Abstract
Mouse virus-like 30S RNAs (VL30m) constitute a family of retrotransposons, present at 100 to 200 copies, dispersed in the mouse genome. They display little sequence homology to Moloney murine leukemia virus (MoMLV), do not encode virus-like proteins, and have not been implicated in retroviral carcinogenesis. However, VL30 RNAs are efficiently packaged into MLV particles that are propagated in cell culture. In this study, we addressed whether the 5′ region of VL30m could replace the 5′ leader of MoMLV functionally in a recombinant vector construct. Our data confirm that the putative packaging sequence of VL30 is located within the 5′ region (nucleotides 362 to 1149 with respect to the cap structure) and that it can replace the packaging sequence of MoMLV. We also show that VL30m contains an internal ribosome entry segment (IRES) in the 5′ region, as do MoMLV, Friend murine leukemia virus, Harvey murine sarcoma virus, and avian reticuloendotheliosis virus type A. Our data show that both the packaging and IRES functions of the 5′ region of VL30m RNA can be efficiently used to develop retrotransposon-based vectors.
The mouse virus-like 30S RNA (VL30m) elements constitute a family of retrotransposons, present at 100 to 200 copies, dispersed in the mouse genome (25, 27, 46). A typical element is 5 to 6 kb in length and contains long terminal repeats (LTRs) of about 500 nucleotides (nt) with an organization resembling that of retroviral LTRs, namely U3-R-U5 (16, 27, 37, 47). The major 30S VL30 RNA transcript is expressed at high levels and in a variety of cell types (10, 16, 22, 34). VL30m elements do not encode virus-like proteins, have little sequence homology to Moloney murine leukemia virus (MoMLV) and have not been implicated in retroviral carcinogenesis (16, 27). However, VL30 RNA possesses the unusual property of being packaged into MLV particles that are propagated in cell culture (17, 19, 33, 34, 67, 78, 79). As a consequence, VL30 elements are able to retrotranspose from cell to cell at high frequency via MLV particles (17, 27).
Packaging of viral RNA is an essential process of retroviral assembly (75). Selection of viral RNA during assembly is governed by interactions between the nucleocapsid (NC) domain of gag and the packaging signal in the viral RNA (24, 75). Packaging signals have been identified in many retroviruses, and in most, if not all, known packaging signals, major determinants are located in the 5′ leader of the viral RNA between the primer binding sites and gag (75). The specificity of genomic RNA recognition and the ability of VL30m RNA to be packaged in type C retroviral particles prompted us to examine whether the 5′ region of VL30m could replace functions known to be directed by the 5′ leader of MoMLV (23).
An interesting feature of the MoMLV, Friend murine leukemia virus (FrMLV), Harvey murine sarcoma virus (HaMSV), and avian reticuloendotheliosis virus type A (REV-A) 5′ leaders is the presence of an internal ribosome entry segment (IRES), which is able to drive translation of the Gag polyprotein in a cap-independent fashion (7, 8, 54, 87). Moreover, in FrMLV, HaMSV, and MoMLV, the IRES was found to be within the packaging signal (7, 8, 23, 87). Therefore, in spite of the fact that no translated VL30 gene products have been identified to date, it was tempting to speculate that VL30m might also have conserved a functional IRES.
Translation initiation is a multistep biochemical pathway aimed at positioning the ribosome at the initiator AUG codon of the mRNA (for a review, see reference 66). In eukaryotes, translation initiation usually takes place by a mechanism dependent on the cap structure at the 5′ end of the mRNA but in certain cases has been shown to proceed in a cap-independent fashion (66, 76). Cap-dependent initiation involves attachment of the initiator methionine tRNA (Met-tRNAi) to the 40S ribosomal subunit, binding of this initiation complex to the 5′ m7GpppG cap structure of the mRNA, and migration of the initiation complex along the mRNA in a 5′→3′ direction until the appropriate AUG codon is encountered to initiate protein synthesis (49). Cap-independent initiation (internal initiation) is mediated by a segment of secondary structure located within the 5′ untranslated region of certain mRNAs, the IRES, which is able to directly recruit the 43S initiation complex (40, 57, 76). Putative IRESes can be functionally discriminated from other 5′ mRNA secondary structures by their ability to mediate translation of the downstream open reading frame (ORF) of a bicistronic reporter mRNA, independent of the translational status of the first ORF (42, 44, 70).
Internal initiation was described first in poliovirus and subsequently in cardiovirus, aphthovirus, rhinovirus, and hepatitis A virus (14, 40, 41, 43, 44, 69). IRESes were later found in other types of viruses, including hepatitis C virus, bovine viral diarrhea virus, and members of the Retroviridae family (4, 7, 8, 39, 54, 85, 87). Cap-independent initiation has also been observed in cellular mRNAs encoding the immunoglobulin heavy-chain binding protein (55), human fibroblast growth factor 2 (86), human insulin-like growth factor (83), translational initiation factor eIF4G (28), platelet-derived growth factor (9), c-myc (61, 82), vascular endothelial growth factor (3, 35, 81), the products of the Drosophila homeotic genes antennapedia and ultrabithorax (63), yeast transcription factors TFIID and HAP4 (36), and the Kv1.4 voltage-gated potassium channel (62).
Based on the criteria of secondary-structure homology between VL30m and MoMLV and sequence homology between VL30m and rat VL30 (38, 48), different regions downstream of the mouse VL30 primer binding site were chosen as putative packaging sequences, and their abilities to replace the MLV 5′ leader in a recombinant construct were examined. Here, we show that the 5′ region of the mouse VL30 retrotransposon can direct packaging of a recombinant RNA and that it contains a functional IRES that is able to promote translation of a downstream ORF in a bicistronic RNA construct. These observations allowed the development of novel retrotransposon-based dicistronic retroviral vectors.
MATERIALS AND METHODS
Plasmid DNA construction and amplification.
Standard procedures were used for restriction nuclease digestion and plasmid DNA construction (77). Escherichia coli HB101 strain 1035 (a recA mutant) was used for plasmid DNA amplification. Details of the constructions are given below.
DNA constructs.
In all cases, nucleotide numbering is with respect to the genomic RNA cap site (position +1).
(i) pKT403.
This plasmid contains the mouse VL30 1.9-kb HindIII fragment in pSP64 (NLV-3; kindly provided by S. Adams) (2).
(ii) pVL30m bicistronic RNAs.
The different VL30 DNA fragments (positions 362 to 1144, 362 to 461, 362 to 575, 576 to 1144, and 462 to 1144) were generated by PCR, digested with NheI, and inserted between the neomycin phosphotransferase gene (neo) and the lacZ gene of NheI-digested pMLV-CB28 (7). The neo-VL30-lacZ sequences are under the control of the T7 RNA polymerase promoter for in vitro expression and the cytomegalovirus early promoter for expression in eukaryotic cells. In these constructs, the initiation of β-galactosidase (β-Gal) translation was from an AUG codon in a favorable context (GCCAUGG) which was generated by PCR (49). pEMCV-CBD260–837 was used as a positive control, while pEMCV-D837–260, containing the same encephalomyocarditis virus (EMCV) IRES fragment but in the reverse orientation, was the negative control (8).
Retroviral vectors.
Retroviral vectors were prepared with pBR322 as the DNA backbone.
(i) pVL30m-SJE1.
The VL30 DNA (positions 362 to 575) generated by PCR (pKT403 template) was digested with BalI and NcoI and cloned into BalI (cutting at position 800)- and NcoI (cutting at position 3449)-digested pLNPOZ (1) containing the lacZ gene and the two MLV LTRs. β-Gal expression was promoted by an AUG codon in a favorable context as for pVLD362–575.
(ii) pVL30m-SJE2.
The VL30 DNA (positions 362 to 1149) generated by PCR was digested with BalI and NcoI and cloned into pLNPOZ as described for pVLEL362–575.
(iii) pVL-SJE3.
The oligonucleotide CCAGCTGAAGCTTGCC was cloned into pLNPOZ which had been digested with BalI (which cuts at position 800) and NcoI (which cuts at position 3449). In this construct, which served as a negative control for RNA packaging, β-Gal expression was promoted by an AUG codon in a favorable context (GCCAUGG), generated by PCR.
(iv) pMLV-LacZ+.
The two LTRs of pMLVK and the MLV Psi+ packaging sequence to position +1035 (5) were inserted into pBluescript KS. The fragment of pCH110 (Pharmacia) containing the lacZ gene was also inserted into this construct. β-Gal expression was promoted by an AUG codon in a favorable context. This plasmid served as a positive control for RNA packaging into MLV virions.
(v) pVL30m-SU8.
VL30 DNA (positions 362 to 575) was generated by PCR, digested with NheI, and cloned between the human placental alkaline phosphatase (PLAP) gene and neo of pMLV-CB71 (7).
(vi) pVL30m-SU9.
VL30 DNA (positions 362 to 1149) was generated by PCR, digested with NheI, and cloned between the PLAP gene and neo of pMLV-CB71 (7).
(vii) pVL30m-SU11.
The EcoRI fragment of pEMCV-CBT4 (83) containing the MLV 5′ LTR and MLV E+ packaging sequence was cloned into pVL30m-SU9/EcoRI.
(viii) pVL30m-SU12.
VL30 DNA (positions 362 to 1149) was generated by PCR, digested with EcoRI, and cloned 5′ of the PLAP gene in pVL-CBT2, which contains rat VL30 between the PLAP gene and neo (8). pEMCV-CBT4 was used as an EMCV-positive control (86). pRev-HW3, containing MoMLV and REV-A IRESes, was described by López-Lastra et al. (54).
In vitro RNA synthesis.
Capped and uncapped RNAs were synthesized by using a DNA template and T7 RNA polymerase (mMessage mMachine or MEGAscript; Ambion) according to the manufacturer’s protocol. Plasmid DNA (1 to 2 μg) digested with SspI (which cuts at position 1240, in the lacZ gene) was used for RNA synthesis in a 20-μl final reaction volume. Transcription was terminated by digestion of the template DNA with DNase I, and RNA was precipitated with lithium chloride. RNA was resuspended in 50 μl of Tris-EDTA buffer and further purified and desalted by application to MicroSpin S-300 columns (Pharmacia Biotech) according to the manufacturer’s protocol. The integrity of RNAs was monitored by 0.7% agarose gel electrophoresis, and RNA concentrations were determined spectrophotometrically.
Translation in a nuclease-treated rabbit reticulocyte lysate (RRL) system.
Capped and uncapped RNAs were translated in the Flex-RRL system (Promega) at 50% concentration with 25 μg of RNA/ml and 0.6 mCi of [35S]methionine (Amersham)/ml at 31°C for 1 h. The assay mixture was supplemented with potassium chloride to a final concentration of 80 mM. For EMCV RNA, 0.5 mM magnesium acetate was added. The effect of foot-and-mouth disease virus (FMDV) leader (L) protease on translation of the capped bicistronic RNAs was assayed as previously described. Briefly, the Flexi-RRL was pretreated with 1.2 μg of purified recombinant L protease (kindly provided by S. J. Morley, Department of Biochemistry, The University of Sussex, Brighton, Sussex, United Kingdom)/ml (64). After translation, samples were heated at 96°C for 3 min in a solution containing 62.5 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol, 5% β-mercaptoethanol, and 0.02% bromophenol blue, and labelled proteins were analyzed by SDS–15% polyacrylamide gel electrophoresis (PAGE). Bands were quantified by using a PhosphorImager (Molecular Dynamics).
Cell culture.
Murine NIH 3T3 cells and the ecotropic packaging cell line GP+E-86 (56) were cultured in Dulbecco’s modified Eagle’s medium (Gibco BRL) with 10% newborn calf serum at 37°C in a 5% CO2 atmosphere.
Transfection, infection, and titration.
Ecotropic GPE-86 cells were seeded at a density of 5 × 105 per 100-mm-diameter plate 24 h prior to transfection with 20 μg of DNA by the calcium phosphate method (21). Infection and titration were performed by adding virus-containing medium to cells. NIH 3T3 cells were seeded at a density of 5 × 105 per 100-mm-diameter plate 24 h prior to infection or at 2 × 104 cells per well in a 24-well plate for titrations. Freshly harvested viruses were filtered (0.45-mm-pore-size filter). Diluted virus-containing supernatants were overlaid on cells in the presence of Polybrene, added to a concentration of 8 mg/ml. Cells were then incubated for 24 h, after which the medium was replaced. Infected cells were grown for a total of 48 h and either subjected to G418 selection at 1 mg/ml or stained for determination of levels of β-Gal or PLAP expression. After 2 months of selection, the G418 concentration was increased to 1.5 mg/ml. The recombinant-virus titer was determined by counting the number of LacZ- or PLAP-positive NIH 3T3 cells 48 h postinfection in limiting-dilution infections. Titers, in transducing units (TU) per milliliter, were calculated as follows: (number of colonies) × (dilution of infecting retrovirus)/(total volume [in milliliters] of diluted vector overlaid on cells).
To analyze long-term virus production of a vector which did not express a selection gene, 2 μg of pSV2neo (80), encoding neomycin phosphotransferase under the control of the simian virus 40 promoter, was cotransfected with 18 μg of the pVL plasmids into GP+E-86 cells. Seventy-two hours after transfection, GP+E-86 cells were diluted and placed under selection conditions in a medium supplemented with G418 at 0.8 mg/ml. After 3 weeks of selection, harvested virus was used to infect NIH 3T3 cells as described above.
Histochemical staining.
Cells were fixed in phosphate-buffered saline (PBS) containing 2% formaldehyde and 0.2% glutaraldehyde. For LacZ staining, after two washes in PBS, cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). For PLAP histochemical staining, after two washes in PBS, cells were incubated at 65°C for 30 min in 1× PBS. Cells were washed twice with AP buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, and 50 mM MgCl2 in double-distilled water) and stained with 0.1-mg/ml 5-bromo-4-chloro-3-indolylphosphate (BCIP), 1-mg/ml nitroblue tetrazolium salt, and 1 mM levamisole in 1× AP buffer.
Enzymatic activities.
Cell extracts were used as substrates for subsequent enzymatic assays. Cells were washed twice with cold 1× PBS, scraped from plates by using a rubber policeman, collected by centrifugation at 600 × g, and resuspended in NP-40 buffer (0.5% NP-40, 140 mM NaCl, 30 mM Tris-HCl [pH 7.5]). Nuclei were removed by a 10-min centrifugation at 14,000 × g. Protein concentrations were determined by using the Micro BCA* protein assay reagent (Pierce). PLAP activity in cell extracts was determined spectrophotometrically (alkaline phosphatase substrate kit; Bio-Rad), using commercial calf intestine alkaline phosphatase (Boehringer Mannheim) as an activity standard. Neomycin phosphotransferase activity was determined by measuring [γ-32P]ATP phosphate transfer to neomycin as previously described (73).
Western blotting.
Cells were washed twice with PBS, trypsinized, and collected by centrifugation at 600 × g. Cells were directly resuspended in NP-40 buffer; this was followed by a 10-min centrifugation at 14,000 × g. The supernatant was transferred to a new tube, and the protein concentration was determined by using the Micro BCA* protein assay reagent (Pierce). Once quantified, 10 μg of total protein was subjected to SDS–15% PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (Boehringer Mannheim) by semidry transfer in a 30% methanol–Tris-glycine buffer. The filter was blocked with 5% fat-free dried milk in TBST (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.05% Tween 20). The membrane was incubated for 1 h at room temperature in a 1:800 dilution of rabbit anti-neomycin phosphotransferase II antibody (5 Prime→3Prime, Inc.) in blocking buffer; after two 15-min washes in TBST, the membrane was incubated as before in a 1:800 dilution of biotinylated anti-rabbit immunoglobulin G antibody (BioSys; BIOSYS S.A., Compiègne, France). After two washes with TBST, the membrane was incubated for 30 min in VECTRASTAIN Elite ABC avidin-peroxidase solution (Vector Laboratories) and developed by enhanced chemiluminescence (ECL; Amersham) according to the manufacturer’s protocol.
Effect of rapamycin on protein synthesis in murine cells.
Stably transduced NIH 3T3 cells were grown to 70 to 80% confluency. Cells were serum starved for 48 h prior to the addition of Dulbecco’s modified Eagle’s medium containing 10% newborn calf serum and either 50 ng of rapamycin (Sigma)/ml or vehicle alone. Four hours after serum stimulation, protein extracts were prepared (see previous section). As previously described (54), the level of reporter gene expression, determined by measuring enzymatic activity in the presence and in the absence of rapamycin, was used to calculate the effect of the drug as a percentage increase or decrease relative to untreated cells.
RESULTS
Recombinant MLV-VL30m vectors allow expression of LacZ in transduced NIH 3T3 cells.
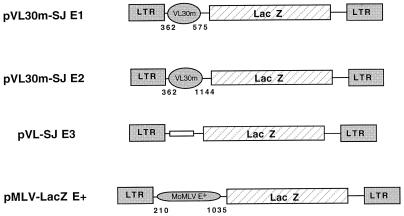
To evaluate whether the 5′ region of VL30m can replace the 5′ leader of MoMLV in a recombinant construct, two regions downstream of the VL30m primer binding site were chosen as putative packaging sequences. These regions were placed upstream of lacZ in a monocistronic MLV-based retroviral vector (Fig. 1A). pMLV-LacZ+, containing the MLV E+ packaging sequence, was used as a positive control, while pVL-SJE3, containing a random sequence in place of a packaging signal, was used as a negative control. Vectors were transfected into GP+E-86 cells, and virus-containing medium was later recovered and used to transduce NIH 3T3 cells. The number of NIH 3T3 LacZ-positive cells obtained after transduction with vectors pVL30m-SJE1 and pVL30m-SJE2 was of the same order of magnitude as that for pMLV-LacZ+, the control vector (data not shown). This preliminary observation prompted us to determine the recombinant-virus titer of the stably producing helper cell line, set up by cotransfecting GP+E-86 cells with pSV2neo and the different vector constructs (80). As shown in Table 1, upon G418 selection, the titers obtained with the MLV-VL30m vectors were of the same order of magnitude as those of the control MLV vector. These data not only confirm the work of Chakraborty et al. (20), which suggested that the 5′ region of VL30m has the ability to drive packaging of a recombinant RNA, but because LacZ is expressed, they also show that VL30m can direct translation of a 3′ cistron in the context of a monocistronic retroviral vector.
FIG. 1.
Schematic representation of MLV-VL30m-lacZ monocistronic retroviral vectors. All MLV-VL30m vectors contain the 5′ LTR and the primer binding site of MLV. In vectors pVL30m-SJ E1 and pVL30m-SJ E2, different segments of the 5′ region of NLV-3 VL30 sequences were inserted upstream of the lacZ reporter cistron. The pMLV-LacZ+ vector, used as a positive control, contains the MLV Psi+ encapsidation sequence. pVL-SJE3, used as a negative control, contains no encapsidation sequence. Numbering is with respect to the VL30m RNA cap site (position +1).
TABLE 1.
Recombinant monocistronic MLV-VL30m-lacZ retroviral titera
| Vector | Retroviral titer (TU/ml) |
|---|---|
| pVL30m-SJ E1 | 0.3 × 105 |
| pVL30m-SJ E2 | 1.3 × 105 |
| pVL-SJ E3 | <1 × 102 |
| pMLV-LacZ E+ | 2 × 105 |
Viral titers were determined by performing serial dilutions followed by LacZ histochemical staining. NIH 3T3 cells were seeded 24 h prior to infection with virus-containing medium harvested from stably transformed ecotropic GP+E-86 cells (56). The viral titers are reported as LacZ+ TU per milliliter of collected medium and correspond to the means of triplicate experiments. The standard deviations did not exceed 10% of the mean values.
The 5′ region of VL30m directs expression of a 3′ cistron of a bicistronic RNA in RRL.
Based on published data for MoMLV (87), HaMSV (8), FrMLV (7), and REV-A (54), we hypothesized that VL30m contains a functional IRES. As a first approach in the characterization of the putative VL30m IRES, capped and uncapped monocistronic RNAs, with different segments of VL30m 5′ RNA upstream from lacZ, were translated in an RRL system. Results suggested that translation of RNAs containing VL30m 5′ sequence proceeded independently of the cap (data not shown). These data prompted us to test the translational ability of these sequences when contained within a bicistronic mRNA (Fig. 2A) (42, 44, 70). In pVL30m bicistronic RNAs, the VL30m sequences from position 362 to 1144, 362 to 461, 362 to 575, 576 to 1144, or 462 to 1144 were inserted between neo and the lacZ gene, as previously described (Fig. 2A) (54). In all constructs, the first cistron lies downstream of a short 5′ capped or uncapped untranslated region (54 nt), and the 3′ cistron (lacZ) would be expressed only if the VL30m sequence has IRES activity or through a termination reinitiation mechanism (42, 66, 70). As a positive control for cap-independent translation, we used pEMCV-CBD260–837 RNA (Fig. 2A), containing the EMCV IRES, while pEMCV-D837–260, with the complete EMCV leader in the reverse orientation, was used as a negative control.
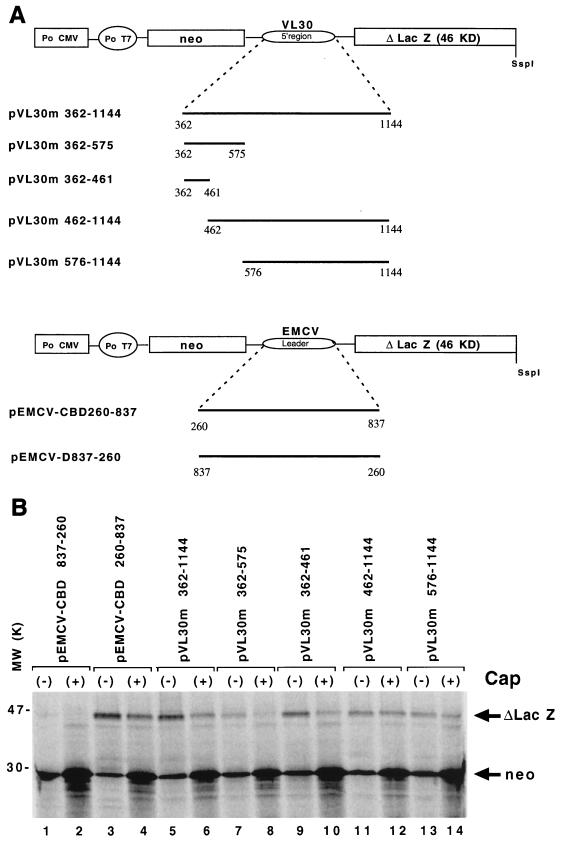
FIG. 2.
Translation of VL30m bicistronic RNA in messenger-dependent RRL. (A) Schematic representation of the bicistronic plasmid constructs containing different portions of the VL30 5′ RNA located between the neo and lacZ genes under the control of the T7 promoter (Po T7) for in vitro expression. Numbering is with respect to the genomic RNA cap site (position +1). Po CMV, cytomegalovirus early promoter; KD, kilodaltons. (B) Translation of uncapped (−) and capped (+) bicistronic RNA in the Flexi-RRL system (Promega). After heat denaturation, 35S-labelled proteins were analyzed by SDS–15% PAGE. The positions of neomycin phosphotransferase (28 kDa) and the C-terminally truncated β-Gal protein (46 kDa) are indicated. Lanes 1 to 4, control RNAs containing the EMCV IRES (see Materials and Methods); lanes 7 to 10 RNAs containing different 3′ deletions in the putative VL30m IRES; lanes 5, 6, and 11 to 14, RNAs containing the 5′ region VL30m RNA or 5′ deletions of this sequence.
The results show that in uncapped pVL30m RNAs (Fig. 2B, lanes 5, 7, 9, 11, and 12) the putative VL30m IRES was capable of promoting synthesis of β-Gal. With capped RNAs, the putative VL30m IRES activities of pVL30m 362–575 and pVL30m 362–461 were reduced to very low levels of β-Gal protein (Fig. 2B, lanes 8 and 10). The addition of cap also reduced, though less drastically, translation of the 3′ cistron in RNA pVL30m 362–1144 (lanes 5 and 6). Interestingly, cap had little effect on the level of β-Gal synthesis with RNAs pVL30m 462–1144 (lanes 11 and 12) and pVL30m 576–1144 (lanes 13 and 14) or with pEMCV-CBD260–823, the control RNA (lanes 3 and 4). As expected, with all RNAs cap enhanced translation of neo, the 5′ cistron. These data indicate that translation initiation promoted by the 5′ region of VL30m is probably cap independent, since a termination-reinitiation mechanism would not explain why 3′ deletions caused a reduction of translation promoted by VL30 sequences (lanes 7 to 10). The above data also suggest that (i) the 3′ region (nt 461 to 1144) of the putative VL30m IRES is required for its optimal activity (lanes 5 and 6 and lanes 11 and 12) and (ii) when present on the same biscistronic RNA, 5′ cap and the IRES compete for the recruitment of translation, as indicated by the shutoff of suboptimal IRES activity of RNAs with 3′ deletions in the VL30 IRES (Δ576–1144 and Δ462–1144) (lanes 7 to 10).
Influence of FMDV L protease on in vitro translation of bicistronic VL30m RNAs.
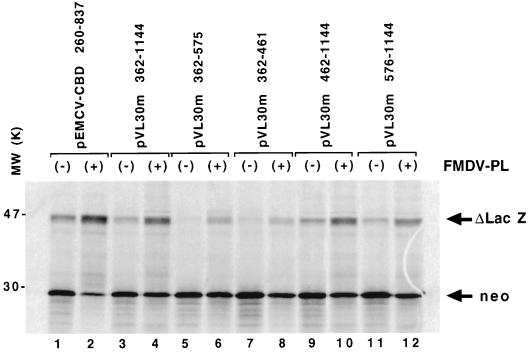
We next examined the effect of the L protease of FMDV on VL30m bicistronic RNA translation (Fig. 3; Table 2). Translation of capped RNA is disrupted when the initiation factor eIF4G is cleaved by viral proteases such as 2A of poliovirus, coxsackievirus, and human rhinovirus or the L protease of FMDV (11, 65, 89, 90). Previous studies using L protease-treated RRL revealed the ability of this protease to partially inhibit translation of capped cellular RNA while internal initiation remained unaffected (54, 64, 65). When capped VL30m bicistronic RNAs or capped pEMCV-CBD260–837 (Fig. 2A) was translated in L protease-treated RRL, the level of β-Gal synthesis was enhanced whereas the level of neomysin phosphotransferase expression decreased (Fig. 3, lanes 1 and 2 [for EMCV] and lanes 9 to 12 [for VL30m]). In confirmation of data presented in Fig. 2B (lanes 8 and 10), β-Gal was poorly expressed by capped pVL30m 362–461 and pVL30m 362–575 RNAs (Fig. 3, lanes 5 and 7). However, when cap-dependent translation was inhibited by L protease, translation from the 3′ cistron was partially restored (Fig. 3; compare lanes 5 and 6 and lanes 7 and 8). This confirms that the 3′ region of the VL30m 5′ sequences is important for optimal IRES activity (Fig. 3, lanes 5 to 8). The effect of L protease on expression of neomycin phosphotransferase and β-Gal was quantitated by scanning densitometry, and data are summarized in Table 2. The contrasting effect of L protease on the synthesis of neomycin phosphotransferase and LacZ is in agreement with previously published data (32, 54, 64, 65, 89, 90) and confirms the presence of a functional IRES within the 5′ region of VL30m RNA. Our in vitro assays showed that cap is able to shut down IRES activity from RNA pVL30m 362–575, an effect that can be abolished by treatment with FMDV L protease (Fig. 2B, lanes 7 and 8; Fig. 3, lanes 5 and 6). It should also be pointed out that ex vivo (Fig. 1; Table 1), monocistronic retroviral vector pVL30m-SJ E1, containing the 5′ VL30m region from nt 362 to 575, was able to promote synthesis of β-Gal in murine cells.
FIG. 3.
Effect of FMDV L protease on bicistronic-RNA translation. Bicistronic capped RNA was translated in the Flexi-RRL system (Promega) with (+) or without (−) L protease (PL). After heat denaturation, 35S-labelled proteins were analyzed by SDS–15% PAGE. The positions of neomycin phosphotransferase (28 kDa) and the C-terminally truncated β-Gal protein (46 kDa) are indicated. Lanes 1 and 2, control RNAs containing the EMCV IRES (see Materials and Methods); lanes 5 to 8, RNAs containing different 3′ deletions in the putative VL30m IRES; lanes 3, 4, and 9 to 12, RNAs containing the 5′ region VL30m RNA or 5′ deletions of this sequence.
TABLE 2.
Relative change in reporter gene expression caused by FMDV L proteasea
| Construct | Relative change (%) in expression of:
|
|
|---|---|---|
| Neomycin phosphotransferase | β-Gal | |
| pEMCV-CBD 260–837 | −85.5 | +145 |
| pVL30m 362–1144 | −35.5 | +281.5 |
| pVL30m 362–575 | −29 | +359 |
| pVL30m 362–461 | −71 | +102.5 |
| pVL30m 462–1144 | −59.5 | +302.5 |
| pVL30m 576–1144 | −63.5 | +154.5 |
The gel shown in Fig. 3 was scanned, and data were used to calculate the effect of L protease on the percentage increase or decrease relative to translation in untreated Flexi-RRL preparations [100% − (level of reporter gene in the presence of L protease) × 100%/(level of reporter gene in the absence of L protease)] (54).
Construction of MLV-based bicistronic retroviral vectors, using the 5′ region of VL30m.
Retroviral vectors incorporating mouse VL30 sequences have been proposed to have potential use in gene therapy (18, 20, 26). To evaluate the use of VL30m 5′ region (E/IRES) in gene transfer and to test its function in cells, we constructed bicistronic retroviral vectors.
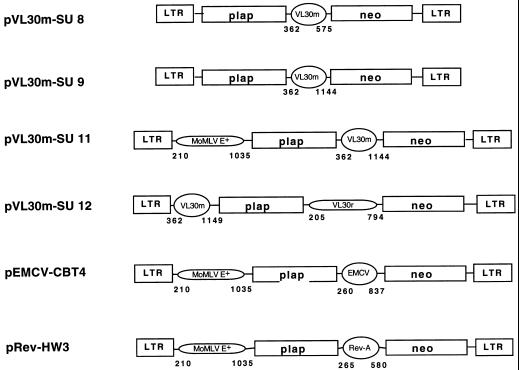
The pVL30m-SU vectors and control vectors pEMCV-CBT4 and pRev-HW3 (54, 84) are shown in Fig. 4. In all constructs, the first cistron encodes PLAP while the second codes for neomycin phosphotransferase. In vectors pVL30m-SU8 and pVL30m-SU9, the 5′ MoMLV E sequence (positions 362 to 575) has been deleted and the putative VL30m E/IRES alone (positions 362 to 1149) was expected to promote both packaging of the recombinant RNA and cap-independent translation of the second cistron in a position-independent manner. In vector pVL30m-SU11, the first cistron is preceded by the MoMLV E+ packaging sequence (positions 210 to 1035), previously shown to contain an IRES (7, 87), while the second cistron is preceded by the putative VL30m IRES (positions 362 to 1149). In the construct pVL30m-SU12, the first cistron is preceded by the putative VL30m E/IRES (positions 362 to 1149) and the second cistron is preceded by the previously described rat VL30 E/IRES (positions 205 to 794) (8).
FIG. 4.
Schematic representation of the bicistronic retroviral vectors. The VL30m-MLV-based retroviral vectors are built on a pBR322 backbone. VL30m corresponds to the 5′ RNA region of the mouse VL30 retrotransposon, VL30r corresponds to the 5′ untranslated region of HaMSV (8), and MLV E+ corresponds to the extended packaging region of MLV (5). Placental alkaline phosphatase (plap) and neomycin phosphotransferase (neo) were used as marker genes. The control vectors pEMCV-CBT4 and pRev-HW3 possess two IRESes (53, 84), the first from MLV (7, 87), which also directs packaging (E+), and the second from EMCV or REV-A (53). In all cases, numbering is with respect to the genomic RNA cap site (position +1).
Vectors were used to transfect ecotropic helper cells (GP+E-86), and neomycin-resistant clones were selected. All transfected GP+E-86 cells were found to stably express both genes (neo and that encoding PLAP) for at least 2 months. Once the integrity of the polycistronic RNA was confirmed by Northern blotting (data not shown), recombinant-virus titers were determined by transducing NIH 3T3 cells with virus-containing medium. The vector titers showed a high degree of variation depending on the position and number of packaging sequences (E) within the same recombinant RNA (Table 3). In these assays, the titers obtained with both control vectors, pEMCV-CBT4 and pREV-HW3, are in agreement with those previously published (108 to 109) (54). These data clearly show that all recombinant RNA can be packaged, with the exception of pVL30-SU8, and that at least the first cistron is expressed in transduced NIH 3T3 cells, allowing their identification by PLAP histochemical staining. However, comparisons between the titers of monocistronic vectors pVL30m-SJE1 and pVL30m-SJE2 (Fig. 1 and Table 1) and those of the bicistronic vectors pVL30m-SU8 and pVL30m-SU9 suggest that in contrast to what has been observed for MoMLV E+, VL30m E seems to act in a position-dependent manner.
TABLE 3.
Recombinant bicistronic retroviral titera
| Vector | Retroviral titer (TU/ml) |
|---|---|
| pVL30m-SU 8 | 2 |
| pVL30m-SU 9 | 1.4 × 103 |
| pVL30m-SU 11 | 2.0 × 107 |
| pVL30m-SU 12 | 1.0 × 105 |
| pRev-HW 3 | 9 × 108 |
| pEMCV-CBT4 | 3.3 × 108 |
Viral titers were determined by performing serial dilutions followed by PLAP histochemical staining (54, 83). NIH 3T3 cells were seeded 24 h prior to transduction with virus-containing medium harvested from stably transformed ecotropic GP+E-86 cells (56). One-milliliter volumes of virus-containing medium were directly used to transduce 8 × 105 NIH 3T3 cells, PLAP-positive colonies were counted after 15 days of G418 selection. The viral titers are reported in PLAP-positive TU per milliliter of collected medium and correspond to the means of triplicate experiments. standard deviations did not exceed 15% of mean values.
To examine the expression of the 3′ neo cistron upon transduction, cells were selected (using G418) as indicated in Materials and Methods. For all vectors, we obtained neomycin-resistant clones positive for PLAP by histochemical staining. This observation suggests that in contrast to packaging ability, the IRES function is position independent. To further confirm these data, expression of both proteins was examined in cellular extracts from transduced cells. The level of PLAP gene expression was determined by a biochemical assay, while the level of neo expression was determined by Western blotting (Fig. 5). In agreement with histochemical staining and drug resistance analyses, both proteins were detected. Interestingly, and despite the fact that 100% of the cells were PLAP positive and G418 resistant, expression of each cistron varied depending on the vector. These types of variation in gene translation have been previously reported (54) and may be due to the general vector context and/or competition for the recruitment of factors necessary for translation. These data suggest that both packaging and IRES functions of the 5′ VL30 region can be efficiently used in the development of retroviral vectors. However, the efficiencies of packaging and protein expression depend on the position of the E sequence and on the combination of IRESes used in the vector construct.
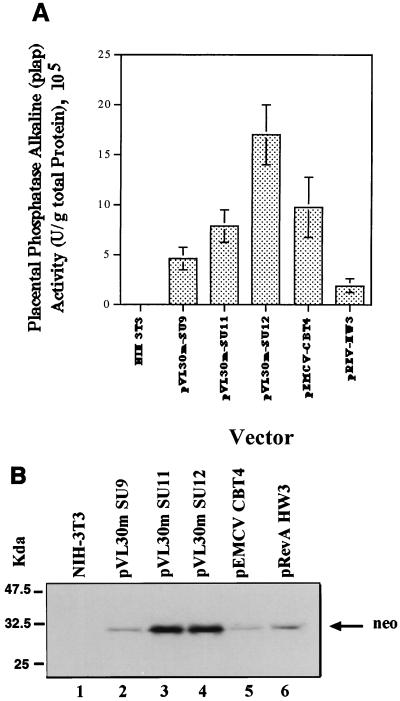
FIG. 5.
Monitoring double transgene expression. Proteins extracted from transduced PLAP-positive neomycin-resistant NIH 3T3 cells were used to determine the level of expression of each transgene by vector constructs. (A) PLAP enzymatic activities were determined as described in Materials and Methods (53, 84). The mean values of alkaline phosphatase specific activities as well as the standard deviation for each set of experiments are shown. Data are the averages of values from three independent experiments. (B) Ten micrograms of total protein was loaded per lane and subjected to SDS–15% PAGE. Proteins were transferred to a polyvinylidene difluoride membrane and probed with a rabbit anti-neomycin phosphotransferase II antibody. The membrane was then incubated with a biotinylated anti-rabbit immunoglobulin G antibody and an avidin-peroxidase solution and, finally, developed by enhanced chemiluminescence. Lane 1, negative control (protein extract from nontransduced NIH 3T3 cells); lanes 2 through 6, protein extracts from cells transduced with the different retroviral vectors. The positions of molecular mass standards are shown on the left.
To further confirm that the 5′ region of VL30m RNA contains a functional IRES, the effect of rapamycin on transgene expression was examined. Rapamycin has been shown to block phosphorylation of the negative regulator of cap binding protein 4E-BP1, PHAS-I. In its dephosphorylated form, PHAS-I acts as a natural repressor of the cap binding protein eIF4E (6, 31, 53), whose nonsequestered levels are probably rate limiting during cap-dependent translation initiation (31, 74). Phosphorylation of PHAS-I results in the release of eIF4E and increased translation activity (68). Beretta et al. (6) have shown that rapamycin blocks PHAS-I phosphorylation in NIH 3T3 cells, specifically attenuating cap-dependent translation. As before (54), we used the protocol of Beretta et al. (6) to determine the effect of rapamycin on the enzymatic activity of the proteins PLAP and neomycin phosphotransferase in cell extracts of transduced NIH 3T3 cells expressing both of these proteins. In these experiments, we used vector pVL30m SU9 as a control, since in this vector the 5′ packaging (E)/IRES region of MLV has been deleted. It is expected that PLAP expression is cap dependent while expression of neomycin phosphotransferase is cap independent. pVL30m-SU11 and pVL30-SU12 are expected to be double IRES vectors; therefore we predicted that translation of both cistrons should be cap independent. Metabolic labelling, as described by Beretta et al. (6) and Morley and McKendrick (59), was used to control the effect of rapamycin on total protein synthesis (data not shown).
As expected, and in agreement with the data of Beretta et al. (6) and López-Lastra et al. (54), in NIH 3T3 cells transduced with vector pVL30m-SU9, rapamycin treatment decreased PLAP enzymatic activity by 13% while increasing neomycin phosphotransferase activity by 57%. In pVL30m-SU11, rapamycin enhanced PLAP activity by 47% and neomycin phosphotransferase activity by 28%. With pVL30m-SU12, in which PLAP expression is directed by the VL30m 5′ region, it was enhanced by 16%, and expression of neomycin phosphotransferase, driven by the VL30 rat IRES, was enhanced by 64%. These ex vivo results are in agreement with the in vitro data obtained by using L protease (Fig. 3 and Table 2) and sustain the conclusion that the 5′ region of VL30m RNA contains a functional IRES that can be efficiently used in the development of retrotransposon-based bicistronic vectors such as pVL30m SU12.
DISCUSSION
A characteristic feature of VL30 retrotransposons is that they can be packaged into MLV virions, leading to their horizontal spread during retrovirus dissemination (17, 19, 33, 34, 67, 78, 79). Based on this observation, we asked whether the 5′ region of VL30m RNA could replace the MLV E+ sequence functionally in a recombinant viral RNA. For this, monocistronic vectors were constructed in which the MLV E+ packaging sequence was replaced by the 5′ region of VL30m. Upon transfection of GP+E-86 helper cells, the packaging potential of the 5′ region of VL30m RNA was determined by titrating the recombinant lacZ vectors. These initial experiments showed that the 5′ region of VL30m RNA can indeed replace the E sequence of MoMLV in a recombinant retroviral construct (Fig. 1; Table 1). Moreover, and contrary to what is predicted by the scanning model of translation initiation, the length and secondary structure of the VL30m 5′ region did not prevent lacZ expression (50–52). This prompted us to determine the mechanism of translation initiation of VL30m 5′ RNA. Since the leaders of MoMLV (87), HaMSV (7), FrMLV (7), and REV-A (54) genomic RNAs have each been described as containing an IRES, we hypothesized that VL30m RNA also has an IRES.
To characterize the VL30m IRES, we used the canonical strategy of bicistronic constructs, first described by Pelletier and Sonenberg (70) and Jang et al. (44) (Fig. 2). The results, presented in Fig. 2B, show that the VL30m 5′ region allows translation of the downstream cistron in a bicistronic construct independently from that of the first cistron (Fig. 2). Moreover, FMDV L protease, which is known to specifically shut off cap-dependent translation initiation, enhanced translation initiation driven by the VL30m 5′ region while reducing expression of neo, the cap-dependent gene (Fig. 3). These data suggest that as for other IRESes, the cap binding protein of the translation preinitiation complex, eIF4E, may not be required for VL30m-driven translation initiation (65, 71). This idea was further sustained by the results obtained with capped RNAs pVL30m 362–461 and pVL30m 362–575, for which a 3′ deletion was able to impair translation of the 3′ cistron. In addition, when cap-dependent translation initiation was inhibited by L protease, translation of the 3′ cistron was enhanced. Together, these data confirm that the 5′ region of VL30m is able to drive translation initiation independently of the 5′ cistron; therefore, this region is capable of recruiting ribosomes by an internal mechanism. The decrease in translation of RNAs pVL30m 362–461 and pVL30m 362–575 as well as the increase in cap-independent translation in the presence of L protease is most probably due to an active competition between the 5′ cap structure and the IRES for the recruitment of canonical translation initiation factors (71, 72, 89, 90).
To confirm the data in a cellular context, a series of retroviral vectors containing the 5′ region of VL30m RNA has been constructed. Retroviral vectors not only allow testing of the functionality of IRESes ex vivo but also permit their exploitation in gene transfer and therapy. In this respect, IRESes represent an efficient means of expressing two transgenes in cells without the need for two promoters or a regulated splicing mechanism (1, 29, 30, 54, 58, 60, 76, 84). In MLV-based vectors, the packaging signal comprising the extended (E+) region of MLV encompasses sequences encoding Gag and glyco-Gag. These sequences, which might be the cause of homologous recombinations possibly generating replication-competent retroviruses, cannot be deleted without destroying the ability of the recombinant RNA to be encapsidated into virions. Our results show that the VL30 5′ region is capable of directing both packaging (Fig. 1 and Table 1) and transgene expression (Fig. 2 and 3 and Table 2). We therefore constructed novel MLV-VL30 vectors (Fig. 4) to determine whether the VL30m E/IRES characterized in vitro could be used in the development of bicistronic vectors.
Upon transduction of murine cells, the VL30m IRES was found to be functional, since it allowed expression of a 3′ cistron of a bicistronic retroviral vector. Moreover, by combining VL30m and rat VL30, we were able to develop an efficient IRES-based vector, pVL30-SU12, that contains only the LTRs of MLV, thus reducing to a minimum the sequences that can potentially recombine with those encoding Gag and glyco-Gag. This improvement, together with those of others (18, 20, 26), will allow the development of safer retrotransposon-based vectors that are less likely to undergo homologous recombination.
Several lines of evidence, such as sequence analysis indicating that the VL30m 5′ region contains stop codons in all reading frames (2), suggest that VL30m has no translational activity. This is supported by the fact that to date no VL30m-encoded polypeptides have been identified (27). Nevertheless, there exist numerous blocks of amino acid homology between VL30 and retroviral genes, suggesting that VL30m RNA contains the remnants of ancestral gag and pol genes (2). This, together with the presence of an IRES, might reflect an ancient translational activity, which could explain why VL30m RNA has been found associated with polyribosomes (25, 45). Moreover, it should be pointed out that in Mus dunni endogenous virus, which appears to be a chimera between VL30m and a virus similar to gibbon ape leukemia virus, genomic RNA packaging and Gag polyprotein expression are most probably driven by the VL30m 5′ region herein reported to contain a functional IRES (88).
It is tempting to speculate about the conservation of these retroviral IRESes. Due to the compact size of retrotransposon genomes, it is not surprising that the long 5′ region participates in multiple transposon life cycle functions. It is clear that higher-order RNA structures can persist through evolution despite substantial changes in primary nucleotide sequence. Thus, it is a distinct possibility that as for MoMLV, FrMLV, and HaMSV, the VL30m IRES is contained within the region which determines dimerization and subsequent packaging of its RNA. Hence, its maintenance would be required for the preservation of the retrotransposon. To date, no specific biological role has been described for the VL30m retrotransposon. VL30s, despite their close association with oncogenic MLVs, have not been directly linked to oncogene activation (27). Since mouse VL30 sequences are actively transcribed under a variety of stimuli, they might act as insertional mutagens via gene activation, as with murine retroviruses (27). On the other hand, the variety of VL30 LTR promoters found in the mouse, together with their mobility, may also provide possible mechanisms for evolutionary changes in gene regulation as well as for the spread of genes across species (15). Although the data presented herein do not allow us to directly address the question of the biological role of this family of retroelements, they do support the hypothesis that VL30m not only activates genes at the level of transcription but also has an effect on translation due to its IRES activity.
ACKNOWLEDGMENTS
Marcelo López-Lastra and Sandrine Ulrici contributed equally to this work.
We thank S. J. Morley, Department of Biochemistry, The University of Sussex, Brighton, Sussex, United Kingdom, for kindly providing purified recombinant FMDV L protease; S. Adams for the gift of pKT403; and M. Rau and E. A. Derrington for critical reading of the manuscript.
This work was supported by grants from ANRS, MGEN, and the European Community (BMH4-CT96-0675). M. López-Lastra presently is supported by a fellowship from the Région Rhône-Alpes.
REFERENCES
- 1.Adam M A, Ramesh N, Miller A D, Osborne W R A. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J Virol. 1991;65:4985–4990. doi: 10.1128/jvi.65.9.4985-4990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams S E, Rathjen P D, Stanway C A, Fulton S M, Malim M H, Wilson W, Ogden J, King L, Kingsman S M, Kingsman A J. Complete nucleotide sequence of a mouse VL30 retroelement. Mol Cell Biol. 1988;8:2989–2998. doi: 10.1128/mcb.8.8.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiri G, Nahari D, Finkelstein Y, Le S Y, Elroy-Stein O, Levi B Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 4.Attal J, Theron M C, Taboit F, Cajero-Juarez M, Kann G, Bolifraud P, Houdebine L M. The RU5 (′R′) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS LETT. 1996;392:220–224. doi: 10.1016/0014-5793(96)00815-0. [DOI] [PubMed] [Google Scholar]
- 5.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 7.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlioz C, Torrent C, Darlix J-L. An internal ribosomal entry signal in the rat VL 30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 10.Besmer P, Olshevsky U, Baltimore D, Dolberg D, Fan H. Virus-like 30S RNA in mouse cells. J Virol. 1979;29:1168–1176. doi: 10.1128/jvi.29.3.1168-1176.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 12.Borman A M, Le Mercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 14.Brown E A, Zajac A J, Lemon S M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bultman S J, Klebig M L, Michaud E J, Sweet H O, Davisson M T, Woychik R P. Molecular analysis of reverse mutations from nonagouti (a) to black-and-tan (a(t)) and white-bellied agouti (Aw) reveals alternative forms of agouti transcripts. Genes Dev. 1994;8:481–490. doi: 10.1101/gad.8.4.481. [DOI] [PubMed] [Google Scholar]
- 16.Carter A T, Norton J D, Avery R J. The genomic DNA organisation and evolution of a retrovirus-transmissible family of mouse (VL30) genetic elements. Biochim Biophys Acta. 1988;951:130–138. doi: 10.1016/0167-4781(88)90033-4. [DOI] [PubMed] [Google Scholar]
- 17.Carter A T, Norton J D, Gibson Y, Avery R J. Expression and transmission of a rodent retrovirus-like (VL30) gene family. J Mol Biol. 1986;188:105–108. doi: 10.1016/0022-2836(86)90485-7. [DOI] [PubMed] [Google Scholar]
- 18.Chakraborty A K, Zink M A, Boman B M, Hodgson C P. Synthetic retrotransposon vectors for gene therapy. FASEB J. 1993;7:971–977. doi: 10.1096/fasebj.7.10.8393821. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty A K, Zink M A, Hodgson C P. Transmission of endogenous VL30 retrotransposons by helper cells used in gene therapy. Cancer Gene Ther. 1994;1:113–118. [PubMed] [Google Scholar]
- 20.Chakraborty A K, Zink M A, Hodgson C P. Expression of VL30 vectors in human cells that are targets for gene therapy. Biochem Biophys Res Commun. 1995;209:677–683. doi: 10.1006/bbrc.1995.1552. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–637. [PubMed] [Google Scholar]
- 22.Clewley J P, Avery R J. The virion RNA species of the Kirsten murine sarcoma-leukemia virus complex released from a clonally related series of mouse cells. Arch Virol. 1982;72:35–46. doi: 10.1007/BF01314448. [DOI] [PubMed] [Google Scholar]
- 23.Corbin A, Darlix J L. Functions of the 5′ leader of murine leukemia virus genomic RNA in virion structure, viral replication and pathogenesis, and MLV-derived vectors. Biochimie. 1996;78:632–638. doi: 10.1016/s0300-9084(96)80009-5. [DOI] [PubMed] [Google Scholar]
- 24.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 25.Fan H, Mueller-Lantzsch H. RNA metabolism of murine leukemia virus. III. Identification and quantitation of endogenous virus-specific mRNA in uninfected BALB/c cell line JLS-V9. J Virol. 1976;18:401–410. doi: 10.1128/jvi.18.2.401-410.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French N S, Norton J D. Construction of a retroviral vector incorporating mouse VL30 retrotransposon-derived, transcriptional regulatory sequences. Anal Biochem. 1995;228:354–355. doi: 10.1006/abio.1995.1364. [DOI] [PubMed] [Google Scholar]
- 27.French N S, Norton J D. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 28.Gan W, Celle M L, Rhoads R E. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 29.Ghattas I R, Sanes J R, Majors J E. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurtu V, Yan G, Zhang G. IRES bicistronic expression vectors for efficient creation of stable mammalian cell lines. Biochem Biophys Res Commun. 1996;229:295–298. doi: 10.1006/bbrc.1996.1795. [DOI] [PubMed] [Google Scholar]
- 31.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatzoglou M, Hodgson C P, Mularo F, Hanson R W. Efficient packaging of a specific VL30 retroelement by psi 2 cells which produce MoMLV recombinant retroviruses. Hum Gene Ther. 1990;1:385–397. doi: 10.1089/hum.1990.1.4-385. [DOI] [PubMed] [Google Scholar]
- 34.Howk R S, Troxler D M, Lowy D, Duesberg P H, Scolnick E M. Identification of a 30S RNA with properties of a defective type C virus in murine cells. J Virol. 1978;25:115–123. doi: 10.1128/jvi.25.1.115-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huez I, Créancier L, Audigier S, Gensac M-C, Prats A-C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNA in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itin A, Keshet E. Nucleotide sequence analysis of the long terminal repeat of murine virus-like DNA (VL30) and its adjacent sequences: resemblance to retrovirus proviruses. J Virol. 1983;47:656–659. doi: 10.1128/jvi.47.3.656-659.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itin A, Rotman G, Keshet E. Conservation patterns of mouse “virus-like” (VL30) DNA sequences. Virology. 1983;127:374–384. doi: 10.1016/0042-6822(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov P A, Karpova O V, Skulachev M V, Tomashevskaya O L, Rodionova N P, Dorokhov Y, Atabekov J G. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- 40.Jackson R J, Howell M T, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 41.Jackson R J, Hunt S L, Gibbs C L, Kaminski A. Internal initiation of translation of picornavirus RNAs. Mol Biol Rep. 1994;19:147–159. doi: 10.1007/BF00986957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 43.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 44.Jang S K, Kraüsslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keshet E, Itin A. Patterns of genomic distribution and sequence heterogeneity of a murine “retrovirus-like” multigene family. J Virol. 1982;43:50–58. doi: 10.1128/jvi.43.1.50-58.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keshet E, Shaul Y. Terminal direct repeats in a retrovirus-like repeated mouse gene family. Nature. 1981;289:83–85. doi: 10.1038/289083a0. [DOI] [PubMed] [Google Scholar]
- 48.Konings D A M, Nash M A, Maizel J V, Arlinghaus R B. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozak M. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1991;1:117–125. [PMC free article] [PubMed] [Google Scholar]
- 51.Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- 52.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 53.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 54.López-Lastra M, Gabus C, Darlix J-L. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum Gene Ther. 1997;7:603–611. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 55.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 56.Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBratney S, Chen C Y, Sarnow P. Internal initiation of translation. Curr Opin Cell Biol. 1993;5:961–965. doi: 10.1016/0955-0674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 58.Morgan R A, Couture L, Elroy-Stein O, Ragheb J, Moss B, Anderson W F. Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Res. 1992;20:1293–1299. doi: 10.1093/nar/20.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morley S J, McKendrick L. Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem. 1997;272:17887–17893. doi: 10.1074/jbc.272.28.17887. [DOI] [PubMed] [Google Scholar]
- 60.Mountford P S, Smith A G. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 62.Negulescu D, Leong L E, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 63.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains a 5′-noncoding sequence that confers translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 64.Ohlmann T, Rau M, Morley S J, Pain V M. Proteolytic cleavage of initiation factor eIF-4 gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acid Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohlmann T, Rau M, Pain V, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 66.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 67.Patience C, Takeuchi Y, Cosset F-L, Weiss R A. Packaging of endogenous retroviral sequences in retroviral vectors produced by murine and human packaging cells. J Virol. 1998;72:2671–2676. doi: 10.1128/jvi.72.4.2671-2676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 69.Pelletier J, Kaplan G, Racaniello V R, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 71.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramesh N, Osborne W R. Assay of neomycin phosphotransferase activity in cell extracts. Anal Biochem. 1991;193:316–318. doi: 10.1016/0003-2697(91)90028-r. [DOI] [PubMed] [Google Scholar]
- 74.Rau M, Ohlmann T, Morley S J, Pain V M. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 75.Rein A. Retroviral RNA packaging: a review. Arch Virol Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 76.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 77.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 78.Scolnick E M, Vass W C, Howk R S, Duesberg P H. Defective retrovirus-like 30S RNA species of rat and mouse cells are infectious if packed by type C helper virus. J Virol. 1979;29:964–972. doi: 10.1128/jvi.29.3.964-972.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sherwin S A, Rapp U R, Benveniste R E, Sen A, Todaro G J. Rescue of endogenous 30S retroviral sequences from mouse cells by baboon type C virus. J Virol. 1978;26:257–264. doi: 10.1128/jvi.26.2.257-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327. [PubMed] [Google Scholar]
- 81.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 83.Teerink H, Voorma H O, Thomas A A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 84.Torrent C, Berlioz C, Darlix J-L. Stable MLV-VL30 dicistronic retroviral vectors with a VL30 or MoMLV sequence promoting both packaging of genomic RNA and expression of the 3′ cistron. Hum Gene Ther. 1996;7:603–611. doi: 10.1089/hum.1996.7.5-603. [DOI] [PubMed] [Google Scholar]
- 85.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 88.Wolgamot G, Bonham L, Miller A D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ziegler E, Borman A M, Deliat F G, Liebig H D, Jugovic D, Kean K M, Skern T, Kuechler E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/s0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 90.Ziegler E, Borman A M, Kirchweger R, Skern T, Kean K M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]