Abstract
Sleeping on the back after 28 weeks of pregnancy has recently been associated with giving birth to a small-for-gestational-age infant and late stillbirth, but whether a causal relationship exists is currently unknown and difficult to study prospectively. This study was conducted to build a computer vision model that can automatically detect sleeping position in pregnancy under real-world conditions. Real-world overnight video recordings were collected from an ongoing, Canada-wide, prospective, four-night, home sleep apnea study and controlled-setting video recordings were used from a previous study. Images were extracted from the videos and body positions were annotated. Five-fold cross validation was used to train, validate, and test a model using state-of-the-art deep convolutional neural networks. The dataset contained 39 pregnant participants, 13 bed partners, 12,930 images, and 47,001 annotations. The model was trained to detect pillows, twelve sleeping positions, and a sitting position in both the pregnant person and their bed partner simultaneously. The model significantly outperformed a previous similar model for the three most commonly occurring natural sleeping positions in pregnant and non-pregnant adults, with an 82-to-89% average probability of correctly detecting them and a 15-to-19% chance of failing to detect them when any one of them is present.
Subject terms: Reproductive disorders, Biomedical engineering, Computer science, Blood flow
Introduction
The supine going-to-sleep position, when adopted after 28 weeks of pregnancy, is associated with giving birth to a small-for-gestational-age infant and late stillbirth1,2. While some professional associations, including the Royal College of Obstetricians and Gynaecologists, have incorporated this evidence into clinical practice guidelines3, the National Institute for Health and Care Excellence has pointed out that the evidence underlying this association is based on retrospective studies of self-reported going-to-sleep position, which may be limited by inaccuracies and recall bias4 and does not account for the variability in sleeping position following sleep onset. As such, the potential impact of sleeping position from 28 weeks through birth on pregnancy outcomes has not yet been prospectively verified with objective measurements.
In clinical practice, sleeping position is either determined by manual review of overnight video recordings or by automated scoring of data from a position sensor that is attached to the participant’s body5. The former method is more common in laboratory-based polysomnography studies, which are typically attended overnight by a registered polysomnographic technician who scores the body position in real-time, and the latter method is almost exclusively employed in home-based sleep studies. In clinical research, in addition to accelerometers and smartphone apps6–9, which require a device to be attached to the body, state-of-the-art non-contact sleeping posture detection methods include use of load cells10,11, pressure mats12–16, radiofrequency signals17, and a combination of these with machine learning18, as in computer vision19–26. One major limitation of many of these methods are that they require specialised sensing equipment, which may incur additional costs, setup time, and risks of device misuse or failure. Another limitation, in the context of pregnancy, is that all of these methods focus on the position of the head and thorax and none of them detect the position of the pelvis, which is critically important owing to its impact on maternal and uteroplacental hemodynamics27. Computer vision can address both of these limitations because it uses equipment that is already in use in clinical and research sleep studies (video), and it is able to be trained to detect the position of each part of the body simultaneously without any additional sensors. There are few quantitative comparisons of performance of computer vision to other methods. In most computer vision studies, performance of the model is compared against the human-applied ground truth labels21–24 or against other computer vision algorithms20. That said, in a study of 78 patients over 94 nights in a polysomnography laboratory, Grimm et al.’s computer vision model achieved an accuracy of 94.0% and outperformed the laboratory accelerometer (91.9% accuracy)19.
We previously built a computer vision model (“SLeeP AIDePt-1”; Sleep in Late Pregnancy: Artificial Intelligence for the Detection of Position) to automatically detect sleeping positions during pregnancy from video recordings. This model was built using a video dataset captured from pregnant individuals in their third trimester simulating a range of sleeping positions in a controlled setting28. However, real-world generalizability and ecological validity was severely limited because the video dataset only contained a single person in the bed, used thin and pattern-free bed sheets, did not allow any objects on the bed besides head pillows, did not include prone posture, and, overall, did not capture the complexities of real-world scenarios.
In an attempt to transition to real-world settings (the present study), our objective was to expand our controlled-setting dataset to include real-world videos and develop a new model (“SLeeP AIDePt-2”) for automated, unobtrusive, and non-contact detection and measurement of the sleeping position of a pregnant individual and their bed partner simultaneously overnight throughout the third trimester in the home setting. We aim to equip researchers with a tool employing this model to enable them to either confirm or disprove the associations between supine sleeping position in late pregnancy and adverse outcomes.
Methods
Design
This study leverages real-world, overnight, video recordings collected from an ongoing, prospective, observational, four-night, home sleep apnea study (ClinicalTrials.gov Identifier: NCT05376475) and controlled-setting video recordings from a previously published cross-sectional, in-home, simulation study, which is open-access and can be found online28. A core outcome set was not used in the design of this study.
Participants
Participants were recruited Canada-wide by the research team via various social media platforms. All data collection was completed within the participants’ own homes.
Participants for the real-world study were eligible to take part if their American Society of Anesthesiologists Physical Status (ASA PS) class was II or lower, they had a low-risk singleton pregnancy, were in the third trimester (between 28 weeks and 0 days through 40 weeks and 6 days gestation, inclusive, determined by first-trimester ultrasound), were aged 18–50 years, had a 2.4 GHz Wi-Fi network in their home, and slept in a bed at night (i.e., not a reclining chair or similar). Participants’ bed partners were eligible to participate if their ASA PS class was II or lower and they slept in the same bed as the pregnant participant. Exclusion criteria for both the participant and their bed partner included non-English speaking, reading, or writing and ASA PS class III or higher.
Participants in the controlled-setting study were eligible to take part if they were healthy, had a low-risk singleton pregnancy, were in the third trimester (between 28 weeks and 0 days through 40 weeks and 6 days gestation, inclusive, determined by first-trimester ultrasound), were aged 18–50 years, had a 2.4 GHz Wi-Fi network in their home, had access to a Android or iPhone device. Participants were excluded if they were non-English speaking, reading, or writing or had a musculoskeletal condition preventing simulation of sleeping positions28.
Interventions
For the real-world study, eligible participants and their bed partners provided voluntary, written, informed consent to participate. Basic demographic data was collected from each participant and their participating bed partner. Each participant in the real-world study was mailed a study kit containing the study equipment. A virtual meeting was held with each participant to describe the study protocol and assist with equipment setup. Each participant installed an app-enabled home-surveillance camera (Wyze Cam V2 by Wyze Labs, Inc., Seattle, USA) by attaching it to the wall, centred at the head of their bed and 1.6–1.7 m above the sleeping surface, to clearly capture all pertinent data. The participant used the Wyze App on an included study iPhone to control the video recordings, which were recorded directly to the iPhone harddrive. Immediately prior to going to sleep each night, the participant started the video recording, and immediately upon waking up for the day, the participant stopped the video recording. Prior to going to sleep each night, the participant and their bed partner (if participating) also donned two home sleep apnea test sensors (NightOwl by Ectosense NV, Leuven, Belgium)—one placed on the fingertip and one placed on the abdomen, centred, and just below the xiphoid process. The sensor on the fingertip recorded peripheral oxygen saturation, photoplethysmography, pulse rate, and actigraphy, and the sensor on the abdomen recorded body position via accelerometry. The participant and bed partner completed the above data collection process each night for four nights and were asked to sleep as they would normally sleep, which included their usual blanket(s), pillow use, and sleeping attire.
For the controlled-setting study, eligible participants gave voluntary, written, informed consent to participate. Basic demographic data was collected from each participant. Each participant used a home-surveillance camera to record a video of themselves simulating a series of twelve pre-defined body positions, first without bedsheets covering their body, and then with bedsheets covering their body. The video feed was shared in real-time with a researcher, and a researcher provided instructions and feedback in real-time to the participant while the video was recorded28.
Outcomes
The main outcomes for this study were the precision and recall of the model for detecting thirteen pre-defined body positions (see “Dataset Development”, below). The outcome of the real-world study pertaining to the present study was the sleeping position of the pregnant participant and bed partner in the overnight video recordings. Note that the sleep physiology data from the NightOwl sensor on the fingertip in the real-world study are beyond the scope of the present study. The accelerometry data from the NightOwl sensor on the abdomen was used to determine the body position during the image annotation process in the rare occasion that blankets densely obscured the participant’s and/or bed partner’s body and position determination was not possible by manual review of the video (see “Dataset Development”, below). The outcome of the controlled-setting study pertaining to the present study was the body position of the participant in the simulated video recordings28.
Sample size
For the real-world study, we selected a target sample size of N = 60 couples (60 pregnant participants and 60 bed partners) based on an approximation of ten events per variable (EPV), of which six were pre-identified and one of which was maternal sleeping position. This approximation of ten EPV in logistic regression analyses is based on guidelines developed by Peduzzi et al.29 and Concato et al.30 No formal power calculation for the selected sample size was performed because the nature of the real-world study is predictive, not inferential31. Recruitment for the real-world study is ongoing, and the present study uses a subset of this data. In the controlled-setting study, the target sample size of twenty was informed by the study budget and timeline, and a formal sample size calculation was not performed28.
Statistical methods
We assessed the normality of continuous variables via the Shapiro–Wilk test at a 0.05 significance level and indicated deviations from normality in our results. Statistical analyses were conducted using the R Statistical Software package (Version 4.2.2)32.
Dataset development
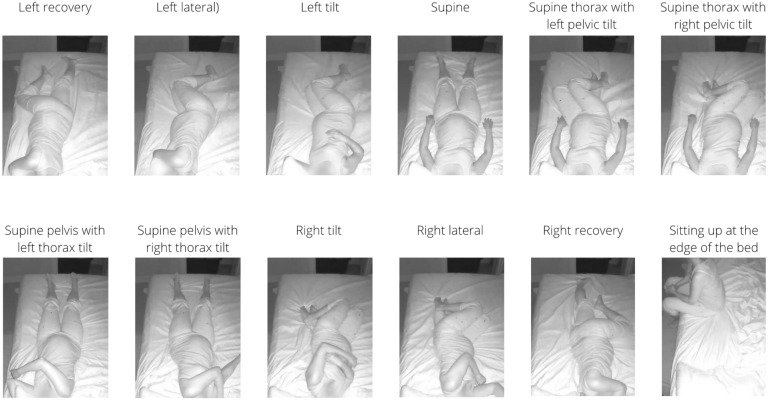
All real-world video recordings were reviewed manually by two reviewers and frames (images) were extracted at specific time points as follows: the participant and/or bed partner changed to a new sleeping position, the participant and/or bed partner remained in the same sleeping position but moved their body and/or limb(s) significantly to yield a variation of that position, and/or the participant and/or bed partner exited or returned to the bed. Low quality frames and blurred frames during sleeping position transitions, which composed less than 1% of the video recordings, were not extracted. Next, the extracted frames were annotated manually by two trained annotators, working independently, according to thirteen pre-defined sleeping positions (prone, left recovery, left lateral, left tilt, supine, supine thorax with left pelvic tilt, supine thorax with right pelvic tilt, supine pelvis with left thorax tilt, supine pelvis with right thorax tilt, right tilt, right lateral, right recovery, and sitting up at the edge of the bed, see Fig. 1), and using an open-source annotation tool, LabelImg (https://github.com/heartexlabs/labelImg)33. Note that because the position of the pelvis impacts maternal and uteroplacental hemodynamics27, we deliberately included detection of its position in our model. For frames with sleeping positions that were difficult to classify or with objects that were difficult to localise, the raw video was manually reviewed by the annotator for clarification. For body positions that could not be classified after manual review of the video, accelerometry data from the NightOwl sensor attached to the abdomen were reviewed and used to ascertain the body position. All frames containing body positions that remained difficult to classify despite following the above process were subsequently reviewed by both annotators and a final classification was agreed upon.
Figure 1.
Sleeping position classification system demonstrating examples of twelve positions previously defined in the SLeeP AIDePt study. Note that the prone position is not shown. This figure was adapted with permission from the SLeeP AIDePt study28. Permission was granted by the author (AJK).
The controlled-setting dataset was originally collected in the SLeeP AIDePt study28. The position classification system used in building the SLeeP AIDePt-1 model delineated between the presence and absence of bed sheets covering the person’s body (e.g., left lateral “without bed sheets” was classified as a separate class than left lateral “with bed sheets”)28. However, for SLeeP AIDePt-2, we wanted to classify sleeping position regardless of bedsheets, so code was written and executed upon the annotated controlled-setting dataset to collapse the “without bed sheets” and “with bed sheets” classes for each position into a single class.
Since pillows can impact sleeping positions, our model (SLeeP AIDePt-2) was designed to detect pillows. This involved identifying pillows—such as head pillows, body pillows, pregnancy pillows, and wedge pillows—in various locations on the bed. As a result, we thoroughly reviewed the controlled-setting dataset and annotated the presence of pillows in those frames as well because they were not annotated in the SLeeP AIDePt-1 model28. During the review of the controlled-setting dataset to annotate the pillows, a single annotator (AJK) also reviewed all the body position classifications and localizations to ensure they followed the classification system developed and used for SLeeP AIDePt-2.
Model development and evaluation
We framed the detection of sleeping positions and pillows as a multi-participant, multi-class, classification problem where state-of-the-art deep Convolutional Neural Networks were used. In this regard, we used YOLOv5s (You Only Look Once, version 5 s) with 7.5 million parameters (https://github.com/ultralytics/yolov5, https://docs.ultralytics.com/), including pre-trained weights from the COCO (Common Objects in Context) dataset and fine-tuned it on our annotated dataset to make predictions of classes. The experiment was implemented using the PyTorch framework. The model was trained using stochastic gradient descent with momentum, an initial learning rate of 0.01, a batch size of 32, and early stopping criteria (https://www.nvidia.com/en-us/data-center/tesla-p100/, https://research.google.com/colaboratory/faq.html). The training was conducted under the Linux virtual machine running on Google Colab with an NVIDIA A100 (SXM4) 40 GB GPU.
Our dataset was split into five non-overlapping folds, and we then trained, validated, and tested SLeeP AIDePt-2 by five-fold cross validation. In each loop of the cross-validation, three folds served as the training set, one fold was the validation set, and one fold was the testing set. Five loops were completed so each fold was given a turn to be the validation set and testing set. The weights that achieved the highest mean average precision (mAP) on the validation set were saved for each loop. Finally, performance evaluation was completed using the best weights on the testing set for each loop, and the following performance measures were calculated for each class (described below): precision, recall, and average precision (AP), which is the area under the precision-recall curve, at standard intersection over union (IoU) values. We used these metrics because they are widely accepted and utilised by the computer vision community for object detection tasks, and their use allows comparison to other models and field benchmarks.
Precision for a given class refers to the likelihood of the model’s prediction being true when it predicts that particular class. This concept is akin to the “positive predictive value” in the clinical domain. Recall for a given class refers to the model’s ability to identify all the positives for that class (“true positives”), which is analogous to “sensitivity” in the clinical realm. Note that “specificity” and “negative predictive value” are not used in YOLO (described below) because YOLO is a detector and negative predictions are not generated (and thus “true negatives” cannot be derived, while “false negatives” can be derived allowing for calculation of recall or “sensitivity”).
The AP for a class is the precision across all recall values between 0 and 1 at various IoU. The IoU is the area of overlap between the predicted object location and the ground truth location divided by the area of union (the total area of both the prediction and ground truth combined). During object detection, the IoU is calculated to determine whether the object is correctly detected or not. By interpolating across all points, the AP can be interpreted as the area under the curve of the precision-recall curve, which is a standard measure of performance in computer vision, similar to how AUROC is the area under the receiver-operator curve in the clinical realm. For each class, we also report the AP@0.50 and the AP@0.50-0.95. The 0.50 in “AP@0.50” is the IoU threshold value. If the predicted IoU is greater than or equal to the threshold, it is considered a true positive. As such, the AP@0.50 means that the AP is calculated for the objects predicted with an IoU value of 0.50 or greater. An IoU of 0.5 is typically selected as the minimum threshold for identifying a true positive, hence our use of AP@0.50. An average of AP at ten different IoU’s starting from 0.50, increasing by 0.05, up to 0.95 is the “AP@0.50-0.95”, which is what the Microscoft COCO dataset (a benchmark dataset for performance evaluation of computer vision object detection models)34 uses and which we also chose to use so that our model could be compared to other vision models for sleeping posture detection models. For a given class, a very high value for AP@0.50-0.95 indicates that the model is more confident about its predictions. The mean average precision (mAP) across all classes is calculated by taking the mean of the AP’s across all classes being predicted by the model.
We selected the YOLO (You Only Look Once) framework (https://pjreddie.com/darknet/yolo/) because of its optimised balance of speed and accuracy, customizability, and popularity as one of the most used, well supported, and famous algorithms for object detection35,36. Speed and customizability attributes enable efficient and adaptable performance, ensuring our models are well-suited for practical deployment which is our ultimate intention. In this study, we trained our model with an implementation of YOLO by Glenn Jocher called YOLOv5 (https://github.com/ultralytics/yolov5, https://docs.ultralytics.com/). YOLOv5 uses the New CSP DarkNet53 as its backbone network, the Spatial Pyramid Pooling and New Cross-Stage Partial Networks as its neck, and YOLOv3’s head as its head. YOLOv5 has different models based on the number of parameters. We used YOLOv5s, which has a total of 7.5 M parameters. The reason for choosing this model is because of its good performance on the standard Common Objects in Context (COCO) dataset and various other advantages including small size, speed, and implementation in the Pytorch framework37–39.
We disabled flip augmentation in the YOLO configuration file to enable SLeeP AIDePt-2 to distinguish leftward and rightward sleeping positions as separate classes. The clinical rationale for distinguishing between left and right as separate classes is that while the human body looks symmetrical on the outside, it is not anatomically symmetrical on the inside, which has physiologic implications. On the inside, the relatively thick-walled and high-pressure aorta runs down the left side of the spinal column whereas the relatively thin-walled and low-pressure inferior vena cava (IVC) runs up the left side of the spinal column. As such, the left lateral and tilt positions are not likely hemodynamically equivalent to the right lateral and tilt positions, and tilting from the supine position to the right tilt position may actually worsen the compression of the IVC (rather than relieving it like tilting to the left does) and reduces cardiac output, and this effect is supported by several studies40–45.
If a model is only fit to a dataset once, it may be better or worse off from a performance perspective depending on what observations in the dataset were used for training and validation. As such, resampling methods are an indispensable tool in machine learning. Resampling involves repeatedly drawing samples from a dataset and refitting a model of interest on the drawn subset (also referred to as a “training set”) in order to obtain additional information about the fitted model46. Cross validation is one of the most commonly used resampling methods in machine learning.
In k-fold cross validation, the dataset is randomly divided into k groups (folds) of approximately equal size46. One fold is “held out” and not used in training or validation. The held out fold is also known as the testing set. Another fold is treated as the validation set, and the model is fit (trained) on the remaining k-2 folds (also known as the training set). The number of misclassified observations, MCO1, is then computed on the observations in the held out fold. Then, this procedure is repeated k times such that each fold gets a turn to be the validation set and testing set and results in k estimates of the number of misclassified observations, MCO1, MCO2, …, MCOk. The k-fold cross validation estimate is computed by averaging these values46. There is a bias-variance trade-off associated with the choice of k in k-fold cross-validation; however, a k = 5 or k = 10 has been demonstrated to yield test error rate estimates that suffer neither from excessively high bias nor very high variance46. We used k-fold cross validation to train SLeeP AIDePt-2 and set k = 5 (see Supplementary File 1 for more details regarding assignment of the testing set and validation set for each loop of the cross validation as well as how the participants were allocated to the five folds).
Overfitting refers to a phenomenon in machine learning where the trained model, instead of predicting the data of interest, follows the errors, or noise, in the data too closely, and thus fails to generalize47. Overfitting is disadvantageous because the trained model will not accurately predict the outcome of interest on new observations that were not part of the original training dataset47. In the overfitting situation, the training error will be very low, but the test error (on unseen data) will be very high. Overfitting occurs when, during training, the machine learning procedure is working too hard to find patterns in the training data and may be picking up data noise and errors in the data that have occurred by random chance instead of the data of interest itself47.
In order to avoid overfitting, we used early stopping criteria to interrupt the model training prior to overfitting occurring. The early stopping patience was set to 20 epochs, that is, training of the model was stopped if the model did not show any improvement in training performance (validation mAP) in the previous 20 epochs. One epoch was the number of iterations it takes for the training process to “see” all the training frames in the training dataset. YOLO does not “see” all the training frames in each iteration; rather, in each iteration, YOLO only takes a “batch” from the total number of training frames, and we set our batch size as 32.
To attenuate the impact of class imbalance on the model’s performance (see the “Dataset” subsection in “Results”), we implemented a weighted random sampler for the training dataloader (https://www.maskaravivek.com/post/pytorch-weighted-random-sampler/) in SLeeP AIDePt 2, which is a necessary and innovative contribution over SLeeP AIDePt-1 from a methodological standpoint. The weighted random sampler assigns a weight to each frame in the training set, representing the probability of selecting that frame during each iteration of sampling. Higher weights correspond to a higher probability of being selected. The weight of each frame is calculated based on the accumulation of classes that appear in the frame. If a class has a lower number of instances in the overall dataset, a frame containing that class is assigned a relatively higher weight, which increases the probability of selecting the frame during sampling.
Patient and public involvement
Patients and public were not involved in the design of this study; however, the rationale for this study was informed by the experiences of pregnancy loss parents, who have brought maternal sleeping position to the attention of researchers and clinicians internationally48.
Ethical approval
The data and procedures underlying this study came from two studies approved by the University of Toronto Health Sciences Research Ethics Board (Protocol No. 40985 approved 18JUN2021; Protocol No. 41612 approved 09MAY2022). These studies were performed in accordance with The Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans. Written informed consent was obtained from all participants in these studies.
Results
From July 2022 through April 2023, 51 people expressed interest in participating in the real-world study. Of these, 34 (67%) were not assessed for eligibility (four decided against participating after learning more about the study, 25 did not respond after their initial expression of interest, and five gave birth prior to screening)—we did not collect any data from these. Of the seventeen (33%) participants that were screened, all met the eligibility criteria and gave written informed consent. All seventeen had bed partners, but only fifteen bed partners gave informed consent and participated. Two participants (and their bed partners) installed the camera incorrectly, so their data was excluded from the model building. As such, fifteen participants (and thirteen bed partners) successfully completed the study.
Demographic characteristics
Demographic characteristics of the pregnant participants and their bed partners (if applicable) are shown in Table 1. Ethnic backgrounds from the real-world dataset included representation from Northern European, Latino, Greek, South Asian, East Asian, Armenian/Turkish, Italian, and Hungarian ancestries. We did not collect ethnicity, gravida, or parity data from the participants in the controlled-setting dataset.
Table 1.
Demographic characteristics of participants in the combined dataset (controlled-setting and real-world), controlled-setting position dataset, and real-world dataseta.
| Characteristic | Pregnant participants | Real-world dataset bed partner (n = 13) | ||
|---|---|---|---|---|
| Combined controlled-setting and real-world datasets (n = 39) | Controlled-setting dataset (n = 24) | Real-world dataset (n = 15) | ||
| Age (years) |
30.6 ± 4.7 30 (27.5–32.5) 21, 44 |
29.5 ± 4.3 29.0 (26.8–31.3) 24, 44 |
32.3 ± 5.1 32 (29.5–36.5) 21, 40 |
32.7 ± 4.6 32 (30.0–36.0) 24, 41 |
| Gravida | NC | NC |
1 (1–2) 1, 3 |
NA |
| Parity | NC | NC |
0 (0–1) 0, 2 |
NA |
| At time of data collectionb | ||||
| Gestational age (days) |
234 ± 23.1 227 (217–255)c 196, 277 |
236 ± 24 229 (218–259) 196, 277 |
231 ± 23 225 (215–245) 196, 272 |
NA |
| Height (meters) |
1.64 ± 0.07 1.64 (1.61–1.67) 1.45, 1.80 |
1.65 ± 0.05 1.64 (1.63–1.65)c 1.55, 1.79 |
1.64 ± 0.09 1.63 (1.60–1.70) 1.45, 1.80 |
1.77 ± 0.07 1.78 (1.73–1.80) 1.63, 1.88 |
| Weight (kilograms) |
79.2 ± 13.9 76.0 (70.3–86.5) 52.4, 108.6 |
77.7 ± 14.7 75.2 (68.8–85.2) 52.4, 106.6 |
81.5 ± 12.6 77.4 (72.9–90.0) 66.8, 108.6 |
88.4 ± 25.0 85.0 (70.5–97.7) 61.4, 151.4 |
| Body mass index (kg/m2) |
29.4 ± 5.7 28.7 (26.0–31.9)c 21.0, 51.8 |
28.7 ± 5.1 27.9 (25.1–31.3) 21.0, 39.6 |
30.6 ± 6.6 28.7 (27.1–32.3)c 23.8, 51.8 |
28.5 ± 9.7 26.2 (22.5–30.2)c 19.4, 57.3 |
Abbreviations: NA, not applicable; NC, not collected.
aFor continuous variables, the mean ± standard deviation is presented on the first line of each cell, the median (25th–75th percentile, i.e., interquartile range) is presented on the second line of each cell, and the minimum and maximum are presented on the third line of each cell. For categorical variables, the median (25th–75th percentile, i.e., interquartile range) is presented on the first line of each cell, and the minimum and maximum are presented on the second line of each cell.
bFirst night of video recording for real-world dataset.
cIndicates that the continuous variable is non-normally distributed.
Dataset
In the real-world study, we collected 29,253 min (487.6 h) of infrared video from which we extracted and annotated 6960 unique frames. Of these, 6514 were multi-participant frames, and 446 were single-participant frames. The majority of the frames were infrared (greyscale), with some frames being in colour (red–green–blue) scale, which happened when the lights in the bedroom were turned on or due to daylight entering the room from the sunrise. All participants and bed partners used bedsheets, which usually covered most of the body. Four participants and bed partners (2775 frames) used patterned, striped, or textured bed sheets. Interestingly, several participants had patterned bedsheets in colour scale, but these patterns disappeared in the infrared scale. All pregnant participants and three bed partners used pillows in addition to a head pillow. For example, one pregnant participant used six pillows (two head pillows, a pillow under the abdomen, a pillow behind the back, a pregnancy pillow, and a pillow between the legs) and their bed partner used two pillows (a head pillow and a long body pillow). Three pregnant participants and one bed partner used a sleep mask during sleeping. Two participants and bed partners (1811 frames) had a child who occasionally entered the bedroom and/or bed during the late morning hours—we did not train the model to detect children. Four participants (2143 frames) had household pets that appeared in the bedroom and on the bed during the videos and in the extracted frames. Of these four, one (979 frames) had multiple pets (two dogs and one cat).
These data from our real-world dataset were combined with our controlled-setting dataset (5970 frames) for a total of 12,930 annotated frames, which contained 47,001 annotations, and comprised our dataset for building SLeeP AIDePt-2. See Table 2 for class-wise information about the datasets.
Table 2.
Total number of annotations containing each class in the real-world dataset, controlled-setting dataset, and combined (controlled-setting and real-world) dataset on which SLeeP AIDePt-2 was traineda.
| Class description | Real-world datasetb | Controlled-setting datasetb | Combined datasetb |
|---|---|---|---|
| Left recovery | 576 (1.7) | 418 (3.4) | 994 (2.1) |
| Left lateral | 3426 (9.9) | 530 (4.3) | 3956 (8.4) |
| Left tilt | 475 (1.4) | 410 (3.3) | 885 (1.9) |
| Supine | 3142 (9) | 702 (5.7) | 3844 (8.2) |
| Supine thorax with left pelvic tilt | 343 (1) | 533 (4.3) | 876 (1.9) |
| Supine thorax with right pelvic tilt | 351 (1) | 574 (4.7) | 925 (2) |
| Supine pelvis with left thorax tilt | 51 (0.1) | 362 (2.9) | 413 (0.9) |
| Supine pelvis with right thorax tilt | 44 (0.1) | 343 (2.8) | 387 (0.8) |
| Right tilt | 455 (1.3) | 345 (2.8) | 800 (1.7) |
| Right lateral | 3113 (9) | 540 (4.4) | 3653 (7.8) |
| Right recovery | 695 (2) | 421 (3.4) | 1116 (2.4) |
| Prone | 536 (1.5) | 0 (0) | 536 (1.1) |
| Sitting up at the edge of the bed | 347 (1) | 459 (3.7) | 806 (1.7) |
| Pillow | 21175 (61) | 6635 (54.1) | 27810 (59.2) |
| Total number of annotations | 34,729 | 12,272 | 47,001 |
| Average # of annotations per class | 2481 | 877 | 3357 |
aBecause of rounding, percentages may not total 100.
bData in the column is displayed as Number (percent).
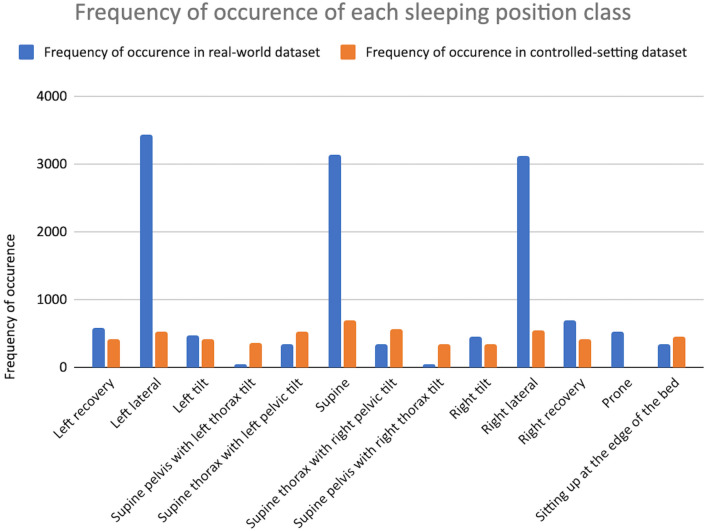
A bar chart of the frequency of occurrences of each position class in the real-world dataset and controlled-setting dataset are shown in Fig. 2. The sleeping positions have been rearranged on the x-axis of the bar chart to demonstrate the progression of the positions starting from left recovery and proceeding leftward, rolling across the back (supine), until right recovery and, finally, prone and sitting.
Figure 2.
Bar chart of the frequency of occurrences of each sleeping position class and the sitting class in the real-world dataset and controlled-setting dataset. Legend: Real-world dataset shown in blue, and controlled-setting dataset shown in orange.
Models
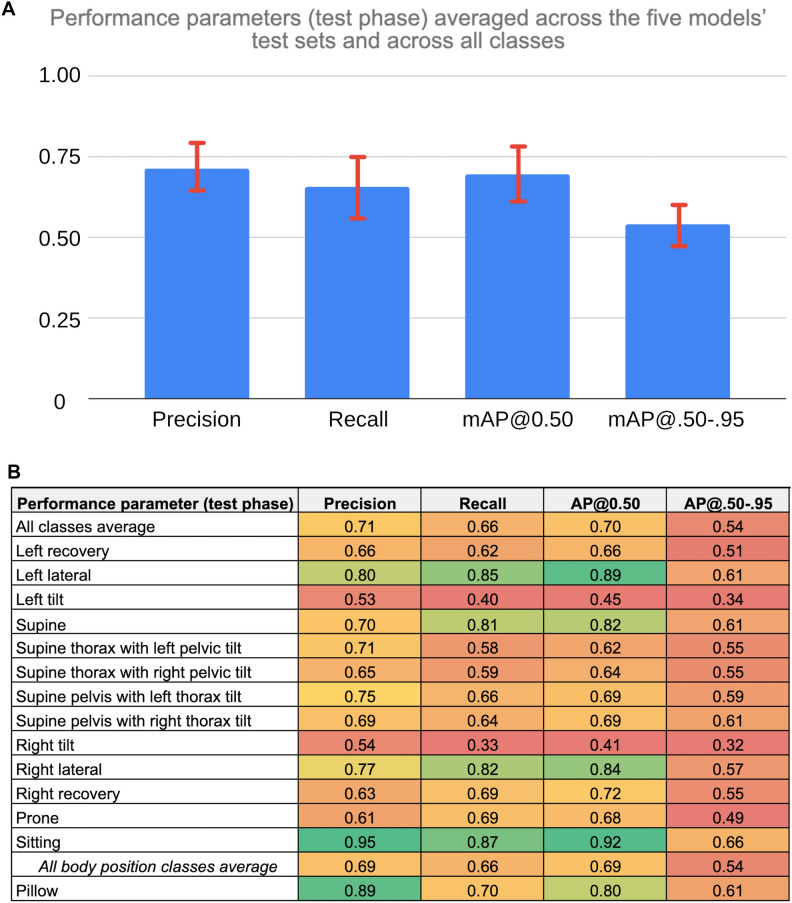
In Fig. 3, class-wise results (averaged across all five loops) are shown using a bar chart and heat map. The bar chart in Fig. 3A shows the four performance metrics from the testing phase averaged across the five models’ (one model per loop of the cross-validation) testing sets and across all classes. The error bars on the bar chart represent one standard deviation of the respective value across all measures, reflecting the variability across models (n = 5) and classes (n = 14). The heatmap in Fig. 3B shows the four performance parameters (columns) from the testing phase averaged across the five models’ test sets for each of the predicted classes (rows).
Figure 3.
(A) Bar Chart of SLeeP AIDePt-2 Performance Metrics From the Testing Phase Averaged Across the Five Models’ Test Sets and Across All Classes. Legend: mAP@0.50 indicates the mean average precision at an intersection of union of 0.50. mAP@.50-.95 indicates the mean average precision at intersections of unions between 0.50 and 0.95. The error bars represent one standard deviation of the respective value across all measures, which reflects the variability both across models and classes. The y-axis does not have units because precision, recall, mAP@0.50, and mAP@.50-.95 are dimensionless values. (B) Heatmap of SLeeP AIDePt-2 Performance Metrics (Columns) From the Testing Phase Averaged Across the Five Models’ Test Sets For Each of the Predicted Classes (Rows). Legend: AP@0.50 indicates the average precision at an intersection of union of 0.50. AP@.50-.95 indicates the average precision at intersections of unions between 0.50 and 0.95. The value of the respective performance metric is mapped to a colour spectrum from red to yellow to green where values of 0.50 or less are represented by red at the lower end of the spectrum, values around 0.75 are shades around yellow (oranger if lower than 0.75; greener if higher than 0.75), and values of 0.90 or more are represented by green at the higher end of the spectrum. The “all body position classes average” is provided as the averaged value of the respective performance metric across the five models’ test sets and the 13 body position classes. For the “all body position classes average” row, the value in the AP@0.50 column is the mean AP@0.50, and the value in the AP@.50-.95 column is the mean AP@.50-.95 since these values represent averages across multiple classes.
On a per-class basis and averaged across the five models, the sitting class had the highest AP@0.50 (0.92). The left lateral, right lateral, and supine classes also had high values of AP@0.50 (0.82–0.89), whereas recovery, prone, and twisted/hybrid positions generally had intermediate values of AP@0.50 (0.62–0.72). The non-hybrid tilted positions (left tilt and right tilt) had the lowest values of AP@0.50 (< 0.50). As for the pillow class, the AP@0.50 was intermediate-to-high (0.80).
See Table S2.1 and Table S2.2 in Supplementary File 2 for a loop-wise summary of the training, validation, and performance testing of the cross-validation of SLeeP AIDePt-2.
Harms
There were no known or identified harms related to this study.
Discussion
Main findings
We collected a heterogeneous video dataset containing instances of single participants and multiple participants, sleeping in or simulating twelve unique sleeping positions along with a sitting position, differing in sleeping environments and pregnancy status (third trimester, non-pregnant), with varying usage of pillows (head, body, pregnancy, wedge) and bed sheets (none, thin, thick; and pattern-free, patterned, striped, textured), both in infrared scale and colour scale video. We trained, validated, and tested a model (SLeeP AIDePt-2) on this dataset to detect the body positions and pillow use of a pregnant person and their bed partner (if any) appearing in an overnight video recording of sleep. The model best detects (high AP@0.50) pillows and the sitting, left lateral, right lateral, and supine positions and has a relatively low false negative rate (high recall) for these detections. The model detects less frequently occurring positions (prone, left/right recovery, supine thorax with left/right pelvic tilt, supine pelvis with left/right thorax tilt) less accurately and is particularly challenged by left/right tilt on which its performance is poorest.
Strengths and limitations
This study describes the transition of a vision-based sleeping position detection model in preparation for real-world use in pregnancy research. The SLeeP AIDePt-2 model has many strengths. Notably, its real-world deployment does not require specialised equipment and takes into account low-lighting, bedsheets, and entry/exit events. It has learned factors unique to pregnancy anatomy and physiology such as determination of the pelvis position (supine, tilt, lateral, recovery) and direction (left, right), which impacts maternal and uteroplacental hemodynamics. It is trained to detect multiple participants and other objects such as pillows in bed simultaneously, enabling it to not only account for more natural occurrences and frequencies of sleeping positions but also more natural sleeping contexts, behaviours, objects, and environments. Prone sleeping in late pregnancy has never been reported in the literature as naturally occurring and, as such, we believe it is exceedingly rare. However, we observed in our real-world dataset that prone sleeping is common in non-pregnant adults. SLeeP AIDePt-2 is trained to detect this. Compared to our previous work28, SLeeP AIDePt-2 is built on an expanded dataset containing a total of 52 participants (39 pregnant, 13 bed partners), including fifteen additional sleeping environments with no restrictions on bedsheets (thickness or patterns) and pillows.
This study has some limitations. One limitation is that SLeeP AIDePt-2 does not account for naturally occurring sleeping positions other than the twelve that we predefined and annotated. We defined “thorax supine with pelvis tilt” as a sleeping position with the persons’ shoulders touching the bed but their legs and pelvis tilted off to one side such that their spine was rotated about 45 degrees in the axial plane. We estimated that any further rotation of the spine would be uncomfortable and would not occur frequently during natural sleep; however, in our real-world dataset, we observed, albeit rarely, more extreme variations of this position such as “thorax supine with pelvis lateral” (90 degrees of spine rotation in the axial plane) and equally extreme but shifted variations of this position such as “thorax tilted (about 45 degrees) with pelvis lateral”. In these events, since the body was twisted with the pelvis being lateral and the thorax being supine or toward supine (tilted), we annotated the position as “thorax supine with pelvis tilt”, which is not 100% accurate but captures, at least in part, the potential physiologic implications of a twisted spine/body and incongruence between the pelvis and thorax position.
Overnight video recordings may not be widely acceptable. While it was not an outcome of this study, in our ongoing, prospective, observational, four-night, home sleep apnea study (ClinicalTrials.gov Identifier: NCT05376475) from which the real-world video dataset was sourced for the current study, we complete an exit interview with each participant and their bed partner and ask them a series of questions about acceptability of overnight video recording. Thus far, acceptability is surprisingly high. We are cautious, however, in applying this acceptability to a population external to our study because our study includes a biassed sample. That is, the participants in our study are likely to be highly accepting of overnight video recordings because they signed up to participate in our study knowing that overnight video recordings would be made and shared with the researchers. Given privacy concerns inherent to the recording of overnight videos, it is likely that other methods of sleeping posture detection (e.g., accelerometer) may have higher acceptance in the general population.
The presence of household pets (e.g., cats, dogs) while sleeping, especially in bed, have recently been implicated in affecting the sleep of their owners49,50, so we also annotated household pets appearing on the bed or in the room in our controlled-setting and real-world datasets. However, this class (household pet), while present in 1266 annotations, appeared in only five participants’ datasets (one controlled-setting and four real-world) and, as such, did not have enough representation across the five folds for training, validation, and testing SLeeP AIDePt-2 to detect this class confidently. As such, SLeeP AIDePt-2 as currently trained cannot detect household pets.
The overall sample size on which SLeeP AIDePt-2 is trained, particularly the real-world dataset, is small and could benefit from further increases in the number of participants, bed partners, and sleeping environments. Finally, the performance of SLeeP AIDePt-2 is sensitive to camera placement, that is, SLeeP AIDePt-2 is trained on frames extracted from video recordings achieved by attaching the camera to the wall at the head of the bed, centred, and 1.6–1.7 m above the sleeping surface. This placement, however, is not possible in some circumstances (e.g., low ceiling height or wall shelving). When this placement is not achieved, the camera orientation and perspective may differ from the predominant orientation and perspective in our underlying training dataset, which is an orientation such that participants’ bodies are oriented vertically in the image with their feet at the top and head at the bottom and a perspective where the camera is looking down from almost directly above the participants’ heads and centred between them. See Supplementary File 3 for a “challenge test” on SLeeP AIDePt-2 by testing its performance on a dataset from a real-world dataset participant and bed partner who inadvertently placed the camera incorrectly (“challenge dataset”) in an effort to assess how model performance is impacted by camera placement.
Interpretation
The SLeeP AIDePt-2 test set was a more challenging test set than the SLeeP AIDePt-1 test set. While the SLeeP AIDePt-1 test set contained the presence of bedsheets in about half of the frames in total, the bedsheets were thin, non-patterned, non-striped, and non-textured. The SLeeP AIDePt-1 test set also contained head pillows only due to restrictions that we imposed on the controlled-setting dataset28. This relative simplicity is in contrast to the test set for SLeeP AIDePt-2, which contained the same frames from the SLeeP AIDePt-1 test set in addition to frames from our real-world dataset, the majority of which contained bed sheets that were thick (e.g., duvets), patterned, striped, and textured, and that were sometimes partially obscured or deformed with multiple pillows above or below the bed sheets. Furthermore, the SLeeP AIDePt-2 test set was more challenging because it contained two people (pregnant participant and bed partner) in most frames in contrast to a single participant. Despite the current model (SLeeP AIDePt-2) being tested on a more challenging test set than our previous model (SLeeP AIDePt-1), it significantly outperformed SLeeP AIDePt-1 for the left lateral, supine, and right lateral positions with AP@0.50’s of 0.89 (vs. 0.72), 0.82 (vs. 0.68), and 0.84 (vs. 0.64), respectively. In contrast, for almost all other positions (except sitting and supine pelvis with right thorax tilt), SLeeP AIDePt-2 performed slightly worse than SLeeP AIDePt-1. The explanation for this difference lies in the underlying datasets. The frequencies of occurrence of the sleeping positions in the controlled-setting dataset on which SLeeP AIDePt-1 was built were approximately equal (Fig. 1); however, this was not so for the sleeping positions in the real-world dataset because people do not spend equal time sleeping in every possible position but, instead, shift between their two or three most comfortable positions. As such, despite our efforts to mitigate class imbalance during training by implementing a weighted random sampler in SLeeP AIDePt-2, it is biassed to best detect the most frequent naturally occurring sleeping positions in pregnancy and in non-pregnant adults, which are left lateral, supine, and right lateral.
Our results are comparable to those from other vision-based sleeping position models in non-pregnant adults19–24. Both Li et al.23 and Mohammadi et al.21 used video recordings from a home-surveillance camera, leveraged controlled-setting data, accounted for bed sheets, and used a similar training methodology to us. Li et al., also combined real-world data with their controlled-setting data, as we did, to build their model23. Unfortunately, comparison of Li et al.’s23 testing performance to ours is not possible because they only present classification accuracy (not recall, precision, nor mAP) but a direct comparison of accuracy is not possible because we cannot compute it for our results—accuracy computation requires the number of true negatives51, which is not computed by YOLO, which is a detector and does not generate negative predictions. Comparing our model to Mohammadi et al.’s model, however, which was trained and validated on approximately 10,000 frames, we calculated their average recall (sensitivity) in the presence of bed sheets to be 0.64 (standard deviation 0.10), which is similar to ours (0.66, standard deviation 0.20). Beyond this, direct comparison of SLeeP AIDePt-2 to other vision-based models and position sensors is challenging because, as far as we are aware, no other measurement tool accounts for the positions of the pelvis and thorax simultaneously.
Conclusion
A computer vision algorithm for prospective measurement of sleeping position using video under real-world conditions across the third trimester in the home setting was built and may hold potential to solve yet unanswered research and clinical questions regarding the relationship between sleeping position and adverse pregnancy outcomes.
While it is currently unknown whether the association between maternal supine sleeping position and adverse pregnancy outcomes is causal, there is currently some controversy and uncertainty regarding the association itself (see Supplementary File 3 for more details)4,52–55. The primary source of scepticism and uncertainty is that the data underlying this association, which is based on subjective, self-reported recollection of sleeping position and does not account for intra-and inter-night variability in sleeping position1,2,4. As such, the current study makes a significant stride toward enabling the study of sleeping positions in the third trimester of pregnancy using more objective methods that were developed with the unique anatomy and physiology of pregnancy in mind (in that it accounts for the position of the pelvis) and can be deployed using ubiquitous equipment without the need for the researcher to visit the participant’s home. Using a model like SLeeP AIDePt-2, researchers could automate measurement of the time spent in each sleeping position across the third trimester, linking it to pregnancy outcomes. As is standard practice in machine learning and software applications, SLeeP AIDePt-2 should be continually audited, iterated, and updated by adding new data to the dataset and re-training, re-validating, and re-testing the model56.
For future research, we propose, “The DOSAGE Study: Dose Of Supine sleep Affects fetal Growth? an Exposure–response Study.” The DOSAGE Study will be an international, prospective, cohort study aiming at gathering objective evidence that will either lend support to, or detract support from, a causal pathway between supine sleeping position after 28 weeks’ gestation, foetal growth, and late stillbirth, and to quantify the safe “dose” of nightly supine sleeping time, if any. See Supplementary File 3 for further details.
Supplementary Information
Acknowledgements
This study was funded by Mitacs through the Mitacs Accelerate Program (Grant No. IT26263) and Mitacs E-Accelerate Program (Grant No. IT26655). The Mitacs Entrepreneur-Accelerate Program funds student and postdoctoral entrepreneurs to further develop the research or technology at the core of their start-up business by way of internships in collaboration with a university, professor and approved incubator. In this study, the total study funding was $15,000 CAD and was provided through Mitacs to the university (University of Toronto) for the professor (Author ED) to administer for the student/intern/entrepreneur (Author AJK) and the study expenses related to the company’s (SBI) technology under the supervision of the incubator (Health Innovation Hub). Mitacs provided 50% of the study funding. Mitacs’ contribution to the grant was matched by SBI, which provided the remaining 50%. Mitacs had no role in the study design; collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication data. However, SBI with the oversight of the University of Toronto had a role in all these aspects via Author’s AJK and ED. This publication was supported by the Canadian Institutes of Health Research (CIHR), Funding Reference Number 192124.
Author contributions
A.J.K.: conceptualization, methodology, software, validation, formal analysis, data curation, writing—original draft, visualization, project administration, funding acquisition. M.E.H.: methodology, software, validation, formal analysis, investigation, data curation, writing—original draft, visualization. H.Z.: formal analysis, investigation, data curation, writing—review & editing. P.E.: investigation, data curation, writing—review & editing. R.A.: investigation, data curation, writing—review & editing. I.L.: methodology, software, formal analysis, data curation, writing—review & editing. L.R.: investigation, data curation, writing—review & editing, project administration. S.A.: conceptualization, methodology, writing—review & editing. B.T.: conceptualization, methodology, writing—review & editing, supervision. S.R.H.: conceptualization, methodology, writing—review & editing, supervision. E.D.: conceptualization, methodology, resources, writing—review & editing, supervision, funding acquisition. All authors read and approved the final manuscript. Written informed consent was obtained to publish identifying images in an online open access publication.
Code availability
The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
Data availability
The datasets generated and analysed during the current study are not publicly available due to restrictions set by the University of Toronto Health Sciences Research Ethics Board because the data from this study (videos and photos containing the participants’ identifiable information, e.g., faces, birthmarks, scars, tattoos) are not easily anonymized and de-identified.
Competing interests
Author AJK is the majority shareholder and the volunteer (unpaid) Chief Executive Officer and President of a startup company, Shiphrah Biomedical Inc. (SBI), which contributed to the study funding through the Mitacs E-Accelerate Program. Author AJK received financial payment from Mitacs through an entrepreneurship internship as part of his graduate studies at the University of Toronto (UofT). Author MEH received financial payment from Mitacs and UofT to complete an international exchange internship with SBI as part of his graduate studies at National Cheng Kung University. The other authors report no financial or non-financial competing interests.
Footnotes
The original online version of this Article was revised: The Acknowledgements section in the original version of this Article was incomplete. Full information regarding the corrections made can be found in the correction for this Article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/17/2024
A Correction to this paper has been published: 10.1038/s41598-024-75418-w
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68472-x.
References
- 1.Cronin, R. S. et al. An individual participant data meta-analysis of maternal going-to-sleep position, interactions with fetal vulnerability, and the risk of late stillbirth. EClinicalMedicine10, 49–57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, N. H. et al. Association of supine going-to-sleep position in late pregnancy with reduced birth weight. JAMA Netw. Open2, e1912614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overview | Antenatal care | Guidance | NICE. https://www.nice.org.uk/guidance/ng201
- 4.National Institute for Health and Care Excellence, National Guideline Alliance & Royal College of Obstetricians and Gynaecologists. Antenatal Care: [W] Maternal Sleep Position during Pregnancy - NICE Guideline NG201 - Evidence Reviews Underpinning Recommendations 1.3.24 to 1.3.25. (2021).
- 5.The AASM Manual for the Scoring of Sleep and Associated Events. American Academy of Sleep Medicine (2020).
- 6.Ferrer-Lluis, I., Castillo-Escario, Y., Montserrat, J. M. & Jané, R. SleepPos app: An automated smartphone application for angle based high resolution sleep position monitoring and treatment. Sensors21, 4531 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrer-Lluis, I., Castillo-Escario, Y., Montserrat, J. M. & Jané, R. Enhanced monitoring of sleep position in sleep apnea patients: Smartphone triaxial accelerometry compared with video-validated position from polysomnography. Sensors21, 3689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer-Lluis, I., Castillo-Escario, Y., Montserrat, J. M. & Jané, R. Analysis of smartphone triaxial accelerometry for monitoring sleep-disordered breathing and sleep position at home. IEEE Access8, 71231–71244 (2020). [Google Scholar]
- 9.Castillo-Escario, Y., Ferrer-Lluis, I., Montserrat, J. M. & Jané, R. Entropy analysis of acoustic signals recorded with a smartphone for detecting apneas and hypopneas: A comparison with a commercial system for home sleep apnea diagnosis. IEEE Access7, 128224–128241 (2019). [Google Scholar]
- 10.Beattie, Z. T., Hagen, C. C. & Hayes, T. L. Classification of lying position using load cells under the bed. Conf. Proc.2011, 474–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahradka, N., Jeong, I. C. & Searson, P. C. Distinguishing positions and movements in bed from load cell signals. Physiol. Meas.39, 125001 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Stern, L. & Roshan Fekr, A. In-bed posture classification using deep neural network. Sensors23, 2430 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadeem, M., Tang, K. & Kumar, A. CNN-based smart sleep posture recognition system. IoT2, 119–139 (2021). [Google Scholar]
- 14.Tang, K., Kumar, A., Nadeem, M. & Maaz, I. CNN-based smart sleep posture recognition system. IoT2, 119–139 (2021). [Google Scholar]
- 15.Chao, Y., Liu, T. & Shen, L.-M. Method of recognizing sleep postures based on air pressure sensor and convolutional neural network: For an air spring mattress. Eng. Appl. Artif. Intell.121, 106009 (2023). [Google Scholar]
- 16.Li, Z., Zhou, Y. & Zhou, G. A dual fusion recognition model for sleep posture based on air mattress pressure detection. Sci. Rep.14, 11084 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue, S., Yang, Y., Wang, H., Rahul, H. & Katabi, D. BodyCompass: Monitoring sleep posture with wireless signals. Proc. ACM Interact. Mob. Wearable Ubiquitous Technol.4, 66:1-66:25 (2020). [Google Scholar]
- 18.Li, X., Gong, Y., Jin, X. & Shang, P. Sleep posture recognition based on machine learning: A systematic review. Pervasive Mob. Comput.90, 101752 (2023). [Google Scholar]
- 19.Grimm, T., Martinez, M., Benz, A. & Stiefelhagen, R. Sleep position classification from a depth camera using Bed Aligned Maps. In 2016 23rd International Conference on Pattern Recognition (ICPR), 319–324. 10.1109/ICPR.2016.7899653 (2016)
- 20.Liu, S., Yin, Y. & Ostadabbas, S. In-bed pose estimation: Deep learning with shallow dataset. IEEE J. Transl. Eng. Health Med.7, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammadi, S. M. et al. Sleep posture classification using a convolutional neural network. In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 1–4 (2018). [DOI] [PubMed]
- 22.Wang, Y. K., Chen, H. Y. & Chen, J. R. Unobtrusive sleep monitoring using movement activity by video analysis. Electronics8, 812 (2019). [Google Scholar]
- 23.Li, Y. Y., Wang, S. J. & Hung, Y. P. A vision-based system for in-sleep upper-body and head pose classification. Sensors22, 2014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbarian, S., Delfi, G., Zhu, K., Yadollahi, A. & Taati, B. Automated non-contact detection of head and body positions during sleep. IEEE Access7, 72826–72834 (2019). [Google Scholar]
- 25.Akbarian, S., Ghahjaverestan, N. M., Yadollahi, A. & Taati, B. Noncontact sleep monitoring with infrared video data to estimate sleep apnea severity and distinguish between positional and nonpositional sleep apnea: Model development and experimental validation. J. Med. Internet Res.23, e26524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam, A. Y. C. et al. A blanket accommodative sleep posture classification system using an infrared depth camera: A deep learning approach with synthetic augmentation of blanket conditions. Sensors21, 5553 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kember, A. J. et al. Impact of maternal posture on fetal physiology in human pregnancy: A narrative review. Front. Physiol.15, 1394707 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kember, A. J. et al. Vision-based detection and quantification of maternal sleeping position in the third trimester of pregnancy in the home setting–building the dataset and model. PLOS Digit. Health2, e0000353 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peduzzi, P., Concato, J., Kemper, E., Holford, T. R. & Feinstein, A. R. A simulation study of the number of events per variable in logistic regression analysis. J. Clin. Epidemiol.49, 1373–1379 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Concato, J., Peduzzi, P., Holford, T. R. & Feinstein, A. R. Importance of events per independent variable in proportional hazards analysis. I. Background, goals, and general strategy. J. Clin. Epidemiol.48, 1495–1501 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Dienes, Z. Understanding Psychology as a Science (Palgrave Macmillan, 2008). [Google Scholar]
- 32.The R Foundation for Statistical Computing. R Statistical Software. (2023).
- 33.Tzutalin. LabelImg Free Software. (2015).
- 34.Lin, T. Y. et al. Microsoft COCO: Common Objects in Context. In Computer Vision—ECCV 2014 (eds Fleet, D. et al.) 740–755 (Springer International Publishing, Cham, 2014). [Google Scholar]
- 35.Redmon, J., Divvala, S., Girshick, R. & Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. http://arxiv.org/abs/1506.02640 (2016).
- 36.Bochkovskiy, A., Wang, C. Y. & Liao, H. Y. M. YOLOv4: Optimal Speed and Accuracy of Object Detection. http://arxiv.org/abs/2004.10934 (2020).
- 37.Jocher, G. et al. ultralytics/yolov5: v6.1—TensorRT, TensorFlow Edge TPU and OpenVINO export and inference. Zenodo 10.5281/zenodo.6222936 (2022).
- 38.Nepal, U. & Eslamiat, H. Comparing YOLOv3, YOLOv4 and YOLOv5 for autonomous landing spot detection in faulty UAVs. Sensors22, 464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karthi, M., Muthulakshmi, V., Priscilla, R., Praveen, P. & Vanisri, K. Evolution of YOLO-V5 algorithm for object detection: Automated detection of library books and performace validation of dataset. In 2021 International Conference on Innovative Computing, Intelligent Communication and Smart Electrical Systems (ICSES) 1–6. 10.1109/ICSES52305.2021.9633834 (2021).
- 40.Fujita, N. et al. Effect of right-lateral versus left-lateral tilt position on compression of the inferior vena cava in pregnant women determined by magnetic resonance imaging. Anesth. Analg.128, 1217–1222 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Bamber, J. H. & Dresner, M. Aortocaval compression in pregnancy: The effect of changing the degree and direction of lateral tilt on maternal cardiac output. Anesth. Analg.97, 256–258 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Saravanakumar, K., Hendrie, M., Smith, F. & Danielian, P. Influence of reverse Trendelenburg position on aortocaval compression in obese pregnant women. Int. J. Obstet. Anesth.26, 15–18 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Humphries, A., Thompson, J. M. D., Stone, P. & Mirjalili, S. A. The effect of positioning on maternal anatomy and hemodynamics during late pregnancy. Clin. Anat.33, 943–949 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Cluver, C., Novikova, N., Hofmeyr, G. J. & Hall, D. R. Maternal position during caesarean section for preventing maternal and neonatal complications. Cochrane Database Syst. Rev.10.1002/14651858.CD007623.pub3 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Fields, J. M. et al. Resuscitation of the pregnant patient: What is the effect of patient positioning on inferior vena cava diameter?. Resuscitation84, 304–308 (2013). [DOI] [PubMed] [Google Scholar]
- 46.James, G., Witten, D., Hastie, T. & Tibshirani, R. Resampling Methods in An Introduction to Statistical Learning, 175–201 (Springer, 2013).
- 47.James, G., Witten, D., Hastie, T. & Tibshirani, R. Statistical Learning in An Introduction to Statistical Learning, 15–57 (Springer, 2013).
- 48.Warland, J., O’Brien, L. M., Heazell, A. E. P., Mitchell, E. A., STARS Consortium. An international internet survey of the experiences of 1,714 mothers with a late stillbirth: The STARS cohort study. BMC Pregnancy Childbirth15, 172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medlin, K. & Wisnieski, L. The association of pet ownership and sleep quality and sleep disorders in United States adults. Hum.-Anim. Interact.2023, 1–8 (2023). [Google Scholar]
- 50.Hoffman, C. L., Browne, M. & Smith, B. P. Human-animal co-sleeping: An actigraphy-based assessment of dogs’ impacts on women’s nighttime movements. Anim. Open Access J. MDPI10, 278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baratloo, A., Hosseini, M., Negida, A. & El Ashal, G. Part 1: Simple definition and calculation of accuracy sensitivity and specificity. Emergency3, 48–49 (2015). [PMC free article] [PubMed] [Google Scholar]
- 52.Silver, R. M. et al. Prospective evaluation of maternal sleep position through 30 weeks of gestation and adverse pregnancy outcomes. Obstet. Gynecol.134, 667–676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCowan, L. M. E. et al. Prospective evaluation of maternal sleep position through 30 weeks of gestation and adverse pregnancy outcomes. Obstet. Gynecol.135, 218 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Silver, R. M., Reddy, U. M. & Gibbins, K. J. Response to letter. Obstet. Gynecol.135, 218–219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fox, N. S. & Oster, E. F. The advice we give to pregnant women: Sleep on it. Obstet. Gynecol.134, 665 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Granlund, T., Stirbu, V. & Mikkonen, T. Towards regulatory-compliant MLOps: Oravizio’s journey from a machine learning experiment to a deployed certified medical product. SN Comput. Sci.2, 342 (2021). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Jocher, G. et al. ultralytics/yolov5: v6.1—TensorRT, TensorFlow Edge TPU and OpenVINO export and inference. Zenodo 10.5281/zenodo.6222936 (2022).
Supplementary Materials
Data Availability Statement
The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
The datasets generated and analysed during the current study are not publicly available due to restrictions set by the University of Toronto Health Sciences Research Ethics Board because the data from this study (videos and photos containing the participants’ identifiable information, e.g., faces, birthmarks, scars, tattoos) are not easily anonymized and de-identified.