Abstract
The major site of in vitro phosphorylation by casein kinase 2 (CK2) was the conserved Ser232 in the P proteins of human, bovine, and ovine strains of respiratory syncytial virus (RSV). Enzymatic removal of this phosphate group from the P protein instantly halted transcription elongation in vitro. Transcription reconstituted in the absence of P protein or in the presence of phosphate-free P protein produced abortive initiation products but no full-length transcripts. A recombinant P protein in which Ser232 was mutated to Asp exhibited about half of the transcriptional activity of the wild-type phosphorylated protein, suggesting that the negative charge of the phosphate groups is an important contributor to P protein function. Use of a temperature-sensitive CK2 mutant yeast revealed that in yeast, phosphorylation of recombinant P by non-CK2 kinase(s) occurs mainly at Ser215. In vitro, P protein could be phosphorylated by purified CK1 at Ser215 but this phosphorylation did not result in transcriptionally active P protein. A triple mutant P protein in which Ser215, Ser232, and Ser237 were all mutated to Ala was completely defective in phosphorylation in vitro as well as ex vivo. The xanthate compound D609 inhibited CK2 but not CK1 in vitro and had a very modest effect on P protein phosphorylation and RSV yield ex vivo. Together, these results suggest a role for CK2-mediated phosphorylation of the P protein in the promoter clearance and elongation properties of the viral RNA-dependent RNA polymerase.
Respiratory syncytial virus (RSV) is a major pediatric pathogen that claims roughly 3 million lives annually throughout the world (19). It is a Pneumovirus belonging to the Paramyxoviridae family and contains a nonsegmented negative-strand RNA genome a little over 15 kb in length (13). The genome is wrapped with the nucleocapsid protein N, and the resultant N-RNA complex serves as the functional template for the viral RNA-dependent RNA polymerase. In vitro (7, 30) and ex vivo reconstitution studies (17, 44) have shown that the minimal RNA polymerase of RSV consists of the large viral protein L and the accessory phosphoprotein P. Optimal transcription additionally requires viral antitermination protein M2 (14, 20), cellular actin (10, 22), and possibly additional cellular proteins (10).
Our laboratory has been investigating the role of phosphorylation of the RSV P protein in viral transcription over the last few years. Using a transcription system reconstituted with human RSV (HRSV, Long serotype) macromolecules in vitro, we have shown that phosphorylation of recombinant P protein by casein kinase 2 (CK2) occurs mainly at Ser232 and that this phosphorylation is essential for the transcription function of P (7). Phosphorylation at Ser237 may additionally stimulate transcription to a small extent, thus playing a modest accessory role (7, 30).
Although most of our knowledge of the molecular biology and biochemistry of RSV has originated from studies of the human strains, there are other strains of RSV that infect nonhuman animals and therefore cause significant damage to the livestock industry and farming. Such strains include bovine RSV (BRSV), ovine RSV (ORSV), and caprine (goat) RSV that readily infect their respective host animals (1, 28, 37, 40, 45). Rapid advancement in gene sequencing and cloning in the recent past has led to the finding that in spite of their natural host preferences, these viruses are identical in genome organization and the predicted primary structures of their proteins are highly similar. For example, the amino acid identities between the N, P, and L proteins of HRSV and BRSV are 93, 77, and 84%, respectively (28, 37, 45). Thus, the transcriptional proteins of these viruses may all have domains in common and, by corollary, the structure and function of their transcription complexes may emulate those of the human prototype. Recently, use of a BRSV ex vivo complementation system based on a minigenome construct has, in fact, shown that BRSV transcription requires the N, P, and L proteins and is stimulated by the BRSV M2 protein (45), a situation identical to that of HRSV (13, 14, 17). A complementation system for ORSV is not available. To determine the functional homology between the transcriptional macromolecules of HRSV and nonhuman RSV in detail, we have used the P protein as a model and have demonstrated the ability of the BRSV and ORSV P proteins to substitute for HRSV P in an otherwise HRSV-based transcription reaction. In addition, we have mapped the major phosphorylation site of the BRSV and ORSV P proteins to Ser232 and provided evidence that the protein kinase involved has properties of CK2.
Although recombinant HRSV P can be phosphorylated by purified and crude CK2 in vitro (7, 29, 30), the exact nature of the in vivo kinase for P remains elusive (7, 38). Drugs such as heparin, that inhibit CK2 in vitro, do not enter cultured cells (ex vivo). In an attempt to identify the intracellular kinase for P, we have therefore adopted two fundamentally different approaches. In the first, we expressed the P proteins and their CK2 site mutant forms in the available temperature-sensitive yeast CK2 mutant, which represents the only known CK2 mutation in any cell (12). In the second approach, we re-evaluated an antiviral xanthate compound, D609, which was previously shown to inhibit RSV P protein phosphorylation, as well as RSV growth in cell culture (42). During the course of these studies, we discovered that the RSV P protein is also an excellent substrate for purified CK1 in vitro and perhaps for a CK1-like activity in yeast. We then mapped the exact site of the CK1-mediated phosphorylation to Ser215 and demonstrated a lack of its role in the transcriptional activity of the P protein.
The precise role of the phosphate groups in the activation of the RSV P protein remains unknown. We now show that the RSV L protein alone cannot initiate transcription and that the unphosphorylated P protein can aid in this process. However, transcription in the presence of the unphosphorylated P protein results in the production of short, discrete oligonucleotide transcripts from within the leader region of RSV. Use of phosphorylated P resulted in promoter clearance of RSV RNA polymerase and production of full-length transcripts. In addition, we have recently shown that the phosphate groups of P protein are susceptible to hydrolysis by the prokaryotic protein phosphatase encoded by phage lambda (PPλ) in vitro (2, 4). We have now successfully used this phosphatase to demonstrate a role of the phosphate groups of RSV P in the elongation phase of transcription. The RSV phosphoprotein, therefore, acts as an elongation factor for the viral RNA-dependent RNA polymerase. Together, our results present a comprehensive picture of the different phosphate groups of the RSV P protein in vitro, as well as ex vivo, and an evaluation of their roles, or lack thereof, in viral transcription.
(This work was submitted in partial fulfillment of the requirements for a Ph.D. at the University of South Alabama by L. C. Dupuy.)
MATERIALS AND METHODS
Bacterial expression of BRSV and ORSV P proteins.
BRSV strain A51908 (28) was purchased from the American Type Culture Collection. ORSV strain WSU 83-1578 was a kind gift from Howard D. Lehmkuhl. The P-encoding genes of BRSV and ORSV were cloned in bacterial expression vector pET-3a by using the same procedures that were adopted for the human strain (29). Briefly, the P-encoding genes were amplified by reverse transcription (RT) and PCR using specific primers containing NdeI and BamHI sites and cloned into the same sites in pET-3a. The recombinant proteins were purified through phosphocellulose and DEAE-cellulose chromatography, during which the proteins behaved essentially like their human counterpart (29). Site-directed mutagenesis was also carried out as described previously (5).
Expression and 32P labeling of RSV P proteins in yeast.
The HRSV P-encoding gene and its mutant forms were first cloned into pET-3a essentially as previously described (29) and then subcloned between BamHI and AvrII sites of the inducible yeast vector YEP-PGAL1 (a kind gift from James Wang; 16). The wild-type and mutant YEP-PGAL1-RSVP constructs and the YEP-PGAL1 vector alone were each introduced into both wild-type yeast and temperature-sensitive CK2 mutant yeast (kind gifts of Claiborne V. C. Glover, University of Georgia; 12). The growth of the transformants at various temperatures, induction of P protein synthesis by galactose, metabolic labeling of cells with [32P]orthophosphate, lysis of cells, and immunoprecipitation of P protein were carried out essentially as previously described (12). CK1 and CK2 activities in yeast extracts were determined by using specific peptide substrates, RRREEESEEE for CK2 and KRRRALS(P)VASLPGL for CK1 in standard in vitro reactions (8, 24). Where mentioned, immunoblot (Western blot) analysis of P protein was performed by using anti-P antibody (29) and Ultra-signal chemiluminescence detection (Pierce, Rockford, Ill.).
Phosphorylation of recombinant P proteins by protein kinases (or HEp-2 cell extracts) and reconstituted RSV transcription reactions were carried out as described previously (7). The CK1 substrate peptide and recombinant CK1 and CK2 were purchased from New England Biolabs (Beverly, Mass.).
Reconstituted RSV transcription.
Transcription reactions based on unfractionated ribonucleoprotein were carried out essentially as described previously (3, 30). Reconstituted reactions (10 μl) were also performed as previously described (7, 30). In brief, 20-μl reaction mixtures contained 50 mM Tris-acetate (pH 7.5), 100 mM Na-acetate, 10% glycerol, 2 μg of actinomycin D per ml, 1 mM dithiothreitol; 120 ng of template N-RNA, 10 ng of L protein (both fractionated from ribonucleoprotein), 50 to 200 ng of purified recombinant (bacterial) P protein, 50 ng of HEp-2 extract containing actin, 50 μCi (50 μM) of [α-32P]UTP, and 500 μM each ATP, CTP, and GTP. The L preparation and the N-RNA complex each contributed about 2 to 4 ng of the M2 protein (22K protein), detectable by immunoblot analysis (data not shown). When phosphorylated P protein was used in a transcription, the bacterial P was phosphorylated by CK2 present in total HEp-2 cell extract (or purified CK1, where mentioned) and then purified away from kinases by phosphocellulose chromatography as previously described (7). HEp-2 extract, pretreated with excess anti-CK2 antibody, was used as the source of actin as previously described (7, 30). 32P-labeled transcripts were quantitated by the DEAE (DE81) paper binding described previously (3).
For some reactions (see Fig. 4), all four α-32P-labeled ribonucleoside triphosphates were used at a final concentration of 100 μM (50 μCi) each. To analyze the RNA products by gel electrophoresis, 100 μl of each transcription reaction mixture was deproteinized with phenol-chloroform, precipitated with ethanol, dried, and dissolved in 5 μl of 90% formamide. Half of the RNA was analyzed by electrophoresis on a 25% polyacrylamide gel containing 6 M urea to analyze small oligoribonuclotide products essentially as previously described (6). The other half of the RNA was electrophoresed on acidic agarose-urea gels as described previously (3). Following electrophoresis, both gels were dried and exposed to X-ray films.
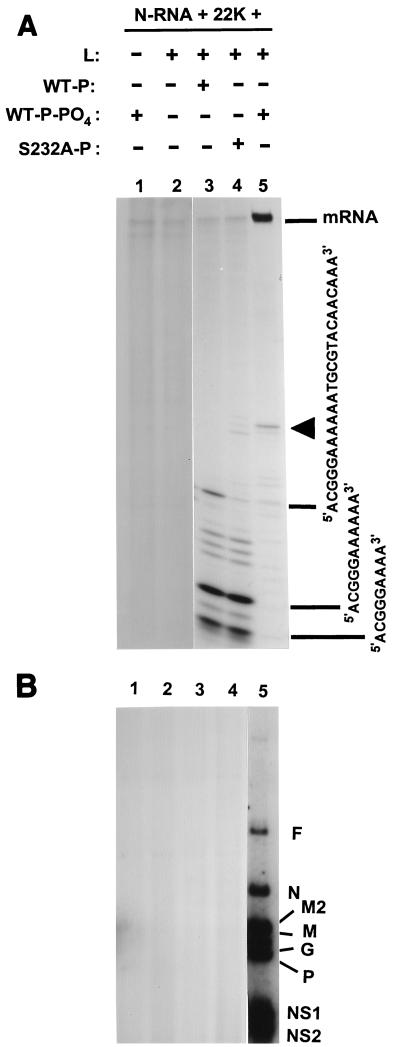
FIG. 4.
Role of phosphorylation of P protein in a postinitiation step of transcription. HRSV transcription was reconstituted essentially as described in Materials and Methods, using all four α-32P-labeled ribonucleoside triphosphates. The following P proteins were used: wild-type, phosphate-free recombinant P (in the presence of 5 μg of anti-CK2 antibody and 200 ng of PPλ per 160-μl reaction mixture) (WT-P; lane 3); wild-type, recombinant P prephosphorylated with total HEp-2 extract (WT-P-PO4; lanes 1 and 5); and Ser232Ala mutant recombinant P (S232A-P; lane 4). Following a 2-h transcription at 30°C, the 32P-labeled RNA was deproteinized and analyzed by electrophoresis on a 25% polyacrylamide gel (A) and an acid-agarose-urea gel (B) as described in Materials and Methods. Autoradiographs of the gels are shown. Each lane contains the equivalent of a 160-μl reaction mixture. In panel A, the arrowhead indicates the putative leader RNA of RSV; in this panel, the mRNAs did not enter the gel and remained in the loading slots, as indicated. The major labeled RNA bands, as shown, were excised from the gel and sequenced as described in Materials and Methods. Panel B shows the mRNAs for the various RSV genes (F, N, etc.), as indicated. Identical lane numbers in the two panels indicate identical reactions. In panel B, lanes 1 to 4 were slightly overexposed to detect any RNA band corresponding to lane 5 and none was found.
Effect of the xanthate compound D609.
To explore the effect of D609 (BIOMOL Research Laboratories, Plymouth Meeting, Pa.) on viral growth, HEp-2 cells grown in 10-cm-diameter dishes to 80% confluency were infected with RSV at a multiplicity of infection of 3. All of the media used in these experiments were brought to pH 7.0 by the addition of 1 M HCl (42). After 2 h of adsorption of the virus, the media were replaced with fresh, prewarmed media containing 0, 15, 30, 45, and 100 μM D609. To label P proteins, the media were removed 24 h later, the infected monolayers were washed twice with phosphate-free media, incubated with phosphate-free media for 2 h, and finally incubated with phosphate-free media plus [32P]orthophosphate (1 mCi per dish) for 4 h. The cells were then processed for immunoprecipitation by using antibodies against RSV P proteins as described previously (29). To determine viral titers, parallel cultures were grown without the 32P label and the supernatant titer was determined by serial dilution on CV-1 cells.
VSV (vesicular stomatitis virus) infection was carried out in an essentially identical manner, except that BHK-21 cells were infected, the infected cells were 32P labeled at 12 h postinfection (because of the faster growth rate of VSV), and the labeled P protein was immunoprecipitated with a polyclonal rabbit antibody against VSV.
To determine the effect of D609 on kinases, standard phosphorylation reactions containing RSV P protein as a substrate, [γ-32P]ATP as a phosphate donor, and purified kinases or total HEp-2 extract as a source of kinase were carried out (7, 29) in the presence of a range of D609 concentrations. Reaction mixtures lacking the substrate P protein but otherwise complete were incubated in ice for 5 min. Reactions were then initiated by addition of the P protein. Incubations were carried out at 37°C for 5 min and then terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer as previously described (29). In parallel reactions it was ensured that the reaction rate was linear at the time of termination. The CK1 inhibitor CK1-7 (Seikagaku America, Ijamsville, Md.) was also used in a similar manner at a final concentration of 40 μM (8).
Sequencing of RNA.
The details of the sequencing of the short transcripts produced in RSV transcription reactions in vitro (see Fig. 4) will be described elsewhere. In brief, the labeled RNAs were located by autoradiography and the bands were excised. RNA was eluted by overnight incubation in the presence of 1% SDS. A 2-μg sample of carrier tRNA was added to it, and the RNA was subjected to phenol-chloroform extraction, followed by precipitation with ethanol and ether. The triphosphate group at the 5′ end was removed by calf intestinal alkaline phosphatase as previously described (25); the 5′ end was then phosphorylated by using T4 polynucleotide kinase. The RNA was sequenced by two different methods. (i) In the ligation-RT-PCR method, the synthetic RNA, G8, was first ligated to the 5′ end of the transcript by using T4 RNA ligase. The 3′ end of the product was then ligated to the phosphorylated 5′ end of another synthetic RNA with the sequence GGU12 to generate the chimeric RNA with the final sequence G8-(RSV RNA)-GGU12. This chimeric RNA was then amplified by RT followed by PCR using avian myeloblastosis virus reverse transcriptase, Pfu polymerase, and primers G8 and A12CC (complementary to the two synthetic termini). The product was cloned into an SmaI-cut vector by blunt-end ligation and then sequenced by using primers in the vector regions flanking the insert. (ii) In the enzymatic sequencing method, corresponding RNAs from a nonradioactive transcription reaction were gel purified and used in enzymatic analysis essentially as previously described (25). The nucleotide sequence at the 3′ end was determined by ligation with 5′-[32P]pCp, followed by digestion with RNase T2. The nucleotide sequence at the 5′ end was determined by labeling of the 5′ end of the RNA with polynucleotide kinase and [γ-32P]ATP, followed by digestion with nuclease P1. For both the 5′ and 3′ nucleotides, the RNase-digested material was analyzed by two-dimensional thin-layer chromatography (25).
RESULTS
Ser232 is the major site of phosphorylation in P proteins of all strains of RSV.
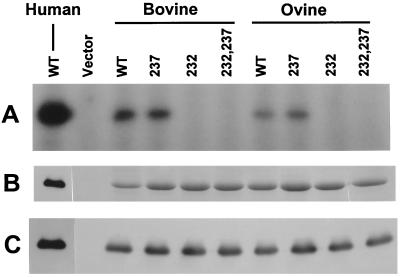
We have previously shown that Ser232 is the major site of phosphorylation of the P protein of HRSV ex vivo (expressed in HEp-2 cells) and in vitro by a cellular protein kinase activity that had the hallmarks of CK2 (7, 29, 38). Using an in vitro transcription assay, we also showed that the Ser232 phosphate is the primary determinant of the transcriptional activity of the P protein while the phosphate at Ser237 makes a much smaller contribution (7). Since this offers an important mechanism of regulation of P function, we wanted to test whether this phenomenon is unique to HRSV or operational in the nonhuman strains also. To this end, we expressed the P proteins of the BRSV and ORSV strains and their S232A, S237A, and S232,237A mutants in bacteria. It should be recalled that the P proteins of the HRSV, BRSV, and ORSV strains are highly similar and have 77% amino acid identity with one another in pairwise alignment (1, 28). Particularly similar are the C-terminal ends of all three, and this area includes Ser232 and Ser237 (7). We therefore expressed the wild-type and mutant P proteins of ORSV BRSV and tested them as substrates for phosphorylation by total HEp-2 cell extract in vitro. Results presented in Fig. 1 show that as in the case of HRSV P, mutation of Ser232 also abrogated phosphorylation in the BRSV and ORSV P proteins. The inhibition of this phosphorylation by heparin and anti-CK2 antibody confirmed that the protein kinase involved is identical to or resembles CK2 (data not shown). Thus, Ser232 appears to be the universal site of phosphorylation in the P proteins of all RSV strains.
FIG. 1.
In vitro phosphorylation of recombinant BRSV and ORSV P proteins. The following purified proteins were phosphorylated by using total HEp-2 cell extract as described in Materials and Methods (lane designations are in parentheses): wild-type HRSV (WT), used as a standard marker and a positive control; an equivalent fraction from a pET-3a vector-only strain (Vector); wild-type BRSV P (WT); S237A mutant BRSV P (237); S232A mutant BRSV P (232); S232, 237A double mutant BRSV P (232,237); wild-type ORSV P (WT); S237A mutant ORSV P (237); S232A mutant ORSV P (232); and S232, 237A double mutant ORSV P (232,237). The phosphorylation reactions were analyzed by SDS-polyacrylamide gel electrophoresis followed by staining (B) or autoradiography (A). The identity of the bands as P was confirmed by immunoblot analysis (C) using an antibody against HRSV P.
Essential role of CK2-mediated phosphorylation of Ser232 in BRSV and ORSV P protein transcriptional activity.
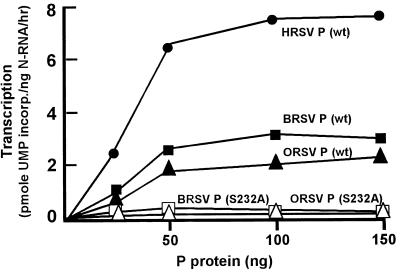
Due to the absence of a reconstituted in vitro transcription system for BRSV and ORSV, we were not able to test the role of Ser232 phosphorylation of their P proteins in homologous viral transcription. Nevertheless, since the nonhuman proteins were highly similar to the human protein, we reasoned that they might be able to replace human P in the reconstituted transcription reaction of HRSV. Thus, standard P protein-responsive HRSV transcription reactions containing template N-RNA, L protein, 22K protein, and uninfected HEp-2 cell extract were reconstituted in which human P was replaced with recombinant bovine or ovine P. Results in Fig. 2 show that at optimal concentrations, transcription achieved by the bovine and ovine P proteins was about 40 and 25% of that achieved by the human counterpart. It is therefore obvious that these two heterologous P proteins are capable of functionally interacting with the transcriptional macromolecules of HRSV.
FIG. 2.
Transcriptional activity of BRSV and ORSV P proteins. Recombinant P proteins of BRSV and ORSV were expressed in Escherichia coli and purified as described in Materials and Methods. Transcription reactions (10 μl) were reconstituted with the HRSV N-RNA template, HRSV L protein, HEp-2 cell extract (as a source of actin and CK2), and various amounts (25, 50, 100, and 150 ng) of recombinant wild-type (wt) and mutant P proteins, as indicated. incorp., incorporated.
The ability of the bovine and ovine P proteins to activate transcription in the human system in effect provided us with an assay for these P proteins. By using this assay, we then tested the effect of the S232A mutation in these proteins. As shown in Fig. 2, just as in human P (7), mutation of Ser232 to Ala in the ovine and bovine P proteins destroyed their transcription activation property. Taken together, results of Fig. 1 and 2 and earlier results obtained with human P (7, 38) prove a critical role of CK2-mediated phosphorylation of Ser232 in all RSV P proteins.
Ser232 phosphates of P protein are essential for RSV transcription elongation.
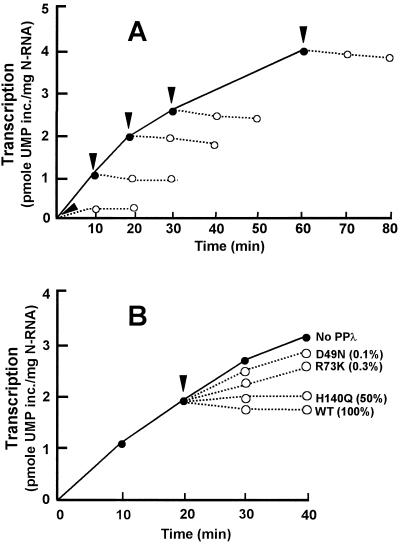
The P protein is the only protein whose phosphorylation is known to be important in RSV transcription and is sensitive to lambda phosphatase (PPλ) (2, 7, 29, 30). There is no evidence that the L or N proteins of RSV are phosphorylated. We have recently shown that actin and its accessory proteins are also required for RSV transcription (10). Although the phosphorylation status of actin is uncertain, pretreatment of either actin or the accessory factor(s) with PPλ did not destroy its transcriptional activity (10). The susceptibility of the HRSV P phosphates to PPλ therefore provided us with a tool to remove the phosphates of P midway in transcription and determine the effect of their removal on transcription. As shown in Fig. 3A, addition of purified PPλ to an RSV transcription reaction completely inhibited transcription regardless of the time at which it was added. The bulk of the transcription measured as shown in Fig. 3 was transcription elongation, based on the following reasoning. (i) The transcripts were labeled by using α-32P-labeled UTP (Materials and Methods), and there is no known RSV transcript that starts with a U. In fact, U is extremely rare within the first 15 nucleotides of all RSV transcripts (for example, see Fig. 4A), A and G being predominant in this region. (ii) The DEAE-cellulose paper (DE81) that we used to measure RNA synthesis (Fig. 3) efficiently binds RNA molecules greater than 15 to 20-nucleotides long. Shorter RNA molecules bind much less efficiently and tend to get dissociated under our washing conditions (3; data not shown). (iii) In standard reconstituted RSV transcription reaction mixtures, full-length transcripts could be detected as early as 5 min (data not shown). In other words, a drastic shutdown of total UMP incorporation by PPλ, such as in Fig. 3A, would not be possible without cessation of RNA elongation. Thus, we conclude that phosphorylation of P is essential for the RNA elongation step of RSV transcription.
FIG. 3.
Effects of wild-type (A) and mutant (B) PPλ on RSV transcription. In vitro transcription reactions (160 μl) were reconstituted as described in Materials and Methods by using wild-type (WT) recombinant P phosphorylated by total HEp-2 extract (●, solid line). At 0, 10, 20, 30, and 60 min after the start of transcription (arrowheads), a 30-μl reaction volume was taken out in a fresh tube to which 40 ng of PPλ (1 μl) was added and incubation was continued. At 0, 10, and 20 min after the addition of PPλ (○, dotted line), a 10-μl reaction volume was spotted onto DE81 paper. The paper was then processed for quantitation of labeled transcripts as described earlier (3). The various mutant forms of PPλ were similarly added at 20 min of transcription, and the reaction was monitored for another 20 min (B). The residual phosphatase activity of each mutant form of PPλ is shown as a percentage of that of the wild type (2). inc., incorporated.
As mentioned above, due to the unreliable binding of small RNA products to DEAE-paper, measurement of total counts (Fig. 3) could not answer whether PPλ additionally inhibited the initiation of RSV transcription. To address this question, therefore, we directly examined the RNA initiation products as described later (see Fig. 4).
It should be mentioned that the amount of PPλ used in these experiments was known to be large enough to achieve nearly instantaneous and complete dephosphorylation of the amount of P protein present in the reaction mixture (2; data not shown). To validate that the transcription inhibitory effect of PPλ is truly due to its phosphatase activity and not due to any other inhibitory activity present in the preparation, we took advantage of the catalytically defective mutant forms of PPλ that we had constructed earlier through site-directed mutagenesis. The mutations altered critical amino acid residues (His140, Arg73, and Asp49) that are invariant in all Ser/Thr phosphatases and play essential roles in the catalytic mechanism (2). Three such mutant enzymes (and their phosphatase activities relative to the wild-type enzyme) were H140Q (50%), R73K (0.3%), and D49N (0.1%). These mutant enzymes were expressed in bacteria and purified in the same manner as the wild type. When these mutant enzymes were added to RSV transcription reaction mixtures, their inhibitory effects were roughly proportional to their residual phosphatase activity (Fig. 3B). Thus, the H140Q mutant was essentially as inhibitory as wild-type PPλ while the R73K mutant was pronouncedly less inhibitory and D49N was almost without effect. These results confirm that the inhibitory effect of PPλ on RSV transcription is truly due to its phosphatase activity and not due to any nonspecific inhibitor present in the preparation.
The negative charge of the phosphate group at Ser232 contributes to P function.
The exact mechanism by which the phosphate groups activate the elongation function of the P protein is not known. Since a predominant feature of a phosphate group is its highly negative charge, we tested the possibility that a negatively charged amino acid, viz., Asp, may be able to functionally substitute for Ser at position 232 to produce a mutant P protein that will be constitutively active without CK2. Ser232 was therefore mutated to Asp by site-directed mutagenesis, and the mutant was expressed in bacteria, purified, and employed in a reconstituted RSV transcription reaction. As shown in Table 1, the S232D mutant exhibited about 40% of wild-type transcription activity (line 3) that was not affected by the addition of either CK2 (line 4) or PPλ (line 5) to the reaction, suggesting that at least part of the transcriptional activity of the Ser232 phosphate group is due to its negative charge. At the same time, these results suggest that the phosphate group itself, and not just its net negative charge, makes a large contribution to the total transcriptional activity of P.
TABLE 1.
Transcriptional activities of RSV P proteinsa
| P protein + treatment | Transcriptional activity (nmol of UMP incorporated/mg of N-RNA/h) |
|---|---|
| Wild type + CK2 | 5.2 |
| Wild type (CK2) + PPλ | 0.3 |
| Ser232Asp | 2.1 |
| Ser232Asp + CK2 | 2.2 |
| Ser232Asp + PPλ | 1.9 |
| Wild type + CK1 | 0.2 |
| Ser215Ala | 5.0 |
RSV transcription reaction mixtures (10 μl) were reconstituted by using 200 ng of the indicated (wild-type or mutant) recombinant HRSV P proteins as described in Materials and Methods. Where indicated, the P protein was prephosphorylated by CK2 (of HEp-2 extract) or by purified CK1. PPλ indicates the presence of 40 ng of PPλ per 20 μl of the transcription reaction mixture (as in Fig. 4A).
Phosphate-free P protein can promote transcription initiation, but phosphorylation is required for promoter clearance.
Since phosphorylation of P was found to be important for transcription elongation, we sought to determine whether it is also important for initiation through direct analysis of the initiated transcripts. RSV transcription was reconstituted under two conditions, both of which would eliminate CK2-mediated phosphorylation of the P protein, i.e., by using S232A mutant P or by using wild-type bacterial (phosphate-free) P in the presence of anti-CK2 antibody and PPλ. The anti-CK2 antibody and PPλ were included to rule out any phosphorylation of the wild-type P protein during transcription by the HEp-2 extract that was needed to provide actin, an essential cellular factor required for RSV transcription. Results are shown in Fig. 4, where panel A depicts the short initiation products resolved on a 24% polyacrylamide gel containing 6 M urea and panel B depicts full-length mRNAs resolved on an acidified agarose-urea gel. Reactions devoid of either L or P produced no detectable transcript, suggesting that the P and L proteins alone are incapable of any aspect of transcription (lanes 1 and 2 in panels A and B). When unphosphorylated P protein was added together with L, however, a heterogeneous population of oligoribonucleotides was produced that represented abortive initiation products (panel A, lane 3). No full-length transcripts were still detectable (panel B, lane 3). An essentially identical RNA profile was obtained by using the S232A mutant P protein, which is nonphosphorylatable by CK2 (lane 4). When the sequences of the three major oligoribonucleotide products from lane 3 were determined, they were found to have identical 5′ termini complementary to the 3′ terminus of the negative-sense genomic RNA (13). The largest species was 24 nucleotides long and had the sequence 5′ACGGGAAAAAATGCGTACAACAAA3′; the two smaller transcripts (9 and 11 nucleotides long) also had the same sequence and were only shorter at the 3′ end. All transcripts were devoid of a 5′ cap and a poly(A) tail (data not shown).
Standard transcription reactions employing phosphorylated P protein (panel B, lane 5) produced mostly larger mRNAs, confirming our previous results (30), but also produced very small amounts of apparently similar short transcripts, which further supported our assertion that the short transcripts may be bona fide intermediates of normal transcription and not degradation products. This is consistent with a model of RSV transcription in which the negative-strand genomic RNA encodes a single promoter at its 3′ end, from which the viral RNA polymerase initiates transcription. Additionally, these results demonstrated that the viral RNA polymerase made up of nonphosphorylated P protein is initiation proficient but does not travel beyond about 24 nucleotides of the leader region, suggesting a possible defect in promoter clearance.
To reiterate, the results in Fig. 3 and 4 strongly suggest that phosphorylation of P is required for the elongation activity of the RNA polymerase, including promoter clearance.
Xanthate compound D609 inhibits CK2 and P phosphorylation in vitro.
D609 has been shown to inhibit the growth of a number of viruses, including VSV and RSV, in cell culture (33, 42). Subsequently, it was shown to inhibit the intracellular phosphorylation of RSV P protein and synthesis of RSV N protein; however, replication of viral genomic RNA was not affected. Although the exact target of D609 remained undefined, based on the foregoing, it was logical to test whether D609 inhibits P protein phosphorylation by CK2 in vitro. As shown in Fig. 5, D609 inhibited the phosphorylation of recombinant RSV P by purified CK2 with a 50% inhibitory concentration of about 30 μM. Similar inhibition of phosphorylation was also seen when the total HEp-2 extract was used as the source of CK2 (data not shown). However, in contrast to a previous study in which 30 μM D609 inhibited intracellular phosphorylation of RSV P or growth of RSV in cell culture (ex vivo), we failed to obtain appreciable inhibition of either of these (Fig. 5). We do not have any explanation for this apparent discrepancy of the ex vivo activity of D609. It is possible that minor differences in cell culture conditions or medium composition have large effects on the potency or cellular uptake of D609. In apparent contrast, under the same conditions (45 μM D609), growth of VSV (on BHK-21 cells) and intracellular phosphorylation of VSV P protein were inhibited by 60 and 95%, respectively (33; data not shown). Thus, we presume that inhibition of RSV growth by D609 is only observed under conditions in which RSV P protein phosphorylation is also inhibited, indicating a possible relationship between the two (42). Since we failed to reproduce ex vivo inhibition of either process, we could not study its mechanism further.
FIG. 5.
Effect of D609 on RSV growth and P protein phosphorylation. The various concentrations of D609 used in vitro and ex vivo are indicated. The effect of D609 on CK2 activity in vitro (○) was tested by using recombinant RSV P as a substrate and total HEp-2 extract as the source of CK2 in a standard kinase reaction; use of purified CK2 produced essentially similar results. Ex vivo effects of D609 were determined by monitoring progeny viral titer (▵) and metabolic labeling of RSV P protein (●) by 32Pi. All data are presented as percentages of values obtained with samples not treated with D609.
RSV P can be a substrate for CK1.
Our investigations of a non-CK2 kinase for RSV P protein were based on two lines of evidence obtained earlier. First, when total HEp-2 extract was the source of the kinase, about 5% of the P protein phosphorylation remained resistant to D609 (data not shown). Second, P protein in which the two CK2 sites—Ser232 and Ser237—were both mutated still contained about 5% of the wild-type phosphates ex vivo (7, 38). To identify the kinases involved in this residual phosphorylation, we tested two multipotent protein kinases, viz., CK1 and protein kinase A, for the ability to phosphorylate purified recombinant RSV P in vitro. CK1 was found to phosphorylate P protein efficiently, whereas protein kinase A did not (data not shown). The stoichiometry of CK1-mediated phosphorylation was found to be about 0.5, suggesting the existence of at least one phosphorylation site in the polypeptide.
In an attempt to understand the significance of CK1-mediated phosphorylation, we first decided to map the site of this phosphorylation (data not shown). Use of a variety of deletion and substitution mutant forms of recombinant RSV P protein in vitro eventually led to the identification of Ser215 as the single site for CK1-mediated phosphorylation. The S215A mutant RSV P protein was completely devoid of phosphorylation by purified CK1 in vitro. Studies of a number of natural and synthetic substrates have shown that the consensus phosphorylation site for CK1 is a Ser or Thr at position n preceded by an acidic amino acid or a phosphorylated Ser/Thr at n − 3 (24). Ser215, belonging to the sequence DEVS215, indeed conformed to the CK1 motif very well.
Relative contributions of CK2 and CK1 to the intracellular phosphorylation states of RSV P protein.
To investigate whether CK1 may also contribute to the phosphorylation of P protein ex vivo, we undertook a series of studies, the results of which will be summarized here without any data shown. First, we analyzed the phosphorylation status of recombinant mutant P proteins expressed in human cells (ex vivo) by metabolic labeling with 32P followed by immunoprecipitation. The S232,237A double mutant was found to contain about 5 to 8% of the wild-type phosphates, confirming earlier findings (7, 38). This residual phosphorylation was found to occur at Ser215, since the 32P content of the S215,232,237A triple mutant was undetectable. Essentially similar results were obtained in vitro by using bacterially expressed recombinant P proteins as substrates for phosphorylation by a total HEp-2 cell extract. Phosphorylation of the S232,237A double mutant was also completely inhibited by including a CK1 inhibitor, CK1-7 (8), in the in vitro reaction mixture, further corroborating the finding that the small amounts (5 to 8%) of residual phosphorylation were indeed due to a CK1-like activity in the HEp-2 cells.
In our second approach, we investigated the phosphorylation status of P following abrogation of CK2. Although CK2-mediated phosphorylation of RSV P can be inhibited by chemical inhibitors such as heparin in vitro (7, 29), these inhibitors either do not enter cells or are not completely specific for CK2, which has precluded studies of intracellular inhibition of CK2 in HEp-2 cells. Thus, we took advantage of the only conditional lethal CK2 mutant of Saccharomyces cerevisiae (yeast) available, which has a temperature-sensitive growth phenotype (12). It has previously been shown that at the nonpermissive temperature (37°C), the CK2 in this mutant is completely inactivated (12). We expressed the wild-type P and its various Ser→Ala mutant forms in the CK2 mutant yeast and in the otherwise isogenic wild-type yeast strain at different temperatures and analyzed their phosphorylation status as described in Materials and Methods. The wild-type and the S232A and S237A mutant P proteins were equally phosphorylated in both yeast strains and at both nonpermissive (37°C) and permissive (25°C) temperatures (data not shown). The S215A mutant protein, to the contrary, was defective in phosphorylation in both yeast strains and at both temperatures. These results strongly suggest that CK1, and not CK2, is the major kinase for RSV P protein in yeast. The inactivation of CK2 in the temperature-sensitive yeast strain at 37°C was confirmed by assaying the CK2 activity of cell extracts in vitro by using a specific peptide substrate as described in Materials and Methods; under the same conditions, CK1 activity remained unaffected (data not shown).
Taken together, results in this section and those published earlier (7, 38) demonstrate that, in vitro as well as ex vivo, serine residues 232, 237, and 215 share all of the phosphates of RSV P. While Ser232 is the major site for CK2-mediated phosphorylation, small amounts of CK1-mediated phosphorylation occur at Ser215 in HEp-2 cells.
CK1-mediated phosphorylation of P is irrelevant for RSV transcription in vitro.
To address the role of the newly discovered, albeit minor, phosphorylation of P by CK1, we phosphorylated recombinant RSV P protein by purified CK1 in vitro and then assayed its transcriptional activity in RSV transcription reconstituted in the presence of anti-CK2 antibody. The anti-CK2 antibody was previously shown to be effective in inhibiting the CK2 activity present in the HEp-2 extract that was needed to provide actin, which is essential for RSV transcription (7, 10, 30). As shown in Table 1, CK1-catalyzed phosphorylation did not bestow transcriptional function on the P protein (line 6). Furthermore, the S215A mutant P was transcriptionally as active as the wild type (Table 1, line 7). These results suggest that phosphorylation of RSV P by CK1 has no discernible role in RSV transcription in vitro. Its lack of biological relevance is further underscored by the fact that this Ser is not conserved in the BRSV and ORSV P proteins and is replaced with Lys and Asn, respectively. The possible role of this phosphorylation in HRSV P ex vivo could not be investigated due to the lack of a conditional lethal CK1 mutant host cell line or a highly specific cell-permeable inhibitor of CK1.
DISCUSSION
In this communication, we have provided a comprehensive account of the phosphate groups of the RSV P protein, the responsible kinases, and their potential role in RSV transcription. To sum up, we have shown the following. (i) Whereas Ser232 is the primary site for CK2-mediated phosphorylation (7; also Fig. 1 and 2), the only site for CK1-mediated phosphorylation is Ser215 both in vitro and ex vivo. These two residues and the minor CK2 site Ser237 account for all of the phosphates of the RSV P protein. (ii) L protein alone, in the absence of P, cannot initiate transcription (Fig. 4). (iii) Initiation of transcription per se does not require phosphorylation of P (Fig. 4 and 6). (iv) CK2-mediated phosphorylation of P is essential for productive exit of the polymerase from the promoter and for continued transcription elongation (Fig. 4 and 6). (v) Minor amounts of phosphorylation by CK1 occur at Ser215 but are not required for transcription in vitro (Table 1).
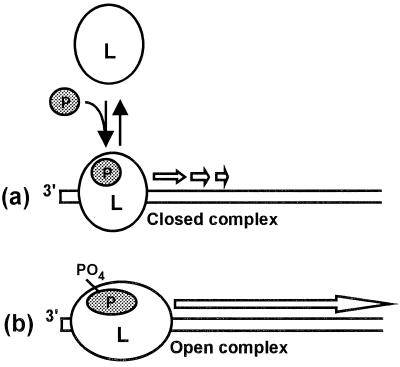
FIG. 6.
Model for the role of P protein in transcription initiation and elongation. (a) Even unphosphorylated P can stimulate loading of the L-P complex at the promoter sequence of the N-RNA template; i.e., phosphorylation is not required for this step. This complex generates abortive transcripts within the leader region. (b) Phosphorylation of P, likely resulting in allosteric conformational changes in the P and L proteins, leads to suppression of the abortive transcription, promoter clearance of the polymerase, and production of full-length transcripts (for details, see the text). The exact stoichiometry of L and P in the functional polymerase is not known.
Although the functional RNA polymerase of RSV requires both the L and P proteins, the relative contributions of the two proteins to polymerase function remain unknown. The essential role of both proteins derives from two kinds of evidence, viz., analysis of conditional lethal mutations and reconstitution experiments in vitro and ex vivo. First, the L protein was found to contain multiple mutations in a cold-sensitive RSV strain (23) and a T736A substitution in another attenuated strain (39). Also, the temperature sensitivity of the tsN19 mutant strain of RSV was due to a G172S mutation in the P protein (11). In an ex vivo cDNA-based reconstitution system, transfection of both L and P cDNAs was required for the synthesis of full-length viral mRNAs and genomic RNA (17, 44). Finally, in vitro, both L and P were absolutely required to reconstitute functional transcription that generated full-length mRNA (7, 30). These studies, however, did not address the role of these proteins or the role of phosphorylation of P in the various steps of the transcription process. Our in vitro results (Fig. 4) showed that the P protein, in the absence of L, did not produce any detectable transcript. The L protein alone was also incapable of transcription. These results suggest that the basic promoter-binding and ribonucleotide polymerase activities of the viral RNA-dependent RNA polymerase require participation of both L and P. In this regard, RSV appears to be similar to VSV and Sendai virus, two nonsegmented negative-strand RNA viruses in both of which the P protein appears to be required for the binding of L to the template N-RNA (15, 21, 32).
When phosphate-free RSV P protein was added along with L, the resultant RSV transcription complex was capable of activating transcription although the transcripts were still short (Fig. 4). This was shown by using either unphosphorylated P (lane 3) or conducting transcription in the presence of PPλ (lane 4). The lack of a need for phosphorylation of RSV P in transcription initiation is also supported by our finding that phosphorylated and unphosphorylated P proteins bound equally well to the template N-RNA in a simple binding assay (30). The sequencing of the short transcripts indicated that they were uncapped, unpolyadenylated, and complementary to the 3′ end of the genomic RNA and thus may be analogous to the leader RNAs that have been found in a number of other negative-strand RNA viruses, such as VSV and Sendai virus (26). Although the minimal promoter of RSV remains undefined, recent studies with VSV have shown that about 24 nucleotides of the terminal bases are absolutely essential for transcription (27, 43). Full transcriptional activity requires additional sequence elements within the leader at positions 19 to 29 and 34 to 46, and a separate element at nucleotides 47 to 50, in the nontranscribed leader-N gene junction (43). The longest abortive RSV transcript, of 24 nucleotides, detected by us in vitro (Fig. 4) is thus reminiscent of the minimal promoter length of VSV and may, in fact, define the minimal functional RSV promoter. To our knowledge, this is the first direct demonstration of free transcripts originating from the leader region of the RSV genome. Since the leader RNA of negative-strand RNA viruses is believed to play an important role in the initiation of encapsidation (binding of N protein to the genomic RNA) and hence in the regulation of the transcription-to-replication switch (9, 35), further characterization of the RSV leader RNAs will be an important breakthrough in RSV gene expression. This is currently in progress.
Full-length mRNAs were only produced when L was used together with phosphorylated P protein (30), suggesting that a major role of phosphorylation of P is to provide the L protein the initial processivity required for promoter clearance (Fig. 4, lanes 5). It should be pointed out that this is fundamentally different from the processivity imparted by the RSV M2 (22K) protein that results in the suppression of intra- as well as intergenic pause and termination throughout the genome (14, 20). Increasing amounts of M2, in fact, generated progressively higher readthrough of adjacent genes and resultant unnatural, chimeric transcripts (20). Phosphoprotein P, in contrast, only suppresses abortive transcription within the leader region at the promoter and does not unduly suppress the natural intergenic termination signals downstream, even at a relatively high concentration (7, 30; data not shown).
The finding that CK1 can phosphorylate recombinant HRSV P is intriguing, since this phosphorylation appears to occur primarily in yeast cells or by use of purified CK1 in vitro and is not important for transcription (Table 1). We do not known if it plays any role in replication. In HEp-2, which is the host cell for HRSV growth and therefore more relevant in our studies, CK2 is responsible for the vast majority of phosphates that are located mostly at Ser232 of the P protein (7, 38) with only about a 5% contribution from Ser215, the site of CK1-mediated phosphorylation. It is hard to ascertain why yeast and HEp-2 cells phosphorylate HSRV P on different residues and through CK1 and CK2, respectively, especially because the exact locations and nature of CK1 and CK2 in any cell are not definitively known (18, 36). Both enzymes are considered ubiquitous based on the fact that their activities can be measured in extracts of virtually all kinds of cells in vitro by using artificial and physiological substrates. However, one cannot rule out the possibility that they associate with various regulatory subunits in cells of different evolutionary origin in vivo (18, 36), which results in their differential accessibility to P protein. Alternatively, the catalytic subunits of CK1 and CK2 of yeast may have substrate recognition properties that differ from those of their HEp-2 counterparts. In any case, until new results are obtained to the contrary, we will consider it unlikely that the minor CK1-mediated phosphorylation of P has any significance in the HRSV life cycle.
Although the various steps of RSV transcription are still to be characterized, we suggest a working hypothesis of the P protein function modeled after our knowledge of eukaryotic transcription (Fig. 6). We believe that although L may encode many transcription-related domains, it cannot by itself recognize the promoter sequence at the 3′ terminus of the N-RNA template (the leader region). P protein, in analogy to the eukaryotic transcription factors, may stabilize promoter-L interactions and thus lead to an active promoter-polymerase complex (46). This function of P does not require phosphorylation and results in the generation of the short leader-like transcripts free of caps and poly(A) tails. The RNA polymerase assembled with phosphate-free P is perhaps akin to a “closed” complex that is still in the initiation mode and fails to proceed beyond the promoter region (31). Phosphorylation of P allows this complex to attain an “open” conformation committed to elongation and necessary for promoter clearance and movement along the template. The transcripts produced in the absence of phosphorylation of P are therefore essentially analogous to the abortive transcription products of other, DNA-dependent, RNA polymerases (31, 41). Since dephosphorylation of P produced immediate loss of RNA elongation at all time points, we further postulate that the open conformation is essential for elongation throughout transcription and that the continued association of phosphorylated P is needed for the maintenance of this open conformation. Recently, use of reconstituted chromatin templates in vitro has led to the discovery of a novel protein factor, FACT (facilitates chromatin transcription), which acts subsequent to transcription initiation to release RNA polymerase II from a nucleosome-induced block to productive transcription (34). We postulate that the P protein acts in an analogous manner for RNA-dependent RNA polymerase, initiating transcription on RNA-based chromatin, i.e., RNA templates wrapped with N protein.
ACKNOWLEDGMENTS
This research was supported in part by a grant from NIAID (AI37938 to S.B.).
We are indebted to James C. Wang (Harvard University) for the gift of the pYEP-PGAL1 plasmid, to Claiborne V. C. Glover (University of Georgia) for the yeast strains, and to Howard D. Lehmkuhl (USDA-ARS, Ames, Iowa) for ovine RSV strain WSU 83-1578.
REFERENCES
- 1.Alansari H, Potgieter L N D. Molecular cloning and sequence analysis of the phosphoprotein, nucleocapsid protein, matrix protein and 22K (M2) protein of the ovine respiratory syncytial virus. J Gen Virol. 1994;75:3597–3601. doi: 10.1099/0022-1317-75-12-3597. [DOI] [PubMed] [Google Scholar]
- 2.Ansai T, Dupuy L C, Barik S. Interaction between a minimal protein serine/threonine phosphatase and its phosphopeptide substrate sequence. J Biol Chem. 1996;271:24401–24407. doi: 10.1074/jbc.271.40.24401. [DOI] [PubMed] [Google Scholar]
- 3.Barik S. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s) J Virol. 1992;66:6813–6818. doi: 10.1128/jvi.66.11.6813-6818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barik S. Expression and biochemical properties of a protein serine/threonine phosphatase encoded by bacteriophage λ. Proc Natl Acad Sci USA. 1993;15:10633–10637. doi: 10.1073/pnas.90.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik S. Mutagenesis and gene fusion by megaprimer PCR. Methods Mol Biol. 1997;67:173–182. doi: 10.1385/0-89603-483-6:173. [DOI] [PubMed] [Google Scholar]
- 6.Barik S, Das A. An analysis of the role of host factors in transcription antitermination in vitro by the Q protein of coliphage lambda. Mol Gen Genet. 1990;222:152–156. doi: 10.1007/BF00283037. [DOI] [PubMed] [Google Scholar]
- 7.Barik S, McLean T, Dupuy L C. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology. 1995;213:405–412. doi: 10.1006/viro.1995.0013. [DOI] [PubMed] [Google Scholar]
- 8.Barik S, Taylor R E, Chakrabarti D. Identification, cloning, and mutational analysis of the casein kinase 1 cDNA of the malaria parasite, Plasmodium falciparum. Stage-specific expression of the gene. J Biol Chem. 1997;272:26132–26138. doi: 10.1074/jbc.272.42.26132. [DOI] [PubMed] [Google Scholar]
- 9.Blumberg B M, Leppert M, Kolakofsky D. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell. 1981;23:837–845. doi: 10.1016/0092-8674(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 10.Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- 11.Caravokyri C, Zajac A J, Pringle C R. Assignment of mutant tsN19 (complementation group E) of respiratory syncytial virus to the P protein gene. J Gen Virol. 1992;73:865–873. doi: 10.1099/0022-1317-73-4-865. [DOI] [PubMed] [Google Scholar]
- 12.Cardenas M E, Dang Q, Glover C V, Gasser S M. Casein kinase II phosphorylates the eukaryote-specific C-terminal domain of topoisomerase II in vivo. EMBO J. 1992;11:1785–1796. doi: 10.1002/j.1460-2075.1992.tb05230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson S U, Schubert M. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1987;84:5655–5659. doi: 10.1073/pnas.84.16.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giaever G N, Snyder L, Wang J C. DNA supercoiling in vivo. Biophys Chem. 1988;29:7–15. doi: 10.1016/0301-4622(88)87020-0. [DOI] [PubMed] [Google Scholar]
- 17.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross S D, Anderson R A. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- 19.Hall C B. Prospects for a respiratory syncytial virus vaccine. Science. 1994;265:1393–1394. doi: 10.1126/science.7915433. [DOI] [PubMed] [Google Scholar]
- 20.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y T, Romito R R, De B P, Banerjee A K. Characterization of the in vitro system for the synthesis of mRNA from human respiratory syncytial virus. Virology. 1993;193:862–867. doi: 10.1006/viro.1993.1195. [DOI] [PubMed] [Google Scholar]
- 23.Juhasz K, Whitehead S S, Bui P T, Biggs J M, Crowe J E, Boulanger C A, Collins P L, Murphy B R. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J Virol. 1997;71:5814–5819. doi: 10.1128/jvi.71.8.5814-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennelly P J, Krebs E G. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 25.Kuchino Y, Nishimura S. Enzymatic RNA sequencing. Methods Enzymol. 1989;180:154–163. doi: 10.1016/0076-6879(89)80099-0. [DOI] [PubMed] [Google Scholar]
- 26.Leppert M, Rittenhouse L, Perrault J, Summers D F, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Pattnaik A K. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J Virol. 1999;73:444–452. doi: 10.1128/jvi.73.1.444-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallipeddi S K, Samal S K. Sequence comparison between the phosphoprotein mRNAs of human and bovine respiratory syncytial viruses identifies a divergent domain in the predicted protein. J Gen Virol. 1992;73:2441–2444. doi: 10.1099/0022-1317-73-9-2441. [DOI] [PubMed] [Google Scholar]
- 29.Mazumder B, Adhikary G, Barik S. Bacterial expression of human respiratory syncytial virus phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology. 1994;205:93–103. doi: 10.1006/viro.1994.1623. [DOI] [PubMed] [Google Scholar]
- 30.Mazumder B, Barik S. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 31.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 32.Mellon M G, Emerson S U. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27:560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller-Decker K, Amtmann E, Sauer G. Inhibition of the phosphorylation of the regulatory non-structural protein of vesicular stomatitis virus by an antiviral xanthate compound. J Gen Virol. 1987;68:3045–3056. doi: 10.1099/0022-1317-68-12-3045. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides G, LeRoy G, Chang C H, Luse D S, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 35.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinna L A, Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- 37.Samal S K, Zamora M, McPhillips T H, Mohanty S B. Molecular cloning and sequence analysis of bovine respiratory syncytial virus mRNA encoding the major nucleocapsid protein. Virology. 1991;180:453–456. doi: 10.1016/0042-6822(91)90057-i. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Seco M P, Navarro J, Martinez R, Villanueva N. C-terminal phosphorylation of human respiratory syncytial virus P protein occurs mainly at serine residue 232. J Gen Virol. 1995;76:425–430. doi: 10.1099/0022-1317-76-2-425. [DOI] [PubMed] [Google Scholar]
- 39.Tolley K P, Marriott A C, Simpson A, Plows D J, Matthews D A, Longhurst S J, Evans J E, Johnson J L, Cane P A, Randolph V B, Easton A J, Pringle C R. Identification of mutations contributing to the reduced virulence of a modified strain of respiratory syncytial virus. Vaccine. 1996;14:1637–1646. doi: 10.1016/s0264-410x(96)00136-3. [DOI] [PubMed] [Google Scholar]
- 40.Trudel M, Nadon F, Simard C, Belanger F, Alain R, Seguin C, Lussier G. Comparison of caprine, human and bovine strains of respiratory syncytial virus. Arch Virol. 1989;107:141–149. doi: 10.1007/BF01313886. [DOI] [PubMed] [Google Scholar]
- 41.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 42.Villanueva N, Navarro J, Cubero E. Antiviral effects of xanthate D609 on the human respiratory syncytial virus growth cycle. Virology. 1991;181:101–108. doi: 10.1016/0042-6822(91)90474-p. [DOI] [PubMed] [Google Scholar]
- 43.Whelan S P J, Wertz G W. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yunus A S, Collins P L, Samal S K. Sequence analysis of a functional polymerase (L) gene of bovine respiratory syncytial virus: determination of minimal trans-acting requirements for RNA replication. J Gen Virol. 1998;79:2231–2238. doi: 10.1099/0022-1317-79-9-2231. [DOI] [PubMed] [Google Scholar]
- 46.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]