Abstract
We previously showed that a didanosine-selected mutation in pNL4-3 background conferred a replication disadvantage on human immunodeficiency virus type 1, resulting in a loss of replication fitness. This work has been extended by showing that a recombinant virus with the HXBc2 backbone and reverse transcriptase (RT) fragments from pNL4-3 containing the Leu74Val mutation produce decreasing amounts of p24 antigen over a 3-week period. The HXBc2 recombinant containing the wild-type RT from pNL4-3 replicated efficiently. When the virion-associated RT containing the Leu74Val mutation was used in an RT processivity assay with homopolymer RNA template-primer, poly(A), and oligo(dT), the RT with altered Leu74Val mutation was less processive, generating fewer cDNA products in comparison to wild-type pNL4-3 RT. The replication kinetics and RT processivity of the mutant with the Leu74Val mutation were compared to those of a lamivudine-selected mutant Met184Val. In replication kinetics assays, mutant Leu74Val replicated slower than the mutant Met184Val. In a processivity assay, the mutant RTs from both viruses show comparable decreases in processivity. These observations provide biochemical evidence of decreased processivity to support the decrease in replication fitness observed with the Leu74Val or Met184Val mutations.
The nucleoside inhibitors of the reverse transcriptase (RT) of human immunodeficiency virus 1 (HIV-1) are converted to the triphosphates, and the nucleoside triphosphates (dNTPs) are incorporated into elongating DNA to greatly inhibit HIV replication (18). The presence of the NTPs in cells produces strong selective pressure on viral replication, causing the wild-type (WT) virus to be rapidly replaced with a drug-resistant variant which is probably present as a minor population in the absence of drugs. The drug-resistant virus is able to replicate to high levels in the presence of drug, but in the absence of drug, the drug-resistant viruses have been suggested to be less fit than the WT population (11). An exception to this general hypothesis are mutants resistant to zidovudine (AZT) containing mutations in the HIV RT gene. The mutants which contain the AZT-related mutation changing threonine to tyrosine at codon 215 and a compensatory mutation lysine to glutamine at codon 219 have been shown to possess an RT with increased processivity compared to the wild-type virus, and viruses containing several AZT mutations replicate to a higher titer in peripheral blood mononuclear cells (PBMC) than the WT virus in the absence of drug (2, 8). Another report has shown contrasting results, i.e., that AZT-resistant HXB2 variants of HIV-1 may replicate more poorly than wild-type virus (25). In contrast, the lamivudine (3TC)-selected mutation Met184Val in the YMDD catalytic region of HIV RT confers a replication disadvantage to HIV in the absence of drug, and the presence of this mutation in purified or virion-associated RT results in an enzyme which exhibits diminished processivity (3, 5). This has also been extended to 3TC-resistant polymerase mutants of hepatitis B virus (HBV), where mutations in the exactly analogous regions of the HBV RT active site (Tyr-Met-Asp-Asp [YMDD]) changing Met 552 to Val results in HBV with a diminished replication capacity in tissue culture (34).
We have previously shown that the didanosine-related mutation Leu74Val in a pNL4-3 background confers a replication disadvantage to the virus and a significant loss in fitness compared to the wild-type virus in a drug-free medium (41). Several clinical trials have shown that didanosine therapy is associated with a persistent low HIV RNA load (15, 24, 32). The detection of mutation Leu74Val in clinical isolates during clinical trials with didanosine monotherapy in AZT-experienced population has been reported to occur frequently in some studies (16, 43). In one study where AZT-experienced individuals were treated with didanosine monotherapy for 12 months, 65% of the individuals developed Leu74Val mutation (43). However, the percentage of clinical isolates that develop Leu74Val mutation is smaller during clinical trials with combination therapy (28, 39, 40). The use of hydroxyurea in combination with didanosine has been suggested as a method to enhance the antiviral affect of didanosine by decreasing intracellular levels of dATP, the natural competitor of ddATP (13). In a clinical trial comparing hydroxyurea and didanosine to didanosine alone, the combination of hydroxyurea and didanosine was associated with a lower HIV-1 RNA level and the appearance of more Leu74Val mutations in patients receiving both drugs (13).
The present study sought to determine the biochemical mechanism for inefficient replication of viruses with Leu74Val mutation and to compare the replication kinetics and RT processivity of didanosine-selected Leu74Val and 3TC selected Met184Val variants. We found that the Leu74Val mutation results in an RT with a decreased processivity on a homopolymer template, poly(A). This finding suggests that the attenuated replication of RT variant Leu74Val is due in part to the decreased processivity of RTs with Leu74Val mutation, providing clear biochemical evidence to support the attenuated viral replication conferred by the didanosine-selected mutation Leu74Val. The in vitro processivities of RTs with Leu74Val and Met184Val mutations were comparable, and RT variant Leu74Val replicated slower than Met184Val variant during replication kinetic analysis.
(This study was presented in part at 5th and 6th Conferences on Retroviruses and Opportunistic Infections, 31 January to 4 February 1998 and 1 to 5 February 1999, Chicago, Ill.)
MATERIALS AND METHODS
Chemicals and medium.
Radionucleotides [methyl-3H]dTTP and [α-32P]dTTP were purchased from NEN, Boston, Mass. Polynucleotides poly(rA), and primer oligo(dT)12–18 were purchased from Boehringer Mannheim, Ind., and poly(rC)-poly(dG)12–18 was purchased from Amersham Pharmacia Biotech, Piscataway, N.J. The oligonucleotides used for mutagenesis were synthesized and high-pressure liquid chromatography purified by Operon, Alameda, Calif. RPMI 1640 medium was used in all cell growth and viral culture assays.
Cells and virus.
Healthy HIV-seronegative donors were routinely bled, and peripheral blood mononuclear cells (PBMC) were separated by Ficoll-Hypaque (Histopaque 1077; Sigma) density gradient centrifugation and stimulated with phytohemagglutinin (PHA; 2 μg/ml) in RPMI 1640 supplemented with 20% heat-inactivated fetal bovine serum, 5% purified human interleukin-2, 250 U of penicillin/ml 250 μg of streptomycin/ml and 2 mM l-glutamine for 48 to 72 h prior to virological study (31). The proviral clone pNL4-3 (contributed by M. Martin) and recombinant RT (HIV-1 BH10) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Site-specific mutagenesis and generation of recombinant viruses.
To insert specific point mutations in RT coding sequences, the Altered Sites in vitro mutagenesis system (Promega, Madison, Wis.) was used according to our previous protocols (41, 42) and directions provided by the manufacturer. Briefly, a 4.3-kb SphI-SalI fragment of proviral clone pNL4-3 was cloned into mutagenesis phagemid pALTER−1. Single-stranded DNA from this recombinant phagemid was used to create specific mutations in RT by using mutagenic oligonucleotides (Table 1). To generate a full-length proviral clone, the 4.3-kb SphI-SalI fragment from pNL4-3 was replaced with the identical fragment of mutagenic vector pALTER-1 carrying various mutations in the RT gene.
TABLE 1.
Oligonucleotides used in this study
| Primer | Primer sequencea | Codon
|
|
|---|---|---|---|
| WT | Mutant | ||
| With mutated RT codon | |||
| Lys70→Thr | 5′TTTTCTCCATGTAGTACTGTC3′ (2768–2748) | AAA | ACA |
| Leu74→Val | 5′GAAATCTACTACTTTTCTCCAT3′ (2780–2759) | TTA | GTA |
| Met184→Val | 5′AATCATCCACGTATTGATAG3′ (3108–3089) | ATG | GTG |
| For sequencing of the RT gene | |||
| 5′CTGCACTTTAAATTTTCCCATT3′ (2534–2555) | |||
| 5′ACAATGGCCATTGACAGAAG3′ (2615–2634) | |||
| 5′AAGTATACTGCATTTACCATACCTAGTATA3′ (2925–2954) | |||
| 5′CCAGCAATATTCCAGTGTAGCATG3′ (3018–3041) | |||
| For PCR amplification of the RT gene | |||
| 5′TCAGATTGGCTGCACTTTGAATTCTCCCATTAGTCCT3′ (2525–2561) | |||
| 5′CTTGGGCCTTATCTATTCCATCTAGAAATAGTAC3′ (4257–4224) | |||
To ensure that the attenuation of viral replication by the Leu74Val mutation in pNL4-3 (41) did not reflect merely the plasmid background, we also analyzed this property in a different plasmid backbone. The wild-type RT fragment (SphI-SalI) of proviral clone HXBc2 was replaced with the equivalent WT or mutated (Leu74Val) RT fragment of the pNL4-3 clone, and the recombinant virus was designated HXPNRT or HXPNRT74, respectively. PBMC (5 × 106) were transfected with 5 μg of each proviral clone, and antigen p24 was measured at different time points.
Transfection, infections, and virus propagation.
PHA-stimulated PBMC (5 × 106) were harvested, washed twice with cold phosphate-buffered saline, and suspended in 250 μl of cold medium (RPMI 1640) supplemented with fetal bovine serum and antibiotics; 5 to ten μg of plasmid DNA was added to the cells in a cuvette (0.4 μm) and kept on ice, and the cells were transfected by electroporation at 250 V and 960 μF with a Bio-Rad Gene Pulser. The electroporated cells were kept on ice, resuspended in 5 ml of RPMI 1640 supplemented with fetal bovine serum and antibiotics, and incubated at 37°C under 5% CO2. Virus production was monitored by determining the presence of antigen p24 with an enzyme-linked immunosorbent assay (ELISA) kit (NEN) and by assaying the RT activity by an in vitro RT assay which measures the incorporation of [methyl-3H]TTP into cDNA of a poly(rA) template in the presence of virion-associated RT and oligo(dT) (41). The cell-free supernatants from these cultures were saved in aliquots for viral propagation. The AIDS Clinical Trials Group (ACTG)-Department of Defense consensus protocols were followed for virus propagation and stock titration (31). Once sufficient virus production (>60 ng of p24 antigen/ml) was achieved, the cell-free virus was stored at −70°C for various analyses. The 50% tissue culture infective dose (TCID50) was calculated by the Spearman-Karber method as discussed elsewhere (31).
Replication kinetic and growth competition assays.
Replication kinetic assays and growth competition assays were carried out as described previously (41). The infections were done with 1,000 TCID50/106 PBMC. In each case, 2 × 106 PBMC were infected for 2 h. Aliquots of culture supernatants were collected everyday until day 7 to monitor viral replication, and cultures were replaced with equivalent amount of fresh RPMI 1640. Cell free virus cultures in duplicate were saved to measure RT activity and antigen p24 concentration, respectively.
Growth competition assays were performed by coinfecting PBMC with equivalent amounts (TCID50) of two viruses and then comparing the fitness of one virus in relation to other over a period of time. This was done by monitoring the extent of virus replication as determined by the presence of the mixture of nucleotides (codons) at a single locus. RT codon 184 was monitored for the presence of the mixture of mutant (G) and WT (A) nucleotides. The less fit virus will show a decrease in peak height over a period of time. To calculate the loss in fitness, the percentage of relative peak heights for both nucleotides at various time points were determined and the selection coefficient (s = fitness difference) was determined by the following mathematical model: s = l/t ln[q(t)p(o)/p(t)q(o)], where t = total time of passage, q(t) = more fit population, p(t = less fit population, and p(0) = q (0) = 0.5 (19, 35).
Quantitation of cDNA product generated during endogenous RT assay.
Virion-associated RT lysates containing endogenous template-primer and RT were analyzed for cDNA synthesis in a time point assay (38). The reaction mixture for this assay contained 50 mM Tris (pH 7.8), 5 mM MgCl2, 60 mM KCl, 10 mM dithiothreitol, 10 mM NaCl, 1 mM EGTA, 0.1% NP-40, 0.4 mM of dNTPs (dCTP, dGTP, and dTTP), 10 μCi of [α-32P]dATP, and equal amounts of RT lysates, normalized on the basis of equivalent RT activity. All reaction mixtures were incubated at 37°C for 1, 5, 10, 20, 40, and 60 min. The reactions were stopped by adding equal volume of stop buffer (1% sodium dodecyl sulfate, 50 mM EDTA, 0.2 M NaCl). Prior to loading on a 1% denaturing agarose gel (20 mM NaOH, 1 mM EDTA), the reaction mixtures were boiled, subjected to digestion with 1 U of pronase (Pharmacia) at 56°C for 30 min, and extracted with phenol-chloroform followed by precipitation with ethanol. After separation on the gel, the products were visualized by autoradiography and scanned for the intensity of α-32P by using the ImageQuant (Molecular Dynamics) software.
In vitro RT processivity assay.
Processivity assays were carried out according to published protocols (3). Briefly, enzymes were released from the virions by NP-40 treatment (0.5%), and RT activity was determined in a poly(rA)-oligo(dT) assay (42). RT lysates of WT, Leu74Val, and Met184Val containing equivalent amounts of RT activity and assay mixtures containing 60 mM Tris (pH 7.8), 75 mM KCl, 5 mM MgCl2, 0.1% NP-40, 1 mM EDTA, 5 μg of poly(rA)3000/ml, 0.16 μg of oligo(dT)15/ml, 4 mM dithiothreitol, and 50 μCi of [α-32P]dTTP/ml were incubated at 37°C water bath for 3 h. Five to 10 μl of reaction mixture was spotted onto DEAE ion-exchange paper (DE81; Whatman), and RT activity was determined. To determine the cDNA length distribution, the reaction mixture was precipitated after extraction with phenol-chloroform, suspended in 5 μl of sterile water, and run on a 6% polyacrylamide sequencing gel. After separation, the gel was dried and exposed to autoradiography. The products were visualized and scanned for α-32P by using the ImageQuant (Molecular Dynamics) software.
RT processivity was also measured in the presence of a 50-fold molar excess of a trap, poly(rC)-oligo(dG). In the preincubated RT assay mixture, trap was added before addition of [α-32P] dTTP. The assay conditions and the protocols to measure RT activity and processivity were identical to those described above.
Confirmation of mutations by nucleotide sequence analysis.
The clones generated by site-specific mutagenesis of the RT region were always confirmed for the desired mutation by an automated DNA sequencer (model 373A; Applied Biosystems). To ensure that the mutation is retained in provirus after transfection and virus production and no other mutations are present, infected genomic DNA was amplified by PCR and the nucleotide sequence was determined from RT codons 10 to 250. Oligonucleotides used during mutagenesis, PCR, and sequencing are listed in Table 1.
Statistical analysis.
Statistical analysis was carried out to compare the processivity of WT RT with mutant RTs containing Leu74Val and Met184Val mutations. The analysis was designed to test the hypothesis that for three RTs, cDNA density decreases at the same rate as DNA band number increases. Across five processivity assays, statistical values were obtained that include mean, median, standard deviation, and minimum and maximum. A nonparametric (Kruskal-Wallis) test was performed to compare the smaller cDNA products. Slopes of regression on DNA density were generated for three RTs, and a t test was performed to compare the mean slopes of band number over five assays.
RESULTS
Attenuated replication of HIV-1 containing RT mutation Leu74Val in the HXBc2 background.
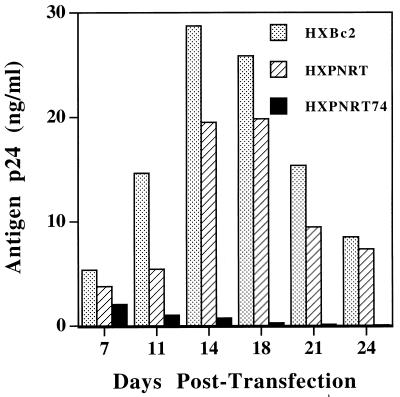
We have earlier shown that didanosine-selected mutation Leu74Val confers a replication disadvantage to the virus in cloned viruses with a pNL4-3 background (41). To determine the effect of this mutation in a different background, recombinant viruses were generated by cloning the WT and mutated RT (Leu74Val) from pNL4-3 plasmid into the HXBc2 proviral clone. It should be noted that the 3′ halves of vectors pNL4-3 and HXBc2 are derived from the same virus isolate (LAV). Since the 4.3-kb SphI-SalI fragment of pNL4-3 was exchanged with the WT fragment of vector HXBc2, the major difference in pNL4-3 virus and recombinant virus is the 1.5-kb upstream sequence that includes 5′ long terminal repeat of the HXBc2 vector. Equivalent amounts (5 μg) of plasmids HXBc2 (WT), HXPNRT (HXBc2 with WT RT of pNL4-3), and HXPNRT74V (HXBc2 with 74ValRT of pNL4-3) were transfected into PHA-stimulated PBMC, and virus replication was monitored by determining HIV antigen p24 production. Relative viral replication is shown in Fig. 1. While a small difference in antigen p24 production was observed between HXPNRT and HXPNRT74V viruses on day 7, there was a marked continuous decrease in the level of antigen p24 production over a period of 3 weeks by the HXPNRT74V virus. These observations show that the didanosine-selected mutation Leu74Val also confers a replication disadvantage in the HXBc2 background.
FIG. 1.
Attenuated replication of recombinant HXPNRT74 virus. Equivalent amounts of DNAs were transfected in PBMC in duplicate, and the production of antigen p24 was monitored until day 24. Aliquots of culture supernatants were collected at various time points. Cultures were fed with fresh PBMC on days 7, 14, and 21. Samples were diluted, and antigen p24 concentration was determined by ELISA. The bar diagram shows the production of antigen p24 in relation to time.
Decreased cDNA synthesis in virion-associated RT lysates of Leu74Val.
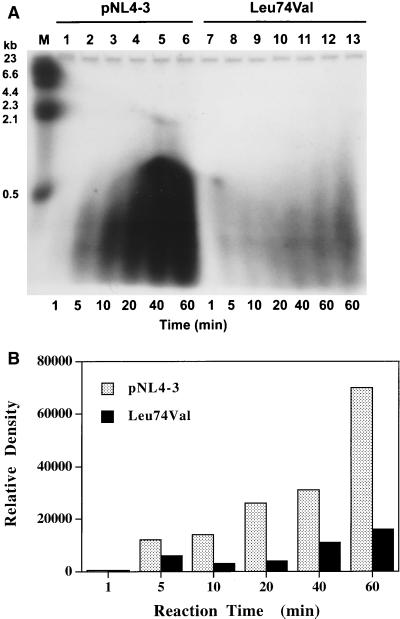
Since our PBMC-based in vivo replication kinetics assays have clearly shown that the Leu74Val mutation confers a replication disadvantage to the virus in two different vectors, pNL4-3 (41) and HXBc2 (as shown above), we compared the extents of cDNA synthesized by the RT from cell-free virions of Leu74Val and WT viruses. RT lysates of WT pNL4-3 and Leu74Val viruses that contained equivalent amounts of RT activity (counts per minute) were incubated with constant amounts of dNTPs, and reactions were stopped at various time points. The DNA generated was subjected to denaturing agarose gel electrophoresis and quantitated by scanning (Fig. 2). Visual examination of the autoradiograph clearly shows that significantly fewer cDNA molecules were generated by the lysates of mutant virus Leu74Val than by WT virus. Densitometer scanning of the autoradiograph revealed that there was a sevenfold decrease in the amount of total DNA product generated in 60 min by mutant RT in comparison to WT RT (Fig. 2B). The DNA products generated from mutant RT appear to be shorter than product formed by WT RT. Although this assay system was not a conventional system to determine the processivity of RT, the generation of fewer DNA molecules suggested that the mutant RT is defective in the ability to bind to template and increase the length of cDNA molecules compared to the longer time period that the WT RT is associated with the template.
FIG. 2.
Decreased synthesis of cDNA products during endogenous RT assay with Leu74Val RT. Virion-associated RT lysates with equivalent RT activity (2.5 × 104 methyl-3H cpm) were incubated at 37°C in the presence of dCTP, dGTP, dTTP, and [α-32P]dATP, and reactions were stopped at 1, 5, 10, 20, 40, and 60 min. Endogenous synthesized DNA products were digested with pronase, extracted with a phenol-chloroform mixture, and precipitated with ethanol. DNA products were boiled and separated on an alkaline agarose gel. The gel was dried and exposed to autoradiography. (A) Autoradiograph. Lanes: 1 to 6, RT lysates of pNL4-3 (WT) virus; 7 to 12, RT lysates of Leu74Val virus; 13, RT lysate of Leu74Val virus containing 1.5-fold-higher RT activity (4.0 × 104 methyl-3H cpm). (B) Quantitation of DNA products. The intensity of the total cDNA synthesized in each lane was quantified with ImageQuant (Molecular Dynamics) software and plotted in relation to time.
Decreased processivity with mutant Leu74Val RT.
Earlier studies have shown that the HIV replication efficiency is related at least in part to the processivity of RT. Combinations of multiple AZT-selected mutations have been shown to increase processivity (2, 8), and 3TC-selected mutation Met184Val has been shown to decrease RT processivity in in vitro processivity assays (3, 5). We determined whether the RT with didanosine-selected mutation Leu74Val also affects the processivity of RT.
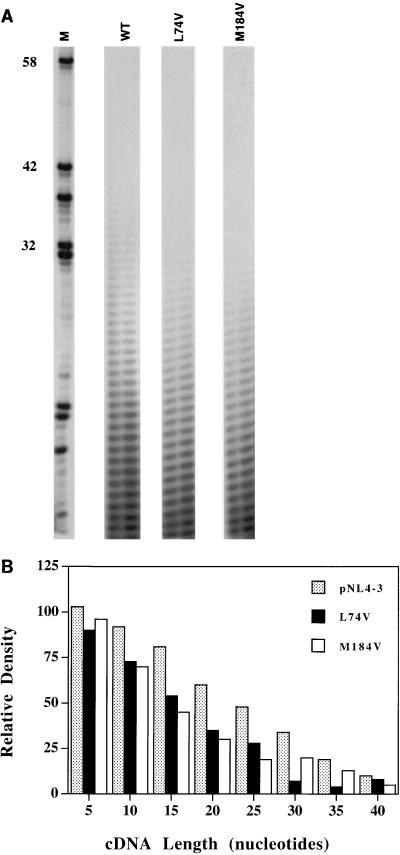
To control the synthesis of cDNA molecules in a way such that each synthesized cDNA molecule results from a single processive cycle, processivity was determined under conditions that use a great template excess and limiting polymerase. To determine the optimal ratio of template-primer and RT, pilot experiments were done at various time points with a range of template-primer concentrations. The processivity assays were performed under conditions that allowed the synthesis of increased numbers of various lengths of cDNA molecules with an increase in the amount of RT lysates. Virion-associated RT lysates of WT (pNL4-3), Leu74Val, and Met184Val that contain equivalent RT activity were incubated with template homopolymer poly(A) and primer oligo(dT). Since virion-associated RT of Met184Val virus has been shown to have a decreased processivity (3), this virus was used as a positive control in our assay system. Moreover, this allowed us to compare the processivity of Leu74Val and Met184Val RTs in the same assay system. Reaction products were subjected to a polyacrylamide gel electrophoresis, and the gel was exposed to autoradiography. Similar results were observed in five independent processivity assays using RT lysates from different virus supernatant stocks, and Fig. 3A shows representative results. The processivity of recombinantly purified RT (HIV-1 BH10) and virion-associated RT of pNL4-3 virus were similar when RT assays were normalized with equivalent RT activity (data not shown). Analysis of cDNA bands on the autoradiograph revealed that RT with the Leu74Val mutation has a decreased processivity, as fewer cDNA molecules were synthesized than with WT RT (Fig. 3A).
FIG. 3.
Decreased processivity of RT containing the didanosine-selected Leu74Val mutation. Various amounts of WT and mutant RT lysates were assayed for RT activity with the poly(rA)-oligo(dT) system (see Materials and Methods), and the reaction products were analyzed on a 6.0% polyacrylamide-7.1 M urea sequencing gel. Reaction products with approximately equivalent amounts of RT activity are shown on a representative gel (A). M, size markers from a dideoxy sequencing reaction with a pBluescript plasmid and M13/pUC reverse sequencing primer (T-track only). RT activities for WT, Leu74Val, and Met184Val viruses were 3.0 × 104, 3.1 × 104, and 3.5 × 104 cpm of α-32P, respectively. Nucleotide lengths of cDNA synthesized for WT, Leu74Val, and Met184Val are 43, 33, and 33, respectively, in this assay. Numbers on the left indicate the nucleotide lengths. (B) Size distribution and relative amounts of cDNA synthesized by WT and mutant RTs. The median values for relative cDNA density from five independent assays were generated by using a statistical program and plotted with respect to sizes of cDNA (Table 2). Groups of five cDNA bands starting at the bottom of gel in individual lanes were quantified at one time, and the intensity of the entire lane was determined with ImageQuant (Molecular Dynamics) software. The bar diagram shows the relative amounts of cDNA synthesized by WT and mutant enzymes.
We measured the density of cDNA bands with ImageQuant analysis and performed statistical analysis on cDNA density data obtained from five independent processivity assays. The median values obtained across five assays are summarized in Table 2, and a bar diagram generated from median values of three viruses is presented in Fig. 3B. A nonparametric (Kruskal-Wallis) test was performed to compare the first five band values for the three viruses, and no significant difference was observed (P = 0.11). We compared the slopes of regression on DNA density by band number for the three viral RTs. Regression lines were fit for each assay, and graphs clearly indicate a quadratic trend (data not shown). A t test was performed to compare the mean slopes of band number over the five assays for WT versus Leu74Val virus and WT versus Met184Val virus. The test indicated that DNA density decreases at a significantly higher rate for virus 74Val compared to WT virus (P = 0.03). Mean slopes for WT and Leu74Val viruses were −0.02 and −0.05, respectively. Similar result were obtained for WT and 184Val viruses (P = 0.02). Mean slopes for WT and 184Val viruses were −0.02 and −0.07. Densitometer scanning of the cDNA products generated by WT and mutant RTs revealed that while the densities of cDNA bands were similar in shorter cDNA products (5 to 10 nucleotides), the differences were striking in the larger cDNA products (>15 nucleotides) of WT and mutant RTs. Under our assay conditions, while the WT RT generated bands up to 43 nucleotides in length, the visible cDNA bands generated with mutant RTs of Leu74Val and Met184Val were around 33 nucleotides in length (Fig. 3A). This indicates that mutant RTs have decreased processivity and are unable to bind to template-primer complex for a prolonged period of time as the WT RT does. The processivity of RT with the Leu74Val mutation was comparable to that of RT with the Met184Val mutation. These observations clearly show that the RTs containing the Leu74Val and Met184Val mutations are less processive than the WT RT enzyme.
TABLE 2.
Summary of relative cDNA synthesis during processivity assaysa
| Group of cDNA bands | Median cDNA density (minimum/maximum)
|
||
|---|---|---|---|
| pNL4-3 | Leu74Val | Met184Val | |
| 5 | 103.5 (91.0/115.0) | 90.0 (88.0/98.0) | 96.5 (92.0/99.0) |
| 10 | 92.5 (87.0/102.0) | 73.0 (68.0/85.0) | 70.0 (55.0/79.0) |
| 15 | 81.5 (71.0/104.0) | 54.0 (39.0/70.0) | 45.0 (21.0/57.0) |
| 20 | 60.5 (57.0/92.0) | 35.0 (23.0/54.0) | 30.0 (13.0/45.0) |
| 25 | 48.5 (36.0/65.0) | 28.0 (12.0/41.0) | 19.0 (5.0/36.0) |
| 30 | 34.0 (26.0/69.0) | 7.0 (5.0/29.0) | 20.0 (6.0/23.0) |
| 35 | 19.0 (12.0/27.0) | 4.0 (2.0/15.0) | 13.0 (2.0/13.0) |
| 40 | 10.0 (6.0/18.0) | 8.0 (8.0/8.0) | 5.0 (5.0/5.0) |
Across the five processivity assays, descriptive statistics of relative density were generated separately by virus and cDNA length (nucleotides). RT activities for WT and mutant viruses were similar in independent processivity assays and within ±10% among different assays.
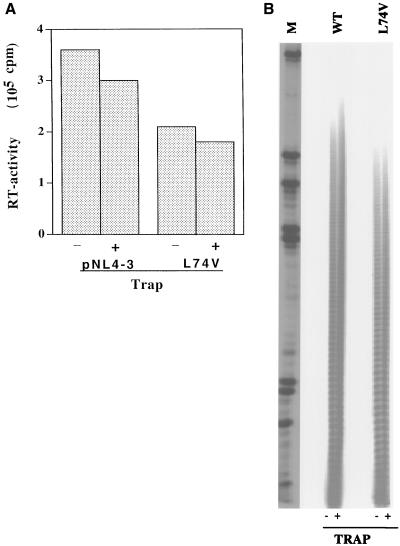
It has previously been shown that the presence or absence of the trap poly(rC)-oligo(dG) did not effect the length or the amount of cDNA products generated under the assay conditions used to measure the processivity of virion-associated RT Met184Val (3). We determined if such a trap could affect the processivity of RT with the Leu74Val mutation. The addition of a 50-fold molar excess of poly(rC)-oligo(dG) to the RT assay resulted in a little effect on RT activity and no effect on processivity of both WT and Leu74Val enzymes (Fig. 4). This showed that we were actually measuring the processivity in a single cycle of reverse transcription.
FIG. 4.
Processivity of WT and Leu74Val RT in the presence of trap. RT assays were done in the absence or in the presence of a 50-fold molar excess of trap poly(rC)-oligo(dG). One-tenth of the reaction mixture was analyzed for RT activity (A), and remaining cDNA products were analyzed for cDNA distribution on a 6% polyacrylamide gel (B).
We also compared the RT activities of WT, Leu74Val, and Met184Val RTs at assay temperatures of 30, 37, and 40°C. No significant difference in RT activity was observed for any of these enzymes at this range of temperatures (data not shown).
Decreased processivity of RTs containing Leu74Val and Met184Val mutations at various MgCl2 concentrations.
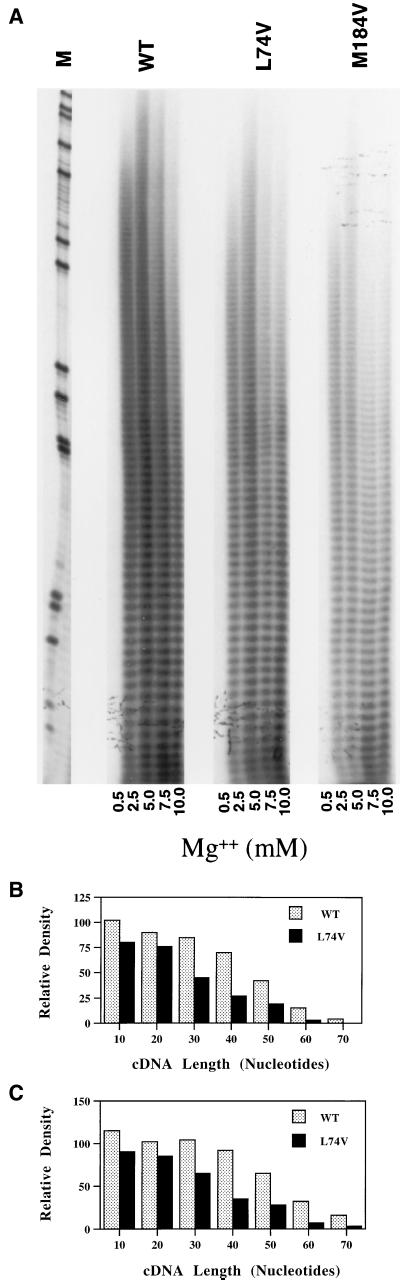
It has been postulated that the aspartic acid residues of the YMDD motif play a role in the metal ion coordination at the catalytic active site of RT. We have analyzed the effect of a range of MgCl2 concentrations on processivity of three RTs (Fig. 5A). The optimal RT processivity for WT, Leu74Val, and Met184Val was measured at 5 mM MgCl2. In a comparison of the WT and Leu74Val RTs, the processivity defect was obvious at MgCl2 concentrations of 2.5, 5.0, and 7.5 mM. In the presence of 10.0 mM MgCl2, the differences between RT processivity of WT and 74Val RTs were smaller. The input RT activities for WT and Leu74Val were 4.5 × 104 and 4.9 × 104 cpm, respectively, in this assay (Fig. 5A). We compared the amounts of cDNA generated by WT and Leu74Val RTs in the presence of 2.5 mM (Fig. 5B) and 5.0 mM (Fig. 5C) MgCl2. Similar to our processivity data (Fig. 3A), the differences in processivity between the WT and Leu74Val RTs were more pronounced for larger cDNA products than the shorter cDNA products. The Leu74Val and Met184Val mutant RTs exhibited very similar decreases in processivity at MgCl2 concentrations of 2.5, 5.0, and 7.5 mM. In contrast to a previous study (3) where shorter cDNA products were observed with the Met184Val RT in the presence of various concentrations of MgCl2, we found approximately similar length cDNA products with all three enzymes, WT, Leu74Val, and Met184Val. It should be noted that the actual input RT activity of Met184Val (1.5 × 104 cpm) was threefold lower than that of the WT and Leu74Val RTs, and therefore the length of cDNA products generated in this assay is not dependent on the RT activity of the three enzymes. Since no major differences were observed in processivity of WT and Leu74Val RT under various concentrations of Mg2+, we conclude that RT residue Leu74 does not play a role in Mg2+ coordination.
FIG. 5.
Analysis of RT processivity under suboptimal concentrations of MgCl2. WT and Leu74Val RT lysates containing RT activities of 4.5 × 104 and 4.9 × 104 cpm of α-32P, respectively, were compared for RT processivity in the presence of various concentrations of MgCl2. RT assays were performed with the template-primer poly(rA)-oligo(dT) and increasing concentrations of MgCl2 (0.5, 2.5, 5.0, 7.5, and 10.0 mM). RT lysates of Met184Val that contain a threefold less RT activity (1.5 × 104 cpm) than WT or Leu74Val were included as a control. cDNA products were run on the gel and visualized by autoradiography (A). M, size markers from a dideoxy sequencing reaction with a pBluescript plasmid and M13/pUC reverse sequencing primer (T-track only). Relative amounts of cDNA synthesized with WT and Leu74Val RT in the presence of 2.5 mM (B) and 5 mM (C) MgCl2 were quantified by ImageQuant (Molecular Dynamics) software.
RT variant Leu74Val replicates slower than Met184Val.
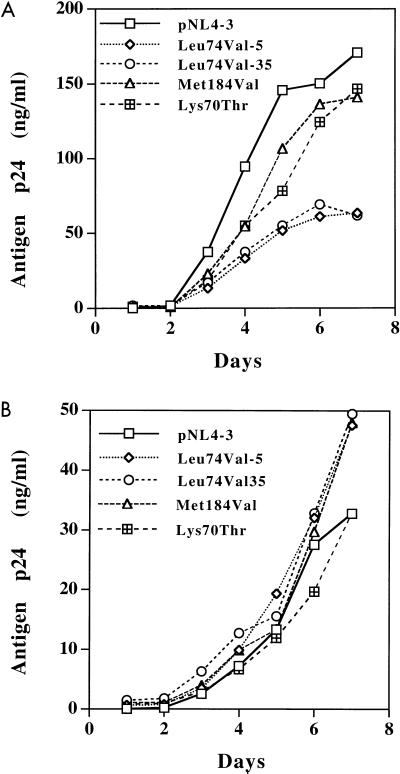
Since our RT processivity analysis showed that RTs with Leu74Val and Met184Val mutations have similar processivities, we examined the replication kinetics in the absence of the drug. To generate RT variants, point mutations were introduced in the RT gene of proviral clone pNL4-3 by site-directed mutagenesis. Two independent clones containing Leu74Val mutations were used in this study. Clone Leu74Val-35 was used in our previous study (41, 42), and a second clone, Leu74Val-5, was included to control for variation among clones. A previously analyzed clone, Lys70Thr, that confers a replication disadvantage to the virus was used as a negative control (41). PHA-stimulated PBMC were infected with equivalent infectious amounts of WT pNL4-3 and RT variants Leu74Val-5, Leu74Val-35, Met184Val, and Lys70Thr in the presence or absence of 10 μM didanosine. Culture supernatants were collected every day until day 7, and replication kinetics were determined by monitoring antigen p24 production (Fig. 6).
FIG. 6.
Comparison of replication kinetics of RT variants. PHA-stimulated PBMC were infected with 1,000 TCID50 of WT Leu74Val-5, Leu74Val-35, Met184Val, and Lys70Thr viruses in the absence (A) or presence (B) of 10 μM didanosine. Culture supernatants were collected every day until day 7, and antigen p24 production was monitored by ELISA. Plots show the production of antigen p24 in relation to time.
Both RT variants Leu74Val and Met184Val replicated less efficiently than WT virus, confirming the previous observations (3, 41). However, Leu74Val variant replicated two- to threefold slower than the variant Met184Val, and the amounts of antigen p24 produced over a period of 7 days were 60 and 140 ng/ml, respectively (Fig. 6A). The statistical significance of a twofold decreased replication of Leu74Val virus in comparison to Met184Val virus is not clear. In view of the amount of antigen p24 production during a 7-day period, the relative replication efficiencies of these viruses were WT > Met184Val > Leu74Val. In three independent replication kinetics assays, the replication disadvantage difference between WT virus and Leu74Val was statistically significant (P = 0.007) by Student t-test paired analysis.
We also compared the replication kinetics of RT variants in the presence of 10 μM didanosine. The assay conditions and viral inoculum were identical to those described above. Both of the variants Leu74Val and Met184Val replicated slightly better than WT virus in the presence of drug between days 1 and 6, except that a rapid increase in antigen p24 production was observed on day 7 (Fig. 6B). Surprisingly, the amounts of p24 antigen produced (50 to 60 ng/ml) by Leu74Val variant on day 7 were similar in both the presence and the absence of the drug, suggesting that the didanosine-selected Leu74Val variant replicates inefficiently but better than the WT virus even in the presence of the drug. However, the plot in Fig. 5B shows that while a plateau is reached for antigen p24 production on day 7 in case of WT virus, there is evidence of increasing viral replication for RT variants Leu74Val and Met184Val. To better understand the statistical significance of the replication differences between the WT and mutant viruses, a detailed evaluation is warranted. Nevertheless, the comparison of replication kinetics in the absence and presence of didanosine suggests that both RT variants containing Leu74Val and Met184Val mutations also replicated slowly in the presence of drug. This suggests that certain drug-selected variants, irrespective of the level of the resistance they confer, may result in an increase in lag phase during replication and a decrease in total viral load, even in the presence of drug(s).
Relative fitness of variants Leu74Val and Met184Val.
We have shown above that the RT variant Leu74Val replicates slower than the 3TC-selected variant Met184Val in drug-free cultures, although in vitro processivities of RTs of the variants are similar. We have previously shown that RT mutation Leu74Val results in a 11% loss of fitness in replication in comparison to WT virus (41). To compare the relative fitness of these variants, we coinfected PHA-stimulated PBMC with equivalent amount of antigen p24 and collected infected cell pellets on days 2, 4, and 7. Genomic DNA was isolated from all cell pellets; the RT gene was amplified and sequenced with a ABI model 373A automated DNA sequencer. Comparison of viral fitness with a mathematical model revealed that the RT variant Met184Val has a 2.4% of loss of fitness in comparison to WT virus. The loss of fitness for Met184Val virus was similar (3%) when the percentage differences in antigen p24 production between WT and Met184Val viruses on day 7 (Fig. 4A) were used to calculate the fitness difference. The loss of fitness for Leu74Val variant was more pronounced (5.7%) in comparison to Met184Val variant. Similar to differences observed in replication kinetic assays, the relative fitness of WT virus and RT variants was WT > Met184Val > Leu74Val.
DISCUSSION
The role of viral fitness in HIV pathogenesis has been appreciated only recently, and few studies have addressed this issue for cloned viruses as well as clinical HIV RT and protease variants (12, 19, 20, 25, 33, 46). Viral fitness may contribute to the optimal antiviral effect of suppressing viral replication and HIV RNA levels in patients treated with antiretroviral therapy. In this study, we have shown that the RT containing the Leu74Val mutation exhibits decreased processivity similar to the processivity of Met184Val RT. Additionally, in vivo PBMC-based replication kinetic assays demonstrate that the Leu74Val variant replicates two- to threefold slower than the Met184Val virus, and there was a 2.4% loss of fitness for Met184Val in comparison to WT virus and a 5.7% loss of fitness for Leu74Val virus compared to Met184Val virus in growth competition assays.
We have shown previously that a didanosine-selected mutation Leu74Val in pNL4-3 background confers a replication disadvantage to the virus that results in a significant loss of fitness to the virus (41). These results were extended to show that the recombinant HXBc2 with a pNL4-3 RT containing a Leu74Val mutation is also attenuated for replication.
Comparison of virion-associated RT of Leu74Val variant and the WT RT showed that the RT of Leu74Val was less processive than the WT RT. This provides evidence that attenuated replication of Leu74Val variant is due to an RT which exhibits decreased processivity, similar to the decreased processivity seen with Met184Val RT (4). Our observations are in contrast to two previous reports where the authors claimed that the WT and Leu74Val RTs have similar processivities (7, 8). A detailed evaluation of the differences between the intensities of various DNA bands of WT and mutant RTs in one of the two reports (8) indicates that the Leu74Val RT appears to be two- to threefold less processive than the WT RT. Since the major focus of that study was comparison of the processivities of RTs containing multiple AZT-selected mutations, it appears that the authors were not impressed with the small differences in the processivity of Leu74Val RT. The assay system used in both of those studies was different in several ways from the one used in the present study. In the previous publications, a heteropolymer RNA template was used and processivity was dependent on the binding of the excess RT to a heparin trap or to excess poly(rC)-oligo(dG) trap. The processivity is limited in these systems due to polymerase stalling at specific pause/stop sites along the template, and as a result enzymes terminated with different efficiencies at these preferred stops. In both of the previous reports, several cDNA products that are synthesized with Leu74Val RT appear to have a density lower than that of WT RT (7, 8). Our processivity assay was optimized on the basis of the presence of excess template and limiting RT activity. We also determined that the addition of trap in a 3-h RT assay effects the RT activity very little and has no effect on processivity of WT or Leu74Val enzymes. The Met184Val RT has been shown convincingly to have a decreased processivity with the template-primer homopolymer poly(A) and oligo(dT), and we used this assay system for our comparisons of Leu74Val and Met184Val RTs. The processivities of RTs with Leu74Val and Met184Val mutations were comparable in this system. This observation was surprising, since the Leu74Val variant replicated slower during replication kinetic analysis and showed a 5.7% loss of fitness in growth competition assays compared to the Met184Val mutant. This suggests that other viral or cellular factors may contribute to viral replication in PBMC cultures or in vivo in humans.
In the ACTG 306 study, no significant benefit was observed with the initial combination of 3TC and didanosine, and when 3TC is present at the same time as didanosine, 3TC cannot confer a further replication disadvantage. Following 24 weeks of didanosine monotherapy, however, the addition of 3TC appeared to result in a further decrease in HIV RNA levels (32). The possibility of the development of 3TC-selected Met184Val mutation that confers cross-resistance to didanosine could not be ruled out in the ACTG 306 study. However, the overall outcome of ACTG 306 study suggests the role of impaired viruses in decreased HIV RNA levels. Further pilot clinical trials are needed to evaluate the relative order of the drugs to be given to achieve an optimal reduction in viral load.
The recent description of a covalently trapped catalytic complex of WT HIV-1 RT provides additional important information on the structural implications of the Leu74Val mutation. The authors determined the three-dimensional crystal structure at a resolution of 3.2 Å of the HIV-1 RT complexed with the DNA template-primer and a dNTP (29). They show that the Leu74Val mutation affects a residue which they state “locks the templating base tightly in place.” The mutation is also likely to influence dNTP binding directly, since Leu74 contacts the side chains on Arg72 and Gln151, which stack on the base of the dNTP. The authors speculate that small changes in the contacting residues can markedly influence the rate of nucleotide incorporation. This provides additional structural information to support the decrease in processivity and slowing of viral replication observed with the Leu74Val mutation. The above study also provided evidence that the direct interaction of DNA duplex with bases occurs in the minor groove at positions n to n − 3, the van der Waals contacts with Pro157 and Met184 (base pair n) and with Ile94 (base pairs n − 2 and n − 3), and hydrogen bonds between Tyr183 and G(n − 1) (29). Ile94 has been shown to be a part of significant structural element called the minor groove binding track (MGBT) that comprises five amino acids, four of them (Gln258, Gly262, Trp266, and Gln269) in α-helix H and one (Ile94) in β-sheet 5b (4). In this study, the authors have shown that the processive synthesis by HIV RT involves the interactions between the minor groove of the template-primer and MGBT. On the basis of the above two structural studies, it can be assumed that the mutation at codon 184 will result in an altered interaction with DNA duplex in the minor groove and indirectly may affect the translocation due to MGBT and thus result in a reduced processivity. The Leu74Val mutation does not appear to directly alter interaction with duplex DNA in the minor groove, and the decrease in processivity observed with Leu74Val suggests that other mechanisms can contribute to a decrease in processivity.
It was shown previously that the WT RT failed to incorporate dideoxynucleotides when the template extension was less than three nucleotides in length, and when the template extension was greater than three nucleotides, WT RT began to incorporate dideoxynucleotides. However, HIV-1 RT variants containing the Leu74Val or Glu89Gly mutations did not readily incorporate dideoxynucleotides even with the presence of the long template extension (6). It has been proposed that these mutations alter the position of the template-primer, which in turn alters the geometry at the polymerase active site (6). Based on our observations in this study, we speculate that these alterations affect not only the incorporation of dideoxynucleotides but also the incorporation of inherent dNTPs in growing cDNA chain resulting in a decreased processivity of Leu74Val RT.
Our observations on the attenuated replication and decreased processivity of RT containing the Leu74Val mutation suggest a mechanism for the clinical benefits observed with didanosine monotherapy. It was shown previously that during HIV infection, extremely high viral loads that correlate with a loss of HIV-specific cytotoxic T lymphocytes can be achieved (9, 21, 27, 36). Viral dynamics studies and the use of mathematical projections have shown that HIV turnover in infected patients is extremely high (26, 37, 44). Combination therapy for HIV treatment has become attractive because of the ability to sustain suppression of viral load (14, 23). However, the presence of a latent reservoir of HIV-1 has been observed in patients on highly active antiretroviral therapy (10, 17, 30, 45). To reduce the latent HIV-1 pool, a combination of drugs that select for HIV variants that are impaired for viral replication or have a significant loss of fitness would be more promising.
ACKNOWLEDGMENTS
Support for this work was provided by NIH grants AI27659 and AI38858 to C.S.C. and training grant 5T32 AI07387 to P.L.S.
We thank Xiaochuan Zhou for performing antigen p24 ELISA and Sita Srinivasan for fractionating cDNA products on sequencing gels. We are extremely thankful to Meredith Regan and Liping Chen of the Biometrics Department for assistance with the statistical analysis. We thank Lin Zhang for careful reading of the manuscript.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arion D, Kaushik N, McCormick S, Borkow G, Parnaik M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 3.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Bebenek K, Beard W A, Darden T A, Li L, Prasad R, Luxon B A, Gorenstein D G, Wilson S A, Kunkel T A. A minor groove binding track in reverse transcriptase. Nat Struct Biol. 1998;4:194–197. doi: 10.1038/nsb0397-194. [DOI] [PubMed] [Google Scholar]
- 5.Boyer P L, Hughes S H. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1995;39:1624–1628. doi: 10.1128/aac.39.7.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer P L, Tantillo C, Jacoba-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer P L, Gao H Q, Hughes S H. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob Agents Chemother. 1998;42:447–452. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caliendo A M, Savara A, An D, DeVore K, Kaplan J C, D’Aquila R T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmichael A, Jin X, Sissons P, Borysiewicz L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1)-specific cytotoxic T lymphocytes (CTL) response at different stages of HIV-1 infection: differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 12.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Antoni A, Foli A, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 14.de Jong M D, Boucher C A B, Galasso G J, Hirsch M S, Kern E R, Lange J M A, Richman D D. Consensus symposium on combined antiviral therapy. Antiviral Res. 1996;29:5–29. doi: 10.1016/0166-3542(95)00910-8. [DOI] [PubMed] [Google Scholar]
- 15.Englund J A, Baker C J, Raskino C, McKinney R E, Petrie B, Fowler M G, Pearson D, Gershon A, McSherry G D, Abrams E J, Schliozberg J, Sullivan J L. Zidovudine, didanosine, or both as the initial treatment for symptomatic HIV-infected children. AIDS Clinical Trials Group (ACTG) study 152 Team. N Engl J Med. 1997;336:1704–1712. doi: 10.1056/NEJM199706123362403. [DOI] [PubMed] [Google Scholar]
- 16.Eron J J, Chow Y K, Caliendo A M, Videler J, Devore K M, Cooley T P, Liebman H A, Kaplan J C, Hirsch M S, D’Aquila R T. pol mutations conferring zidovudine and didanosine resistance with different effects in vitro yield multiply resistant human immunodeficiency virus type 1 isolates in vivo. Antimicrob Agents Chemother. 1993;37:1480–1487. doi: 10.1128/aac.37.7.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 18.Furman P A, Fyfe J A, St. Clair M H, Weinhold K, Rideout J L, Freeman G A, Lehman S N, Bolognesi D P, Broder S, Mitsuya H, Barry D W. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc Natl Acad Sci USA. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goudsmit J, Ronde A D, Ho D D, Perelson A S. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5664. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gougeon M-L, Lecoeur H, Dulioust A, Enouf M-G, Crouvoisier M, Goujard C, Debord T, Montagnier L. Programmed cell death in peripheral lymphocytes from HIV-infected persons: increased susceptibility to apoptosis of CD4 and CD8 T cells correlates with lymphocyte activation and with disease progression. J Immunol. 1996;156:3509–3520. [PubMed] [Google Scholar]
- 22.Gu Z, Gao Q, Li X, Parnaik M A, Wainberg M A. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 24.Hammer S, Katzenstein D A, Hughes M D, Gundacker H, Schooley R T, Haubrich R H, Henry W K, Lederman M M, Phair J P, Niu M, Hirsch M S, Merigan T C for the ACTG 175 Study Team. A trial comparing nucleoside monotherapy with combination therapy in HIV-infected adults with CD4 cell counts from 200 to 500 per cubic millimeter. N Engl J Med. 1996;335:1081–1090. doi: 10.1056/NEJM199610103351501. [DOI] [PubMed] [Google Scholar]
- 25.Harrigan P R, Bloor S, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature (London) 1995;373:123–126.26. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 27.Hoffenbach A, Langlade-Demoyen P, Dadaglio G, Vilmer E, Michel F, Mayaud C, Autran B, Plata F. Unusually high frequencies of HIV-specific cytotoxic T lymphocytes in humans. J Immunol. 1989;142:452–462. [PubMed] [Google Scholar]
- 28.Holodniy M, Mole L, Margolis D, Moss J, Dong H, Boyer E, Urdea M, Kolberg J, Eastman S. Determination of human immunodeficiency virus RNA in plasma and cellular viral DNA genotypic zidovudine resistance and viral load during zidovudine-didanosine combination therapy. J Virol. 1995;69:3510–3516. doi: 10.1128/jvi.69.6.3510-3516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 30.Ikuta K, Kameoka M, Luftig R B. AIDS pathogenesis: the role of accessory gene mutations, leading to formation of long-lived persistently infected cells and/or apoptosis-inducing HIV-1 particles. Virus Res. 1997;52:145–156. doi: 10.1016/s0168-1702(97)00125-1. [DOI] [PubMed] [Google Scholar]
- 31.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpacker C S The AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuritzkes D R, Marschner I, Johnson V A, Bassett R, Eron J J, Fischl M A, Murphy R L, Fife K, Maenza J, Rosandich M E, Bell D, Wood K, Sommadossi J-P, Pettinelli C the National Institute of Allergy and Infectious Disease AIDS Clinical Trials Group Protocol 306 Investigators. Lamivudine in combination with zidovudine, stavudine, or didanosine in patients with HIV-1 infection. A randomized, double-blind, placebo-controlled trial. National Institute of Allergy and Infectious Disease AIDS Clinical Trials Group Protocol 306 Investigators. AIDS. 1999;13:685–694. doi: 10.1097/00002030-199904160-00009. [DOI] [PubMed] [Google Scholar]
- 33.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melegari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 35.Nagylaki T. Introduction to theoretical population genetics. Berlin, Germany: Springer-Verlag KG; 1992. pp. 25–27. [Google Scholar]
- 36.Pantaleo G, De Maria A, Koenig S, Butini L, Moss B, Baseler M, Lane H C, Fauci A S. CD8+ T lymphocytes of patients with AIDS maintain normal broad cytolytic function despite the loss of human immunodeficiency virus-specific cytotoxicity. Proc Natl Acad Sci USA. 1990;87:4818–4822. doi: 10.1073/pnas.87.12.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 38.Quan Y, Gu Z, Li X, Li Z, Morrow C D, Wainberg M A. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (−)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J Virol. 1996;70:5642–5645. doi: 10.1128/jvi.70.8.5642-5645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schooley R T, Ramirez-Ronda C, Lange J M A, Cooper D A, Lavelle J, Lefkowitz L, Moore M, Larder B A, St. Clair M, Mulder J W, McKinnan R, Pennington K, Harrigan P R, Kinghorn I, Skol M, Rooney J F Wellcome Resistance Study Collaborative Group. Virologic and immunologic benefits of initial combination therapy with zidovudine and zalactabine or didanosine compared to zidovudine monotherapy. J Infect Dis. 1996;173:1354–1366. doi: 10.1093/infdis/173.6.1354. [DOI] [PubMed] [Google Scholar]
- 40.Shafer R W, Iversen A K N, Winters M A, Aguiniga E, Katzenstein D A, Merigan T C the AIDS Clinical Trials Group 143 Virology Team. Drug resistance and heterogenous long-term virologic response of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J Infect Dis. 1995;172:70–78. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- 41.Sharma P L, Crumpacker C S. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J Virol. 1997;71:8846–8851. doi: 10.1128/jvi.71.11.8846-8851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma P L, Chatis P A, Dogon A L, Mayers D L, McCutchan F E, Page C, Crumpacker C S. AZT related mutation Lys70Arg in reverse transcriptase of human immunodeficiency virus confers decrease in susceptibility to ddATP in in vitro RT assay. Virology. 1996;223:365–369. doi: 10.1006/viro.1996.0488. [DOI] [PubMed] [Google Scholar]
- 43.St. Clair M, Martin J, Tudor-Williams G, Bach M, Vavro C, King D, Kellam P, Kemp S, Larder B. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 44.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Han B S, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 45.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 46.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple gag and gag-pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]