Abstract
Aim
Severe functional tricuspid regurgitation (FTR) is associated with high risk of cardiovascular events, particularly heart failure (HF) and mortality. MicroRNAs (miRNAs) have been recently identified as novel biomarkers in different cardiovascular conditions, but no studies have focused on FTR. We sought to (1) to identify and validate circulating miRNAs as regulators of FTR and (2) to test association of miRNA with heart failure and mortality in FTR.
Methods and results
Consecutive patients with isolated severe FTR (n = 100) evaluated in the outpatient Heart Valve Clinic and age‐ and gender‐matched subjects with no TR (controls, n = 50) were prospectively recruited. The experimental design included (1) a screening phase to identify candidate miRNA differentially expressed in FTR (n = 8) compared with controls (n = 8) through miRNA array profiling of 192 miRNAs using quantitative reverse transcription PCR arrays [qRT‐PCR]) and (2) a validation phase in which candidate miRNAs identified in the initial screening were selected for further validation by qRT‐PCR in a prospectively recruited cohort of FTR (n = 92) and controls (n = 42). Bioinformatics analysis was used to predict their potential target genes and functional pathways elicited. A combined endpoint of hospital admission due to heart failure (HF) and all‐cause mortality was defined. Initial screening identified 16 differentially expressed miRNAs in FTR compared with controls, subsequently confirmed in the validation phase (n = 16 were excluded due to significant haemolysis). miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p identified FTR with high predictive value [AUC of 0.93 (0.88–0.97), 0.83 (0.75–0.91) and 0.84 (0.76–0.92), respectively]. During a median follow‐up of 20.4 months (IQR 8–35 months), 32% of FTR patients reached the combined endpoint. Patients with low relative expression of miR‐15a‐5p, miR‐92a‐3p, miR101‐3p, and miR‐363‐3p, miR‐324‐3p, and miR‐22‐3p showed significantly higher rates of events (log‐rank test for all P < 0.01). Both miR‐15a‐5p [hazard ratio: 0.21 (0.06–0.649, P = 0.007) and miR‐92a‐3p (0.27 (0.09–0.76), P = 0.01] were associated with outcomes after adjusting for age, gender, and New York Heart Association functional class.
Conclusions
Circulating miRNAs are novel diagnostic and prognostic biomarkers in severe FTR. The quantification of miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p held strong diagnostic value, and the quantification of miR‐15a‐5p and miR‐92a‐3p are independently associated with outcomes. The recognition of specific miRNAs offers a novel perspective for TR evaluation.
Keywords: Biomarker, Heart failure, Micro‐RNA, Tricuspid regurgitation
Introduction
With the robust evidence of the impact of tricuspid regurgitation (TR) on patients' outcomes, interest in understanding its pathophysiology and mechanism has grown. Functional tricuspid regurgitation (FTR) is the most common cause of TR, accounting for more than 80% of the cases. 1 , 2 , 3 However, FTR is not a single pathophysiological condition but includes several phenotypes and aetiologies with subsequent different outcomes. 3 , 4 , 5
Long‐term persistent atrial fibrillation (AF) has been associated with the development of atrial FTR (A‐FTR) in patients with structurally standard valve leaflets due to progressive dilatation and dysfunction of the tricuspid annulus (TA). The underlying pathophysiological factors behind TA enlargement in combination with the development of A‐FTR are still unclear, given that despite RA enlargement, a significant fraction of patients presenting long‐term persistent AF will fail to develop substantial TR during follow‐up. 3 , 4 , 5 , 6 On the other side, ventricular TR is caused by tethering of the leaflets due to adverse RV remodelling secondary to pulmonary hypertension, RV myopathy, or mitral or aortic valve disease. 3 , 7 Similarly, predisposing pathophysiologic factors may interplay with RV remodelling, which may justify the lack of TR progression in all patients. To date, the underlying molecular mechanisms that lead to the progression of both A‐FTR and V‐FTR and the differences in the outcome of patients have yet to be identified.
MicroRNAs (miRNAs) are small (20–25 nucleotides), non‐coding endogenous regulatory RNA molecules that post‐transcriptionally regulate gene expression. miRNAs have recently attracted increasing interest as regulators of valvular disease, pathogenesis, diagnostic biomarkers, and therapeutical targets, with the advantage of being easily measured in accessible blood samples. 8 In vivo and in vitro studies have demonstrated stimulatory or inhibitory roles in mitral valve prolapse development, aortic leaflet fusion, and calcification pathways. However, no study has been designed to target specific miRNAs as brand‐new biomarkers of FTR.
The aims of this study were (1) to identify and validate a panel of serum miRNA differentially expressed in FTR and (2) to test miRNA's ability to stratify the risk of HF hospitalization and all‐cause mortality.
Methods
Study population and subject enrolment
Consecutive patients with isolated functional severe TR evaluated in the outpatient Heart Valve Clinic were prospectively included. Age‐ and gender‐matched subjects with normal biventricular function, absence of valvular heart disease and adjusted by the presence non‐valvular permanent atrial fibrillation (AF) and bi‐atrial enlargement were enrolled and included as a control group.
Patients with FTR were divided into two groups based on the aetiology of FTR: atrial and ventricular FTR (A‐FTR and V‐FTR, respectively). 3 , 5 , 7 A‐FTR was defined by the absence of any leaflet abnormality, left ventricular (LV) dysfunction (LVEF <60%), left‐sided valve disease, pulmonary hypertension (pulmonary artery systolic pressure >50 mmHg by echocardiography or mean pulmonary arterial pressure >20 mmHg by right heart catheterization) or cardiac implantable electronic device, and supported by the evidence of longstanding or permanent AF. V‐FTR was defined by the absence of any leaflet abnormality, presence of leaflet tethering in the context of previously corrected left heart valve diseases or LV dysfunction resulting in pulmonary hypertension or secondary pulmonary hypertension (e.g. chronic lung disease, pulmonary thromboembolism, and left‐to‐right shunt) detected by echocardiography or right heart catheterization as previously described.
Exclusion criteria for all subjects included significant (> mild) uncorrected left heart valve disease, <18 years old, pregnancy, and active tumour disease. All patients underwent a detailed protocol, including clinical evaluation, comprehensive echocardiogram, and blood testing for established and novel biomarkers. 9
Blood samples for determination of serum miRNAs were extracted on the first visit to the Heart Valve Clinic (TR group) and during an outpatient visit (control group) and stored at the local Biobank. Samples from patients included in this study were provided by the BioBank Hospital Ramón y Cajal‐IRYCIS (National Registry of Biobanks B.0000678), integrated in the Biobanks and Biomodels Platform of the ISCIII (PT20/00045), and they were processed following standard operating procedures with the appropriate approval of the Ethical and Scientific Committees.
Blood samples were collected in serum tubes with separating gel, and serum tubes were allowed to clot for 30 min and centrifuged for 10 min at 1500× g room temperature. Three aliquots of 500 μL were obtained and stored at −80°C.
The study protocol was reviewed and approved by the local institutional ethics committees (internal code 249/17). Written informed consent to perform the protocol was obtained from all patients. All procedures were carried out in accordance with the Declaration of Helsinki (2000).
Experimental design
Figure 1 demonstrates the two‐phase design of this study (screening and validation phase).
Figure 1.
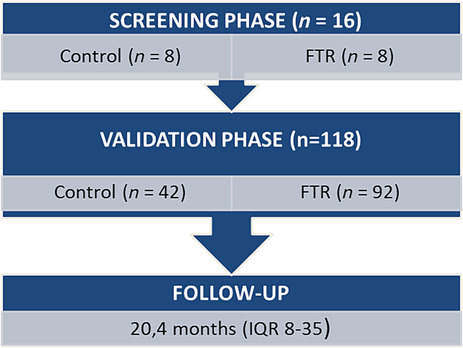
Flow chart: study design.
Screening phase: MicroRNA array profiling
To identify serum miRNAs differentially expressed in patients with TR, a screening assay for 192 miRNAs using quantitative reverse transcription PCR arrays was performed in eight patients with FTR and eight controls adjusted for AF, age, sex, and cardiovascular (CV) risk factors (Table 1 ). The miRCURY LNA miRNA PCR Panel (QIAGEN, Cat. No. YAHS‐106Y), combined with a miRCURY LNA SYBR Green Master Mix, was used. Data were analysed using the QIAGEN data analysis web portal (http://www.qiagen.com/geneglobe). Array data were normalized using the 2−ΔΔCt method.
TABLE 1.
Identification stage
| miRNA | ID fold regulation | P‐value |
|---|---|---|
| hsa‐miR‐409‐3p | 1.59 | 0.048 |
| hsa‐miR‐375 | 1.74 | 0.034750 |
| hsa‐miR‐222‐3p | −2.26 | 0.003783 |
| hsa‐miR‐15a‐5p | −2.45 | 0.009925 |
| hsa‐miR‐29b‐3p | −2.30 | 0.007424 |
| hsa‐miR‐152‐3p | −1.59 | 0.043350 |
| hsa‐miR‐30e‐5p | −2.10 | 0.019476 |
| hsa‐miR‐186‐5p | −2.10 | 0.006234 |
| hsa‐miR‐101–3p | −1.95 | 0.016495 |
| hsa‐miR‐126‐5p | −2.73 | 0.003620 |
| hsa‐miR‐92a‐3p | −2.22 | 0.028454 |
| hsa‐miR‐363‐3p | −2.47 | 0.007119 |
| hsa‐miR‐148a‐3p | −1.93 | 0.037182 |
| hsa‐miR‐324‐3p | −1.54 | 0.046501 |
| hsa‐miR‐22‐3p | −2.02 | 0.005555 |
| hsa‐miR‐29c‐3p | −1.85 | 0.029347 |
Differential miRNA expression between FTR versus controls. Controls were defined as age and gender subjects adjusted by the presence of persistent AF fold‐change (2−ΔΔCt) is the normalized miRNA expression (2−ΔΔCt) in a test sample divided the normalized miRNA expression (2−ΔΔCt) in the control sample. Fold‐regulation represents fold‐change results in a biologically meaningful way. Fold‐change values greater than 1.5 indicates a positive‐ or an up‐regulation, and the fold‐regulation is equal to the fold‐change. Fold‐change values less than 1.5 indicate a negative or down‐regulation, and the fold‐regulation is the negative inverse of the fold‐change.
Validation phase
Candidate miRNAs identified in the initial screening were selected for further validation by individual quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis in a prospectively recruited cohort of FTR (n = 92) and controls (n = 42). Blood samples from 16 subjects (15 TR and 1 control) were excluded due to significant haemolysis.
MicroRNAs extraction from serum and retrotranscription reaction
The total RNA enrichment for miRNAs was performed from 200 μL of serum using the miRNeasy Serum/Plasma Advanced Kit (Ref. 217204, Qiagen), following the manufacturer's instructions. During the extraction process, UniSp2 is added for subsequent amplification as technical control of RNA isolation, using the RNA Spike‐in Kit (Ref. 339390, Qiagen). cDNA synthesis is performed from 7 μL of RNA using the miRCURY LNA RT Kit (Reference 339340, Qiagen) in a final volume of 35 μL, following the manufacturer's instructions. During the reverse transcription process, cel‐miR‐39‐3p is added for subsequent amplification as technical control of cDNA synthesis using the RNA Spike‐in Kit (Ref. 339390, Qiagen).
Validation of selected microRNAs by individual quantitative reverse transcription PCR
miRCURY LNA™ miRNA PCR Assay and miRCURY SYBR Green PCR Kit (Ref. 339347, Qiagen) were used, following the manufacturer's instructions. The exogenous UniSp2 is also amplified as technical control of extraction.
The specific references for each studied miRNA are shown in Table S1 . For data normalization, the expression of two miRNAs, hsa‐miR‐103a‐3p and hsa‐miR‐191‐5p, was analysed in all samples. Then the stability of each was evaluated using the Normfinder (miR‐191) and Bestkeeper software, demonstrating that the expression of miR‐191 or the average expression of miR‐103 and miR‐191 presents the most stable values among all samples. The results were finally normalized using the 2−ΔΔCt method.
Bioinformatics analysis
Bioinformatics analysis predicted target genes of selected miRNAs using predictive analysis across three different databases: FunRich, 10 miRDB, 11 and TargetScan. 12
For the visual representation of predictive analysis results, FunRich software was applied to generate a Venn diagram. The diagram facilitated the selection of targets that exhibited concordance in at least two databases. FunRich open software is a stand‐alone software tool for gene target prediction, functional enrichment, and interaction network analysis of genes and proteins.
Furthermore, enrichment analyses were conducted utilizing the ShinyGo 13 and Enrichr 14 web tools. Specifically, the ShinyGo tool was employed for analysing Gene Ontology (GO) molecular function, GO biological process, and GO cellular component, while Enrichr was used for WikiPathway 2021, KEGG 2021, and Reactome 2022. Subsequently, an interaction analysis of predicted miRNA targets was executed using FunRich software and involved two distinct databases: Uniprot and FunRich.
Clinical outcomes
Clinical data were obtained prospectively from hospital records in the Heart Valve Clinic and direct communication with the patients. A combined endpoint of hospital admission due to heart failure (HF) and all‐cause mortality at follow‐up was defined. HF admission was based on the universal definition of HF as a clinical syndrome with HF symptoms and/or signs corroborated by elevated natriuretic peptide and/or objective evidence of pulmonary or systemic congestion. 15
Statistical analysis
GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA) was used for descriptive statistics, comparative analysis, and graphic representation. The D'Agostino & Pearson, Shapiro–Wilk, and Kolmogorov–Smirnov tests were used to determine the normality of the sample distribution. Subsequently, the Student's t‐test or Mann–Whitney test was used to compare two groups, and the one‐way ANOVA or Kruskal–Wallis test for three subgroups, as appropriate. For comparison of categorical variables, the χ 2 test was used. Categorical data were expressed as percentages and continuous variables as mean ± SD or median (interquartile range). SPSS (version 21.0; SPSS, Chicago, IL, USA) and Stata Statistical Software (version 14.1, Stata Corporation, College Station, TX, USA) were used for linear regression and Kaplan–Meier analysis. Medians were used to define the miRNA cut‐off point in the survival analysis. All results with a P‐value ≤0.05 were considered significant.
Results
Study population characteristics
Identification cohort
Sixteen subjects were included in the screening stage (8 with FTR and 8 controls). Mean age was 78 ± 4 years in FTR and 79 ± 6 in controls (P = 0.64). In both groups, there were a 62.5% (five in each group) of females, and all patients had permanent AF. No significant differences were found in the presence of CV risk factors (50% vs. 62%, P = 0.24).
Validation cohort
Demographic data, baseline characteristics, and imaging parameters of FTR patients and controls are shown in Table 2 . Most of subjects were females (65% in FTR and 61% in controls) with no significant difference in age or CV risk factors. Regarding the aetiology of FTR, 52% had functional atrial TR and 48% functional ventricular TR. According to echocardiographic parameters, FTR patients showed higher right ventricle (RV) and right atrium area and lower values of RV and RA strain. Mean values of the different parameters of RV and LV function are described in Table 2 . Baseline values of laboratory data for FTR are also shown in Table 2 .
TABLE 2.
Demographic and characteristics in FTR and controls
| Variable | FTR (N = 77) | Controls (N = 41) | P‐value |
|---|---|---|---|
| Age, years | 77 ± 8 | 77 ± 11 | 0.73 |
| Female, n (%) | 50 (65) | 25 (61) | 0.67 |
| CV risk factors | |||
| Type 2 diabetes mellitus, n (%) | 13 (17) | 8 (19) | 0.74 |
| Hypertension, n (%) | 40 (52) | 24 (58) | 0.32 |
| Hypercholesterolaemia, n (%) | 34 (44) | 24 (58) | 0.16 |
| Smoker, n (%) | 10 (13) | 5 (12) | 0.63 |
| Atrial fibrillation, n (%) | 73 (95) | 41 (100) | 0.91 |
| Coronary artery disease, n (%) | 11 (14) | 5 (12) | 0.42 |
| COPD or asthma, n (%) | 7 (9) | 4 (9) | 0.32 |
| Renal disease, n (%) | 7 (9) | 3 (7) | 0.62 |
| Previous left valve surgery | 37 (48) | 0 (0) | NA |
| TR aetiology | |||
| Atrial TR, n (%) | 40 (52) | NA | NA |
| Ventricular TR, n (%) | 37 (48) | NA | NA |
| NYHA class | <0.001 | ||
| I/II | 63 (81) | 41 (100) | |
| III | 13 (17) | 0 | |
| IV | 1 (1) | 0 | |
| Diuretic treatment | NA | ||
| At least one type of diuretic treatment | 48 (62) | 0 | |
| Loop diuretics | 33 (43) | 0 | |
| Potassium‐sparing diuretics | 27 (35) | 0 | |
| Biochemistry | |||
| Creatinine, mg/dL | 0.9 ± 0.22 | 0.97 ± 0.8 | 0.5 |
| Haemoglobin, g/dL | 12.6 ± 2 | 13.2 ± 1.6 | <0.001 |
| Adjusted BNP (BNP/upper limit of normal for age/gender) | 1.2 (0.7–1.7) | 0.9 (0.6–1.4) | 0.001 |
| Total bilirubin, mmol/L | 0.89 (0.6–1.3) | 0.76 (0.6–1.2) | 0.78 |
| ASAT, U/L | 23 (18–28) | 21 (18–26) | 0.65 |
| ALAT, U/L | 16 (14–22) | 16 (13–21) | 0.79 |
| GGT, U/L, med | 57 (33–110) | 45 (27–67) | 0.002 |
| LDH, U/L | 247 (191–247) | 225 (188–278) | 0.39 |
| ALP, U/L | 84 (74–111) | 76 (66–90) | 0.005 |
| Echocardiographic parameters | |||
| LV ejection fraction, % | 60 ± 8 | 64 ± 9 | 0.94 |
| LA volume, mL/m2 | 106 ± 60 | 99 ± 28 | 0.006 |
| Average E/e′ | 11 ± 4 | 11.3 ± 4 | 0.35 |
| RV end‐diastolic area, cm2 | 23 ± 8 | 16 ± 5 | 0.004 |
| RV end‐systolic area, cm2 | 13 ± 5 | 9 ± 6 | 0.005 |
| RA area, cm2 | 31 ± 14 | 26 ± 9 | 0.02 |
| RA volume, mL | 111 (70–165) | 86 (60–103) | 0.012 |
| RV FAC, % | 44.1 ± 9 | 44 ± 7 | 0.78 |
| TAPSE, mm | 20 ± 4 | 22 ± 3 | 0.01 |
| S′ wave TDI, cm/s | 10.4 ± 2 | 11.6 ± 2 | 0.07 |
| RV‐free wall longitudinal strain | −21.7 ± 6 | −25.9 ± 3 | <0.001 |
| RA reservoir strain | 12 ± 7 | 22 ± 8 | <0.001 |
| TR max velocity, cm/s | 263 ± 51 | NA | NA |
ALAT, alanine aminotransferase; ALP, alkaline phosphatase; ASAT, aspartate aminotransferase; BNP, brain natriuretic peptide; CV, cardiovascular; COPD, chronic obstructive pulmonary disease; GGT, gamma‐glutamyl transpeptidase; LDH, serum lactate dehydrogenase; LV, left ventricular; NYHA, New York Heart Association; RV, right ventricular; TAPSE, tricuspid annular plane systolic excursion; TDI: tissue Doppler imaging; TR, tricuspid regurgitation.
Identification and validation cohorts did not significantly differ in terms of age, gender, CV risk factors (P > 0.05 for all).
Identification stage: Functional tricuspid regurgitation is associated with differential miRNA expression
We first performed an initial screening in 16 subjects (eight FTR and eight controls) to test whether FTR patients differentially expressed selected serum microRNAs when compared with controls, resulting in the identification of 16 serum candidates (Table 1 , Figure 2 ). A heat map diagram shows the expression organized into a two‐way hierarchical clustering by miRNA and samples, indicating that FTR exhibited a different pattern of miRNA expression compared with subjects with similar atrial dimensions and matched by age, gender, and the presence of AF (Figure 3).
Figure 2.
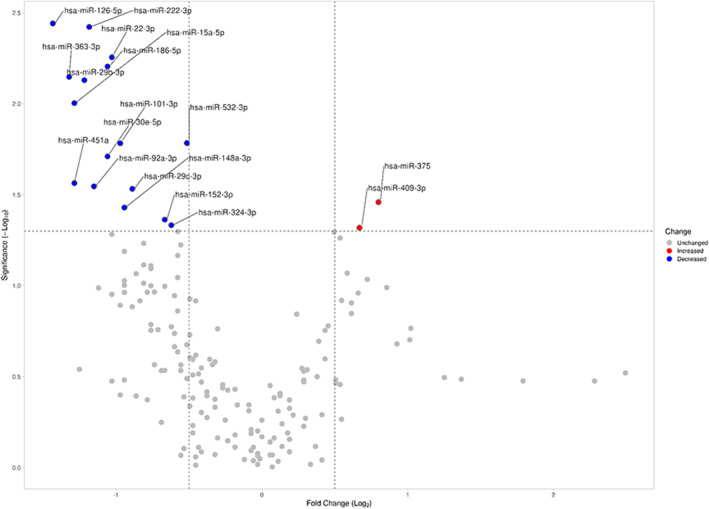
Graphic representation (Volcano plot) of significant miRNA expression changes obtained in the miRCURY LNA miRNA PCR Panels. Data are expressed as the log2 of the fold change on the x‐axis and the log10 of the P‐value on the y‐axis. The dotted lines delimit the limit of statistically significant values (P‐value <0.05) and log2 of fold change >0.5. All miRNAs with statistically significant results, represented in red (increased) or blue (decreased) in the volcano plot, were selected for further validation.
Figure 3.
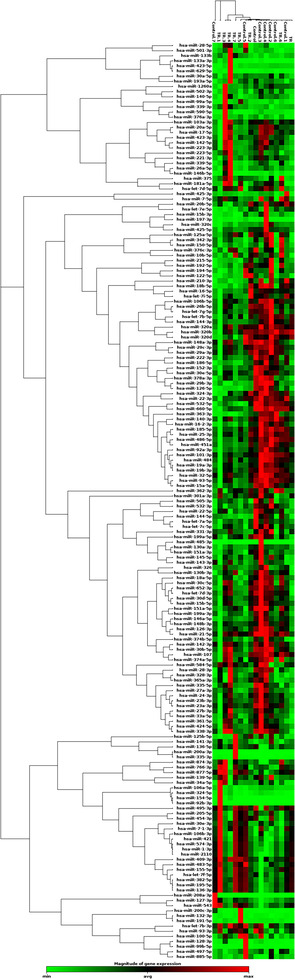
miRNA expression clustergram (miRNA expression signature). The hierarchical clustering represented by the dendrograms can be used to identify groups of miRNAs with related expression levels. Samples re‐classification based on miRNAs expression could be also observed.
Validation stage: Validation of miRNAs in a selected cohort of patients with tricuspid regurgitation
The 16‐candidate miRNA identified in the initial screening were selected for further validation by individual quantitative reverse transcription polymerase chain reaction (qRT‐PCR) analysis in 118 samples collected from TR patients and controls. Demographic data, baseline characteristics, and imaging parameters of selected patients are presented in Table 2 . No differences were found in age, gender, or CV risk factors between TR and controls. No sex‐specific differences in miRNA expression were found in patients with TR.
Our results show differential miRNA expression in FTR compared with controls. In particular, A‐FTR exhibited significant different levels of microRNAs: miR‐29b‐3p, miR‐152‐3p, miR‐30e‐5p, miR‐186‐5p, miR‐126‐5p, and miR‐148a‐3p (P < 0.05), whereas V‐FTR preferentially expressed microRNAs: miR‐15a‐5p, miR‐152‐3p, miR‐30e‐5p, miR‐186‐5p, miR‐22‐3p, and miR324‐3p, respectively (P < 0.05; Table 3 and Figure 4 ). Further analysis showed that miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p yielded a highly significant predictive value for the detection of FTR, as showing areas under the curve of 0.93 (0.88–0.97), 0.83 (0.75–0.91) and 0.84 (0.76–0.92), respectively (Figure 5 ). Among all the data, miR‐324‐3p and miR‐29c‐3p were differentially expressed in A‐FTR compared with V‐FTR (P < 0.05 for both).
TABLE 3.
Comparative analysis of relative miRNA expression between the control group and patients with atrial TR and ventricular TR
| miRNA | Control | Atrial TR | Ventricular TR |
|---|---|---|---|
| hsa‐miR‐409‐3p | 1.00 (0.72–1.37) | 1.22 (0.86–1.72) | 1.35 (1.01–1.81) |
| hsa‐miR‐375 | 1.00 (0.73–1.36) | 1.12 (0.80–1.57) | 0.95 (0.70–1.29) |
| hsa‐miR‐222‐3p | 1.00 (0.90–1.11) | 1.08 (0.96–1.21) | 1.13 (1.00–1.28) |
| hsa‐miR‐15a‐5p | 1.00 (0.88–1.13) | 1.07 (0.89–1.28) | 1.21 (1.05–1.39) * |
| hsa‐miR‐29b‐3p | 1.00 (0.92–1.09) | 0.83 (0.71–0.98) * | 0.97 (0.85–1.11) |
| hsa‐miR‐152‐3p | 1.00 (0.90–1.11) | 0.58 (0.49–0.67) **** | 0.66 (0.59–0.73) **** |
| hsa‐miR‐30e‐5p | 1.00 (0.83–1.20) | 0.43 (0.36–0.50) **** | 0.48 (0.42–0.54) **** |
| hsa‐miR‐186‐5p | 1.00 (0.89–1.12) | 0.08 (0.03–0.16) **** | 0.13 (0.07–0.26) **** |
| hsa‐miR‐101‐3p | 1.00 (0.89–1.12) | 0.94 (0.77–1.13) | 1.14 (0.92–1.42) |
| hsa‐miR‐126‐5p | 1.00 (0.89–1.12) | 0.82 (0.75–0.97) * | 0.95 (0.83–1.09) |
| hsa‐miR‐92a‐3p | 1.00 (0.86–1.16) | 1.03 (0.84–1.27) | 1.19 (1.02–1.38) |
| hsa‐miR‐363‐3p | 1.00 (0.84–1.19) | 1.09 (0.85–1.39) | 1.23 (1.02–1.48) |
| hsa‐miR‐148a‐3p | 1.00 (0.89–1.12) | 0.84 (0.72–0.98) * | 0.92 (0.80–1.05) |
| hsa‐miR‐324‐3p | 1.00 (0.90–1.11) | 0.99 (0.86–1.13) # | 1.17 (1.04–1.31) * , # |
| hsa‐miR‐22‐3p | 1.00 (0.87–1.14) | 1.05 (0.88–1.25) | 1.24 (1.09–1.42) * |
| hsa‐miR‐29c‐3p | 1.00 (0.89–1.13) | 0.90 (0.78–1.03) # | 1.03 (0.92–1.17) # |
Data represented as geometric mean (95% confidence interval). The relative expression of the miRNAs analysed by qPCR is expressed as the normalized value of 2−ΔΔCt.
TR, tricuspid regurgitation.
P‐value ≤0.05, compared with the control group.
P‐value ≤0.0001, compared with the control group.
P‐value ≤0.05, comparing atrial TR versus ventricular TR patients.
Figure 4.
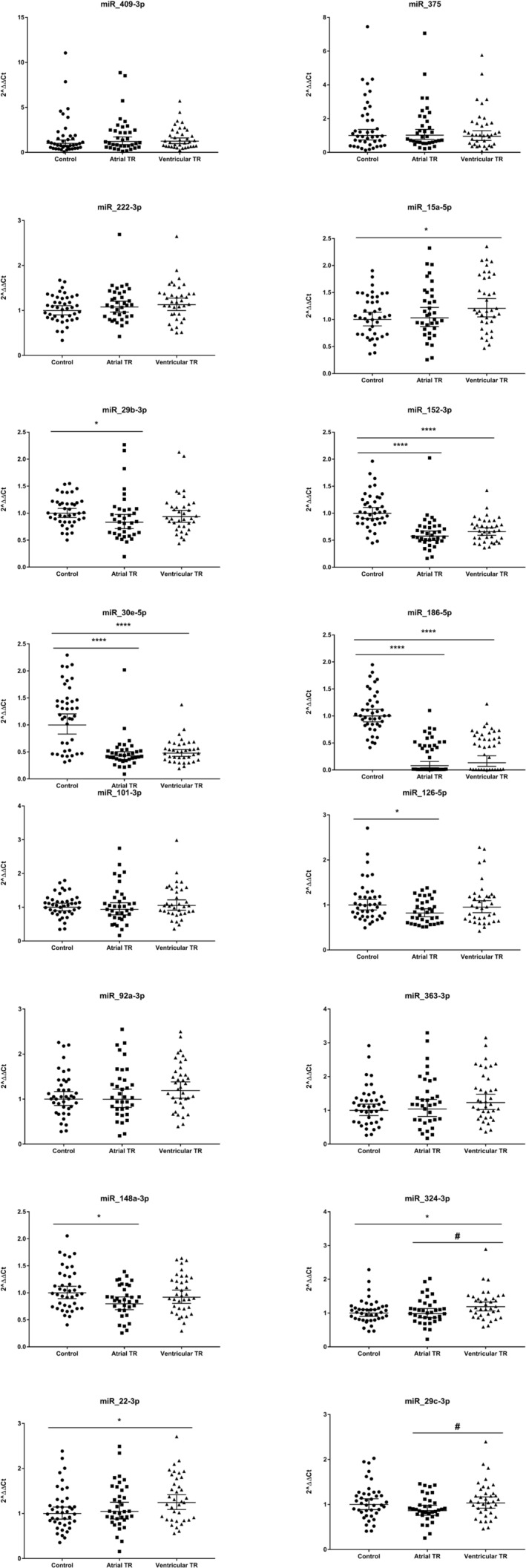
Comparative analysis of the qPCR validation study of miRNA expression between the control, A‐FTR, and V‐FTR groups. The normalized values of 2−ΔΔCt were used for the graphical representation. Data were represented as the geometric mean (95% confidence interval). *P‐value ≤0.05 compared with its respective control group. ****P‐value ≤0.0001 compared with its respective control group. # P‐value ≤0.05, comparing atrial TR versus ventricular TR patients.
Figure 5.
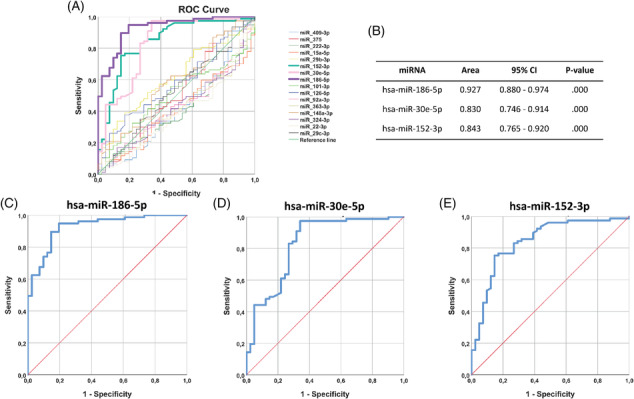
ROC curves of the statistically significant miRNAs. (A) Graphic representation of the set of all selected miRNAs. Note the pattern of the miRNAs hsa‐miR‐186‐5p, hsa‐miR‐30e‐5p, and hsa‐miR‐152‐3p. (B) Area under the curve and 95% confidence interval of the three miRNAs with significant results in the ROC analysis. (C) ROC curve of hsa‐miR‐186‐5p. (D) ROC curve of hsa‐miR‐30e‐5p. (E) ROC curve of hsa‐miR‐152‐3p.
Association of selected microRNAs with cardiac imaging parameters
Further analysis demonstrated the association between a group of serum miRNAs with cardiac imaging parameters of RV dimension, RV systolic function, and TR severity (Table 4 ). Linear regression analysis demonstrated that expression of specific miRNAs was associated with right chamber size/function [RV‐end‐diastolic/end‐systolic area (EDA, ESA) RA area, RA reservoir strain], together with different measures of RV systolic function (TAPSE, systolic excursion velocity by Doppler Tissue Imaging [S′ by TDI)] and RV free‐wall longitudinal strain [RV‐FWLS]) and TR severity (TR vena contracta). miR‐15a‐5p was associated with RV‐EDA, RV‐ESA, RA reservoir strain, TAPSE, and TR vena contracta. miR‐363‐3p was associated with RV‐EDA and RV‐ESA. miR‐22‐3p was associated with RV‐ESA, RV‐FWLS, RA reservoir strain, and TAPSE. miR‐152‐3p was associated with RA area and RA volume, RA reservoir strain, TAPSE, and S′ by TDI. miR‐101‐3p was associated with RV‐FWLS, and miR‐30e‐5p was associated with TAPSE and S´ by TDI (Table 4 ).
TABLE 4.
Linear regression analysis
| Parameter | miRNA | Beta | B (95% CI) | P‐value |
|---|---|---|---|---|
| RV end‐diastolic area | miR‐15a‐5p | −0.345 | −0.024 (−0.039, −0.008) | 0.003 |
| miR‐363‐3p | 0.242 | −0.026 (−0.050, −0.001) | 0.041 | |
| RV‐end‐systolic area | miR‐15a‐5p | −0.329 | −0.035 (−0.059, −0.011) | 0.005 |
| miR‐363‐3p | −0.233 | −0.038 (−0.076, 0.000) | 0.048 | |
| miR‐22‐3p | −0.259 | −0.028 (−0.052, −0.003) | 0.028 | |
| RA area | miR‐152‐3p | −0.204 | −0.005 (−0.010, 0.000) | 0.036 |
| RA volume | miR‐152‐3p | −0.197 | −0.001 (−0.001, 0.000) | 0.046 |
| RV‐free wall longitudinal strain | miR‐101–3p | −0.394 | −0.083 (−0.133, −0.033) | 0.002 |
| miR‐22‐3p | 0.265 | 0.011 (0.001, 0.022) | 0.037 | |
| RA reservoir strain | miR‐222‐3p | 0.262 | 0.016 (0.000, 0.031) | 0.043 |
| miR‐15a‐5p | 0.262 | 0.019 (0.001, 0.038) | 0.043 | |
| miR‐152‐3p | 0.322 | 0.012 (0.003, 0.022) | 0.012 | |
| miR‐22‐3p | 0.288 | 0.023 (0.003, 0.042) | 0.026 | |
| TAPSE | miR‐15a‐5p | 0.204 | 0.012 (0.001, 0.023) | 0.038 |
| miR‐152‐3p | −0.440 | −0.017 (−0.023, −0.010) | <0.001 | |
| miR‐30e‐5p | −0.556 | −0.030 (−0.038, −0.021) | <0.001 | |
| miR‐186‐5p | −0.646 | −0.033 (−0.041, −0.025) | <0.001 | |
| miR‐22‐3p | 0.223 | 0.014 (0.002, 0.025) | 0.023 | |
| TDI | miR‐152‐3p | 0.362 | 0.060 (0.022, 0.099) | 0.002 |
| miR‐30e‐5p | 0.298 | 0.065 (0.014, 0.116) | 0.014 | |
| miR‐148a‐3p | 0.248 | 0.050 (0.002, 0.099) | 0.042 | |
| miR‐29c‐3p | 0.265 | 0.058 (0.006, 0.111) | 0.029 | |
| Vena contracta | miR‐92a‐3p | −0.227 | −0.099 (−0.196, −0.001) | 0.047 |
| miR‐15a‐5p | −0.314 | −0.426 (−0.775, −0.078) | 0.017 |
Data in bold indicate P < 0.01.
TR, tricuspid regurgitation.
Follow‐up and outcomes
During a median follow‐up of 20.4 months (IQR 8–35 months), 25 (32%) patients reached the combined endpoint. Twenty‐four patients were referred for HF, and 15 died [11 were CV deaths].
Table 5 shows the clinical, biomarker, and imaging parameter differences between patients presenting with and without events. Patients with events were older, mostly men, with reduced haemoglobin, elevated basal brain natriuretic peptide (BNP), and gamma‐glutamyl transpeptidase (GGT) levels. In addition, patients with events showed more severe TR and larger RV dimensions. Both RA and RV‐free wall longitudinal strains were impaired in patients with events.
TABLE 5.
Comparisons between patients with and without events
| Variable | Patients with events, n = 25 | Patients without events, n = 52 | P‐value |
|---|---|---|---|
| Basal characteristics | |||
| Age, years | 80 ± 8 | 76 ± 8 | 0.037 |
| Female, n (%) | 10 (40) | 40 (77) | 0.002 |
| At least one CV risk factor, n (%) | 20 (80) | 35 (67) | 0.25 |
| Coronary artery disease, n (%) | 5 (20) | 6 (11) | 0.28 |
| COPD or asthma, n (%) | 3 (12) | 5 (9) | 0.10 |
| Renal disease, n (%) | 5 (20) | 5 (10) | 0.14 |
| Biochemistry | |||
| Creatinine, mg/dL | 1.13 ± 0.3 | 1.08 ± 0.8 | 0.83 |
| Haemoglobin, g/dL | 11.8 ± 2 | 13.0 ± 2 | 0.016 |
| Adjusted BNP (BNP/upper limit of normal for age/gender) | 3.05 ± 2.1 | 1.4 ± 1.2 | <0.001 |
| Total bilirubin, mmol/L | 1.33 ± 0.6 | 0.96 ± 0.5 | 0.014 |
| ASAT, U/L, median (IQR) | 24 (20–34) | 22 (18–26) | 0.14 |
| ALAT, U/L, median (IQR) | 17 (14–22) | 16 (13–23) | 0.78 |
| GGT, U/L, median (IQR) | 112 (84–155) | 44 (28–73) | <0.001 |
| LDH, U/L, median (IQR) | 249 (206–324) | 246 (189–31) | 0.59 |
| ALP, U/L, median (IQR) | 104 (74–146) | 84 (73–99) | 0.113 |
| Imaging parameters | |||
| Atrial TR, n (%) | 14 (56) | 26 (50) | 0.28 |
| LV ejection fraction, % | 58 ± 10 | 61 ± 8 | 0.24 |
| LA volume, mL/m2 | 103 ± 60 | 107 ± 63 | 0.83 |
| Average E/e′ | 10 ± 4 | 11 ± 4 | 0.53 |
| RV end‐diastolic area, cm2 | 29 ± 7 | 20 ± 7 | <0.001 |
| RV end‐systolic area, cm2 | 16 ± 6 | 11 ± 4 | <0.001 |
| RA volume, mL/m2 | 163 ± 80 | 127 ± 90 | 0.18 |
| RV FAC, % | 44 ± 10 | 44 ± 9 | 0.73 |
| TAPSE, mm | 19.6 ± 4 | 21 ± 4 | 0.16 |
| S′ wave TDI, cm/s | 10.1 ± 2 | 10.4 ± 2 | 0.60 |
| RV‐free wall longitudinal strain | −16 ± 5 | −24 ± 6 | 0.004 |
| RA reservoir strain | 8 ± 4 | 14 ± 7 | <0.001 |
| TR biplane vena contracta, mm | 1.2 ± 0.4 | 0.8 ± 0.3 | 0.004 |
| ERO, median (IQR), cm2 | 0.99 ± 0.8 | 0.49 ± 0.5 | 0.023 |
| TR max velocity, cm/s | 254 ± 65 | 268 ± 43 | 0.28 |
| Relative miRNA expression | |||
| hsa‐miR‐409‐3p | 1.11 (0.76–2.47) | 1.3 (0.59–2.66) | 0.93 |
| hsa‐miR‐375 | 1.57 (0.69–2.89) | 0.93 (0.54–1.41) | 0.054 |
| hsa‐miR‐222‐3p | 1.10 (0.84–1.40) | 1.19 (0.92–1.39) | 0.57 |
| hsa‐miR‐15a‐5p | 0.94 (0.66–1.24) | 1.28 (1.01–1.84) | <0.001 |
| hsa‐miR‐29b‐3p | 0.82 (0.62–1.07) | 0.94 (0.72–1.12) | 0.11 |
| hsa‐miR‐152‐3p | 0.54 (0.39–0.75) | 0.68 (0.53–0.86) | 0.037 |
| hsa‐miR‐30e‐5p | 0.39 (0.02–0.55) | 0.47 (0.40–0.60) | 0.133 |
| hsa‐miR‐186‐5p | 0.40 (0.02–0.55) | 0.43 (0.01–0.65) | 0.78 |
| hsa‐miR‐101–3p | 0.84 (0.53–1.04) | 1.10 (0.88–1.59) | <0.001 |
| hsa‐miR‐126‐5p | 0.84 (0.60–1.16) | 0.93 (0.68–1.22) | 0.258 |
| hsa‐miR‐92a‐3p | 0.80 (0.56–1.32) | 1.38 (1.03–1.74) | 0.001 |
| hsa‐miR‐363‐3p | 0.78 (0.45–1.38) | 1.33 (0.98–2.07) | 0.002 |
| hsa‐miR‐148a‐3p | 1.06 (0.64–1.22) | 0.87 (0.69–1.20) | 0.594 |
| hsa‐miR‐324‐3p | 0.89 (0.78–1.14) | 1.17 (0.94–1.47) | 0.040 |
| hsa‐miR‐22‐3p | 0.91 (0.80–1.03) | 1.35 (1.04–1.79) | 0.002 |
| hsa‐miR‐29c‐3p | 0.89 (0.76–1.33) | 1.00 (0.82–1.25) | 0.49 |
Clinical and imaging parameters and relative miRNA expression.
BNP, brain natriuretic peptide; CV, cardiovascular; COPD, chronic obstructive pulmonary disease; ERO, effective regurgitant orifice; GGT, gamma‐glutamyl transpeptidase; LV, left ventricular; NYHA, New York Heart Association; RA, right atrium; RV, right ventricular; TR, tricuspid regurgitation.
Patients with events showed significant different levels of miR‐15a‐5p, miR‐152‐3p, miR101‐3p, miR‐92a‐3p, miR‐363‐3p, miR‐324‐3p, and miR‐22‐3p (P < 0.05 for all). Based on the high or low relative expression of selected miRNAs (below or above the median value) patients were stratified in two groups of risk; those patients with low relative expression of miR‐15a‐5p, miR‐92a‐3p, miR101‐3p, and miR‐363‐3p, miR‐324‐3p, and miR‐22‐3p, showed significantly higher rates of HF admission and all‐cause mortality (log rank test for all P < 0.01, Figure 6 ).
Figure 6.
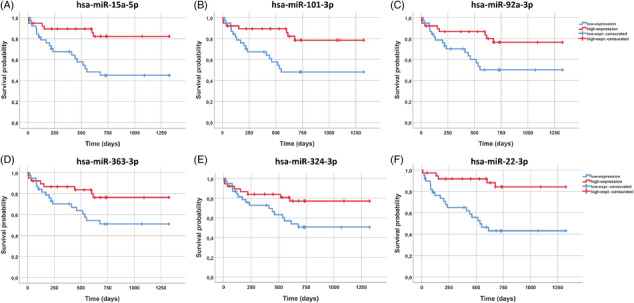
Kaplan–Meier survival graphs of the miRNAs statistically related to survival (or IC). The miRNA cut‐off was defined based on the median value to subdivide the IT population into high or low miRNA. (A) miR‐15a‐5p Kaplan–Meier graph. P‐value (log rank) = 0.002; (B) miR101‐3p Kaplan–Meier graph. P‐value (log rank) = 0.006; (C) miR‐92a‐3p Kaplan–Meier graph. P‐value (log rank) = 0.020; (D) miR‐363‐3p Kaplan–Meier graph. P‐value (log rank) = 0.034; (E) miR‐324‐3p Kaplan–Meier graph. P‐value (log rank) = 0.041; (F) miR‐22‐3p Kaplan–Meier graph. P‐value (log rank) < 0.0001.
Cox regression analyses are shown in Table 6 . The level of expression of miR‐15a‐5p, miR‐92a‐3p, miR101‐3p, and miR‐363‐3p, and miR‐22‐3p were predictors of HF and all‐cause mortality in univariate analysis (Table 3 ; LR χ 2: 57.8, P < 0.001). Among all, miR‐15a‐5p and miR‐92a‐3p were analysed in a multivariate model (due to higher AUC and the most significant association with outcomes). Both miR‐15a‐5p [hazard ratio: 0.21 (0.06–0.649, P = 0.007) and miR‐92a‐3p (0.27 (0.09–0.76), P = 0.01] were associated with outcomes after adjusting for age, gender, and New York Heart Association functional class.
Table 6.
Results of univariate and multivariate analyses in prediction of the outcome endpoints
| Univariate | Multivariate model 1 (LR χ 2: 27.15) | Multivariate MODEL 2 (LR χ 2: 25.73) | ||||
|---|---|---|---|---|---|---|
| Unadj HR (95% CI) | P | Adj HR (95% CI) | P | Adj HR (95% CI) | P | |
| Age (years) | 1.08 (1.02–1.15) | 0.01 | 1.07 (1.01–1.14) | 0.014 | 1.06 (1.00–1.13) | 0.04 |
| Gender (female) | 0.35 (0.15–0.81) | 0.02 | 0.24 (0.09–0.63) | 0.004 | 0.29 (0.11–0.73) | 0.009 |
| NYHA class | 2.69 (1.57–4.60) | <0.001 | 1.68 (0.95–2.96) | 0.07 | 1.61 (0.86–3.01) | 0.13 |
| LVEF, % | 0.97 (0.92–1.01) | 0.14 | — | — | — | — |
| RV‐basal diameter, mm | 1.02 (0.97–1.07) | 0.46 | — | — | — | — |
| RV end‐diastolic area, cm2 | 1.09 (1.05–1.14) | <0.001 | — | — | — | — |
| TR velocity, mm/s | 0.99 (0.98–1.01) | 0.60 | — | — | — | — |
| TAPSE, mm | 0.89 (0.79–1.01) | 0.06 | — | — | — | — |
| S′ wave TDI, cm/s | 0.95 (0.76–1.17) | 0.62 | — | — | — | — |
| RV‐FWLS, % | 1.16 (1.10–1.23) | 0.001 | — | — | — | — |
| Biplane VC, mm | 3.42 (1.30–9.01) | 0.013 | — | — | — | — |
| ERO, median (IQR), cm2 | 1.51 (1.06–2.12) | 0.02 | — | — | — | — |
| hsa‐miR‐15a‐5p | 0.21 (0.08–0.55) | 0.001 | 0.21 (0.06–0.64) | 0.007 | — | — |
| hsa‐miR‐101–3p | 0.20 (0.06–0.59) | 0.004 | — | — | — | — |
| hsa‐miR‐92a‐3p | 0.22 (0.09–0.54) | 0.001 | — | — | 0.27 (0.09–0.76) | 0.01 |
| hsa‐miR‐363‐3p | 0.34 (0.17–0.71) | 0.004 | — | — | — | — |
| hsa‐miR‐324‐3p | 0.31 (0.09–1.06) | 0.062 | — | — | — | — |
| hsa‐miR‐22‐3p | 0.23 (0.09–0.58) | 0.002 | — | — | — | — |
For univariate analyses, results are presented with unadjusted hazard ratios (unadj HR) with 95% confidence intervals (95% CI). Model 1: hsa‐miR‐15a‐5p, age, gender, NYHA class. Model 2: hsa‐miR‐92a‐3p, age, gender, NYHA class.
ERO, effective regurgitant orifice; LVEF, left ventricular ejection fraction; NYHA, New York Heart Association; RA, right atrium; RV, right ventricular; TR, tricuspid regurgitation.
MicroRNAs functional significance
The biological significance of the circulating miRNAs with diagnostic capability was explored through bioinformatic studies. A predictive analysis of the potential targets of miRNAs with diagnostic capacity (hsa‐miR‐186‐5p, hsa‐miR‐30e‐5p, and hsa‐miR‐152‐3p) revealed interesting connections with genes potentially involved in CV pathologies. As depicted in Figure 7 , EGFR and ESR1 exhibited the highest links within the network of targets potentially related to miRNAs. Subsequently, the enrichment analysis showed biological, structural or molecular pathways statistically associated with the set of targets. The enrichment analysis results unveiled noteworthy outcomes, particularly in pathways such as FoxO, ErbB, Nrf2, and MECP2 signalling, which were associated with the proper functioning of CV activity (Figure S1 ).
Figure 7.
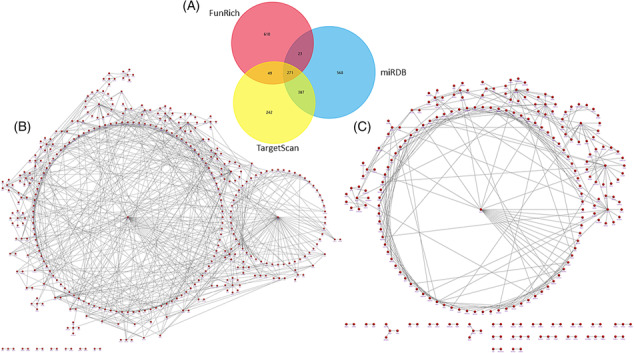
Biological pathways for predicted targets of miRNAs related to TR diagnosis (miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p). (A) Representative Venn diagram of the target prediction analysis methodology. (B) Graphical representation of the interaction of targets performed with the Uniprot database. (C) Graphical representation of the interaction of targets using the Funrich database.
Discussion
The current study is the first report of miRNA profile in patients with severe FTR. We demonstrated differential miRNA expression in severe A‐FTR compared with controls. V‐FTR also exhibits differential miRNA expression compared with controls and A‐FTR. In particular, the following miRNAs were identified as diagnostic markers of FTR: miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p. Our results describe a group of miRNAs that could stratify the CV risk in severe FTR, defined as HF admission or all‐cause mortality.
FTR is the most common cause of TR, representing the underlying aetiology in 80–90% of cases. FTR is distinguished by the structural integrity of the tricuspid valve leaflets, in which TR results in tricuspid annulus (TA) dilatation. 3 In V‐FTR, annular dilatation is commonly assumed to result from adverse RV remodelling secondary to different factors leading to papillary muscle displacement, RV leaflet anchoring, and TR. Despite the original aetiology, FTR has a negative impact on the outcome and predicts poor survival, heart failure, and impaired functional capacity. 3 , 16 Although different predictors of V‐FTR have been suggested in left valvular heart disease or pulmonary hypertension, the in‐depth underlying mechanisms are not fully understood. 17 , 18
A‐FTR is a relatively new phenotype of FTR secondary to TA dilatation in response to RA remodelling and dilatation with leaflet separation and diminished leaflet coaptation. 19 Patients with A‐FTR usually show normal/mildly dilated right ventricular (RV) size and preserved RV systolic function, at least in the early stages. Although RA size has been demonstrated to play a crucial role in determining the changes in the tricuspid annulus area, this study demonstrates different levels of miRNAs expression in A‐FTR compared with subjects with AF despite similar RA volumes. A group of these miRNAs was subsequently validated in a larger cohort, suggesting a potential role in identifying significant TR development in the context of longstanding or permanent AF. We further validated a new group of differentially expressed miRNAs in V‐FTR compared with controls, including miR‐186‐5p, miR‐30e‐5p, and miR‐152‐3p. Lastly, this study demonstrated the association of a selected group of miRNAs with specific imaging parameters of RV dimension, RV systolic function, and TR severity.
Echocardiography helps to differentiate between A‐FTR vs. V‐FTR, yet both entities may show an overlapping phenotypic picture in the advanced stages of valvular disease. 7 In addition, long‐standing V‐FTR may also evolve with AF or may become present in early stages in patients with concomitant left valvular heart disease; consequently, diagnosis of the primary cause in advanced stages of FTR may result quite challenging, and hence, the development of new tools that enable better identification would be of great help for a more accurate diagnosis. Here, we found differential miRNA expression in A‐FTR vs. V‐FTR with the potential to serve as a differential fingerprint biomarkers.
Beyond clinical status and imaging parameters, robust predictive biomarkers in tricuspid valvular heart disease still need to be improved. In this regard, our data emphasized the potential role of specific miRNAs, particularly the analysis of miR‐15a‐5p, miR‐92a‐3p, miR101‐3p, and miR‐363‐3p, miR‐324‐3p, and miR‐22‐3p, useful in the risk stratification of patients with FTR. Taken together, our data point towards selected miRNAs as novel prognostic biomarkers with the potential to complement current clinical algorithms.
A predictive analysis of putative gene targets on the three miRNAs hsa‐miR‐186‐5p, hsa‐miR‐30e‐5p, and hsa‐miR‐152‐3p has revealed a potential link to EGFR, and ESR1 genes, implicated in several CV diseases. EGFR signalling is crucial to cardiomyocyte function and survival, 20 , 21 and its activation is associated with endothelial dysfunction, neointimal hyperplasia, atherogenesis, cardiac remodelling, and valve development. 22 Oestrogen‐induced cardiac protection 23 , 24 is thought to be mediated through the activation of nuclear receptors ERα and ERβ, encoded by the ESR1 and ESR2 genes, respectively. 23 On the other hand, a wealth of evidence underscores the crucial roles of three members within the FoxO subfamily—FoxO1, FoxO3, and FoxO4—in preserving cardiac function and responding to cardiac stresses in adults. 25 Meanwhile, the ErbB family of receptor tyrosine kinases (RTKs) serves as a group of receptors facilitating cellular interactions with the extracellular environment. These receptors transduce signals to the nucleus, promoting processes such as differentiation, migration, and proliferation vital for proper heart morphogenesis and function. 26 Additionally, recent findings highlight the significance of Nrf2 and its target genes as critical regulators in maintaining CV homeostasis. Their role involves the suppression of oxidative stress, a key contributor to the development and progression of heart failure. 27 Lastly, MeCP2 emerges as a pivotal player in various CV activities, encompassing angiogenesis, heart development, blood pressure regulation, myocardial contractility, cardiac conduction, and the modulation of myocardial hypertrophy and fibrosis. 28 Our study is the first report connecting the possibility of selected miRNAs targeting FTR by regulating specific candidate gene expression.
TA dilatation and the degree of leaflet adaptation or growth may ultimately determine the degree of valve dysfunction leading to the development of TR in FTR. Whether the proposed targets of three predictive miRNAs of tricuspid pathology in our study are related to leaflet growth or adaptation has to be explored in future studies. This hypothesis would open a new exciting pathway in the pathophysiology of FTR with potential new targets.
Limitations
This was an observational single centre study. Despite the relative low number of patients, the study demonstrated the diagnostic and prognostic value of different miRNA as novel promising biomarkers. The exploratory nature of the study and the small sample size did not allow adjusting for additional imaging prognostic parameters. Lastly, an established gold standard for bioinformatic analysis has not been defined. In this study, we used the FunRich software, which combines different databases, allowing a first approximation of potential targets of selected miRNAs expression. Future prospective studies are needed to confirm our results.
In conclusion, the present study demonstrates, for the first time, the capability of several miRNAs determined in blood samples to identify FTR with the potential to stratify the risk of mortality and heart failure hospitalizations, providing new putative target genes for further investigations on this pathology.
Conflict of interest
No conflict of interest to disclosure
Funding
This study was supported by the Instituto de Salud Carlos III, PI20/01206.
Supporting information
Figure S1. Enrichment analysis using the main databases: A) KEGG 2021 Human; B) WikiPathway 2021 Human; C) Reactome 2022; D) GO Molecular function; E) GO Cellular Component; F) GO Biological Process.
Table S1. Supporting information.
Hinojar, R. , Moreno‐Gómez‐Toledano, R. , Conde, E. , Gonzalez‐Gomez, A. , García‐Martin, A. , González‐Portilla, P. , Fernández‐Golfín, C. , García‐Bermejo, M. L. , Zaragoza, C. , and Zamorano, J. L. (2024) Circulating miRNA in functional tricuspid regurgitation. Unveiling novel links to heart failure: A pilot study. ESC Heart Failure, 11: 2272–2286. 10.1002/ehf2.14765.
References
- 1. Vahanian A, Beyersdorf F, Praz F, Milojevic M, Baldus S, Bauersachs J, et al. ESC/EACTS Scientific Document Group, 2021 ESC/EACTS Guidelines for the management of valvular heart disease: Developed by the Task Force for the management of valvular heart disease of the European Society of Cardiology (ESC) and the European Association for Cardio‐Thoracic Surgery (EACTS). Eur Heart J 2021;ehab395:561‐632. doi: 10.1093/eurheartj/ehab395 [DOI] [Google Scholar]
- 2. Otto CM, Nishimura RA, Bonow RO, Carabello BA, Erwin JP 3rd, Gentile F, et al. 2020 ACC/AHA guideline for the management of patients with valvular heart disease: A report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Thorac Cardiovasc Surg 2021;162:e183‐e353. [DOI] [PubMed] [Google Scholar]
- 3. Hahn RT, Badano LP, Bartko PE, Muraru D, Maisano F, Zamorano JL, et al. Tricuspid regurgitation: Recent advances in understanding pathophysiology, severity grading and outcome. Eur Heart J Cardiovasc Imaging 2022;23:913‐929. doi: 10.1093/ehjci/jeac009 [DOI] [PubMed] [Google Scholar]
- 4. Muraru D, Guta AC, Ochoa‐Jimenez RC, Bartos D, Aruta P, Mihaila S, et al. Functional regurgitation of atrioventricular valves and atrial fibrillation: An elusive pathophysiological link deserving further attention. J Am Soc Echocardiogr 2020. Jan;33:42‐53. doi: 10.1016/j.echo.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 5. Schlotter F, Dietz MF, Stolz L, Kresoja KP, Besler C, Sannino A, et al. Atrial functional tricuspid regurgitation: Novel definition and impact on prognosis. Circ Cardiovasc Interv. 2022;15:e011958. doi: 10.1161/CIRCINTERVENTIONS.122.011958 [DOI] [PubMed] [Google Scholar]
- 6. Guta AC, Badano LP, Tomaselli M, Mihalcea D, Bartos D, Parati G, et al. The pathophysiological link between right atrial remodeling and functional tricuspid regurgitation in patients with atrial fibrillation: A three‐dimensional echocardiography study. J Am Soc Echocardiogr 2021. Jun;34:585‐594.e1. doi: 10.1016/j.echo.2021.01.004 [DOI] [PubMed] [Google Scholar]
- 7. Florescu DR, Muraru D, Florescu C, Volpato V, Caravita S, Perger E, et al. Right heart chambers geometry and function in patients with the atrial and the ventricular phenotypes of functional tricuspid regurgitation. Eur Heart J Cardiovasc Imaging 2022;23:930‐940. doi: 10.1093/ehjci/jeab211 [DOI] [PubMed] [Google Scholar]
- 8. Nappi F, Iervolino A, Avtaar Singh SS, Chello M. MicroRNAs in valvular heart diseases: Biological regulators, prognostic markers and Therapeutical targets. Int J Mol Sci 2021;22:12132. doi: 10.3390/ijms222212132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hinojar R, Zamorano JL, González Gómez A, García‐Martin A, Monteagudo JM, García Lunar I, et al. Prognostic impact of right ventricular strain in isolated severe tricuspid regurgitation. J Am Soc Echocardiogr 2023;S0894‐7317:94‐99. doi: 10.1016/j.echo.2023.02.009 [DOI] [PubMed] [Google Scholar]
- 10. Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol 2021;433:166747. doi: 10.1016/j.jmb.2020.166747 [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Wang X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127‐D131. doi: 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed‐pairing stability and high target‐site abundance decrease the proficiency of lsy‐6 and other microRNAs. Nat Struct Mol Biol 2011;18:1139‐1146. doi: 10.1038/nsmb.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ge SX, Jung D, Jung D, Yao R. ShinyGO: A graphical gene‐set enrichment tool for animals and plants. Bioinformatics 2020;36:2628‐2629. doi: 10.1093/BIOINFORMATICS/BTZ931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90‐W97. doi: 10.1093/NAR/GKW377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bozkurt B, Coats AJS, Tsutsui H, Abdelhamid CM, Adamopoulos S, Albert N, et al. Universal definition and classification of heart failure: a report of the Heart Failure Society of America, Heart Failure Association of the European Society of Cardiology, Japanese Heart Failure Society and Writing Committee of the Universal Definition of Heart Failure: Endorsed by the Canadian Heart Failure Society, Heart Failure Association of India, Cardiac Society of Australia and New Zealand, and Chinese Heart Failure Association. Eur J Heart Fail 2021;23:352‐380. doi: 10.1002/ejhf.2115 [DOI] [PubMed] [Google Scholar]
- 16. Praz F, Muraru D, Kreidel F, Lurz P, Hahn RT, Delgado V, Senni M, von Bardeleben RS, Nickenig G, Hausleiter J, Mangieri A, Zamorano JL, Prendergast BD, Maisano F Transcatheter treatment for tricuspid valve disease. EuroIntervention 2021. 17:791–808. doi: 10.4244/EIJ-D-21-00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shiran A, Sagie A. Tricuspid regurgitation in mitral valve disease incidence, prognostic implications, mechanism, and management. J Am Coll Cardiol 2009;53:401‐408. doi: 10.1016/j.jacc.2008.09.048 [DOI] [PubMed] [Google Scholar]
- 18. Medvedofsky D, Aronson D, Gomberg‐Maitland M, Thomeas V, Rich S, Spencer K, et al. Tricuspid regurgitation progression and regression in pulmonary arterial hypertension: Implications for right ventricular and tricuspid valve apparatus geometry and patients outcome. Eur Heart J Cardiovasc Imaging 2017;18:86‐94. doi: 10.1093/ehjci/jew010 [DOI] [PubMed] [Google Scholar]
- 19. Muraru D, Addetia K, Guta AC, Ochoa‐Jimenez RC, Genovese D, Veronesi F, et al. Right atrial volume is a major determinant of tricuspid annulus area in functional tricuspid regurgitation: A three‐dimensional echocardiographic study. Eur Heart J Cardiovasc Imaging 2021;22:660‐669. [DOI] [PubMed] [Google Scholar]
- 20. Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, et al. Architecture and membrane interactions of the EGF receptor. Cell 2013;152:557‐569. doi: 10.1016/j.cell.2012.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holowka D, Baird B. Mechanisms of epidermal growth factor receptor signaling as characterized by patterned ligand activation and mutational analysis. Biochim Biophys Acta Biomembr 2017;1859:1430‐1435. doi: 10.1016/j.bbamem.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kokubo H, Miyagawa‐Tomita S, Tomimatsu H, Nakashima Y, Nakazawa M, Saga Y, et al. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res 2004;95:540‐547. doi: 10.1161/01.RES.0000141136.85194.f0 [DOI] [PubMed] [Google Scholar]
- 23. Puzianowska‐Kuznicka M. ESR1 in myocardial infarction. Clin Chim Acta 2012;413:81‐87. doi: 10.1016/J.CCA.2011.10.028 [DOI] [PubMed] [Google Scholar]
- 24. Zheng Z, Yu S, Zhang W, Peng Y, Pu M, Kang T, et al. Genistein attenuates Monocrotaline‐induced pulmonary arterial hypertension in rats by activating PI3K/Akt/ENOS signaling. Histol Histopathol 2017;32:35‐41. doi: 10.14670/HH-11-768 [DOI] [PubMed] [Google Scholar]
- 25. Ronnebaum SM, Patterson C. The FoxO family in cardiac function and dysfunction. Annu Rev Physiol 2010;72:81‐94. doi: 10.1146/annurev-physiol-021909-135931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanchez‐Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Semin Cell Dev Biol 2010;21:929‐935. doi: 10.1016/j.semcdb.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou S, Sun W, Zhang Z, Zheng Y. The role of Nrf2‐mediated pathway in cardiac remodeling and heart failure. Oxid Med Cell Longev 2014;2014:260429. doi: 10.1155/2014/260429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang C, Wang F, Cao Q, Li Z, Huang L, Chen S. The effect of Mecp2 on heart failure. Cell Physiol Biochem 2018;47:2380‐2387. doi: 10.1159/000491610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Enrichment analysis using the main databases: A) KEGG 2021 Human; B) WikiPathway 2021 Human; C) Reactome 2022; D) GO Molecular function; E) GO Cellular Component; F) GO Biological Process.
Table S1. Supporting information.


