Abstract
Oncogenes or tumor suppressor genes are rarely mutated in several pediatric tumors and some early stage adult cancers. This suggests that an aberrant epigenetic reprogramming may crucially affect the tumorigenesis of these tumors. Compelling evidence support the hypothesis that cancer stem cells (CSCs), a cell subpopulation within the tumor bulk characterized by self-renewal capacity, metastatic potential and chemo-resistance, may derive from normal stem cells (NSCs) upon an epigenetic deregulation. Thus, a better understanding of the specific epigenetic alterations driving the transformation from NSCs into CSCs may help to identify efficacious treatments to target this aggressive subpopulation. Moreover, deepening the knowledge about these alterations may represent the framework to design novel therapeutic approaches also in the field of regenerative medicine in which bioengineering of NSCs has been evaluated. Here, we provide a broad overview about: 1) the role of aberrant epigenetic modifications contributing to CSC initiation, formation and maintenance, 2) the epigenetic inhibitors in clinical trial able to specifically target the CSC subpopulation, and 3) epigenetic drugs and stem cells used in regenerative medicine for cancer and diseases.
Keywords: cancer stem cells, epigenetic alterations, epigenetic inhibitors, regenerative medicine, normal stem cells
Epigenetics and normal vs. cancer stem cells
What’s epigenetics and how important is it in normal and cancer stem cell biology?
Although in the eukaryotic organism all cells contain the same DNA sequence, different cell types in distinct tissues perform different functions. This potential is determined by the regulation of gene expression. Epigenetics is a regulatory mechanism of gene expression that does not lead to alterations of DNA sequence but operates on the physical structure of DNA or histones, the DNA associated proteins. In eukaryotes, octameric histones compact DNA in a higher order and dynamic 3D structure, called chromatin, which is acquired in a progressive manner through covalent modifications and is therefore reversible. At the basal level chromatin is formed by nucleosomes, where two turns of DNA (146 base pairs) are wrapped on histone octamers formed by two copies of each core histone H3, H4, H2A and H2B. Histone H1 binds nucleosomes to each other, further compacting the DNA molecule into a 30 nm fiber. The dynamism of epigenetic regulation allows cells to adapt and respond to external stimuli and has a central role in different cellular processes, including transcription, DNA repair, replication and cellular differentiation. The epigenetic information not contained in the DNA sequences is heritable and pass on from mother to daughter cells. Alterations in epigenetic regulation are hallmarks of senescence and many diseases, including fragile X syndrome and cancer (Okano et al., 1999; Bond et al., 2015; Gupta et al., 2019). Of note, in cancer despite the advanced knowledge and continuous research on driver mutations, recent studies on the non-genetic determinants confirmed that these epigenetic mechanisms are involved in chromosomal instability, oncogene activation, silencing of oncosuppressor genes and also in the development of tumor heterogeneity. Interestingly, several aberrant epigenetic alterations have been associated and linked to the determination and formation of a subpopulation of cells known as cancer stem cells (CSCs), promoting tumor initiation (Vicente-Duenas et al., 2018; Kumar et al., 2022). Indeed, tumor tissues seem organized hierarchically, at the top of which there are CSCs (Sato et al., 2003; Kreso and Dick, 2014). This population has been identified in many tumor types such as breast, colon, thyroid, brain, ovary, prostate, pancreas, liver, skin and lung cancers, and show many features, tumor-initiating capability, self-renewal, DNA repair, high density of drug transporter and thus therapy resistant mechanisms (Todaro et al., 2007; Turdo et al., 2019; Turdo et al., 2020). In addition, CSCs are considered responsible for the metastatic process (Medema, 2013; Todaro et al., 2014; Veschi et al., 2020; Gaggianesi et al., 2021). Each epigenetic mechanism has its purpose in lesser or greater compactness of the chromatin, determining accessibility (euchromatin) to the RNA polymerase and to all transcriptional factors. In particular, epigenetic mechanisms include events that modify the elements of chromatin, DNA and histone proteins. DNA methylation is the best-known epigenetic process, which regulates chromatin remodeling and gene expression, thus determining several biological processes such as differentiation and genomic stability (Atlasi and Stunnenberg, 2017; Park et al., 2022). Differentiated cells showed a stable and unique DNA methylation pattern (Moore et al., 2013). The addition and maintenance of methyl groups on cytosine residues to form 5-methylcytosine (5mC) in CpG dinucleotides, called CpG sites, are orchestrated by a family of DNA methyltransferases (DNMTs). In particular, DNMT3A and DNMT3B are responsible for the de novo methylation of DNA, a heightened process in embryonic cells and development (Okano et al., 1999). It has been shown that altered methylation due to the lack of these enzymes is also associated with an alteration of the OCT4 and NANOG stem genes (Li et al., 2007; Bibikova et al., 2008). The characterization of ten-eleven translocation protein 1 (TET1) and its role shed new lights on the complexity of DNA methylation and in particular how it could be erased (Tahiliani et al., 2009), although until recently it was considered an irreversible process (Rasmussen and Helin, 2016). Additionally two enzymes from the same family, TET2 and TET3, were identified. All these three enzymes catalyze the oxidation of 5 mC, thus inducing the formation of 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Jiang, 2020). It has been demonstrated that alterations on gene expression profiles of these enzymes appear to be involved in many cancer types (Pan et al., 2015; Xu et al., 2019).
Histone modifications include phosphorylation, acetylation (Ac), crotonylation, methylation (Me), sumoylation, and mono-ubiquitination (Ub) on the selected amino-terminal tail, lysines (K), arginines (R), and serines (S) that are catalyzed by proteins engaged by transcription machinery (Berger, 2007). Histone changes occur following a precise order, which is referred to as the “histone code” as it regulates the chromatin condensation and the accessibility of transcription systems (Strahl and Allis, 2000). In particular, histone methyltransferases (HMTs) and histone acetyltransferases (HATs) add methyl or acetyl groups to the histone tails, respectively, whereas histone demethylases (HDMs) and histone deacetylases (HDACs) remove these groups. Modifications affecting histone H3 and its biological significance have been extensively studied in CSCs compared to histone H2 and histone H4 (Chi et al., 2010). In particular, the post-transcriptional modifications of histone H3 such as H3K27me3, H3K27ac and H3K4me3, are responsible for plastic and dynamic chromatin maintenance in CSCs. To support these data, it was demonstrated that during the cellular differentiation, stem cells (SCs) lose H3K4me3 mark and acquire a compact chromatin profile (Suva et al., 2013; Yamazaki et al., 2013). While acetylation is usually linked to a chromatin opening and gene activation, the role of histone methylation depends on the histone in which it occurs, in fact, both events could lead to the induction or repression of gene transcription. For example, mono-methylation of lysine 4 on histone H3 (H3K4me1) in the promoter region is associated with a limited recruitment of specific enzymes involved in chromatin remodeling (Cheng et al., 2014), while di- and tri-methylation (H3K4me2, H3K4me3) are generally linked to a transcriptionally active form of chromatin (Pekowska et al., 2011). Trimethylation of lysine 9 or 27 on histone H3 (H3K9me3 or H3K27me3), are associated with regions of inactive gene transcription (Barski et al., 2007; Eissenberg and Shilatifard, 2010). Of note, many histone modifying enzymes display non histone targets which may play a crucial role in modulating the epigenetic reprogramming of pediatric cancers derived from neuroblasts, supporting their potential role in affecting also the plasticity of the stem cell compartment (Veschi et al., 2017; Veschi and Thiele, 2017; Veschi et al., 2019).
Another important change affecting histones is the incorporation of histone variants. The histone variants can totally replace the canonical histones or form heterotypic nucleosomes with them. Histone genes can be classified as replication dependent histones (S phase), which are known as canonical histones, tissue specific histones and cell cycle replication independent histones. The latter two types are known as histone variants, which differ from canonical in the moment of deposition on chromatin and the amino acid sequence. For example, human embryonic stem cells (hESCs) exhibit higher levels of H1.1, H1.3, and H1.5 compared with specialized cells while the variant H1.0 is involved in cell differentiation (Terme et al., 2011). One of the most common variant of H2A histone is macroH2A, which is correlate with a repression of gene transcription, that replaces the canonical H2A in inactivated X chromosomes of female cells and senescence process (Zhang et al., 2005). Furthermore, macroH2A is upregulated during differentiation processes of embryonic and adult SCs (Barrero et al., 2013).
In 2012, H2A.Z-H2A hybrid couple has been shown as highly expressed in mouse SCs in particular localized at the transcriptional start sites (TSS) of expressed genes (Nekrasov et al., 2012). In addition, the variant H2A.Z has been studied in cancer progression and it has been associated to increased risk of metastasis in ERα-positive breast cancer patients (Hua et al., 2008). Compelling evidence highlighted the fundamental role of H2A.Z in ESC development and in regulating the gene expression patterns of specialized cells (Subramanian et al., 2013).
Non-coding RNA (ncRNA) are RNA sequences that are transcribed but not translated into proteins. They can be classified according to their length in small RNA and long ncRNA, of about 20–22 or 200 nucleotides, respectively. NcRNAs have recently been reported to interact with nucleic acids and proteins. Small ncRNAs usually result in downregulation/silencing of gene expression, while long ncRNAs (lncRNAs) may have structural and regulatory functions, and they can act both in nucleus and in cytosol, interacting with DNA, RNA or proteins, leading to different effects on the regulation of biological processes (Statello et al., 2021). Interestingly, lncRNAs are cell-type specific and play important roles in SC maintenance and differentiation (Statello et al., 2021).
All this evidence points out an urgent need to better understand the epigenetic mechanisms, that influence the transition of normal versus cancer SCs and their characteristic features including their tumorigenic capacity and metastatic potential.
Normal versus cancer stem cells
The above-described mechanisms of epigenetic regulation are essential and crucial for the future of SCs. Normal stem cells (NSCs) are responsible for different biological processes such as embryonic development and tissue homeostasis (Rossi et al., 2020), and they are characterized by limited self-renewal. These cells can undergo a particular cell division giving rise to two SCs with the same characteristics, to maintain the SC pool, and another division that leads to one SC and one progenitor cell to continue the differentiation process (Fuchs and Chen, 2013). Regenerating and repairing organs, as well as maintaining normal tissue homeostasis and differentiating into specialized cells under specific signals, represent additional functions of NSCs. NSCs display a diploid genome, are generally quiescent and characterized by a restricted proliferation potential (Voog and Jones, 2010). Tissue SCs create continuous crosstalk with their microenvironment, called niche. The released niche factors regulate, at the epigenetic level, the fate of SCs (Wu and Sun, 2006). SCs are classified based on their differentiation potential, into totipotent SCs that are able to generate all cell types, including embryonic and extraembryonic tissue (for example morula), pluripotent SCs generate all body cells excluding extraembryonic tissue (such as blastocyst), multipotent SCs that have the ability to develop specific cell types (such as hematopoietic cells), and unipotent SCs that have an aptitude to generate exclusively single cell types (for example hepatic cells) (Mardanpour et al., 2008). The stemness potential and the differentiation status of progenitor cells are determined by the epigenetic changes, which include DNA methylation and histone modifications. The pluripotent cells have a different chromatin configuration from the other cells they present the chromatin lightly packed and permissive for an active gene transcription. During the differentiation process, stem genes turn off, while only specialized genes of committed cells became transcriptionally active (Stergachis et al., 2013; Atlasi and Stunnenberg, 2017).
A study regarding the genome-wide distribution of DNA methylation highlighted that the methylation of CpG islands undergoes modifications during cellular differentiation. This event drives SCs towards their differentiation fate (Meissner et al., 2008). Generally, CpG islands remain unmethylated in NSCs, however, there are differentially methylated regions (DMRs) such as those present in the inactivated X chromosome. The differentiation process leads to an increase in methylated areas with a reduction of the multipotent capacity and self-renewal through gene silencing (Bibikova et al., 2008; Fuchs and Blau, 2020). It is known that the expression pattern of pluripotency is regulated by the transcription factors Oct4, Nanog and Sox2, when these factors are reintroduced into specialized cells these cells can reprogram themselves (Li et al., 2012). Accordingly, it has recently been shown that TET1 and TET2, enzymes that demethylate DNA, are expressed in mouse ESCs (Ito et al., 2010) and are Oct4-regulated, supporting the pluripotent state (Koh et al., 2011). Since the discovery of stem cell-like cells in patients affected by leukemia, a novel theory of tumor initiation was postulated regarding the presence within the tumor of CSCs derived from NSCs, that have underwent abnormal epigenetic alterations. Therefore, upon the epigenetic reprogramming CSC subpopulation, defined by uncontrolled proliferation and self-renewal capacity, has been considered responsible for tumor initiation, progression and therapy resistance (Turdo et al., 2019). In summary, in the majority of cases scientists believe that CSCs, derive from NSCs that have lost the control of proliferation processes, present deregulation of common Wnt/β-catenin, JAK-STAT, TGF-β and hedgehog (HH) signaling pathways, or have undergone mutation and/or abnormal epigenetic alteration (Rycaj and Tang, 2015).
An important characteristic to distinguish NSCs versus CSCs is represented by the chromatin accessibility. Nowadays, despite the gold standard methods used in the past decades, multiple complex biochemical techniques have been developed to study chromatin accessibility and to design maps of the chromatin status across a large number of tissues and cell types in several diseases, including cancer. Specifically, high-throughput technology, through the evaluation of cis-regulatory sequences analysis, the adoption of single-cell methods together with in silico and bioinformatic approaches allow the exploration of chromatin accessibility profiles both in bulk population and at a single-cell level. Here, we will discuss the innovative high-throughput technologies applied for the study of chromatin accessibility in NSCs and CSCs. RNA expression with sequencing (SHARE-seq) is used for single or several measures of chromatin accessibility at a single-cell level and the relative gene expression profile. Of note, this technology allows the study of the trajectory lineage for each cell. Ma et al. (2020), studied the chromatin potential state of a single cell to its future RNA states, identifying the cell fate which may follow during developmental transitions by SHARE-seq analysis application. Additionally, transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) has been used to investigate chromatin states transition in early human pre-implantation development. Particularly, ATAC-seq, through integrative analysis allows studying the conservation or divergence in regulatory circuitry during early stage of embryonic development both in human and mouse and, between human pluripotent stem cells in vivo and hESCs (Wu et al., 2018). Single-cell chromatin accessibility methods allow also the identification of a stem-like cell subpopulation in primary tumors starting from the bulk population. Specifically, by the application of ATAC-seq technique, Guilhamon et al. demonstrated that the transcription factor SP1 promotes stemness and invasion in glioblastoma and that FOXD1, a pluripotency transcription factor, controls the expression of the aldehyde dehydrogenase ALDH1A3, a marker of invasive potential in glioblastoma SCs (Guilhamon et al., 2021). Finally, formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq) is an emerging technology applied to identify regulatory elements distant from their gene targets. FAIRE-seq consists of a silico approach to predict transcription factors binding sites and a high-throughput sequencing of a plethora of cell types, aimed to evaluate the complexity of several transcriptional networks focusing on biological processes such as epithelial–mesenchymal transition (EMT) and CSC formation. The FAIRE-seq application in breast cancer models highlighted regions with increased accessibility. Specifically, FOX and AP-1 regions are associated with increased expression of CSC-associated genes. On the other hand, the transcriptional repression of two FOX family members, FOXN2 and FOXQ1, impairs CSC formation and reduces the expression of stemness-related genes (Hardy et al., 2016). Elucidating the differences in chromatin accessibility between NSCs and CSCs by the above-mentioned high-throughput techniques will help to identify novel potential biomarkers/therapeutic targets specific for this aggressive subpopulation.
In the following paragraphs we will review the principal epigenetic mechanisms and/or alterations related to the initiation and maintenance of CSCs.
Epigenetic alterations critical for cancer stem cell initiation
Despite the effort employed in the study of cancer biology, the specific trigger that induces cell transformation and tumor initiation is still unclear (Vicente-Duenas et al., 2013). In the past, the study of biological processes related to the biology of cancer has been focused on finding molecular pathways and genetic alterations, at the SC compartment level, trying to shed light on the molecular basis responsible for initiation, promotion and progression of cancer. In the recent years there has been an increasing emphasis on the characterization of the epigenetic mechanisms associated with cancer initiation and evolution (Kumar et al., 2022). Many works have shown that epigenetic alteration are implicated in numerous steps during cancer initiation, being responsible for abnormal expression of oncogenes and tumor suppressor genes, thus leading to tumor transformation (Cheng et al., 2019). In particular, SCs and progenitors are characterized by specific epigenetic profiles, which make them more susceptible to acquire DNA mutations than differentiated cells (Beerman and Rossi, 2015). Both cell-intrinsic (i.e., mutations) and cell-extrinsic (i.e., environmental cues) factors control the epigenetic status of cancer cells. Specifically, epigenetic alterations occurring in the SC compartment may be amplified within the cell compartment, bringing a selective advantage on cell growth and maintenance. During this process, further epigenetic changes or additional alterations at the genetic level that lead to cellular transformation may also occur. These changes are inherited from the daughter cells, resulting in a pool of cells that support tumor initiation (Beerman and Rossi, 2015). Gene expression is dramatically altered in CSCs compared to healthy cells resulting from an aberrant modulation of the epigenetic machinery. Hypermethylation of CpG islands and silencing of tumor suppressor genes and/or pro-differentiation factors are characteristic epigenetic traits of CSCs. Of note, the epigenetic status of malignant ESCs is significantly different regarding the methylation profile compared with CSCs in adult cancers (Ohm et al., 2007).
Among several theories proposed in order to explain the origin of CSC, further hypotheses have been postulated about alternative mechanisms contributing to CSC formation or other precursor cells from which CSC may derive, beyond NSCs. According to the “cell reversal theory” (CRT), a somatic cell can dedifferentiate after a specific perturbation (a potential carcinogenic event), in this way the cell switches to a different epigenetic state which could activate an uncontrolled proliferation. A cell can enter on the pathological\cancerous epigenetic program and lead to the formation of what is labeled as a CSC (Carvalho, 2020). Similarly to CRT, Friedmann-Morvinski and Verma presented a theory about the origin of CSC resulting from tumor progression. Specifically, they explained that upon reprogramming into a pluripotent state that could drive tumor progression, the dedifferentiation of tumor cells could determine CSC formation via epigenetic resetting (Friedmann-Morvinski and Verma, 2014). Moreover, Nimmakayalaa et al. (2019) hypothesized that CSC origin could be driven by cell fusion, horizontal gene transfer, exposure to hypoxia and toxic agents, metabolic reprogramming or mutations in differentiated cells that guide the reprogramming of them into CSC.
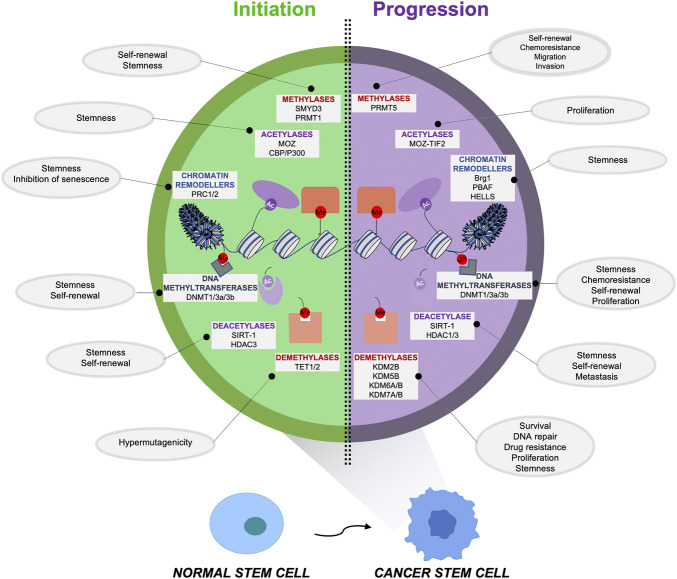
In the following paragraphs we will summarize the principal epigenetic mechanisms responsible for CSC initiation, formation and maintenance, which are shown in Figure 1 and described in Tables 1, 2, focusing on the aberrant epigenetic reprogramming driving the transformation process of a NSC to a CSC.
FIGURE 1.
Epigenetic alterations responsible for the initiation and maintenance of cancer stem cells (CSCs). An aberrant epigenetic reprogramming plays a critical role in CSC initiation and maintenance, driving the transformation process of a normal stem cell (NSC) versus a cancer stem cell (CSC). The principal alterations involving the epigenetic regulators that sustain the formation (left) and the maintenance (right) of this aggressive subpopulation are indicated along with the functions/properties of CSCs that these enzymes are promoting.
TABLE 1.
Epigenetic alterations in cancer stem cell (CSC) initiation.
| Epigenetic enzymes/LncRNAs | Cancers | Modifications | Functions | References |
|---|---|---|---|---|
| Methylases | ||||
| PRMT1 | Leukemia | Upregulation | CSC initiation | Chi et al. (2010) |
| SMYD3 | Gastric cancer | Upregulation | Self renewal, stemness | Wang et al. (2018) |
| Demethylases | ||||
| KDM1A | Prostate and breast cancer, leukemia | Overexpression | CSC initiation | Lim et al. (2010), Crea et al. (2012), Vu et al. (2013) |
| KDM1B | Glioblastoma | Overexpression | CSC initiation | Hu et al. (2016) |
| KDM3A | Colorectal cancer | mRNA stabilization | Wnt/β-catenin pathway upregulation | Wang et al. (2019) |
| Acetylases | ||||
| CBP/p300 | Leukemia | c-Myb-CBP/p300 interaction | AML1-ETO and MLL fusion-protein upregulation | Pattabiraman et al. (2014) |
| MOZ | Leukemia | MOZ/TIF2 fusion protein | CSC initiation | Aikawa et al. (2010) |
| Deacetylases | ||||
| HDAC3 | Hepatocellular carcinoma | Overexpression | Self renewal, stemness | Liu et al. (2013) |
| SIRT-1 | Colorectal and neuronal cancer | Overexpression | Self renewal, stemness | Chen et al. (2014), Lee et al. (2015a) |
| Chromatin remodellers | ||||
| PRC1 | Prostate cancer | Overexpression | Senescence inhibition | Yap et al. (2010) |
| PRC2 | Hematopoietic cancer | Downregulation | CSC initiation | Beerman and Rossi, (2015) |
| DNA methyltransferases | ||||
| DNMT1 | Breast cancer | Downregulation | CSC initiation | Pathania et al. (2015) |
| DNMT3A | Leukemia, myelofibrosis | Downregulation | CSC initiation | Mayle et al. (2015) |
| DNMT3B | Squamocellular carcinoma | Downregulation | CSC initiation | Rinaldi et al. (2017) |
| DNA demethylases | ||||
| TET1 | Non-Hodgking B cell lymphoma | Detection | 5-hydroxymethylcytosine loss and DNA hypermethylation | Cimmino et al. (2015) |
| TET2 | Leukemia | Downregulation | Stem cell hypermutagenicity | Pan et al. (2017) |
| Long non coding RNAs | ||||
| HOTAIR | Breast cancer | Overexpression | Self renewal, stemness, CSC initiation | Deng et al. (2017) |
| HOTTIP | Pancreatic cancer | Overexpression | Self renewal, stemness | Melendez-Zajgla and Maldonado, (2021) |
| LnchPVT1 | Hepatocellular carcinoma | Upregulation | Self renewal, stemness, CSC initiation | Wang et al. (2014) |
| Linc00617 | Breast cancer | Upregulation | Self renewal, stemness, CSC initiation | Li et al. (2017) |
| MALAT-1 | Hepatocellular carcinoma, pancreatic cancer | Upregulation | Self renewal, stemness, CSC initiation | Wang et al. (2015), Zeng et al. (2018) |
| RNA methyl transferases | ||||
| METTL3 | Glioblastoma | ncRNA m6A-modification | CSC initiation | Cui et al. (2017) |
| METTL14 | Glioblastoma | ncRNA m6A-modification | Self renewal, stemness, CSC initiation | Cui et al. (2017) |
TABLE 2.
Epigenetic alterations in cancer stem cell (CSC) maintenance.
| Epigenetic enzymes/LncRNAs | Cancers | Modifications | Functions | References |
|---|---|---|---|---|
| Methylases | ||||
| PRMT5 | Breast cancer | Overexpression | CSC maintenance | Chiang et al. (2017) |
| Demethylases | ||||
| KDM2B | Leukemia, glioblastoma | Overexpression | CSC maintenance, survival and chemoresistance | Frescas et al. (2007), He et al. (2011), Staberg et al. (2018) |
| KDM6A/B | Glioblastoma | Overexpression | CSC proliferation, drug resistance | Liau et al. (2017) |
| KDM7A/B | Glioblastoma | NR | CSC survival and DNA repair | Mallm et al. (2020) |
| Acetylases | ||||
| MOZ-TIF2 | Leukemia | Overexpression | CSC proliferation | Aikawa et al. (2010) |
| Deacetylases | ||||
| HDAC1 | NSCLC | NR | Self renewal and CSC proliferation | Wang et al. (2017) |
| HDAC3 | Hepatocellular carcinoma | NR | Self renewal | Liu et al. (2013) |
| SIRT-1 | Pancreatic cancer | Overexpression | Cancer metastasis and stem cell properties | Leng et al. (2021) |
| Chromatin remodellers | ||||
| Brg1 | Colorectal cancer | NR | CSC maintenance | Yoshikawa et al. (2021) |
| HELLS | Glioblastoma | NR | CSC maintenance | Zhang et al. (2019) |
| PBAF | Prostate cancer | NR | CSC maintenance | Hagiwara et al. (2021) |
| DNA methyltransferases | ||||
| DNMT1 | Hepatocellular carcinoma, breast cancer | NR, overexpression | Self renewal, CSC maintenance | Pathania et al. (2015), Wang et al. (2021) |
| DNMT3A | NR | NR | CSC proliferation and chemoresistance | Wainwright and Scaffidi, (2017) |
| DNMT3B | NR, hepatocellular carcinoma | NR | CSC maintenance | Shukla and Meeran, (2014), Lai et al. (2019) |
| Long non coding RNAs | ||||
| H19 | Breast cancer, papillary thyroid carcinoma | NR, upregulation, NR, NR | CSC maintenance, proliferation and self renewal | Li et al. (2018), Peng et al. (2018), Peperstraete et al. (2020), Singh et al. (2020) |
| HOTAIR | Breast, colon and gastric cancer | NR, overexpression, NR | Prometastatic, CSC maintenance, tumor growth | Zhang et al. (2014a), Wang et al. (2016), Deng et al. (2017) |
| Lnc34a | Colorectal cancer | Overexpression | CSC proliferation | Amirkhah et al. (2019) |
| MALAT-1 | Glioma, breast and pancreatic cancer | Upregulation | CSC proliferation, self renewal, colony formation and invasion | Jiao et al. (2014), Jiao et al. (2015), Han et al. (2016), Zeng et al. (2018) |
| NORAD | Pancreatic cancer | Upregulation | Self renewal, proliveration and tumorigenesis | Ma et al. (2021) |
NSCLC, non small cell lung cancer; NR, not reported.
Epigenetic changes in tumor initiation: Theories about epigenetic switch of normal stem cells into cancer stem cells
CSCs are characterized by abnormalities in genes that play a pivotal role in normal differentiation. The alterations of these genes mainly involve a hypermethylated profile that characterized CSCs since their premalignant state (Mayle et al., 2015; Pan et al., 2017). The hypermethylated status in these cells is due and maintained by the activity of several epigenetic regulators such as TET1 and TET2, the proto-oncogene IDH1 and IDH2, Polycomb group proteins, that tightly regulate the epigenetic profile of CSCs (Nguyen et al., 2002; Kondo et al., 2003; Cimmino et al., 2015; Turcan et al., 2018). Specifically, the aforementioned epigenetic status is characterized by a bivalent chromatin pattern: histone H3 trimethylated at Lys27 (H3K27me3), a repressive trait, plus dimethylated H3K4 that is active mark (Ohm et al., 2007). On the basis of these observations, several theories regarding tumor initiation were described postulating a crucial role of epigenetics, such as the “Epigenetic Priming model” reported by C. Vicente-Duenas et al. (2018), based on the presence of a cause/effect ratio between the epigenome and genetic alterations. Depending on the SC pre-existing epigenome, the oncogenic hit is a fundamental occurrence that is necessary only in the early stages of tumor initiation reshaping the epigenetic profile of stem cells without inducing any phenotypic modification. This theory is based on numerous observations that suggest that these changes, regarding the epigenetic modification on target genes, remain in a primed state and downstream effects persist even when the oncogene is no longer expressed by the cells during later stages of the tumor pathology. The epigenetic changes thus introduced give rise to an aberrant differentiation program, which ultimately leads to tumor formation. Moreover, by comparing mutational background and epigenetic profiles of healthy vs. tumor tissues, Direna Alonso-Curbelo et al. (2021) demonstrated that tissue damage and oncogene mutations determine a series of epigenetic changes which drive cancer initiation only in tumor tissues while they are not occuring during the physiological processes of tissue regeneration in healthy counterparts. For instance, this epigenetic state early following tissue damage has been reported to induce a dysregulation of survival-associated signaling pathways, thus leading to tumorigenesis in pancreatic cancer patients.
Epigenetic changes in tumor initiation: Histone modifications and chromatin remodeling
The aim of this paragraph is to highlight the pivotal role of epigenetics, above the variations at the genetic level, in CSC determination and the subsequent tumor initiation. The epigenetic switch, observed in the previously reported studies, is characterized by significant variations in the level of chromatin and genes that regulate the structure and accessibility of chromatin, and in the methylation profile of specific genes. All these changes are fundamental for the understanding of the mechanisms underlying tumor initiation. However, the epigenetic alterations that determine CSC profile are still to be completely defined and represent a challenge in understanding cancer biology. In the recent years, epigenetic mechanisms underlying the switch between NSCs to CSCs have been reported as the basis for CSC cellular plasticity, representing an important focus of study to determine the origins of tumor heterogeneity and the possibility of making reversible the acquisition of a transformed phenotype (Kumar et al., 2022).
Several works, through the use of advanced techniques, focused the attention on both genomic and epigenomic landscape, genes related to the regulation, and the structural alteration of epigenetics. The presence of mutations that give stem cells self-renewal aberrant properties is related to the presence of chromosomal rearrangements. Histone modification represents the most studied epigenetic modification. Histone modification induces structural and functional alteration of cellular phenotype. Histone proteins governed DNA tridimensional structure and each of these modifications is implicated and covers pivotal steps of tumor formation and evolution (Feinberg, 2018). In particular, numerous histone modifications generate epigenetic alterations implicated in tumorigenesis, mainly modulating the gene expression pattern of the SC compartment. Two are the main methylase enzymes involved in the early step of tumorigenesis: PRMT1, especially involved in leukemia SC transformation (Pereira et al., 2010), and SMYD3, that maintains self-renewal and tumorigenicity ability of gastric CSCs (Wang et al., 2018). Likewise, aberrant histone demethylation is responsible for tumor initiation process. KDM3A responsible for demethylation of histone H3 lysine 9, when overexpressed, increases the expression of the Wnt/β-catenin pathway inducing colorectal CSC determination and colorectal cancer tumorigenesis (Wang et al., 2019). Moreover, KDM1A and KDM1B enzymes were found overexpressed in several solid and hematopoietic tumors collaborating in the early step of the tumorigenesis process (Lim et al., 2010; Crea et al., 2012; Vu et al., 2013; Hu et al., 2016).
Of particular importance seems to be the gene KMT2A/MLL encoding for a methyl transferase that regulates the changes at the chromatin level. The formation of fusion proteins, as a result of chromosomal rearrangements at the level of the aforementioned gene, induces the formation of SCs that have “degenerated” into numerous solid tumors and acute myeloid leukemia. Cohesins represent a family of proteins that regulate the definition of three-dimensional structure of chromatin. The degeneration of this family of proteins determines, through significant chromatin changes, the transcriptional profile definition related to neoplastic transformation. Mutation on this protein machinery that govern the tridimensional chromatin structure, have been positively correlated with a degenerated remodeling at the chromatin level and with an increased expression of stemness-related genes in SCs. All these changes are linked to the leukemia SCs arising (Mazumdar et al., 2015) and the degenerated activation of signaling pathways, linked with tumor initiation, such as SFN5-Nanog and SWI/SNF (Wilson and Roberts, 2011). These events have been mostly characterized in liquid tumors, in which CSC model represents a fundamental paradigm. However, many epigenetic alterations have also been observed in solid tumors. Histone modification is strongly represented in cancers that have been found particularly enriched in the CSC compartment. The loss of H3K27me3 induces the expression of stem-related genes such as SOX9, LGR5, ASCL2, OLFM4 and EPHB in colorectal cancer (Lu et al., 2020). Glioblastoma is a tumor specifically studied for the rich stem component, about one-third of glioblastomas of pediatric type are characterized by gain of function at the histone H3 level. In particular, the most altered gene appears to be H3F3A, which causes Polycomb repressive complex 2 (PRC2) inhibition. The result is the reduction of the trimethylated form of histone, which makes the chromatin less accessible. This observation clearly illustrates how chromatin plays a fundamental role in defining a transformed profile of CSCs (Wainwright and Scaffidi, 2017).
Likewise, PRC1 induces histone modification by a different mechanism involving monoubiquitination of histone H2A at lysine 119 (H2AK119Ub1). The result of this epigenetic alteration is the inhibition of senescence and the promotion of tumorigenesis through INK4a/ARF repression (Yap et al., 2010). Polycomb group proteins, including PRC1 and PRC2, tightly regulate the methylation profile of CSCs in colorectal and bladder cancers (Nguyen et al., 2002; Kondo et al., 2003). Proteins involved in establishing and maintaining DNA methylation have also been identified as drivers for CSC formation. Compelling evidence suggests that hypermethylation of CpG islands is an early event in cancer development and, in some cases, may occur at SC level contributing to neoplastic transformation. Hypermethylated CpG islands leave a molecular imprinting on cancer cells and can be used as molecular marker to study the evolution of the different epigenetic profiles during tumor growth and progression. DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) and methylcytosine dioxygenases (TET1 and TET2) regulate the methylation status of CpG regions and the occurrence of mutations in these genes may ultimately interfere with protein activity. Several studies show the correlation between the specific amino acid alteration in DNMT3A and a specific gene expression profile (Cancer Genome Atlas Research et al., 2013). For example, most DNMT3A inactivating mutations which are responsible for the malignant transformation of leukemia stem cells (LSCs) have been identified in hematopoietic tumors. Decreased expression of TET proteins have been found in many tumors, suggesting their role in epigenetic stability. Loss of function of TET2 leads to a hypermethylation of target genes and increase the mutation rate of hematopoietic stem cells (HSCs) (Pan et al., 2017). TET1 is mainly expressed in HSCs and plays an important role in SC epigenetic profile maintenance, avoiding DNA hypermethylation. Mutations in TET1 and the subsequential TET1 protein downregulation lead to 5hmC loss and to malignant transformation of HSCs (Cimmino et al., 2015). IDH1 and IDH2 retain a dehydrogenase activity that has been found altered in many solid tumors resulting in a pro-tumorigenic function. Mutations in proto-oncogene IDH1 and IDH2, are usually found in glioma and acute myeloid leukemia (AML), resulting in a hypermethylated profile that confer a selective advantage to the mutated subpopulation of cells, in term of growth rate and stemness capability (Turcan et al., 2018). Abundant genes that normally suppress tumor growth in normal cells are hypermethylated, such as RASSF10 in kidney cancer, SIX3 in glioblastoma, CDKN2A and PTEN in melanoma. Hypomethylation of several genes may also represent another fundamental step in tumor initiation, including LY6K in glioblastoma, SLC34A2 in papillary thyroid carcinoma (PTC), and RBBP6 in colorectal cancer.
Of note, the addition of acetyl group on H3/H4 modifying chromatin configuration allows the interaction with transcription factors (Morrison and Thakur, 2021). Multiple enzymes are responsible for catalyzing the addition and removal of acetyl groups, including HATs and HDACs respectively (Lu et al., 2020). The mechanism of histone acetylation on H3/H4 facilitates a tight packaging of chromatin structure (Liu et al., 2017a). For leukemia SCs malignant transformation modification on the acetylation profile is strictly related to the activity of CBP/p300 by the interaction with c-Myb resulting in the induction of acute myeloid leukemia (Pattabiraman et al., 2014). Moreover, leukemia SCs are characterized by the acetylation status of AML1-ETO fusion protein by p300 enzyme (Wang et al., 2011). Another HAT enzyme involved in the transformation process and in the maintenance of malignant phenotype is the fusion protein MOZ-TIF2 increasing the expression of CSF1R (Aikawa et al., 2010). Similarly, HDAC collaborates in the initiation step of several solid tumors. In liver CSCs, HDAC3 is overexpressed compared to normal stem compartment and study conducted by genetic ablation of this enzyme showed its critical role for CSC renewal ability. In fact, HDAC3 knock-out impairs sphere and clone formation and reduces the expression of stem markers (Liu et al., 2013). Furthermore, in colorectal and neural SCs SIRT-1 is required for the maintenance of cell survival and stem phenotype. SIRT-1 deficiency was demonstrated to impair the tumorigenic ability of the aforementioned CSCs (Chen et al., 2014; Lee et al., 2015a).
Epigenetic changes in cancer initiation: Role of long non-coding RNAs and RNA modifications
LncRNAs are involved in the regulation of stem-like phenotype in several tumors. Many of these ncRNAs play a pivotal role in determining a malignant transformation on progenitor/stem cell tissue, determining the CSCs typical properties (Schwerdtfeger et al., 2021). Lnc-PVT1 is particularly correlated with HCC development and has been found to induce CSC transformation, by stabilizing NOP2 protein (Wang et al., 2014). Linc00617 is a nuclear lncRNA that binds the promoter of SOX2 and, by epigenetic reprogramming, increases the transcription rate, enhancing self-renewal and promoting stemness in breast cancer cells (Li et al., 2017). Compelling evidence suggests that lncRNAs play a pivotal role in the molecular processes required for determining the stem phenotype (Castro-Oropeza et al., 2018). Moreover, lncRNAs play a key role in the initiation and progression of pancreatic ductal adenocarcinoma (PDAC). Recent studies have shown that these RNAs play a relevant role in the maintenance of CSC phenotype. MALAT-1, a known oncogenic lncRNA, was able to promote stemness and increase the number of pancreatic CSCs, which lead to increased tumorigenicity in vivo, probably through the regulation of SOX2/SOX9 (Zeng et al., 2018). In HCC, lncTCF7 is significantly upregulated, increases CSC survival, recruits and activates Wnt pathway components (Wang et al., 2015). Furthermore, HOTAIR is another lncRNA involved in the establishment of CSC properties (Deng et al., 2017). HOTAIR interacts mainly with miRNAs influencing tumor initiation and development (Cantile et al., 2021). In particular, HOTAIR blocks the inhibitory effect of mir-34 on the JAK/STAT pathway, thus activating this crucial stemness-related signaling pathway (Deng et al., 2021). Moreover, HOTTIP is an important epigenetic regulator of CSC phenotype acquisition and stabilization. HOTTIP is a RNA binding protein that activates the HOXA9 transcription factor, which in turn leads to Myc signaling activation and stemness profile determination in PCSCs (Melendez-Zajgla and Maldonado, 2021). HOTTIP drives HOXA9 transcriptional profile even in Leukemia CSCs, activating downstream targets mostly related to Wnt/β-catenin signaling (Luo et al., 2022).
In addition to the role of lncRNAs in the initiation step of tumorigenesis, further epigenetic alterations involve structural and functional RNA alteration through several biochemical-related modification processes (Zhao et al., 2020). The methyltransferases and demethylases recognize, bind and change RNA methylation status influencing the stability, the post-translational modification and the splicing variants reflecting on protein expression (Xie et al., 2020). For example, down-regulation of METTL3 and METTL14 by increasing RNA N6—methyladenosine promotes the survival and the self-renewal ability of glioblastoma SCs thus leading to the acquisition of a transformed phenotype (Cui et al., 2017).
Role of epigenetic alterations induced by mechanotransduction on cancer stem cells
Mechanotransduction is a biological process through which cells are able to convert mechanical stimuli in biochemical signals. Of note, altered biophysical forces derived from surrounding tumor microenvironment (TME) could promote EMT process and cancer stemness through the alteration of epigenetic signatures. Accordingly, epigenetic enzymes such as histone modifiers have been reported to be regulated by an altered mechanotransduction (Veerasubramanian et al., 2020). The origin and maintenance of CSCs is finely regulated by the mechanotransduction proprieties of extracellular matrix (ECM) through a great variety of cell surface receptors and the physical interaction of CSCs with their surrounding ECMs. Several emerging evidence support the idea that mechanical inputs could epigenetically regulate CSC origin, maintenance and behavior. Tan and colleagues showed that matrix softness regulates CSC maintenance via H3K9 demethylation resulting in increased expression of Sox2 in melanoma CSCs. These findings confirmed the link between matrix softness-induced epigenetic alterations with self-renewal and survival properties of CSCs (Tan et al., 2014). Moreover, the methylation in the promoter region of the oncosuppressor RASSF1A is associated with the constitutive nuclear accumulation of YAP1 and high expression levels of prolyl 4-hydroxylase alpha-2 (P4HA2) which sustain collagen deposition. The elevated collagen deposition induces ECM stiffness, which drives a stem-like reprogramming and promotes the metastatic potential of CSCs in lung adenocarcinoma (Pankova et al., 2019).
Epigenetic alterations critical for cancer stem cell maintenance
CSCs derive from NSCs in a complex multistep process characterized by both genetic and epigenetic alterations. Epigenetic lesions are a multitude and different in their own nature: they are linked to a modification of the structure and function of the genome or can be associated with limitless uncontrolled cell growth and, generally, to the acquisition of the phenotypic hallmarks of the malignant CSCs. Basically, epigenetic regulations are a primary mechanism which define the SC identity, notably studying CSCs, they show how epigenetically different they are from the healthy counterpart.
Alteration of the epigenetic code, particularly regarding the expression of proteins involved in writing or reading post-translational modifications represent two mechanisms that induce cancer formation. Moreover, carcinogenesis is characterized by epigenetic modifications including change in methylation patterns of cytosines in DNA, modifications of the proteins that bind to DNA, and the nucleosome positioning along DNA. The epigenetic status follows a well-balanced homeostasis in normal cells, but it results strongly altered in many ways in CSCs. The principal epigenetic alterations responsible for CSC maintenance are described in Table 2.
Alterations in epigenetic regulators as histone modifying enzymes in cancer stem cells
The variety in histone modifications showed a deep complex scenario: Several coexisting histone modifications induce activation, and some of them induce repression. Importantly, these modification patterns are not static but they are a fluid and dynamic changing in cellular context. Furthermore, the activation and repression induced by histone modifications are not necessary mutually exclusive, as announced by ‘‘bivalent domains.’’ The resulting influence that one or more histone modifications display on cell fate is termed ‘‘histone crosstalk,’’ and recent evidence would suggest that crosstalk have a great biological significance, particularly in cancer (Esteller, 2007; Lee et al., 2010). Histone methylation involves mainly the K and R residues and these methylations represent chemical marks that serve as binding sites for histone readers (Stallcup, 2001). K and R methylation can occur on both histones and non-histone proteins. Lysine methylation is directly associated to gene activation or repression. For instance, while histone H3 lysine 4 (H3K4), histone H3 lysine 36 (H3K36), and histone H3 lysine 79 (H3K79) induce gene activation, histone H3 lysine 9 (H3K9) and histone H3 lysine 27 (H3K27) lead to gene repression. In many cancers, epigenetic modifications are associated not only with the malignant transformation of NSCs into CSCs, but also influence their stemness phenotype and promote tumor progression. Accordingly, PRMT5 is an arginine methyltransferase, which guides both in vitro and in vivo breast CSCs (BCSCs) proliferation and self-renewal via the transcription factor FOXP1. Mechanistically, PRMT5 is recruited on the FOXP1 promoter and catalyzes Histone H3 R2 Dimethyl Symmetric (H3R2me2s), which in turn induce the recruitment of SET1 and favors the H3K4me3 with subsequent stemness genes expression (Chiang et al., 2017). Similarly, brother of the regulator of the imprinted site (BORIS) regulates CSC-like properties in liver cancer via OCT4 expression. Particularly, BORIS overexpression facilitates its binding on the OCT4 promoter and it has been associated with high levels of H3K4me2 (Liu et al., 2017b). Interestingly, methylation alteration was found in inflammatory disease model, particularly EZH2 catalyzes trimethylation of histone 3 lysine 27 (H3K27me3) is critical for ameliorate the intestinal immune regulation during inflammatory bowel disease (IBD) (Zhou et al., 2019). Accordingly, Kelly et al. (2018) identified a gene signature showing iperactivation of pathways involving immunoregulation, cell survival and metabolism associated to altered levels of H3K4me3, that correlated with worst prognosis in IBD patients.
The initial idea that histone lysine methylation was a highly durable, static modification has now been confuted by the identification of eight classes of lysine demethylases (KDM1-8). Depending on which lysine residue is modified, KDMs can regulate transcriptionally, by activating or repressing, both oncogenes and tumor suppressors. The KDM7 subfamily has catalytic activity at lysine residues on histone tails. KDM7A and KDM7B demethylase H3K9me2/me1, H3K27me2/me1 and H4K20me1, while KDM7C only demethylases H3K9me2 (Lee et al., 2015b; Pappa et al., 2019). KDM7A maintains low levels of H3K9 and H3K27 methylation by guaranteeing viability of glioblastoma SCs (Mallm et al., 2020). Accordingly, some studies show how KDM2B is characterized by demethylase activity at H3K4me3 KDM2B has an important role in leukemia SC maintenance and proliferation of glioblastoma SCs (Frescas et al., 2007; He et al., 2011). Of note, the knockdown of KDM2B in glioblastoma cells decreases the levels of H3K36me2, reduces the number of proliferating cells and increase DNA damage accumulation, supporting KDM2B role in glioblastoma SC maintenance (Staberg et al., 2018). The histone demethylases KDM6A/B are highly expressed in glioblastoma SCs and via NOTCH pathway induce plasticity in CSCs contributing to tumor progression and relapse (Liau et al., 2017).
The lysine residue acetylation represents a histone modification involved in DNA repair machinery, transcription and chromatin structure modeling. Biologically, acetylation induces a neutral charge on lysine’s positive residues by leading to a reduction of electrostatic interaction between positive histones and negatively charged DNA. For this reason, histone acetylation is linked to a more ‘‘open’’ chromatin conformation. Acetylation is regulated by the competing activities of two families of epigenetic enzymes, the histone lysine acetyltransferases (KATs) and HDACs. Specifically, KATs are divided in two groups: 1) type-B, which are predominantly cytoplasmic, and 2) type-A, which are primarily nuclear. The expression of KATs and HDACs impairs the structure and integrity of the genome of NSCs, so these two groups of histone modifiers drive CSC transformation in several cancers.
The constitutively expressed acetyltransferase fusion protein MOZ-TIF2 is able to interact with transcription factor PU.1 by inducing the transcriptional activation of CSF1R. In an in vivo model, PU.1\MOZ-TIF2 interaction drives the upregulation of CSF1R and it leads to maintenance of LSCs (Aikawa et al., 2010). Upon chronic Cr(VI) exposure, lung cancer cells display a glycolytic shift, which is directly associated with an upregulation of the proto oncogene c-Myc expression. Moreover, this glycolytic shift in Cr(VI)-transformed cells promotes an increased production of acetyl-CoA and induces histone acetylation, thus enhancing CSC-like properties and their tumorigenic capacity (Clementino et al., 2020).
A pro-tumorigenic role of HDACs has been demonstrated in a model of non-small cell lung cancer (NSCLC), resistant to cisplatin (CDDP)-based therapy. In vivo studies confirmed that CDDP resistant tumors display high expression levels of CSC-associated transcription factors. CDDP-enriched CSCs show an aberrant activation of the C/EBP-β/TRIB1/HDAC1/p53 axis. Specifically, Wang et al. (2017) hypothesized that C/EBP-β induces the transcription of the protein kinase TRIB1, which cooperates with HDAC1 promoting p53 acetylation and activation. This axis is involved in CSC enrichment and chemotherapy resistance induced by CDDP in NSCLC. Several studies showed high expression levels of HDACs in CSCs. In particular, increased HDAC3 levels has been observed in liver CSCs and positively correlated with Nanog and CD133 expression levels (Liu et al., 2013). The HDAC SIRT-1 is highly overexpressed in CD133+ colorectal stem-like cells (Chen et al., 2014). Moreover, SIRT-1 was found to maintain, in cooperation with CRL4B complex, the stemness features of pancreatic CSCs (Leng et al., 2021).
Alterations in epigenetic regulators as chromatin remodelers in cancer stem cells
Basic biological events such as transcription, replication and DNA repair depend on DNA accessibility to the enzyme complex implicated in each process. ATP-dependent chromatin remodelers are able to define DNA accessibility by acting on nucleosomes for repositioning, ejecting, or modifying their structure. Eukaryotic cells are characterized by four families of chromatin remodelers, which are classified on the similarities and differences of the ATPase subunits, including SWI/SNF, imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), and INOsitol requiring 80 (INO80). The epigenetic deregulation can derive from chromatin remodeling that results altered in various ways in cancer: redistribution or mistargeting; down or over expression of subunits; loss-of-function mutations in SWI/SNF-subfamily remodelers which is linked to reduction of DNA accessibility at promoters and enhancers of tumor-suppressor or other genes; or gain-of-function mutations in SWI/SNF-subfamily remodelers which cause high dynamic nucleosome mobility and DNA accessibility at oncogenes or genome-wide. This scenario is frequently observed in CSCs and other diseases. For example, Brg1 is a chromatin-remodeling regulator for maintenance of intestinal CSCs. Specifically, Brg1 plays a crucial role in intestinal CSCs in mice by affecting apoptosis and improving cell survival and SC subpopulation in human colorectal cancer cells (Yoshikawa et al., 2021). Similarly it has been demonstrated that MUC1-C, an oncogenic protein, interacts with SWI\SNF family complex PBAF by balancing ROS levels and pluripotency gene expression in prostate CSCs (Hagiwara et al., 2021). Therefore in silico analysis, the chromatin remodeler HELLS was able to form a complex with the transcription factors Myc and E2F3 by promoting stemness in glioblastoma cells (Zhang et al., 2019). Besides, it has been found how the SWI/SNF complex activates transcriptionally the oncogenes AR/FOXA1 expression in prostate cancer (Xiao et al., 2022).
Taken together this data indicate that both histone modifications and chromatin remodeling alter the gene expression of oncogenes and/or tumour suppressor genes thus affecting genome integrity and unbalancing NSCs homeostasis in favor of CSCs transformation.
DNA methylation in cancer stem cells maintenance
DNA methylation is a covalent chromatin modification regulating genome stability and gene expression. The lack of DNA methylation regulation mechanisms causes several diseases, including cancer. CSCs present an aberrant DNA methylation, mainly occurring in CpG islands. Accordingly, several tumor suppressor genes (TSGs) are hypermethylated in cancer and, for this reason, silenced by promoting cancer progression. Changing of DNA methylation pattern (hypomethylation or hypermethylation) in cancer is often associated with an aberrant expression of DNMTs (1, 3A, and 3B). Specifically, DNMT family has a crucial role for maintaining the CSC statement (Wainwright and Scaffidi, 2017). It has been shown that DNMT1 ablation could decrease proliferation and tumorigenesis in lung CSCs, besides it leads to the extinguished of CSCs from tumor bulk by enhancing apoptosis and differentiation (Toh et al., 2017; Gao et al., 2018). Accordingly, DNMT1 is able to regulate BEX1 in liver cancer, by controlling self-renewal and maintenance of liver CSCs via Wnt/β-catenin signaling pathway activation (Wang et al., 2021). It has been found that DNMT1 expression is overexpressed in breast cancer, and mammary gland-specific DNMT1 deletion protects in vivo mouse model from breast cancer tumorigenesis by reducing BCSC pool. Through genome-scale methylation approach, it has been identified ISL1 as a direct DNMT1 methylation target. Specifically, ISL1 is hypermethylated and downregulated in breast tumors and CSCs subpopulation (Pathania et al., 2015).
DNMT3B is involved in the increasing methylation of CSCs guaranteeing an undifferentiated state (Shukla and Meeran, 2014). Therefore, mutations in DNMT3A also induce the maintenance of CSC subpopulation in cancers. These DNMT3A mutations directly activate CSC proliferation pathways and increase chemoresistance (Wainwright and Scaffidi, 2017). In accordance, DNA methylation controls the expression of surface markers of CSCs such as CD44 and CD133 for CSC maintenance, as well as ABC transporters, involving drug efflux into CSCs promoting chemotherapy resistance (Crea et al., 2009). Moreover, it has been found that DNMT3B/OCT4 axis expression induces sorafenib resistance and stem-like properties in HCC via IL-6/STAT3 pathway regulation (Lai et al., 2019).
A deeper analysis of DNA methylation patterns in CSC population is crucial for a better understanding of the molecular mechanisms CSC-driven associated to tumor relapse and poor prognosis, and will lead to the identification of more specific DNA methylation inhibitors.
Long non-coding RNA role in cancer stem cell maintenance
NcRNAs are a subgroup of RNAs that do not translate into protein but control gene expression at the post-transcriptional level, by considering them as an important epigenetic regulator. Particularly, lncRNAs (ncRNAs over 200 nucleotides in length) have a critical function in maintaining CSC populations through stemness genes regulations and, generally, by activating pathways related to SCs (Chen et al., 2017).
The expression of HOTAIR is closely linked with advanced tumor stage, metastasis, and poor prognosis in a variety of human cancers (Min et al., 2017). HOTAIR’ pro-metastatic role is developed by the targeting of polycomb repressive complex 2 (PRC2) and by the downregulation of metastasis repressor genes (Deng et al., 2017). Moreover, HOTAIR is upregulated in CSC populations of breast and colon cancer. Particularly, in colon CSC subpopulation (CD133+/CD44+) shows overexpressed levels of HOTAIR, suggesting that HOTAIR regulates the acquisition of stemness. In breast cancer, HOTAIR is able to suppress the tumor inhibitor miR-7, by upregulating the expression levels of c-Myc, TWIST and miR-9 and maintaining the BCSC pool (Zhang et al., 2014a). In addition, HOTAIR was found to unpair the association of P300, CREB and RNA pol II to the SETD2 promoter region by promoting the growth of human liver CSC (Wang et al., 2016).
Additionally, Lnc34a has been found overexpressed in colorectal CSCs. Mechanistically, Lnc34a recruits both DNMT3A and HDAC1 on MIR34A promoter, by inducing the methylation and deacetylation of the promoter. Both the epigenetic perturbations blocked the MIR34A gene expression and enhanced CSC proliferation (Amirkhah et al., 2019).
MALAT-1 is a lncRNA upregulated in glioma SCs, BCSCs and PCSCs. It enhances CSC properties, such as proliferation, self-renewal, colony formation and invasion in vitro (Jiao et al., 2014; Jiao et al., 2015; Han et al., 2016; Zeng et al., 2018; Amirkhah et al., 2019). Mechanistically, MALAT-1 may represent a molecular sponge for miRNAs. Particularly, MALAT-1 binds miR-200c, which induces an upregulation of ZEB1, an important epithelial–mesenchymal transition (EMT) transcription factor (Korpal et al., 2008; Pa et al., 2017).
Similarly, the lncRNA NORAD was found to be upregulated in pancreatic cancer tissues and cells by acting as a molecular sponge. Accordingly, the lncRNA NORAD could selectively bound to miR-202-5p, by inducing the expression of the miR202-5p target gene ANP32E, which is linked to self-renewal of PCSCs and proliferation in vitro, as well as enhancing tumorigenesis of PCSCs in vivo (Ma et al., 2021).
The lncRNA H19 controls the maintenance of the CSC pool. Accordingly, microarray analysis showed that high levels of H19 are positively correlated with the overexpression of a cluster of transcriptional factors stemness-related such as Sox2, Oct4, and Nanog in acute lymphoblastic leukemia cells (Singh et al., 2020). In addition, BCSCs (ALDH1A1+; CD44+/CD24−) showed high H19 expression levels, induced by miR-675, supporting a role for both H19 and miR-675 in the enrichment of BCSC compartment (Peperstraete et al., 2020). LncRNA H19 plays a role in regulating CSC maintenance also through its function as a miRNA sponge. Of note, it has been found that H19 and LIN28 cooperate in promoting CSC self-renewal. Increasing levels of LIN28 are regulated by H19 through the sponging of miR-196b (Ren et al., 2018). Similarly, H19 can also sponge miR-3126-5p to increase the expression of ERβ receptor in PTC by promoting CSC-like properties in PTC (Li et al., 2018). Furthermore, H19 regulates miR-let-7 by controlling self-renewal in BCSCs (Peng et al., 2018).
The novel studies of lncRNA biology in human disorders are opening new scenarios for the use of lncRNA as disease biomarkers and/or therapeutic targets. However, to increase our current knowledge, more methodological improvements are necessary in order to study lncRNA structure and unravel the spatial, developmental specificities, and biological networking of lncRNAs.
Epigenetic reprogramming during inflammation in cancer stem cells
Chronic inflammation has been correlated to several cancers and it contributes to different steps during carcinogenesis (Di Franco et al., 2021). A plethora of inflammatory cytokines, interleukins, interferons, transforming growth factors, chemokines, and adhesion molecules have been associated with chronic inflammation. These inflammatory mediators are directly reported to regulate aberrantly the transcriptomic and the epigenetic machinery, particularly DNA methylation and histone modifications in cancers, driving the pathogenesis of tumor and by fueling CSC pool. For instance, TGF-β regulates DNTMs transcription and activity, leading to radical changes in DNA methylation for the acquisition of stemness phenotype by ovarian cancer cells (Cardenas et al., 2014). Similarly, TGFβ regulates the recruitment of epigenetic enzymes such as DNMTs and HMTs (EHMT2 and SUV39H1) to the CDH1 gene promoter by inducing endothelial cell transformation, which seems to contribute to the development of BCSCs (David and Massague, 2018). The TGFβ pathway through Smad2/3 induces EMT, which is directly linked to CSC generation. Specifically, the arginine methylation of Smad7 by PRMT1 induces TGFβ-induced EMT and CSC generation (Katsuno et al., 2018). An alternative important mechanism, which involves the TGFβ/Smad-induced EMT in CSC generation, is characterized by two double-negative feedback loops: Zeb/miR-200 and Snail-miR-34. The two double negative loops drive both EMT and CSC generation (Brabletz and Brabletz, 2010; Siemens et al., 2011; Zhang et al., 2014b). Therefore, the Akt/miR-200/E-cadherin axis controlled by TGFβ pathway induces both EMT in mammary epithelial cells and CSC generation (Iliopoulos et al., 2009).
IL-6 mediated inflammation regulates cancer cell stemness. Mechanistically, it exists a specific loop in which p53 deletion causes demethylation of the IL-6 promoter by activating IL-6 signaling. Subsequently, the hyperactivation of IL-6 induces the overexpression of DNMT1, which, in turn, methylates the promoter of the p53 gene by initiating IL-6\p53\DNMT1 autocrine loop (Hodge et al., 2005; D'Anello et al., 2010; Liu et al., 2015). This autocrine loop drives cancer cells to a stem-like phenotype acquisition via epigenetic reprogramming (D'Anello et al., 2010; Liu et al., 2015). Similarly, IL-6 induces hypermethylation of the promoter of the miR142-3p gene by repressing its expression and promoting cell stemness and invasiveness in glioblastoma (Chiou et al., 2013). Interestingly, IL-6 has also a critical role in the conversion of non-CSCs into CSCs (Iliopoulos et al., 2011). Moreover, the IL-6/STAT3/PTEN/NF-κB inflammatory axis is preferentially activated in CD44+CD24− stem-like breast cancer cells compared with other tumor cell types such as cancer associated fibroblasts and inflammatory cells (Iliopoulos et al., 2011; Marotta et al., 2011).
Epigenetic therapy using stem cells for regenerative medicine
A clear understanding of epigenetic processes could be crucial for the full control of SCs and their use in regenerative medicine. Excellent progress in the biomedical field leads to the use of SCs for the regeneration of organs and tissue that have lost their physiologic functions and mechanisms. Epigenetic mechanisms may drive the cellular reprogramming to convert somatic cells into induced pluripotent stem cells (iPSCs), which can be directed to differentiate in specific cell types.
The employment of SCs for patients that are affected by diseases such as type 1 diabetes mellitus, hematological malignancies or cancer represents a new effective therapeutic strategy. Although pharmacological therapy in type 1 diabetes mellitus was excellent to restore the chronic disease and avoid complications, the possibility to re-generate new islets by using embryonic SCs (ESCs) is a promising approach (Chen et al., 2020). Legoy et al. (2020) demonstrated that the human- iPSCs (h-iPSCs) are able to generate insulin-producing β-cells. Proteomic analysis of the transplanted cells revealed that in vivo microenvironment is predominant to drive to islet profile, selecting specific hormones through regulation of epigenetic factors and generating human pancreatic progenitors. This study highlighted the capacity of hiPSCs, under particular conditions, to differentiate in β-cells.
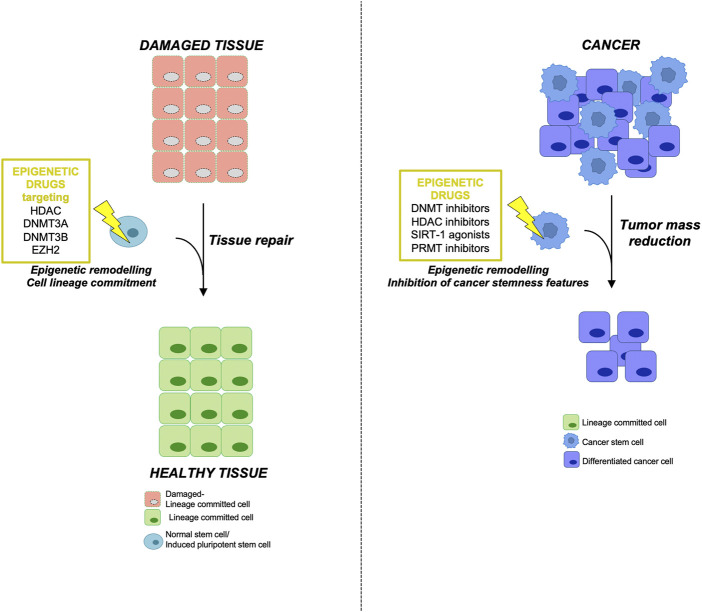
Although the use of SCs is wrapped by various problems such as the isolation and expansion in vitro along with ethical issues, there are promising and compelling results on the use of SCs as a possible therapeutic approach for regenerative medicine (Figure 2).
FIGURE 2.
Epigenetic therapy using NSCs in regenerative medicine and targeting CSCs in tumors (left) Epigenetically engineered adult normal stem cells (NSCs) or induced pluripotent stem cells (iPSCs) represent an innovative and promising tool in regenerative medicine. Adult stem cells deriving from damaged tissue or iPSCs could be reprogrammed using the indicated epigenetic drugs in order to obtain a specific lineage commitment, thus allowing tissue repair and regeneration. (right) On the other hand, the use of epigenetic inhibitors in cancer therapy targeting the aberrant epigenetic modifications responsible for CSC initiation and maintenance may lead to a reduction of tumor mass by inhibiting the stemness features of CSC subpopulation.
Regenerative medicine is used to repair or replace damaged human tissue by engineering adult and embryonic SCs. The concepts of regenerative medicine hold the potential to help in healing previously irreparable tissues or correcting genetic defects responsible for diseases. Bioengineering in vitro human SCs, towards evaluating genetic variation responsible for the disease and identifying new targets for therapy, strikes a new direction for regenerative medicine as a potential approach for cell-based therapies (Bailey et al., 2014; Mendelson and Frenette, 2014). Epigenetic control of gene expression is a trait heritable during cell divisions and has emerged as a key mechanism defining the cell lineage commitment and influencing the evolution of many diseases (Choi and Friso, 2010; Hirabayashi and Gotoh, 2010). Adult and embryonic SCs represent an excellent tool to understand human development and organogenesis and may be used to treat several diseases by manipulation of environmental signals and intracellular pathways influencing cell proliferation, self-renewal ability, and cell lineage differentiation (Audet, 2004). The phenotypic and functional changes associated with the commitment of SCs into progenitor precursors and then to terminally differentiated cells are the results of remarkable changes in gene expression patterns. To a large extent, a unique epigenetic program orchestrates the promotion and maintenance of the gene expression profile during differentiation involving the silencing of self-renewal genes and the activation of cell type-specific genes (Wu and Sun, 2006).
Engineered SCs could be used to favour tissue repair upon several pathological conditions and for treating multiple inherited and degenerative diseases such as hematopoietic and immune system disorders, diabetes, Parkinson’s and Alzheimer’s diseases. Moreover, bioengineered SCs are exploited also as a cell-based therapy for the treatment of aggressive and recurrent cancers (Mimeault et al., 2007). Yamanaka and colleagues demonstrated that terminally differentiated cells could undergo unnatural conversion into iPSCs (Takahashi and Yamanaka, 2006). Oct4, Sox2, Klf4, and cMyc, ectopically expressed in terminally differentiated cells, cooperate with PRC2 proteins silencing lineage-specific genes and inducing reprogramming of cell identity and function (Pereira et al., 2010). A characteristic trait of this epigenetic reprogramming is the methylation of H3 histone (Mansour et al., 2012). This discovery breaks up the paradigm according to the cellular destiny that may only follow the natural lineage direction recapitulating embryonic development (Cherry and Daley, 2012). Moreover, iPSCs are easier to obtain from patients compared to SCs and for this reason represent an important source for regenerative medicine applications.
Epigenetic mechanisms cooperate in defining the fate of every cell. DNA methylation, histone modifications, and ncRNAs are responsible for gene expression control during embryonic development, and physiological and pathological processes in adults (Wu and Sun, 2006). Engineering SCs or iPSCs, by inducing an epigenetic reprogramming and modifying downstream gene regulatory networks, is a strategy exploited in regenerative medicine to drive cell destiny. Many epigenetic labels have been found as character traits of many neoplastic and pathological tissue. Furthermore, different epigenetics characteristics are strongly correlated to the differentiation status of a particular tissue or cells. Recent publications highlighted the different roles of each specific DNMT during the transition of SCs from quiescence to proliferating state up to differentiation status (Naito et al., 2016). For example, early-stage steps in normal hematopoietic development are associated with hypermethylation and the loss of function of the methylation-responsible enzymes (Gore and Weinstein, 2016).
Interestingly, histone modification, especially the inactivation of the EZH2 subunit, impairs the self-renewal ability of SCs reducing their regenerative potential (Juan et al., 2011). In many adult tissues, epigenetic modifications regulate cellular plasticity in committed cells, contributing to tissue repair machinery. As reported in the literature, differentiated hepatocytes, in response to tissue damage undergo a rewiring of genomic methylome/hydroxymethylome landscapes reverting lineage-committed and contributing to the regeneration of liver parenchyma (Aloia et al., 2019). Epigenetic modifications influence cell signaling but can be tightly controlled by intra and extracellular factors. As an example, Notch signaling is precisely regulated in a time- and space-restricted manner regulating the quiescence status of stellate cells that represent the staminal compartment of muscle tissue (Bjornson et al., 2012). The myogenesis process is characterized by high cellular plasticity and is strictly controlled by the epigenetic process. Hypomethylation and/or hydroxymethylation of some intragenic or intergenic regions of Notch receptors interfere with the terminal differentiation of myoblasts into mature myofibers.
DNA methylation is one of the major repressive systems for the muscle gene. DNMT inhibitor delivery has been proved to be an alternative strategy for patients affected by dystrophic muscles restoring the regenerative ability of stellate cells (Sincennes et al., 2016). Moreover, epigenetic mechanisms are the main regulator of gene expression patterns during the differentiation of MSCs (Ghorbaninejad et al., 2020). Modulation of DNMT3A and DNMT3B activity influences the whole process of underlying bone tissue development. Mouse depleted for DNMT3B die during embryonic development since it was demonstrated to be necessary for proper lengthening and mineralization of both axial and appendicular bones. DMNT3B is reactivated during the regeneration of bone tissue decreasing at the final stages (Xu et al., 2018). Moreover, the alteration of the methylation status in immune cells is involved in the pathogenesis of autoimmune disease. The association between the methylation status of methylation-sensitive genes and the development of the immune thrombocytopenia (ITP) is currently under evaluation in a clinical trial (NCT04100876). In particular, many single nucleotide polymorphisms (SNPs) in the DNMT3A and in the DNMT3B gene may influence catalytic activity of these enzymes and may be used as prognostic markers (NCT04100876).
The growing interest in the role of epigenetics as cause-and/or-effect of several diseases and as a key regulator of human tissue organogenesis and differentiation has led to intensifying research for the development of new more specific drugs. Targeting epigenetic machinery appears to be a promising therapeutic strategy for several diseases such as neurodegenerative disorders and oncology pathologies. One of the most used epigenetic modulators in the clinic for the treatment of epilepsy patients and bipolar disorders is valproic acid (VPA), a HDAC inhibitor that has been prescribed for several years without any severe side effects (Amitai et al., 2015). VPA enhances the activity of the inhibitory transmitter gamma-aminobutyric acid (GABA) reducing its degradation and promoting its synthesis. Nowadays, several epigenetic drugs are under pre-clinical and clinical evaluation (NCT01021449; NCT04608448; NCT02284477). Epigenetic drugs act on chromatin structure by the inhibition of DNMTs and HDACs influencing the time and space of gene expression (Rodriguez-Paredes and Esteller, 2011; Altucci and Rots, 2016). In orthopedic regenerative medicine, epigenetic drugs represent a promising therapeutic approach (van Wijnen and Westendorf, 2019). Several EZH2 inhibitors such as GSK126, UNC 1999, and EPZ005687, inhibit osteoclastogenesis and promote bone regeneration, and are currently in clinical trials to treat patients with osteoarthritis (van Wijnen and Westendorf, 2019; Ball et al., 2022).
Epigenetic drugs in clinical trial targeting the epigenetic modifications sustaining cancer stem cells
Epigenetics is crucial for the organism’s development and is highly responsive to environmental cues (Baylin and Jones, 2011). To date, a link between the microenvironment (diet, exposure to chemicals) and epigenetic alterations associated with pathological conditions has not been assessed. In this regard, epigenetic alterations become attractive targets for the treatment of different cancers and diseases. Moreover, epigenetic modifications, due to their reversible nature, represent potential biomarkers useful for clinical purposes (Nebbioso et al., 2012).
A better knowledge of the exact pattern of epigenetic modifications could help clinicians to identify a personalized therapy. In glioma patients, in fact, the expression of O6-methylguanine-DNA methyltransferase (MGMT), an enzyme involved in alkylating agent-induced DNA damage repair, is predictive of response to the treatment with carmustine and temozolomide. Indeed, in presence of a hypermethylated MGMT promoter, the ability to repair the damage induced by alkylating agents is consequently decreased, resulting in an increased sensitivity to these chemotherapeutic drugs (Yu et al., 2019). Heritable or environmental-induced epigenetic abnormalities are associated with several diseases, and the differences between healthy and pathological tissues allowed the identification of diagnostic and/or prognostic biomarkers. In this regard, FDA approved an early colorectal cancer-screening program, based on the identification of specific DNA methylation patterns of selected gene promoters in non-invasive tissue (e.g., vimentin, or BMP3, septin and NDRG4). Moreover, DNA methylation at the promoter of brain-derived neurotrophic factor (BDNF) is currently being tested for the treatment of autism or depression (Delgado-Morales et al., 2017). In addition to DNA methylation altered profiles, aberrant epigenetic modifications can derive from somatic mutations occurring in the genes codifying for histone-modifying enzymes and/or chromatin remodelers, altering their expression levels and activity, thus predisposing to cancer and diseases (Han et al., 2019). Importantly, the different families of epigenetic enzymes such as readers, writers, and erasers, can be targeted by epigenetic probes/inhibitors to treat different cancer types.
In the following paragraphs we will revise the epigenetic inhibitors in clinical trial for several cancers focusing on the compounds targeting the epigenetic modifications sustaining the initiation and maintenance of CSCs (Figure 2; Table 3).
TABLE 3.
Epigenetic inhibitors targeting epigenetic alterations of cancer stem cells (CSCs).
| Drugs | Target | Cancer | References/NTC |
|---|---|---|---|
| DNMT inhibitors | |||
| Azacitydine | DNMT1 | AML, MDS | NCT01074047 |
| Decitabine | DNMT1 | AML, MDS | NCT00043381, NCT00866073 |
| Disulfiram | DNMT1 | Recurrent glioblastoma, prostate and metastatic breast cancer | Lin et al. (2011), NCT03323346, NCT02678975 |
| Hydralazine | DNMT1 | Ovarian, testis, lung, breast and cervix cancers | NCT00404508 |
| EGCG | DNMT1 and DNMT3A/B | Colorectal, esophageal and prostate cancers | Fang et al. (2003) |
| Guadecitabine (SGI-110) | DNMT1/3 | HCC, AML, MDS, CMML | NCT01752933, NCT02348489, NCT02920008, NCT02907359 |
| SGI-1027 | DNMT1 and DNMT3A/B | Histiocytic lymphoma, Burkitt’s lymphoma, breast and prostate cancer | Datta et al. (2009) |
| HDAC inhibitors | |||
| Tefinostat | Pan HDAC | CTCL, PTCL | NCT02759601 |
| Vorinostat (SAHA) | Pan HDAC | Breast cancer | NCT 04190056 |
| Romidepsin | Class I HDAC | HCC | Barbarotta and Hurley, (2015) |
| SIRT-1 agonists | |||
| EX-527 (SEN0014196 or selisistat) | SIRT-1 | HCC, brast cancer | Gollavilli et al. (2015), Ceballos et al. (2018) |
| Nicotinammide | SIRT-1 | CLL, NMSC | NCT04844528 |
| suramin | SIRT-1 | NSCLC, hormone-Refractory Prostate, adrenocortical cancer | NCT01038752, NCT00002723, NCT00002921 |
| PRMT inhibitors | |||
| GSK3326595 | PMRT5 | Solid tumors, non-Hodgkin’s lymphoma | NCT02783300 |
| LLY-283 | PMRT5 | Breast, gastric, lung, skin, ovarian and hematological cancers | Bonday et al. (2018) |
CSC, cancer stem cell; NTC, national clinical trial; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; HCC, hepatocellular carcinoma; CMML, chronic myelomonocytic leukemia; CTLT, cutaneous T-cell lymphoma; PTCL, peripheral T-cell lymphoma; NSCLC, non small cell lung cancer; CLL, chronic lymphocytic leukemia; NMSC, non-melanoma skin cancer.
DNA methyltransferase inhibitors
An aberrant methylation status has been associated with tumorigenesis, and metastasis formation. Moreover, altered DNA methylation patterns have been correlated with poor prognosis and therapy resistance in cancer patients (Romero-Garcia et al., 2020). Different DNA methylation patterns were observed in colon, prostate, lung, ovarian and breast cancers, particularly represented in NSCs versus CSCs (Pechalrieu et al., 2017).
DNMT inhibitors can reprogram cancer cells by inducing cell death and growth arrest. Decitabine (5-aza-2′-deoxycytidine) and azacitydine (5-azacytidine) are nucleoside inhibitors used for the treatment of AML and myelodysplastic syndrome (MDS) (NCT01074047, NCT00043381, NCT00866073). The FDA-approved guadecitabine, 5-Fluoro-2′-deoxycytidine (FdCyd), and zebularine belong to the next-generation hypomethylating agents (Nepali and Liou, 2021). In particular, guadecitabine (known as SGI-110) promotes enhanced uptake of the active metabolite, decitabine, into the DNA of quickly dividing cancer cells, and is resistant to the action of cytidine deaminase (Issa et al., 2015). SGI-110 is currently being tested for the treatment of advanced HCC (NCT 01752933) and the treatment of AML, musculoskeletal disorders (MSDs) and chronic myelomonocytic leukemia (CMML) in patients refractory to standard therapies (Global phase 3 ASTRAL-1, NCT02348489, ASTRAL-2 NCT02920008, and ASTRAL-3 study NCT02907359). The use of nucleoside DNMT inhibitors is associated with genomic instability and risk of mutagenicity, for this reason, non-nucleoside analogs were developed. Mainly, these inhibitors consist of small molecule agents derived from natural and chemical sources, targeting specifically the catalytic region of the methyltransferases. Among the natural non-nucleoside DNMT inhibitors, EGCG (epigallocatechin-3 gallate), a polyphenol derived from green tea demethylates the promoter of several genes such as p16 (INK4a), RAR (retinoic acid receptor beta), human mutL homologue 1 (hMLH1) in cancer cell lines derived from colon, prostate and esophageal tumors (Fang et al., 2003). Hydralazine is an antihypertensive and vasodilator drug, recently known as a non-nucleoside DNMT inhibitor. This compound can induce cell death in p53-mutant leukemic T cells by a caspase-dependent mechanism (Ruiz-Magana et al., 2016). Hydralazine has been tested in different clinical cancer trials and in combination with valproate and chemotherapy showed clinical benefit in patients with refractory solid tumors (NCT00404508). Another non-nucleoside inhibitor is disulfiram, which can decrease genomic 5-methyl cytosine content, reactivating tumor suppressor genes silenced by hypermethylation, thus leading to a growth arrest of prostate cancer cells (Lin et al., 2011). Disulfiram has been tested in clinical trials for treatment of metastatic breast tumors (NCT03323346) and recurrent glioblastoma (NCT02678975). SGI-021 is a non-nucleoside DNMT inhibitor that exerts a potent anti-proliferative effect against cancer cell lines derived from breast cancer, histiocytic lymphoma, Burkitt’s lymphoma and prostate cancer (Datta et al., 2009).
Histone deacetylases inhibitors
HDACs are considered targets for epigenetic cancer therapy and multiple diseases (Tang et al., 2013; West and Johnstone, 2014). FDA approved four categories of HDAC inhibitors: hydroxamic acids such as SAHA; cyclic peptides; benzamides and short-chain fatty acids (Nepali and Liou, 2021). SAHA, romidepsin, and tefinostat are HDAC inhibitors for class I HDAC and are in several clinical trials in breast (NCT 04190056); CTCL, PTCL (Barbarotta and Hurley, 2015), and HCC patients (NCT 02759601). Citarinostat (ACY-241), is a selective inhibitor of HDAC6, currently in phase Ib study for the treatment of NSLC patients (Awad et al., 2021). SIRT-1 is a HDAC classIII, known as sir-2-like proteins (sirtuin) has been identified in several tumors (breast, colorectal, prostate liver, pancreatic and lung cancer) and promotes cell migration, proliferation and chemoresistance of CSCs (Sun et al., 2013; Santolla et al., 2015; Pinho et al., 2016). A preclinical study demonstrated that SIRT-1-agonists (STACs) could represent a new therapeutic approach for the treatment of pancreatic cancer. Chini et al. (2016), in fact showed that use of STACs induced pancreatic cancer cells death through a process dependent by SIRT-1 and lysosomes. EX-527 (SEN0014196 or selisistat) is the most potent inhibitor of SIRT-1, tested in Huntigton’s disease patients in phase II clinical trial (Sussmuth et al., 2015). This inhibitor displays different biological effects on several tumor cells (Broussy et al., 2020). Broussy et al. showed that combination of two SIRT-1 inhibitors (EX-527 and nicotinamide) on lymphoma cells, induced apoptosis and cell cycle arrest (Broussy et al., 2020). Additional SIRT-1 inhibitors are nicotinamide, thioacyllysine, sirtinol, cambinol, splitomicin, suramin and tenovin (Hu et al., 2014). A clinical trial is ongoing to evaluate the effect of nicotinamide in patients with chronic lymphocytic leukemia (CLL) (NCT 04844528). Different clinical trials tested the effect of suramin in NSCLC (NCT01038752), hormone-refractory prostate cancer (NCT00002723) and adrenocortical cancer (NCT 00002921).
Protein arginine methyltransferases inhibitors
Protein arginine methyltransferases (PRMTs) are enzymes that physiologically catalyze the methylation of R residues on histones. Dysfunction of PRMT is associated with the occurrence and progression of cancer (Mohammad et al., 2019). PRMT5 is a methyltransferase of class II that can catalyze symmetrical dimethylation and monomethylation, and is involved in signal transduction, cellular differentiation and RNA metabolism (Dacwag et al., 2007; Jansson et al., 2008; Bezzi et al., 2013). In cancer, PRMT5 is known to interact with cancer-associated deregulated pathways such as ErbB, FGF/FGFR signaling pathways and, for this reason, is considered a promising therapeutic target for the treatment of cancer (Sheng and Wang, 2016). PRMT5 is highly expressed in gastric cancer, breast cancer, glioblastoma and lymphoma. Different types of PRMT5 inhibitors have been identified. Bonday et al. (2018) showed that LLY-283, a PRMT5 inhibitor, exerted an anti-proliferative effect on several tumor cell lines. To date, a phase I study is ongoing to evaluate the effect of a selective PRMT5 inhibitor for the treatment of solid tumors and non-Hodgkin’s lymphoma (NCT 02783300).
Demethylase inhibitors
Aberrant expression of different demethylases (LSDs or KDM1) and jumonji C (JmjC) groups of N-methyl-lysine demethylases (JmjC KDMs, KDM2–7) were identified in different tumors (McAllister et al., 2016). In particular, JmjC-KDMs depend on 2-oxoglutarate and oxygen availability (Tsai and Wu, 2014). It was shown that JmjC-KDMs are upregulated in hypoxia (Hancock et al., 2015), a common feature of solid tumors. Hoffmann et al. (2012) highlighted the presence of specific JmjC-KDMs, KDM3A, and KDM6B, in hypoxic tumors their expression has been correlated with cancer aggressiveness and development. To date, some compounds are currently been tested in clinical trials to inhibit LSD1 such as INCB059872 (NCT03514407) IMG-7289 (IMG) (NCT02273102), GSK2879552 (NCT02929498), CC90011 (NCT03850067), ORY-1001 (NCT02913443), while the development of JmjC-KDM specific inhibitors is more difficult due to their structural similarity and poor permeability (Morera et al., 2016).
Concluding remarks
Aberrant epigenetic changes contribute to the transformation process of NSCs into CSCs. Specifically, epigenetic modifications sustain and maintain the self-renewal and stemness properties of NSCs, thus ultimately promoting the initiation and maintenance of CSCs by supporting their proliferation, invasiveness, metastatic potential and chemo-resistance capabilities.
Deepening the knowledge about the above-mentioned epigenetic mechanisms, thus identifying the most promising epigenetic druggable targets, would be useful for both the regenerative medicine and oncological settings.
Acknowledgments
We apologize to our colleagues whose work we have not been able to include due to space constraints.
Funding Statement
This work is supported by grants from AIRC (21445), PRIN 2017WNKSLR and POFESR (G78I1800086007) to Giorgio Stassi, and by EUROPEAN SOCIAL FUND (ESF) PON AIM 1892002, n.2. LM, MC, TA, and VV are research fellows funded by European Union-FESR FSE, PON Ricerca e Innovazione 2014–2020.
Author contributions
VF, PV, VV and SG conceptualized and wrote the manuscript. MC, LM, DC, PG, CD, TA, GM, DS, and TM contributed to draft the manuscript. TA and CD provided graphical contribution.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aikawa Y., Katsumoto T., Zhang P., Shima H., Shino M., Terui K., et al. (2010). PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat. Med. 16 (5), 580–585. 10.1038/nm.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L., McKie M. A., Vernaz G., Cordero-Espinoza L., Aleksieva N., van den Ameele J., et al. (2019). Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 21 (11), 1321–1333. 10.1038/s41556-019-0402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Curbelo D., Ho Y. J., Burdziak C., Maag J. L. V., Morris J. P. t., Chandwani R., et al. (2021). A gene-environment-induced epigenetic program initiates tumorigenesis. Nature 590 (7847), 642–648. 10.1038/s41586-020-03147-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altucci L., Rots M. G. (2016). Epigenetic drugs: From chemistry via biology to medicine and back. Clin. Epigenetics 8, 56. 10.1186/s13148-016-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirkhah R., Naderi-Meshkin H., Shah J. S., Dunne P. D., Schmitz U. (2019). The intricate interplay between epigenetic events, alternative splicing and noncoding RNA deregulation in colorectal cancer. Cells 8 (8), E929. 10.3390/cells8080929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai M., Sachs E., Zivony A., Remez R., Ben Baruch R., Amit B. H., et al. (2015). Effects of long-term valproic acid treatment on hematological and biochemical parameters in adolescent psychiatric inpatients: A retrospective naturalistic study. Int. Clin. Psychopharmacol. 30 (5), 241–248. 10.1097/YIC.0000000000000084 [DOI] [PubMed] [Google Scholar]
- Atlasi Y., Stunnenberg H. G. (2017). The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18 (11), 643–658. 10.1038/nrg.2017.57 [DOI] [PubMed] [Google Scholar]
- Audet J. (2004). Stem cell bioengineering for regenerative medicine. Expert Opin. Biol. Ther. 4 (5), 631–644. 10.1517/14712598.4.5.631 [DOI] [PubMed] [Google Scholar]
- Awad M. M., Le Bruchec Y., Lu B., Ye J., Miller J., Lizotte P. H., et al. (2021). Selective histone deacetylase inhibitor ACY-241 (citarinostat) plus nivolumab in advanced non-small cell lung cancer: Results from a phase Ib study. Front. Oncol. 11, 696512. 10.3389/fonc.2021.696512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. M., Mendicino M., Au P. (2014). An FDA perspective on preclinical development of cell-based regenerative medicine products. Nat. Biotechnol. 32 (8), 721–723. 10.1038/nbt.2971 [DOI] [PubMed] [Google Scholar]
- Ball H. C., Alejo A. L., Samson T. K., Alejo A. M., Safadi F. F. (2022). Epigenetic regulation of chondrocytes and subchondral bone in osteoarthritis. Life (Basel) 12 (4), 582. 10.3390/life12040582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarotta L., Hurley K. (2015). Romidepsin for the treatment of peripheral T-cell lymphoma. J. Adv. Pract. Oncol. 6 (1), 22–36. [PMC free article] [PubMed] [Google Scholar]
- Barrero M. J., Sese B., Marti M., Izpisua Belmonte J. C. (2013). Macro histone variants are critical for the differentiation of human pluripotent cells. J. Biol. Chem. 288 (22), 16110–16116. 10.1074/jbc.M113.466144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129 (4), 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Baylin S. B., Jones P. A. (2011). A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11 (10), 726–734. 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Rossi D. J. (2015). Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell 16 (6), 613–625. 10.1016/j.stem.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 (7143), 407–412. 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Bezzi M., Teo S. X., Muller J., Mok W. C., Sahu S. K., Vardy L. A., et al. (2013). Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27 (17), 1903–1916. 10.1101/gad.219899.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Laurent L. C., Ren B., Loring J. F., Fan J. B. (2008). Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell 2 (2), 123–134. 10.1016/j.stem.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Bjornson C. R., Cheung T. H., Liu L., Tripathi P. V., Steeper K. M., Rando T. A. (2012). Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30 (2), 232–242. 10.1002/stem.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A. M., Ming G. L., Song H. (2015). Adult mammalian neural stem cells and neurogenesis: Five decades later. Cell Stem Cell 17 (4), 385–395. 10.1016/j.stem.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonday Z. Q., Cortez G. S., Grogan M. J., Antonysamy S., Weichert K., Bocchinfuso W. P., et al. (2018). LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 9 (7), 612–617. 10.1021/acsmedchemlett.8b00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S., Brabletz T. (2010). The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 11 (9), 670–677. 10.1038/embor.2010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussy S., Laaroussi H., Vidal M. (2020). Biochemical mechanism and biological effects of the inhibition of silent information regulator 1 (SIRT1) by EX-527 (SEN0014196 or selisistat). J. Enzyme Inhib. Med. Chem. 35 (1), 1124–1136. 10.1080/14756366.2020.1758691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N., Ley T. J., Miller C., Ding L., Raphael B. J., Mungall A. J., et al. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368 (22), 2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantile M., Di Bonito M., Tracey De Bellis M., Botti G. (2021). Functional interaction among lncRNA HOTAIR and MicroRNAs in cancer and other human diseases. Cancers (Basel) 13 (3), 570. 10.3390/cancers13030570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas H., Vieth E., Lee J., Segar M., Liu Y., Nephew K. P., et al. (2014). TGF-beta induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics 9 (11), 1461–1472. 10.4161/15592294.2014.971608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J. (2020). Cell reversal from a differentiated to a stem-like state at cancer initiation. Front. Oncol. 10, 541. 10.3389/fonc.2020.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Oropeza R., Melendez-Zajgla J., Maldonado V., Vazquez-Santillan K. (2018). The emerging role of lncRNAs in the regulation of cancer stem cells. Cell. Oncol. 41 (6), 585–603. 10.1007/s13402-018-0406-4 [DOI] [PubMed] [Google Scholar]
- Ceballos M. P., Decandido G., Quiroga A. D., Comanzo C. G., Livore V. I., Lorenzetti F., et al. (2018). Inhibition of sirtuins 1 and 2 impairs cell survival and migration and modulates the expression of P-glycoprotein and MRP3 in hepatocellular carcinoma cell lines. Toxicol. Lett. 289, 63–74. 10.1016/j.toxlet.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Chen S., Du K., Zou C. (2020). Current progress in stem cell therapy for type 1 diabetes mellitus. Stem Cell Res. Ther. 11 (1), 275. 10.1186/s13287-020-01793-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhu J., Wang F., Guan Z., Ge Y., Yang X., et al. (2017). LncRNAs and their role in cancer stem cells. Oncotarget 8 (66), 110685–110692. 10.18632/oncotarget.22161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Sun K., Jiao S., Cai N., Zhao X., Zou H., et al. (2014). High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci. Rep. 4, 7481. 10.1038/srep07481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Blum R., Bowman C., Hu D., Shilatifard A., Shen S., et al. (2014). A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell 53 (6), 979–992. 10.1016/j.molcel.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., et al. (2019). Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 4, 62. 10.1038/s41392-019-0095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry A. B., Daley G. Q. (2012). Reprogramming cellular identity for regenerative medicine. Cell 148 (6), 1110–1122. 10.1016/j.cell.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P., Allis C. D., Wang G. G. (2010). Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10 (7), 457–469. 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K., Zielinska A. E., Shaaban A. M., Sanchez-Bailon M. P., Jarrold J., Clarke T. L., et al. (2017). PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 21 (12), 3498–3513. 10.1016/j.celrep.2017.11.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini C. C., Espindola-Netto J. M., Mondal G., Guerrico A. M., Nin V., Escande C., et al. (2016). SIRT1-Activating compounds (STAC) negatively regulate pancreatic cancer cell growth and viability through a SIRT1 lysosomal-dependent pathway. Clin. Cancer Res. 22 (10), 2496–2507. 10.1158/1078-0432.CCR-15-1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou G. Y., Chien C. S., Wang M. L., Chen M. T., Yang Y. P., Yu Y. L., et al. (2013). Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol. Cell 52 (5), 693–706. 10.1016/j.molcel.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Choi S. W., Friso S. (2010). Epigenetics: A new bridge between nutrition and health. Adv. Nutr. 1 (1), 8–16. 10.3945/an.110.1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L., Dawlaty M. M., Ndiaye-Lobry D., Yap Y. S., Bakogianni S., Yu Y., et al. (2015). TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 16 (6), 653–662. 10.1038/ni.3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementino M., Xie J., Yang P., Li Y., Lin H. P., Fenske W. K., et al. (2020). A positive feedback loop between c-myc upregulation, glycolytic shift, and histone acetylation enhances cancer stem cell-like property and tumorigenicity of Cr(VI)-transformed cells. Toxicol. Sci. 177 (1), 71–83. 10.1093/toxsci/kfaa086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F., Danesi R., Farrar W. L. (2009). Cancer stem cell epigenetics and chemoresistance. Epigenomics 1 (1), 63–79. 10.2217/epi.09.4 [DOI] [PubMed] [Google Scholar]
- Crea F., Sun L., Mai A., Chiang Y. T., Farrar W. L., Danesi R., et al. (2012). The emerging role of histone lysine demethylases in prostate cancer. Mol. Cancer 11, 52. 10.1186/1476-4598-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 (11), 2622–2634. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Anello L., Sansone P., Storci G., Mitrugno V., D'Uva G., Chieco P., et al. (2010). Epigenetic control of the basal-like gene expression profile via Interleukin-6 in breast cancer cells. Mol. Cancer 9, 300. 10.1186/1476-4598-9-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. (2007). The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol. 27 (1), 384–394. 10.1128/MCB.01528-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J., Ghoshal K., Denny W. A., Gamage S. A., Brooke D. G., Phiasivongsa P., et al. (2009). A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 69 (10), 4277–4285. 10.1158/0008-5472.CAN-08-3669 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- David C. J., Massague J. (2018). Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19 (7), 419–435. 10.1038/s41580-018-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Morales R., Agis-Balboa R. C., Esteller M., Berdasco M. (2017). Epigenetic mechanisms during ageing and neurogenesis as novel therapeutic avenues in human brain disorders. Clin. Epigenetics 9, 67. 10.1186/s13148-017-0365-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Yang M., Jiang R., An N., Wang X., Liu B. (2017). Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (CSC) subpopulation enriched from breast cancer cells. PLoS One 12 (1), e0170860. 10.1371/journal.pone.0170860 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Deng S., Wang J., Zhang L., Li J., Jin Y. (2021). LncRNA HOTAIR promotes cancer stem-like cells properties by sponging miR-34a to activate the JAK2/STAT3 pathway in pancreatic ductal adenocarcinoma. Onco. Targets. Ther. 14, 1883–1893. 10.2147/OTT.S286666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Franco S., Bianca P., Sardina D. S., Turdo A., Gaggianesi M., Veschi V., et al. (2021). Adipose stem cell niche reprograms the colorectal cancer stem cell metastatic machinery. Nat. Commun. 12 (1), 5006. 10.1038/s41467-021-25333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Shilatifard A. (2010). Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339 (2), 240–249. 10.1016/j.ydbio.2009.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2007). Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8 (4), 286–298. 10.1038/nrg2005 [DOI] [PubMed] [Google Scholar]
- Fang M. Z., Wang Y., Ai N., Hou Z., Sun Y., Lu H., et al. (2003). Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 63 (22), 7563–7570. [PubMed] [Google Scholar]
- Feinberg A. P. (2018). The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378 (14), 1323–1334. 10.1056/NEJMra1402513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. (2007). JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450 (7167), 309–313. 10.1038/nature06255 [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D., Verma I. M. (2014). Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 15 (3), 244–253. 10.1002/embr.201338254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Blau H. M. (2020). Tissue stem cells: Architects of their niches. Cell Stem Cell 27 (4), 532–556. 10.1016/j.stem.2020.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Chen T. (2013). A matter of life and death: Self-renewal in stem cells. EMBO Rep. 14 (1), 39–48. 10.1038/embor.2012.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggianesi M., Di Franco S., Pantina V. D., Porcelli G., D'Accardo C., Verona F., et al. (2021). Messing up the cancer stem cell chemoresistance mechanisms supported by tumor microenvironment. Front. Oncol. 11, 702642. 10.3389/fonc.2021.702642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Sheng Y., Yang J., Wang C., Zhang R., Zhu Y., et al. (2018). Osteopontin alters DNA methylation through up-regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 37 (1), 179. 10.1186/s13046-018-0832-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbaninejad M., Khademi-Shirvan M., Hosseini S., Baghaban Eslaminejad M. (2020). Epidrugs: Novel epigenetic regulators that open a new window for targeting osteoblast differentiation. Stem Cell Res. Ther. 11 (1), 456. 10.1186/s13287-020-01966-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollavilli P. N., Kanugula A. K., Koyyada R., Karnewar S., Neeli P. K., Kotamraju S. (2015). AMPK inhibits MTDH expression via GSK3β and SIRT1 activation: Potential role in triple negative breast cancer cell proliferation. FEBS J. 282 (20), 3971–3985. 10.1111/febs.13391 [DOI] [PubMed] [Google Scholar]
- Gore A. V., Weinstein B. M. (2016). DNA methylation in hematopoietic development and disease. Exp. Hematol. 44 (9), 783–790. 10.1016/j.exphem.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhamon P., Chesnelong C., Kushida M. M., Nikolic A., Singhal D., MacLeod G., et al. (2021). Single-cell chromatin accessibility profiling of glioblastoma identifies an invasive cancer stem cell population associated with lower survival. Elife 10, e64090. 10.7554/eLife.64090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. B., Pastushenko I., Skibinski A., Blanpain C., Kuperwasser C. (2019). Phenotypic plasticity: Driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 24 (1), 65–78. 10.1016/j.stem.2018.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M., Fushimi A., Yamashita N., Bhattacharya A., Rajabi H., Long M. D., et al. (2021). MUC1-C activates the PBAF chromatin remodeling complex in integrating redox balance with progression of human prostate cancer stem cells. Oncogene 40 (30), 4930–4940. 10.1038/s41388-021-01899-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Jia L., Lv W., Wang L., Cui W. (2019). Epigenetic enzyme mutations: Role in tumorigenesis and molecular inhibitors. Front. Oncol. 9, 194. 10.3389/fonc.2019.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhou L., Wu T., Huang Y., Cheng Z., Li X., et al. (2016). Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell. Mol. Neurobiol. 36 (7), 1097–1107. 10.1007/s10571-015-0303-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. L., Dunne K., Walport L. J., Flashman E., Kawamura A. (2015). Epigenetic regulation by histone demethylases in hypoxia. Epigenomics 7 (5), 791–811. 10.2217/epi.15.24 [DOI] [PubMed] [Google Scholar]
- Hardy K., Wu F., Tu W., Zafar A., Boulding T., McCuaig R., et al. (2016). Identification of chromatin accessibility domains in human breast cancer stem cells. Nucleus 7 (1), 50–67. 10.1080/19491034.2016.1150392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Nguyen A. T., Zhang Y. (2011). KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117 (14), 3869–3880. 10.1182/blood-2010-10-312736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Gotoh Y. (2010). Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11 (6), 377–388. 10.1038/nrn2810 [DOI] [PubMed] [Google Scholar]
- Hodge D. R., Hurt E. M., Farrar W. L. (2005). The role of IL-6 and STAT3 in inflammation and cancer. Eur. J. Cancer 41 (16), 2502–2512. 10.1016/j.ejca.2005.08.016 [DOI] [PubMed] [Google Scholar]
- Hoffmann I., Roatsch M., Schmitt M. L., Carlino L., Pippel M., Sippl W., et al. (2012). The role of histone demethylases in cancer therapy. Mol. Oncol. 6 (6), 683–703. 10.1016/j.molonc.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Jing H., Lin H. (2014). Sirtuin inhibitors as anticancer agents. Future Med. Chem. 6 (8), 945–966. 10.4155/fmc.14.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Sun T., Wang H., Chen Z., Wang S., Yuan L., et al. (2016). MiR-215 is induced post-transcriptionally via HIF-drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 29 (1), 49–60. 10.1016/j.ccell.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Kallen C. B., Dhar R., Baquero M. T., Mason C. E., Russell B. A., et al. (2008). Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 4, 188. 10.1038/msb.2008.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Hirsch H. A., Wang G., Struhl K. (2011). Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc. Natl. Acad. Sci. U. S. A. 108 (4), 1397–1402. 10.1073/pnas.1018898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Polytarchou C., Hatziapostolou M., Kottakis F., Maroulakou I. G., Struhl K., et al. (2009). MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci. Signal. 2 (92), ra62. 10.1126/scisignal.2000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa J. J., Roboz G., Rizzieri D., Jabbour E., Stock W., O'Connell C., et al. (2015). Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet. Oncol. 16 (9), 1099–1110. 10.1016/S1470-2045(15)00038-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466 (7310), 1129–1133. 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., et al. (2008). Arginine methylation regulates the p53 response. Nat. Cell Biol. 10 (12), 1431–1439. 10.1038/ncb1802 [DOI] [PubMed] [Google Scholar]
- Jiang S. (2020). Tet2 at the interface between cancer and immunity. Commun. Biol. 3 (1), 667. 10.1038/s42003-020-01391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao F., Hu H., Han T., Yuan C., Wang L., Jin Z., et al. (2015). Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 16 (4), 6677–6693. 10.3390/ijms16046677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao F., Hu H., Yuan C., Wang L., Jiang W., Jin Z., et al. (2014). Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 32 (6), 2485–2492. 10.3892/or.2014.3518 [DOI] [PubMed] [Google Scholar]
- Juan A. H., Derfoul A., Feng X., Ryall J. G., Dell'Orso S., Pasut A., et al. (2011). Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 25 (8), 789–794. 10.1101/gad.2027911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y., Qin J., Oses-Prieto J., Wang H., Jackson-Weaver O., Zhang T., et al. (2018). Arginine methylation of SMAD7 by PRMT1 in TGF-beta-induced epithelial-mesenchymal transition and epithelial stem-cell generation. J. Biol. Chem. 293 (34), 13059–13072. 10.1074/jbc.RA118.002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D., Kotliar M., Woo V., Jagannathan S., Whitt J., Moncivaiz J., et al. (2018). Microbiota-sensitive epigenetic signature predicts inflammation in Crohn's disease. JCI Insight 3 (18), 122104. 10.1172/jci.insight.122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K. P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8 (2), 200–213. 10.1016/j.stem.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Shen L., Issa J. P. (2003). Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell. Biol. 23 (1), 206–215. 10.1128/MCB.23.1.206-215.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Lee E. S., Hu G., Kang Y. (2008). The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283 (22), 14910–14914. 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., Dick J. E. (2014). Evolution of the cancer stem cell model. Cell Stem Cell 14 (3), 275–291. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Kumar V. E., Nambiar R., De Souza C., Nguyen A., Chien J., Lam K. S. (2022). Targeting epigenetic modifiers of tumor plasticity and cancer stem cell behavior. Cells 11 (9), 1403. 10.3390/cells11091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S. C., Su Y. T., Chi C. C., Kuo Y. C., Lee K. F., Wu Y. C., et al. (2019). DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J. Exp. Clin. Cancer Res. 38 (1), 474. 10.1186/s13046-019-1442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Park J. R., Kwon O. S., Lee T. H., Nakano I., Miyoshi H., et al. (2015a). SIRT1 is required for oncogenic transformation of neural stem cells and for the survival of "cancer cells with neural stemness" in a p53-dependent manner. Neuro. Oncol. 17 (1), 95–106. 10.1093/neuonc/nou145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Smith E., Shilatifard A. (2010). The language of histone crosstalk. Cell 142 (5), 682–685. 10.1016/j.cell.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Park J. W., Sung H. S., Choi Y. J., Kim W. H., Lee H. S., et al. (2015b). PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene 34 (22), 2897–2909. 10.1038/onc.2014.219 [DOI] [PubMed] [Google Scholar]
- Legoy T. A., Mathisen A. F., Salim Z., Vethe H., Bjorlykke Y., Abadpour S., et al. (2020). In vivo environment swiftly restricts human pancreatic progenitors toward mono-hormonal identity via a HNF1A/HNF4A mechanism. Front. Cell Dev. Biol. 8, 109. 10.3389/fcell.2020.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng S., Huang W., Chen Y., Yang Y., Feng D., Liu W., et al. (2021). SIRT1 coordinates with the CRL4B complex to regulate pancreatic cancer stem cells to promote tumorigenesis. Cell Death Differ. 28 (12), 3329–3343. 10.1038/s41418-021-00821-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu L., Xu L., Qin K., Liu C., Yu Y., et al. (2017). Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol. Carcinog. 56 (1), 3–17. 10.1002/mc.22338 [DOI] [PubMed] [Google Scholar]
- Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., et al. (2007). Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 27 (24), 8748–8759. 10.1128/MCB.01380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chai H. F., Peng F., Meng Y. T., Zhang L. Z., Zhang L., et al. (2018). Estrogen receptor beta upregulated by lncRNA-H19 to promote cancer stem-like properties in papillary thyroid carcinoma. Cell Death Dis. 9 (11), 1120. 10.1038/s41419-018-1077-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Tian E., Chen Z. X., Sun G., Ye P., Yang S., et al. (2012). Identification of Oct4-activating compounds that enhance reprogramming efficiency. Proc. Natl. Acad. Sci. U. S. A. 109 (51), 20853–20858. 10.1073/pnas.1219181110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau B. B., Sievers C., Donohue L. K., Gillespie S. M., Flavahan W. A., Miller T. E., et al. (2017). Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 20 (2), 233–246. 10.1016/j.stem.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Janzer A., Becker A., Zimmer A., Schule R., Buettner R., et al. (2010). Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31 (3), 512–520. 10.1093/carcin/bgp324 [DOI] [PubMed] [Google Scholar]
- Lin J., Haffner M. C., Zhang Y., Lee B. H., Brennen W. N., Britton J., et al. (2011). Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate 71 (4), 333–343. 10.1002/pros.21247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Lin J. H., Hsu T. W., Su K., Li A. F., Hsu H. S., et al. (2015). IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. Int. J. Cancer 136 (3), 547–559. 10.1002/ijc.29033 [DOI] [PubMed] [Google Scholar]
- Liu C., Liu L., Shan J., Shen J., Xu Y., Zhang Q., et al. (2013). Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Lett. 339 (1), 60–69. 10.1016/j.canlet.2013.07.022 [DOI] [PubMed] [Google Scholar]
- Liu N., Li S., Wu N., Cho K. S. (2017a). Acetylation and deacetylation in cancer stem-like cells. Oncotarget 8 (51), 89315–89325. 10.18632/oncotarget.19167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chen K., Liu Z., Huang Y., Zhao R., Wei L., et al. (2017b). BORIS up-regulates OCT4 via histone methylation to promote cancer stem cell-like properties in human liver cancer cells. Cancer Lett. 403, 165–174. 10.1016/j.canlet.2017.06.017 [DOI] [PubMed] [Google Scholar]
- Lu Y., Chan Y. T., Tan H. Y., Li S., Wang N., Feng Y. (2020). Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 19 (1), 79. 10.1186/s12943-020-01197-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Zhu G., Eshelman M. A., Fung T. K., Lai Q., Wang F., et al. (2022). HOTTIP-dependent R-loop formation regulates CTCF boundary activity and TAD integrity in leukemia. Mol. Cell 82 (4), 833–851. 10.1016/j.molcel.2022.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Zhang B., LaFave L. M., Earl A. S., Chiang Z., Hu Y., et al. (2020). Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183 (4), 1103–1116. 10.1016/j.cell.2020.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. S., Yang X. L., Liu Y. S., Ding H., Wu J. J., Shi Y., et al. (2021). Long non-coding RNA NORAD promotes pancreatic cancer stem cell proliferation and self-renewal by blocking microRNA-202-5p-mediated ANP32E inhibition. J. Transl. Med. 19 (1), 400. 10.1186/s12967-021-03052-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallm J. P., Windisch P., Biran A., Gal Z., Schumacher S., Glass R., et al. (2020). Glioblastoma initiating cells are sensitive to histone demethylase inhibition due to epigenetic deregulation. Int. J. Cancer 146 (5), 1281–1292. 10.1002/ijc.32649 [DOI] [PubMed] [Google Scholar]
- Mansour A. A., Gafni O., Weinberger L., Zviran A., Ayyash M., Rais Y., et al. (2012). The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488 (7411), 409–413. 10.1038/nature11272 [DOI] [PubMed] [Google Scholar]
- Mardanpour P., Guan K., Nolte J., Lee J. H., Hasenfuss G., Engel W., et al. (2008). Potency of germ cells and its relevance for regenerative medicine. J. Anat. 213 (1), 26–29. 10.1111/j.1469-7580.2008.00930.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta L. L., Almendro V., Marusyk A., Shipitsin M., Schemme J., Walker S. R., et al. (2011). The JAK2/STAT3 signaling pathway is required for growth of CD44⁺CD24⁻ stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 121 (7), 2723–2735. 10.1172/JCI44745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., Curry C. V., et al. (2015). Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125 (4), 629–638. 10.1182/blood-2014-08-594648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar C., Shen Y., Xavy S., Zhao F., Reinisch A., Li R., et al. (2015). Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 17 (6), 675–688. 10.1016/j.stem.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister T. E., England K. S., Hopkinson R. J., Brennan P. E., Kawamura A., Schofield C. J. (2016). Recent progress in histone demethylase inhibitors. J. Med. Chem. 59 (4), 1308–1329. 10.1021/acs.jmedchem.5b01758 [DOI] [PubMed] [Google Scholar]
- Medema J. P. (2013). Cancer stem cells: The challenges ahead. Nat. Cell Biol. 15 (4), 338–344. 10.1038/ncb2717 [DOI] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454 (7205), 766–770. 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez-Zajgla J., Maldonado V. (2021). The role of lncRNAs in the stem phenotype of pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 22 (12), 6374. 10.3390/ijms22126374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson A., Frenette P. S. (2014). Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20 (8), 833–846. 10.1038/nm.3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M., Hauke R., Batra S. K. (2007). Stem cells: A revolution in therapeutics-recent advances in stem cell biology and their therapeutic applications in regenerative medicine and cancer therapies. Clin. Pharmacol. Ther. 82 (3), 252–264. 10.1038/sj.clpt.6100301 [DOI] [PubMed] [Google Scholar]
- Min S. N., Wei T., Wang X. T., Wu L. L., Yu G. Y. (2017). Clinicopathological and prognostic significance of homeobox transcript antisense RNA expression in various cancers: A meta-analysis. Med. Baltim. 96 (23), e7084. 10.1097/MD.0000000000007084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad H. P., Barbash O., Creasy C. L. (2019). Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 25 (3), 403–418. 10.1038/s41591-019-0376-8 [DOI] [PubMed] [Google Scholar]
- Moore L. D., Le T., Fan G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 (1), 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera L., Lubbert M., Jung M. (2016). Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics 8, 57. 10.1186/s13148-016-0223-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison O., Thakur J. (2021). Molecular complexes at euchromatin, heterochromatin and centromeric chromatin. Int. J. Mol. Sci. 22 (13), 6922. 10.3390/ijms22136922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Mori M., Inagawa M., Miyata K., Hashimoto N., Tanaka S., et al. (2016). Dnmt3a regulates proliferation of muscle satellite cells via p57Kip2. PLoS Genet. 12 (7), e1006167. 10.1371/journal.pgen.1006167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A., Carafa V., Benedetti R., Altucci L. (2012). Trials with 'epigenetic' drugs: An update. Mol. Oncol. 6 (6), 657–682. 10.1016/j.molonc.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M., Amrichova J., Parker B. J., Soboleva T. A., Jack C., Williams R., et al. (2012). Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 19 (11), 1076–1083. 10.1038/nsmb.2424 [DOI] [PubMed] [Google Scholar]
- Nepali K., Liou J. P. (2021). Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 28 (1), 27. 10.1186/s12929-021-00721-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. T., Weisenberger D. J., Velicescu M., Gonzales F. A., Lin J. C., Liang G., et al. (2002). Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 62 (22), 6456–6461. [PubMed] [Google Scholar]
- Nimmakayala R. K., Batra S. K., Ponnusamy M. P. (2019). Unraveling the journey of cancer stem cells from origin to metastasis. Biochim. Biophys. Acta. Rev. Cancer 1871 (1), 50–63. 10.1016/j.bbcan.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm J. E., McGarvey K. M., Yu X., Cheng L., Schuebel K. E., Cope L., et al. (2007). A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39 (2), 237–242. 10.1038/ng1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 (3), 247–257. 10.1016/s0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Pa M., Naizaer G., Seyiti A., Kuerbang G. (2017). Long noncoding RNA MALAT1 functions as a sponge of MiR-200c in ovarian cancer. Oncol. Res. [Epub ahead of print]. 10.3727/096504017X15049198963076 [DOI] [PubMed] [Google Scholar]
- Pan F., Weeks O., Yang F. C., Xu M. (2015). The TET2 interactors and their links to hematological malignancies. IUBMB Life 67 (6), 438–445. 10.1002/iub.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Wingo T. S., Zhao Z., Gao R., Makishima H., Qu G., et al. (2017). Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat. Commun. 8, 15102. 10.1038/ncomms15102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankova D., Jiang Y., Chatzifrangkeskou M., Vendrell I., Buzzelli J., Ryan A., et al. (2019). RASSF1A controls tissue stiffness and cancer stem-like cells in lung adenocarcinoma. EMBO J. 38 (13), e100532. 10.15252/embj.2018100532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa S., Padilla N., Iacobucci S., Vicioso M., Alvarez de la Campa E., Navarro C., et al. (2019). PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. U. S. A. 116 (39), 19464–19473. 10.1073/pnas.1903188116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee D. H., Ham S., Oh J., Noh J. R., Lee Y. K., et al. (2022). Targeted erasure of DNA methylation by TET3 drives adipogenic reprogramming and differentiation. Nat. Metab. 4, 918–931. 10.1038/s42255-022-00597-7 [DOI] [PubMed] [Google Scholar]
- Pathania R., Ramachandran S., Elangovan S., Padia R., Yang P., Cinghu S., et al. (2015). DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat. Commun. 6, 6910. 10.1038/ncomms7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D. R., McGirr C., Shakhbazov K., Barbier V., Krishnan K., Mukhopadhyay P., et al. (2014). Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes. Blood 123 (17), 2682–2690. 10.1182/blood-2012-02-413187 [DOI] [PubMed] [Google Scholar]
- Pechalrieu D., Etievant C., Arimondo P. B. (2017). DNA methyltransferase inhibitors in cancer: From pharmacology to translational studies. Biochem. Pharmacol. 129, 1–13. 10.1016/j.bcp.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., et al. (2011). H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30 (20), 4198–4210. 10.1038/emboj.2011.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F., Wang J. H., Fan W. J., Meng Y. T., Li M. M., Li T. T., et al. (2018). Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 37 (8), 1062–1074. 10.1038/onc.2017.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peperstraete E., Lecerf C., Collette J., Vennin C., Raby L., Volkel P., et al. (2020). Enhancement of breast cancer cell aggressiveness by lncRNA H19 and its mir-675 derivative: Insight into shared and different actions. Cancers (Basel) 12 (7), E1730. 10.3390/cancers12071730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. F., Piccolo F. M., Tsubouchi T., Sauer S., Ryan N. K., Bruno L., et al. (2010). ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6 (6), 547–556. 10.1016/j.stem.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Pinho A. V., Mawson A., Gill A., Arshi M., Warmerdam M., Giry-Laterriere M., et al. (2016). Sirtuin 1 stimulates the proliferation and the expression of glycolysis genes in pancreatic neoplastic lesions. Oncotarget 7 (46), 74768–74778. 10.18632/oncotarget.11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K. D., Helin K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30 (7), 733–750. 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Fu J., Ma T., Yan B., Gao R., An Z., et al. (2018). LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle 17 (11), 1372–1380. 10.1080/15384101.2018.1482137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L., Avgustinova A., Martin M., Datta D., Solanas G., Prats N., et al. (2017). Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-γ. Elife 6, e21697. 10.7554/eLife.21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M., Esteller M. (2011). Cancer epigenetics reaches mainstream oncology. Nat. Med. 17 (3), 330–339. 10.1038/nm.2305 [DOI] [PubMed] [Google Scholar]
- Romero-Garcia S., Prado-Garcia H., Carlos-Reyes A. (2020). Role of DNA methylation in the resistance to therapy in solid tumors. Front. Oncol. 10, 1152. 10.3389/fonc.2020.01152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F., Noren H., Jove R., Beljanski V., Grinnemo K. H. (2020). Differences and similarities between cancer and somatic stem cells: Therapeutic implications. Stem Cell Res. Ther. 11 (1), 489. 10.1186/s13287-020-02018-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Magana M. J., Martinez-Aguilar R., Lucendo E., Campillo-Davo D., Schulze-Osthoff K., Ruiz-Ruiz C. (2016). The antihypertensive drug hydralazine activates the intrinsic pathway of apoptosis and causes DNA damage in leukemic T cells. Oncotarget 7 (16), 21875–21886. 10.18632/oncotarget.7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycaj K., Tang D. G. (2015). Cell-of-Origin of cancer versus cancer stem cells: Assays and interpretations. Cancer Res. 75 (19), 4003–4011. 10.1158/0008-5472.CAN-15-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolla M. F., Avino S., Pellegrino M., De Francesco E. M., De Marco P., Lappano R., et al. (2015). SIRT1 is involved in oncogenic signaling mediated by GPER in breast cancer. Cell Death Dis. 6, e1834. 10.1038/cddis.2015.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Maitra A., Fukushima N., van Heek N. T., Matsubayashi H., Iacobuzio-Donahue C. A., et al. (2003). Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 63 (14), 4158–4166. [PubMed] [Google Scholar]
- Schwerdtfeger M., Desiderio V., Kobold S., Regad T., Zappavigna S., Caraglia M. (2021). Long non-coding RNAs in cancer stem cells. Transl. Oncol. 14 (8), 101134. 10.1016/j.tranon.2021.101134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X., Wang Z. (2016). Protein arginine methyltransferase 5 regulates multiple signaling pathways to promote lung cancer cell proliferation. BMC Cancer 16, 567. 10.1186/s12885-016-2632-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Meeran S. M. (2014). Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim. Biophys. Acta 1840 (12), 3494–3502. 10.1016/j.bbagen.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Siemens H., Jackstadt R., Hunten S., Kaller M., Menssen A., Gotz U., et al. (2011). miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10 (24), 4256–4271. 10.4161/cc.10.24.18552 [DOI] [PubMed] [Google Scholar]
- Sincennes M. C., Brun C. E., Rudnicki M. A. (2016). Concise review: Epigenetic regulation of myogenesis in health and disease. Stem Cells Transl. Med. 5 (3), 282–290. 10.5966/sctm.2015-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Padi S. K. R., Bearss J. J., Pandey R., Okumura K., Beltran H., et al. (2020). PIM protein kinases regulate the level of the long noncoding RNA H19 to control stem cell gene transcription and modulate tumor growth. Mol. Oncol. 14 (5), 974–990. 10.1002/1878-0261.12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staberg M., Rasmussen R. D., Michaelsen S. R., Pedersen H., Jensen K. E., Villingshoj M., et al. (2018). Targeting glioma stem-like cell survival and chemoresistance through inhibition of lysine-specific histone demethylase KDM2B. Mol. Oncol. 12 (3), 406–420. 10.1002/1878-0261.12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup M. R. (2001). Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20 (24), 3014–3020. 10.1038/sj.onc.1204325 [DOI] [PubMed] [Google Scholar]
- Statello L., Guo C. J., Chen L. L., Huarte M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis A. B., Neph S., Reynolds A., Humbert R., Miller B., Paige S. L., et al. (2013). Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154 (4), 888–903. 10.1016/j.cell.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403 (6765), 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Subramanian V., Mazumder A., Surface L. E., Butty V. L., Fields P. A., Alwan A., et al. (2013). H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet. 9 (8), e1003725. 10.1371/journal.pgen.1003725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Li H., Chen J., Dehennaut V., Zhao Y., Yang Y., et al. (2013). A SUMOylation-dependent pathway regulates SIRT1 transcription and lung cancer metastasis. J. Natl. Cancer Inst. 105 (12), 887–898. 10.1093/jnci/djt118 [DOI] [PubMed] [Google Scholar]
- Sussmuth S. D., Haider S., Landwehrmeyer G. B., Farmer R., Frost C., Tripepi G., et al. (2015). An exploratory double-blind, randomized clinical trial with selisistat, a SirT1 inhibitor, in patients with Huntington's disease. Br. J. Clin. Pharmacol. 79 (3), 465–476. 10.1111/bcp.12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M. L., Riggi N., Bernstein B. E. (2013). Epigenetic reprogramming in cancer. Science 339 (6127), 1567–1570. 10.1126/science.1230184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324 (5929), 930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tan Y., Tajik A., Chen J., Jia Q., Chowdhury F., Wang L., et al. (2014). Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat. Commun. 5, 4619. 10.1038/ncomms5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Yan H., Zhuang S. (2013). Histone deacetylases as targets for treatment of multiple diseases. Clin. Sci. 124 (11), 651–662. 10.1042/CS20120504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terme J. M., Sese B., Millan-Arino L., Mayor R., Belmonte J. C. I., Barrero M. J., et al. (2011). Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J. Biol. Chem. 286 (41), 35347–35357. 10.1074/jbc.M111.281923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M., Alea M. P., Di Stefano A. B., Cammareri P., Vermeulen L., Iovino F., et al. (2007). Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1 (4), 389–402. 10.1016/j.stem.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., et al. (2014). CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 14 (3), 342–356. 10.1016/j.stem.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Toh T. B., Lim J. J., Chow E. K. (2017). Epigenetics in cancer stem cells. Mol. Cancer 16 (1), 29. 10.1186/s12943-017-0596-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. P., Wu K. J. (2014). Epigenetic regulation of hypoxia-responsive gene expression: Focusing on chromatin and DNA modifications. Int. J. Cancer 134 (2), 249–256. 10.1002/ijc.28190 [DOI] [PubMed] [Google Scholar]
- Turcan S., Makarov V., Taranda J., Wang Y., Fabius A. W. M., Wu W., et al. (2018). Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat. Genet. 50 (1), 62–72. 10.1038/s41588-017-0001-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turdo A., Porcelli G., D'Accardo C., Franco S. D., Verona F., Forte S., et al. (2020). Metabolic escape routes of cancer stem cells and therapeutic opportunities. Cancers (Basel) 12 (6), E1436. 10.3390/cancers12061436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turdo A., Veschi V., Gaggianesi M., Chinnici A., Bianca P., Todaro M., et al. (2019). Meeting the challenge of targeting cancer stem cells. Front. Cell Dev. Biol. 7, 16. 10.3389/fcell.2019.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen A. J., Westendorf J. J. (2019). Epigenetics as a new frontier in orthopedic regenerative medicine and oncology. J. Orthop. Res. 37 (7), 1465–1474. 10.1002/jor.24305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerasubramanian P. K., Trinh A., Akhtar N., Liu W. F., Downing T. L. (2020). Biophysical and epigenetic regulation of cancer stemness, invasiveness and immune action. Curr. Tissue Microenviron. Rep. 1 (4), 277–300. 10.1007/s43152-020-00021-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veschi V., Liu Z., Voss T. C., Ozbun L., Gryder B., Yan C., et al. (2017). Epigenetic siRNA and chemical screens identify SETD8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell 31 (1), 50–63. 10.1016/j.ccell.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veschi V., Thiele C. J. (2017). High-SETD8 inactivates p53 in neuroblastoma. Oncoscience 4 (3-4), 21–22. 10.18632/oncoscience.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veschi V., Verona F., Lo Iacono M., D'Accardo C., Porcelli G., Turdo A., et al. (2020). Cancer stem cells in thyroid tumors: From the origin to metastasis. Front. Endocrinol. 11, 566. 10.3389/fendo.2020.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veschi V., Verona F., Thiele C. J. (2019). Cancer stem cells and neuroblastoma: Characteristics and therapeutic targeting options. Front. Endocrinol. 10, 782. 10.3389/fendo.2019.00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Duenas C., Hauer J., Cobaleda C., Borkhardt A., Sanchez-Garcia I. (2018). Epigenetic priming in cancer initiation. Trends Cancer 4 (6), 408–417. 10.1016/j.trecan.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Vicente-Duenas C., Romero-Camarero I., Cobaleda C., Sanchez-Garcia I. (2013). Function of oncogenes in cancer development: A changing paradigm. EMBO J. 32 (11), 1502–1513. 10.1038/emboj.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voog J., Jones D. L. (2010). Stem cells and the niche: A dynamic duo. Cell Stem Cell 6 (2), 103–115. 10.1016/j.stem.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L. P., Luciani L., Nimer S. D. (2013). Histone-modifying enzymes: Their role in the pathogenesis of acute leukemia and their therapeutic potential. Int. J. Hematol. 97 (2), 198–209. 10.1007/s12185-012-1247-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright E. N., Scaffidi P. (2017). Epigenetics and cancer stem cells: Unleashing, hijacking, and restricting cellular plasticity. Trends Cancer 3 (5), 372–386. 10.1016/j.trecan.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Yuan J. H., Wang S. B., Yang F., Yuan S. X., Ye C., et al. (2014). Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 60 (4), 1278–1290. 10.1002/hep.27239 [DOI] [PubMed] [Google Scholar]
- Wang L., Gural A., Sun X. J., Zhao X., Perna F., Huang G., et al. (2011). The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 333 (6043), 765–769. 10.1126/science.1201662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu X., Ren Y., Zhang J., Chen J., Zhou W., et al. (2017). Cisplatin-enriching cancer stem cells confer multidrug resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis. 8 (4), e2746. 10.1038/cddis.2016.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liang N., Yang T., Li Y., Li J., Huang Q., et al. (2021). DNMT1-mediated methylation of BEX1 regulates stemness and tumorigenicity in liver cancer. J. Hepatol. 75 (5), 1142–1153. 10.1016/j.jhep.2021.06.025 [DOI] [PubMed] [Google Scholar]
- Wang S., Liu F., Deng J., Cai X., Han J., Liu Q. (2016). Long noncoding RNA ROR regulates proliferation, invasion, and stemness of gastric cancer stem cell. Cell. Reprogr. 18 (5), 319–326. 10.1089/cell.2016.0001 [DOI] [PubMed] [Google Scholar]
- Wang T., Wu H., Liu S., Lei Z., Qin Z., Wen L., et al. (2018). SMYD3 controls a Wnt-responsive epigenetic switch for ASCL2 activation and cancer stem cell maintenance. Cancer Lett. 430, 11–24. 10.1016/j.canlet.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., et al. (2015). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16 (4), 413–425. 10.1016/j.stem.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yang X., Liu C., Li X., Zhang B., Wang B., et al. (2019). Acetylation of PHF5A modulates stress responses and colorectal carcinogenesis through alternative splicing-mediated upregulation of KDM3A. Mol. Cell 74 (6), 1250–1263. e1256. 10.1016/j.molcel.2019.04.009 [DOI] [PubMed] [Google Scholar]
- West A. C., Johnstone R. W. (2014). New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124 (1), 30–39. 10.1172/JCI69738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. G., Roberts C. W. (2011). SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11 (7), 481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- Wu H., Sun Y. E. (2006). Epigenetic regulation of stem cell differentiation. Pediatr. Res. 59 (4), 21R–25R. 10.1203/01.pdr.0000203565.76028.2a [DOI] [PubMed] [Google Scholar]
- Wu J., Xu J., Liu B., Yao G., Wang P., Lin Z., et al. (2018). Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557 (7704), 256–260. 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- Xiao L., Parolia A., Qiao Y., Bawa P., Eyunni S., Mannan R., et al. (2022). Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 601 (7893), 434–439. 10.1038/s41586-021-04246-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Chen W., Chen K., Chang Y., Yang F., Lin A., et al. (2020). Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int. 20 (1), 585. 10.1186/s12935-020-01679-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wang C., Shen J., Tong P., O'Keefe R. (2018). Ablation of Dnmt3b in chondrocytes suppresses cell maturation during embryonic development. J. Cell. Biochem. 119 (7), 5852–5863. 10.1002/jcb.26775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. P., Lv L., Liu Y., Smith M. D., Li W. C., Tan X. M., et al. (2019). Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J. Clin. Invest. 129 (10), 4316–4331. 10.1172/JCI129317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J., Estecio M. R., Lu Y., Long H., Malouf G. G., Graber D., et al. (2013). The epigenome of AML stem and progenitor cells. Epigenetics 8 (1), 92–104. 10.4161/epi.23243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. L., Li S., Munoz-Cabello A. M., Raguz S., Zeng L., Mujtaba S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38 (5), 662–674. 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Fukuda A., Omatsu M., Namikawa M., Sono M., Fukunaga Y., et al. (2021). Brg1 is required to maintain colorectal cancer stem cells. J. Pathol. 255 (3), 257–269. 10.1002/path.5759 [DOI] [PubMed] [Google Scholar]
- Yu W., Zhang L., Wei Q., Shao A. (2019). O(6)-Methylguanine-DNA methyltransferase (MGMT): Challenges and new opportunities in glioma chemotherapy. Front. Oncol. 9, 1547. 10.3389/fonc.2019.01547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Cen Y., Chen J. (2018). Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 15 (2), 2117–2122. 10.3892/ol.2017.7557 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang G., Dong Z., Prager B. C., Kim L. J., Wu Q., Gimple R. C., et al. (2019). Chromatin remodeler HELLS maintains glioma stem cells through E2F3 and MYC. JCI Insight 4 (7), 126140. 10.1172/jci.insight.126140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Cai K., Wang J., Wang X., Cheng K., Shi F., et al. (2014a). MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 32 (11), 2858–2868. 10.1002/stem.1795 [DOI] [PubMed] [Google Scholar]
- Zhang J., Tian X. J., Zhang H., Teng Y., Li R., Bai F., et al. (2014b). TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 7 (345), ra91. 10.1126/scisignal.2005304 [DOI] [PubMed] [Google Scholar]
- Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., et al. (2005). Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8 (1), 19–30. 10.1016/j.devcel.2004.10.019 [DOI] [PubMed] [Google Scholar]
- Zhao L. Y., Song J., Liu Y., Song C. X., Yi C. (2020). Mapping the epigenetic modifications of DNA and RNA. Protein Cell 11 (11), 792–808. 10.1007/s13238-020-00733-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Huang S., Wang Z., Huang J., Xu L., Tang X., et al. (2019). Targeting EZH2 histone methyltransferase activity alleviates experimental intestinal inflammation. Nat. Commun. 10 (1), 2427. 10.1038/s41467-019-10176-2 [DOI] [PMC free article] [PubMed] [Google Scholar]