Summary
Background
The ubiquitin regulatory X (UBX) domain-containing proteins (UBXNs) are putative adaptors for ubiquitin ligases and valosin-containing protein; however, their in vivo physiological functions remain poorly characterised. We recently showed that UBXN3B is essential for activating innate immunity to DNA viruses and controlling DNA/RNA virus infection. Herein, we investigate its role in adaptive immunity.
Methods
We evaluated the antibody responses to multiple viruses and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza in tamoxifen-inducible global and constitutive B cell-specific Ubxn3b knockout mice; quantified various immune populations, B lineage progenitors/precursors, B cell receptor (BCR) signalling and apoptosis by flow cytometry, immunoblotting and immunofluorescence microscopy. We also performed bone marrow transfer, single-cell and bulk RNA sequencing.
Findings
Both global and B cell-specific Ubxn3b knockout mice present a marked reduction in small precursor B-II (>60%), immature (>70%) and mature B (>95%) cell numbers. Transfer of wildtype bone marrow to irradiated global Ubxn3b knockouts restores normal B lymphopoiesis, while reverse transplantation does not. The mature B population shrinks rapidly with apoptosis and higher pro and activated caspase-3 protein levels were observed following induction of Ubxn3b knockout. Mechanistically, Ubxn3b deficiency leads to impaired pre-BCR signalling and cell cycle arrest. Ubxn3b knockout mice are highly vulnerable to respiratory viruses, with increased viral loads and prolonged immunopathology in the lung, and reduced production of virus-specific IgM/IgG.
Interpretation
UBXN3B is essential for B lymphopoiesis by maintaining constitutive pre-BCR signalling and cell survival in a cell-intrinsic manner.
Funding
United States National Institutes of Health grants, R01AI132526 and R21AI155820.
Keywords: UBXN, Haematopoiesis, Lymphopoiesis, B cell, COVID-19
Research in context.
Evidence before this study
We recently reported that UBXN3B is essential for controlling DNA virus infection using a tamoxifen-inducible global Ubxn3b−/− mouse model. Mechanistically, UBXN3B is required for activating the cGAS (cyclic GMP-AMP synthase)-STING (stimulator-of-interferon-genes) signalling pathway and antiviral innate immune responses, e.g., type I IFNs.1 Therefore, it is necessary to examine if UBXN3B regulates adaptive viral immunity.
Added value of this study
We have demonstrated that UBXN3B is essential for B lymphopoiesis from the precursor B stage in the bone marrow and production of virus-specific IgM/IgG. UBXN3B plays a crucial role in clearing respiratory virus infection and limiting immunopathology in the lung. These functions are independent of the cGAS-STING signalling pathway. Rather, UBXN3B maintains constitutive pre-BCR signalling in large precursor B and promotes cell survival in mature B cells.
Implications of all the available evidence
The UBXN3B protein sequence is highly conserved between humans and rodents (98% identity), and so its function in murine B lymphopoiesis may be conserved in humans also. Because UBXN3B is essential for pre-B through mature B survival, it may be a potential therapeutic target for B-cell related diseases such as B lymphomas and autoimmunity.
Introduction
The immune system is comprised of various cell types that coordinate responses to infection, and maintain tissue and immune homeostasis. Peripheral immune cells, with the exception of a few cell types such as long-lived memory T cells and some tissue macrophages, are constantly replenished from bone marrow stem cells through progenitor cells.2 For instance, approximately 0.5–1 × 1011 granulocytes are generated daily in adult human individuals.3 The haematopoietic system is a hierarchically organised, somatic stem cell-maintained organ system, with long-lived and self-renewing pluripotent haematopoietic stem cells (LT-HSCs) at its apex.2 LT-HSCs differentiate into short-term multipotent progenitors (MPPs or ST-HSCs) and lineage-committed haematopoietic progenitors, which in turn will eventually differentiate into the numerous mature blood cell lineages.4 While at the apex of the haematopoietic hierarchy, LT-HSCs are largely quiescent, and the highly proliferative MPPs are the primary contributor to steady-state haematopoiesis.5,6 MPPs can differentiate into lineage-committed progenitors, e.g., common lymphoid progenitors (CLPs) and common myeloid progenitors (CMP), which turn into blast cells leading to specific cell types. Among haematopoiesis, B cell development is the best studied, with several stages clearly defined, including pre-progenitor (pre-pro) B, precursor B I (pre-BI), large pre-BII, small pre-BII and immature B inside bone marrow. In bone marrow, the pre–BI transition to large pre-BII is considered an essential checkpoint, involving rearrangement of variable (V)/diversity (D)/joining (J) gene segments by recombination activating genes (RAG1/2) and assembly of pre-B cell receptor (pre-BCR) with a surrogate light chain (SLC) and immunoglobulin (Ig) μ heavy chain (μH) on the cell surface. Pre-BI cells without a functional pre-BCR undergo apoptosis. Once passing the first quality check, pre-BI becomes large pre-BII, which proliferates for several rounds and differentiates into small pre-BII. At this stage, small pre-BII no longer expresses SLC, but begins expressing an Ig light chain κ or λ that forms a BCR together with μH and becomes immature B (checkpoint 2). Immature B cells exit the bone marrow and mature in the peripheral immune organs such as the spleen and lymph node.7 This process is controlled by a unique set of cell-intrinsic transcription factors and cell-extrinsic factors such as cytokines, chemokines and growth factors in its bone marrow niche.4
The human genome encodes 13 ubiquitin regulatory X (UBX) domain-containing proteins, designated UBXNs. The UBX domain shares weak homology with ubiquitin but adopts the same three dimensional fold as ubiquitin.8 Many UBXNs are capable of binding multiple E3 ubiquitin ligases and p97 (also known as VCP), an ATPase associated with various cellular activities (AAA ATPase).9,10 However, the physiological functions of UBXNs remain poorly characterised. We and other research groups have recently shown that several UBXNs regulate pattern recognition receptor11, 12, 13 and nuclear factor-kappa B (NF-κB) signalling pathways.14,15 Of note, we recently reported that UBXN3B positively regulates cGAS (cyclic GMP-AMP synthase)-STING (stimulator-of-interferon-genes) signalling and innate immunity to DNA viruses.1 Herein, we report the essential role of UBXN3B in B lymphopoiesis independent of STING signalling. This function is B-cell intrinsic because transfer of wild type bone marrow to irradiated global Ubxn3b knockout restores the normal B population, while reverse transplantation does not. Mechanistically, UBXN3B deficiency leads to impaired constitutive pre-BCR signalling and cell cycle gene expression in both pre-B and mature B cells. Ubxn3b deficiency renders animals highly vulnerable to respiratory viruses such SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) and influenza virus, typified by reduced production of virus-specific antibodies, increased viral replication and immunopathology in the lung. This study reveals the cell-intrinsic essential role of UBXN3B in pre-B cell signalling, B cell survival, and UBXN3B as a potential therapeutic target for B lymphomas and autoimmunity.
Methods
Mouse models
The mouse line with the exon 1 of Ubxn3b flanked by two LoxP sites (Ubxn3bflox/flox) were generated via homologous recombination by Dr. Fujimoto at Nagoya University.16 The homozygous Ubxn3bflox/flox were then crossed with homozygous tamoxifen-inducible Cre recombinase-oestrogen receptor T2 mice (The Jackson Laboratory, Stock # 008463) to generate Cre+ Ubxn3bflox/flox littermates. To induce Ubxn3b deletion, > 6-week old mice were injected with 100 μL of tamoxifen (10 mg/mL in corn oil) (Sigma, #T5648) via intraperitoneal (i.p.) injection every 2 days for a total duration of 8 days (4 doses). Successful deletion of Ubxn3b was confirmed in our recent study.13 Half of Cre+ Ubxn3bflox/flox litters were treated with tamoxifen and designated Ubxn3b−/−; the other half were treated with corn oil only and designated Ubxn3b+/+. Mice were allowed to purge tamoxifen for at least 4 weeks (unless specified) before any infection or analyses was performed. B6.SJL-Ptprca Pepcb/BoyJ (Stock No. # 002014) is a congenic strain used widely in transplant studies because it carries the differential pan-leukocyte marker Ptprca, commonly known as CD45.1 or Ly5.1. The Mb1-Cre knock-in/knock-out mice (Stock No. # 020505) carry an allele in which exons 2 and 3 of the Mb1 gene (encoding CD79α) have been replaced with a codon optimised Cre recombinase gene and the ATG codon of exon 1 is deleted; both abolishing endogenous Mb1 gene function and placing Cre expression under the control of the endogenous Mb1 promoter/enhancer elements. Homozygous mice are also useful for studying B cell-deficiency. To generate the B-cell specific Ubxn3b knockout, Ubxn3bflox/flox mice were crossed with Mb1-Cre+/+, resulting in Mb1-Cre+/−/Ubxn3bflox/WT mice (Mb1HetUbxn3bHet) which were mated to each other. Five genotypes were analysed: Mb1-Cre−/− Ubxn3BWT/WT (Mb1WTUbxn3bWT), Mb1-Cre−/+ Ubxn3BWT/WT (Mb1HetUbxn3bWT), Mb1-Cre−/+ Ubxn3Bflox/WT (Mb1HetUbxn3bHet), Mb1-Cre+/+ Ubxn3BWT/WT (Mb1KOUbxn3bWT), and Mb1-Cre−/+ Ubxn3Bflox/flox (Mb1HetUbxn3bKO). The main comparison was between Mb1HetUbxn3bWT and Mb1HetUbxn3bKO. The Sting−/− (Stock # 025805) and cGas−/− (Stock # 026554) were purchased from the Jackson Laboratory. All experiments were performed in accordance with relevant guidelines and regulations approved by the Institutional Animal Care and Use Committee at the UConn Health and Yale University. Both sexes were included in the study design and results reporting.
Antibodies, cell lines and viruses
The rabbit anti-UBXN3B (Clone D8H6D, Cat #34945), anti-BLNK mAb (Clone D3P2H, Cat #36438), anti-ß-Actin mAb (Clone 8H10D10, Cat # 3700), anti-phospho-BLNK mAb (Thr152) (Clone E4P2P, Cat #62144), anti-GAPDH (Clone D16H11, Cat # 5174), anti-BTK mAb (Clone D3H5, Cat # 8547), anti-phospho-BTK mAb (Tyr223) (Clone D9T6H, Cat # 87141), anti-MEK1/2 mAb (Clone D1A5, Cat # 8727), and anti-phospho-MEK1/2 mAb (Ser221) (Clone 166F8, Cat # 2338), anti-PLCγ2 mAb (Clone D9H10, Cat #3872), anti-phospho-PLCγ2 Ab (Tyr1217) (Cat #3871), anti-FoxO1 mAb (Clone C29H4, Cat # 2880), ant-phospho-FoxO1 mAb (Ser256) (Clone E1F7T, Cat # 84192), anti-Akt mAb (Clone C67E7, Cat # 4691), anti-phospho-Akt mAb (Ser473) (Clone D9E, Cat # 4060), anti-Stat5 mAb (Clone D2O6Y, Cat # 94205), anti-phospho-Stat5 mAb (Tyr694) (Clone D47E7, Cat # 4322), anti-Stat1 Ab (Cat # 9172) and anti-phospho-Stat1 mAb (Tyr701) (Clone 58D6, Cat # 9167) were purchased from Cell Signaling Technology (Danvers, MA 01923, USA).
Human embryonic kidney 293 cells transformed with T antigen of SV40 (HEK293T, # CRL-3216), human pre-B cell from acute lymphoblastic leukaemia (NALM6, # CRL-3273), and Vero cells (monkey kidney epithelial cells, # CCL-81) were purchased from American Type Culture Collection (ATCC) (Manassas, VA20110, USA). These cell lines are not listed in the database of commonly misidentified cell lines maintained by ICLAC. Cells were grown in DMEM or RPMI-1640 medium supplemented with 10% foetal bovine serum (FBS) and antibiotics/antimycotics (Life Technologies, Grand Island, NY 14072 USA). We routinely added MycoZAP (Lonza Group, Basel, Switzerland) to cell cultures to prevent mycoplasma contamination.
SARS-CoV-2 (Cat# NR-52281, Strain/Isolate hCov-19/USA-WA1/2020) was provided by BEI Resources (funded by National Institute of Allergy and Infectious Diseases and managed by ATCC, United States). The full-length human ACE2 [Accession No: NM_021804] cDNA was inserted into pAV-EGFP-CMV/FLAG and Ad5 viruses were prepared by Vector Builder Inc. (Chicago, IL 60609, USA). Herpes simplex virus (HSV-1) (Cat# VR1493) were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). These viruses were amplified in Vero cells.
Validation of all the antibodies and cell lines are available on the manufacturer's website and can be found in Reagent Validation File.
Generation of UBXN3B knockout by CRISPR-Cas9
The procedures and guide RNA were described in our published study.1 Briefly, a guide RNA specific for UBXN3B (TACGTTCCCTGGTAGAAGAC common for both mouse and human UBXN3B) was cloned into lentiCRISPR-v2 vector and co-transfected into HEK293T cells with the packaging plasmids pVSVg and psPAX2. The wild type was a non-targeting gRNA from Integrated DNA technology. Lentiviral particles in the cell culture supernatants were collected and cleared by brief centrifugation. The supernatants were centrifuged at 28,000×g for 2 h to concentrate viral particles. The virus pellets were resuspended in a small volume of cell culture medium and added together with polybrene (final 8 μg/mL) to target cells such as NALM6. Twenty-four hours after incubation, the transduction medium was replaced with fresh growth medium. Cells were incubated for 24 h and then selected with 0.75 μg/mL of puromycin for 5 days. Knockout efficiency was confirmed by immunoblotting.
Induction of UBXN3B deletion in splenic B cells ex vivo
Splenocytes from WT mice (Cre+ Ubxn3bflox/flox) were treated with 2 μM of 4-hydroxyl tamoxifen or DMSO for 5 days in IMDM medium supplemented with 10% FBS, 4 ng/mL BAFF (Novus, Cat # 2149-BF) and antibiotics/antimycotics to generate Ubxn3b−/− or Ubxn3b+/+ cells respectively. CD19+ B cells were isolated from splenocytes using the CD19 MicroBeads and autoMACS Pro Separator (Miltenyi Biotec).
Concentration of SARS-CoV-2
The virus was grown in Vero cells for 72 h, and the culture medium was cleared by brief centrifugation. A PEG-it Virus Precipitation Solution (Cat# LV810A-1, System Biosciences, Palo Alto, CA 94303, USA) was added to 40 mL of virus culture at a 1:4 ratio, incubated overnight at 4 °C. The mixture was centrifuged at 1500×g for 30 min, and the resulting pellet was suspended in 1 mL of DMEM medium. In parallel, Vero cell culture medium without virus was processed in the same way and used for mock infection.
Plaque-forming assay
Quantification of infectious viral particles in sera or homogenised tissues was performed on Vero cell monolayer.17 Briefly, viral samples were incubated with 90% confluent Vero cells (6-well plate) at 37 °C for 4 h. The inoculum was then removed and replaced with 2 mL of DMEM complete medium with 1% SeaPlaque agarose (Cat. # 50100, Lonza). The cells were incubated at 37 °C, 5% CO2 for 3 days, and on the fourth day the cells were stained with Neutral Red (Sigma–Aldrich) overnight.
Mouse infection/immunisation and monitoring
Mice were administered 2 × 108 plaque forming units (PFU) of Ad5-hACE2 intranasally, then after 5 days intranasally inoculated with 2 × 105 PFU of SARS-CoV-2 or mock. Three hundred and fifty CCID50 (cell culture infectious dose 50% assay) of Influenza A PR8/34H1N1 strain was administered to mice by intranasal instillation in 40 μL of sterile phosphate buffered saline. The body mass of individual mice was weighed on the day of infection (Day 0) as a baseline. The percentage change in an animal was calculated as 100× (Day n-Day 0)/Day 0, where n defines the length of infection (in days). 1 × 104 PFU of HSV-1 was injected intraperitoneally. For immunisation, each mouse was injected with 10 μg of chicken ovalbumin (OVA) intramuscularly on Day 1 and 10 respectively.
Bone marrow transplantation
Eight weeks or older wild type (WT, B6, CD45.1, recipient) male mice were irradiated at a lethal dose (900 rad) with a Gammacell-40 irradiator once and transplanted with ERT2-Cre+-Ubxn3bflox/flox bone marrow (BM) cells (donor, CD45.2) intravenously. Thirty days after transplantation, half of the mice were administered 100 μL of tamoxifen (10 mg/mL in corn oil) (Sigma, #T5648) via intraperitoneal injection (i.p.) every 2 days for a total duration of 8 days (4 doses) (designated Ubxn3b−/− BM–WT). The other half was treated with corn oil in the same way (designated Ubxn3b+/+ BM–WT). Conversely, Ubxn3bflox/flox or ERT2-Cre+ Ubxn3bflox/flox mice were irradiated and transplanted with WT BM. Thirty days after transplantation, all the recipient mice were treated with tamoxifen, resulting in chimeric WT BM–Ubxn3b+/+ and WT BM-Ubxn3b−/− mice. Fifteen to forty-five days after the last dose of tamoxifen, immune cells were analysed by flow cytometry and/or mice were infected with SARS-CoV-2.
Tissue histology
Tissues were fixed in 4% paraformaldehyde (PFA), embedded in paraffin, cut into 4 μM thick sections, immobilised to glass slides, decalcified, and processed for haematoxylin and eosin staining. Arbitrary arthritic disease scores (on a 1–5 scale with 1 being the slightest, 5 the worst) were assessed using a combination of histological parameters, including exudation of fibrin and inflammatory cells into the joints, alteration in the thickness of tendons or ligament sheaths, and hypertrophy and hyperlexia of the synovium18 in a double-blinded manner.
Haemosiderosis was evaluated by iron staining (Prussian Blue stain) (Cat. # ab150674, from Abcam, Cambridge, CB2 0AX, UK). Lungs were fixed in 4% PFA, embedded in paraffin, cut into 4 μM thick sections, immobilised to glass slides, deparaffinised in xylene, rinsed with 100% ethanol, and hydrated progressively in 95%, 70% ethanol and distilled water. The slides were incubated in Iron Stain Solution (1:1 of potassium ferrocyanide solution to hydrochloric acid solution) for 3 min at ambient temperature, rinsed thoroughly in distilled water, stained in Nuclear Fast Red Solution for 5 min, rinsed again with distilled water 4 times, dehydrated in 95% alcohol followed by absolute alcohol, and finally mounted in synthetic resin. The slides were assessed with an Accu-Scope microscope EXI-310 and images were acquired by an Infinity II camera and software. Haematoxylin and eosin (H&E) staining was performed with paraffin-embedded 4 μM thick tissue sections following standard procedures.
TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labelling) was performed with frozen spleen sections. Briefly, spleens were surgically removed, embedded in OCT (Fisher, Cat# 23-730-571), frozen at −80 °C, cut into 4 μm sections and mounted on glass slides. The sections were fixed with 95% ethanol for 10 min and washed twice with PBS for 5 min each time. The slides were post-fixed in 4% PFA for 15 min, then washed twice with PBS. The sections were subsequently saturated with 50 μL of equilibration buffer (Promega, Cat#: G3250) for 10 min and incubated with the TUNEL reaction mixture at 37 °C for 1 h. The slides were then washed in 2× SSC for 15 min and with PBS three times. For double immunofluorescence labelling, the slides were blocked with 5% FBS in PBS and then incubated overnight at 4 °C with 100 μL of an AF647-conjugated anti-mouse CD19 antibody (1:50 in PBS with 5% FPS, BioLegend, Cat#: 115522) or 100 μL of an AF594-conjugated anti-mouse CD3 antibody (1:50 in PBS with 5% FPS, BioLegend, Cat#:100240). After incubation, the slides were washed and mounted using Vectashield Mounting Medium (Vector Laboratories, Cat#: H-1900). Images were acquired using a Zeiss 880 confocal microscope.
Flow cytometry and fluorescence-activated cell sorting (FACS)
Flow and FACS was performed according to our published study.19 Mouse tissues were minced with fine scissors and digested in 4 mL of digestion medium [20 mg/mL collagenase IV (Sigma–Aldrich, St. Louis, MO, USA), 5 U/mL dispase (StemCell, Cambridge, MA, USA), and 50 mg/mL DNase I mix (Qiagen, Germantown, MD, USA) in complete RPMI1640 medium] at 37 °C for 4 h. The lysate was filtrated with a 40 μm cell strainer. Cells were then pelleted down by centrifugation at 500×g for 5 min. The red blood cells in the cell pellet were lysed three times with a lysis buffer (Cat. # 420301 from BioLegend, San Diego, CA 92121, USA). Cells were suspended in FACS buffer and stained for 30 min at 4 °C with the desired antibody cocktails (BioLegend, San Diego, CA, US) of APC-Fire 750-anti CD11b (Cat. # 101261, clone M1/70), Alexa Fluor 700-anti Ly-6G (Cat. # 127621, clone 1A8), Brilliant Violet 421-anti CD11c (Cat. # 117343, clone N418), PerCP-Cy5.5-anti MHC II (Cat. # 107625, clone M5/114.15.2), PE-anti Tetherin (PCDA1) (Cat. # 127103, clone 10C2), Brilliant Violet 510-anti F4/80 (Cat. # 123135, clone BM8), APC-anti CD68 (Cat. # 137007, clone FA-11), PE-Dazzle 594-anti CD3 epsilon (Cat. # 100347, clone 145-2C11), Brilliant Violet 711-anti CD4 (Cat. # 100557, clone RM4-5), Brilliant Violet 570-anti CD8a (Cat. # 100739, clone 53–6.7), Brilliant Violet 650 anti-CD161 (NK1.1) (Cat. # 108735, clone PK136), FITC anti-CD117 (cKit) (Cat. # 105805, clone 2B8), PE anti-erythroid cells (Cat. # 116207, clone TER-119), Brilliant Violet 711-anti CD115 (Cat. # 135515, clone AFS98), FITC-anti CD25 (Cat. # 102005, clone PC61), Zombie UV (Cat. # 423107), PE-Cy7-anti CD45 (Cat. # 103113, clone 30-F11), TruStain FcX-anti CD16/32 (Cat. # 101319, clone 93), APC anti-CD127 (IL-7Ra) (Cat. # 135011, clone A7R34), PE-Dazzle 594 anti-Sca-1 (Ly-6A/6E) (Cat. # 108137, clone D7), PE-Cy5 anti-Flt3 (CD135) (Cat. # 135311, clone A2F10), Brilliant Violet 421-anti CD34 (Cat. # 119321, clone MEC14.7), PE anti-CD16/32 (Cat. # 101307, clone 93), Brilliant Violet 711-anti IgM (Cat. # 406539, clone RMM-1), Brilliant Violet 421-anti CD45R (B220) (Cat. # 103239, clone RA3-6B2), Alexa Fluor 700-anti CD19 (Cat. # 115527, clone 6D5), PE-Cy7 anti-CD93 (Cat. # 136505, clone AA4.1) and Lin- (anti-CD4, CD8, CD11b, CD11c, Gr1, NK1.1, TER119, Siglec-F, FceRIa, CD19, B220 cocktail). Lin-cocktail contains FITC anti-CD4 (Cat.# 100405, clone GK1.5), FITC anti-CD8 (Cat.# 100705, clone 53-6.7), FITC anti-CD11b(Mac1) (Cat.# 101205, clone M1/70), FITC anti-CD11c (Cat.# 117305, clone N418), FITC anti-Gr1(Ly-6G/C) (Cat.# 108405, clone RB6-8C5), FITC anti-NK1.1 (Cat.# 108705, clone PK136), FITC anti-Ter119 (Cat.# 116205, clone TER-119), FITC anti-B220 (Cat.# 103205, clone RA3-6B2), FITC anti-CD19 (Cat.# 115505, clone 6D5), FITC anti-Siglec-F (Cat.# 155503, clone S17007L) and FITC anti-FcεRIα (Cat.# 134305, clone MAR-1). After staining and washing, the cells were fixed with 4% PFA and analysed on a Becton–Dickinson FACS ARIA II, CyAn advanced digital processor (ADP). Data were analysed using the FlowJo software. Among CD45+ cells, CD11b+ Ly6G+ cells were classified as neutrophils, Ly6G− CD11b+ F4/80+ as monocytes/macrophages, Ly6G− CD11b+ CD115+ as monocytes, Ly6G− CD11c+ MHC II+ as dendritic cells (DC), CD3+ as total T cells, CD3+ CD4+ as CD4 T cells, CD3+ CD8+ as CD8 T cells, CD19+ as B cells. Lin− Sca+ cKit+ cells were identified as total HSCs, which were subdivided into Flt3high (short-term HSC or MPP) and Flt3low (long-term HSC). Lin− Sca− cKit+ cells were subdivided into CD34low CD16/32low (MEP), CD34high CD16/32low (CMP) and CD34low CD16/32high (GMP). The Lin− CD127+ cKit+ cells were identified as CLP.
To analyse B lineage fractions, non-B cells (after lysis of red blood cells) were first dumped with FITC-CD3, -TER119, -LY6G, -LY6C, -CD11b, and -NK1.1. The remaining cells were then sequentially gated on BV650 anti-B220 (Cat. # 103241, clone RA3-6B2), APC anti-CD43 (Cat. # 143207, clone S11), PerCP-Cy5 anti-CD24 (Cat. # 101824, clone M1/69), PE anti-BP1/CD249/Ly51 (Cat. # 108307, clone 6C3), BV421 anti-IgM (Cat. # 406517, clone RMM-1), APC-Cy7 anti-IgD (Cat. # 405715, clone 11-26c.2a), PE-Cy7 anti-CD93 (Cat. # 136505, clone RAA4.1), PE-Dazzle 594 anti-CD19 (Cat. # 115553, clone 6D5). The phospho-specific antibodies were PerCP/Cy5.5 anti-MEK1 Phospho (Ser298) (Cat. # 610605, clone A16117B), PE/Dazzle 594 anti-ERK1/2- Phospho Thr202/Tyr204 (Cat. # 369517, clone 6B8B69), PE anti-BTK Phospho Tyr223 (Cat. # 601703, clone A16128B), Alexa 647 anti-AKT Phospho Ser473 (Cat. # 606555, clone A21001C), PE anti-PLCγ1 Phospho Tyr783 (Cat. # 612403, clone A17025A). All the above antibodies were from BioLegend. PE anti-NF-κB Phospho Ser529 (Cat. # MA537412, clone NFkBp65S529-H3) was purchased from Invitrogen.
Single cell RNA sequencing (scRNA-seq)
Bone marrow was pooled from three mice per genotype, and red blood cells were lysed. HSCs/progenitors and B subsets (excluding mature B) were sorted as described above. In order to obtain an even coverage of each cell compartment/subset, we mixed approximately 1/6 of pre-B, ¼ of immature B (which are much more abundant than the others are) with all the HSCs/progenitors (including B progenitors). About 5 × 104 live cells were subjected to a droplet-based 10× Genomics chromium single cell RNA-Seq on a NovaSeq 6000 sequencer and analysed with a Cell Ranger pipeline. Approximately 11,042 Ubxn3b+/+ and 9269 Ubxn3b−/− cells were sequenced and ∼1600 gene/cell and 3800 unique molecular identifier (UMI) counts/cell were obtained. Clustering cells, annotating cell clusters and analysing differentially expressed genes were performed with Loupe Browser 5.0.1. Statistical significance of differential gene expression between two groups was calculated using an exact negative binomial test (sSeq method). For annotating cell clusters, we did not merely rely on the surface markers for flow cytometry, rather we referred to a database, Bloodspot, which provides transcript expression profiles of genes and gene signatures in healthy and malignant haematopoiesis and includes data from both humans and mice.20 The B subsets were readily identified by a medium to high level of B-restricted transcription factors (Pax5, Ebf1) and surface markers (Cd19, Cd79a), while a barely detectable level of Fcgr3 and Cebpa. Sequences are available from the GEO (accession number GSE266190). Bioinformatics analyses were performed using the Reactome (https://reactome.org/).
Bulk RNA-seq
ERT2-Cre+/Ubxn3bflox/flox mice were treated with tamoxifen (Ubxn3b−/−) or corn oil (Ubxn3b+/+) every two days for 4 doses. Five days after the last dose of tamoxifen treatment, bone marrow was collected from both femurs and tibias. Various B cell subsets from bone marrow were sorted as described above. Total RNA was extracted from about 1 × 105 the mature B subset using the RNAeasy Mini Plus kit (Qiagen Cat.No. 74134). mRNA was purified from 400 ng total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesised using random hexamer primers followed by the second strand cDNA synthesis. After end repair, A-tailing, adapter ligation, size selection, amplification, and purification, library quality was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on NovaSeq X Plus platform (PE150). The total clean reads were >48.7 M, clean bases >7.3 Gb. The total mapping ratio >92%. Sequences are available from the GEO (accession number GSE264055). High-quality sequencing reads were aligned to the mouse reference genome (GRCm39, Release M34 https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M34/GRCm39.genome.fa.gz) using Hisat2 (version 2.2.1). Default parameters for Hisat2 were employed. Aligned reads were quantified at the gene level using the featureCounts function from the Subread package (version 2.0.3). The parameters -t exon -g gene_id was specified to count reads mapping to exons and aggregate them by gene identifier, respectively. The annotation gtf file (https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M34/gencode.vM34.basic.annotation.gtf.gz) corresponding to the GRCm39 genome release was utilised to guide the quantification process. Differential gene expression analysis was conducted using the DESeq2 package (version 1.38.3) in R(4.2.3). Raw counts obtained from featureCounts were input into DESeq2, where normalisation and statistical testing were performed to identify differentially expressed genes between conditions using the Wald test. Bioinformatics analyses were performed using the Reactome (https://reactome.org/).
Calcium flux assay
Calcium influx was assayed with a Fluo-4 Direct™ Calcium Assay Kit (ThermoFisher cat# F10471). Briefly cells were incubated with Fluo-4 Direct™ at 37 °C for 1 h and room temperature for 1 h. The cells were prewarmed at 37 °C in a water bath for 1 min and stimulated with 20 μg/mL of a purified F(ab')2 goat anti-mouse IgM (μ chain) antibody (BioLegend, Clone Poly21571, Cat# 157102) for primary mouse B fractions or an anti-human IgM (BioLegend, Clone Poly23973, Cat# 397302) for NALM6, at 37 °C for 10 s and were immediately analysed by flow cytometry on a Becton–Dickinson FACS ARIA II, CyAn advanced digital processor (ADP). The results were presented as the ratio of the mean fluorescence intensity at any given time point [F] subtracted by the fluorescence at time point zero [F0] (before stimulation) and divided by F0, i.e., ΔF/F0.
Quantification of serum IgG by Enzyme-Linked ImmunoSorbent Assay (ELISA)
Anti-SARS-CoV-2 Spike IgG titres in sera were measured with a commercial ELISA kit (Acro Biosystems, Cat # RAS-T018). 100 μL of each diluted serum specimens (500-fold) was added to a well and incubated at room temperature for 1 h, then the unbound serum was washed off three times with the wash solution. 100 μL of diluted horseradish peroxidase-conjugated goat anti-mouse IgG was added to each well and incubated at room temperature for 1 h. After stringency wash, 100 μL of substrate 3,3′,5,5′–tetramethylbenzidine (TMB) was added to each well and incubated at room temperature for 5–30 min for colour development and terminated by 100 μL of 0.16 M sulfuric acid. The absorption at wavelength 450 nm (A450nm) was read on a Cytation 1 plate reader (BioTek, Winooski, VT, USA). For quantification of OVA/HSV-1 IgGs, 100 ng of OVA/1 × 106 PFU of HSV-1 was coated into a 96-well microplate in 100 μL of antigen coating buffer (0.1 M pH 9.5 bicarbonate/carbonate) overnight at 4 °C. The antigen-coated wells were blocked with 200 μL of blocking solution (2% bovine serum albumin in phosphate buffered saline PBS) at room temperature for 2 h. The dilution of OVA sera (1:4000) and HSV-1 sera (1:2000) in blocking solution was pre-optimised. The following steps were the same as for SARS-CoV-2 IgG ELISA.
Immunoblotting
To prepare B cell fractions for immunoblotting, bone marrow B cells were sorted by flow cytometry as described above (∼104–105 cells in each fraction), pelleted down by brief centrifugation, suspended in 50 μL of 2 × SDS-PAGE sample buffer, boiled at 95 °C for 5 min, and centrifuged at 13,000×g for 10 min. Immunoblotting was performed using standard procedures. Briefly, protein samples were resolved by SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis, 4–20% gradient) and transferred to a nitrocellulose membrane. The membrane was blocked in 5% fat-free milk at room temperature for 1 h, incubated with a primary antibody over night at 4 °C, washed briefly and incubated with an HRP-conjugated secondary antibody for 1 h at room temperature. An ultra-sensitive or regular enhanced chemiluminescence (ECL) substrate was used for detection (ThermoFisher, Cat# 34095, 32106). For immunoblotting of proteins from an extremely low number of sorted bone marrow B cells, a Lumigen ECL substrate ought to be used (Southfield, Michigan 48033, USA).
Reverse transcription and quantitative (q) PCR
Up to 1 × 106 cells or 10 mg, tissues were collected in 350 μL of RLT buffer (QIAGEN RNeasy Mini Kit). RNA was extracted following the QIAGEN RNeasy manufacturer's instructions. Reverse transcription of RNA into complementary DNA (cDNA) was performed using the BIO-RAD iScript™ cDNA Synthesis Kit. Quantitative PCR (qPCR) was performed with gene-specific primers and SYBR Green PCR master mix. Results were calculated using the –ΔΔCt method and a housekeeping gene, beta actin, as an internal control. The qPCR primers and probes for immune genes were reported in our previous studies.1,21,22 The new primers are listed in Supplemental Table S1.
Data acquisition and statistical analysis
The sample size chosen for our animal experiments in this study was estimated according to our prior experience in similar sets of experiments and power analysis calculations. All animals were included in analysis, and no method of randomisation/blinding was applied. GraphPad Prism version 10.1.1 was employed for graphing and statistical analysis. All the statistical tests satisfied the data independency assumption other than repeated measures analyses. p-values ≤ 0.05 were considered significant. The sample sizes (biological replicates), sample types, statistical tests, and p-values are specified in the figure legends. The data were first assessed for normal distribution using the Shapiro–Wilk test and, for the assumption of homogeneity of variance, by F-test or Brown–Forsythe test. For the two grouped datasets with normal distribution, a standard two-tailed, unpaired t-test without or with Welch's correction was applied to the same or different homogeneity of variance, e.g., Supplemental Fig. S1d and Fig. 1b. The nonparametric Mann–Whitney test was conducted on datasets that did not satisfy normal distribution, e.g., Fig. 1a. The Holm-Šídák test was simultaneously applied to the above multiple comparisons. The ordinary two-way ANOVA with two-stage step-up method of Benjamini, Krieger and Yekutieli multiple comparisons test was used for normal residuals and equal SDs in three or more group variables, e.g., Fig. 1e. Log transformation was applied to satisfy a normal distribution, e.g., Fig. 1e. In the analyses of repeated measures, the two-way ANOVA followed by Bonferroni's multiple comparisons test was used for time-course datasets, e.g., Fig. 5e. The Greenhouse-Geisser correction was performed if equal variability of differences was not satisfied, e.g., Fig. 4b. The mixed-effects model was applied if there were missing values, e.g., Supplemental Fig. S15f. Survival curves were analysed using the Log–Rank (Mantel–Cox) test and Gehan-Breslow-Wilcoxon test with no censoring, e.g., Supplemental Fig. S15h.
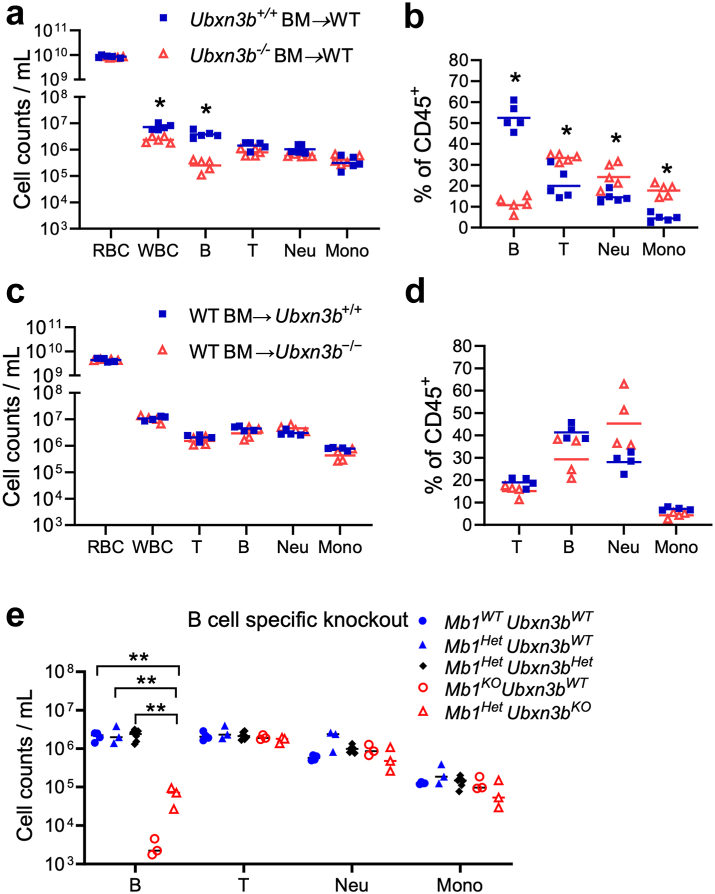
Fig. 1.
UBXN3B controls B cell development in a B cell-intrinsic manner. a, b) Irradiated wild type (WT, CD45.1) recipient mice were transplanted with ERT2-Cre+Ubxn3bflox/flox bone marrow (CD45.2). The mice were then treated with tamoxifen (TMX) to delete Ubxn3b in haematopoietic cells (designated Ubxn3b−/− BM–WT) or corn oil (designated Ubxn3b+/+ BM–WT). c, d) Irradiated Cre+Ubxn3bflox/flox and Ubxn3bflox/flox (CD45.2) mice were transplanted with wild type (WT, CD45.1) bone marrow. The mice were then treated with TMX to delete Ubxn3b (designated WT BM–Ubxn3b−/−) or not (designated WT BM–Ubxn3b+/+) in non-haematopoietic cells. Data shown in a-d) are the cellularity of major blood immune populations at 45 days after TMX. e) The cellularity of blood immune cells in Mb1-Cre−/−Ubxn3BWT/WT (Mb1WTUbxn3bWT), Mb1-Cre−/+Ubxn3BWT/WT (Mb1HetUbxn3bWT), Mb1-Cre−/+Ubxn3Bflox/WT (Mb1HetUbxn3bHet), Mb1-Cre+/+Ubxn3BWT/WT (Mb1KOUbxn3bWT), and Mb1-Cre−/+Ubxn3Bflox/flox (Mb1HetUbxn3bKO) mice. RBC: red blood cells, WBC: white blood cells. The horizontal line indicates the median of the result. Each symbol = one mouse. In a, b) N = 5, c, d) N = 4, e) N = 3 mice/genotype. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 [multiple unpaired Mann–Whitney tests with Holm-Šídák correction for a; multiple unpaired Welch t-tests with Holm-Šídák correction for b-d; two-way ANOVA with two-stage step-up method of Benjamini, Krieger and Yekutieli multiple comparisons test after log-transformation for e].
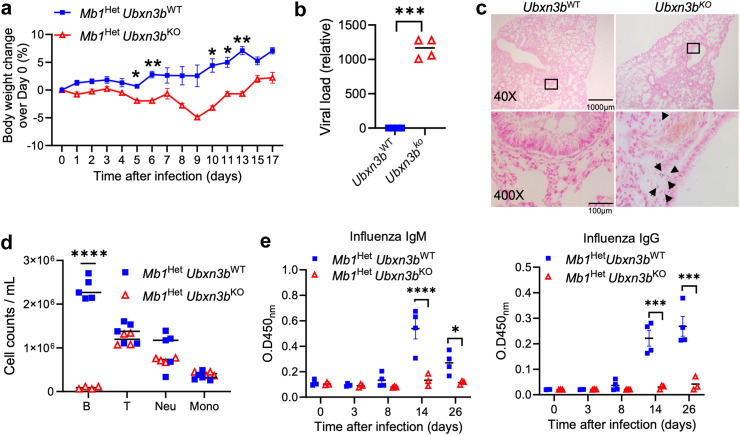
Fig. 5.
UBXN3B limits influenza pathogenesis. Sex- and age-matched B cell specific Ubxn3bKO (Mb1HetUbxn3bKO) and matched Ubxn3bWT (Mb1HetUbxn3bWT) were infected with 350 CCID50 (cell culture infectious dose 50% assay) influenza A PR/8/34H1N1 intranasally. a) The body weight changes over day 0. N = 5 Ubxn3bWT, 3 Ubxn3bKO. b) The viral RNA loads in lungs at 9 days after infection. N = 3 mice per genotype. c) Representative microscopic images of iron-staining (blue) of lung sections at 24 days p.i. (n = 3 mice per genotype). The arrow heads indicate blue cells with haemosiderosis. d) The cell counts of various immune cell populations in the blood at 7 days p.i. N = 5 Ubxn3bWT, 4 Ubxn3bKO. e) The concentrations of IAV-specific serum IgM and IgG presented as optical density (OD) at λ450nm. N = 4 Ubxn3bWT, 3 Ubxn3bKO. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 [repeated measures two-way ANOVA Bonferroni's multiple comparisons test a; unpaired Welch t-test for b; multiple unpaired Welch t-tests with Holm-Šídák correction for d; repeated measures two-way ANOVA with Bonferroni's multiple comparisons test for e].
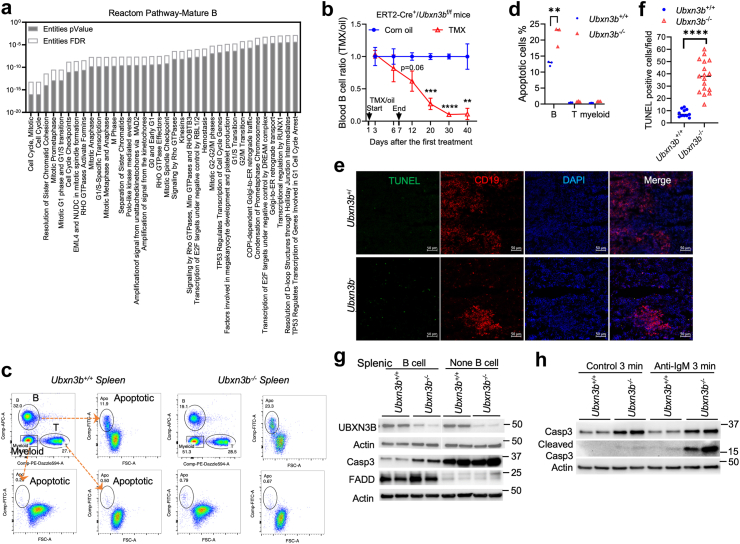
Fig. 4.
UBXN3B is crucial for mature B cell survival. ERT2-Cre+Ubxn3bflox/flox mice were treated with tamoxifen (TMX) (Ubxn3b−/−) or corn oil (Ubxn3+/+) every two days for 7 days. a) At 14 days after the first treatment, the whole transcriptomes in the bone marrow mature B fraction (N = 2 Ubxn3+/+, 3 Ubxn3b−/−) are analysed by bulk RNA-seq. Shown are the top pathways enriched from significantly downregulated genes in Ubxn3b−/−. The statistical method for p-values is Binomial Test, and for FDR (false discovery rate) is Benjamini-Hochberg. b) The blood B cell ratios of TMX to corn oil-treated ERT2-Cre+Ubxn3bflox/flox mice at various timepoints. Data point: mean ± SD, N = 4 mice/genotype, ∗∗p < 0.01, ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001 [repeated measures two-way ANOVA Bonferroni's multiple comparison test with Greenhouse-Geisser correction]. c) The flow cytometry gating strategy and d) percentage of apoptosis (Annexin V positivity) in the splenic B, T, or myeloid populations at 14 days after the first TMX/oil treatment. N = 3 mice/genotype, ∗p < 0.05 [multiple unpaired Welch t-tests with Holm-Šídák correction]. e) Immunofluorescence staining for apoptotic (TUNEL) and B cells (CD19) in the spleens of mice at 14 days after the first TMX/oil treatment. Objective: 20×. Scale bar: 50 μM. DNA is stained with DAPI. f) The numbers of TUNEL-positive cells per microscopic field. Each symbol = one field. N = 11 fields from 3 Ubxn3+/+, N = 17 fields from 3 Ubxn3b−/− spleens. ∗∗∗∗p < 0.0001 [unpaired Welch t-test]. g, h) The immunoblots of indicated proteins in g) splenic B and non-B immune cells, h) splenic B cells stimulated with an anti-mouse IgM antibody or isotype control. Spleens are from the mice at 14 days after the first TMX/oil treatment.
Ethics
All experiments were performed in accordance with relevant guidelines and regulations approved by the Institutional Animal Care and Use Committee at UConn Health (Protocol: AP-200417) and Yale University (Protocol: 10404), adhering to federal and state laws. This research is reported in adherence to the ARRIVE guidelines.
Role of funders
Funders played no role in study design, data collection, data analysis, interpretation, or writing of the manuscript.
Results
UBXN3B is essential for B lymphopoiesis
We have long been interested in UBXNs because of their potential function in ubiquitination and immune regulation. Using a tamoxifen-inducible Cre-LoxP system, we successfully deleted an embryonically essential gene, UBXN3B, in adult mice and demonstrated that UBXN3B positively regulates the cGAS-STING-mediated the type I IFN response to a DNA virus, herpes simplex virus 1 (HSV-1).1 We now asked if UBXN3B regulates the adaptive immune response to HSV-1. To this end, we infected mice with a sublethal dose of HSV-1 (10,000 virions) and monitored the kinetics of HSV-1-specific immunoglobulin G (IgG) production over 30 days. Of note, the serum HSV-1 IgG levels at days 14 through 30 post infection (p.i.) in Ubxn3b−/− mice were significantly lower than those in Ubxn3b+/+ littermates (Supplemental Fig. S1a, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test). Next, we asked if this UBXN3B function requires cGAS-STING. However, Sting−/− and cGAS−/− animals presented even higher HSV-1 IgG levels than their wild type (WT) controls did (Supplemental Fig. S1b and c, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test), likely a result of increased viral replication due to deficient innate immunity. Nonetheless, these results suggest that UBXN3B regulates the IgG response to HSV-1 independently of cGAS-STING. To further confirm this, we examined the IgG response to a nonviral antigen, chicken ovalbumin (OVA). Indeed, the OVA IgG levels at 20 days after immunisation were significantly reduced in Ubxn3b−/− when compared to Ubxn3b+/+ littermates (Supplemental Fig. S1d, two tailed unpaired t-test). We next asked if reduced IgG production is because of fewer B cells in Ubxn3b−/−. Indeed, the B cell counts in both the spleen and blood of Ubxn3b−/− mice were over 10-fold lower than those of Ubxn3b+/+ littermates; while there was a moderate increase in myeloid counts (neutrophils, monocytes, macrophages) in Ubxn3b−/− mice. The total cluster of differentiation (CD) 45+ and T cell counts were moderately lower in Ubxn3b−/− than Ubxn3b+/+ littermates too (Supplemental Fig. S2a–c). Conversely, the neutrophil-to-lymphocyte ratios were significantly higher in Ubxn3b−/− mice (Supplemental Fig. S2b and c, multiple unpaired Welch t-tests with Holm-Šídák correction). Of note, we also observed reduced cell density of the white pulp and dispersed small clusters in the red pulp in the spleen of Ubxn3b−/− (Supplemental Fig. S2d). These results suggest that UBXN3B is essential for B lymphopoiesis. We then asked if STING has also a role in steady state haematopoiesis. However, the blood immune cell compositions were similar between Sting+/+ and Sting−/− littermates (Supplemental Fig. S3), suggesting a STING-independent role of UBXN3B in haematopoiesis.
Haematopoiesis involves a global change of gene expression controlled by cell-intrinsic transcription factors and epigenetic modifiers, and cell-extrinsic factors such as cytokines, chemokines, growth factors, and interactions with osteoblasts, endothelial cells, reticular cells and stromal cells in its bone marrow niche.4 To investigate if the B cell defect in Ubxn3b−/− mice is cell-intrinsic or extrinsic, we performed reciprocal bone marrow transplantation. Firstly, we transferred Cre+ Ubxn3b flox/flox bone marrow (CD45.2) to irradiated wild type (WT, CD45.1) recipient mice. Thirty days after transplantation, the recipient WT mice were treated with either corn oil (designated Ubxn3b+/+ BM–WT) or tamoxifen (dissolved in corn oil) to induce Ubxn3b deletion in the bone marrow (BM) (designated Ubxn3b−/− BM–WT). We confirmed that >99% of blood B cells/neutrophils/monocytes, and >82% of T cells were derived from the CD45.2 donor in both mouse groups at 45 days after transplantation (Supplemental Fig. S4a), indicating successful irradiation and immune reconstitution. The B cell cellularity and total CD45+ cell counts in the chimeric Ubxn3b−/− BM–WT mice were significantly lower than that in Ubxn3b+/+ BM–WT mice (Fig. 1a, multiple unpaired Mann–Whitney tests with Holm-Šídák correction; b, multiple unpaired Welch t-tests with Holm-Šídák correction). The reciprocal bone marrow transplantation [Ubxn3bflox/flox or Cre+ Ubxn3bflox/flox mice (CD45.2) into WT (CD45.1) bone marrow] was successful (Supplemental Fig. S4b) and all the immune cell compartments were comparable between the two groups (Fig. 1c and d, multiple unpaired Welch t-tests with Holm-Šídák correction). These results suggest that UBXN3B controls B cell production in a cell-intrinsic manner. To strengthen this concept, we generated B cell specific Ubxn3b knockout by crossing Ubxn3bflox/flox with an Mb1-driven Cre mouse line. The Mb1-Cre allele abolishes endogenous Mb1 gene (that encodes CD79α) function and places Cre expression under the control of the endogenous Cd79a gene promoter/enhancer, a gene exclusively expressed in the B lineage. B cell-specific Ubxn3b knockout mice should carry a wild type Mb1 and Mb1-Cre allele (Mb1Het), and homologous Ubxn3bflox/flox (Mb1Het Ubxn3bKO). Indeed, mice with Mb1-Cre+/+ alleles resulted in an Mb1KO phenotype, i.e., nearly complete depletion of mature B cells, while having normal T/neutrophils/monocytes (open red circle, Fig. 1e, two-way ANOVA with Benjamini, Krieger and Yekutieli multiple comparisons test), when compared to either Mb1Het or Mb1WT (closed blue circle or triangle, respectively; Fig. 1e). The B cell numbers were reduced by >20-fold in Mb1Het Ubxn3bKO compared to that in Mb1Het Ubxn3bWT (closed blue triangle versus open red triangle, Fig. 1e). Mb1Het Ubxn3bHet demonstrated a similar phenotype as Mb1Het Ubxn3bWT, suggesting that one copy of Ubxn3b is sufficient.
UBXN3B is crucial for B lymphopoiesis beginning from the pre-B stage
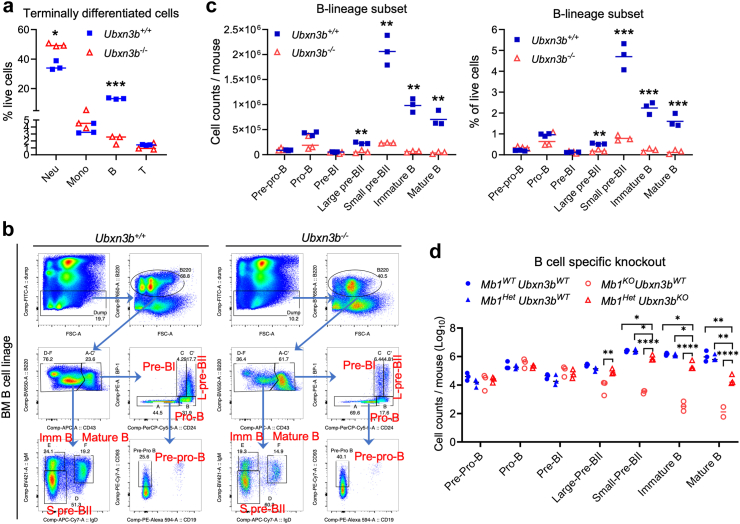
The aforementioned results have demonstrated that UBXN3B primary controls B lymphopoiesis. To this end, we quantitated the terminally differentiated immune cells in bone marrow. Among all live cell types (after lysis of red blood cells), neutrophil was the most abundant, then B cell (recirculating mature B cells). The percentage of B cells was ∼6-fold lower, while the frequency of neutrophils was moderately higher, in Ubxn3b−/− than Ubxn3b+/+ bone marrow (Fig. 2a, multiple unpaired Welch t-tests with Holm-Šídák correction). These results suggest that dysregulated haematopoiesis in Ubxn3b−/− is due to a defect in B lymphopoiesis, which we tested by assessing all the stages of B cell development. Of note, the percentage and cellularity of large precursor BII (pre-BII), small pre-BII, immature B (imm-B) and mature B (recirculating B) fractions were significantly lower in Ubxn3b−/− than Ubxn3b+/+ (∼4, 9, 16, and 18 times respectively by cell number) (Fig. 2b and c, multiple unpaired Welch t-tests with Holm-Šídák correction). Next, we examined stem cells and other lineage progenitors. Total HSCs (Lin− Sca+ Kit+) (LSK) contains two populations, long-term HSCs, which are capable of self-renewal but are quiescent at steady state, and short/mid-term multipotent HSCs (also known as MPPs), which can differentiate into lineage-committed progenitors. The LSK and MPP percentage was modestly decreased in Ubxn3b−/− when compared to Ubxn3b+/+. The frequency of lineage-committed common lymphoid progenitors (CLPs) was also reduced, while the common myeloid progenitors (CMPs) trended higher in Ubxn3b−/− (Supplemental Fig. S5). Largely consistent with the results from the global knockout mice, the numbers of small pre-BII, imm-B and recirculating mature B fractions were significantly smaller (1.9, 3.4, 30 times respectively) in the Mb1Het Ubxn3bKO (open red triangle) mice than those in Mb1Het Ubxn3bWT mice (closed blue triangle). Although the counts of large pre-BII trended lower too (9.4 × 104 versus 1.3 × 105), these values did not reach statistical significance (Fig. 2d, two-way ANOVA with Benjamini, Krieger and Yekutieli multiple comparisons test). Of note, the Mb1KO animals demonstrated defective B cell development as early as in the large pre-BII stage, consistent with its essential role in pre-BCR signalling (Fig. 2d). Overall, these results demonstrate that UBXN3B primarily regulates B lymphopoiesis beginning at the pre-B stage.
Fig. 2.
UBXN3B is essential for the proliferation of large pre-BII. a) The cellularity of terminally differentiated immune cells, b) the flow cytometry gating strategy, c) cellularity and percentage of B lineage fractions in the bone marrow of inducible global Ubxn3b knockout and wildtype littermates derived from ERT2-Cre+Ubxn3bflox/flox. d) The cellularity of B lineage fractions in the bone marrow of B cell specific knockout mice derived from Mb1-Cre−/+/Ubxn3bflox/+. Neu: neutrophil, Mono: monocyte, pre-pro-B: pre-progenitor B, Fraction B: progenitor B (pro-B), C: precursor BI (pre-BI), C’: large (L) pre-BII, D: small (S) pre-BII, E: immature B (Imm-B), F: mature B. The horizontal line indicates the median of the result. Each symbol = one mouse. N = 3 mice/genotype. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 [multiple unpaired Welch t-tests with Holm-Šídák correction for a, c; two-way ANOVA with two-stage step-up method of Benjamini, Krieger and Yekutieli multiple comparisons test after log-transformation for d].
B lymphopoiesis involves a global change of gene expression controlled by cell-intrinsic transcription factors. Therefore, we sorted early bone marrow B fractions and quantified the mRNA expression of well-established transcription factors for haematopoiesis, several of which are B lineage-specific/dominant transcription factors, including early B cell factor (EBF1), paired box protein 5 (PAX5), myocyte enhancer factor (MEF2C) and Ikaros family zinc finger 1/3 (IKZF1/3).23 Ebf1, Pax5 and Ikzf3 (encoding Aiolos) mRNA levels were dramatically induced (>100 fold) in pro-B, pre-B and mature B, when compared to those in pre-pro-B cells. Of note, Ikzf3 was decreased by 5-fold and Ikzf1 (encoding Ikaros) was modestly reduced but only transiently in Ubxn3b−/− pro-B cells, when compared to that in Ubxn3b+/+ pro-B cells (Supplemental Fig. S6a, multiple unpaired Welch t-tests with Holm-Šídák correction). We observed no difference in B cell surface marker genes (Cd19, Cd79a and Il7ra) and a moderate reduction in immunoglobulin V-D-J recombinase Rag1 (Supplemental Fig. S6b, multiple unpaired Welch t-tests with Holm-Šídák correction). These data demonstrate that UBXN3B is largely dispensable for transcription factor expression.
UBXN3B maintains constitutive pre-BCR signalling
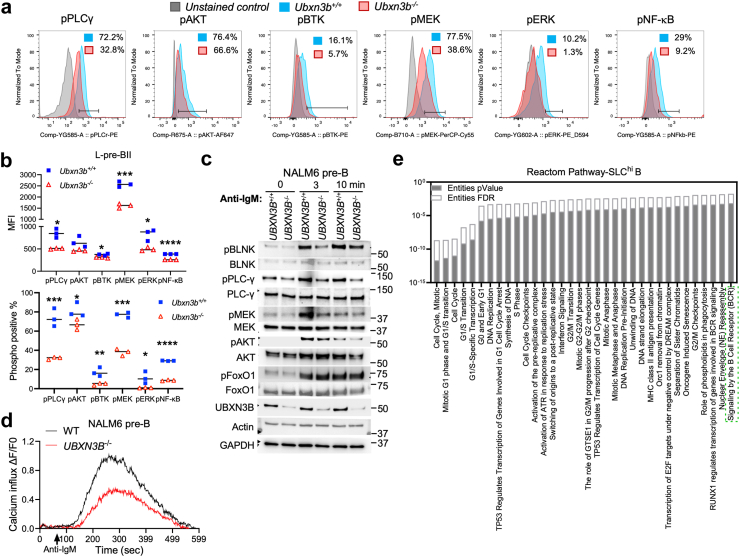
The above results demonstrate that UBXN3B is involved in pre-BI transition to and proliferation of large pre-BII. These steps are monitored by two checkpoints involving pre-BCR signalling,7 which plays several important roles, including allelic exclusion, negative/positive selection, proliferation of large pre-BII, and termination of SLC transcription etc.24 A pre-BCR comprises an Ig μ heavy chain (μH) and a surrogate light chain (SLC); the latter is transiently and robustly induced in pro-B and pre-BI, but rapidly downregulated in pre-BII to allow for BCR recombination and expression.25 However, the endogenous ligands for pre-BCR remain poorly defined. Anti-μH antibodies are known to cross-link and activate BCR signalling. Indeed, an anti-μH antibody stimulated BCR signalling robustly in mouse primary mature B cells, slightly in small pre-BII and immature B, but not at all in earlier fractions in terms of phosphorylation of the major BCR signalling components including BTK (Bruton tyrosine kinase), AKT (RAC serine/threonine-protein kinase), MEK (mitogen-activated protein kinase kinase), ERK (extracellular signal-regulated kinase), PLC-γ (phospholipase C gamma) and NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) (Supplemental Fig. S7). In addition to ligand-dependent activation, auto-crossing of surface pre-BCR or CD79α/β and intracellular pre-BCR complex (tonic) can activate downstream signalling essential for B cell development in the pro-B and pre-B stages.26,27 Highly conserved regions within the Igμ CH1 domain and λ5 (surrogate light chain) induce the aggregation of multiple pre-BCR complexes and downstream signalling.28, 29, 30, 31 Therefore, we assessed constitutive BCR signalling in the bone marrow B fractions by flow cytometry. The phosphorylation of BTK, MEK, ERK, PLC-γ and NF-κB was reduced in Ubxn3b−/− large pre-BII. Moreover, pPLC-γ, pMEK and pERK trended lower in the Ubxn3b−/− pre-BI. The phosphorylation of all these proteins was normal in the other fractions including mature B (Fig. 3a and b, multiple unpaired Welch t-tests with Holm-Šídák correction; Supplemental Fig. S8b, multiple unpaired Welch t-tests with Holm-Šídák correction). The total BTK, PLC-γ and MEK protein levels were not affected (Supplemental Fig. S8c), indicating that UBXN3B regulates their activation rather than protein turnover. We next asked if the UBXN3B function is conserved in humans using NALM6. We knocked UBXN3B out by CRISPR-Cas9 as we did previously,1 and stimulated BCR signalling with an anti-human IgM antibody. We noted a modest decrease in the basal phosphorylation of BLNK, PLC-γ and MEK in UBXN3B−/− cells. Importantly, an anti-human IgM antibody enhanced BLNK, PLC-γ, MEK and AKT phosphorylation, which was impaired in UBXN3B−/− cells. The reduction in proximal BCR signalling components further impaired calcium influx in UBXN3B−/− cells (Fig. 3c and d). Next, we investigated if UBXN3B is required for activating mature BCR signalling or not. We collected ERT-Cre+ Ubxn3bflox/flox splenocytes and used one half to induce Ubxn3b deletion ex vivo with 4-OH tamoxifen for 5 days, while the other half was treated with the solvent, dimethyl sulfoxide, resulting in WT. Mature B cells were then positively purified with an anti-CD19 antibody. This ex vivo approach ensured maximum homogeneity of WT/KO cells except for the Ubxn3b gene. We observed no significant differences in anti-IgM-induced calcium influx and activation (phosphorylation) of key BCR signalling molecules (Supplemental Fig. S9a and b). Consistently, activation of the same BCR signalling proteins was comparable in mature B cells isolated directly from Ubxn3b+/+ and Ubxn3b−/− mice (in vivo tamoxifen treatment) (Supplemental Fig. S9c) demonstrating that UBXN3B is critical for autonomous pre-BCR signalling but dispensable for tonic and induced mature BCR signalling. These results also suggest that UBXN3B targets the early stage of pre-BCR signalling, likely the pre-BCR complex. Therefore, we checked if UBXN3B colocalised with the BCR complex in the plasma membrane. We noted that in the mouse spleen UBXN3B was localised to the cell membrane, and some colocalisations between CD79 and UBXN3B. Similarly, in NALM6 cells, UBXN3B was also localised to the cell periphery (Supplemental Fig. S9d).
Fig. 3.
Pre-BCR signalling is impaired in large pre-BII. a-b) ERT2-Cre+Ubxn3bflox/flox mice were treated with tamoxifen (TMX) (Ubxn3b−/−) or corn oil (Ubxn3+/+) every two days for 7 days. At 14 days after the first dose of TMX, bone marrow B cells were analysed by flow cytometry. a) Representative histograms of the indicated phosphorylated proteins in the large (L) pre-BII fraction. X-axis: fluorescence intensity, Y-axis: normalised to mode. b) The mean fluorescence intensity (MFI) of indicated phosphorylated proteins and percentage of positive L-pre-BII cells (%). Each symbol = one animal. N = 3 mice/genotype. The horizontal line indicates the mean of the results. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 [multiple unpaired Welch t-tests with Holm-Šídák correction]. c) The immunoblots of phosphorylated (p) and total proteins in human NALM6 pre-B cells stimulated with an anti-human IgM. d) The ratio of ΔF (the difference of calcium load between any a given time after anti-IgM treatment and time point zero F0) to F0 in NALM6. Each dot represents the ratio of mean ΔF/F0 of all the cells recorded at a given time (every second). The results are representative of two independent experiments or mice (c, d). e) The top pathways enriched from significantly downregulated genes in the Ubxn3b−/− B fraction with high expression of pre-BCR surrogate light chains (SCLhi). The whole transcriptomes (pooled from 3 mice/genotype) are analysed by single cell (sc) RNA-seq. The statistical method for p-values is Binomial Test, and for FDR (false discovery rate) is Benjamini-Hochberg.
In addition to pre-BCR, interleukin 7(IL-7)/IL-7R signalling is required for B cell development in the bone marrow.32 In mice, IL-7/IL-7R signalling is essential for the transition from MPP to CLP and pro-/pre-B to pre-BI cells.33,34 To clarify if UBXN3B regulates IL-7R signalling, we stimulated NALM6 pre-B cells with a recombinant IL-7 protein and examined STAT5 (signal transducer and activator of transcription 5), AKT, MEK and FoxO1 (Forkhead box protein O1) activation. No significant differences were observed between WT and UBXN3B−/− cells (Supplemental Fig. S9e).
To validate the pre-BCR signalling in an unbiased manner, we performed single cell RNA sequencing (scRNA-seq) on the bone marrow B fractions after removing all the other terminally differentiated haematopoietic cells with an antibody cocktail (FITC-conjugated CD3, TER119, LY6G, -LY6C, CD11b, and NK1.1). We also removed most immature B (IgM+ IgD−) and mature B cells (IgM+ IgD+) from cell preparations for scRNA-seq. We divided cells into the B and non-B lineages and clustered the B lineage based on SLC expression levels (Supplemental Fig. S10) into SLC-high (SLChi), SLC-intermediate (SLCin) and SLC-low (SLClo) populations. We observed a significant reduction in Ubxn3b (alias: Faf2) expression in all the Ubxn3b−/− when compared to Ubxn3b+/+ fractions, validating the tamoxifen-inducible Ubxn3b knockout and scRNA-seq quality (Supplemental Tables S2–S4, exact negative binomial test). In SLChi Ubxn3b−/− cells, 263 differentially expressed genes (DEGs) were significantly downregulated (p < 0.05, Log2 fold <−0.6), and these genes were primarily enriched in the cell cycle (34 genes) and the BCR signalling pathway (11 genes, p < 0.05, FDR <0.05) (Fig. 3e). There were 806 upregulated genes (Log2 fold >0.6, p < 0.05), with an enrichment in the response of EIF2AK1 and type I IFN signalling (Supplemental Table S2). In SLCin Ubxn3b−/− cells, 76 genes were downregulated, and these genes were enriched in the cell cycle (15 genes) and BCR signalling (5 genes) (p < 0.05, FDR <0.05); while top two enriched pathways of the 787 upregulated DEGs were ATF6-activated chaperon and response of EIF2AK1 (p < 0.05, FDR <0.05) (Supplemental Table S3). In SLClo Ubxn3b−/− B cells, only 38 genes were downregulated, including 5 immunoglobulin heavy/light chain genes. Nineteen genes were upregulated with 5 genes related to response of EIF2AK1/EIF2AK4 (Supplemental Table S4). Consistent with a previous RNA-seq analysis of pre-BCR-deficient pre-B fractions,35 we observed decreased expression of genes involved in BCR signalling-regulated genes, such as Il2ra, Ikzf3, Blk, Iglc2, Iglc1, Rasgrp1, Cd22 and Cd38 in SLChi Ubxn3b−/− cells (Log2 fold <−1, p < 0.05). In contrast, SLC genes (Vpreb1/2, Igll1) and Dntt (DNA nucleotidyl transferase) were upregulated (Log2 fold >1, p < 0.05) (Supplemental Table S2), consistent with the failure of pre-BCR signalling to suppress their expression in this stage.25,35 We noted the similar set of down and up-regulated DEGs except Ikzf3 in SLCin Ubxn3b−/− cells (Supplemental Table S3). We validated the mRNA expression of Vpreb1/2 by quantitative RT-PCR. Vpreb expression was barely detectable in pre-pro-B cells but was drastically upregulated in pro-B and further in pre-BI cells, then was suppressed in large pre-BII, further negligible in recirculating mature B and other fully differentiated lineages. Of note, the Ubxn3b expression kinetic was coincident with that of Vpreb. In agreement with scRNA-seq data, Vpreb expression in Ubxn3b−/− was higher than Ubxn3b+/+ large pre-BII (Supplemental Fig. S10b). Lastly, there were 1002 downregulated DEGs in the non-B cells (Log2 fold <−0.6, p < 0.05), however, no significant enrichment in any pathways. On the other hand, 394 genes were upregulated (Log2 fold >0.6, p < 0.05), with enrichment in response of EIF2AK1 (Supplemental Table S5, exact negative binomial test). Overall, these results suggest that UBXN3B positively regulates pre-BCR signalling and cell cycle.
UBXN3B is crucial to mature B cell survival
Next, we performed bulk RNA-seq to test if UBXN3B regulates cell cycle in mature B cells as in SLChi/in cells. Indeed, 414 genes were downregulated (Log2 fold <−0.6, p < 0.05), among which 65 were related to the cell cycle (p < 0.05, FDR <0.05) (Fig. 4a, Supplemental Table S6, Wald test). The upregulated 202 genes (Log2 fold >0.6, p < 0.05) were enriched in the SREBP regulation of cholesterol biosynthesis pathways (p < 0.05, FDR <0.05) and response of EIF2AK1 (p < 0.05, FDR = 0.08) (Supplemental Table S6). Of note, 72% of these cell cycle genes are indirectly repressed by tumour protein 53 (TP53)-DREAM,36 indicating hyperactivation of TP53 signalling in Ubxn3b−/− cells (Supplemental Table S6). Indeed, nearly half of the cell cycle genes were TP53 targets in SLChi and SLCin Ubxn3b−/− B cells (Supplemental Tables S2 and S3). Considering the potential role of TP53 signalling in driving apoptosis, we thus examined if UBXN3B maintains steady-state mature B cell survival. The peripheral B cell population remains largely constant, achieved by the balance between apoptosis and continuous input from the bone marrow. Indeed, in Ubxn3b+/+ animals, the blood B cell counts were ∼ 4 × 106/mL during the 40-day longitudinal study. However, the B cell numbers dropped by 40% on Day 12 after administration of tamoxifen (significant gene deletion took place on ∼ Day 8, after 3 doses of tamoxifen), 73% on Day 20, 90% on Day 30 (Fig. 4b, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test and Greenhouse-Geisser correction). The estimated half-life of Ubxn3b−/− B cells was ∼2 weeks, much shorter than the normal half-life (i.e., 7–8 weeks) of naïve murine mature B cells from WT mice. These results demonstrate that UBXN3B is critical for maintaining B cell survival. At steady state, B cell apoptosis ensures that the size of the peripheral B cell compartment remains stable despite the continuous input of B cells from the bone marrow. Therefore, we quantified Annexin V positive apoptotic blood immune cells by flow cytometry (Fig. 4c). Likely due to high vulnerability of B cells to in vitro processing (red blood cell lysis etc.), we observed a high rate of apoptosis (∼13%) in Ubxn3b+/+ B cells, while negligible T and myeloid death. Of note, there were more apoptotic B cells in Ubxn3b−/− mice (∼23%) (Fig. 4d, multiple unpaired Welch t-tests with Holm-Šídák correction), which was confirmed by TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labelling) staining of the spleen (Fig. 4e and f, unpaired Welch t-test). These TUNEL positive cells were largely negative for CD3+ T cell (Supplemental Fig. S11a). Furthermore, the B cell density was greatly reduced in the Ubxn3b−/− spleen (Supplemental Fig. S11b), consistent with H&E result (Supplemental Fig. S2d). Consistent with increased apoptosis, the pro-caspase-3 protein level was higher in splenic Ubxn3b−/− than Ubxn3b+/+ B cells, as was cleaved caspase-3 before and after BCR stimulation (Fig. 4g and h). While the level of pro-caspase-3 protein was higher in non-B than B cells, its expression was similar between the two genotypes in non-B cells.
UBXN3B limits immunopathogenesis of respiratory viruses
The aforementioned data have demonstrated the essential role of UBXN3B in steady-state B lymphopoiesis and haematopoietic homeostasis. We then asked if this holds true during emergency haematopoiesis triggered by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, the etiological agent of life-threatening COVID-19. It has been clear that COVID-19 mortality is strongly associated with an imbalance in immune cell compartmentalisation, characterised by neutrophilia and lymphopenia,37 reminiscent of Ubxn3b deficiency in mice. Because mice are not permissive to clinical isolates of SARS-CoV-2, we delivered human angiotensin-converting enzyme 2 (ACE2, the cellular entry receptor for SARS-CoV)-expressing Ad5 vector (replication-defective adenovirus vector) intranasally to the mouse lung before infection.38,39 We observed a slight drop in body mass a few days post SARS-CoV-2 infection and rapid recovery of Ubxn3b+/+ mice, while a ∼10% reduction in the body weight of Ubxn3b−/− littermates at days 3–4 p.i. and a significant delay in recovery (Supplemental Fig. S12a, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test). Histopathological analyses by H&E staining revealed more immune infiltrates in the lung of both Ubxn3b+/+ and Ubxn3b−/− mice at 3 days p.i., compared to uninfected mice (day 0). Notably at 10 days p.i., many clusters of brown cells were noted in all the H&E sections from Ubxn3b−/− lungs but were completely absent in Ubxn3b+/+ lungs (Supplemental Fig. S12b). We reasoned that these brownish cells were representative of haemosiderosis, a form of iron overload disorder resulting in the accumulation of haemosiderin, an iron–storage complex. In the lung, macrophages phagocytose red blood cells following vascular leakage, leading to iron overload. Using iron staining we tested this hypothesis and detected a few lightly iron-laden cells at 3 days p.i., but many clusters of heavily iron-laden cells by 10 days p.i. in all the Ubxn3b−/− lungs compared to Ubxn3b+/+. Lung haemosiderosis even persisted up to day 35 p.i. in a few knockout mice (Supplemental Fig. S12c and d). We next examined viral loads and innate immune responses. The viral loads in the lungs trended higher in Ubxn3b−/− when compared to Ubxn3b+/+ mice, though they varied dramatically among individuals at 3 days p.i. By 10 days p.i. the virus was almost cleared from the lung in both mouse genotypes (Supplemental Fig. S12e). The serum cytokines IL-6, TNF-α, IL-10 and granulocyte-macrophage colony-stimulating factor (GM-CSF) were higher in Ubxn3b−/− than those in Ubxn3b+/+ on Day 3 p.i., which is consistent with clinical observations in patients with severe COVID-19. The concentrations of serum interferon alpha (IFN-α), C-X-C motif chemokine ligand 10 (CXCL10) and IFN-γ were modestly upregulated, but equally between Ubxn3b−/− and Ubxn3b+/+ mice after SARS-CoV-2 infection, suggesting normal type I/II IFN responses in Ubxn3b−/− (Supplemental Fig. S13, multiple unpaired Mann–Whitney tests with Holm-Šídák correction). We next examined various immune cell compartments in the lung and peripheral blood, and the longer impact of SARS-CoV-2 infection on immune cells. We noted a ∼3-fold increase in neutrophil and 10-fold decrease in B cell frequencies (Supplemental Figs. S14 and S15a, multiple unpaired Welch t-tests with Holm-Šídák correction); the N/L ratio was also much higher in the blood of Ubxn3b−/− than Ubxn3b+/+ mice at 3 days p.i. (Supplemental Fig. S15b, multiple unpaired Welch t-tests with Holm-Šídák correction). By 35 days p.i., the total immune cells and T cell number were moderately lower, while neutrophils, macrophages/monocytes and N/L ratios were higher in Ubxn3b−/− than Ubxn3b+/+ lungs. Remarkably, the B cell counts were decreased by 5-fold in the lung of Ubxn3b−/− mice (Supplemental Fig. S15c–e, multiple unpaired Welch t-tests with Holm-Šídák correction). Because of the dramatic defect in B cell numbers that might lead to a weak antibody response, we thus measured the concentrations of serum anti-SARS-CoV-2 Spike IgM/IgG. Indeed, both the IgM and IgG concentrations in Ubxn3b−/− were significantly lower than those in Ubxn3b+/+ mice (Supplemental Fig. S15f and g, mixed effect model with Bonferroni's multiple comparisons test).
We next asked if these observations could be extended to other respiratory viruses, such as influenza. Of note, 70% of the global Ubxn3b−/− mice succumbed to a dose of H1N1 influenza A (IAV) that was only sublethal to Ubxn3b+/+ mice (Supplemental Fig. S15h, Log–Rank test). The B cell specific Ubxn3b KO mice did not succumb to the same dose of IAV, consistent with more defects in the host antiviral defence associated with the global Ubxn3b knockout. Mb1Het Ubxn3bKO mice lost more body weight and presented higher viral loads at 9 days p.i. in the lung than Mb1Het Ubxn3bWT (Fig. 5a, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test; b, unpaired Welch t-test). Although the viral RNA was barely detectable in either genotype at 24 days p.i., all the Ubxn3bKO mice (n = 3) presented moderate haemosiderosis, while this disorder was not present in Ubxn3bWT (n = 3) (Fig. 5c). The blood B cell counts at 7 days p.i. and concentrations of serum anti- IAV IgM/IgG were significantly lower in Mb1Het Ubxn3bKO mice (Fig. 5d, multiple unpaired Welch t-tests with Holm-Šídák correction; e, repeated measures two-way ANOVA with Bonferroni's multiple comparisons test).
Discussion
The role of UBXN3B in B lymphopoiesis
The HSC differentiation cascade must be finely regulated to enable timely production of the right number and type of mature cells, i.e., homeostasis, disruption of which may lead to a pathological state, such as autoimmunity, immunodeficiency, cancer, etc. As an example, lymphopenia and a skewed myeloid-to-lymphoid ratio in the elderly may contribute to inflammageing and impaired anti-microbial immunity.40 In this study, we have discovered the essential role of UBXN3B in the maintenance of haematopoietic homeostasis, especially, B lymphopoiesis (Supplemental Fig. S2). This function is primarily B-cell intrinsic as evidenced by the bone marrow transplantation studies and B cell specific Ubxn3b knockout mouse phenotype (Fig. 1). Of note, consistent with the role of pre-BCR signalling,41 UBXN3B is required for the proliferation of large pre-BII (Fig. 2). This stage requires a transient yet essential pre-BCR signalling to drive VDJ recombination and proliferation of large pre-BII.24 Indeed, constitutive activation of the major molecules of pre-BCR signalling is impaired in the Ubxn3b−/− large pre-BII fraction, culminating in deficient B cell development, IgM and IgG antiviral antibody response in knockout mice (Fig. 3a and b; Supplemental Fig. S15f and g; Fig. 5e). Moreover, RNA-seq has confirmed that BCR signalling is one of the significantly downregulated pathways in the SLChi/in Ubxn3b−/− subsets (Fig. 3e, Supplemental Tables S2 and S3). However, in mature B cells, UBXN3B is dispensable for tonic and induced BCR signalling (Supplemental Fig. S8 and S9), which shares the same major components with the pre-BCR pathway except the light chains of BCR. This is unlikely to be due to UBXN3B expression, which is relatively stable during B cell development (Supplemental Fig. S10b). The pre-BCR-associated UBXN3B function could be attributed to different cellular regulations for pre-BCR and BCR respectively, which are unfortunately still poorly defined. Co-receptors that negatively regulate the activity of resting pre-BCR and BCR are developmentally regulated,42 such as CD22, which is not expressed until after the pre-BCR-dependent stage.43 Thus, it is plausible that UBXN3B may require a pre-B specific cellular factor to influence pre-BCR signalling. Alternatively, UBXN3B could target intracellular pre-BCR complex or associated proteins that activate tonic signalling in the pre-B stage.27,42 Indeed, our results have demonstrated that the phosphorylation of BTK is reduced in the steady-state Ubxn3b−/− large pre-BII (Fig. 3a and b; Supplemental Fig. S8b), while the surface Vpreb1 abundance is normal (not shown).
Ubiquitination is essential for BCR signalling, particularly for the signalling molecules downstream of the BCR complex, e.g., NF-kB.44 Degradative ubiquitination of pre-BCR surrogate light chains mediated by the E3 ligase Synoviolin 1 (SYVN1, also known as HRD1) promotes the transition of large pre-BII to small pre-BII.45 In mature B, antigen binding to BCR triggers endocytosis of the BCR complex and sorting into the MHC class II antigen-presenting compartment, which is dependent on ubiquitinated CD79β.45 As a putative adaptor for multiple E3 ligases and p97, UBXN3B might regulate the pre-BCR complex via ubiquitination. Indeed, our recent work showed that UBXN3B interacts with an E3 ligase TRIM56 via its coiled-coil domain to mediate non-degradative K63-linked ubiquitination of STING.1 Alternatively, UBXN3B may play a ubiquitination-independent role in the pre-BCR complex. For example, the ER-associated pool of UBXN3B senses cholesterol and interacts with the mechanistic target of rapamycin complex 1 (mTORC1) to drive non-alcoholic fatty liver disease to hepatocellular carcinoma progression.46
Notably, the whole genome transcriptome analyses have revealed that the primary downregulated pathway is the cell cycle specifically in SLChi/in precursor and mature B cells, but not in the non-B lineage precursors/progenitors (Figs. 3e and 4a, Supplemental Tables S5 and S6). This may be owed in part to deficient pre-BCR signalling, which is essential for cell proliferation and survival.24 However, tonic mature BCR signalling is normal (Supplemental Figs. S8 and S9), suggesting that UBXN3B employs multiple mechanisms to maintain normal cell cycle and survival in mature B cells. Interestingly, >40% of the downregulated cell cycle genes are also direct or indirect TP53-DREAM targets (Supplemental Tables S2, S3 and S6), which suppresses more than 250 cell cycle-associated genes.36 Consistent with the hyperactivation of TP53 signalling, increased apoptosis leads to a striking reduction in the half-life of peripheral B cells when UBXN3B is deleted (Fig. 4b–e). These results imply that UBXN3B might regulate cell cycle directly or via TP53-DREAM. Furthermore, Ubxn3b-deficient B cells have higher pro and activated caspase-3 levels (Fig. 4g and h), suggesting that UBXN3B may alternatively target apoptotic pathways. In favour of this, a recent study found that together with p97 and endoplasmic reticulum (ER)-localised E3 ubiquitin ligases, UBXN3B inhibits apoptosis by degrading proapoptotic factors, phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1, also known as NOXA) and BCL2 interacting killer (BIK) in HeLa cells (cervical carcinoma).47 Our results further imply that UBXN3B keeps the caspase-3 protein level in check as B cells are more sensitive to death than the other immune cells (Fig. 4d), evidenced by significantly lower caspase-3 expression in B than non-B cells (Fig. 4g). Consistent with this concept, loss of apoptosis or caspase-3 leads to expansion of the B cell compartment and autoimmunity.48,49 In addition to eliciting apoptosis, caspase-3 also negatively regulates cell cycle by cleaving several cell cycle regulators, leading to cell cycle arrest, DNA damage response, and activation of TP53 signalling.50 Future studies intend to identify the putative binding partners of UBXN3B in B-cells.
The role of UBXN3B in the immunopathogenesis of respiratory viruses
Mechanistically, UBXN3B may control RNA virus infection by regulating STING signalling and the type I IFN response.1,22 However, both STING and UBXN3B are dispensable for the innate immune response to many RNA viruses including Chikungunya,22,51 West Nile viruses,52 and SARS-CoV-253 (Supplemental Fig. S13). These results suggest that the primary function of UBXN3B during RNA virus infection is independent of STING and that dysregulated haematopoiesis may be the main driver of prolonged immunopathology in Ubxn3b−/− mice. In particular, the B-cell deficiency leads to a striking reduction in virus-specific IgM/IgGs (Fig. 5e, Supplemental Fig. S15f and g) that are essential for early control and clearance of numerous viruses including influenza54,55 and SARS-CoV-2.56,57 Indeed, IAV infection persisted much longer in the lung of B cell specific Ubxn3b knockout mice (Fig. 5b). The same was true during SARS-CoV-2 infection whereby Ubxn3b-deficient mice have higher viral load at 3 days p.i., however, the virus is eventually cleared by 10 days p.i. (Supplemental Fig. S12e). This discrepancy is due to the SARS-CoV-2 isolate employed for mouse infection. The clinical strain USA/WA1/2020 is unable to infect mice because of the lack of its cognate receptor, human ACE2. To overcome this, mice need to be sensitised with a viral vector carrying human ACE2. However, Ad5-vectored human ACE2 expression in the lung is transient, which peaks at 5 days but disappearing by 9 days after intranasal delivery (equivalent to 4 days after SARS-CoV-2 challenge).39 Thus, clinical SARS-CoV-2 is unable to sustain a prolonged infection in the lung. Nonetheless, the lung damage lasts beyond 10 days in the global Ubxn3b−/− mice (Supplemental Fig. S12c and d), suggesting that virus-elicited immunopathology is the main driver of lung damage. Consistent with our results, both SARS-CoV-2 and IAV elicit immunopathology in the lung, including massive immune infiltrates and elevated levels of systemic pro-inflammatory mediators.58 In particular, COVID-19 fatality is strongly associated with neutrophilia and lymphopenia,37 which is partly recapitulated in Ubxn3b−/− mice. This hyperinflammatory phase of COVID-19 generally occurs after the peak of virus replication when only a few infectious particles remain.52 Again, Ubxn3b−/− mice had sustained immunopathology (e.g., lung haemosiderosis) whilst viral load returned to baseline by 10 days p.i. (Supplemental Fig. S12). At this post clearance stage, the high N/L ratio in Ubxn3b−/− mice may sustain inflammation, which is also the most reliable biomarkers of chronic inflammatory conditions, such as type II diabetes,59 cardiovascular disease,60 and ageing61 etc. These conditions are predictive of severe COVID-19 morbidity and risk factors for mortality.37
In summary, using two murine models, we have rigorously demonstrated the essential role of UBXN3B in B lymphopoiesis during steady state and viral infections. Mechanistically, UBXN3B is crucial for pre-BCR signalling and B cell survival. However, we recognise a few limitations of this study. First, the in-depth molecular mechanism of UBXN3B action remains unclear; significant future efforts are required to pinpoint the exact target in the pre-BCR pathway and how UBXN3B controls caspase-3 protein turnover. Second, even though the UBXN3B protein sequence is highly conserved in mammals, its function in human B lymphopoiesis needs validating. Third, we employed a clinical SARS-CoV-2 strain and transiently human ACE2-sensitised global UBXN3B knockout mice, which could not address the specific, long-term impact of B cell defect on SARS-CoV-2 replication and pathogenesis. Fourth, the increase in myeloid populations, though moderate, when UBXN3B is depleted globally in mice, may lead to chronic inflammation. This should be considered a potential risk if UBXN3B is explored as a B-cell depleting therapeutic target.
Contributors
T.G designed and performed most of the experimental procedures and data analyses. D.Y. helped T.G. with most of the experimental procedures. T.L. and A.G.H. contributed to some of the experimental procedures. B.W. contributed to flow cytometry analysis. Z.C. and Z.F. performed the bulk RNA-seq analysis. B.T. and L.H. helped to acquire the influenza data. K.W provided guidance to bone marrow transplantation experiments. Y.W., L.Y., G.C., L.H., J.P., A.T.V., R.A.F. and E.F. contributed to discussion, data interpretations and/or helped improve writing. P.W. conceived and oversaw the study. T.G. and P.W. wrote the paper. All the authors read and approved the final version of the manuscript. T.G. and P.W. have accessed and verified the underlying data.
Data sharing statement
All data generated or analysed during this study are included in this published article (and its supplemental information files). The raw RNA-seq data are publicly available at https://www.ncbi.nlm.nih.gov/geo/, GEO # (GSE264055 & GSE266190). All unique materials used are readily available from the authors. However, the availability of live animals may change over time.
Declaration of interests
No financial or non-financial interest to disclose.
Acknowledgements
This project was funded in part by National Institutes of Health grants R01AI132526 and R21AI155820 to P.W., R01CA262430 to K.W., and R01AI113040 to J. P. P., and UConn Health Startup Fund and UConn COVID-RSF to P.W. The authors thank Dr. Evan Jellison, Director of the Flow Cytometry Facility for his tremendous technical support.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105248.
Appendix A. Supplementary data
References
- 1.Yang L., Wang L., Ketkar H., et al. UBXN3B positively regulates STING-mediated antiviral immune responses. Nat Commun. 2018;9(1):2329. doi: 10.1038/s41467-018-04759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boettcher S., Manz M.G. Regulation of inflammation- and infection-driven Hematopoiesis. Trends Immunol. 2017;38(5):345–357. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Dancey J.T., Deubelbeiss K.A., Harker L.A., Finch C.A. Neutrophil kinetics in man. J Clin Invest. 1976;58(3):705–715. doi: 10.1172/JCI108517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieger M.A., Schroeder T. Hematopoiesis. Cold Spring Harb Perspect Biol. 2012;4(12) doi: 10.1101/cshperspect.a008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busch K., Klapproth K., Barile M., et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518(7540):542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Fraticelli A.E., Wolock S.L., Weinreb C.S., et al. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553(7687):212–216. doi: 10.1038/nature25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melchers F. Checkpoints that control B cell development. J Clin Invest. 2015;125(6):2203–2210. doi: 10.1172/JCI78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuberth C., Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65(15):2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo H., Rabouille C., Newman R., et al. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388(6637):75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- 10.Alexandru G., Graumann J., Smith G.T., Kolawa N.J., Fang R., Deshaies R.J. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134(5):804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P., Yang L., Cheng G., et al. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013;3(4):1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J.H., Park M.E., Nikapitiya C., et al. FAS-associated factor-1 positively regulates type I interferon response to RNA virus infection by targeting NLRX1. PLoS Pathog. 2017;13(5) doi: 10.1371/journal.ppat.1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai T., Wu L., Wang S., et al. FAF1 regulates antiviral immunity by inhibiting MAVS but is antagonized by phosphorylation upon viral infection. Cell Host Microbe. 2018;24(6):776–790.e5. doi: 10.1016/j.chom.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y.B., Tan B., Mu R., et al. Ubiquitin-associated domain-containing UBX protein UBXN1 is a negative regulator of NF-kappaB signaling. J Biol Chem. 2015;290(16):10395–10405. doi: 10.1074/jbc.M114.631689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y., O'Boyle K., Auer J., et al. Multiple UBXN family members inhibit retrovirus and lentivirus production and canonical NFkappaBeta signaling by stabilizing IkappaBalpha. PLoS Pathog. 2017;13(2) doi: 10.1371/journal.ppat.1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai N., Suzuki M., Hayashi K., et al. Hepatocyte-specific depletion of UBXD8 induces periportal steatosis in mice fed a high-fat diet. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P., Arjona A., Zhang Y., et al. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat Immunol. 2010;11(10):912–919. doi: 10.1038/ni.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin T., Geng T., Harrison A.G., et al. CXCL10 signaling contributes to the pathogenesis of arthritogenic alphaviruses. Viruses. 2020;12(11):1252. doi: 10.3390/v12111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagger F.O., Sasivarevic D., Sohi S.H., et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–D924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L., Geng T., Yang G., et al. Macrophage scavenger receptor 1 controls Chikungunya virus infection through autophagy in mice. Commun Biol. 2020;3(1):556. doi: 10.1038/s42003-020-01285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng T., Lin T., Yang D., et al. A critical role for STING signaling in limiting pathogenesis of Chikungunya virus. J Infect Dis. 2020;223(12):2186–2196. doi: 10.1093/infdis/jiaa694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choukrallah M.A., Matthias P. The interplay between chromatin and transcription factor networks during B cell development: who pulls the trigger first? Front Immunol. 2014;5:156. doi: 10.3389/fimmu.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martensson I.L., Almqvist N., Grimsholm O., Bernardi A.I. The pre-B cell receptor checkpoint. FEBS Lett. 2010;584(12):2572–2579. doi: 10.1016/j.febslet.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 25.Dul J.L., Argon Y., Winkler T., ten Boekel E., Melchers F., Martensson I.L. The murine VpreB1 and VpreB2 genes both encode a protein of the surrogate light chain and are co-expressed during B cell development. Eur J Immunol. 1996;26(4):906–913. doi: 10.1002/eji.1830260428. [DOI] [PubMed] [Google Scholar]
- 26.Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 27.Guloglu F.B., Roman C.A. Precursor B cell receptor signaling activity can be uncoupled from surface expression. J Immunol. 2006;176(11):6862–6872. doi: 10.4049/jimmunol.176.11.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzog S., Hug E., Meixlsperger S., et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9(6):623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi K., Melchers F. The nonimmunoglobulin portion of lambda5 mediates cell-autonomous pre-B cell receptor signaling. Nat Immunol. 2003;4(9):849–856. doi: 10.1038/ni959. [DOI] [PubMed] [Google Scholar]
- 30.Ubelhart R., Bach M.P., Eschbach C., Wossning T., Reth M., Jumaa H. N-linked glycosylation selectively regulates autonomous precursor BCR function. Nat Immunol. 2010;11(8):759–765. doi: 10.1038/ni.1903. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda T., Sanjo H., Pages G., et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity. 2008;28(4):499–508. doi: 10.1016/j.immuni.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Chen D., Tang T.X., Deng H., Yang X.P., Tang Z.H. Interleukin-7 biology and its effects on immune cells: mediator of generation, differentiation, survival, and homeostasis. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.747324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peschon J.J., Morrissey P.J., Grabstein K.H., et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180(5):1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Freeden-Jeffry U., Vieira P., Lucian L.A., McNeil T., Burdach S.E., Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181(4):1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadhouders R., de Bruijn M.J., Rother M.B., et al. Pre-B cell receptor signaling induces immunoglobulin kappa locus accessibility by functional redistribution of enhancer-mediated chromatin interactions. PLoS Biol. 2014;12(2) doi: 10.1371/journal.pbio.1001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25(1):114–132. doi: 10.1038/cdd.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J., Zhuang Z., Zheng J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3):734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan A.O., Case J.B., Winkler E.S., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182(3):744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groarke E.M., Young N.S. Aging and hematopoiesis. Clin Geriatr Med. 2019;35(3):285–293. doi: 10.1016/j.cger.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu T., Mundt C., Licence S., Melchers F., Martensson I.L. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol. 2002;168(12):6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- 42.Monroe J.G. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6(4):283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 43.Fuentes-Panana E.M., Bannish G., Shah N., Monroe J.G. Basal Igalpha/Igbeta signals trigger the coordinated initiation of pre-B cell antigen receptor-dependent processes. J Immunol. 2004;173(2):1000–1011. doi: 10.4049/jimmunol.173.2.1000. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T., Sun J., Cheng J., et al. The role of ubiquitinase in B cell development and function. J Leukoc Biol. 2021;109(2):395–405. doi: 10.1002/JLB.1MR0720-185RR. [DOI] [PubMed] [Google Scholar]
- 45.Veselits M., Tanaka A., Chen Y., et al. Igbeta ubiquitination activates PI3K signals required for endosomal sorting. J Exp Med. 2017;214(12):3775–3790. doi: 10.1084/jem.20161868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F., Tian T., Zhang Z., et al. Publisher Correction: long non-coding RNA SNHG6 couples cholesterol sensing with mTORC1 activation in hepatocellular carcinoma. Nat Metab. 2022;4(9):1215. doi: 10.1038/s42255-022-00650-5. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J., Cao Y., Yang J., Jiang H. UBXD8 mediates mitochondria-associated degradation to restrain apoptosis and mitophagy. EMBO Rep. 2022;23(10) doi: 10.15252/embr.202254859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo M., Hakem R., Furlonger C., et al. Caspase-3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol. 2003;4(10):1016–1022. doi: 10.1038/ni976. [DOI] [PubMed] [Google Scholar]
- 49.Wright J.A., Bazile C., Clark E.S., et al. Impaired B cell apoptosis results in autoimmunity that is alleviated by ablation of Btk. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.705307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Connolly P., Garcia-Carpio I., Villunger A. Cell-cycle cross talk with caspases and their substrates. Cold Spring Harb Perspect Biol. 2020;12(6) doi: 10.1101/cshperspect.a036475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng T., Yang D., Lin T., Cahoon J.G., Wang P. UBXN3B controls immunopathogenesis of arthritogenic alphaviruses by maintaining hematopoietic homeostasis. mBio. 2022;13(6) doi: 10.1128/mbio.02687-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He X., Lau E.H.Y., Wu P., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 53.Liu J., Liu Y., Xiang P., et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med. 2020;18(1):206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopf M., Brombacher F., Bachmann M.F. Role of IgM antibodies versus B cells in influenza virus-specific immunity. Eur J Immunol. 2002;32(8):2229–2236. doi: 10.1002/1521-4141(200208)32:8<2229::AID-IMMU2229>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 55.Skountzou I., Satyabhama L., Stavropoulou A., et al. Influenza virus-specific neutralizing IgM antibodies persist for a lifetime. Clin Vaccine Immunol. 2014;21(11):1481–1489. doi: 10.1128/CVI.00374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gasser R., Cloutier M., Prevost J., et al. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34(9) doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klingler J., Weiss S., Itri V., et al. Role of immunoglobulin M and A antibodies in the neutralization of severe acute respiratory syndrome coronavirus 2. J Infect Dis. 2021;223(6):957–970. doi: 10.1093/infdis/jiaa784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai Y., Tao X. Comparison of COVID-19 and influenza characteristics. J Zhejiang Univ Sci B. 2021;22(2):87–98. doi: 10.1631/jzus.B2000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo X., Zhang S., Zhang Q., et al. Neutrophil: lymphocyte ratio is positively related to type 2 diabetes in a large-scale adult population: a Tianjin chronic low-grade systemic inflammation and health cohort study. Eur J Endocrinol. 2015;173(2):217–225. doi: 10.1530/EJE-15-0176. [DOI] [PubMed] [Google Scholar]
- 60.Angkananard T., Anothaisintawee T., McEvoy M., Attia J., Thakkinstian A. Neutrophil lymphocyte ratio and cardiovascular disease risk: a systematic review and meta-analysis. Biomed Res Int. 2018;2018 doi: 10.1155/2018/2703518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Chen Q., Luo X., et al. Neutrophil-to-Lymphocyte ratio positively correlates to age in healthy population. J Clin Lab Anal. 2015;29(6):437–443. doi: 10.1002/jcla.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.