Abstract
Herpes simplex virus type 1 (KOS)ΔgK is a mutant virus which lacks glycoprotein K (gK) and exhibits defects in virion egress (S. Jayachandra, A. Baghian, and K. G. Kousoulas, J. Virol. 69:5401–5413, 1997). To further understand the role of gK in virus egress, we constructed recombinant viruses, ΔgKhpd-1, -2, -3, and -4, that specified gK amino-terminal portions of 139, 239, 268, and 326 amino acids, respectively, corresponding to truncations immediately after each of the four putative membrane-spanning domains of gK. ΔgKhpd-1 and ΔgKhpd-2 viruses produced lower yields and smaller plaques than ΔgK. Numerous ΔgKhpd-1 capsids accumulated predominately within large double-membrane vesicles of which the inner membrane appeared to be derived from viral envelopes while the outer membrane appeared to originate from the outer nuclear membrane. The mutant virus ΔgKhpd-3 produced higher yields and larger plaques than the ΔgK virus. The mutant virus ΔgKhpd-4 produced yields and plaques similar to those of the wild-type virus strain KOS, indicating that deletion of the carboxy-terminal 12 amino acids did not adversely affect virus replication and egress. Comparisons of the gK primary sequences specified by alphaherpesviruses revealed the presence of a cysteine-rich motif (CXXCC), located within domain III in the lumen side of gK, and a tyrosine-based motif, YTKΦ (where Φ is any bulky hydrophobic amino acid), located between the second and third hydrophobic domains (domain II) in the cytoplasmic side of gK. The mutant virus gK/Y183S, which was constructed to specify gK with a single-amino-acid change (Y to S) within the YTKΦ motif, replicated less efficiently than the ΔgK virus. The mutant virus gK/C304S-C307S, which was constructed to specify two serine instead of cysteine residues within the cysteine-rich motif (CXXCC changed to SXXSC) of gK domain III, replicated more efficiently than the ΔgK virus. Our data suggests that gK contains domains in its amino-terminal portion that promote aberrant nucleocapsid envelopment and/or membrane fusion between different virion envelopes and contains domains within its domains II and III that function in virus replication and egress.
Glycoproteins specified by herpes simplex virus (HSV) are synthesized in the endoplasmic reticulum (ER) and are thought to be transported to the plasma membrane via the Golgi apparatus (38, 43, 44), presumably following cellular vesicular transport pathways (16, 33, 34, 39, 41). Herpes virions assemble their nucleocapsids within the nucleus and acquire an envelope containing viral glycoproteins by budding through the inner nuclear lamellae (31, 38, 42, 48). The process by which enveloped virions are transported from the perinuclear spaces through the cytoplasm to extracellular spaces and adjacent cells is not completely understood. According to one hypothesis, virions are transported as enveloped particles from the ER to the Golgi within vesicles derived from the outer nuclear lamellae and from the Golgi to extracellular spaces via Golgi-derived vesicles in a manner analogous to vesicular transport of viral and cellular glycoproteins. In this model, viral glycoproteins are modified in situ during transport before they are released into extracellular spaces (5, 10, 22, 38, 43, 44, 48). Nucleocapsids devoid of viral envelopes, which are frequently found in the cytoplasm of infected cells, are thought to represent a nonproductive population of viruses (5). An alternative model suggests that after virions acquire their envelopes from the inner nuclear membrane, they fuse with the outer nuclear lamellae, releasing unenveloped nucleocapsids into the cytoplasm of infected cells (4, 14, 15, 24, 50). In contrast to the previous model, free nucleocapsids in the cytoplasm represent a prerequisite step for subsequent budding into Golgi-derived vacuoles, generating enveloped virions containing fully processed glycoproteins. In both egress models, virions are released from the infected cells via fusion of vacuoles containing enveloped virions with the plasma membranes (25, 38, 45).
At least three genes, UL11, UL20, and UL53 (glycoprotein K [gK]), code for proteins that are known to be involved in HSV type 1 (HSV-1) virion egress (2, 3, 20, 21). Deletion of the UL20 gene caused accumulation of enveloped virions within the perinuclear spaces (3), while deletion of the gK gene led to accumulation of enveloped virions within the cytoplasm (21). Both UL20 and gK null mutant viruses were partially complemented by cellular factors, because the UL20 null mutant virus replicated in 143 TK− cells and the replication of the gK null mutant virus was enhanced in actively replicating cells (3, 21).
HSV-1 gK is encoded by the UL53 open reading frame (9, 28), and it has characteristics of a membrane protein, including an N-terminal signal sequence, and two potential sites for N-glycosylation 10 amino acids apart (9, 35). It exists as a single 40-kDa protein species in infected cells (19), while gK translated in vitro had an apparent molecular mass of 36 kDa, and N-linked glycosylation occurred in the first 112 residues of the protein consistent with glycosylation at residues 48 and 58 (36). Initially, gK was predicted to have four membrane-spanning regions (9); however, recent, experiments with in vitro-translated gK in the presence of microsomal membranes have suggested that gK contains three instead of four membrane-spanning regions (30).
To investigate the role of gK in infectious-virus production and virion egress, we constructed mutant viruses containing stop codons within the gK gene immediately after gene segments coding for each of the four putative hydrophobic domains (hpd) predicted by Debroy et al. (9) as well as mutant viruses containing mutated codons causing single-amino-acid changes within gK amino acid motifs conserved among all alphaherpesviruses. Characterization of these viruses in cell culture revealed structural features of gK that are important for infectious virus production and egress.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Manassas, Va.). The cells were propagated and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma Chemical Co., St. Louis, Mo.) containing sodium bicarbonate and 15 mM HEPES and supplemented with 7% heat-inactivated fetal bovine serum (FBS). V27 cells carry a stably integrated copy of the HSV-1 (KOS) ICP27 gene and were kindly provided by D. M. Knipe, Harvard Medical School. These cells were propagated in DMEM supplemented with 7% FBS and 500 μg of G418/ml (37). The gK-transformed cell line VK302 was obtained from D. C. Johnson, Oregon Health Sciences University, and was maintained in DMEM lacking histidine (GIBCO Laboratories, Grand Island, N.Y.) supplemented with 7% FBS and 0.3 mM histidinol (Sigma Chemical Co.). All cells were passed once in DMEM plus 7% FBS without selection prior to infection with virus (20). HSV-1 (KOS), the parental wild-type strain used in this study, was originally obtained from P. A. Schaffer (University of Pennsylvania, Philadelphia). HSV-1 (KOS) d27-1, which has a 1.6-kb BamHI-StuI deletion of the ICP27 gene, was kindly provided by D. M. Knipe and was propagated in V27 cells (37). The ΔgK virus was propagated on VK302 cells and was described previously (21).
Reagents.
Restriction enzymes and DNA modification enzymes were obtained from New England Biolabs (Beverly, Mass.). RNase and proteinase K were purchased from Boehringer Mannheim (Indianapolis, Ind.). Gel fragment purification matrix and buffers (Prep-a-gene) were obtained from Bio-Rad (Hercules, Calif.). Sequencing grade 35S-dATP was obtained from DuPont NEN (Wilmington, Del.). AmpliTaq, XL Polymerase, and deoxynucleoside triphosphates were purchased from PE Biosystems (Foster City, Calif.). All synthetic oligonucleotide primers were synthesized by the Louisiana State University Gene Probes and Expression Systems Laboratory “GeneLab” by using phosphoamidite chemistry on an Applied Biosystems 394 DNA/RNA synthesizer with PE Biosystems Inc. reagents.
Plasmids.
Truncations in gK were generated by the insertion of stop codons in the gK gene by PCR. Antisense oligonucleotides which contained a stop codon and a BamHI restriction site at their 5′ termini were synthesized. Antisense primers were ΔgKhpd-1 (5′-TCGGGATCCTCAGAGGGCGACGAACG-3′); ΔgKhpd-2 (5′-CAGGATCCTCAGGATATGAAAGCGG-3′); ΔgKhpd-3 (5′-GCCGGGATCCTCAATACAGCTCTGTCAGGC-3′), and ΔgKhpd-4 (5′-CCTGGGATCCTCAGTGAAGCGCCACGAGC-3′). The sense primer for all PCRs was UL52KpnI (5′-TAGTCGCGTGCATCGAAACCC-3′). PCR was performed as described previously with HSV-1 (KOS) viral DNA as a template (7, 21). Reaction conditions for the XL PCR were as follows: 98°C for 3 s and 72°C for 2 min for 30 cycles. The PCR products were precipitated, restricted with KpnI and BamHI, and gel purified. Plasmid pSJ1723, containing the UL52, UL53, and UL54 genes, was described previously (21). PCR-derived DNA fragments containing stop codons within the gK gene were cloned into the unique KpnI and BamHI sites of plasmid pSJ1723 to produce plasmids pTF9101, pTF9102, pTF9103, and pTF9104 coding for gK truncations after the first, second, third, and fourth putative hydrophobic domains, respectively (Fig. 1). Single-codon changes within the gK gene were produced by splice-overlap extension with synthetic oligonucleotides, and the PCR-derived gK genes containing single-codon changes were cloned into the acceptor plasmid pSJ1723. Plasmid pTF9120 specified gK with two C-to-S mutations within the CXXCC gK motif. Plasmid pTF9121 specified gK with a C-to-S change at amino acid position 269. Plasmid pTF9122 specified gK with a Y-to-S change within the YTKΦ motif. Plasmid pTF9105 was derived by insertion of a PCR-amplified wild-type KOS gK DNA fragment into pSJ1723.
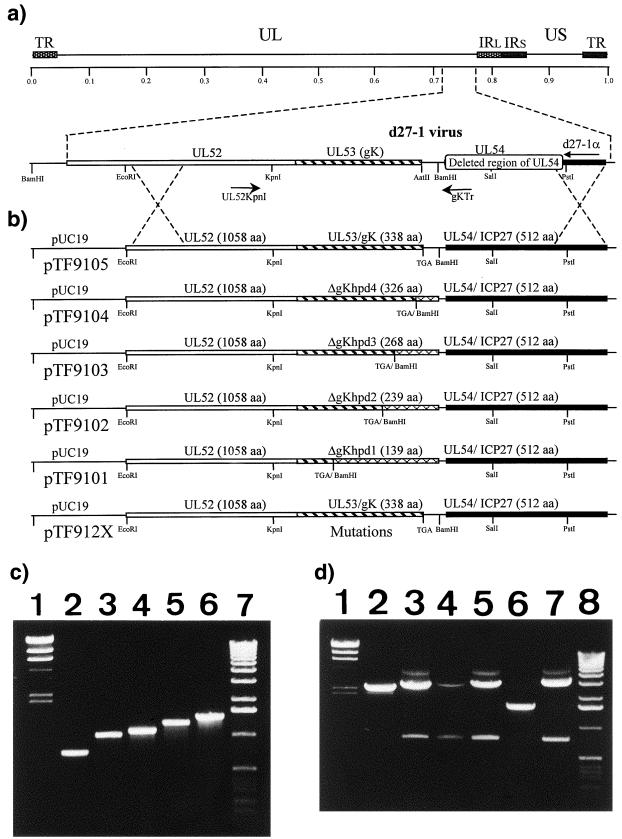
FIG. 1.
Strategy for the isolation and PCR detection of HSV-1 mutants specifying gK truncations. (a) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. Shown below is the region of the mutant virus HSV-1 d27-1 genome (between map units 0.7 and 0.8) containing the UL52, the UL53, and the partially deleted UL54 open reading frames with relevant restriction endonuclease sites. (b) Plasmid constructs containing each of the truncated gK genes used to generate the ΔgKhpd-1, -2, -3, and -4 mutant viruses. Represented on the HSV-1 (KOS) genome are the relative positions of the PCR primers UL52KpnI/gKTr used to detect the truncated gK genes. The hatched segments represent the portions of genes that are expressed after truncation, while segments 3′ to the TGA stop codon are portions of the gK genes that are deleted. aa, amino acids. (c) Agarose gel electrophoresis of double-stranded DNA PCR products with the UL52KpnI/gKTr primer pair used to detect the truncated gK genes. Lane 1, lambda phage DNA digested with HindIII (marker); lanes 2 to 6, Viral DNA of ΔgKhpd-1, ΔgKhpd-2, ΔgKhpd-3, ΔgKhpd-4, and KOS viruses, respectively, amplified with the PCR primer pair UL52KpnI/gKTr; lane 7, molecular size marker (1-kbp ladder). (d) Agarose gel electrophoresis of double-stranded DNA PCR products with the UL52KpnI/d27-1α primer pair used to confirm the purity of the recombinant viruses after extensive plaque purification. Lane 1, lambda phage DNA digested with HindIII (marker); Lanes 2 to 7, viral DNA of ΔgKhpd-1, ΔgKhpd-2, ΔgKhpd-3, ΔgKhpd-4, d27-1, and KOS viruses, respectively, amplified with the PCR primer pair UL52KpnI and d27-1α; lane 8, molecular size marker (1-kbp ladder).
Construction and purification of gK mutant viruses.
Plasmids specifying truncations in the UL53 gK gene were transfected into 50% confluent VK302 cells with Lipofectamine (GIBCO-BRL, Gaithersburg, Md.) according to the manufacturer’s instructions. Twenty-four hours posttransfection, the cells were infected at a multiplicity of infection (MOI) of 10 with the d27-1 virus (ICP27 null) as described previously for the generation of the ΔgK virus (21). At 48 h postinfection (p.i.), the cells were lysed by three freeze-thaw cycles, and the resultant virus stocks were plated onto VK302 cells and overlaid with agarose. Virus plaques were picked and plaque purified five times on VK302 cells. Viruses specifying amino acid changes in gK were produced following a similar protocol with the exception that Vero cells were used instead of VK302 cells to eliminate the possibility of revertants arising from the rescue of mutants by the resident wild-type gK gene.
Viral DNA.
Viral DNA was prepared from infected Vero cells as described previously (26). Briefly, Vero cells were infected with plaque-purified virus at an MOI of 5. At 2 days p.i., the cells were lysed with 1% NP-40, 0.5% deoxycholate in 10 mM Tris, and 1 mM EDTA. The lysates were treated with RNase (10 μg/ml) for 10 min at 37°C, followed by the addition of sodium dodecyl sulfate (1% final concentration) and proteinase K (10 μg/ml) at 55°C overnight. DNA was purified with two phenol-ether extractions and precipitated with 3 M sodium acetate and ice-cold ethanol.
PCR confirmation and sequencing of gK mutant viruses.
The UL53 region encoding the truncated gK was PCR amplified from plaque-purified recombinant viral DNA with the UL52KpnI oligonucleotide as the sense primer and the gKTr oligonucleotide (5′-CATACCCCGTTCCGCTTCC-3′), which binds downstream of the UL53 gene, as the antisense primer. The size differences of each gK gene truncation were determined by agarose gel electrophoresis followed by ethidium bromide staining. Truncated gK genes as well as specific codon changes were confirmed by direct sequencing with the fmol DNA cycle sequencing system (Promega, Madison, Wis.) according to the manufacturer’s directions. Sequencing reactions were resolved in a Gel-Mix 6 polyacrylamide gel (GIBCO-BRL). Mutant viruses were tested for the absence of any contaminating d27-1 virus by diagnostic PCR with primers UL52KpnI and d27-1α. These PCRs were performed under long-PCR conditions with XL Polymerase (Perkin-Elmer, Inc.) essentially as described previously (7, 12, 13).
Rescue and complementation of gK mutant viruses.
Isolated viruses specifying truncations or mutations in the UL53 gene were rescued by transfecting plasmid pTF9105 into Vero cells and superinfecting them with each of the truncated virus isolates at 24 h posttransfection. At 48 h p.i., the cells were freeze-thawed three times and plated to confluent Vero cells, and the number of wild-type plaques in relation to ΔgK-like plaques was determined. The wild-type plaques were picked, plaque purified, and tested by PCR and sequencing to determine whether the wild-type UL53 gene was present. All gK mutant viruses were tested for their ability to form KOS-like plaques on the complementing cell line VK302, which complements the ΔgK virus (21).
Electron microscopy of gK-truncated virions in Vero cells.
Vero cells were infected with truncated viruses prepared from VK302 cells or KOS at an MOI of 5 and incubated at 37°C for the time designated. The infected cells were prepared for negative staining electron microscopy as described previously (21). All sections were examined with a Phillips 410 transmission electron microscope.
Production of infectious virions.
Different subconfluent Vero monolayers containing approximately 8 × 105 cells per 9.62-cm2 well at the time of infection were infected with each mutant virus at an MOI of 5. After adsorption for 2 h at 4°C, the viruses were removed and the cultures were washed with medium. Fresh prewarmed medium was added to the cells, and the cultures were incubated at 37°C for the duration of the study. At 12 and 24 h p.i., the combined cells and supernatant fluid samples were frozen and thawed three times and sonicated, and the number of infectious virions was determined by standard endpoint plaque assays on VK302 cells.
RESULTS
Construction and genetic characterization of HSV-1 (KOS) mutant viruses containing stop codons within the gK gene.
Specific portions of the gK gene were generated by PCR with a 3′ oligonucleotide primer containing an in-frame stop codon. Stop codons were inserted immediately after the gK gene sequences coding for each of the four putative hydrophobic domains of gK, resulting in predicted truncated gKs of 139, 239, 268, and 326 amino acids. Plasmids pTF9101, pTF9102, pTF9103, and pTF9104 contained the truncated gK genes between the UL52 and UL54 genes, reconstructing the prototypic sequence of the genes (UL52-UL53-UL54). Each plasmid was used to rescue the mutant virus d27-1(KOS), which has a lethal deletion within the UL54 gene specifying the immediate-early protein ICP27, as shown in Fig. 1 and described previously for the construction of the ΔgK virus (21). Putative recombinant virus isolates were plaque purified and tested by PCR for both the presence of contaminating d27-1 virus and the engineered gK truncations. Primers UL52KpnI/gKTr, which flank the gK coding region, were used to amplify the gK genes. This set of primers will not produce a PCR product against the d27-1 virus, because primer gKTr is located within the deleted ICP27 gene portion of the d27-1 viral genome (Fig. 1). Amplification of the gK gene specified by each of the four different mutant viruses with the UL52KpnI/gKTr primer set generated the predicted DNA fragments of 730, 1,030, 1,117, and 1,291 bp, confirming the presence of the predicted truncated gK genes (Fig. 1c). The presence of the engineered stop codons was confirmed by DNA sequencing (not shown). To confirm that there was no contaminating d27-1 virus present, additional diagnostic PCR was performed with the primer pair UL52KpnI/d27-1α. The parental d27-1 virus generated a single PCR product of 1,613 bp (Fig. 1d, lane 6), while the wild-type strain KOS generated the predicted 3,241-bp DNA fragment (Fig. 1d, lane 7). The mutant viruses ΔgKhpd-1, -2, -3, and -4 produced the predicted PCR-amplified DNA fragments of 2,358, 2,658, 2,745, and 2,919 bp, respectively (Fig. 1d, lanes 2 to 5). None of the mutant viruses generated the d27-1-specific DNA fragment of 1,613 bp, indicating that there was no contaminating viral DNA in the mutant virus stocks (Fig. 1d, lanes 2 to 5).
Plaque morphologies of virus isolates and virus yields.
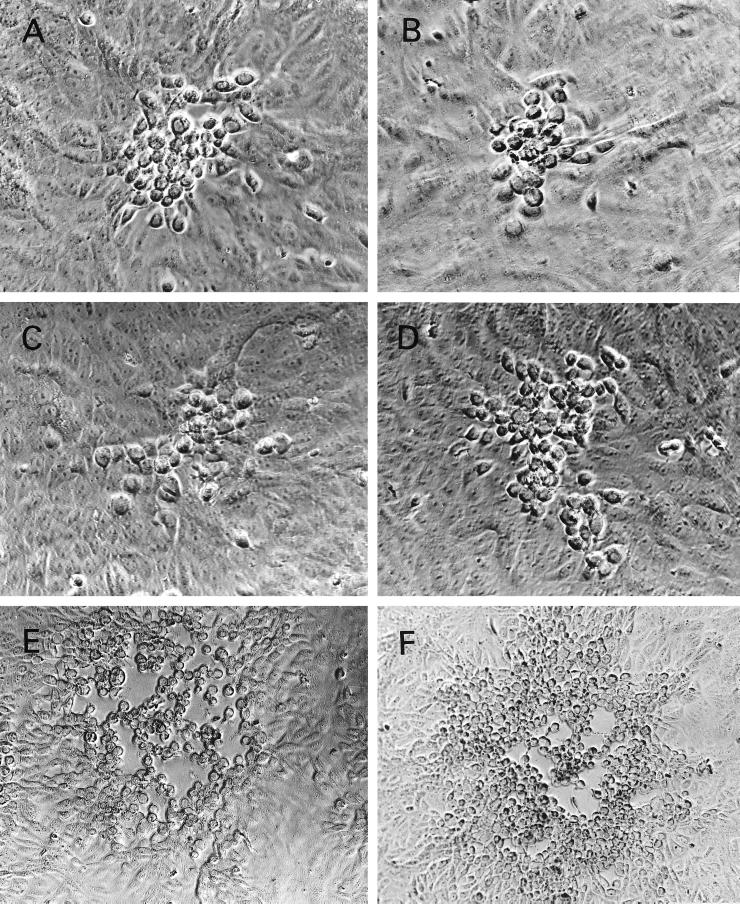
The plaque morphologies of mutant viruses were compared to those of the ΔgK and KOS viruses on Vero cell monolayers. The ΔgKhpd-1 and ΔgKhpd-2 viruses produced plaques that were reproducibly smaller (on average, approximately 20 to 30% fewer cells per plaque) than those of the ΔgK virus, while the ΔgKhpd-3 virus produced plaques which were slightly larger (on average, approximately 40 to 50% more cells per plaque) than those of the ΔgK virus. ΔgKhpd-4 viral plaques were similar in size to those of the wild-type KOS virus (Fig. 2). The ΔgK virus produces wild-type-KOS-like plaques on the complementing cell line VK302 (21). Similarly, all four gK mutant viruses produced KOS-like plaques on the complementing cell line VK302. Rescue of the hpd-1, -2, and -3 mutant viruses by plasmid pTF9105 containing the wild-type KOS gK gene produced yields similar to those of the KOS virus (not shown).
FIG. 2.
Plaque morphology of KOS, ΔgK, and ΔgKhpd-1, -2, -3, and -4 on Vero cells. (A) ΔgK; (B) ΔgKhpd-1; (C) ΔgKhpd-2; (D) ΔgKhpd-3; (E) ΔgKhpd-4; (F) KOS. The cells were infected at an MOI of 0.01 PFU/cell and photographed with a phase-contrast microscope at 48 h p.i.
Subconfluent Vero cell monolayers were infected in parallel with the viruses KOS, ΔgK, ΔgKhpd-1, ΔgKhpd-2, ΔgKhpd-3, and ΔgKhpd-4, and the total number of infectious virions (intracellular and extracellular) was determined at 12 and 24 h p.i. The yields of ΔgKhpd-1 and ΔgKhpd-2 viruses were similar to that of ΔgK virus at 24 h p.i. The yield of the ΔgKhpd-3 virus was approximately fivefold higher than those of the ΔgK, ΔgKhpd-1, and ΔgKhpd-2 viruses, while the yield of the ΔgKhpd-4 virus was approximately 10-fold higher than that of ΔgKhpd-3 and identical to that of KOS virus. The yields of all four gK mutant viruses were substantially higher in VK302 cells, approaching those of KOS virus. The ratios of extracellular to intracellular virus at 24 h p.i. were similar for ΔgK, ΔgKhpd-1, ΔgKhpd-2, and ΔgKhpd-3, while the ΔgKhpd-4 ratio was identical to that of KOS virus (Table 1).
TABLE 1.
Yields of KOS and gK-truncated mutant virusesa
| Virus | Total yield
|
Out/in (24 h p.i.) | |
|---|---|---|---|
| 12 h p.i. | 24 h p.i. | ||
| ΔgK | 3.2 × 105 | 3.7 × 106 | 0.043 |
| ΔgKhpd-1 | 8.6 × 105 | 2.1 × 106 | 0.028 |
| ΔgKhpd-2 | 1.6 × 106 | 2.6 × 106 | 0.056 |
| ΔgKhpd-3 | 2.1 × 106 | 1.6 × 107 | 0.047 |
| ΔgKhpd-4 | 3.0 × 106 | 3.0 × 108 | 0.5 |
| KOS | 3.5 × 106 | 3.0 × 108 | 0.5 |
Subconfluent Vero cell monolayers (approximately 8 × 105 cells) were infected with each virus at an MOI of 5, and at 12 and 24 h p.i. the total number of infectious virions was determined, as well as the number of infectious virions within cells and extracellular fluids. The ratio of extracellular (out) to intracellular (in) virions at 24 h p.i. is also shown. The virus yields represent one of three experiments in which individual numbers varied by less than twofold.
Electron microscopy.
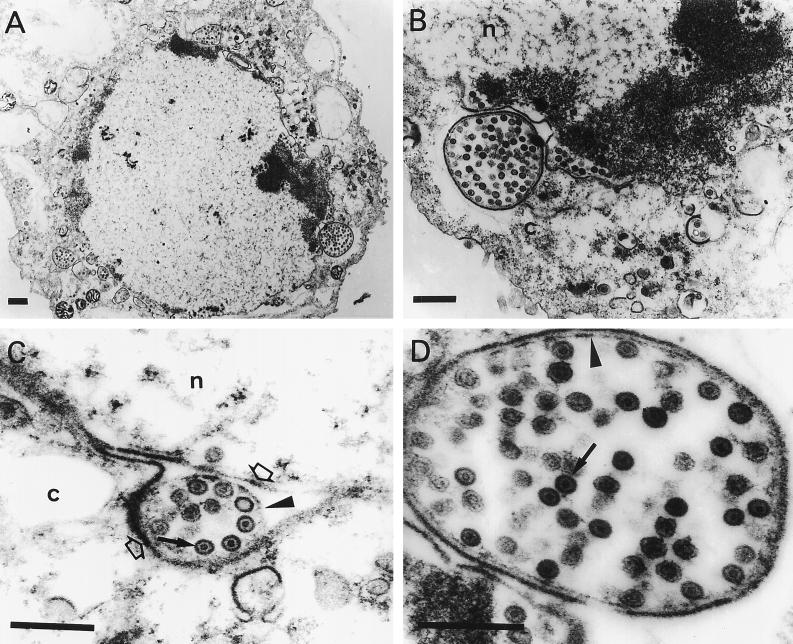
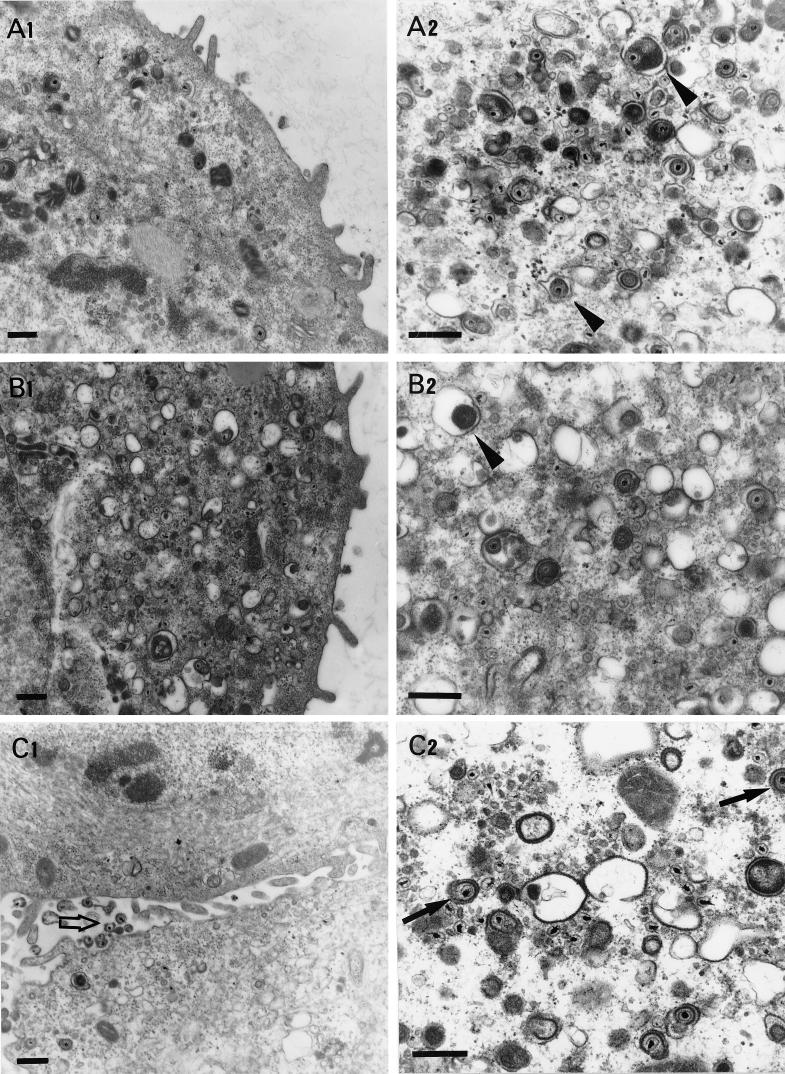
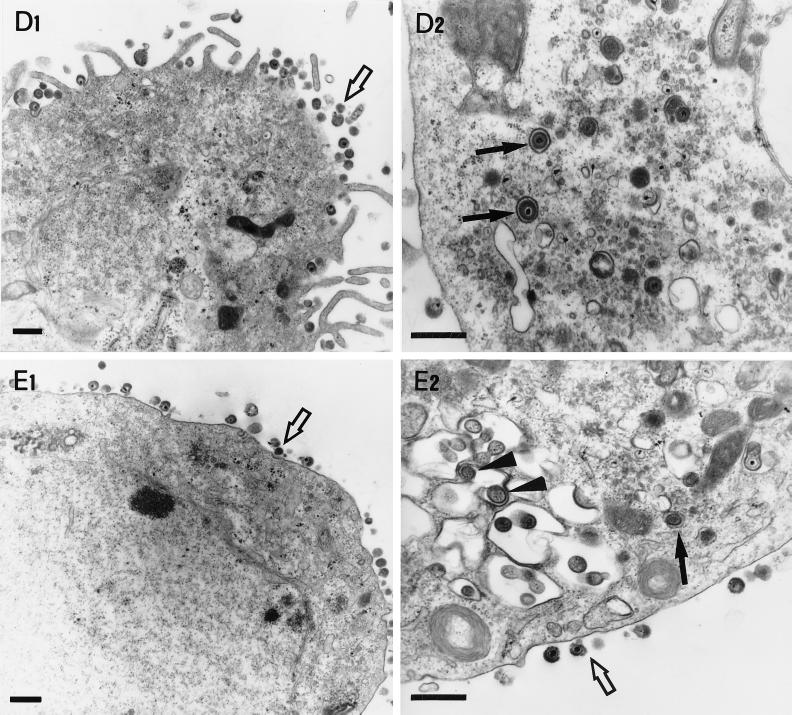
Conventional fixation-embedding electron microscopic analysis was undertaken to examine the intracellular localization of ΔgK mutant viruses in Vero cells as described previously (21). Examination of Vero cells infected with the ΔgKhpd-1 mutant virus revealed the presence of large double-membrane vesicles containing tens to hundreds of nucleocapsids in the cytoplasm located proximal to the nuclear membrane (Fig. 3A, B, and D). Single-membrane vesicles containing numerous nucleocapsids were also visualized within the perinuclear space in the process of budding through the outer nuclear lamella at regions of high electron density (Fig. 3C). In these micrographs it appeared that the outer membrane of the cytoplasmic vesicles was derived from the outer nuclear lamellae. Herpes virions are thought to acquire their envelopes by budding through the inner nuclear lamellae; therefore, it is hypothesized that the internal membrane of the double-membrane cytoplasmic vesicles must be derived from the inner nuclear lamellae (Fig. 3C and D). In contrast to ΔgKhpd-1, only a few membrane vesicles were observed with ΔgKhpd-2, while no such vesicles were found in ΔgKhpd-3-, ΔgKhpd-4-, ΔgK-, or KOS-infected cells. The ΔgKhpd-2 and ΔgKhpd-3 viruses accumulated virion particles in the cytoplasm of infected Vero cells (Fig. 4B2 and C2). ΔgKhpd-1 (Fig. 3A and B), ΔgK (Fig. 4A2), and ΔgKhpd-2 (Fig. 4B2)-infected cells contained enveloped virion particles within cytoplasmic vacuoles, while ΔgKhpd-3 and -4 and KOS contained single enveloped virions within well-defined, electron-dense vesicles (Fig. 4C2, D2, and E2, respectively). The outside surfaces of ΔgKhpd-1 (Fig. 3A)- and ΔgKhpd-2 (Fig. 4B1)-infected cells were devoid of virion particles, while only a few viruses per cell were detected on ΔgKhpd-3-infected cell surfaces (Fig. 4C1). In contrast, a high number of virions were detected on the outside surfaces of ΔgKhpd-4- and wild-type-KOS-infected cells (Fig. 4D1 and E1, respectively).
FIG. 3.
Electron micrographs of Vero cells infected with the ΔgKhpd-1 mutant virus. Subconfluent Vero cell monolayers were infected at an MOI of 5 PFU/cell, incubated at 37°C for 36 h, and prepared for electron microscopy. The solid arrows in panels C and D mark nucleocapsids. The open arrows in panel C mark the outer and inner nuclear membranes in the cytoplasmic (c) and nuclear (n) compartments. The arrowheads mark membranes surrounding nucleocapsids within the perinuclear space in panels C and D. Bars, 0.5 μm.
FIG. 4.
Electron micrographs of Vero cells infected with different gK mutant viruses. Subconfluent Vero cells were infected with ΔgK virus (A1 and A2), ΔgKhpd-2 (B1 and B2), ΔgKhpd-3 (C1 and C2), ΔgKhpd-4 (D1 and D2), and KOS (E1 and E2). All cells were infected at an MOI of 5 PFU/cell, incubated at 37°C for 36 h, and prepared for electron microscopy. The arrowheads in panels A2 and B2 mark enveloped virions within cytoplasmic vacuoles. The open arrows in panels C1, D1, E1, and E2 mark extracellular virions. The solid arrows in panels C2, D2, and E2 mark enveloped virions within electron-dense vesicles. Bars = 0.5 μm.
Alignment of gK specified by alphaherpesviruses.
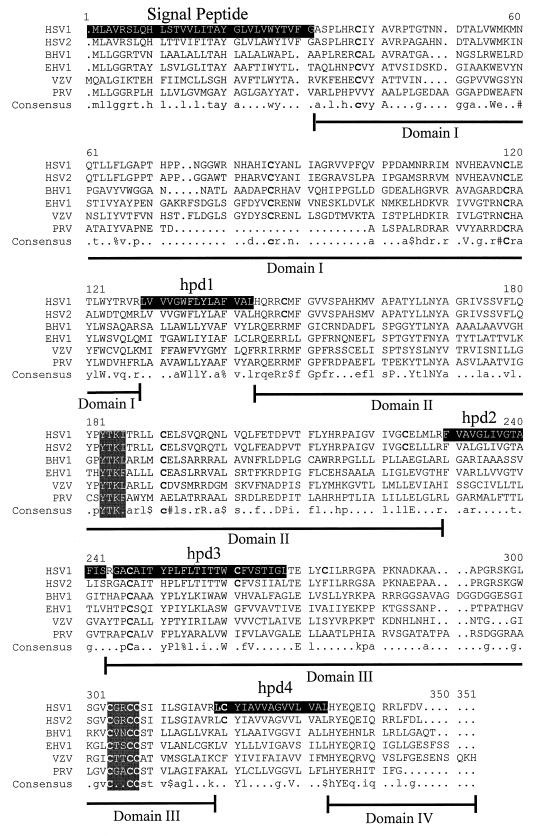
The gK gene is highly conserved among different herpesviruses. Motivated by the hypothesis that domains important in the structure and function of gK should be conserved among different herpesviruses, we investigated whether there are conserved amino acid motifs within gK domains II and III. The gK primary structures of six alphaherpesviruses were aligned with the MultiAlign program (8) (Fig. 5). The HSV-1 gK amino acid sequence contains 13 cysteine residues at positions 37, 82, 114, 144, 187, 220, 243, 257, 269, 296, 299, 300, 312. Cysteines 114, 243, 296, 299, and 300 were conserved among all gK sequences. Cysteine 114 is located within domain I, while cysteines, 243, 296, 299, and 300 are located within domain III, and both domains face the lumen or extracellular side (see Fig. 7). The alignment revealed conservation of two short amino acid sequences. In the predicted lumen side of gK, domain III contained the cysteine-rich motif (CXXCC). Domain II, predicted to be oriented toward the cytoplasm, was the most conserved among different herpesviruses and contained a conserved tyrosine-based amino acid motif (YXXΦ), where X denotes any amino acid and Φ denotes a bulky hydrophobic amino acid. The tyrosine-based and cysteine-rich motifs were also conserved in the alphaherpesviruses Marek’s disease virus and gallid herpesvirus 1 (not shown).
FIG. 5.
Alignment of gK amino acid sequences specified by alphaherpesviruses. Alignment was performed with the MultiAlign program (8). Hydrophobic domains (signal peptide, hpd1, hpd2, hpd3, and hpd4) on the HSV-1 gK amino acid sequence are shaded. The two other shaded areas contain conserved amino acid motifs. YXXΦ is a tyrosine-based motif known to function in vesicular transport of membrane-embedded glycoproteins. X denotes any amino acid, and Φ denotes a bulky hydrophobic amino acid. CXXCC is a cysteine-rich motif. The last line depicts the consensus gK sequence, with conserved residues indicated by capital letters. $, either L or M amino acids; %, either F or Y residues; #, D or N; !, I or V.
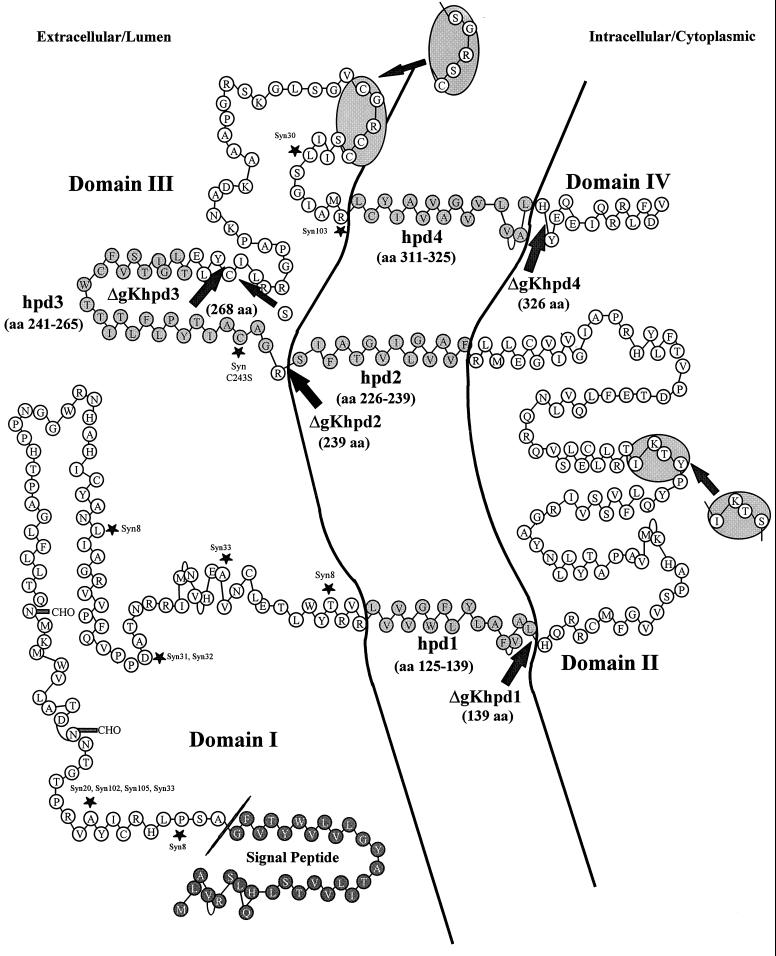
FIG. 7.
Schematic model of the predicted secondary structure of gK. The gK model of Debroy et al. (9) was modified to have three instead of four membrane-spanning domains, as suggested by Mo and Holland (30) and as predicted by computer-based predictions by the PSORT (32), Tmpred (18), and SOSUI (17) algorithms. The predicted putative hydrophobic domains (hpd) (lightly shaded circles) of gK which transverse the membrane (lines) are shown as embedded within the membrane. The arrows indicate the termination sites for truncated gKs specified by the designated viruses. Syncytial mutations are marked by asterisks. Amino acid motifs that are conserved among alphaherpesviruses are contained within shaded oval areas. The darkly shaded circles represent a signal peptide.
Construction of recombinant viruses specifying amino acid changes within the CXXCC and YXXΦ motifs.
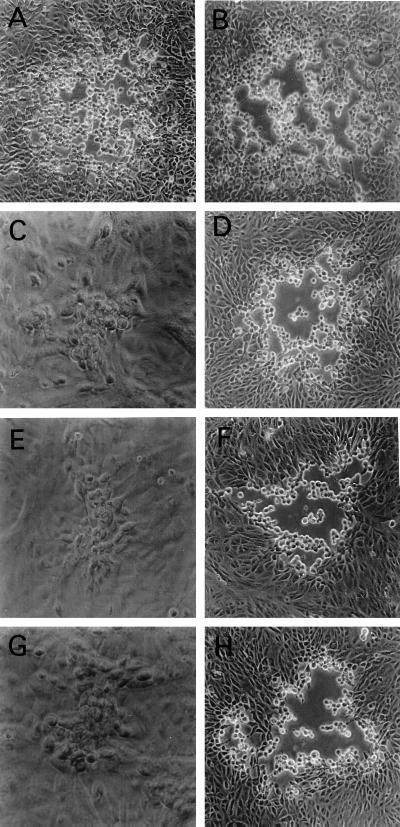
To investigate the role of the conserved amino acid motifs in the structure and function of gK, mutant viruses were constructed specifying single- and double-amino-acid changes within these two motifs. The mutant virus gK/Y183S (gK/YS) specified gK with a single-amino-acid change (Y to S) within the YTKΦ motif, and the mutant virus gK/C304S-C307S (gK/CSCS) specified gK with two cysteines changed to serine residues within the cysteine-rich motif (CXXCC changed to SXXSC) of domain III. The mutant virus gK/CSCS produced small plaques (Fig. 6C) similar to those of the ΔgK virus (Fig. 6G); however, gK/CSCS yields were similar to ΔgKhpd3 yields. Similarly, the mutant virus gK/YS (Fig. 6E) formed plaques that appeared to be similar in size to ΔgK plaques (Fig. 6G). In contrast to all other gK mutant viruses, yields of the gK/YS mutant were consistently lower by 10- to 100-fold than those of the ΔgK virus (Table 1). The mutant virus gK/C269S (Fig. 6B), specifying a C-to-S amino acid change, exhibited plaque morphology and egress characteristics similar to those of the wild-type KOS strain (Fig. 6A). All of the gK mutant viruses described above produced substantially larger plaques in VK302 cells (Fig. 6D, F, and H), approaching the KOS plaque in size (Fig. 6A), and their yields in VK302 approached those of the ΔgK virus on VK302 cells.
FIG. 6.
Plaque morphology of gK mutant viruses with amino acid changes within conserved gK motifs. Vero cells (A, B, C, E, and G) or VK302 cells (D, F, and H) were infected at an MOI of 0.01 PFU/cell and photographed with a phase-contrast microscope at 48 h p.i. (A) KOS; (B) gK/C269S; (C and D) gK/C304S-C307S; (E and F) gK/Y183S; (G and H) ΔgK.
DISCUSSION
Previously, we showed that the mutant virus ΔgK, which lacked the entire gK gene, replicated inefficiently and was unable to egress from infected cells (21). To improve our understanding of the role of gK in virus replication and egress, we engineered either stop codons at different sites of the gK gene or mutations specifying single-amino-acid changes altering amino acid motifs that are conserved among all alphaherpesviruses. Our results support and extend previous observations that gK plays an important role in virus replication and egress, and furthermore, they suggest that gK is a multifunctional protein involved in virion envelopment, intracellular virion transport, and egress.
The initial prediction of gK secondary structure indicated that gK may possess four hydrophobic domains that transverse cellular membranes (9). Recently, differential protection experiments with gK expressed in vitro in the presence of microsomal membranes suggested that gK may have three membrane-spanning regions (30). In support of this model, computer-assisted predictions with different computer algorithms available through the Internet, including PSORT (32), Tmpred (18), and SOSUI (17), revealed that the third hydrophobic domain predicted by Debroy et al. (9) has a low probability for transversing cellular membranes (not shown). Based on these considerations, and to facilitate the discussion of results, we present a modified version of the secondary-structure model initially proposed by Debroy et al. (9), with the third hydrophobic domain of gK located extracellularly (Fig. 7). A striking consequence of having three instead of four membrane-spanning domains is that a substantial portion of gK is predicted to lie within the lumen of cellular organelles that is functionally equivalent to the outside of the cell. Based on this gK model, we have subdivided the gK primary structure into four domains: domain I is the amino-terminal portion of gK terminating with the last amino acid of the putative hydrophobic domain hpd1; domain II includes the entire intracellular portion of gK and terminates with the last amino acid of the putative hydrophobic domain hpd2; domain III starts with the first hydrophilic amino acid immediately after hpd2 and terminates with the last hydrophobic amino acid of the putative hydrophobic domain 4; domain IV includes the carboxyl-terminal 13 amino acids of gK (Fig. 7). Characteristically, this model predicts that all syncytial mutations are on the external portion of gK located within either domain I or domain III, suggesting that these domains may cooperate in virus-induced cell fusion (11, 30).
The mutant viruses ΔgKhpd1 and ΔgKhpd2 produced substantially lower yields than the ΔgK virus, indicating that gK truncations specified by these viruses interfered with infectious virus production. In contrast, the yield of the gK mutant virus ΔgKhpd3 was approximately 10-fold higher than that of the ΔgK virus, indicating that this truncated gK retained partial function in the production of infectious virus. Expression of the entire domain III in gK specified by mutant virus ΔgKhpd4 restored entirely wild-type viral replication and plaque morphology. Collectively, these results suggest that domain III is important for the structure and function of gK. Furthermore, the first 28 amino acids of domain III must contain gK elements that contribute to the structure and function of gK, since the yields of ΔgKhpd3 were higher than those of ΔgK. Deletion of the terminal 12 amino acids in gK specified by the hpd4 mutant virus indicated that the carboxyl terminus of gK (domain IV) is not required for virus replication and virus spread.
Electron microscopic examination of over 100 Vero cells infected with ΔgKhpd1 revealed the presence of many large, double-membrane vesicles containing numerous capsids. Considering that each electron micrograph represented a cross section of an infected cell, it was estimated that each double-membrane vesicle contained hundreds of capsids. These vesicles were found adjacent to nuclear membranes as well as throughout the cytoplasm. The outer membrane of each vesicle appeared electron dense and morphologically similar to the outer nuclear lamellae, while an inner membrane of each vesicle appeared less electron dense and otherwise morphologically similar to the inner nuclear membrane. Additional vesicles containing numerous capsids were found within the perinuclear spaces of infected cells. The overall appearance of these vesicles suggested that they constituted precursor forms of the double-membrane vesicles found in the cytoplasm of infected cells. It is not clear from the electron microscopic data how these vesicles were formed. One possibility is that simultaneous budding of multiple nucleocapsids may be responsible for the production of the large vesicles, indicating that the expression of truncated gKs (ΔgKhpd1 and ΔgKhpd2) interferes with viral envelopment mechanisms. Alternatively, the inner membrane of the double-membrane vesicles may be derived from the fusion of virion envelopes after budding into the perinuclear space. In this scenario, fusion of enveloped virions within the perinuclear space must occur rapidly, because we could not find single enveloped virions within perinuclear spaces.
Based on the assumption that amino acid sequences conserved among all herpesviruses may represent functional domains of gK, we aligned gK sequences specified by alphaherpesviruses. This analysis revealed that the five cysteine residues which are conserved in all alphaherpesviruses are located in either domain I or III, suggesting that these cysteine residues may be involved in cooperative interactions between domains I and III. Domain III contained a CXXCC motif that was conserved among all alphaherpesviruses. Mutating the CXXCC motif to SXXSC caused the appearance of small plaques, while yields were similar to those of the ΔgKhpd3 virus. In contrast, a single C-to-S change at position 269 did not adversely affect plaque formation and virus yield. These results indicate that both the CXXCC motif and the first 28 amino acids of domain III contribute to the structure and function of gK, while Cys269 does not affect gK functions. Domain II, predicted to be oriented toward the cytoplasm, was the most conserved among different herpesviruses and contained the tyrosine-based amino acid motif (YXXΦ). Similar motifs are known to serve as putative signals for post-Golgi glycoprotein transport and receptor-specific endocytosis (6, 23, 27, 29, 49). Proper subcellular localization of the varicella-zoster virus glycoprotein I (gI) was shown to depend on two different determinants, one of which is a tyrosine-containing tetrapeptide related to endocytosis-sorting signals (1). Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE inhibited endocytosis of gE and caused a small-plaque phenotype, while it did not alter in vivo virulence (47). A single-amino-acid change of Y to S within this motif caused the formation of small viral plaques and reduced virus yields drastically in comparison to those of the ΔgK virus, indicating that the mutated gK interfered with virus replication.
It is conceivable that gK truncations and the other mutations described here may destabilize gK, causing its rapid degradation. Attempts to detect truncated gK in infected cellular extracts by radioimmunoprecipitation with rabbit antibodies raised against gK peptide antigens were inconclusive due to the high background reactivity of these sera. However, the different phenotypic properties and yields of gK mutant viruses strongly suggest that mutated gK proteins are expressed in biologically active forms. Specifically, the severe truncations of ΔgKhpd1 and ΔgKhpd2 reduced virus titers substantially in comparison to those of the ΔgK virus and produced large vesicles containing virions that were readily detected by electron microscopy. Similarly, we noted previously that expression of 112 amino acids by FgKβ resulted in gK-specific virus-induced cell fusion, supporting our hypothesis that this amino-terminal truncation of gK is expressed (21). The tyrosine-to-serine change within gK domain II produced small plaques and reduced virus yields drastically in comparison to those of ΔgK, indicating that this truncated gK exerts a negative effect on infectious virus production. Mutations within the CXXCC motif of gK domain III produced viruses which replicated more efficiently than the ΔgK virus, indicating that the mutant gK protein is expressed in a partially functional form. It is important to note that all gK mutant viruses produced yields and plaques which were similar to those of the wild-type KOS virus when propagated in VK302 cells, indicating that VK302 cells complemented gK mutant viruses in a trans-dominant manner, overcoming the negative effect of the hpd1, hpd2, and gK/Y183S mutations. This cellular complementation indicates that gK mutant viruses do not contain any secondary mutations that may affect their phenotypic and replication properties. It is unclear at this point why VK302 cells complement all gK mutant viruses. One of the possible explanations for the trans-dominant complementation of gK mutations by VK302 cells is that gK expressed by the cellular gene complexes with cellular proteins found in limiting amounts within infected cells. In this scenario, mutated gK expressed by the virus cannot displace preformed gK-cellular protein complexes.
Our electron microscopic data is consistent with the hypothesis that enveloped virions are transported to the Golgi via vesicles which originate from the outer nuclear lamellae. Such vesicles were readily observed in wild-type-KOS-infected cells containing single enveloped virions. Furthermore, expression of truncated gKs (hpd1 and hpd2) resulted in the accumulation of hundreds of virion particles within vesicles which, except for their large size, otherwise appeared to be morphologically similar to those containing single KOS virions. The intracellular transport of various intracellular cargo, including soluble and membrane-bound proteins, is achieved in cells through the use of intricate systems of targeted vesicular transport. These systems dictate a well-orchestrated cascade of molecular events which control the formation of vesicles from the ER and their bidirectional transport to the Golgi, intracellular organelles, and extracellular spaces. The hallmark of this cellular transport system is that specific targeting of vesicles is achieved through the use of pilot and sorter proteins embedded within the transport vesicle membrane and the receiving membrane, respectively (40, 41, 46). It is tempting to consider that herpes simplex virions, which have evolved to use cellular systems masterfully to their advantage, may utilize specific elements of the vesicular transport pathways for intracellular virion transport and virion egress. In this regard, differences in the virion egress pathways of HSV and those of pseudorabies virus and varicella-zoster virus may be due to the differential localization and functions of gK and other viral proteins involved in virion egress.
ACKNOWLEDGMENTS
We acknowledge the expert technical assistance of Laura Younger with electron microscopy.
This work was supported in part by Public Health Service grant AI43000 to K. G. Kousoulas, by the LSU School of Veterinary Medicine, and by a grant from the Louisiana Board of Regents Educational Quality Support Fund to K. G. Kousoulas. T. P. Foster was supported by a Louisiana Board of Regents graduate fellowship.
Footnotes
LSU GeneLab publication no. 200.
REFERENCES
- 1.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne H, Bell S, Minson T, Wilson D W. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canfield W M, Johnson K F, Ye R D, Gregory W, Kornfeld S. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24–29 of the cytoplasmic tail. J Biol Chem. 1991;266:5682–5688. [PubMed] [Google Scholar]
- 7.Chouljenko V, Jayachandra S, Rybachuk G, Kousoulas K G. Efficient long-PCR site-specific mutagenesis of a high GC template. Biotechniques. 1996;21:472–474. doi: 10.2144/96213st05. , 476–478, 480. [DOI] [PubMed] [Google Scholar]
- 8.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 10.Di Lazzaro C, Campadelli-Fiume G, Torrisi M R. Intermediate forms of glycoconjugates are present in the envelope of herpes simplex virus virions during their transport along the exocytic pathway. Virology. 1995;214:619–623. doi: 10.1006/viro.1995.0073. [DOI] [PubMed] [Google Scholar]
- 11.Dolter K E, Ramaswamy R, Holland T C. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster T P, Chouljenko V N, Kousoulas K G. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology. 1999;258:365–374. doi: 10.1006/viro.1999.9743. [DOI] [PubMed] [Google Scholar]
- 13.Foster T P, Rybachuk G V, Kousoulas K G. Expression of the enhanced fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J Virol Methods. 1998;75:151–160. doi: 10.1016/s0166-0934(98)00107-4. [DOI] [PubMed] [Google Scholar]
- 14.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths G, Simons K. The trans Golgi network: sorting at the exit site of the Golgi complex. Science. 1986;234:438–443. doi: 10.1126/science.2945253. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics, in press. [DOI] [PubMed]
- 18.Hofmann K, Stoffel W. TMbase—database of membrane spanning protein segments. Biol Chem. 1993;347:166. [Google Scholar]
- 19.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchhausen T, Bonifacino J S, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 24.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro M, Tajima M, Kato K. Transformation of Golgi membrane into the envelope of herpes simplex virus in rat anterior pituitary cells. Eur J Cell Biol. 1989;50:398–406. [PubMed] [Google Scholar]
- 26.Kousoulas K G, Pellett P E, Pereira L, Roizman B. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology. 1984;135:379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- 27.Marks M S, Woodruff L, Ohno H, Bonifacino J S. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J Cell Biol. 1996;135:341–354. doi: 10.1083/jcb.135.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 29.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 30.Mo C, Holland T C. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K. J Biol Chem. 1997;272:33305–33311. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- 31.Morgan C, Rose H M, Holden M, Jones E P. Electron microscopic observations on the development of herpes simplex virus. J Exp Med. 1959;110:643–656. doi: 10.1084/jem.110.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer S R, Rothman J E. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- 35.Pogue-Geile K L, Spear P G. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 36.Ramaswamy R, Holland T C. In vitro characterization of the HSV-1 UL53 gene product. Virology. 1992;186:579–587. doi: 10.1016/0042-6822(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 37.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 39.Rothman J E. Intracellular membrane fusion. Adv Second Messenger Phosphoprotein Res. 1994;29:81–96. doi: 10.1016/s1040-7952(06)80008-x. [DOI] [PubMed] [Google Scholar]
- 40.Rothman J E. The protein machinery of vesicle budding and fusion. Protein Sci. 1996;5:185–194. doi: 10.1002/pro.5560050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Scinece. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz J, Roizman B. Concerning the egress of herpes simplex virus from infected cells: electron and light microscope observations. Virology. 1969;38:42–49. doi: 10.1016/0042-6822(69)90126-3. [DOI] [PubMed] [Google Scholar]
- 43.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 44.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 201–232. [Google Scholar]
- 45.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teasdale R D, Jackson M R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 47.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torrisi M R, Di Lazzaro C, Pavan A, Pereira L, Campadelli-Fiume G. Herpes simplex virus envelopment and maturation studied by fracture label. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trowbridge I S, Collawn J F, Hopkins C R. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 50.Whealy M E, Robbins A K, Tufaro F, Enquist L W. A cellular function is required for pseudorabies virus envelope glycoprotein processing and virus egress. J Virol. 1992;66:3803–3810. doi: 10.1128/jvi.66.6.3803-3810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]