Abstract
The activation of gene expression by the human cytomegalovirus (HCMV) particle was investigated. The HCMV major immediate-early (IE) promoter was cloned upstream of the Escherichia coli lacZ coding sequences, and the resulting cassette was introduced into the genome of a herpes simplex virus type 1 (HSV-1) mutant lacking functional VP16. Upon infection with the HSV-1 recombinant in the presence of cycloheximide, to block de novo protein synthesis, expression of lacZ-specific transcripts was increased by fivefold when HCMV was included in the inoculum. Accumulation of HSV-1 IE RNAs was also stimulated by coinfection with HCMV, as was expression of the adenovirus 5 VAI transcript when the VAI gene was cloned into the HSV-1 genome. Coinfection with HCMV did not alter mRNA stability or uncoating of the HSV-1 genome. The coding sequences for the HCMV phosphoprotein pp71, controlled by the HCMV IE promoter, were cloned into an HSV-1 recombinant impaired for the production of the three major transactivators (VP16, ICP0, and ICP4) to yield a recombinant (in1324) which expressed pp71 but did not cause significant cytotoxicity. Infection with in1324 resulted in stimulation of HCMV IE, HSV-1 IE, and VAI expression, demonstrating that pp71 is responsible for the effects we observed when using the entire HCMV particle. Therefore, HCMV pp71 exhibits novel properties in its ability to stimulate gene expression from a range of promoters present in a herpesvirus genome.
The program of herpesvirus gene expression in infected cells is controlled at multiple levels, one of which is the activation of immediate-early (IE) transcription by structural proteins of the incoming virion. In the case of herpes simplex virus type 1 (HSV-1), the tegument protein VP16 activates IE transcription by forming a complex with the cellular proteins Oct-1 and HCF at the sequence TAATGARAT (R is a purine nucleotide) (reviewed in reference 26). Similarly, components of the human cytomegalovirus (HCMV) particle stimulate HCMV IE gene expression, but in this case, the proteins responsible and their modes of action are less clear than for HSV-1. The promoter controlling expression of the HCMV major IE locus is very complex, containing an enhancer region with reiterated binding sites for a variety of cellular transcription factors (5, 11, 15, 39). As a result of this complexity, the HCMV IE promoter, and hence viral IE gene expression, is sensitive to modulation by many cellular regulatory pathways. The major IE locus encodes a family of proteins which have positive and negative effects on transcription and combine to regulate the expression of the viral genome.
The HCMV tegument phosphoprotein pp71 (encoded by gene UL82) is the most obvious candidate for a functional counterpart of VP16. In cotransfection assays, this protein activates expression from the HCMV IE promoter, and deletion analysis suggested that the target sequences were the ATF or AP-1 recognition sites present in the repeated 19-bp units of the promoter (16). Cotransfection with plasmids encoding pp71 also enhances the infectivity of HCMV DNA, although this effect may not be mediated through a direct effect on the major IE promoter, since a plasmid encoding pp71 gave a greater stimulation of infectivity than a plasmid encoding the entire IE locus (2). Another tegument protein, encoded by gene UL69, stimulates IE promoter activity in cotransfection assays, especially in concert with pp71 (43), but surprisingly the expression of UL69 antagonizes the enhancement of infectivity by a pp71-encoding plasmid (2). Components of the virion influence cellular signal transduction pathways, and since the HCMV major IE promoter is responsive to manipulations which operate through such pathways, these are additional potential mechanisms for activating IE transcription (4, 11, 13, 35, 40). The cell transcription factors NF-κB and Sp1 are activated by glycoproteins gB and gH of the infecting HCMV particle, thereby altering the expression of a number of cellular genes and possibly that of the IE genes by virtue of the NF-κB sites in the 18-bp repeat units in the major IE promoter (35, 39, 44). The virus also specifies a family of genes with homology to G-coupled receptors, which mediate signal transduction and consequent cellular gene activation (3, 6, 9), and one member (UL33) is known to be a virion component (18). Finally, HCMV particles contain protein kinases and phosphatases which may affect cellular or viral gene expression by altering the phosphorylation of transcription factors, either directly or through effects on other kinases which mediate signal transduction (21). Virion components activate transcription of interferon-responsive genes, although the proteins involved and their relevance to viral IE gene expression are unclear at present (23, 45).
To date, the activation of HCMV IE or cell promoters by virion components has been investigated in assays based on the stimulation of expression from plasmid templates or genes resident in the cellular genome (16, 41). It is not possible at present to determine the responses of promoters within the HCMV genome, because viral mutants defective for virion components are not available and because genetic manipulation of HCMV remains a formidable task. We demonstrated previously that the stimulatory effect of the HCMV particle could be reproduced by coinfection of cells with HCMV and an HSV-1 recombinant, derived from the VP16 mutant in1814, containing the HCMV IE promoter controlling Escherichia coli lacZ (33). Analysis of RNA produced after infection with the HSV-1 recombinant in the presence of cycloheximide revealed a fivefold increase in lacZ-specific transcript levels when HCMV was added, due to activation of the HCMV IE promoter by components of the HCMV virion. This system provides a means of studying the sequence specificity of the effect of HCMV virion proteins at the level of RNA accumulation and with the target promoters in the HSV-1 genome, an environment that may more closely resemble the HCMV genome than do transfected plasmid DNA or transformed cells. We describe here the responses of promoters located in the HSV-1 genome to activation by components of the HCMV particle. In addition, we have investigated the contribution of pp71 to gene activation by the construction of an HSV-1-derived recombinant, in1324, which expresses the protein efficiently in human fibroblasts, a cell type that is permissive for HCMV replication. The recombinant is derived from the HSV-1 mutant in1312, which contains the VP16 mutation from in1814, a deletion of the essential RING domain of ICP0, and the tight temperature-sensitive mutation from tsK which inactivates ICP4 function at the nonpermissive temperature (34). Mutant in1312 therefore fails to produce the major HSV-1-specified activators of gene expression. After infection of cells at 38°C, mutants lacking these three functions do not induce detectable cytopathology at effective multiplicities of up to 5 PFU per cell yet express functional levels of foreign gene products for at least 2 days when the HCMV IE promoter is used to direct transcription (30, 31, 38). We have used in1324 to investigate the properties of pp71 in isolation from other HCMV virion proteins.
MATERIALS AND METHODS
Plasmids.
HindIII fragments of HCMV (strain AD169), cloned into pAT153, were provided by J. Macnab. The HindIII-BamHI fragment of HindIII 1, containing most of the pp71 coding sequences (36), was cloned between the HindIII and BamHI sites of pUC18, to give pCP8327. The HCMV HindIII c fragment was inserted into the HindIII site of pCP8327 to yield plasmid pCP8401, in which the pp71 coding sequences are contiguous. The XbaI site at the 3′ end of the pp71 coding region was changed to an SstI site by insertion of an oligonucleotide, retaining the termination codon. The pp71 open reading frame was cloned as a 1,711-bp FspI-SstI fragment between the SmaI and SstI sites of pJ7Ω (22), giving plasmid pCP8671. The shuttle vector pCP1802 was constructed by replacing the lacZ coding sequences of pMJ101 (14) with a polylinker containing a unique HpaI site which could be used to place open reading frames between the HCMV IE promoter and simian virus 40 polyadenylation signals, all embedded in HSV-1 thymidine kinase (TK) coding sequences to facilitate recombination into the viral genome. The pp71 open reading frame was excised from pCP8671 as a BamHI-SstI fragment, treated with Klenow fragment, and cloned into the unique HpaI site of pCP1802, giving plasmid pCP43937. The green fluorescent protein (GFP) coding protein sequences from plasmid pGFPemd (Packard Instrument Company) were cloned into the HpaI site of pCP1802, giving pAR29, in which expression of GFP is controlled by the HCMV IE promoter.
Cells and viruses.
HSV-1 recombinants were propagated and titrated on BHK-21 (clone 13) cells, with 3 mM hexamethylene bisacetamide present to complement the VP16 mutation derived from in1814 (1, 19). HCMV (strain AD169) was propagated and titrated on human fetal lung (HFL) fibroblasts. For irradiation with UV, the HCMV preparation was diluted 10-fold in Eagle medium lacking serum and phenol red and UV irradiated as described previously (25), with an HSV-1 preparation treated in parallel to monitor the efficacy of irradiation. The titer of HCMV was reduced to undetectable levels, and HSV-1 was reduced from to 2.5 × 109 PFU/ml to <5 × 102 PFU/ml. The HSV-1 recombinants used were derived from mutant in1814 and contained insertions consisting of the E. coli lacZ gene controlled by various herpesvirus promoters or of the adenovirus 5 (Ad5) VAI gene at the TK or UL41 locus. Although known additional mutations were present in the parents of some of the recombinants, these are not relevant to experiments reported here, in which de novo protein synthesis was inhibited by the addition of 50 μg of cycloheximide per ml from the time of infection. The promoter sequences controlling lacZ in the recombinants derived from in1814 are listed in Table 1. Recombinants in1853, in1820K, in1312, in1332, in1382, in1383, and in1389 have been described previously (7, 31, 32, 34). Recombinants in1341 and in1342 were derived from in1820K and contained the HSV-1 ICP0 promoter from an SstI site at −808 to a BbvI site at +48, upstream of lacZ, with deletions from the EagI site at −687 to the BstXI site at −417 (in1341), or from the BstXI site at −417 to the SmaI site at −126 (in1342). Mutant in1373 was derived from in1312 and possessed a cassette of the HCMV major IE promoter (a Sau3AI fragment from −750 to +7) controlling lacZ, inserted between BamHI and BstXI sites in the UL41 coding sequences, thereby deleting 935 bp of UL41. Mutant in1375 was derived from in1373 by insertion of the Ad5 VAI gene (an XhoI-NheI fragment representing nucleotides −326 to +189 with respect to the start site of the 155-nucleotide RNA) cloned into the SstI site in the HSV-1 TK gene. Mutant in1324 was constructed by recombination of ScaI-cleaved pCP43937 with in1312 DNA and selection for TK-deficient viruses, as described previously (31), and in1325 was prepared by cotransfection of in1324 DNA with ScaI-cleaved pAR29 and subsequent selection of fluorescent plaques. Mutant in1329 was constructed by recombination of ScaI-cleaved pTM8 (in which lacZ is controlled by the ICP4 promoter and upstream sequences) (17) with in1312 DNA and selection of lacZ-expressing plaques. The VP16 mutation of in1312 was rescued by cotransfection with pMC1 (1) to give in1330. The structures of recombinant viruses were confirmed by Southern hybridization.
TABLE 1.
Promoter sequences cloned into HSV-1 recombinants
| Mutant | Type of inserted sequencea |
|---|---|
| in1853, in1382, and in1332 | HCMV IE (−750 to +7)-lacZ (TK) |
| in1335 and in1329 | HSV-1 ICP4 (−331 to +29)-lacZ (TK) |
| in1383 | HSV-1 ICP0 (−808 to +48)-lacZ (TK) |
| in1341 | HSV-1 ICP0 (−808 to −687 and −417 to +48)-lacZ (TK) |
| in1342 | HSV-1 ICP0 (−808 to −417 and −126 to +48)-lacZ (TK) |
| in1389 | HSV-1 ICP0 (−106 to +48)-lacZ (TK) |
| in1373 | HCMV IE (−750 to +7)-lacZ (UL41) |
| in1375 | HCMV IE (−750 to +7)-lacZ (UL41), VAI (−326 to +189) (TK) |
| in1324 | HCMV IE (−750 to +7)-pp71 (TK) |
| in1325 | HCMV IE (−750 to +7)-GFP (TK) |
The site of insertion is in parentheses.
Analysis of RNA accumulation.
Monolayers of 107 HFL cells were infected with HSV-1 recombinants at a multiplicity of 5 PFU per cell (or the equivalent amount of in1373, in1375, in1389, in1324, in1325, and in1330, which give lower titers due to the absence of functional ICP0), with 50 μg of cycloheximide per ml present throughout. Where present, HCMV (AD169) was added at 0.3 PFU per cell. After incubation at 38.5°C for 1 h, culture medium containing 50 μg of cycloheximide per ml was added, and incubation was continued at 38.5°C for 5 h. Cells were harvested, polyadenylated RNA was extracted and analyzed on RNA blots, and hybridization with probes specific for γ-actin, lacZ, and the HSV-1 IE mRNAs was carried out, as described previously (1, 24). For analysis of VAI production, total cytoplasmic RNA was prepared from Ad5-infected HFL cells as described by Preston (29). The VAI probe was the 515-bp XhoI-NheI fragment described above. A probe specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Ambion. Radioactive signals in individual bands were quantified by use of a PhosphorImager (Molecular Dynamics).
Analysis of uncoating of viral DNA.
Monolayers of 2 × 106 HFL cells were infected with 0.5 PFU of in1332 per cell, with or without 1 PFU of HCMV per cell, in the presence of 50 μg of cycloheximide per ml, and maintained at 38.5°C for 12 h. Nuclei were incubated, with or without 5 U of DNase I, at 37°C for 20 min, under conditions described previously (14). DNA was extracted, cleaved with BamHI, electrophoresed, blotted, and hybridized with a probe specific for the Moloney murine leukemia virus (Mo-MuLV) long terminal repeat insert in the HSV-1 long repeat, as described previously (14).
Expression of pp71.
Protein blots to detect pp71 were carried out with polyclonal antibody BgL2 (kindly supplied by G. Hensel), as described by Hensel et al. (12).
β-Galactosidase assays.
Cell extracts were prepared and β-galactosidase activities were determined by a fluorometric assay, as described by Preston and Nicholl (33).
Cytotoxicity assay.
Monolayers of Vero cells were infected with in1324 or in1325, and the efficiency of colony formation after trypsinization and dilution was determined as described previously (31).
RESULTS
Activation of gene expression by HCMV particles.
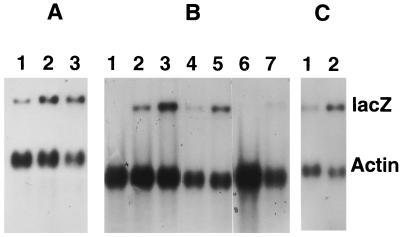
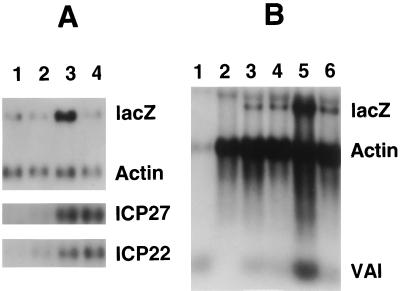
In the protocol used for many of the experiments reported here, monolayers of HFL cells were infected with HSV-1 mutants lacking functional VP16, with or without HCMV, in the presence of cycloheximide to block protein synthesis. At 6 h postinfection, polyadenylated RNA was extracted and analyzed by gel electrophoresis, blotting, and hybridization. Previously, we used this approach to demonstrate that the HCMV particle contains proteins that stimulate expression from the HCMV major IE promoter, when placed upstream of lacZ coding sequences, by approximately fivefold (33), and initial experiments were carried out to confirm and extend these findings. To ensure that the effects were due to HCMV virion components rather than a low level of IE protein synthesis, even in the presence of cycloheximide, HCMV was UV irradiated prior to coinfection with in1332 (Fig. 1A). Expression of lacZ-specific RNA, controlled by the HCMV IE promoter, was observed in the absence of coinfecting virus (lane 1) and was stimulated to the same extent (fivefold, relative to the level of actin-specific RNA) by irradiated (lane 2) and unirradiated (lane 3) virus, demonstrating that the components exerting the effect were not affected by heavy irradiation of the HCMV preparation. This result, together with the observation that increasing the concentration of cycloheximide from 50 μg/ml to 100 μg/ml did not affect the level of stimulation (results not shown), demonstrates that the effect was due to virion components. Our previous studies also showed that expression from the HSV-1 ICP0 IE promoter was also increased by two- to threefold upon coinfection with HCMV in the presence of cycloheximide (33). To investigate the nature of the sequences in this promoter (initially defined as an 856-bp fragment from −808 to +48), HSV-1 mutants containing deleted versions of the promoter upstream of lacZ were tested for responsiveness to HCMV (Fig. 1B). lacZ-specific RNA accumulation was increased by two- to threefold when mutants in1341 (lacking −687 to −417) and in1342 (lacking −417 to −126) were tested (lanes 2 to 5), and even the minimal promoter (from −106 to +48) present in in1389 was activated by coinfection with HCMV (lanes 6 and 7). Therefore, no specific region mediating the response to HCMV was identified in the ICP0 promoter. The properties of another HSV-1 IE promoter, that controlling ICP4, were also investigated (Fig. 1C). Coinfection with HCMV resulted in a fivefold stimulation of lacZ-specific RNA production, demonstrating that this promoter also responds to components of the HCMV virion.
FIG. 1.
Effects of HCMV on promoter activities. HFL cell monolayers were infected in the presence of cycloheximide and incubated at 38.5°C for 6 h. Polyadenylated RNA was hybridized with probes specific for lacZ and actin. (A) Cells were infected with in1332 alone (lane 1), with in1332 plus UV-irradiated HCMV (lane 2), or with in1332 plus unirradiated HCMV (lane 3). (B) Cells were mock infected (lane 1) or infected with in1341 (lanes 2 and 3), in1342 (lanes 4 and 5), or in1389 (lanes 6 and 7), either with no other virus (lanes 1, 2, 4, and 6) or with HCMV (lanes 3, 5 and 7). (C) Cells were infected with in1335 alone (lane 1) or in1335 plus HCMV (lane 2).
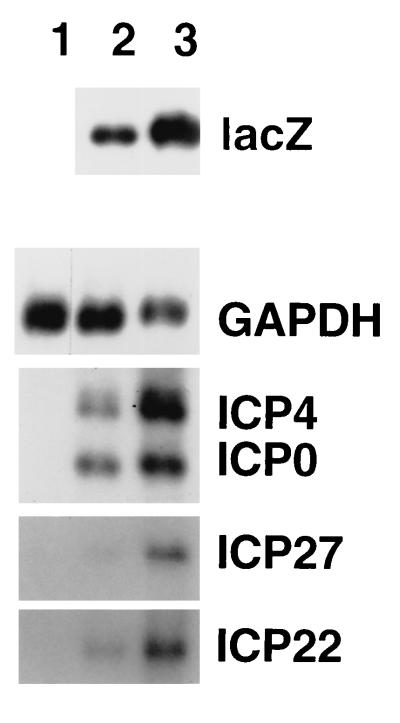
To investigate whether all HSV-1 promoters respond to HCMV, and to confirm that the effects shown in Fig. 1 were not due to lacZ sequences or flanking regions from the HSV-1 TK gene, HFL monolayers were infected with in1853 and coinfected with HCMV in the presence of cycloheximide, and HSV-1 IE RNA accumulation was measured by using gene-specific fragments as hybridization probes (Fig. 2). The amounts of the HSV-1 IE RNAs were increased by two- to threefold (ICP0 and ICP22) or four- to fivefold (ICP4 and ICP27). Therefore, the HSV-1 promoters in their natural locations are activated by components of the HCMV virion. As expected, lacZ-specific RNA (controlled by the HCMV IE promoter) increased by approximately sixfold.
FIG. 2.
Effects of HCMV on HSV-1 IE promoters. HFL cell monolayers were mock infected (lane 1), infected with in1853 alone (lane 2), or infected with in1853 plus HCMV (lane 3) in the presence of cycloheximide and incubated at 38.5°C for 6 h. Polyadenylated RNA was prepared and hybridized with probes specific for lacZ and GAPDH, ICP4 and ICP0, ICP27, and ICP22.
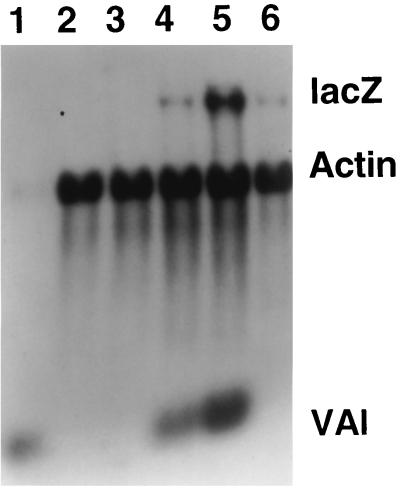
To extend the range of promoters analyzed by the coinfection approach, it would be desirable to clone cellular promoters into the HSV-1 genome and determine their responses to coinfection with HCMV. This approach is problematic, however, since most heterologous promoters are not expressed with an IE pattern of regulation in the context of the HSV-1 genome but are dependent upon IE protein synthesis (27). In searching for promoters that are active in the presence of cycloheximide, the adenovirus VAI gene, which is transcribed by RNA polymerase III, was investigated. HSV-1 mutant in1375 contains lacZ controlled by the HCMV IE promoter inserted into the nonessential UL41 gene (which encodes the virion host shutoff function), and VAI (transcribed sequences plus promoter and terminator elements) at the TK locus. Cells were infected with in1375 or its parent in1373, which contains the UL41 insertion but not VAI, and coinfected with HCMV in the presence of cycloheximide (Fig. 3). Synthesis of VAI was readily detected in cells infected with in1375 but not in1373 (lanes 4 and 6), and coinfection with HCMV increased the level of this transcript (lane 5) by two- to threefold (taking data from three experiments). The level of lacZ-specific RNA was increased by fivefold (lanes 4 and 5). This experiment shows that the effect of HCMV is not restricted to herpesvirus IE promoters or to genes transcribed by RNA polymerase II.
FIG. 3.
Effects of HCMV on VAI expression. HFL monolayers were mock infected (lanes 2 and 3), infected with in1375 (lanes 4 and 5), or infected with in1373 (lane 6), alone (lanes 2, 4 and 6) or with HCMV (lanes 3 and 5). After incubation at 38.5°C for 6 h in the presence of cycloheximide, cytoplasmic RNA was prepared. Cytoplasmic RNA from Ad5-infected cells (24 h postinfection) was analyzed in lane 1. Hybridization was with probes specific for lacZ, actin, and VAI.
Coinfection with HCMV does not affect mRNA stability or uncoating of HSV-1.
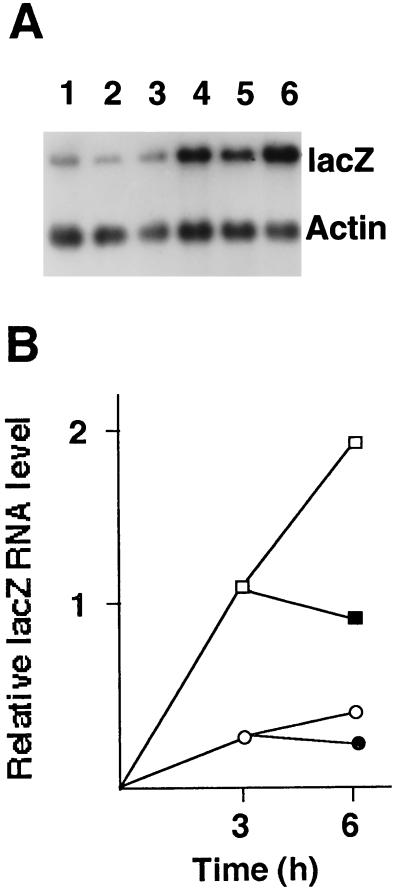
The effects shown in Fig. 1 to 3 could be due to mRNA stabilization by HCMV rather than a true increase in the rate of accumulation; thus, RNA stability was investigated (Fig. 4). Monolayers were infected with in1853, with or without HCMV, and incubated in the presence of cycloheximide for 3 h. At this time, cultures were treated with actinomycin D, to block transcription, and RNA was analyzed after a further 3 h. The level of lacZ-specific RNA, relative to that of actin mRNA, increased between 3 and 6 h postinfection regardless of the presence of HCMV and only decreased by approximately 20% over 3 h in the presence of actinomycin D in both sets of cultures. Therefore, lacZ mRNA was relatively stable over the time course of the experiments described here, demonstrating that mRNA stabilization cannot account for the increase mediated by coinfection with HCMV.
FIG. 4.
HCMV does not affect RNA stability. (A) HFL monolayers were infected with in1853 in the presence of cycloheximide, without (lanes 1 to 3) or with (lanes 4 to 6) HCMV. At 3 h postinfection, samples were taken for RNA preparation (lanes 1 and 4), and the remaining cultures were incubated for a further 3 h in the presence (lanes 2 and 5) or absence (lanes 3 and 6) of actinomycin D. RNA was hybridized with probes specific for lacZ and actin. (B) Amounts of lacZ-specific RNA, relative to actin mRNA levels, in cells infected with in1853 and HCMV (□), infected with HCMV and in1853 and actinomycin D treated (■), infected with in1853 alone (○), or infected with in1853 and actinomycin D treated (●).
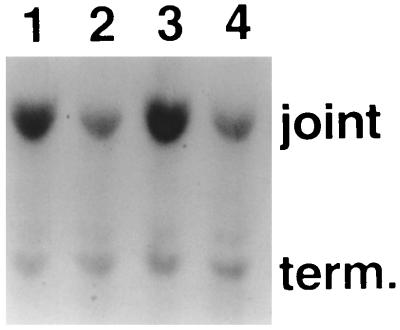
Two other possible explanations for the effects of HCMV were investigated. The incorporation of [3H]uridine into precursor pools and RNA was not affected by infection with HCMV under the experimental protocol reported here (results not shown), ruling out a general stimulation of cellular RNA synthesis. The uptake and uncoating of HSV-1 genomes were also investigated (Fig. 5). HFL monolayers were infected with in1332 in the presence of cycloheximide, with and without HCMV. At 12 h postinfection, nuclei were prepared and digested with DNase I or mock digested. DNA was extracted, cleaved with BamHI, and analyzed by Southern hybridization with a probe which detects a fragment from the long repeat and joint-spanning regions of the HSV-1 genome. Two parameters of uncoating were measured: the overall sensitivity to DNase I digestion was determined, and the ratio of joint to terminal fragment was calculated. It is known that HSV-1 DNA is converted to a circular form, which contains two joints but no termini and is DNase I sensitive, shortly after uncoating; thus, an increase in the ratio of joint to terminus would indicate a greater extent of uncoating irrespective of the absolute recovery of HSV-1 genomes (10, 14, 28). The results of repeated experiments showed, as illustrated in Fig. 5, that coinfection with HCMV did not alter the amount of in1332 DNA in the nucleus, the sensitivity to digestion with DNase I, or the ratio of hybridization to joint and terminal fragments. Therefore, the increase in gene expression upon coinfection with HCMV was not a consequence of greater uptake or uncoating of HSV-1 genomes.
FIG. 5.
HCMV does not affect uptake and uncoating of HSV-1 DNA. HFL monolayers were infected with in1332 without (lanes 1 and 2) or with (lanes 3 and 4) 1 PFU of HCMV per cell in the presence of cycloheximide. After incubation at 38.5°C for 12 h, nuclei were prepared and mock digested (lanes 1 and 3) or digested with DNase I (lanes 2 and 4). DNA was purified, digested with BamHI, and analyzed by hybridization with a probe specific for the Mo-MuLV sequences in the long repeat region. The joint and terminal (term.) fragments are labelled.
Activation of gene expression by HCMV pp71.
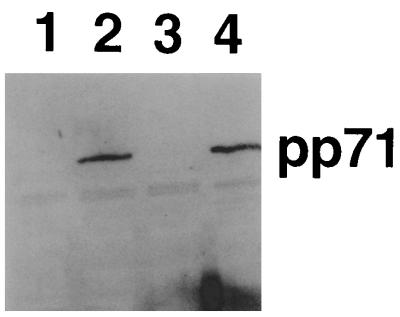
To investigate the contribution of pp71 to the increases in gene expression described above, the pp71 coding sequences were cloned into the TK locus of the HSV-1 multiple mutant in1312 to produce recombinant in1324. Infection with in1324 was expected to result in efficient expression of pp71 in HFL cell monolayers and thus enable the effects of this protein to be analyzed in the absence of other HCMV-specified products and without cytopathic changes to cells. Protein blots confirmed that pp71 was produced after infection of HFL cells with in1324 (Fig. 6). A control virus, in1325, was constructed from in1324 by replacement of the pp71 coding sequences with those of GFP to ensure that the observed effects were due to the presence of pp71 rather than other mutations that had arisen during the construction of in1324. From the results described above, if pp71 alone is functional, it might be expected that in1324 would synthesize greater amounts of ICP27, ICP22, ICP47, the nonfunctional ICP4, and truncated ICP0, and thus pp71 might act indirectly due to the larger amounts of these proteins. To provide a control for this possibility, the VP16 mutation of in1312 was rescued to give in1330, a virus in which IE gene expression of in1312 was elevated by VP16 in the absence of ICP0 or ICP4 function.
FIG. 6.
HSV-1 recombinant in1324 expresses pp71. HFL cell monolayers were mock infected (lane 1), infected with in1324 (lane 2), or infected with in1312 (lane 3), and cell extracts were prepared after incubation at 38.5°C for 6 h. An extract was also prepared from HFL cells 24 h after infection with 0.5 PFU of HCMV per cell (lane 4). Protein blots were probed with antibody BgL2.
Monolayers of HFL cells were infected with in1324, in1325, or in1330; maintained at 38.5°C for 3 h; and infected with in1382 for 5 h in the presence of cycloheximide. RNA was analyzed by hybridization to lacZ- and actin-specific probes and, after sequential stripping of the membrane, to an ICP27-specific probe and an ICP22-specific probe (Fig. 7A). The results show that infection with in1324 followed by a period of 3 h to permit protein synthesis resulted in an increase (approximately sixfold) in the accumulation of in1382-specified lacZ RNA (lane 3), whereas preinfection with in1325 (lane 2) or in1330 (lane 4) caused a small decrease compared with a sample from mock-preinfected cells (lane 1). ICP27- and ICP22-specific RNA levels were also increased by preinfection with in1324 compared with in1325, showing that pp71 specified by in1324 also acted on its promoters. Activation of HSV-1 IE gene expression by VP16 was similar to that achieved by pp71 (compare lanes 3 and 4), confirming that the activation of the HCMV IE promoter was due to the presence of pp71 synthesized by in1324 rather than as an indirect consequence of raised levels of IE proteins. The effects of pp71 on VAI synthesis were also tested (Fig. 7B). The experimental protocol used for the experiment shown in Fig. 7A was followed, except that the second virus added was in1375 instead of in1382. An RNA blot probed for lacZ, actin, and VAI sequences confirmed the activation of the HCMV IE promoter, located in the UL41 locus rather than TK, and also demonstrated that preinfection with in1324, but not in1325 or in1330, resulted in an increase in VAI accumulation (fourfold, from three determinations).
FIG. 7.
Stimulation of gene expression by in1324. (A) HFL cell monolayers were mock preinfected (lane 1); preinfected with in1325 (lane 2), in1324 (lane 3), or in1330 (lane 4); and incubated at 38.5°C for 3 h. Monolayers were then infected with in1382 in the presence of cycloheximide and incubated at 38.5°C for a further 5 h, and polyadenylated RNA was prepared and analyzed. The blot was hybridized to probes specific for lacZ and actin; followed, after stripping, to ICP27; and, after further stripping, to ICP22. (B) HFL cell monolayers were mock preinfected (lane 3) or preinfected with in1325 (lane 4), in1324 (lane 5), or in1330 (lane 6) and incubated at 38.5°C for 3 h. Monolayers were then infected with in1375 in the presence of cycloheximide and incubated for a further 5 h at 38.5°C. Total cytoplasmic RNA was prepared and analyzed by hybridization to probes specific for lacZ, actin, and VAI. Cytoplasmic RNA from Ad5-infected cells (lane 1) and mock-infected cells (lane 2) was also analyzed.
As a further test of the activity of pp71, monolayers were preinfected with in1324 or in1325, and after incubation at 38.5°C for 3 h, infected with in1382, in1383, or in1329 without cycloheximide and maintained at 38.5°C for 5 h. Mutants in1382, in1383, and in1329 are, like in1324 and in1325, devoid of VP16, ICP0, and ICP4 activity at 38.5°C, and thus it is possible to measure the effect of pp71 by carrying out β-galactosidase assays with infected cell extracts (Table 2). Preinfection with in1324 resulted in a four- to fivefold increase in expression from the three promoters tested (HCMV IE, HSV-1 ICP0, and ICP4), similar to the effects on RNA levels after infection with HCMV in the presence of cycloheximide (Fig. 1, 2, and 3).
TABLE 2.
Activation of promoters by preinfection with in1324a
| Preinfection | Second infection | β-Galactosidase activity (arbitrary fluorescence units)b |
|---|---|---|
| None | in1382 | 800 |
| in1325 | in1382 | 700 |
| in1324 | in1382 | 3,850 |
| None | in1383 | 64 |
| in1325 | in1383 | 84 |
| in1324 | in1383 | 245 |
| None | in1329 | 19 |
| in1325 | in1329 | 20 |
| in1324 | in1329 | 82 |
Monolayers of HFL cells were mock preinfected or preinfected with in1324 or in1325. After incubation at 38.5°C for 3 h, cells were infected with in1382, in1383, or in1329 and incubated at 38.5°C for a further 5 h. Cytoplasmic extracts were then assayed for β-galactosidase activity.
The values shown are the means of duplicate determinations which did not vary by more than 20%.
To investigate whether the expression of pp71 was toxic to the host cell, monolayers of Vero cells were infected with in1324 or in1325, with the same amounts of virus as in the experiments shown in Fig. 7 and Table 2. After incubation at 38.5°C for 16 h, monolayers were trypsinized, diluted, and plated at 38.5°C. Cells infected with in1324 or in1325 formed colonies as efficiently as mock-infected cells (140 ± 35 colonies on a 10−4 dilution of the original monolayer), demonstrating that expression of pp71 was not cytotoxic in this assay.
DISCUSSION
In the experiments reported here, we used HSV-1 mutants which encode nonfunctional VP16 as vehicles to investigate the responses of promoters to stimulation by HCMV virion components. This methodology has allowed us to analyze the effects of the entire HCMV particle on gene expression at the RNA level in the absence of protein synthesis, an approach that would be difficult by using transfection of plasmids as reporter constructs due to the low efficiency and variability of transfection in human fibroblasts, the only realistic tissue culture host cells for HCMV. There is currently no system in which the activation of promoters by virion components can be analyzed in the context of the HCMV genome, and the use of the HSV-1 genome provides a closer approximation to the natural situation than the analysis of promoters in transfected plasmids or stably transformed cell lines.
Although many components of the HCMV virion could contribute to promoter activation, the finding that preinfection with in1324 gives analogous responses, both in terms of magnitude and lack of specificity, indicates that pp71 is the virion constituent responsible for the activation of expression from promoters in the HSV-1 genome. It is possible, however, that other virion components may have similar effects or modulate the activity of pp71. The fact that removal of the pp71 open reading frame abolished the effectiveness of in1324 confirms that pp71 is the active component of this virus and that fortuitous mutations have not arisen during its construction. This finding demonstrates that pp71 can act alone and that interaction with other virion proteins is not required for its activity. It was not anticipated that raising the levels of ICP27, -22, and -47 would affect promoter activity, because there is no evidence that these proteins stimulate gene expression in the absence of functional ICP0 or ICP4, but this possibility was checked by construction and analysis of in1330. Preinfection with in1330 did not affect expression from the HCMV IE or VAI promoters, and the use of this virus demonstrated that production of pp71 by preinfection with in1324 was almost as effective as provision of VP16 by in1330 in activating the ICP27 and ICP22 promoters. Comparisons between the activities of the two proteins is complicated by the fact that VP16 is delivered as a single dose, whereas the amounts of pp71 are expected to rise through the early stages of infection, since the HCMV IE promoter controls the expression of pp71, possibly establishing a positive feedback system.
Our results emphasize that pp71 is capable of activating a range of heterologous promoters present in a herpesvirus genome and that the effect does not exhibit a strict sequence specificity. The only elements common to all HSV-1 IE promoters, as revealed by sequence comparisons, are the TAATGARAT motif, which mediates the response to VP16, and the TATA motif. Previous studies have demonstrated that the HCMV IE promoter does not respond to VP16 (33, 41), and, furthermore, the minimal ICP0 promoter present in in1389 has no TAATGARATs; thus, the only element known to be common to the RNA polymerase II-recognized promoters that we have analyzed is the TATA sequence. Since VAI synthesis was also stimulated, the spectrum of responsive sequences appears to be large. It is known, however, that the TATA-binding protein TFIID is required for transcription of VAI as well as for most RNA polymerase II-recognized genes; therefore, this factor may be a target for the action of pp71 (20, 42). A mechanism of this type is consistent with the general stimulation of expression from the HCMV genome which occurs when plasmids encoding pp71 are cotransfected with viral DNA (2). In plasmid cotransfection studies, the cell factors ATF and AP-1 were implicated as mediating the response to pp71 (16). This model is appropriate for the HCMV IE promoter, but it may not be universally applicable, since recognition sites for these factors have not been identified in HSV-1 IE promoters, although it is possible that ATF and AP-1 response elements have been overlooked in previous analyses due to the high degree of degeneracy exhibited by the binding sites. In addition, there were differences in the degrees of response of the promoters tested: expression from the HSV-1 ICP0 and ICP22 promoters and VAI was stimulated by two- to threefold, whereas expression from the HCMV IE and HSV-1 ICP4 and ICP27 promoters was increased by five- to sixfold.
Although the role of pp71 appears to be similar to that of VP16, the broader specificity of the HCMV-specified protein may signify differences between HSV-1 and HCMV in the requirements for virion transactivators. VP16 gives an initial boost to IE gene expression, after which IE proteins maintain transcription of the viral genome and a vigorous, rapid, productive infection. Arguably, the major role of VP16 is to ensure that sufficient ICP0 is produced to prevent conversion of the HSV-1 genome to a quiescent state in which promoters become unresponsive to transactivators (1, 8, 33, 37, 38). It is less apparent why the powerful HCMV major IE promoter should require further activation by a virion component, and once IE proteins have been expressed, they would be expected to control the transcription of the HCMV genome. Possibly, pp71 is important for the efficient expression of other IE loci which lack potent promoters, and it may also function to improve or maintain genome accessibility to transcription factors.
ACKNOWLEDGMENTS
We thank D. J. McGeoch for comments on the manuscript, P. Lomonte for help in purifying GFP-expressing viruses, and G. Hensel for providing antibody to pp71.
REFERENCES
- 1.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billstrom M A, Johnson G L, Avdi N J, Worthen G S. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J Virol. 1998;72:5535–5544. doi: 10.1128/jvi.72.7.5535-5544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldogh I, Abubaker S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 5.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 6.Chee M S, Satchwell S C, Preddie E, Weston K M, Barrell B G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 7.Ecob-Prince M S, Hassan K, Denheen M T, Preston C M. Expression of β-galactosidase in neurons of dorsal root ganglia which are latently infected with herpes simplex virus type 1. J Gen Virol. 1995;76:1527–1532. doi: 10.1099/0022-1317-76-6-1527. [DOI] [PubMed] [Google Scholar]
- 8.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J-L, Murphy P M. Human cytomegalovirus open reading frame UL28 encodes a functional β chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 10.Garber D A, Beverley S M, Coen D M. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 11.Ghazal P, Lubon H, Fleckenstein B, Hennighausen L. Binding of transcription factors and creation of a large nucleoprotein complex on the human cytomegalovirus enhancer. Proc Natl Acad Sci USA. 1987;84:3658–3662. doi: 10.1073/pnas.84.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel G M, Meyer H H, Buchman I, Pommerehne D, Schmolke S, Plachter B, Radsak K, Kern H F. Intracellular localization and expression of the human cytomegalovirus matrix protein pp71 (UL82): evidence for its translocation to the nucleus. J Gen Virol. 1996;77:3087–3097. doi: 10.1099/0022-1317-77-12-3087. [DOI] [PubMed] [Google Scholar]
- 13.Hunninghake G W, Monick M M, Liu B, Stinski M F. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J Virol. 1989;63:3026–3033. doi: 10.1128/jvi.63.7.3026-3033.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamieson D R S, Robinson L H, Daksis J I, Nicholl M J, Preston C M. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus Vmw65 mutants. J Gen Virol. 1995;76:1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 15.Kothari S, Baillie J, Sissons J G P, Sinclair J. The 21bp repeat element of the human cytomegalovirus major immediate early enhancer is a negative regulator of gene expression in undifferentiated cells. Nucleic Acids Res. 1991;19:1767–1771. doi: 10.1093/nar/19.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowenstein P R, Morrison E E, Bain D, Hodge P, Preston C M, Clissold P, Stow N, McKee T A, Castro M G. Use of recombinant vectors derived from herpes simplex 1 mutant tsK for short-term expression of transgenes encoding cytoplasmic and membrane anchored proteins in postmitotic polarized cortical neurons and glial cells in vitro. Neuroscience. 1994;60:1059–1077. doi: 10.1016/0306-4522(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 18.Margulies B J, Bowne H, Gibson W. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology. 1996;225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFarlane M, Daksis J I, Preston C M. Hexamethylene bisacetamide stimulates herpes simplex virus immediate early gene expression in the absence of trans-induction by Vmw65. J Gen Virol. 1992;73:285–292. doi: 10.1099/0022-1317-73-2-285. [DOI] [PubMed] [Google Scholar]
- 20.Meyers R E, Sharp P A. TATA-binding protein and associated factors in polymerase II and polymerase III transcription. Mol Cell Biol. 1993;13:7953–7960. doi: 10.1128/mcb.13.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelson S, Turowski P, Picard L, Goris J, Landini M P, Topilko A, Hemmings B, Bessia C, Garcia A, Virelizier J L. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J Virol. 1996;70:1415–1423. doi: 10.1128/jvi.70.3.1415-1423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern J P, Land H. A series of mammalian expression vectors and characterization of their expression or a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholl M J, Preston C M. Inhibition of herpes simplex virus type 1 immediate-early gene expression by alpha interferon is not VP16 specific. J Virol. 1996;70:6336–6339. doi: 10.1128/jvi.70.9.6336-6339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notarianni E L, Preston C M. Activation of cellular stress protein genes by herpes simplex virus temperature-sensitive mutants which overproduce immediate early polypeptides. Virology. 1982;123:113–122. doi: 10.1016/0042-6822(82)90299-9. [DOI] [PubMed] [Google Scholar]
- 26.O’Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 27.Panning B, Smiley J R. Regulation of cellular genes transduced by herpes simplex virus. J Virol. 1989;63:1929–1937. doi: 10.1128/jvi.63.5.1929-1937.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poffenberger K L, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preston C M. The cell-free synthesis of herpesvirus-induced polypeptides. Virology. 1977;78:344–353. doi: 10.1016/0042-6822(77)90109-x. [DOI] [PubMed] [Google Scholar]
- 30.Preston, C. M. Unpublished observations.
- 31.Preston C M, Mabbs R, Nicholl M J. Construction and characterization of herpes simplex virus type 1 mutants with conditional defects in immediate early gene expression. Virology. 1997;229:228–239. doi: 10.1006/viro.1996.8424. [DOI] [PubMed] [Google Scholar]
- 32.Preston C M, McFarlane M. Cytodifferentiating agents affect the replication of herpes simplex virus type 1 in the absence of functional VP16. Virology. 1998;249:418–426. doi: 10.1006/viro.1998.9314. [DOI] [PubMed] [Google Scholar]
- 33.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston C M, Rinaldi A, Nicholl M J. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J Gen Virol. 1998;79:117–124. doi: 10.1099/0022-1317-79-1-117. [DOI] [PubMed] [Google Scholar]
- 35.Prosch S, Staak K, Stein J, Liebenthal C, Stamminger T, Volk H-D, Kruger D H. Stimulation of human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFα is mediated via induction of NF-κB. Virology. 1995;208:197–206. doi: 10.1006/viro.1995.1143. [DOI] [PubMed] [Google Scholar]
- 36.Ruger B, Klages S, Walla B, Albrecht J, Fleckenstein B, Tomlinson P, Barrell B. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp65 and pp71 of human cytomegalovirus. J Virol. 1987;61:446–453. doi: 10.1128/jvi.61.2.446-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell J, Stow N D, Stow E C, Preston C M. Herpes simplex virus genes involved in latency in vitro. J Gen Virol. 1987;68:3009–3018. doi: 10.1099/0022-1317-68-12-3009. [DOI] [PubMed] [Google Scholar]
- 38.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambucetti L C, Cherrington J M, Wilkinson G W G, Mocarski E S. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;13:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamminger T, Fickenscher H, Fleckenstein B. Cell type-specific induction of the major immediate early enhancer of human cytomegalovirus. J Gen Virol. 1990;71:105–113. doi: 10.1099/0022-1317-71-1-105. [DOI] [PubMed] [Google Scholar]
- 41.Stinski M F, Roehr T J. Activation of the major immediate early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J Virol. 1985;55:431–441. doi: 10.1128/jvi.55.2.431-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White R J, Khoo B C E, Inostroza J A, Reinberg D, Jackson S P. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science. 1994;266:448–450. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- 43.Winkler M, Shmolke S, Plachter B, Stamminger T. The pUL69 protein of human cytomegalovirus (HCMV), a homologue of the herpes simplex virus ICP27, is contained within the tegument of virions and activates the major immediate-early enhancer of HCMV in synergy with the tegument protein pp71 (ppUL82) Scand J Infect Dis Suppl. 1995;99:8–9. [Google Scholar]
- 44.Yorochko A D, Hwang E-S, Rasmussen L, Keay S, Pereira L, Huang E-S. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu H, Cong J-P, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]