Abstract
The availability of nutrients from mosquito blood meals accelerates the development of Plasmodium falciparum laboratory strains in artificially infected Anopheles gambiae mosquitoes. The impact of multiple blood meals on the number of P. falciparum genotypes developing from polyclonal natural human malaria infections (field-isolates) remains unexplored. Here, we experimentally infect An. gambiae with P. falciparum field-isolates and measure the impact of an additional non-infectious blood meal on parasite development. We also assess parasite genetic diversity at the blood stage level of the parasite in the human host and of the sporozoites in the mosquito. Additional blood meals increase the sporozoite infection prevalence and intensity, but do not substantially affect the genetic diversity of sporozoites in the mosquito. The most abundant parasite genotypes in the human blood were transmitted to mosquitoes, suggesting that there was no preferential selection of specific genotypes. This study underlines the importance of additional mosquito blood meals for the development of parasite field-isolates in the mosquito host.
Keywords: Malaria, Plasmodium falciparum, Anopheles gambiae, Parasite development, Oocysts, Sporozoites, Nutritional stress, Genetic diversity, Multiplicity of infection
Subject terms: Microbiology, Parasitology, Genotype, Haplotypes
Introduction
To complete their life cycle, malaria parasites are transmitted from humans to mosquitoes and from mosquitoes to humans. During this cycle, different forms of parasites are involved in the transmission and for survival inside the human host and mosquito vector. Gametocytes, the transmissible forms of the parasite are transmitted from human to mosquito during the blood meal of a female mosquito. In the mosquito, male and female gametocytes undergo sexual reproduction to produce ookinetes that will develop into oocysts, which in turn produce thousands of sporozoites. The sporozoites migrate to the mosquitoes salivary glands to be transmitted to the human host during the next blood-meal1,2.
During its development in the mosquito, the parasites depend on nutrients from the mosquito's blood meals, including lipids3 and free amino acids4,5, to support the massive proliferation of sporozoites in the oocyst. Therefore, the successful development of the parasite is reliant upon mosquito nutritional status6. Female mosquitoes undergo several reproductive cycles over their lifetime, with each necessitating a blood meal to provide nutrients for the production of eggs7,8. In infected mosquitoes, malaria parasites compete for the remaining available resources and the limitation of these resources exposes the developing oocysts to nutritional stress3. Consequently, the parasite has developed strategies to cope with depleted resources. For example, oocysts respond to starvation and nutritional stress by becoming dormant9, a state in which they cease their development and growth9–11. However, multiple studies have shown that additional, non-infectious blood meals will replenish the availability of host-derived nutrients. Experimental mosquito infections with laboratory cultured P. falciparum strains have shown that in single fed Anopheles mosquitoes, oocyst maturation and growth are boosted with post infection blood meals9,11. Sporozoite infection intensity in the mosquito is higher after an additional blood meal12, while the speed of sporozoites development is substantially accelerated10.
Although evidence is accumulating that parasite growth in the mosquito is substantially driven by nutrients from the mosquitoes’ blood meals, current knowledge is restricted to laboratory-cultured parasite strains. These are considerably different from experimental mosquito infections using P. falciparum field-isolates. The absence of environmental exposure and selection pressure to laboratory strains of malaria parasites induces genetic bottlenecking and may have distanced these parasites from their naturally circulating relatives. Therefore, results from experimental mosquito infections using P. falciparum laboratory strains may not apply to field-isolates. While mosquito infections using laboratory-cultured gametocytes usually result in high-intensity infections13–15, the majority of infected mosquitoes contain below five oocysts per midgut15–18 under natural conditions19. Furthermore, experiments using laboratory parasite strains are usually monoclonal, while natural malaria infections are generally polyclonal20 with different parasite clones (henceforth called genotypes) circulating in the bloodstream of the infected human host. The impact of polyclonality on parasite infectiousness has yielded mixed findings, with some studies reporting a negative impact21 while others reported higher mosquito infection rates22.
Several studies that genotyped parasites before and after mosquito-feeding experiments reported that low-intensity genotypes (minority clones) undetected in human blood appeared in the mosquitoes23,24. Successful transmission of minority clones seemingly maintains the genetic diversity of the parasite25 and could have implications in preserving drug resistant genotypes in absence of drug-pressure26. However, these studies only investigated genetic diversity at the oocyst stage of the parasite or did not measure the quantitative transmissibility of distinct clones. To our knowledge, only one study has quantitatively explored the transmission of distinct Plasmodium genotypes with the conclusion that most genotypes detected from the human blood were transmitted to the mosquito27.
Here, we examined the impact of an additional non-infectious blood meal on the infection prevalence, intensity, and genetic diversity of P. falciparum field-isolates in An. gambiae mosquitoes. Two independent series of direct membrane feeding assays (DMFAs) were conducted. In the first series, we investigated the development of parasites from oocyst to sporozoites and the genetic diversity of blood stage parasites and sporozoites, while the second series of DMFAs explored the time to development of sporozoites. We demonstrated that replenishment of nutrients by an additional blood meal positively affects the sporogonic development of P. falciparum field-isolates in Anopheles mosquitoes. However, an additional blood meal did not affect the development of distinct parasite genotypes within the mosquito.
Results
Prevalence and intensity of infection in the first series of DMFAs
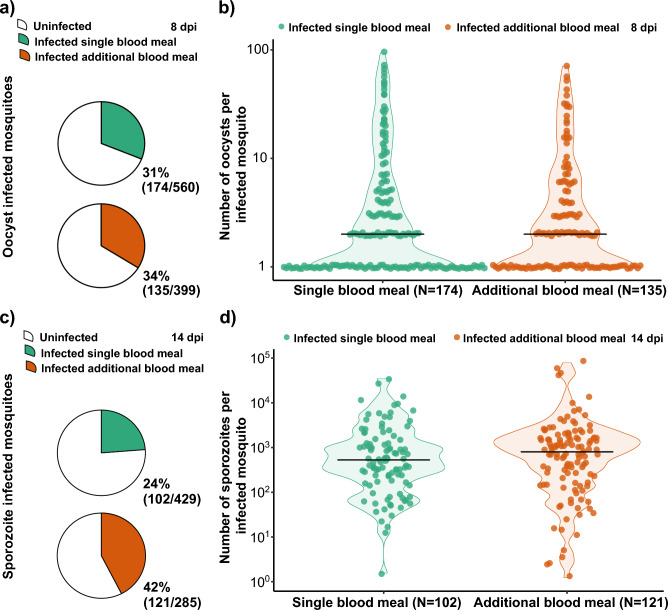
In the first series of DMFAs, we estimated the association between an additional blood meal and infection prevalence of oocysts and sporozoites as well as the intensity of infection at eight and 14 days post infection (DPI) respectively. Prevalence of infection per gametocyte carrier for the first series of DMFAs is displayed in supplementary figures S1. We dissected 959 mosquitoes for oocysts of which 560 obtained a single infectious blood meal and 399 received an additional non-infectious blood meal. Oocyst prevalence in the single blood meal group was 31% (174/560) and in the additional blood meal group 34% (135/399). Both groups had a median oocyst intensity of two oocysts, with a range of 1–94 oocysts per midgut in the single blood meal group and 1–71 oocysts in the additional blood meal group. There was no evidence of a difference in the group with an additional blood meal compared to the single feed group for oocyst infection prevalence (OR: 1.30; 95% CI 0.93–1.80; p = 0.12; Fig. 1A) or intensity (RR: 1.15; CI 0.86–1.54; p = 0.34; Fig. 1B).
Figure 1.
Results of DMFAs 8 DPI by oocyst dissection and 14 DPI by sporozoite detection and quantification by qPCR. (A) The proportion of oocyst infected mosquitoes receiving a single infectious blood meal (31%) or an additional non-infectious blood meal (34%). There was no evidence of a difference for the single blood meal group compared to the additional blood meal group (OR 1.30; CI 0.93–1.80; p = 0.12). (B) Intensity of infection as number of oocysts per individual infected mosquito. There was no evidence of a difference in the median oocyst intensity between single and additional blood meal group (RR: 1.15; CI 0.86–1.54: p = 0.34). (C) The proportion of sporozoite infected mosquitoes receiving a single infectious blood meal (24%) or an additional non-infectious blood meal (42%), with evidence of an increase in prevalence for the additional blood meal group (OR 2.57; CI 1.76–3.67; p < 0.0001). (D) Intensity of infection as number of sporozoites from individual infected mosquitoes with median sporozoite infection intensity in the single blood meal group (2438) and the additional blood meal group (3108) (RR: 2.45; CI 1.14–5.28; p = 0.02).
We analysed 714 individual mosquitoes for sporozoite infection at 14 DPI using qPCR. Of those, 429 obtained only a single infectious blood meal and 285 received an additional non-infectious blood meal. The sporozoite infection prevalence in the single blood meal group was 24% (102/429) with a median sporozoite infection intensity of 530 (1–35′390) sporozoites per infected mosquito. In the additional blood meal group, we found an infection prevalence of 42% (121/285) with median sporozoite infection intensity of 800 (1–79′296) per infected mosquito (Table 1). There was a significant increase with the additional blood meal in both sporozoite prevalence (OR 2.57; 95% CI 1.76–3.67; p < 0.0001; Fig. 1C) and intensity (RR: 2.45; CI 1.14–5.28; p = 0.02; Fig. 1D).
Table 1.
The infection prevalence and intensity of oocysts and sporozoites by time point.
| Day | Parasite stage | Prevalence of infection | Median intensity of infection | ||
|---|---|---|---|---|---|
| Single BM (%) | Additional BM (%) | Single BM | Additional BM | ||
| 1st series of DMFAs | |||||
| 8 | Oocysts | 31 | 34 | 2 (1–94) | 2 (1–74) |
| 14 | Sporozoites | 24 | 42 |
530 (1 -35′390) |
800 (1–79′296) |
| 2nd series of DMFAs | |||||
| 10 | Sporozoites | 0 | 5 | 0 |
362 (2–5′577) |
| 13 | Sporozoites | 5 | 20 |
1′732 (328–2′364) |
999 (1–7′898) |
| 16 | Sporozoites | 12 | 23 |
1364 (5–10′338) |
4852 (4–37′109) |
Lowest and highest parasite counts are in brackets.
BM Blood meal.
Prevalence and intensity of infection in the second series of DMFAs
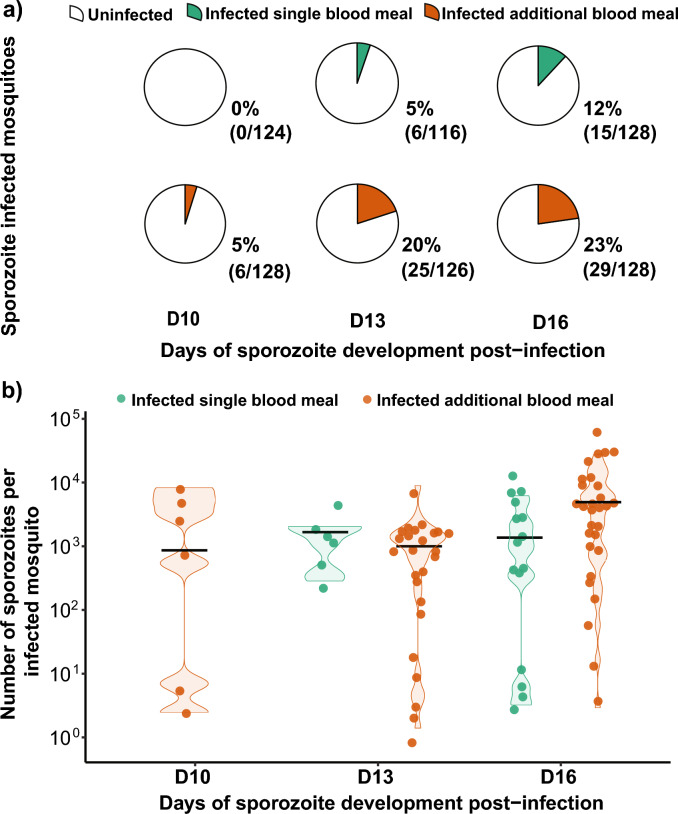
In the second series of DMFAs, we estimated the association between an additional blood meal and sporozoite infection prevalence and intensity (Table 1) over three different time points. Prevalence of infection per gametocyte carrier for the second series of DMFAs is displayed in supplementary figure S1. We included 750 individual mosquitoes of which 368 received a single infectious blood meal and 382 received an additional non-infectious blood meal. Across all time points, the average infection prevalence of sporozoites in the single blood meal group was 6% (21/368) with a median sporozoite infection intensity of 1363 (1–10′338) sporozoites per infected mosquito. In the additional blood meal group, the infection prevalence was 16% (60/382) with a median infection intensity of 1249 (1–37′109) sporozoites per infected mosquito. We found a positive, significant association between the additional blood meal and the prevalence (OR 4.73; 95% CI 2.63–8.52; p < 0.0001, Fig. 2A) and the intensity of infection of sporozoites (RR: 5.67; 95% CI 1.38–23.32; p = 0.02, Fig. 2B).
Figure 2.
Results of DMFAs at three different time points: 10-, 13- and 16-days post infection by sporozoite detection and quantification by qPCR. (A) The proportion of sporozoite infected mosquitoes at different time points as observed in this study. A consistently higher prevalence of sporozoites infected mosquitoes was observed at each time-point in the additional blood meal group compared to single fed mosquitoes. Taking the timepoints together a significant difference was observed (OR 4.73; 95% CI 2.63–8.52; p < 0.0001). (B) Intensity of infection as number of sporozoites from individual infected mosquitoes with median sporozoite infection intensity in the single blood meal group (1363) and the additional blood meal group (1249) (RR: 5.67; 95% CI 1.38–23.32; p = 0.02).
Genetic diversity and multiplicity of infection
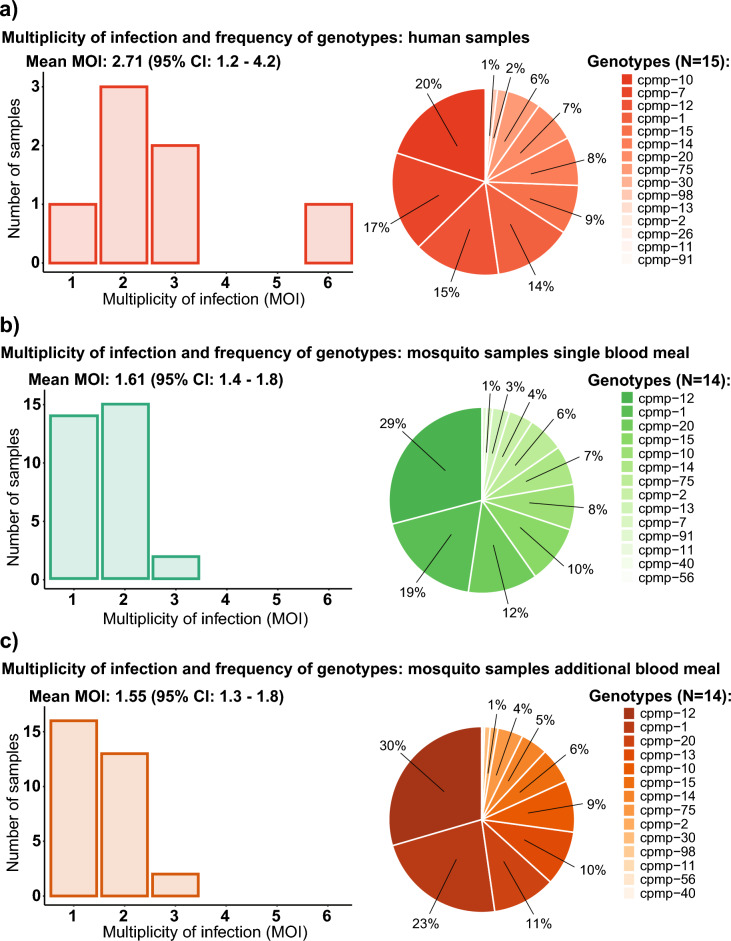
Seven human blood samples from different gametocyte carriers and 62 mosquito samples, qPCR positive for sporozoites on day 14 DPI were genotyped. Thirty-one mosquitoes from each group with median sporozoite infection intensity of 2055 (85–35′390) sporozoites in the single blood meal group and 1808 (433–79′269) sporozoites in the additional blood meal group were included in the analysis. The samples were genotyped in triplicates and sequencing data from all three replicates was merged for analysis. After merging the replicates, we obtained a total number of 12 million reads for all samples and a mean number of 175′850 reads per sample. Four different markers were used for targeted sequencing. From these four markers, the conserved Plasmodium membrane protein (cpmp)28 was found to be the best performing marker with the highest diversity (Supplementary files 1, Fig. S2). Using cpmp, we found 17 unique parasite genotypes in both human and mosquito samples (Fig. 3). Fourteen of those were detected in both the mosquito and human samples, one only in the human samples (cpmp-26) and two only in the mosquito samples (cpmp-40 and cpmp-56). The multiplicity of infection (MOI) in human samples ranged from one to six different genotypes (mean MOI: 2.71) and in mosquito samples from one to three (mean MOI: 1.55). The estimated mean MOI in mosquitoes from the single blood meal group was 1.61 and 1.55 from the additional blood meal group. MOIs of individual participants are displayed in Table 2.
Figure 3.
Multiplicity of infection (MOI) and frequency of genotypes in different sample groups (A) Human samples; (B) Mosquito samples from the single blood meal group; (C) Mosquito samples from the additional blood meal group. The number of samples refers to the total number of individuals (human or mosquito) for each specific MOI. Genotype frequency was calculated as the proportion of reads of a specific genotype from all reads from the specific sample group of all genotypes.
Table 2.
Multiplicity of infection per participant for human and mosquito sample groups.
| Participant Nr | MOI Human blood | MOI Mosquito single blood meal | MOI Mosquito additional blood meal |
|---|---|---|---|
| Participant 01 | 2 | 1.9 (N = 7) | 1.4 (N = 7) |
| Participant 02 | 2 | 1.3 (N = 3) | 1.7 (N = 3) |
| Participant 03 | 3 | 1.5 (N = 2) | 1.5 (N = 2) |
| Participant 04 | 3 | 2 (N = 2) | 1 (N = 2) |
| Participant 05 | 1 | 1.1 (N = 9) | 1.2 (N = 9) |
| Participant 06 | 6 | 2 (N = 3) | 2.5 (N = 4) |
| Participant 07 | 2 | 2 (N = 5) | 1.8 (N = 4) |
Numbers of samples used for calculating mean MOI in brackets.
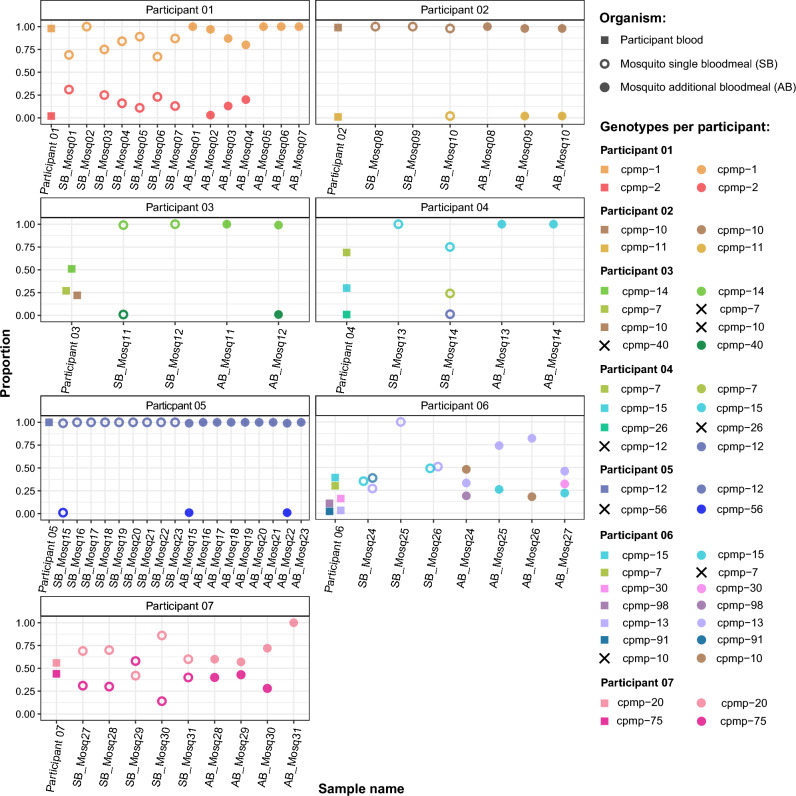
Overall 19 genotypes were detected in the human blood samples from which two (cpmp-10 and cpmp-15) were detected in two participants and cpmp-7 was detected in the blood of three different participants. From these 19 genotypes detected in the human blood 79% (15/19, Fig. 4) were transmitted to mosquitoes. We observed that all genotypes with the highest proportions of reads in the human blood samples were transmitted to the mosquitoes. The different genotypes and their proportions varied among individual participants and in infected mosquitoes (Fig. 4).
Figure 4.
Human-to-mosquito transmission of individual genotypes as detected in human blood samples and mosquito heads and thoraxes. Squares represent parasites detected from human samples while circles represent mosquito samples. Crosses indicate failure of detection either in the human blood samples (left panel) or mosquitoes (right panel). Y-axis represents the frequencies of different genotypes detected as percentage from 0 to 1.0 while 1.0 represents 100%. Each individual genotype is represented by a different colour. AB: Additional blood meal; SB: Single blood meal; HB: Human blood.
Discussion
In this study, we compared infection prevalence and intensity of oocysts and sporozoites in groups of mosquitoes that received a single infectious blood meal and mosquitoes that received an additional blood meal after being infected with P. falciparum field-isolates. We did not observe an impact of an additional blood meal on oocyst prevalence nor intensity and this finding was expected as described by others previously9. It has been shown that an additional blood meal does not affect the prevalence of oocysts in mosquitoes in general but rather affects the speed of their development being larger in size when the mosquitoes receive an additional blood meal10.
To investigate the impact of an additional blood meal on sporogonic development, we have analysed prevalence and intensity of sporozoite infection on four different time points. We found that both prevalence and intensity were higher in the group of mosquitoes that received an additional blood meal compared to single fed mosquitoes. Our results indicate that sporozoites appear earlier in mosquitoes that received an additional blood meal. In this group, the first sporozoites were detected on day 10, whereas no infection was observed in mosquitoes fed only once. A substantial increase in sporozoite-infected mosquitoes was noted between days 13 and 16 in the single blood meal group. In contrast, a significant rise in the prevalence of sporozoite infection occurred earlier, between days 10 and 13, in mosquitoes given an additional blood meal. This suggests that the observed difference in sporozoite infection prevalence is due to the slower development of oocysts in single-fed mosquitoes compared to those receiving an additional blood meal. Indeed, Nutrients limitation affects the general growth of oocysts and consequently the increased availability of nutrients from an additional blood meal accelerates the appearance of sporozoites in the mosquito salivary glands29. The nutrients from additional blood meals reduce the parasite extrinsic incubation period (EIP) rendering infected mosquitoes infectious sooner and therefore increasing the parasite's potential of being transmitted30.
We hypothesized that competition is higher in high-intensity infections with many co-existing oocysts in a single mosquito4 as they often occur in mosquito infections from monoclonal laboratory infections. However, our results show that resource limitation also affects the sporogonic development of parasites in mosquito infections at lower densities as they occur when using field-isolates in DMFAs15,31. While the dependency of monoclonal laboratory strains of P. falciparum on essential resources from blood meals ingested by its vector for sporogonic development is well documented9–12, the need of field isolates of P. falciparum for additional resources to develop into sporozoites was similarly striking in the present study.
The numbers of sporozoites as quantified in the present study are rather an estimation than an exact number, as the use of plasmids and conversion of copy numbers to parasites is of limited precision32. Better quantitative, molecular diagnostic assays to accurately measure sporozoite numbers from mosquito salivary glands or heads and thoraxes of mosquitoes would be desirable33,34. Also, our results are limited as we only quantify total sporozoites from the head and thorax and not salivary gland sporozoites only.
Our findings support efforts to better study parasite growth in mosquitoes and associated nutrients and pathways as potential targets for control interventions4. In addition, the association between additional blood meals, oocyst growth and sporozoite development indicates that additional blood meals should be incorporated into standard laboratory infection assays for the evaluation of transmission blocking interventions35. Furthermore, this study indicates that interventions designed to reduce mosquito blood feeding on human hosts will reduce vectorial capacity by acting on the mosquito through reducing man-vector contact but also acting on the parasite by reducing the probability of the parasite completing its extrinsic incubation period. For example, long lasting insecticide nets that continue to operate as physical barriers even under reduced insecticide bioavailability could be important, as every blood meal can serve as a booster for the development of sporozoites.
We hypothesized that selective pressure in single fed mosquitoes would favour the development of most competitive genotypes by increasing their frequency or by reducing the overall number of genotypes in single fed mosquitoes when compared to mosquitoes receiving an additional blood meal. Our results do not support this hypothesis, as MOI and the frequencies of detected parasite genotypes in both the single and additional blood meal group were similar. While the frequencies of the three most transmitted genotypes in both groups of mosquitoes were nearly identical, other genotypes showed considerable differences such as genotype cpmp-13, which was detected at a threefold higher frequency in the additional blood meal group. Overall we have not detected a striking difference in both the MOI and the frequencies of the distinct genotypes between the two groups of mosquitoes.
The human-to-mosquito transmission of distinct P. falciparum genotypes remains largely unexplored. We could only identify one study using targeted amplicon deep sequencing to track individual parasite genotypes from gametocytes in the human blood to sporozoites in the mosquito. This study reported an efficient transmission of genotypes27, which is in line with our findings showing efficient transmission of distinct genotypes from human to mosquito. As most genotypes detected in the human blood were also detected as sporozoites, we conclude that the vast majority of circulating genotypes in asymptomatic malaria infections produce transmissible gametocytes36.
We found that the most abundant genotypes in the human blood were all transmitted to at least one mosquito, confirming an effect of genotype density on transmission efficiency, and that highly abundant genotypes had all produced gametocytes. Four genotypes in different DMFAs were not transmitted to mosquitoes, this might be due to the fact that these genotypes were not present as gametocytes but only as asexual stages. The sequencing of parasite DNA, does not allow discrimination between sexual and asexual stages of the parasite, which is clearly a limitation of our study.
Finally, we also detected genotypes in the mosquito that were not detected in the human blood samples. These emerging genotypes could be present in the human blood as minority clones occurring at very low densities and thus not be detected by amplicon deep sequencing. Other studies have also reported the emergence of minority clones in the mosquito24,26, arguing that the passage through the mosquito increases the MOI in mosquitoes and promotes transmission of minority clones.
Our data suggest that the abundance of a clone in the human blood is mostly translated to its final abundance in the mosquito as sporozoites. Newly emerged genotypes not abundant in the blood were predominantly present at low proportions in the mosquito, indicating that only a small proportion of all sporozoites belonged to those genotypes. However, the transmission dynamics of minority clone P. falciparum genotypes in infections with higher MOI should be investigated with larger sample sizes from mosquitoes and humans, ideally with a targeted sequencing approach on human-blood samples purified for gametocytes and separated from non-transmissible asexual blood-stages, or directly targeting gametocyte specific targets for sequencing. Finally, our study is limited by the relatively small number of sequenced mosquitoes. Further studies should investigate genotype specific human-to-mosquito transmission including higher numbers of sporozoite infected mosquitoes. Since mosquito infections from field-isolates are often not very efficient, gametocytes could be enriched prior to mosquito feeding to increase infection success in DMFAs37.
The results presented in this study underline the importance of nutrients derived from the mosquito vectors’ blood meal on P. falciparum sporogonic development and the length of the parasite extrinsic incubation period critical to malaria transmission dynamics. Better knowledge of involved nutrients and pathways might lead to novel targets for interventions to control malaria. Finally, our study provides detailed insights into the relative transmission of distinct parasite genotypes, which to date remains a poorly explored field in malaria research.
Methods
Study population and design
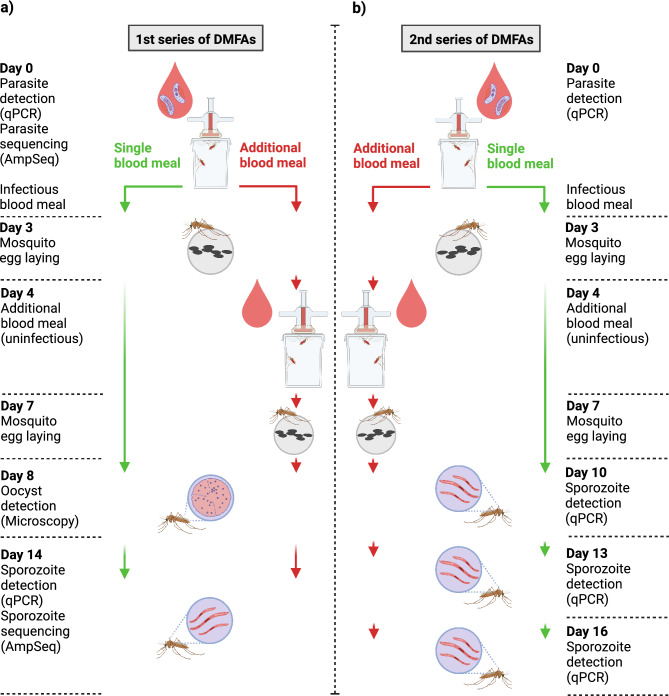
This study was conducted in Bagamoyo, a coastal town located 80 km north of Dar es Salaam in Tanzania. Malaria transmission intensity is moderate and peaks after the long rains from March to June and short rains from October to November38. Two cross sectional-studies were conducted from November 2020 to January 2021 and May to July 2021 to recruit study participants. Gametocytemic participants from the two cross-sectional studies were enrolled into two series of DMFAs (Fig. 5), each series representing an independent experiment.
Figure 5.
Study procedures and experimental set-up. A 1st series of DMFAs B 2nd series of DMFAs (created with BioRender.com).
Ethical considerations
The study received ethical clearance from the ethical commission of Northwestern Switzerland (EKNZ, Req-2018–01019), the Institutional Review Board (IRB) of IHI (IHI/IRB/No: 11 -2019) and by the National Institute for Medical Research (NIMR), Tanzania (NIMR/HQ/R.8a/Vol. IX/3069). From all participants enrolled in this study, written informed consent from parent or guardians was obtained. All children gave oral assent prior to initiate study procedures. All research was performed in accordance to relevant guidelines and regulations.
Screening for asymptomatic gametocyte carriers
School-aged children were screened for malaria infection in primary schools in Bagamoyo District, Tanzania. Only children between 6 and 14 years old with no clinical symptoms of malaria were eligible for enrolment.
Thick smears were prepared for all participants, stained with 10% Giemsa and examined by a certified microscopist under a light-microscope (OLYMPUS, Japan). Gametocytes were counted per 500 white-blood cells (WBC). All children screened positive for infection with asexual or sexual blood stages of Plasmodium were treated with Artemether-Lumefantrine (ALU, Artefan, Ajanta Pharma Limited, India) within 24 h of diagnosis according to Tanzania national guidelines39. Participants with P. falciparum infections and with gametocyte density ≥ 16 gametocytes/ µL were enrolled for blood-drawing to feed mosquitoes in DMFAs.
Mosquito rearing
We used the Anopheles gambiae sensu stricto (Ifakara strain) colony, reared at IHI, Bagamoyo Branch Insectary in Kingani. The colony is maintained at an ambient 12:12 h dark:light cycle, 70 ± 20% relative humidity and 27 ± 2 °C under strict hygiene measures. Mosquito eggs are bleached with 1% bleach solution for 60 s prior to floating. Larvae are fed on TetraMin® fish flakes (Tetra Ltd., UK) and reared at a intensity of 200 larvae (from L2 larval stage) per 1 L of deionized water (ELGA, Purelab, Option R, UK). Approximately 1000 adult mosquitoes are kept in 30 × 30 × 30 cm cages and exclusively membrane-fed using parafilm (Parafilm®, France) on non-infectious human blood 40 (National Blood Transfusion Service, Tanzania). Mosquitoes have access to 10% sucrose solution ad libitum. The solution is autoclaved (121 °C:15 min) to ensure its sterility. Adult mosquitoes are biannually checked for microsporidia infections41,42, and cages with microsporidia infected mosquitoes are discarded.
Direct membrane feeding assays (DMFAs)
Blood for DMFAs was drawn in Lithium-Heparin coated vacutainers (Vacuette®, Greiner, Austria) from P. falciparum positive asymptomatic gametocyte carriers. Autologous serum was replaced with preheated (37 °C) malaria-naïve AB serum43. DMFAs were conducted in the arthropod containment Level 2 (ACL2) transmission facility at IHI Bagamoyo Branch under appropriate biosafety measures44.
Two series of DMFAs were conducted for mosquito infections (Fig. 1). In each DMFA, approximately 240–400 mosquitoes (3–5 days old) in eight batches of 30–50 female mosquitoes were fed with gametocytemic blood. Each batch of mosquitoes was fed on 200 µL of gametocytemic blood via a membrane feeding system with water-jacketed glass feeder covered with Parafilm membrane maintained at 37 °C by a circulating water-bath45. Blood-fed mosquitoes were kept in a paper cup with filter paper covering the bottom of the cup for egg laying. The morning after DMFA, unfed mosquitoes were removed using a mouth aspirator, the remaining ones were provided with 10% sugar solution. Thereafter, cups with mosquitoes were kept in plastic cages (30 × 30 × 30 cm) at 75 ± 2% humidity and 27 ± 1 °C at 12:12 h dark:light cycle, in a climatic chamber (AraLab, Portugal) upon parasite detection.
In the first series of DMFAs (cross-sectional survey November 2020 to January 2021), blood from eleven participants was used to infect mosquitoes. One third of the fed mosquitoes from each cup of each of the eleven DMFAs were dissected (N = 959 individual mosquitoes) for oocysts on day eight post infection (DPI). Eight of the eleven DMFAs had an oocyst prevalence over 10% in all dissected mosquitoes, and were selected for detection of sporozoites by qPCR at 14 DPI mosquitoes. In the second series of DMFAs (cross-sectional survey May to July 2021), mosquitoes were infected with blood from seven participants and analysed for sporozoites using qPCR on 10 DPI, 13 DPI and 16 DPI.
Additional blood meals and egg laying
In the evening of the third day after the infectious DMFA (3 DPI, ~ 72 h), filter papers on the bottom of the cups were wetted with 3 mL of deionized water. This allowed mosquitoes to lay eggs on the moist filter papers overnight. In the evening (4 DPI, ~ 96 h), four cups (containing between 120 and 200 mosquitoes) were randomly selected for the additional uninfectious blood meal using freshly drawn blood of a male voluntary blood donor, tested negative for malaria infection by malaria rapid diagnostic test. Mosquitoes were allowed to feed for 60 min on water-jacketed glass feeders as described before. Finally, at seven DPI (~ 168 h), filter papers of all cups were wetted again to allow mosquitoes to lay their eggs from oogenesis induced by the additional blood meal.
Dissection of mosquitoes and oocyst detection
On eight DPI, one third of the surviving mosquitoes from each cup were separated and killed by spraying ethanol (70%). The remaining mosquitoes were kept for sporozoite development and transferred to a fresh cup. Midguts were dissected under a stereoscope at 10× magnification and stained for 10 min using 1% mercurochrome solution (Merck, Switzerland) before examination under a compound microscope (OLYMPUS, Japan) at 40× magnification for presence of oocysts.
DNA extraction
DNA was extracted from heads and thoraxes of mosquitos in 200 µL DNAzol reagent (Molecular Research Center, AlfaAesar, Germany). Mosquito tissue was ground and incubated at 55 °C for 20 min in a heating block. This was followed by centrifugation at 18′000 relative centrifugal force (rcf) for ten minutes. DNA was precipitated in cold ethanol ( − 20 °C, 100%) and pelleted by centrifugation at 15′000 rcf for eight minutes and washed with 75% ethanol followed by a last centrifugation step at 15′000 rcf for five minutes. Finally, the DNA pellet was air-dried in a heating-block for 15 min at 45 °C and eluted in 50ul of DNAse/RNAse free water.
qPCR for detection and relative quantification of sporozoites
Plasmodium falciparum infection was detected using a multiplex TaqMan based qPCR assay targeting the P. falciparum specific acidic terminal sequence of the var genes (PfvarATS) and the Pan Plasmodium 18S rRNA sequence (Pspp18S) as described previously32,46. We added an Anopheles specific pair of primers and probe targeting the mosquito 28S rRNA sequence as an internal control for DNA extraction (Supplementary Fig. 1). We used the Pspp18S target for detection and quantification of sporozoites. The PfvarATS target was used as confirmatory target for P. falciparum infection. Only mosquitoes positive for both targets were counted as parasite infected.
Sporozoites were quantified using a plasmid containing the 18S rRNA gene (GenBank: AF145334) from P. falciparum (BEI Resources, NIAID, MRA-177). Plasmid copy numbers per µL were calculated as described elsewhere47 and standard curves were prepared by serial dilutions over eight magnitudes assuming an average of six copies of the 18S rRNA gene sequence per parasite genome48, from one million parasites per µL (6′000′000 plasmid copies per µL) to 0.1 parasites per µL (0.6 copies/ µL). Each concentration from the serial dilution (standards) was run in triplicate to determine qPCR efficiency, limit of detection, the slope and y-intercept for relative quantification of Plasmodium DNA in positive mosquito samples.
Samples were run as single qPCR reactions and repeated if the internal qPCR control did not meet the internally defined cut-off for quality control (CT ≥ 21.99). In case of negative repetition of the internal control target, the sample was excluded from analysis. Samples were considered positive if both Plasmodium targets gave positive results below the qPCR cut-off of 40 cycles of amplification.
Molecular detection of Plasmodium species other than P. falciparum from gametocyte carriers (qPCR)
All gametocyte positive children enrolled for blood-drawing were also tested for the presence of P. malariae49 and P. ovale38 that are also found in the study area. This was done to avoid co-infections and interspecific parasite competition when transmitted to mosquitoes.
DNA was extracted from 180 µL whole blood drawn into EDTA tubes (Vacuette®, Greiner, Austria) and preserved in 540 µL 1× DNA/RNA Shield™ (Zymo Research, Irvine, USA). The Quick-DNA™ Miniprep kit (Zymo Research, Irvine, USA) was used for DNA extraction according to the manufacturer’s instructions. DNA was eluted in 50 µL elution buffer and stored at − 20 °C until use.
We used a qPCR assay described by Schindler et al.50 targeting relevant human Plasmodium species different to P. falciparum in the Bagamoyo district (supplementary table 1). All qPCR assays were run in duplicates with appropriate positive controls (PC) for the different Plasmodium species and non-template controls (NTC). Participants positive for Plasmodium species other than P. falciparum were excluded from the study.
Amplicon deep sequencing
Sporozoite infected mosquitoes from the single and additional blood meal group with similar sporozoite numbers were selected for sequencing to avoid an eventual bias of sequencing data due to the number of sporozoites sequenced. Targeted amplicon deep sequencing was performed using four different markers (supplementary Fig. 1). DNA of individual samples was amplified with primary and nested PCR in triplicates. Amplification was confirmed on an automated capillary electrophoresis system (QIAxcel, Qiagen). Then nPCR products were purified as previously described28. Finally, sequence libraries were generated in a final round of PCR adding Illumina sequencing adaptors and sample specific molecular indexes. Final sequencing libraries were quantified, normalised, pooled together, purified and DNA concentration was quantified using a Qubit dsDNA kit (Thermofisher Scientific) to adjust for final sequencing concentration. Library pools were sequenced in triplicates on an Illumina MiSeq platform in paired-end mode (600 cycles; 2× 300 bp v3), with 15% Enterobacteria phage PhiX control (Illumina, PhiX Control v3)51.
Sequence read analysis and haplotype calling
Sequencing reads were analysed using the HaplotypR pipeline (version 0.3.1). Samples yielding fewer than 25 reads were excluded from analysis. Trimmed sequences were compared to the reference sequence P. falciparum strain 3D7 (PlasmoDB) and cut-offs were defined as described elsewhere52.
Code availability
The code of the pipeline used to process the data sequenced here is available under https://github.com/lerch-a/HaplotypR and published by Lerch et al.28.
Statistical analysis
In the first series of DMFAs, we estimated the impact of an additional blood meal on the prevalence and intensity of oocyst infection at 8 DPI and sporozoite infection at 14 DPI. Logistic regression was performed including the infection status of mosquitoes with oocysts or sporozoites as the binary outcome variable, the number of blood meals as a categorical fixed effect and participants donating gametocytemic blood (participant ID) as a random effect to account for multiple mosquitoes being fed on the same participant’s blood. The adjusted odds ratios (aOR) for the odds of infection in the additional blood meal group compared to the odds of infection in the single blood meal group were estimated and presented with 95% confidence intervals (CI).
To estimate the association between the additional blood meal and the intensity of oocyst or sporozoite infection in the mosquitoes, negative binomial regressions were performed. The number of oocysts or sporozoites was incorporated as the outcome variable, the number of blood meals as a categorical variable, and the participants that donated gametocytemic blood (participant ID) were incorporated as a random effect into the regression model. Rate ratios (RR) comparing the number of parasites in the additional blood meal group to the single blood meal group were estimated with 95% CI.
In the second series of DMFAs, we estimated the association between having an additional blood meal and the prevalence of sporozoites infection over the three time points (10, 13 and 16 DPI) using logistic regression. We used logistic regression for prevalence and negative binomial regression for the intensity of sporozoite infections with a random effect for participant. To control for differences between the time points, we included day (of qPCR detection for sporozoite infection) into the model as a fixed effect. We investigated whether the association between infection and blood meal differed by time points using an interaction test. The analysis of sporozoite intensity was not adjusted for day of infection due to the overall small number of sporozoite-infected mosquitoes (Supplementary information).
All data were entered into Excel (Microsoft, 2016) and analysed with the statistical software R (version 4.1.3) and the R studio interface (Version 1.3.1093) using the “lme4” package.
Supplementary Information
Author contributions
MMT, SJM, CN and LMH designed the study; LMH, PAK, RM, RMS conducted the DMFAs and laboratory work for parasite detection; RMS reared the mosquitoes for the DMFAs; MMT led participant recruitment and informed consent procedures; MSC, SLM, MMS and MMT conducted participant screenings, blood-drawings and treatment of participants; SLM managed data entry and data collection; LMH and AS conducted amplicon deep sequencing experiments; LMH, PHHS and MG analysed sequencing data; TH gave technical inputs and initial study design; LMH, SJM and AR analysed the data; LMH, SMJ, MMT, CN and AR wrote the manuscript. All authors reviewed the manuscript.
Funding
LMH was funded trough Rudolf Geigy Foundation, Switzerland; Fieldwork and Labwork were funded trough, Leading House Africa, Switzerland; Novartis Stiftung für Medizinisch-Biologische Forschung, #18C189, Switzerland; Vector Control and Product Testing Unit (VCPTU), Ifakara Health Institute, Bagamoyo, Tanzania.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-67990-y.
References
- 1.Nilsson, S. K., Childs, L. M., Buckee, C. & Marti, M. Targeting human transmission biology for malaria elimination. PLoS Pathog.11, e1004871. 10.1371/journal.ppat.1004871 (2015). 10.1371/journal.ppat.1004871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baton, L. A. & Ranford-Cartwright, L. C. Spreading the seeds of million-murdering death: metamorphoses of malaria in the mosquito. Trends Parasitol.21, 573–580. 10.1016/j.pt.2005.09.012 (2005). 10.1016/j.pt.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 3.Costa, G. et al. Non-competitive resource exploitation within mosquito shapes within-host malaria infectivity and virulence. Nat. Commun.9, 3474. 10.1038/s41467-018-05893-z (2018). 10.1038/s41467-018-05893-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw, W. R., Marcenac, P. & Catteruccia, F. Plasmodium development in Anopheles: a tale of shared resources. Trends Parasitol.38, 124–135. 10.1016/j.pt.2021.08.009 (2022). 10.1016/j.pt.2021.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lampe, L., Jentzsch, M., Kierszniowska, S. & Levashina, E. A. Metabolic balancing by miR-276 shapes the mosquito reproductive cycle and Plasmodium falciparum development. Nat. Commun.10, 5634. 10.1038/s41467-019-13627-y (2019). 10.1038/s41467-019-13627-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takken, W. et al. Larval nutrition differentially affects adult fitness and Plasmodium development in the malaria vectors Anopheles gambiae and Anopheles stephensi. Parasit. Vectors6, 345. 10.1186/1756-3305-6-345 (2013). 10.1186/1756-3305-6-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott, T. W. & Takken, W. Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends Parasitol.28, 114–121. 10.1016/j.pt.2012.01.001 (2012). 10.1016/j.pt.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 8.Klowden, M. J. & Briegel, H. Mosquito gonotrophic cycle and multiple feeding potential: contrasts between Anopheles and Aedes (Diptera: Culicidae). J. Med. Entomol.31, 618–622. 10.1093/jmedent/31.4.618 (1994). 10.1093/jmedent/31.4.618 [DOI] [PubMed] [Google Scholar]
- 9.Habtewold, T. et al. Plasmodium oocysts respond with dormancy to crowding and nutritional stress. Sci. Rep.11, 3090. 10.1038/s41598-021-81574-0 (2021). 10.1038/s41598-021-81574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw, W. R. et al. Multiple blood feeding in mosquitoes shortens the Plasmodium falciparum incubation period and increases malaria transmission potential. PLoS Pathog.16, e1009131. 10.1371/journal.ppat.1009131 (2020). 10.1371/journal.ppat.1009131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon, H., Simões, M. L., Reynolds, R. A., Dimopoulos, G. & Smith, R. C. Additional feeding reveals differences in immune recognition and growth of plasmodium parasites in the mosquito host. mSphere10.1128/mSphere.00136-21 (2021). 10.1128/mSphere.00136-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponnudurai, T. et al. Sporozoite load of mosquitoes infected with plasmodium falciparum. Trans. R Soc Trop. Med. Hyg.83, 67–70. 10.1016/0035-9203(89)90708-6 (1989). 10.1016/0035-9203(89)90708-6 [DOI] [PubMed] [Google Scholar]
- 13.Eldering, M. et al. Comparative assessment of An.gambiae and An.stephensi mosquitoes to determine transmission-reducing activity of antibodies against P.falciparum sexual stage antigens. Parasit Vectors10, 489. 10.1186/s13071-017-2414-z (2017). 10.1186/s13071-017-2414-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponnudurai, T. et al. Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology98(Pt 2), 165–173. 10.1017/s0031182000062065 (1989). 10.1017/s0031182000062065 [DOI] [PubMed] [Google Scholar]
- 15.Stone, W. J. et al. The relevance and applicability of oocyst prevalence as a read-out for mosquito feeding assays. Sci. Reports3, 3418. 10.1038/srep03418 (2013). 10.1038/srep03418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg, R. Malaria: Some considerations regarding parasite productivity. Trends Parasitol.24, 487–491. 10.1016/j.pt.2008.07.009 (2008). 10.1016/j.pt.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 17.Billingsley, P. F., Medley, G. F., Charlwood, D. & Sinden, R. E. Relationship between prevalence and intensity of Plasmodium falciparum infection in natural populations of Anopheles mosquitoes. Am. J. Trop. Med. Hyg.51, 260–270. 10.4269/ajtmh.1994.51.260 (1994). 10.4269/ajtmh.1994.51.260 [DOI] [PubMed] [Google Scholar]
- 18.Graumans, W., Jacobs, E., Bousema, T. & Sinnis, P. When is a plasmodium-infected mosquito an infectious mosquito?. Trends Parasitol.10.1016/j.pt.2020.05.011 (2020). 10.1016/j.pt.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bompard, A. et al. High plasmodium infection intensity in naturally infected malaria vectors in Africa. Int. J. Parasitol.50, 985–996. 10.1016/j.ijpara.2020.05.012 (2020). 10.1016/j.ijpara.2020.05.012 [DOI] [PubMed] [Google Scholar]
- 20.Juliano, J. J. et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc. Natl. Acad. Sci. USA107, 20138–20143. 10.1073/pnas.1007068107 (2010). 10.1073/pnas.1007068107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nsango, S. E. et al. Genetic clonality of Plasmodium falciparum affects the outcome of infection in Anopheles gambiae. Int. J. Parasitol.42, 589–595. 10.1016/j.ijpara.2012.03.008 (2012). 10.1016/j.ijpara.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 22.Barry, A. et al. Higher gametocyte production and mosquito infectivity in chronic compared to incident Plasmodium falciparum infections. Nat. Commun.12, 2443. 10.1038/s41467-021-22573-7 (2021). 10.1038/s41467-021-22573-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nwakanma, D. et al. High gametocyte complexity and mosquito infectivity of Plasmodium falciparum in the Gambia. Int. J. Parasitol.38, 219–227. 10.1016/j.ijpara.2007.07.003 (2008). 10.1016/j.ijpara.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 24.Grignard, L. et al. Transmission of molecularly undetectable circulating parasite clones leads to high infection complexity in mosquitoes post feeding. Int. J. Parasitol.48, 671–677. 10.1016/j.ijpara.2018.02.005 (2018). 10.1016/j.ijpara.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morlais, I. et al. Plasmodium falciparum mating patterns and mosquito infectivity of natural isolates of gametocytes. PLoS One10, e0123777. 10.1371/journal.pone.0123777 (2015). 10.1371/journal.pone.0123777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry, A. et al. The rare, the best: Spread of antimalarial-resistant plasmodium falciparum parasites by anopheles mosquito vectors. Microbiol. Spectr.9, e0085221. 10.1128/Spectrum.00852-21 (2021). 10.1128/Spectrum.00852-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian, S. et al. Efficient transmission of mixed plasmodium falciparum/vivax infections from humans to mosquitoes. J. Infect. Dis.221, 428–437. 10.1093/infdis/jiz388 (2020). 10.1093/infdis/jiz388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerch, A. et al. Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections. BMC Gen.18, 864–864. 10.1186/s12864-017-4260-y (2017). 10.1186/s12864-017-4260-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw, W. R. & Catteruccia, F. Vector biology meets disease control: Using basic research to fight vector-borne diseases. Nat. Microbiol.4, 20–34. 10.1038/s41564-018-0214-7 (2019). 10.1038/s41564-018-0214-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohm, J. R. et al. Rethinking the extrinsic incubation period of malaria parasites. Parasit Vectors11, 178. 10.1186/s13071-018-2761-4 (2018). 10.1186/s13071-018-2761-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura, K. et al. Qualification of standard membrane-feeding assay with Plasmodium falciparum malaria and potential improvements for future assays. PLoS One8, e57909. 10.1371/journal.pone.0057909 (2013). 10.1371/journal.pone.0057909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann, N. et al. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med12, e1001788. 10.1371/journal.pmed.1001788 (2015). 10.1371/journal.pmed.1001788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaumeau, V. et al. Contribution of asymptomatic plasmodium infections to the transmission of malaria in Kayin State, Myanmar. J. Infect. Dis.219, 1499–1509. 10.1093/infdis/jiy686 (2019). 10.1093/infdis/jiy686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, C. Y. T. et al. Assessing Plasmodium falciparum transmission in mosquito-feeding assays using quantitative PCR. Malar J17, 249. 10.1186/s12936-018-2382-6 (2018). 10.1186/s12936-018-2382-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Habtewold, T. et al. Streamlined SMFA and mosquito dark-feeding regime significantly improve malaria transmission-blocking assay robustness and sensitivity. Malar J.18, 24. 10.1186/s12936-019-2663-8 (2019). 10.1186/s12936-019-2663-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann, N. E. et al. Assessment of ultra-sensitive malaria diagnosis versus standard molecular diagnostics for malaria elimination: An in-depth molecular community cross-sectional study. Lancet Infect. Dis.18, 1108–1116. 10.1016/s1473-3099(18)30411-0 (2018). 10.1016/s1473-3099(18)30411-0 [DOI] [PubMed] [Google Scholar]
- 37.Andolina, C. et al. Quantification of sporozoite expelling by Anopheles mosquitoes infected with laboratory and naturally circulating P. falciparum gametocytes. Elife10.7554/eLife.90989 (2024). 10.7554/eLife.90989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarimo, B. B. et al. Seasonality and transmissibility of Plasmodium ovale in Bagamoyo District, Tanzania. Parasit. Vectors15, 56. 10.1186/s13071-022-05181-2 (2022). 10.1186/s13071-022-05181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanzania, N. National guidelines for malaria diagnosis and treatment. Dar es Salaam (2006).
- 40.Emami, S. N., Ranford-Cartwright, L. C. & Ferguson, H. M. The transmission potential of malaria-infected mosquitoes (An gambiae-Keele, An arabiensis-Ifakara) is altered by the vertebrate blood type they consume during parasite development. Sci Rep7, 40520. 10.1038/srep40520 (2017). 10.1038/srep40520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herren, J. K. et al. A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes. Nat. Commun.11, 2187. 10.1038/s41467-020-16121-y (2020). 10.1038/s41467-020-16121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musiime, A. K. et al. Is that a real oocyst? Insectary establishment and identification of Plasmodium falciparum oocysts in midguts of Anopheles mosquitoes fed on infected human blood in Tororo, Uganda. Malaria J.18, 287. 10.1186/s12936-019-2922-8 (2019). 10.1186/s12936-019-2922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sangare, I. et al. Studying fitness cost of Plasmodium falciparum infection in malaria vectors: Validation of an appropriate negative control. Malaria J.12, 2. 10.1186/1475-2875-12-2 (2013). 10.1186/1475-2875-12-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organisation, Laboratory Biosafety Manual. 4th Edition (2020).
- 45.Hofer, L. M. et al. Malaria rapid diagnostic tests reliably detect asymptomatic Plasmodium falciparum infections in school-aged children that are infectious to mosquitoes. Parasit Vectors16, 217. 10.1186/s13071-023-05761-w (2023). 10.1186/s13071-023-05761-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamau, E., Alemayehu, S., Feghali, K. C., Saunders, D. & Ockenhouse, C. F. Multiplex qPCR for detection and absolute quantification of malaria. PLoS One8, e71539. 10.1371/journal.pone.0071539 (2013). 10.1371/journal.pone.0071539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whelan, J. A., Russell, N. B. & Whelan, M. A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods278, 261–269. 10.1016/s0022-1759(03)00223-0 (2003). 10.1016/s0022-1759(03)00223-0 [DOI] [PubMed] [Google Scholar]
- 48.Mercereau-Puijalon, O., Barale, J. C. & Bischoff, E. Three multigene families in Plasmodium parasites: Facts and questions. Int. J. Parasitol.32, 1323–1344. 10.1016/s0020-7519(02)00111-x (2002). 10.1016/s0020-7519(02)00111-x [DOI] [PubMed] [Google Scholar]
- 49.Schindler, T. et al. Two cases of long-lasting, sub-microscopic plasmodium malariae infections in adults from coastal Tanzania. Malar J18, 149. 10.1186/s12936-019-2787-x (2019). 10.1186/s12936-019-2787-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindler, T. et al. Molecular monitoring of the diversity of human pathogenic malaria species in blood donations on Bioko Island, Equatorial Guinea. Malar J.18, 9. 10.1186/s12936-019-2639-8 (2019). 10.1186/s12936-019-2639-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruenberg, M., Lerch, A., Beck, H. P. & Felger, I. Amplicon deep sequencing improves Plasmodium falciparum genotyping in clinical trials of antimalarial drugs. Sci. Report9, 17790. 10.1038/s41598-019-54203-0 (2019). 10.1038/s41598-019-54203-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lerch, A. et al. Longitudinal tracking and quantification of individual Plasmodium falciparum clones in complex infections. Sci. Report9, 3333–3333. 10.1038/s41598-019-39656-7 (2019). 10.1038/s41598-019-39656-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.