Abstract
During retroviral DNA synthesis reverse transcriptase frequently performs nonrequired template switches that can lead to genetic rearrangements or recombination. It has been postulated that template switching occurs after pauses in the action of reverse transcriptase. Hence factors which affect pausing, such as polymerization rate, may affect the frequency of template switching. To address the hypothesis that increasing the time required to complete reverse transcription increases the frequency of template switching, we established conditions that lengthened the time required to complete a single round of intracellular Moloney murine leukemia virus reverse transcription approximately threefold. Under these conditions, which resulted from intracellular nucleotide pool imbalances generated with hydroxyurea, we examined template switching frequency using a lacZ-based tandem repeat deletion assay. We observed that the frequency of deletion during reverse transcription in hydroxyurea-treated cells was approximately threefold higher than that in untreated control cells. These findings suggest that rates of retroviral recombination may vary when the intracellular environment is altered.
Retroviruses can persist in infected cells in the form of double-stranded DNA copies of the viral RNA genome. During reverse transcription, viral reverse transcriptase (RT) must perform two template switches—the first and second strong-stop template switches—to complete the synthesis of integration-competent viral DNA (11). It has been postulated that the requirement to perform these two obligatory template switches confers on RT the tendency to make additional, nonrequired template switches which can lead to viral genetic recombination (6, 32, 37). Paired with RT’s relatively high base substitution rate, such recombination events are thought to introduce much of the genetic variation that is observed in retroviral populations (15, 22, 30, 33). Both genetic alterations to RT and changes to the intracellular environment can affect RT-mediated base substitution rates (21, 24, 29, 38). However, factors that affect template switching during reverse transcription remain largely unknown.
During recombinogenic template switching, RT begins DNA synthesis on one copy of the viral genome and then switches to the other, copackaged genome (intermolecular template switching). The frequency of recombinogenic template switching varies, but experimental evidence suggests that on average, such events happen roughly once per genome per cycle of reverse transcription (19). RT can also perform template switches between two different positions on the same template molecule (intramolecular template switching). Homologous recombination—template switching between regions with high sequence similarity—is far more frequent than recombination between nonhomologous sequences (15, 43, 44). In studies where homology patterns should permit either type of switch, intramolecular template switching was much more common than intermolecular template switching (13).
Template switching during reverse transcription on templates containing direct repeats frequently results in the deletion of one copy of the repeat (3, 8, 13, 20, 22, 31, 32, 35, 36, 40). Spleen necrosis virus-based vectors with direct repeats of 1,333, 788, and 383 bp have been shown to yield deleted products 93%, 85%, and 28 to 40% of the time, respectively (20). Although recombination hot spots have been characterized and template switching frequency does not always reflect the length of sequence homology, direct-repeat deletion rates are generally roughly proportional to the length of the direct repeat (1, 19, 20, 31, 44).
Early models for retroviral template switching during minus strand synthesis evoked the need for a template break to force transfer to the homologous region of the copackaged genome (6). However, the high frequency of tandem repeat deletions and the relatively minor effects on recombination of experimental conditions designed to enhance template lesions suggest that physical breaks in templates—although possibly involved in some template switching—are not required (14, 20, 40). It has also been suggested that kinetic disruptions, such as pausing by RT, may promote departure from one template and association with another. Support for this possibility comes from observations that some template features, such as RNA structures and homopolymeric runs, which may impede RT elongation, are retroviral recombination hot spots (19, 31, 39). For many types of polymerases, slowing of elongation can affect a wide range of elongation properties. For example, there is a correlation between decreased rates of elongation and pausing and/or termination for Escherichia coli RNA polymerase. An RNA polymerase mutant with decreased affinity for the substrates ATP and GTP showed a decrease in elongation rate and increased pausing (18). Additional studies in the E. coli system demonstrated that elongation rate may determine the probability of transcript release and that termination efficiency is indirectly proportional to the elongation rate (17, 25). The authors of those studies noted that small changes in the elongation rate of RNA polymerase can have large effects on termination efficiency.
In this study we sought to address the effects of decreasing the RT elongation rate on template switching events, such as those that lead to retroviral genetic recombination. Previous determinations of rates of retroviral recombination have been performed in transformed cells, but viral replication in its natural setting occurs in cells whose intracellular environments are less metabolically rich than those of actively dividing cultured cells. In this study, hydroxyurea (HU) was used to produce an imbalance of intracellular nucleotide pools and template switching was scored by determining deletion rates of a 117-bp direct repeat in a lacZ reporter gene. We found that HU treatment lengthened the amount of time required to complete retroviral DNA synthesis and that template switching frequency under these conditions was increased threefold compared to that of untreated control cells.
MATERIALS AND METHODS
Plasmid construction.
The gag-pol-puro plasmid, pGPP, was a derivative of an infectious Moloney murine leukemia virus (M-MuLV) provirus plasmid in which the env coding region was replaced with an expression cassette for puromycin resistance. To make pGPP, a BamHI-SpeI fragment containing the simian virus 40 (SV40) promoter, a puromycin resistance gene, long terminal repeats (LTRs), and plasmid backbone portions of pBabepuro (26) was ligated to two restriction fragments, SpeI-SalI and SalI-BamHI, which together contained the gag and pol portions from the infectious M-MuLV provirus plasmid, pNCA (7).
pLacPuro and derivatives were M-MuLV-based vectors in which the viral genes were replaced by the lacZ gene driven by the LTR promoter, followed by a puromycin resistance gene transcribed from the SV40 promoter. pLacPuro contained the lacZ cassette from the BAG vector (34) and the puromycin resistance gene from pBabepuro in the “tipless” M-MuLV provirus backbone of pAM86-5 (23). An XbaI to HindIII fragment including portions of the upstream LTR and the complete lacZ gene from pBAG was combined with XbaI-EcoRI and HindIII-EcoRI pAM86-5 fragments to yield pLacPuro. To make different lengths of direct repeats within lacZ, a restriction fragment containing the LacZ coding region was further cleaved with different blunt-cutting restriction enzymes. Pairwise combinations of upstream and downstream portions of lacZ from different digests were then religated into the parental vector so that 117-, 284-, and 971-bp direct repeats were made (pLaac-117, pLaac-284, and pLaac-971, respectively). The 117-bp direct repeat contained a duplication of sequences between EcoRV and SspI, the 284-bp direct repeat contained a duplication of sequences between HincII and FspI, and the 971-bp direct repeat contained a duplication of sequences between EcoRV and FspI.
The pMLV Ψ− construct was made by deleting sequences between MscI and AatII sites in the packaging signal region of pNCA (7, 10). Further construction details are available on request.
Cell lines and viruses.
NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum (Gibco). 293T cell derivatives (ET, ET pLacPuro, and ET pLaac lines; see below) were grown in DMEM supplemented with 10% fetal bovine serum (HyClone). Transfections were performed by using lipofectamine (Gibco) according to the manufacturer’s instructions, except where noted. Infections were performed in the presence of 0.8 μg of hexadimethrine bromide (polybrene) (Sigma) per ml for either 10 min or 2 h at 37°C, as noted. Puromycin-resistant cells were selected in puromycin (Sigma) at either 1-μg/ml (ET lines) or 6-μg/ml (3T3 cells) concentration.
To make the ET cell line, 293T cells were stably transfected with pAM 178, a plasmid that confers histidinol resistance and contains the ecotropic envelope gene driven by the cytomegalovirus immediate-early promoter. This plasmid was constructed by inserting the his gene from pSV2 His and portions of pNCA that contain ecotropic env into pCI (Promega). Construction details are available on request. Transfectants were single-cell cloned, and several cloned transfectants were functionally tested for the ability to supply ecotropic envelope in trans by transiently transfecting them with pGPP and then testing the resulting virus for the ability to confer puromycin resistance on transduced 3T3 cells. The cell clone that consistently produced the largest number of puroR colonies was used in subsequent experiments as the ET cell line.
gag-pol-puro vector virus was generated by stably transfecting ET cells with pGPP and pooling over 300 puromycin-resistant colonies. Virus used in the timing of reverse transcription experiments was harvested from 80% confluent 100-mm-diameter plates of the stably transfected pool every 12 h. Collected virus was pooled, filtered with 0.45-μm-pore-size filters (Fisher), aliquoted, and frozen at −70°C prior to use.
To make LacPuro and Laac vector virus, pLacPuro and the various pLaac plasmids were first stably transfected into ET cells. Puromycin-resistant single-cell clones from these transfections were chosen as candidate ET-pLac and ET-pLaac vector-expressing cell lines. Candidate cell line genomic DNA was examined by Southern blotting for the presence of undeleted lacZ. Each candidate cell line was further tested for vector integrity and functionality by transiently transfecting it with pMLV Ψ−, harvesting virus, and using the vector virus to transduce fresh 3T3 cells. The resulting puromycin-resistant colonies were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to detect functional LacZ. Suitable ET-derived single-cell-cloned expressors of each vector were then chosen and called ET pLacPuro, ET pLaac-117, ET pLaac-284, and ET pLaac-971.
To generate virus for the assays of the rates of deletion, each line (ET pLaac-117, ET pLaac-284, or ET pLaac-971) was transiently transfected with pMLV Ψ− by using a calcium phosphate precipitation method. Briefly, 5 μg of pMLV Ψ− plasmid in 200 μl of 250 mM CaCl2 was combined with 200 μl of precipitation buffer (250 mM NaCl, 50 mM HEPES–NaOH [pH 7.1], 1.5 mM Na2HPO4–NaH2PO4), mixed well, and incubated for 30 min at room temperature. Fresh serum-containing medium (4 ml) was applied to a 50% confluent 60-mm-diameter plate, and the DNA precipitate was added dropwise. After 24 h, the medium was replaced with fresh DMEM containing fetal serum. Forty-eight hours after transfection, 5 ml of virus was harvested, filtered, aliquoted, and stored at −70°C. The virus samples used for all infections within each experiment were from a single stock of virus.
Reverse transcription assays.
Reverse transcription was timed by using virus from the ET gag-pol-puro cell line. 3T3 cells were infected with ET gag-pol-puro virus according to the “short infection” protocol, which involved washing cells twice with phosphate-buffered saline (PBS) 10 min after 50 μl of virus and 750 μl of complete medium containing 0.8 μg of polybrene per ml were applied to the cells. After the wash step, cells either remained untreated or were treated with 60 μM HU (Sigma). The media on all plates were replaced at 5 h postinfection to remove HU. At different times postinfection (1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 h) cells were treated with 200 μM 3′-azido-3′-deoxythymidine (AZT) (Sigma) to stop reverse transcription. At 48 h postinfection, the media on all plates were replaced with puromycin-containing media. After 2 weeks of puromycin selection the colonies were counted.
For experiments in which the times of HU exposure were varied, cells were infected as described above and 60 μM HU was added after infection. HU-containing media were removed from these plates at 5 or 10 h postinfection. AZT (200 μM) was added to the plates at 2, 3, 4, 5, 7, 9, or 11 h postinfection. A control set of plates received AZT but not HU at the same time points. Again, puromycin was added to the cells at 48 h postinfection and colonies were counted 2 weeks later. Note that AZT was used only in assays of the timing of reverse transcription: in experiments where error or deletion rates were measured, no AZT was added.
Rates of error during reverse transcription were examined by determining rates of lacZ inactivation of the wild-type lacZ vector. The ET pLacPuro cell line was transiently transfected with pMLV Ψ− by the calcium phosphate precipitation method described above. Virus was harvested and used to infect 3T3 cells as described above. After the wash step, cells either remained untreated or were treated with 60 μM HU for 5 h, as described above. After 2 weeks of puromycin selection, colonies were stained with X-Gal.
The deletion frequencies of the different-sized direct repeats in lacZ were analyzed by using virus produced from the ET pLaac cell lines. Assays to examine rates of tandem repeat deletion were performed as follows. Fifty microliters of virus-containing medium harvested from pMLV Ψ−-transfected ET pLaac cells as described above was combined with 750 μl of complete medium containing polybrene, and the mix was added to 20% confluent 3T3 cells in a 60-mm-diameter culture dish. After a 10-min incubation at 37°C, the virus was removed and the cells were washed twice with PBS. Three milliliters of the appropriate medium (with or without HU) was added to each plate, and reverse transcription was allowed to proceed at 37°C. The medium on each plate was replaced at 5 h postinfection to eliminate HU. Forty-eight hours after infection puromycin was added to the plates, and puromycin-resistant colonies were stained with X-Gal 2 weeks later.
Deletion frequencies were adjusted to account for lacZ-inactivating mutations. In our direct-repeat assays, blue colonies resulted when one copy of the vector’s direct repeat was deleted. However, because the mutations that accumulate in intact lacZ during a single cycle of viral replication (see Table 1) presumably accumulate at the same rate regardless of whether a deletion occurs, we assume that some deleted vectors failed to generate blue colonies due to these additional mutations. Therefore, the ratios of determinations of deletion frequencies for the various pLaac vectors for blue colonies to the determinations for total colonies are likely to be underestimated. We corrected for these presumptive inactivating mutations as follows: if the rate of lacZ mutational inactivation was 8.8% and the blue-colony to total-colony ratio for apparent deletion frequency was 5.1%, we calculated that 5.1% corresponded to 91.2% of the total deletions and that the actual deletion rate was 5.6%. Since HU treatment resulted in increased rates of lacZ inactivation, the deletion frequencies for cells treated with HU were normalized by using an inactivation rate different from that used for untreated cells.
TABLE 1.
Effect of HU treatment on lacZ inactivation
| Expt no. | Without HU treatment
|
With HU treatment
|
||||
|---|---|---|---|---|---|---|
| No. of blue colonies | No. of white colonies | % Inactivationa | No. of blue colonies | No. of white colonies | % Inactivationa | |
| 1 | 253 | 16 | 5.9 | 131 | 17 | 11.5 |
| 2 | 194 | 19 | 8.9 | 103 | 24 | 18.9 |
| 3 | 207 | 18 | 8.0 | 92 | 15 | 14.0 |
| 4 | 149 | 21 | 12.4 | 59 | 15 | 20.3 |
| Avg | 8.8 | 16.2 | ||||
| SD | 2.7 | 4.1 | ||||
% Inactivation was determined by dividing the number of white colonies by the total number of colonies and multiplying by 100.
For experiments on cells in serum at low concentration, cells were grown in 0.5 or 10% calf serum-containing media for 48 h prior to infection. Cells were then infected with ET pLaac virus as described above but did not receive HU. Forty-eight hours after infection, fresh medium containing 10% calf serum was added to each plate. Puromycin was added to the cells 48 h after infection, and after 2 weeks of selection colonies were stained with X-Gal.
LacZ staining was performed by using standard protocols (34). Briefly, cells were washed once with PBS. One milliliter of a fixing solution (2% formaldehyde, 0.2% glutaraldehyde in PBS) was added, and cells were incubated for 5 min at 4°C. Cells were then washed in PBS, and 1 ml of a staining solution (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.1% X-Gal [Gibco BRL] in PBS) was added.
RESULTS
Assessing the time required to complete intracellular reverse transcription.
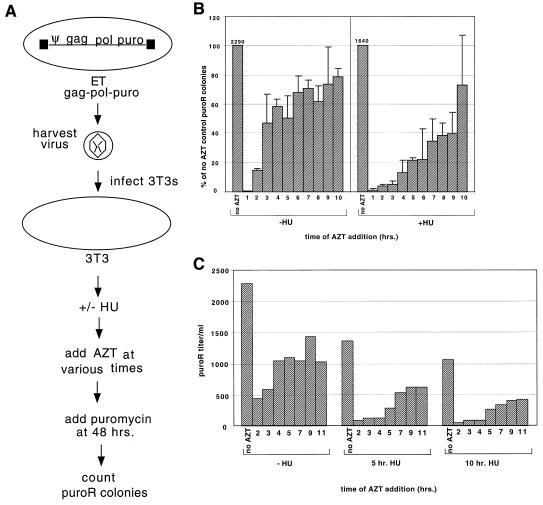
We developed an assay to gauge the length of time required for the completion of a single round of viral DNA synthesis. This assay involved infecting NIH 3T3 cells with replication-defective retroviral vectors that confer puromycin resistance, treating infected cells with an excess of the RT inhibitor AZT at various times postinfection, and then scoring whether viral DNA synthesis was completed prior to AZT addition by determining the puromycin-resistant colony titer of the infected cells. For each of these timing assays, 3T3 cells were infected with gag-pol-puro vector virus harvested as described in the Materials and Methods section (Fig. 1A). Because the cells were washed with PBS at 10 min postinfection and the culture medium was replaced, most infections should have been fairly synchronous and limited to virus that had attached within the first 10 min. Under these conditions, the productive multiplicity of infection was less than 0.01. At different times after infection (1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 h), AZT was added to separate plates. The concentration of AZT used was more than 100-fold higher than that required to inhibit M-MuLV replication (23a), but this amount of AZT does not cause apparent cytopathology. Forty-eight hours after initial infection, the AZT-containing medium was removed and puromycin-containing medium was added. After 2 weeks of selection in puromycin, drug-resistant colonies were counted. In this assay, only cells in which viral DNA synthesis had been completed prior to AZT addition should form puromycin-resistant colonies. In control experiments, where AZT was added prior to infection, no puromycin-resistant colonies formed, thus indicating that the concentration of AZT used in our assays completely inhibited provirus generation.
FIG. 1.
Assay of timing of reverse transcription. (A) Schematic overview of timing of reverse transcription assays. ET cells stably expressing gag-pol-puro were used as a source of virus for the timing assays. (ET cells are 293T cells that stably express ecotropic Env.) 3T3 cells were infected for 10 min with ET gag-pol-puro virus, and the cells were washed to remove most unattached virions. After being washed, cells were treated with 60 μM HU for 5 h or remained untreated. At various times postinfection, 200 μM AZT was added to individual plates. Puromycin was added to the plates 48 h postinfection. Colonies were counted after 2 weeks of selection. (B) Averaged results of two independent experiments measuring timing of reverse transcription. The left half of the graph shows results of experiments carried out in the absence of HU (−HU). The positive control received no AZT, and the negative control (data not shown) had no virus or AZT and no colonies were present. Numbers 1–10 denote the times of AZT addition (hours postinfection). The right half of the graph shows results of experiments done in the presence of 60 μM HU (+HU). The positive control received HU but no AZT. For these experiments, the media on all plates were changed at 5 h postinfection. The average puromycin-resistant (puroR) colony titers for positive controls in experiments conducted in the absence and presence of HU were 2,290 and 1,640 colonies/ml, respectively, as indicated above the bars. (C) Timing of reverse transcription with varying times of HU exposure. Cells were infected as described for panel B except that they were treated with 60 μM HU for 5 or 10 h or remained untreated (−HU). At various times postinfection (2, 3, 4, 5, 7, 9, or 11 h) 200 μM AZT was added to individual plates. AZT was removed and puromycin was added after 48 h, and colonies were counted 2 weeks later.
The left half of Fig. 1B shows a time course of AZT inhibition of provirus synthesis and includes combined data obtained from two independent experiments. A few colonies were present on plates which had received AZT at 2 h postinfection, suggesting that DNA synthesis was completed in less than 2 h for a small percentage of the virus. However, the majority of viral DNAs took at least 3 h to complete. When the time point at which 50% of the positive (no AZT) control titer was formed was set as the mean DNA synthesis completion time, it appeared that the mean reverse transcription time under our standard assay conditions in 3T3 cells was 2 to 4 h.
Altering the intracellular environment to prolong the time required to complete reverse transcription.
We also performed this assay for the timing of reverse transcription in cells treated with HU. HU, an inhibitor of cellular ribonucleotide reductase, has been shown to inhibit reverse transcription by decreasing the levels of substrate deoxynucleoside triphosphates (2, 12, 21). We performed trials to determine a concentration of HU that partially inhibited viral DNA synthesis. In the experiments described below, the same infection protocol as described above was used except that 60 μM HU was included in the culture media used after the postinfection wash step. Again, AZT was added at different times after infection, and at 5 h the HU-containing medium was replaced with fresh medium. Again, colonies were counted after 2 weeks of selection in puromycin. The right half of Fig. 1B shows data obtained from two independent experiments. Time courses of viral DNA completion in cells treated with HU and in untreated cells are shown in this figure. At the 3-h time point, untreated cells had completed synthesis of nearly half of the total viral DNAs, while less than 10% of the total DNAs had been synthesized in HU-treated cells at this time point. Colony counts gradually increased over successive time points of AZT addition for the HU-treated cells, resulting in a mean completion time that was delayed to more than 9 h. The titers of puromycin-resistant colonies in the absence of AZT were only slightly higher for the untreated controls than for the HU-treated cells (2,290 and 1,640 puromycin-resistant colonies/ml, respectively), thus suggesting that the HU treatment employed was not highly cytotoxic. It is possible that some component of this modest titer decrease resulted from increased rates of abortive reverse transcription rather than HU cytotoxicity.
Because the mean completion time determined as described above for cells treated with HU was longer than the duration of HU treatment, it seemed possible that the observed delay may have resulted from a cessation of reverse transcription during HU exposure followed by a restoration of reverse transcription at the normal rate when HU was removed. To address this possibility, experiments measuring the timing of reverse transcription were performed as described above except that infected cells were exposed to HU for 10 h, which is slightly longer than the mean completion time given above. AZT was added at 2, 3, 4, 5, 7, 9, or 11 h postinfection. Figure 1C shows the results of these experiments. Cells not treated with HU showed a pattern for DNA synthesis completion time similar to that for cells not treated with HU in the experiment summarized in Fig. 1B, and the mean DNA synthesis completion time was 2 to 4 h. Although overall colony counts were reduced somewhat when HU treatment time was increased, cells treated with HU for 5 h and those treated with HU for 10 h both showed comparable mean completion times. Since the mean completion time was reached during the period of HU treatment for the cells treated with HU for 10 h, reverse transcription was ongoing during the period of HU treatment. Since the times required to complete DNA synthesis for both HU treatment regimens were increased relative to that for cells not treated with HU, this suggests that on average, the reverse transcription machinery was engaged in the process of DNA synthesis for a longer period before completing viral DNA in cells treated with HU than in untreated cells.
Analysis of reverse transcription error rates under altered intracellular conditions.
We also analyzed the effect of HU treatment on rates of error during reverse transcription by examining rates of LacZ inactivation in a manner similar to that employed in previously reported forward-mutation rate assays (21, 32). To perform these experiments, we used LacPuro (Fig. 2B), a replication-defective retroviral vector that confers puromycin resistance and encodes intact LacZ under the control of the M-MuLV LTR promoter. Mammalian cells which have been transduced with this vector stain blue with X-Gal unless the lacZ gene has been mutationally inactivated. Virions containing LacPuro were harvested as described in the Materials and Methods section and used to infect 3T3 cells, either in the presence or absence of HU treatment for 5 h. After puromycin selection, transduced cells were stained with X-Gal and blue and white colonies were counted (Table 1). When the criterion of LacZ inactivation was employed, untreated cells showed an error rate of 8.8%, while HU-treated cells gave rise to an error rate of 16.2%, which suggests that HU treatment during reverse transcription increased the mutation rate roughly 1.8-fold. This value is similar to a previously reported 2.7-fold increase in error rate observed during the reverse transcription of murine leukemia virus (MLV)-based vectors in 2 mM HU-treated D17 cells (21). Although some of the white colonies in our experiments may have resulted from effects such as the imbalanced expression that is sometimes observed when two genes are coexpressed in a single retroviral vector (9), we assumed that differences in white colony/blue colony ratios in the presence and absence of HU resulted from different levels of LacZ-inactivating errors during reverse transcription.
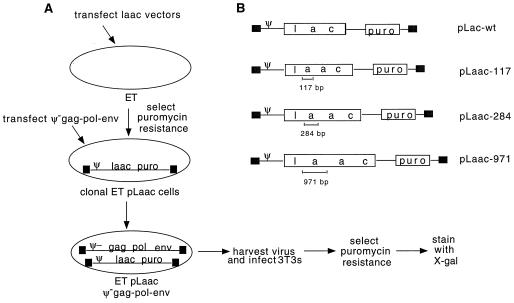
FIG. 2.
Template switching assay and lacZ vectors. (A) Template switching assay using the lacZ vectors described in panel B. 293T cell-derived ET cells were transfected with pLaac-117, pLaac-284, or pLaac-971, and stable clonal transfectants were obtained by puromycin selection. pMLV Ψ− was then transiently transfected into single-cell clones expressing each vector, and virus was harvested and used to infect 3T3 cells. After puromycin selection, the resulting 3T3 cell colonies were stained with X-Gal to determine the deletion frequency. (B) M-MuLV-based vectors containing direct repeats of different lengths within the lacZ gene. The parental vector, pLacPuro, contains the puromycin resistance gene transcribed from the SV40 promoter and the lacZ gene transcribed from the upstream LTR. pLaac-117, pLaac-284, and pLaac-971 are derivatives of pLacPuro which contain 117-, 284-, and 971-bp repeats, respectively, within the lacZ gene. LTRs are represented by black boxes.
Establishing a system to measure rates of tandem repeat deletion.
To assess rates of template switching during reverse transcription, we constructed the Laac series of retroviral vectors. The plasmids which encode these were derivatives of pLacPuro with direct repeats of different lengths within the lacZ reporter gene (Fig. 2B). If the direct repeat within lacZ remained undeleted during reverse transcription, cells transduced by the resulting vector DNA should remain unstained when incubated with X-Gal. However, if precise deletion of the direct repeat occurred during reverse transcription, then the transduced cells should stain blue.
We produced M-MuLV-derived virions containing Laac vector RNAs in human 293T cell-derived cells as described in the Materials and Methods section and used these to transduce fresh 3T3 cells (Fig. 2A). These virions should not reinfect the 293T cell-derived producer cells because the human cells lack the ecotropic receptor. However, the virus can be used to infect cells, such as murine 3T3 cells, which contain the ecotropic receptor. After puromycin selection, transduced 3T3 cells were stained with X-Gal and the numbers of blue and white colonies were counted (Table 2). The Laac vectors with 117-, 284-, and 971-bp direct repeats yielded blue colony/total colony ratios of 5.1, 27.1, and 60.0%, respectively. If the measured LacZ mutational inactivation rate of 8.8% given above is factored in, then these values suggest that the rates of tandem deletion for the 117-, 284-, and 971-bp repeats were roughly 5.6, 30, and 66%, respectively (see the Materials and Methods section). Thus, in agreement with data presented by other groups, our data revealed that deletion rates increased with the length of the direct repeats.
TABLE 2.
Deletion rates of Laac vectors
| Expt no. | Deletion rate ofa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ET Laac-117
|
ET Laac-284
|
ET Laac-971
|
|||||||
| No. of blue colonies | No. of white colonies | % Deletion | No. of blue colonies | No. of white colonies | % Deletion | No. of blue colonies | No. of white colonies | % Deletion | |
| 1 | 6 | 89 | 6.3 | 35 | 154 | 18.5 | 43 | 25 | 63.2 |
| 2 | 10 | 179 | 5.3 | 54 | 124 | 30.3 | 48 | 30 | 61.5 |
| 3 | 5 | 134 | 3.6 | 50 | 104 | 32.5 | 47 | 38 | 55.3 |
| Avg | 5.1 | 27.1 | 60.0 | ||||||
| SD | 1.4 | 7.5 | 4.2 | ||||||
% Deletion was determined by dividing the number of blue colonies by the total number of colonies and multiplying by 100.
Comparing rates of tandem repeat deletion under differing intracellular conditions.
To address whether increasing the duration of reverse transcription affected template switching, rates of tandem repeat deletion in HU-treated and untreated cells were compared. For these experiments, Laac-117 vectors were chosen because their low rate of blue colony formation (5.1%) should readily allow higher deletion rates to be scored. 3T3 cells were infected with Laac-117 virus harvested from 293T cell-derived producer cells as described in the Materials and Methods section. Reverse transcription was allowed to proceed in either the presence or absence of 60 μM HU, the resulting puromycin-resistant transductants were stained with X-Gal, and blue and white colonies were counted. The results of 15 independent experiments are listed in Table 3. In these assays, untreated cells and HU-treated cells showed apparent template switching frequencies of 5.0 and 13.5%, respectively, suggesting that HU treatment increased template switching 2.7-fold. When calculated LacZ inactivation rates were factored in (see the Materials and Methods section), these findings suggest that direct-repeat deletion rates were nearly threefold higher in HU-treated cells than in untreated cells.
TABLE 3.
Effect of HU on Laac deletion rates
| Expt no. | Without HU
|
With HU
|
||||
|---|---|---|---|---|---|---|
| No. of blue colonies | No. of white colonies | % Deletiona | No. of blue colonies | No. of white colonies | % Deletiona | |
| 1 | 10 | 168 | 5.6 | 22 | 146 | 13.1 |
| 2 | 7 | 178 | 3.8 | 14 | 81 | 14.7 |
| 3 | 16 | 348 | 4.4 | 12 | 73 | 14.3 |
| 4 | 8 | 175 | 4.4 | 17 | 89 | 16.0 |
| 5 | 12 | 181 | 6.2 | 20 | 112 | 15.2 |
| 6 | 5 | 164 | 3.0 | 8 | 68 | 10.5 |
| 7 | 11 | 202 | 5.2 | 39 | 206 | 15.9 |
| 8 | 10 | 205 | 4.7 | 27 | 226 | 10.7 |
| 9 | 8 | 182 | 4.2 | 16 | 123 | 11.5 |
| 10 | 13 | 227 | 5.4 | 19 | 145 | 11.6 |
| 11 | 18 | 281 | 6.0 | 14 | 86 | 14.0 |
| 12 | 14 | 207 | 6.3 | 10 | 75 | 11.8 |
| 13 | 19 | 284 | 6.3 | 19 | 107 | 15.1 |
| 14 | 11 | 274 | 3.9 | 19 | 130 | 12.8 |
| 15 | 17 | 304 | 5.3 | 30 | 158 | 16.0 |
| Avg | 5.0 | 13.5 | ||||
| SD | 1.0 | 2.0 | ||||
% Deletion was determined by dividing the number of blue colonies by the total number of colonies and multiplying by 100.
DISCUSSION
We demonstrate here that alterations to the intracellular environment can affect rates of tandem repeat deletion during M-MuLV reverse transcription. Specifically, we determined that treating cells with HU resulted in an increase in tandem repeat deletion rates. The design of this study was based on predictions of models for retroviral genetic recombination and on known elongation properties of polymerases. Viral template switching has been suggested to proceed by a “pause and jump” mechanism, and polymerase elongation rates affect pausing (27, 40). Therefore, we postulated that decreasing the rate of reverse transcription might increase template switching. Substrate limitations can reduce polymerase elongation rates, and HU treatment results in intracellular nucleotide pool imbalances. We thus sought to examine whether HU treatment affects template switching rates. We developed two main assays in the course of this work: an intracellular vector DNA synthesis assay for timing of the duration of reverse transcription and a lacZ-based tandem repeat deletion template switching assay.
The assay for timing of reverse transcription used AZT to terminate vector DNA synthesis. The data suggested that the average time required to complete reverse transcription of an 8.6-kb vector was 2 to 4 h and that some DNA synthesis was completed within the first 1 or 2 h postinfection. These values are fairly consistent with the predicted completion time, 2.4 h, for our vectors if intracellular elongation proceeded at the rate, approximately 1 nucleotide/s, which has been determined by using heteropolymeric primer and/or templates in purified reaction mixtures (4, 16). It should be noted that neither the time required for AZT-triphosphate formation in the cell nor the times required for viral processes such as uncoating and template switches are known precisely. However, we assume that the times required for AZT triphosphorylation and viral entry did not vary significantly among our experimental samples under our experimental conditions and that hence the approach taken allowed us to compare differences in duration of viral DNA synthesis.
Using this timing assay and a concentration of HU empirically determined to partially inhibit viral DNA synthesis, we determined that HU treatment resulted in a significantly prolonged mean completion time of viral DNA synthesis. Whether the additional time required to complete DNA synthesis was due to slowed DNA polymerization per se or if other factors such as damage to template RNAs caused or contributed to increased synthesis times was not explicitly examined. Because the amounts of viral DNAs generated in our experiments were very small, our attempts to monitor the accumulation of specific DNA intermediates over time were not successful. However, our experiments suggest that synthesis was ongoing throughout the period of HU treatment (Fig. 1c) and hence our findings are consistent with the possibility that synthesis of individual proviruses took longer in HU-treated cells than in untreated cells.
The deletion of direct repeats is presumed to be mechanistically related to the intermolecular template switching that results in genetic recombination, and direct-repeat deletion has been used frequently as a measure of intramolecular template switching. Therefore, a direct-repeat deletion assay was developed to score the amounts of template switching in HU-treated and untreated cells. The assay used retroviral vectors containing direct repeats of different lengths in the lacZ gene, such that cells containing deleted vectors stained blue with X-Gal while cells containing undeleted or mutagenized lacZ regions remained unstained. Using this approach, we demonstrated that treatment of cells with HU increased template switching about threefold. These findings are consistent with predictions derived from our hypothesis that decreasing reverse transcription rates might increase template switching rates. However, this increase may have resulted from either the increase in the time required to complete DNA synthesis or other effects, such as a possible increase in the number of broken RNA molecules, which some models postulate to promote retroviral recombination (6). In separate experiments, we altered intracellular conditions by growing 3T3 cells in 0.5% calf serum rather than our standard 10% calf serum. We observed a twofold increase in template switching with the Laac-117 vector in cells fed 0.5% serum compared to that in cells fed 10% serum (data not shown). This suggests that changes to the intracellular environment other than HU treatment can also affect template switching rates and hence that these effects are not specific to HU treatment.
The effects we report here have interesting implications for the potential contributions of recombination to genetic variation in retroviral populations. Rates of retroviral recombination have typically been assessed in cultured transformed cells, which are significantly more metabolically active than most cells that retroviruses are likely to encounter during natural infection of an organism. For example, replication of human immunodeficiency virus type 1 is slower in certain primary cells than in transformed cell lines (28) and an interesting feature of human immunodeficiency virus is its ability to productively infect nondividing cells or to partially reverse transcribe in quiescent cells (5, 41, 42). Our findings suggest that proviruses generated under such conditions may contain relatively high levels of RT-related errors, such as template switch-induced recombination or other genomic rearrangements.
ACKNOWLEDGMENTS
We thank Vicki Larson for help with some early experiments and Michael Imperiale and David Friedman for critical reading of the manuscript.
This work was supported by American Cancer Society grant RPG-95-058-04-MBC to A.T. and NIH training grant T32 GM 07544 to J.K.P.
REFERENCES
- 1.Anderson J A, Bowman E H, Hu W-S. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J Virol. 1998;72:1195–1202. doi: 10.1128/jvi.72.2.1195-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back N K T, Berkhout B. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1997;41:2484–2491. doi: 10.1128/aac.41.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman R R, Hu W-S, Pathak V K. Relative rates of retroviral reverse transcriptase template switching during RNA- and DNA-dependent DNA synthesis. J Virol. 1998;72:5198–5206. doi: 10.1128/jvi.72.6.5198-5206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckle M, Williams R M, Negroni M, Buc H. Real time measurements of elongation by a reverse transcriptase using surface plasmon resonance. Proc Natl Acad Sci USA. 1996;93:889–894. doi: 10.1073/pnas.93.2.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 7.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 8.Delviks K A, Hu W-S, Pathak V K. ψ-Vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J Virol. 1997;71:6218–6224. doi: 10.1128/jvi.71.8.6218-6224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerman M, Temin H M. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984;39:459–467. [PubMed] [Google Scholar]
- 10.Fisher J, Goff S P. Mutual analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 11.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 12.Goulaouic H, Subra F, Mouscadet J F, Carteau S, Auclair C. Exogenous nucleosides promote the completion of MoMLV DNA synthesis in G0-arrested Balb c/3T3 fibroblasts. Virology. 1994;200:87–97. doi: 10.1006/viro.1994.1166. [DOI] [PubMed] [Google Scholar]
- 13.Hu W-S, Bowman E H, Delviks K A, Pathak V K. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol. 1997;71:6028–6036. doi: 10.1128/jvi.71.8.6028-6036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W-S, Temin H M. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W-S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber H E, McCoy J M, Seehra J S, Richardson C C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis and template switching. J Biol Chem. 1989;264:4669–4678. [PubMed] [Google Scholar]
- 17.Jin D J, Burgess R R, Richardson J P, Gross C A. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci USA. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin D J, Gross C A. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J Biol Chem. 1991;266:14478–14485. [PubMed] [Google Scholar]
- 19.Jones J S, Allan R W, Temin H M. One retroviral RNA is sufficient for synthesis of viral DNA. J Virol. 1994;68:207–216. doi: 10.1128/jvi.68.1.207-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julias J G, Hash D, Pathak V K. E− vectors: development of novel self-inactivating and self-activating vectors for safer gene therapy. J Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julias J G, Pathak V K. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 23.Kulpa D, Topping R, Telesnitsky A. Determination of the site of first strand transfer during Moloney murine leukemia virus reverse transcription and identification of strand transfer-associated reverse transcriptase errors. EMBO J. 1997;16:856–865. doi: 10.1093/emboj/16.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Larson, V., and A. Telesnitsky. Unpublished data.
- 24.Martin-Hernandez A M, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell J C, Roberts J W, Jin D J, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien W A, Namazi A, Kalhor H, Mao S H, Zack J A, Chen I S. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 30.Parthasarathi S, Varela-Echavarria A, Ron Y, Preston B D, Dougherty J P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87:6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston B D, Dougherty J P. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 34.Price J, Turner D, Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc Natl Acad Sci USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhode B W, Emerman M, Temin H M. Instability of large direct repeats in retrovirus vectors. J Virol. 1987;61:925–927. doi: 10.1128/jvi.61.3.925-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M A, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 39.Wooley D P, Bircher L A, Smith R A. Retroviral recombination is nonrandom and sequence dependent. Virology. 1998;243:229–234. doi: 10.1006/viro.1998.9052. [DOI] [PubMed] [Google Scholar]
- 40.Xu H, Boeke J D. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:8553–8557. doi: 10.1073/pnas.84.23.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 42.Zack J A, Haislip A M, Krogstad P, Chen I S Y. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Temin H M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Temin H M. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol. 1994;68:2409–2414. doi: 10.1128/jvi.68.4.2409-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]