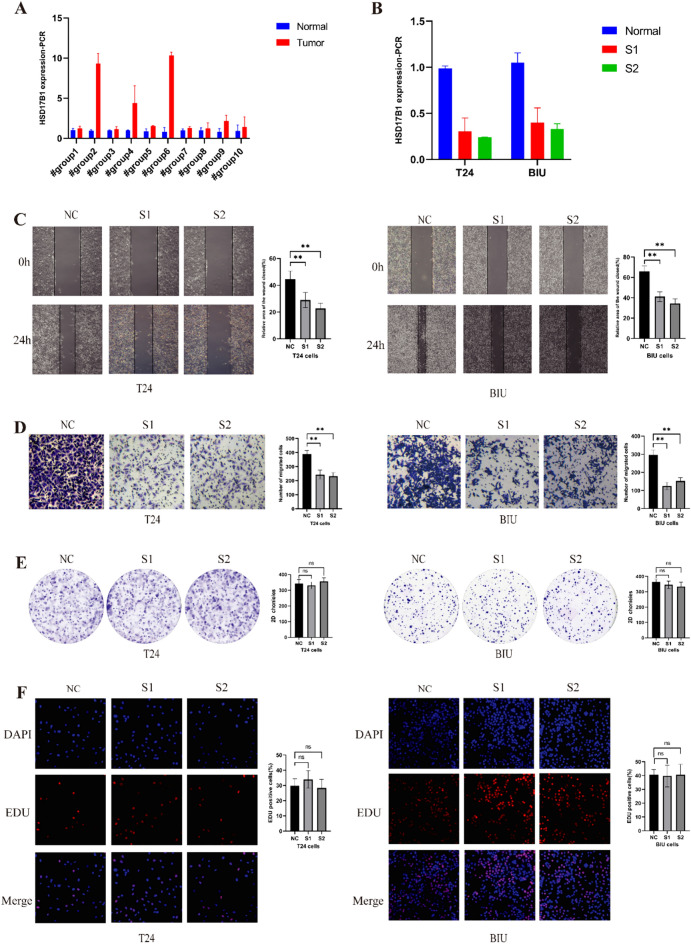
We conducted PCR analysis on BLCA patient tissues, confirming its pronounced overexpression in tumor tissues (Fig. 6A). To verify successful transfection of S1 and S2 in T24 and BIU cells, PCR assessments were performed (Fig. 6B). Results from Transwell and wound-healing assays compellingly demonstrate a significant reduction in the migratory capability of BLCA cells, directly attributed to suppressed HSD17B1 expression (Fig. 6C, D). Interestingly, the knockdown of HSD17B1 did not significantly impact the proliferation of T24 and BIU cells, as evidenced by both colony formation and EdU experiments (Fig. 6E, F). In summary, these findings unequivocally highlight the pivotal role of HSD17B1 in modulating the migratory behavior of BLCA cells.
Fig. 6.
Functional verification of knockdown HSD17B1. HSD17B1 expression in bladder cancer tissues was detected using qPCR, and the interference efficiency of small interfering RNA (siRNA) was also validated (A, B). Effect of HSD17B1 knockdown on the migration of bladder cancer cells (C, D). Knockdown of HSD17B1 was followed by colony (E) and EdU (F) experiments in T24 and BIU cells
Discussion
BLCA is a prevalent malignancy characterized by notable progression, significantly impacting patients’ quality of life [22]. The imperative for practical prognostic tools to steer treatment decisions and facilitate outcomes in BLCA patients cannot be overstated. Over time, various gene predictive models associated with BLCA have emerged, spanning those linked to basement inflammation [23], ferroptosis-related mechanisms [24], telomere maintenance [25], and dendritic cell activity [26]. These models have collectively enriched our understanding of BLCA and its therapeutic landscape. Despite these advancements, exploring propionate metabolism-related biomarkers in BLCA remains relatively uncharted territory. Notably, while predictive models related to propionate metabolism have been established for hepatocellular carcinoma [27], their application in BLCA has been sparse. Our study pioneers a novel initiative to address this gap by introducing a predictive model explicitly tailored to propionate metabolism-related risks in BLCA. Through this endeavor, we delineate essential biomarkers intricately linked to propionate metabolism, paving the way for enhanced prognostic assessments and personalized treatment strategies in BLCA patients.
In this study, we utilized an integrated approach combining both differential expression analysis and univariate Cox regression analysis to identify PMRGs that are closely linked to prognosis in BLCA. Subsequently, we stratified BLCA patients into two distinct subgroups, denoted as Cluster A and Cluster B, with Cluster A exhibiting a notably higher survival rate compared to Cluster B. Notably, nearly all identified PMRGs displayed significantly elevated expression levels in Cluster B. Interestingly, our investigation into immune cell infiltration using ssGSEA revealed significantly elevated levels of T cells and macrophages within Cluster B compared to Cluster A. Among these, CD8+ T cells are pivotal in immune surveillance against infections and cancer, while macrophages, the predominant myeloid cell type in cancer, are believed to exert protumor functions predominantly [28].
In this investigation, we conducted an extensive analysis of 812 PMRGs. This meticulous process involved carefully selecting genes exhibiting distinct expression patterns and significant prognostic relevance, culminating in developing a comprehensive risk signature. The finalized set of PMRGs, comprising SCD, GLP1R, MMP9, HSD17B1, RUNX2, and DMD, was the foundation for constructing the risk signature. Notably, SCD emerged as a pivotal enzyme in lipid metabolism, with its expression intricately linked to malignant transformation, tumor proliferation, and OS [29–31]. Furthermore, in a BLCA xenograft model, the inhibition of SCD resulted in substantial suppression of tumor progression, underscoring its potential as a therapeutic target in BLCA management [32]. The GLP1R gene encodes a seven-transmembrane protein, acting as a receptor for GLP1 [33]. According to Toru Shigeoka et al., upregulation of GLP1R may inhibit prostate carcinoma growth by restraining cell cycle progression [34]. Elevated MMP-9 levels are commonly observed in cancer tissues compared to normal adjacent tissues, promoting cancer cell migration and invasion [35, 36]. Studies suggest increased MMP-9 expression in mouse bladder tissues exposed to MC-LR may raise BLCA risk [37]. RUNX2, a member of the RUNX family, regulates crucial developmental processes, including differentiation, apoptosis, proliferation, and cell lineage specification [38]. Initially identified for its role in osteogenesis, RUNX2’s oncogenic functions have been linked to osteosarcoma progression [39]. In BLCA, downregulation of RUNX2 inhibits cell growth, promotes apoptosis, and reduces migration and invasion. These findings suggest that RUNX2 may serve as a prognostic biomarker and therapeutic target in BLCA [40]. The DMD gene, responsible for encoding the DMD protein, is notably abundant in humans [41]. This protein comprises two subunits, α and β, both of which have been associated with involvement in tumorigenesis [42]. HSD17B1 plays a critical role in various cancers. Research suggests it regulates active estrogen biosynthesis, promoting breast cancer cell proliferation [43]. In non-small cell lung cancer, increased HSD17B1 expression facilitates cancer progression [44]. Clinical studies indicate high HSD17B1 expression is associated with advanced pathologic N stage in BLCA, suggesting its potential as a prognostic biomarker [45]. As studies increasingly highlight the significance of HSD17B1 in various tumors, its exploration in BLCA remains limited. Consequently, we aimed to elucidate the specific role of HSD17B1 in BLCA. Our findings reveal upregulated mRNA expression of HSD17B1 in BLCA tissues, with knockdown experiments demonstrating a notable inhibition of BLCA cell migration and invasion.
Furthermore, we developed a nomogram that combines the risk score associated with propionate metabolism and additional clinical parameters, demonstrating excellent performance in prognostic predictions for BLCA patients. This novel risk signature offers an innovative approach to forecasting the survival outcomes of BLCA patients. Calibration curves across the entire dataset are closely aligned with the 45° angle, confirming the robust performance of the nomogram.
The TME plays a pivotal role in driving the proliferation and progression of cancer cells [46]. Our investigation revealed a significant disparity in the immune cell infiltration pattern within the TME, distinguishing between high-risk and low-risk BLCA patients based on the calculated risk score. Specifically, the high-risk group exhibited a notable increase in the abundance of CD4 memory resting T cells, follicular helper T cells, and macrophages compared to the low-risk group. It is evident that the gut microbiota and short-chain fatty acids such as propionate play crucial roles in shaping the generation of both effector and regulatory T cells through epigenetic and metabolic mechanisms [47]. Dysbiosis of the gut microbiota and subsequent propionate production have promoted cancer progression by inducing autophagy in cancer cells and M2 polarization in macrophages [48]. Our investigation identified significant disparities in the TME between high-risk and low-risk BLCA patients, as determined by the risk score. Previous studies have highlighted the predictive role of TME in immunotherapy outcomes, particularly in non-muscle-invasive BLCA [49]. Our findings underscore the importance of TME profiling in BLCA prognosis and treatment decision-making. Future research should focus on elucidating the intricate interplay between TME characteristics, PMRGs, and immunotherapy to optimize patient outcomes in BLCA. To enhance clinical outcomes, we analyzed drug sensitivity to identify different responses between high and low-risk groups. Our results suggest that high-risk individuals may respond well to conventional chemotherapy drugs like cyclophosphamide but may be resistant to leflunomide and oxaliplatin. This highlights the potential for combination chemotherapy to benefit high-risk BLCA patients, enabling more precise and personalized treatment approaches.
However, our study is not without limitations. Firstly, its reliance on public databases means the absence of clinical cohorts, hindering direct clinical validation of the model’s reliability. Secondly, while we observed the impact of HSD17B1 on BLCA cell proliferation and migration, we did not delve deeper into its regulatory mechanisms. Additionally, the retrospective and indirect nature of predicting immune therapy responses underscores the necessity for prospective trials involving larger patient cohorts to enhance the reliability of the scoring system.
Conclusion
Our study highlights the importance of propionate metabolism in BLCA and introduces a reliable predictive model for predicting patient outcomes and guiding immunotherapy and drug selection. Additionally, we identify HSD17B1 as a promising therapeutic target for BLCA treatment. These findings have meaningful implications and could improve BLCA patients’ treatment outcomes.
Supplementary Information
Supplementary Material 1: Fig. S1. Overview of study design.
Supplementary Material 2: Fig. S2. Establish risk model and nomogram for predicting survival probability in bladder cancer patients.The volcano plot indicates PMRGs.Consensus matrixes were obtained for k = 3, 4, and 5. When k = 2, the CDF curve has the lowest slope.PCA, tSNE, and UAMP identified two subtypes based on the expression of PMRGs.Sankey diagram of the interrelationship between two subtypes and high and low risks.The plot of CIR after risk stratification of the nomogram.
Supplementary Material 3: Fig. S3. Clinical pathological characteristics and immune cell infiltration in risk grouping.Correlation analysis between risk scores and clinicopathological features.The infiltrating levels of different immune cells in the high and low-risk groups.
Supplementary Material 4: Fig. S4.Patients’ response to nine common chemotherapeutic drugs in the low- and high-risk groups.
Supplementary Material 5: Fig. S5. Single-cell analysis and validation of the HSD17B1 from external databases.The cell type of GSE145281_aPDL1 dataset.The principal distributions of HSD17B1 on cell types in the GSE145281_aPDL1 dataset.Expression of HSD17B1 in BLCA patients in the TISCH database.Survival analysis for the subgroup classified by HSD17B1 mRNA expression.Expression of HSD17B1 in pan-cancer patients in the TISCH database.The expression of HSD17B1 between normal and tumor was analyzed by immunohistochemistryfrom the Human Protein Atlasdataset.
Acknowledgements
The authors express their gratitude for the collaborative efforts of the TCGA and GEO databases.
Author contributions
FZ: protocol, data collection, data analysis, manuscript writing; ZW: protocol, data collection, data analysis, manuscript writing; SL: protocol, data collection, data analysis; SX: protocol, manuscript writing; JZ: protocol, data collection; YT: data collection, data analysis; XL: protocol; HX: protocol, data collection; BF: protocol, data collection, data analysis, manuscript writing. All authors read and approved the final manuscript.
Funding
This study was supported by the Jiangxi Provincial “Double Thousand Plan” Fund Project (Grant No. jxsq2019201027), Jiangxi Provincial Natural Science Foundation for Distinguished Young Scholars (No. 20232ACB216014) and Jiangxi Provincial Natural Science Foundation (20232BAB206090).
Availability of data and materials
Data analyzed by the present study can be gained from TCGA (https://portal.gdc.cancer.gov/) databases (under accession: TCGA-BLCA) and GEO (http://www.ncbi.nlm.nih.gov/geo/) (under accession: GSE13507).
Declarations
Ethics approval and consent to participate
All organizations included in this study obtained ethical approval from the Ethics Committee of the First Affiliated Hospital of Nanchang University [Approval ID: (2022) CDYFYYLK (11-031)], and patient participation was contingent upon informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fuchun Zheng, Zhipeng Wang and Sheng Li contributed equally to the work and should be regarded as co-first authors.
Contributor Information
Yifan Tan, Email: yfantan@163.com.
Bin Fu, Email: urofubin@126.com.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Al-Zalabani AH, Stewart KFJ, Wesselius A, Schols AMWJ, Zeegers MP. Modifiable risk factors for the prevention of bladder cancer: a systematic review of meta-analyses. Eur J Epidemiol. 2016;31:811–51. 10.1007/s10654-016-0138-6. 10.1007/s10654-016-0138-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stecca C, Abdeljalil O, Sridhar SS. Metastatic urothelial cancer: a rapidly changing treatment landscape. Ther Adv Med Oncol. 2021;13:17588359211047352. 10.1177/17588359211047352. 10.1177/17588359211047352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tambaro R, Napoli MD, Pisano C, Cecere SC, Attademo L, Rossetti S, et al. From clinical trials to clinical use of checkpoint inhibitors for patients with metastatic urothelial cancer. Immunotherapy. 2021;13:67–77. 10.2217/imt-2020-0128. 10.2217/imt-2020-0128 [DOI] [PubMed] [Google Scholar]
- 5.Santos LPA, Assunção LDP, Lima PDS, Tristão GB, Brock M, Borges CL, et al. Propionate metabolism in a human pathogenic fungus: proteomic and biochemical analyses. IMA Fungus. 2020;11:9. 10.1186/s43008-020-00029-9. 10.1186/s43008-020-00029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia Z, Chen X, Chen J, Zhang L, Oprescu SN, Luo N, et al. ACSS3 in brown fat drives propionate catabolism and its deficiency leads to autophagy and systemic metabolic dysfunction. Clin Transl Med. 2022;12: e665. 10.1002/ctm2.665. 10.1002/ctm2.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham CH, Lee J-E, Yu J, Lee SH, Yu K-R, Hong J, et al. Anticancer effects of propionic acid inducing cell death in cervical cancer cells. Molecules. 2021;26:4951. 10.3390/molecules26164951. 10.3390/molecules26164951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes AP, Ilter D, Low V, Drapela S, Schild T, Mullarky E, et al. Altered propionate metabolism contributes to tumor progression and aggressiveness. Nat Metab. 2022;4:435. 10.1038/s42255-022-00553-5. 10.1038/s42255-022-00553-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim W-J, Kim E-J, Kim S-K, Kim Y-J, Ha Y-S, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. 10.1186/1476-4598-9-3. 10.1186/1476-4598-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43: e47. 10.1093/nar/gkv007. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Therneau TM, Grambsch PM. The cox model. In: Therneau TM, Grambsch PM, editors. Modeling survival data: extending the cox model. New York: Springer; 2000. p. 39–77. 10.1007/978-1-4757-3294-8_3. [Google Scholar]
- 12.Gordon: advanced forest plot using “grid” graphics... https://scholar.google.com/scholar_lookup?hl=en&publication_year=2020&author=M+Gordon&author=T+Lumley&title=Advanced+forest+plot+using+%E2%80%98grid%E2%80%99+graphics+%5BR+package+forestplot+version+1.10.1%5D. Accessed 7 May 2024.
- 13.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. 10.1093/bioinformatics/btq170. 10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Bai X, Wang X, Wang D, Ma G, Zhang F, et al. A novel fatty acid metabolism-associated risk model for prognosis prediction in acute myeloid leukaemia. Curr Oncol. 2023;30:2524–42. 10.3390/curroncol30020193. 10.3390/curroncol30020193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Fang X, Lu L, Liu G. Prognostic significance and immune landscape of a fatty acid metabolism-related gene signature in colon adenocarcinoma. Front Genet. 2022;13: 996625. 10.3389/fgene.2022.996625. 10.3389/fgene.2022.996625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. 10.1186/1472-6947-8-53. 10.1186/1472-6947-8-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. 10.1038/nmeth.3337. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9: e107468. 10.1371/journal.pone.0107468. 10.1371/journal.pone.0107468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun D, Wang J, Han Y, Dong X, Ge J, Zheng R, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–30. 10.1093/nar/gkaa1020. 10.1093/nar/gkaa1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13:2669. 10.1038/s41467-022-30342-3. 10.1038/s41467-022-30342-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–23. 10.3322/caac.21631. 10.3322/caac.21631 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Tang Y, Liu Z, Tan X, Zou Y, Luo S, et al. Identification of an inflammation-related risk signature for prognosis and immunotherapeutic response prediction in bladder cancer. Sci Rep. 2024;14:1216. 10.1038/s41598-024-51158-9. 10.1038/s41598-024-51158-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J, Hu J, Zhao L, Wu Z, Li R, Deng W. Identification of clinical prognostic factors and analysis of ferroptosis-related gene signatures in the bladder cancer immune microenvironment. BMC Urol. 2024;24:6. 10.1186/s12894-023-01354-y. 10.1186/s12894-023-01354-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Xu D, Jiang C, Huili Y, Nie S, Zhu H, et al. Telomere maintenance-related genes are important for survival prediction and subtype identification in bladder cancer. Front Genet. 2022;13:1087246. 10.3389/fgene.2022.1087246. 10.3389/fgene.2022.1087246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An B, Guo Z, Wang J, Zhang C, Zhang G, Yan L. Derivation and external validation of dendritic cell-related gene signatures for predicting prognosis and immunotherapy efficacy in bladder urothelial carcinoma. Front Immunol. 2022;13:1080947. 10.3389/fimmu.2022.1080947. 10.3389/fimmu.2022.1080947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao J, Wang J, Zhou C, Luo J. Development and validation of a propionate metabolism-related gene signature for prognostic prediction of hepatocellular carcinoma. J Hepatocell Carcinoma. 2023;10:1673–87. 10.2147/JHC.S420614. 10.2147/JHC.S420614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Romero P. Metabolic control of CD8+ T cell fate decisions and antitumor immunity. Trends Mol Med. 2018;24:30–48. 10.1016/j.molmed.2017.11.005. 10.1016/j.molmed.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 29.Anand U, Agarwal R, Anand CV. Pulmonary lipid peroxidation in cigarette smokers and lung cancer patients. Chest. 1992;101:290. 10.1378/chest.101.1.290a. 10.1378/chest.101.1.290a [DOI] [PubMed] [Google Scholar]
- 30.Piao C, Cui X, Zhan B, Li J, Li Z, Li Z, et al. Inhibition of stearoyl CoA desaturase-1 activity suppresses tumour progression and improves prognosis in human bladder cancer. J Cell Mol Med. 2019;23:2064–76. 10.1111/jcmm.14114. 10.1111/jcmm.14114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–80. 10.1158/1078-0432.CCR-12-3249. 10.1158/1078-0432.CCR-12-3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du X, Wang Q-R, Chan E, Merchant M, Liu J, French D, et al. FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res. 2012;72:5843–55. 10.1158/0008-5472.CAN-12-1329. 10.1158/0008-5472.CAN-12-1329 [DOI] [PubMed] [Google Scholar]
- 33.Jones B, Buenaventura T, Kanda N, Chabosseau P, Owen BM, Scott R, Goldin R, Angkathunyakul N, Corrêa IR Jr, Bosco D, Johnson PR. Targeting GLP-1 receptor trafficking to improve agonist efficacy. Nat Commun. 2018;9(1):1602. 10.1038/s41467-018-03941-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shigeoka T, Nomiyama T, Kawanami T, Hamaguchi Y, Horikawa T, Tanaka T, et al. Activation of overexpressed glucagon-like peptide-1 receptor attenuates prostate cancer growth by inhibiting cell cycle progression. J Diabetes Investig. 2020;11:1137–49. 10.1111/jdi.13247. 10.1111/jdi.13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gobin E, Bagwell K, Wagner J, Mysona D, Sandirasegarane S, Smith N, Bai S, Sharma A, Schleifer R, She JX. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer. 2019. 10.1186/s12885-019-5768-0. 10.1186/s12885-019-5768-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors. 2018;18(10):3249. 10.3390/s18103249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Wu W, Peng Y, Liu L, Zhang Y, Wang R, et al. Chronic exposure to microcystin-leucine-arginine induces epithelial hyperplasia and inflammation in the mouse bladder. Ecotoxicol Environ Saf. 2022;244: 114033. 10.1016/j.ecoenv.2022.114033. 10.1016/j.ecoenv.2022.114033 [DOI] [PubMed] [Google Scholar]
- 38.Ito Y, Bae S-C, Chuang LSH. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81–95. 10.1038/nrc3877. 10.1038/nrc3877 [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Zhao L, Tian W, Liao Z, Zheng H, Wang G, et al. Correlation of WWOX, RUNX2 and VEGFA protein expression in human osteosarcoma. BMC Med Genom. 2013;6:56. 10.1186/1755-8794-6-56. 10.1186/1755-8794-6-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Y, Huang S, Wang Z. Identify and validate RUNX2 and LAMA2 as novel prognostic signatures and correlate with immune infiltrates in bladder cancer. Front Oncol. 2023;13:1191398. 10.3389/fonc.2023.1191398. 10.3389/fonc.2023.1191398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–28. 10.1016/0092-8674(87)90579-4. 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- 42.Brennan PA, Jing J, Ethunandan M, Górecki D. Dystroglycan complex in cancer. Eur J Surg Oncol. 2004;30:589–92. 10.1016/j.ejso.2004.03.014. 10.1016/j.ejso.2004.03.014 [DOI] [PubMed] [Google Scholar]
- 43.Kristanti AN, Aminah NS, Siswanto I, Manuhara YSW, Abdjan MI, Wardana AP, et al. Anticancer potential of β-sitosterol and oleanolic acid as through inhibition of human estrogenic 17beta-hydroxysteroid dehydrogenase type-1 based on an in silico approach. RSC Adv. 2022;12:20319–29. 10.1039/d2ra03092f. 10.1039/d2ra03092f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drzewiecka H, Gałęcki B, Jarmołowska-Jurczyszyn D, Kluk A, Dyszkiewicz W, Jagodziński PP. Increased expression of 17-beta-hydroxysteroid dehydrogenase type 1 in non-small cell lung cancer. Lung Cancer. 2015;87:107–16. 10.1016/j.lungcan.2014.12.012. 10.1016/j.lungcan.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Xiao Y, Yu Q, Zhang C. HSD17B1 could serve as a prognostic biomarker for urinary bladder patients. Asian J Surg. 2023;46:5379–81. 10.1016/j.asjsur.2023.07.088. 10.1016/j.asjsur.2023.07.088 [DOI] [PubMed] [Google Scholar]
- 46.Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70–9. 10.4161/auto.26534. 10.4161/auto.26534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangan P, Mondino A. Microbial short-chain fatty acids: a strategy to tune adoptive T cell therapy. J Immunother Cancer. 2022;10: e004147. 10.1136/jitc-2021-004147. 10.1136/jitc-2021-004147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duan H, Wang L, Huangfu M, Li H. The impact of microbiota-derived short-chain fatty acids on macrophage activities in disease: mechanisms and therapeutic potentials. Biomed Pharmacother. 2023;165: 115276. 10.1016/j.biopha.2023.115276. 10.1016/j.biopha.2023.115276 [DOI] [PubMed] [Google Scholar]
- 49.Semeniuk-Wojtaś A, Poddębniak-Strama K, Modzelewska M, Baryła M, Dziąg-Dudek E, Syryło T, et al. Tumour microenvironment as a predictive factor for immunotherapy in non-muscle-invasive bladder cancer. Cancer Immunol Immunother. 2023;72:1971–89. 10.1007/s00262-023-03376-9. 10.1007/s00262-023-03376-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Fig. S1. Overview of study design.
Supplementary Material 2: Fig. S2. Establish risk model and nomogram for predicting survival probability in bladder cancer patients.The volcano plot indicates PMRGs.Consensus matrixes were obtained for k = 3, 4, and 5. When k = 2, the CDF curve has the lowest slope.PCA, tSNE, and UAMP identified two subtypes based on the expression of PMRGs.Sankey diagram of the interrelationship between two subtypes and high and low risks.The plot of CIR after risk stratification of the nomogram.
Supplementary Material 3: Fig. S3. Clinical pathological characteristics and immune cell infiltration in risk grouping.Correlation analysis between risk scores and clinicopathological features.The infiltrating levels of different immune cells in the high and low-risk groups.
Supplementary Material 4: Fig. S4.Patients’ response to nine common chemotherapeutic drugs in the low- and high-risk groups.
Supplementary Material 5: Fig. S5. Single-cell analysis and validation of the HSD17B1 from external databases.The cell type of GSE145281_aPDL1 dataset.The principal distributions of HSD17B1 on cell types in the GSE145281_aPDL1 dataset.Expression of HSD17B1 in BLCA patients in the TISCH database.Survival analysis for the subgroup classified by HSD17B1 mRNA expression.Expression of HSD17B1 in pan-cancer patients in the TISCH database.The expression of HSD17B1 between normal and tumor was analyzed by immunohistochemistryfrom the Human Protein Atlasdataset.
Data Availability Statement
Data analyzed by the present study can be gained from TCGA (https://portal.gdc.cancer.gov/) databases (under accession: TCGA-BLCA) and GEO (http://www.ncbi.nlm.nih.gov/geo/) (under accession: GSE13507).