Abstract
In order to map linear B-cell (LBC) epitopes in the major capsid protein of feline calicivirus (FCV), an expression library containing random, short (100- to 200-bp) fragments of the FCV F9 capsid gene was constructed. Analysis of this library showed it to be representative of the region of the capsid gene that encodes the mature capsid protein. The library was screened by using polyclonal antisera from a cat that had been challenged experimentally with F9 to identify immunoreactive clones containing LBC epitopes. Twenty-six clones that reacted positively to feline antisera in immunoblots were identified. FCV-derived sequence from these clones mapped to a region of the capsid that spanned 126 amino acids and included variable regions C and E. An overlapping set of biotinylated peptides corresponding to this region was used to further map LBC epitopes by using F9 antisera. Four principal regions of reactivity were identified. Two fell within the hypervariable region at the 5′ end of region E (amino acids [aa] 445 to 451 [antigenic site {ags} 2] and aa 451 to 457 [ags 3]). However, the other two were in conserved regions (aa 415 to 421 [ags 1; region D] and aa 475 to 479 [ags 4; central region E]). The reactivity of the peptide set with antisera from 11 other cats infected with a range of FCV isolates was also determined. Ten of 11 antisera reacted to conserved ags 4, suggesting that this region may be useful for future recombinant vaccine design.
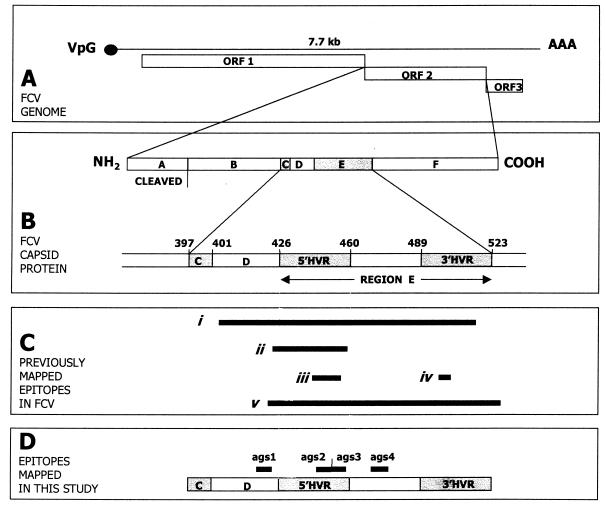
Feline calicivirus (FCV) is an important acute, oral and respiratory pathogen of domestic cats (13) and belongs to the family Caliciviridae (6). It contains a single-stranded, positive-sense RNA genome of approximately 7.7 kb that contains three open reading frames (ORFs) (Fig. 1A) (2, 16, 30, 32, 35, 50, 53). ORF1 is located at the 5′ end of the genome and codes for the nonstructural proteins. ORF3 encodes a putative minor structural protein. ORF2 encodes the major capsid protein and is divided into six regions designated A to F (Fig. 1B). Region A is cleaved to release the mature capsid protein (3). Regions B, D, and F are relatively conserved between FCV isolates, whereas regions C and E are variable between isolates (31, 46–48). Region E has been further divided into 5′ and 3′ hypervariable regions (HVRs) separated by a conserved central domain (Fig. 1B) (48). Whereas isolates of FCV can often be distinguished from each other both antigenically (8, 24, 28, 40) and by sequence analysis (14, 16), FCVs are currently considered to belong to a single serotype (40) and a single genotype (14, 16).
FIG. 1.
Summary of FCV structure. (A) The FCV genome contains three ORFs. (B) ORF2 encodes the major capsid protein, which is divided into conserved regions (regions B, D, and F) and variable regions (regions C and E, including 5′ HVR and 3′ HVR). (C) Antigenic regions previously identified: i, aa 408 to 517 (18); ii, aa 422 to 458 (29); iii, aa 441 to 455 (55); iv, aa 493 to 494 (55); v, aa 420 to 529 (33). (D) Map of region ags 1, 2, 3, and 4 identified in this study is shown. All amino acids are numbered in accordance with the published FCV F9 capsid sequence (3).
Following recovery from clinical disease, cats may develop a persistent, inapparent infection with FCV, and such carriers represent a reservoir of infection for susceptible animals (41, 58). Vaccination against FCV is available (13). The vaccines used are based on whole virus, often an isolate called F9. Although generally effective at preventing clinical disease, cats may develop subclinical and persistent infections under the protection of vaccine-induced immunity (10, 12). Despite vaccination, the prevalence of FCV within the cat population has remained high (5, 19), at levels similar to those reported prior to its introduction (59). Vaccine failures have also been reported. In the majority of cases, these are associated with field virus (8, 42), and it is likely that due to the antigenic variability of FCVs, current vaccines will not induce protection against all field isolates (14, 27). However, vaccine-derived virus has also been implicated as the cause of disease in some vaccine failures (8, 42).
Following infection with FCV, serum virus-neutralizing (VN) antibodies develop by approximately 7 days postinfection (21, 25). The levels of such VN antibodies correlate well with protection against homologous challenge (39). There is also production of immunoglobulin G (IgG)- and IgA-associated mucosal immunity (25) and serum immunofluorescence, complement fixation, complement fixation inhibition, and agar gel precipitation antibodies (15, 34, 57). Although major histocompatibility complex-restricted cytotoxic activity of peripheral blood T lymphocytes has been demonstrated in vaccinated cats, the significance of cytotoxic T lymphocytes to FCV protection is not known (51).
Attempts to characterize the antigenically important regions of FCV are summarized in Fig. 1C. A recombinant peptide corresponding to amino acids (aa) 408 to 517 of FCV F9 induced the formation of neutralizing polyclonal antisera in rabbits, and cats vaccinated with F9 produced a polyclonal antisera that reacted to this peptide (18). The peptide corresponding to aa 422 to 458 contained the epitopes for two neutralizing mouse monoclonal antibodies (N-MAbs) (29). Amino acids 441 to 445 and 493 to 494 are critical to the formation of four linear and three conformational epitopes, respectively (55) (see also Fig. 2). Finally, aa 420 to 529, when transferred from one FCV isolate to another, conveyed neutralization characteristics of the donor virus to the recipient (33). No neutralizing epitopes have currently been mapped in conserved regions of the FCV genome, although one N-MAb to a conformational FCV capsid epitope neutralized all FCV isolates tested (52).
FIG. 2.
Predicted amino acid sequence of the mature capsid coding region of FCV F9 derived from p1BSF9. Regions B, C, D, E, and F (31, 48) are indicated. The underlined sequence (aa 382 to 507) represents the region corresponding to the immunoreactive clones identified by screening λF9CAP.p1 with anti-F9 serum from cat 1. ags 1 (aa 415 to 421), ags 2 (aa 445 to 451), ags 3 (aa 451 to 457), and ags 4 (aa 475 to 479) identified by using the peptide library and the same serum from cat 1 are indicated by asterisks beneath the sequence. ↑ and ▴ indicate sites of mutations that disrupt LBC and conformational B-cell epitopes, respectively, in FCV F4 (55).
Despite these previous studies, no methodical search has been carried out for linear B-cell (LBC) epitopes recognized by feline antibodies in the FCV capsid. The aim of this study was to construct an expression library containing random, short fragments of an FCV capsid gene, suitable for screening with antisera from an experimentally infected cat. The isolate chosen to make the library (F9) is frequently incorporated in FCV vaccines. By using this library, 26 immunoreactive clones were mapped within the capsid. Subsequent fine mapping of LBC epitopes within this region was performed by using an overlapping set of 9-mer peptides.
MATERIALS AND METHODS
Construction of plasmid p1BSF9.
The construction of pF9VAC, which contains the mature capsid protein coding region of FCV F9 (nucleotides 5686 to 7329 [2]), has been described elsewhere (17) and was kindly provided by M. Glenn. To facilitate subsequent manipulation, the F9 capsid gene was excised from pF9VAC and cloned into pBluescript (Stratagene). One clone, designated p1BSF9, was sequenced to confirm that it contained regions B to F of the F9 capsid (nucleotides 5686 to 7329 of the published F9 sequence [2]).
Construction of lambda expression library λF9CAP.p1.
Random overlapping fragments of p1BSF9 of 100 to 200 bp were generated by bovine pancreatic DNase I (Boehringer Mannheim) digestion using standard protocols (45). The correct degree of digestion was achieved by serial 10-fold dilutions of DNase I, and all reactions were performed for 1 min at 14°C. Products of the correct size range were gel purified, blunt ended with the Klenow fragment of DNA polymerase I (Stratagene), and ligated to EcoRI linkers (5′-CCGGAATTCCGG-3′; Stratagene) by using manufacturers’ and standard protocols (45). The products of ligation were digested with EcoRI. Fragments of the appropriate size were gel purified, ligated into the lambda gt11 expression system, and packaged in accordance with the manufacturer’s instructions (lambda gt11 system and Packagene; Promega).
All subsequent manipulations of lambda were performed in accordance with the manufacturers’ protocols unless otherwise stated. The resulting library was designated λF9CAP. Initial screening of λF9CAP was performed by blue-white plaque selection. The library was amplified and aliquoted to produce working stocks designated λF9CAP.p1, which were used in all subsequent experiments.
Characterization of insert size within λF9CAP.
The size of individual inserts within nine randomly picked plaques from λF9CAP was determined by PCR amplification across the λgt11 cloning site using lambda gt11 forward and reverse primers (Promega) in accordance with the manufacturer’s instructions.
Characterization of insert origin within λF9CAP.p1.
In situ hybridization using three probes corresponding to the 5′, middle, and 3′ regions of the mature FCV capsid coding region was used to confirm that λF9CAP.p1 contained DNA of FCV origin and that the library was representative of the entire coding region.
Briefly, probes were generated from p1BSF9 by digestion with the restriction enzyme pairs NheI/NcoI, NcoI/NdeI, and NdeI/BamHI to generate three fragments equivalent to the 5′, middle, and 3′ regions of the mature FCV capsid-coding region. Fragments of the predicted sizes were gel purified (QIAquick gel extraction kit; Qiagen), labelled with [α-32P]dCTP (Prime-it RmT random primer labelling kit; Stratagene), and purified with Sephadex G50 columns (ProbeQuant G-50 microcolumns; Pharmacia Biotech) in accordance with the manufacturers’ instructions.
Target DNA was prepared by plating λF9CAP.p1 in order to produce approximately 300 discrete lambda plaques on a standard 90-mm-diameter petri dish. Plaque blots were prepared in triplicate with a Hybond-N nylon membrane (Amersham). Blots were processed and hybridized essentially in accordance with the manufacturer’s instructions, using stringent wash conditions.
Antisera.
Antisera from specific-pathogen-free cats were prepared as described previously (9), using FCV isolates F9 (1), LS015 and LS027 (25), and Bu13 (7), three commercial vaccine viruses based on F9 (VacA, VacB, and VacC), and a previously undescribed American field isolate (FCVus1).
Immunological screening of λF9CAP.p1.
Hybond-N nylon membrane filters (Amersham) were soaked in 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) and air dried. All wash stages were performed at room temperature with gentle agitation unless otherwise stated.
Approximately 2,000 PFU of λF9CAP.p1 were plated in 90-mm-diameter petri dishes by using standard protocols (Promega). As negative controls, antiserum against F9 was also used to screen recombinant lambda gt11 containing a non-FCV-derived insert as supplied by the manufacturer (Promega). Plates were incubated at 42°C for approximately 4 h until plaques were just visible, overlaid with the IPTG-impregnated membranes, and incubated overnight at 4°C. The membranes were removed from the plates and washed twice in phosphate-buffered saline (PBS; 154 mM NaCl, 3 mM KCl, 9 mM Na2HPO4, 1.65 mM KH2PO4) for 5 min. Membranes were blocked for 1 h in 1% (wt/vol) bovine serum albumin (BSA; Sigma) in PBS and washed twice in 0.05% Tween 20 (Sigma) in PBS (PBS-T).
Anti-F9 serum from cat 1 was diluted 1:200 in 0.5% (wt/vol) BSA in PBS. Membranes were incubated with 10 ml of this primary antiserum for 1 h at 37°C. After the membranes were washed three times in PBS-T for 5 min per wash, they were incubated in 10 ml of mouse monoclonal anti-cat IgG biotin conjugate (Sigma) diluted 1:6,000 in PBS-T containing 0.5% BSA (wt/vol). The membranes were washed three times in PBS-T for 5 min per wash and incubated for 10 min in extravidin peroxidase (Sigma) diluted 1:2,000 in 0.5% (wt/vol) BSA in PBS-T. The membranes were washed for 5 min, twice in PBS-T and twice in PBS, and incubated at room temperature with gentle agitation with fresh 3,3′-diaminobenzadine tetrahydrochloride (DAB; Sigma) prepared in accordance with the manufacturer’s instructions until plaques became visible. The reaction was terminated by washing the membranes twice in tap water and air dried. Positive plaques were stored in 1 ml of phage buffer and subjected to two further rounds of immunological screening prior to sequencing.
Sequencing of DNA from positive plaques.
For sequencing, DNA from reactive plaques was amplified by PCR. Briefly, 10 μl of phage buffer containing each stored plaque was added directly to a 90-μl PCR mix containing 5 U of Taq DNA polymerase and 1× PCR buffer (Advanced Biotechnologies), 100 μM each deoxynucleoside triphosphate, and 200 nM each of primer gt1 (5′-CGGTTTCCATATGGGGATTGGTGGCG-3′) and gt2 (5′-CGCGAAATACGGGCAGACATGGCCTGC-3′) (Kings College, London, United Kingdom). Thermal cycling conditions consisted of 95°C for 2 min, followed by 40 cycles of 95°C (1 min), 50°C (1 min), and 72°C (3 min). A final extension was performed at 72°C for 5 min. A negative control of water was processed simultaneously. Amplicons were purified (Wizard PCR prep DNA purification system; Promega) and sequenced (Prism big dye terminator cycle sequencing ready reaction kit; Perkin-Elmer ABI 377) in accordance with the manufacturers’ instructions.
Peptide mapping.
To span the 126-aa region identified by screening λF9CAP.p1, overlapping 9-mer peptides corresponding to this region were synthesized, each peptide offset from the next one by 2 aa. Peptides were made simultaneously on derivatized polyethylene pins (Chiron Technologies, Clayton, Australia), cleaved, and supplied as lyophilized powders (peptides 3 to 62). Peptides were N-terminally biotinylated and included a 4-aa spacer arm (SGSG). Negative controls consisted of two similar peptides not based on FCV sequence (peptides 1 and 2), and they were supplied by the manufacturer. Two FCV-based peptides that map outside the 126-aa region were also synthesized (RHFDFNQET and QSKIVVFQD; peptides 63 and 64, respectively). The peptide set was screened by using a solid-phase immunoassay in accordance with the manufacturer’s protocol. Briefly, a 1:200 dilution of antisera from FCV-challenged cats was used to detect LBC epitopes. Bound feline IgG was detected by using a 1:1,000 dilution of peroxidase-labelled goat anti-feline IgG (Kirkegaard & Perry Laboratories, Inc.) and ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] (Sigma). Optical density (OD) values (405 nm) for each peptide were determined in duplicate, averaged, and corrected by subtracting the average of the two negative control peptides.
RESULTS
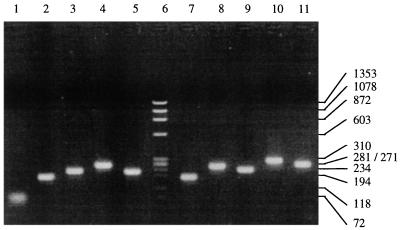
Predicted amino acid sequence of the mature capsid coding region derived from p1BSF9 is shown in Fig. 2. Results of PCR analysis of nine randomly picked plaques from λF9CAP.p1 are shown in Fig. 3. Amplicon sizes range from approximately 180 to 310 bp, which is equivalent to lambda gt11 insert sizes of 90 to 220 bp (PCR amplification across the cloning site of λgt11 adds approximately 90 nucleotides of vector-derived DNA to the insert). Probes representing the 5′, middle, and 3′ regions of the F9 capsid gene each reacted positively with approximately 10% of plaques in λF9CAP.p1 (data not shown). This is in broad agreement with the proportion of p1BSF9 that was FCV derived and suggested that λF9CAP.p1 was equally representative of the mature capsid-coding region.
FIG. 3.
PCR amplification across the cloning site of nine randomly picked plaques (lanes 2 to 5 and 7 to 11) from λF9CAP.p1. Lane 1, negative control; lane 6, HaeIII-digested φX174 molecular weight markers (Boehringer Mannheim) (with values in base pairs indicated on the right). Amplicon sizes range from approximately 180 to 310 bp.
Immunoscreening of λF9CAP.p1 with anti-F9 serum from cat 1 identified approximately 10 immunoreactive plaques per 2,000 PFU. The antiserum did not react positively to any plaques in the lambda negative control. Sequence was obtained from 26 of the immunoreactive plaques after two further rounds of plaque purification. Reactive clones covered 126 aa between aa 382 and aa 507 and included sequence from the 3′ end of region B to the 5′ end of the 3′ HVR of region E (Fig. 2). Several clones did not overlap each other, suggesting that there were at least two regions containing LBC epitopes. No immunoreactive clones were identified outside this region, despite several attempts using duplicate blots and attempting to select immunoreactive clones that did not hybridize in a Southern blot to a probe containing sequence from the 5′ HVR (data not presented).
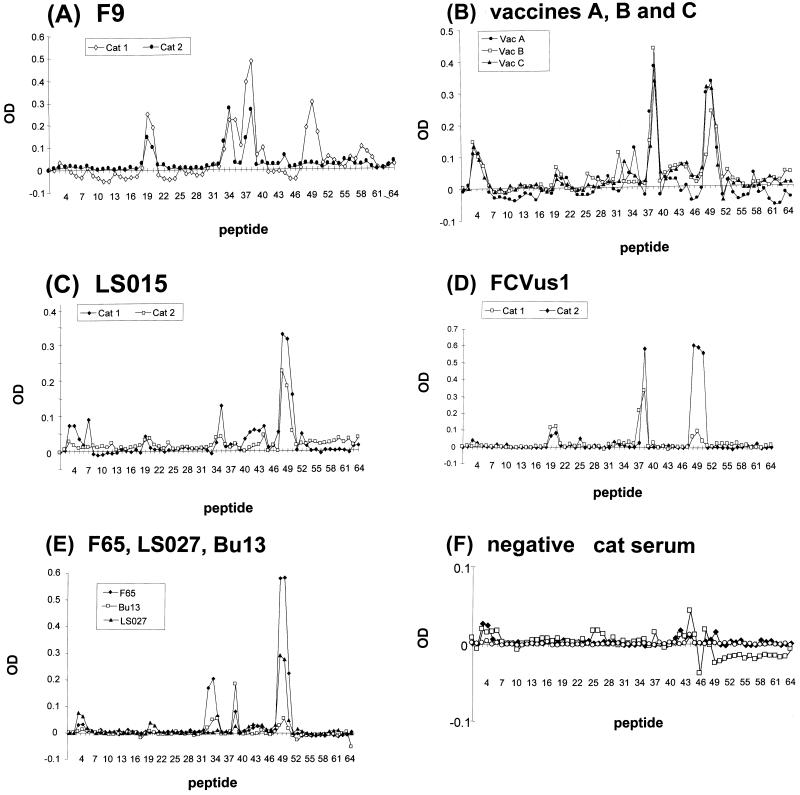
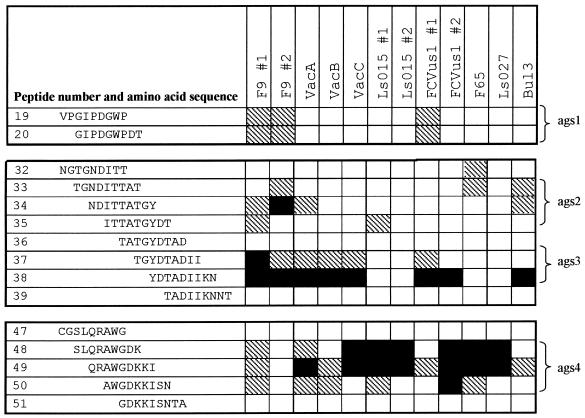
Results of screening the overlapping peptide set using antisera from cats infected with different FCV isolates are shown in Fig. 4 and 5. Anti-F9 serum from cat 1 demonstrated four putative LBC epitope-containing antigenic sites (ags) identified as GIPDGWP (aa 415 to 421; peptides 19 and 20, ags 1), ITTATGY (aa 445 to 451; peptides 34 and 35, ags 2), YDTADII (aa 451 to 457; peptides 37 and 38, ags 3), and AWGDK (aa 475 to 479; peptides 48 to 50, ags 4), with the strongest reactivity against region ags 3. ags 1 was located in conserved region D, ags 2 and ags 3 were located in the 5′ HVR of region E, and ags 4 was located in the conserved central part of region E (Fig. 1D and 2). All immunoreactive clones identified in λF9CAP.p1 encoded at least one of the ags, either ags 1, ags 2, ags 3, or ags 4 (data not presented).
FIG. 4.
OD values for screening the 64 peptides of the peptide library with antisera from cats infected with F9 (cats 1 and 2) (A), vaccine-derived FCV (VacA, VacB, and VacC) (B), LS015 (cats 1 and 2) (C), FCVus1 (cats 1 and 2) (D), F65, LS027, and Bu13 (E), and negative specific-pathogen-free cat serum (cats 1, 2, and 3) (F). Peptides 1 and 2 are negative control peptides. All values represent the average of two tests and have been corrected by subtracting the average of the values for the negative peptides.
FIG. 5.
Reactivity of each antiserum with the peptide library. The average corrected enzyme-linked immunosorbent assay OD result for each peptide with any given antiserum is represented as a percentage of the maximum OD value obtained with that antiserum. Values >25% of the maximum are indicated by hatching. Values within 75% of the maximum are indicated by black shading. Antigenic sites are shown on the right. The start of peptide 19 corresponds to aa 413 of the capsid protein.
When anti-F9 serum from cat 2 was used, the strongest reactivity was to both variable region ags 2 and ags 3. However, ags 2 was 2 aa further towards the NH2 terminus of the protein than that with the antiserum from cat 1 (NDITTAT; aa 443 to 449). There was also some activity against region ags 1, but surprisingly, there was no detectable reactivity in region ags 4.
Antisera raised to vaccine viruses (VacA, VacB, and VacC) reacted most with variable ags 3 and conserved ags 4. Reactivity to other regions was less apparent, only exceeding 25% of maximum for VacA antiserum in ags 2.
Antisera from the remaining cats infected with the field isolates reacted consistently with peptides corresponding to conserved ags 4. For antiserum against LS015, F65, and LS027, reactivity was strongest to this region. However, for the remaining antisera (FCVus1 and Bu13), reactivity was as strong or stronger to region ags 3. Overall, reactivity to ags 1 and ags 2 was less apparent.
A low-level reactivity was frequently observed with peptides 3 and 4, but this was also noted with the negative control antisera.
DISCUSSION
It is important to identify those regions of viruses that closely interact with host proteins since these regions determine the pathogenesis and clearance of the virus. For caliciviruses, such regions are largely unknown, although in FCV, current evidence suggests that the 5′ HVR of region E contains the immunodominant regions of the capsid (18, 29, 49, 55, 56). However, these studies either have used nonfeline antibodies (29, 49, 55) or have not analyzed the whole of the FCV capsid coding region (18). In this study, we have carried out a systematic search of the mature FCV capsid for LBC epitopes with feline antisera. Using an expression library, we have shown that the only immunoreactive region of the capsid appears to be confined to 126 aa spanning variable regions C to E. Within these regions, we have demonstrated the presence of at least four putative LBC epitopes by using an overlapping peptide library. Two of these regions (ags 2 and ags 3) map close to each other in the 5′ HVR. However, the other two (ags 1 and ags 4) map to region D and the middle of region E, respectively, in regions of the FCV capsid that are considered to be conserved on the basis of sequence analysis.
In a previous study using mouse N-MAb escape mutants (55), four neutralizing LBC epitopes clustered in the 5′ HVR of FCV isolate F4 were identified (Fig. 2). Mutations that disrupt these epitopes map very close to the ags 2 and ags 3 identified in our study (55). This confirms the significance of the 5′ HVR as a major antigenic determinant in FCV and as an important target for neutralizing antibodies. Indeed, we have previously shown that a mutation at aa 449 of the capsid allows F9 to escape neutralization by the N-MAb IG9 (4) (data not shown).
In addition to these variable epitopes, our study also identified epitopes in conserved regions on either side the 5′ HVR which have not been reported in previous epitope mapping studies. Such conserved B-cell epitopes may ultimately prove to be good candidates for future vaccine design, although the neutralizing ability of antibodies targeting such regions has not yet been determined. Neutralizing immune responses targeting conserved epitopes should in theory be broadly cross-reactive and reduce the risk of viral escape mutant formation. This may be particularly true for the ags 4 identified in this study, since all the cats that were infected with non-F9 FCV isolates also produced antibodies that reacted with this conserved region.
Although we have identified conserved antigenic sites in this study, one of the two cats infected with F9 failed to produce a detectable antibody response to ags 4, and the majority of cats failed to produce detectable antibodies to conserved ags 1. This may suggest that in the isolates used to produce antisera in this study, there may have been minor sequence variability in these otherwise conserved regions, sufficient to disrupt the epitopes. Unfortunately, capsid sequence is not available for all the isolates used in this study. However, in comparisons of the 22 full capsid sequences available to the authors (16), whereas ags 4 was completely conserved, ags 1 contained three sequences. Nineteen isolates had the sequence GIPDGWPA as in F9, one contained a single substitution (GIQDGWP; FCV 2280), and one contained an insertion and a substitution (GIPDQVWP; FCV LLK). This suggests that ags 1 may tolerate more variability than ags 4 and explain why, in this study, ags 1 reactivity was less than that of ags 4.
Alternatively, there may be variability in the response of individual cats to conserved epitopes. This may simply reflect antibody titer differences between antisera used in this study such that if a higher concentration of antiserum had been used, antibodies to these conserved epitopes may have ultimately been detected. It is also possible that variability in antigen presentation associated with differences in histocompatability type may have meant that some cats responded less efficiently to these epitopes.
The failure of studies based on mouse MAbs to identify conserved epitopes may reflect the unnatural host in which MAbs against FCV were made. It is likely that as the natural host, cats infected with FCV (as in this study) are exposed to a greater repertoire of antigens than the mice used to produce MAbs and in which FCV is considered not to replicate. It is also likely that the populations of mice used to produce MAbs represent inbred populations, and, as such, the repertoire of antigens they respond to is likely to be more restricted. Since the earlier MAb studies selected N-MAbs (54); it is also possible that the conserved epitopes we have identified induce nonneutralizing antibodies. It will therefore be important to further characterize the antibodies that react with these conserved regions, as to their ability to neutralize FCV. The presence of shared and therefore possibly conserved epitopes targeted by neutralizing antibodies is suggested by the observed cross-reactivity between FCV antisera and heterologous FCV isolates in vitro which has led to the single serotype definition of FCVs (40). There has also been an N-MAb described which consistently neutralizes different FCV isolates in vitro and that reacts with a conformational epitope in the capsid (1D7) (52). However, attempts to map the 1D7 epitope have been unsuccessful, since it has not been possible to manufacture an escape mutant to this antibody, suggesting that this epitope forms part of an essential region of the FCV capsid (52).
In this study, no epitopes were mapped to other variable regions of the capsid, namely, region C and the 3′ HVR. This suggests that if the observed variability in these regions is immunologically significant, then these regions may contain either conformational B-cell epitopes or T-cell epitopes. Indeed, the 3′ HVR has been implicated in the formation of conformational epitopes using N-MAb escape mutants (55). Fine structural studies of the calicivirus capsid that would identify secondary conformational interactions between regions of primary capsid protein sequence have not yet been performed.
The colocalization of neutralizing B-cell epitopes with HVRs in viral capsid and envelope proteins is well recognized and is believed to reflect the presence of these domains on the surface of viral proteins. In the envelope gene (env) of human immunodeficiency virus type I, the third variable region contains B- and T-cell epitopes (20, 36, 44). In feline immunodeficiency virus, B-cell epitopes have also been mapped to HVRs in env (37). Equivalent regions have also been identified in HVRs of hepatitis C virus envelope glycoprotein (23, 60).
The inherent variability of such surface-expressed immunodominant regions has important implications to FCV antigenicity and neutralization by the host. First, it may explain the wide spectrum of related but slightly different antigenic profiles seen between FCV isolates (8, 9, 22, 24, 40). Second, FCV isolates have been shown to evolve both antigenically and by sequence analysis of the capsid HVRs in carrier cats, suggesting a possible role for virus evolution in the mechanism of FCV persistence (26, 43). Finally, since most FCV vaccines have traditionally been based upon a single isolate, often F9, it is probable that vaccine-induced antibodies that target variable regions will not protect equally against all FCV isolates (8, 9, 38). Indeed, FCV-related disease in vaccinated cats, although uncommon, is well described (8, 9, 38). Conventional polyvalent and subunit recombinant vaccines have been developed in an attempt to improve the cross-reactivity of vaccine-induced immunity (9, 11). In the latter case, a recombinant polypeptide containing regions C to E from five different FCV isolates was able to induce neutralizing antibodies and some protection from FCV challenge but did not alter the duration of FCV shedding compared to that in controls (11).
In conclusion, we have confirmed that the 5′ HVR of capsid region E is an important immunodominant region of FCV, containing linear B-cell epitopes. However, we have for the first time mapped epitopes to more conserved regions of the FCV capsid. Although the importance of these epitopes to virus neutralization needs to be determined, such epitopes may account for the observed cross-reactivity between FCV strains and may lead to the development of improved recombinant vaccines capable of providing protection against all field isolates.
ACKNOWLEDGMENTS
The authors are especially grateful to Ruth Ryvar for technical assistance.
We thank Margaret Hughes at the Liverpool School of Tropical Medicine for all sequencing and Barry Hodson from Chiron Technologies for assistance with peptide set design and handling. We are also very grateful to Satya Malik for help with the enzyme-linked immunosorbent assays.
REFERENCES
- 1.Bittle J L, York C J, Newberne J W, Martin M. Serological relationship of new feline cytopathogenic viruses. Am J Vet Res. 1960;21:547–550. [Google Scholar]
- 2.Carter M J, Milton I D, Meanger J, Bennett M, Gaskell R M, Turner P C. The complete nucleotide sequence of feline calicivirus. Virology. 1992;190:443–448. doi: 10.1016/0042-6822(92)91231-i. [DOI] [PubMed] [Google Scholar]
- 3.Carter M J, Milton I D, Turner P C, Meanger J, Bennett M, Gaskell R M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch Virol. 1992;122:223–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter M J, Routledge E G, Toms G L. Monoclonal antibodies to feline calicivirus. J Gen Virol. 1989;70:2197–2200. doi: 10.1099/0022-1317-70-8-2197. [DOI] [PubMed] [Google Scholar]
- 5.Coutts A J, Dawson S, Willoughby K, Gaskell R M. Isolation of feline respiratory viruses from clinically healthy cats at UK cat shows. Vet Rec. 1994;135:555–556. [PubMed] [Google Scholar]
- 6.Cubitt D, Bradley D W, Carter M J, Chiba S, Estes M K, Saif L J, Schaffer F L, Smith A W, Studdert M J, Thiel H J. Virus taxonomy. Classification and nomenclature of viruses. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 359–363. [Google Scholar]
- 7.Dawson S. Studies of feline calicivirus and its role in feline disease. Ph.D. thesis. Liverpool, United Kingdom: University of Liverpool; 1991. [Google Scholar]
- 8.Dawson S, McArdle F, Bennett D, Carter S D, Bennett M, Ryvar R, Gaskell R M. Investigation of vaccine reactions and breakdowns after feline calicivirus vaccination. Vet Rec. 1993;132:346–350. doi: 10.1136/vr.132.14.346. [DOI] [PubMed] [Google Scholar]
- 9.Dawson S, McArdle F, Bennett M, Carter M, Milton I P, Turner P, Meanger J, Gaskell R M. Typing of feline calicivirus isolates from different clinical groups by virus neutralisation tests. Vet Rec. 1993;133:13–17. doi: 10.1136/vr.133.1.13. [DOI] [PubMed] [Google Scholar]
- 10.Dawson S, Smyth N R, Bennett M, Gaskell R M, McCracken C M, Brown A, Gaskell C J. Effect of primary-stage feline immunodeficiency virus infection on subsequent feline calicivirus vaccination and challenge in cats. AIDS. 1991;5:747–750. doi: 10.1097/00002030-199106000-00016. [DOI] [PubMed] [Google Scholar]
- 11.DeSilver D A, Guimond P M, Gibson J K, Thomsen D R, Wardley R C, Lowery D E. Expression of the complete capsid and the hypervariable region of feline calicivirus in the baculovirus expression system. In: Chasey D, Gaskell R M, Clarke I N, editors. First International Symposium on Caliciviruses. Proceedings of a European Society for Veterinary Virology Symposium. Reading, United Kingdom: European Society for Veterinary Virology and Central Veterinary Laboratory; 1997. pp. 131–143. [Google Scholar]
- 12.Gaskell C J, Gaskell R M, Dennis P E, Woolridge M J A. Efficacy of an inactivated feline calicivirus (FCV) vaccine against challenge with United Kingdom field strains and its interaction with the FCV carrier state. Res Vet Sci. 1982;32:23–26. [PubMed] [Google Scholar]
- 13.Gaskell R M, Dawson S D. Feline respiratory disease. In: Greene C E, editor. Infectious diseases of the dog and cat. 2nd ed. Philadelphia, Pa: The W. B. Saunders Co.; 1998. [Google Scholar]
- 14.Geissler K, Schneider K, Platzer G, Truyen B, Kaaden O-R, Truyen U. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res. 1997;48:193–206. doi: 10.1016/s0168-1702(97)01440-8. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie J H, Judkins B, Kahn D E. Feline viruses. XIII. The use of the immunofluorescent test for the detection of feline picornaviruses. Cornell Vet. 1971;61:172–179. [PubMed] [Google Scholar]
- 16.Glenn M, Radford A D, Turner P C, Carter M, Lowery D, DeSilver D A, Meanger J, Baulch-Brown C, Bennett M, Gaskell R M. Nucleotide sequence of UK and Australian isolates of feline calicivirus (FCV) and phylogenetic analysis of FCVs. Vet Microbiol. 1999;67:75–193. doi: 10.1016/s0378-1135(99)00043-7. [DOI] [PubMed] [Google Scholar]
- 17.Glenn M A. Molecular and phylogenetic studies on feline calicivirus. Ph.D. thesis. Liverpool, United Kingdom: University of Liverpool; 1997. [Google Scholar]
- 18.Guiver M, Littler E, Caul E O, Fox A J. The cloning, sequencing and expression of a major antigenic region from the feline calicivirus capsid protein. J Gen Virol. 1992;73:2429–2433. doi: 10.1099/0022-1317-73-9-2429. [DOI] [PubMed] [Google Scholar]
- 19.Harbour D A, Howard P E, Gaskell R M. Isolation of feline calicivirus and feline herpesvirus from domestic cats 1980 to 1989. Vet Rec. 1991;128:77–80. doi: 10.1136/vr.128.4.77. [DOI] [PubMed] [Google Scholar]
- 20.Javaherian K, Langlois A F, McDanal C, Ross K L, Eckler L I, Jellis C L, Pofry A T, Rusche J R, Bolognesi D P, Putney S D, Matthews T J. Principle neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahn D E, Hoover E A, Bittle J L. Induction of immunity to feline caliciviral disease. Infect Immun. 1975;11:1003–1009. doi: 10.1128/iai.11.5.1003-1009.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalunda M, Lee K M, Holmes D F, Gillespie J H. Serological classification of feline caliciviruses by plaque-reduction and immunodiffusion. Am J Vet Res. 1975;36:353–356. [PubMed] [Google Scholar]
- 23.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knowles J O, Dawson S, Gaskell R M, Gaskell C J, Harvey C E. Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Vet Rec. 1990;127:125–127. [PubMed] [Google Scholar]
- 25.Knowles J O, McArdle F, Dawson S, Carter S D, Gaskell C J, Gaskell R M. Studies on the role of feline calicivirus in chronic stomatitis in cats. Vet Microbiol. 1991;27:205–219. doi: 10.1016/0378-1135(91)90148-9. [DOI] [PubMed] [Google Scholar]
- 26.Kreutz L C, Johnson R P, Seal B S. Phenotypic and genotypic variation of feline calicivirus during persistent infection of cats. Vet Microbiol. 1998;59:229–236. doi: 10.1016/s0378-1135(97)00158-2. [DOI] [PubMed] [Google Scholar]
- 27.Lauritzen A, Jarrett O, Sabara M. Serological analysis of feline calicivirus isolates from the United States and United Kingdom. Vet Microbiol. 1997;56:55–63. doi: 10.1016/S0378-1135(96)01252-7. [DOI] [PubMed] [Google Scholar]
- 28.McArdle F, Dawson S, Carter M J, Milton I D, Turner P C, Meanger J, Bennett M, Gaskell R M. Feline calicivirus strain differentiation using monoclonal antibody analysis in an enzyme-linked immuno-flow-assay. Vet Microbiol. 1996;51:197–206. doi: 10.1016/0378-1135(96)00017-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milton I D, Turner J, Teelan A, Gaskell R, Turner P C, Carter M J. Location of monoclonal antibody binding sites in the capsid protein of feline calicivirus. J Gen Virol. 1992;73:2435–2439. doi: 10.1099/0022-1317-73-9-2435. [DOI] [PubMed] [Google Scholar]
- 30.Neill J D. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 1990;17:145–160. doi: 10.1016/0168-1702(90)90061-f. [DOI] [PubMed] [Google Scholar]
- 31.Neill J D. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus sequences. Virus Res. 1992;24:211–222. doi: 10.1016/0168-1702(92)90008-w. [DOI] [PubMed] [Google Scholar]
- 32.Neill J D, Reardon I M, Heinrikson R L. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J Virol. 1991;65:5440–5447. doi: 10.1128/jvi.65.10.5440-5447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neill J D, Sosnovtsev S, Green K Y. Structure/function studies of the capsid protein of caliciviruses: domain swaps between different feline calicivirus strains. In: Chasey D, Gaskell R M, Clarke I N, editors. First International Symposium on Caliciviruses. Proceedings of a European Society for Veterinary Virology Symposium. Reading, United Kingdom: European Society for Veterinary Virology and Central Veterinary Laboratory; 1997. pp. 120–124. [Google Scholar]
- 34.Olsen R G, Kahn D E, Hoover E A, Saxe N J, Yohn D S. Differences in acute and convalescent-phase antibodies of cats infected with feline picornaviruses. Infect Immun. 1974;10:375–380. doi: 10.1128/iai.10.2.375-380.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshikamo R, Tohya Y, Kawaguchi Y, Tomonaga K, Maeda K, Takeda N, Utagawa E, Kai C, Mikami T. The molecular cloning and sequencing of an open reading frame encoding non-structural proteins of feline calicivirus F4 strain isolated in Japan. J Vet Med Sci. 1994;56:1093–1099. doi: 10.1292/jvms.56.1093. [DOI] [PubMed] [Google Scholar]
- 36.Palker T J, Clark M E, Langlois A J, Matthews T J, Weinhold K J, Randall R R, Bolognesi D P, Haynes B F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pancino G, Chappey C, Saurin W, Sonigo P. B epitopes and selection pressures in feline immunodeficiency virus envelope glycoproteins. J Virol. 1993;67:664–672. doi: 10.1128/jvi.67.2.664-672.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen N C, Hawkins K F. Mechanisms of persistence of acute and chronic feline calicivirus infections in the face of vaccination. Vet Microbiol. 1995;47:141–156. doi: 10.1016/0378-1135(95)00101-f. [DOI] [PubMed] [Google Scholar]
- 39.Povey C, Ingersoll J. Cross-protection among feline caliciviruses. Infect Immun. 1975;11:877–885. doi: 10.1128/iai.11.5.877-885.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Povey R C. Serological relationships among feline caliciviruses. Infect Immun. 1974;10:1307–1314. doi: 10.1128/iai.10.6.1307-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Povey R C, Wardley R C, Jessen H. Feline picornavirus infection: the in vivo carrier state. Vet Rec. 1973;92:224–229. doi: 10.1136/vr.92.9.224. [DOI] [PubMed] [Google Scholar]
- 42.Radford A D, Bennett M, McArdle F, Dawson S, Turner P C, Glenn M A, Gaskell R M. The use of sequence analysis of a feline calicivirus (FCV) hypervariable region in the epidemiological investigation of FCV related disease and vaccine failures. Vaccine. 1997;15:1451–1458. doi: 10.1016/s0264-410x(97)00059-5. [DOI] [PubMed] [Google Scholar]
- 43.Radford A D, Turner P C, Bennett M, McArdle F, Dawson S, Glenn M A, Williams R A, Gaskell R M. Quasispecies evolution of a hypervariable region of the feline calicivirus capsid gene in cell culture and in persistently infected cats. J Gen Virol. 1998;79:1–10. doi: 10.1099/0022-1317-79-1-1. [DOI] [PubMed] [Google Scholar]
- 44.Rusche J R, Javaherian K, McDanal C, Petro J, Lynn D L, Grimaila R, Langlois A, Gallo R C, Arthur L O, Fischinger P J, Bolognesi D P, Putney S D, Matthews T J. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci USA. 1988;85:3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Seal B S. Analysis of capsid protein gene variation among divergent isolates of feline calicivirus. Virus Res. 1994;33:39–53. doi: 10.1016/0168-1702(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 47.Seal B S, Neill J D. Capsid protein gene sequence of feline calicivirus isolates 255 and LLK: further evidence for capsid protein configuration among feline caliciviruses. Virus Genes. 1994;9:183–187. doi: 10.1007/BF01702662. [DOI] [PubMed] [Google Scholar]
- 48.Seal B S, Ridpath J F, Mengeling W L. Analysis of feline calicivirus capsid protein genes: identification of variable antigenic determinant regions of the protein. J Gen Virol. 1993;74:2519–2524. doi: 10.1099/0022-1317-74-11-2519. [DOI] [PubMed] [Google Scholar]
- 49.Shin Y-S, Tohya Y, Oshikamo R, Kawaguchi Y, Tomonaga K, Miyazawa T, Kai C, Mikami T. Antigenic analysis of feline calicivirus capsid precursor protein and its polypeptides produced in a mammalian cDNA expression system. Virus Res. 1993;30:17–26. doi: 10.1016/0168-1702(93)90012-c. [DOI] [PubMed] [Google Scholar]
- 50.Sosnovtsev S, Green K Y. RNA transcripts derived from a cloned full-length copy of feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 51.Tham K M, Studdert M J. Antibody and cell-mediated immune responses to feline calicivirus following inactivated vaccine and challenge. J Vet Med Ser B. 1987;34:640–654. doi: 10.1111/j.1439-0450.1987.tb00445.x. [DOI] [PubMed] [Google Scholar]
- 52.Tohya Y, Masuoka K, Takahashi E, Mikami T. Neutralizing epitopes of feline calicivirus. Arch Virol. 1991;117:173–181. doi: 10.1007/BF01310763. [DOI] [PubMed] [Google Scholar]
- 53.Tohya Y, Taniguchi Y, Takahashi E, Utagawa E, Takeda N, Miyamura K, Yamazaki S, Mikami T. Sequence analysis of the 3′-end of the feline calicivirus genome. Virology. 1991;183:810–814. doi: 10.1016/0042-6822(91)91016-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tohya Y, Taniguchi Y, Tsubikamoto M, Takahashi E, Mikami T. Preparation and characterization of neutralizing monoclonal antibodies to feline calicivirus. Jap J Vet Sci. 1990;52:521–526. doi: 10.1292/jvms1939.52.251. [DOI] [PubMed] [Google Scholar]
- 55.Tohya Y, Yokoyama N, Maeda K, Kawaguchi Y, Mikami T. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J Gen Virol. 1997;78:303–305. doi: 10.1099/0022-1317-78-2-303. [DOI] [PubMed] [Google Scholar]
- 56.Viaplana E, Plana J, Villaverde A. Antigenicity of VP60 structural protein of rabbit haemorrhagic disease virus. Arch Virol. 1997;142:1843–1848. doi: 10.1007/s007050050201. [DOI] [PubMed] [Google Scholar]
- 57.Wardley R C. Studies on feline calicivirus with particular reference to persistent infections. Ph.D. thesis. Bristol, United Kingdom: University of Bristol; 1974. [Google Scholar]
- 58.Wardley R C. Feline calicivirus carrier state: a study of the host/virus relationship. Arch Virol. 1976;52:243–249. doi: 10.1007/BF01348021. [DOI] [PubMed] [Google Scholar]
- 59.Wardley R C, Gaskell R M, Povey R C. Feline respiratory viruses—their prevalence in clinically healthy cats. J Small Anim Pract. 1974;15:579–586. doi: 10.1111/j.1748-5827.1974.tb06538.x. [DOI] [PubMed] [Google Scholar]
- 60.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]